Abstract
The Mexican population is characterized by high and particular admixture, and the picture of variants associated with disease remains unclear. Here we investigated the distribution of single nucleotide polymorphisms (SNPs) in the Mexican population. We focused on two non-synonymous and three synonymous SNPs in the beta-2 adrenergic receptor gene (ADRB2), which plays key roles in energy balance regulation. These SNPs were genotyped in 2,011 Mexican Amerindians (MAs) belonging to 62 ethnic groups and in 1,980 geographically matched Mexican Mestizos (MEZs). The frequency distribution of all five ADRB2 variants significantly differed between MAs, MEZs, and other continental populations (CPs) from the 1000 Genomes database. Allele frequencies of the three synonymous SNPs rs1042717A, rs1042718A, and rs1042719C were significantly higher in Mexican individuals, particularly among MAs, compared to in the other analyzed populations (P<0.05). The non-synonymous ADRB2 Glu27 allele (rs1042714G), which is associated with several common conditions, showed the lowest frequency in MAs (0.03) compared to other populations worldwide. Among MEZs, this allele showed a frequency of 0.15, intermediate between that in MAs and in Iberians (0.43). Moreover, Glu27 was the only SNP exhibiting a geographic gradient within the MEZ population (from 0.22 to 0.11), reflecting admixed mestizo ancestry across the country. Population differentiation analysis demonstrated that Glu27 had the highest FST value in MAs compared with Europeans (CEU) (0.71), and the lowest between MAs and Japanese (JPT) (0.01), even lower than that observed between MAs and MEZs (0.08). This analysis demonstrated the genetic diversity among Amerindian ethnicities, with the most extreme FST value (0.34) found between the Nahuatls from Morelos and the Seris. This is the first study of ADRB2 genetic variants among MA ethnicities. Our findings add to our understanding of the genetic contribution to variability in disease susceptibility in admixed populations.
Introduction
The Mexican Mestizo (MEZ) population is one of the most genetically diverse populations worldwide due to the admixture between Native American, European, and African populations [1]. In addition to MEZs, the Mexican population also includes a great diversity of Mexican Amerindians (MAs), who were the original settlers of Mexico. The MA people currently constitute 14.9% of the population (15 million), distributed into 68 ethnic groups throughout the Mexican territory [2,3]. Genomic diversity studies reveal vast genetic differences between the MEZ population and most of the continental populations (CPs), as well as between MAs and MEZs [3–5]. Therefore, the Mexican population is characterized by a high and particular admixture.
Recent studies suggest that ethnic diversity may introduce genetic variations that can potentially generate inter-individual differences in disease susceptibility and therapeutic efficacy [6–8]. These findings could be explained within an evolutionary framework, in which the frequencies of specific alleles reflect ancient genetic adaptations that have shifted due to environment and lifestyle differences among human populations [9]. However, most research in this field has been performed among Caucasians [7,10].
The protein encoded by the beta-2 adrenergic receptor gene (ADRB2) plays a key role in energy balance regulation and is a target for many drugs that are commonly used to treat different conditions [11,12]. ADRB2 is an intron–less gene located on chromosome 5q31-32, which is of particular interest due to its impact on the genetic risk for several common illnesses, including obesity, asthma, and cardiovascular disease [13–15]. Notably, ADRB2 shows great inter-population variability in allele frequencies [16,17]. Since ADRB2 may have been subjected to balancing selection during human evolution, it is a particularly interesting candidate for evaluating how the genetic structure of a population affects the inter–individual differences in susceptibility to chronic degenerative diseases and response to therapeutic drugs [17,18].
Among the single nucleotide polymorphisms (SNPs) found in the ADRB2 coding region, the two most studied are the non-synonymous SNPs rs1042713 and rs1042714, which result in amino acid changes at protein positions 16 (Gly16Arg) and 27 (Gln27Glu), respectively. The variant alleles of these SNPs modify the receptor activity at several levels, and may also affect the response to therapies with beta-2 adrenergic receptor (b2-AR) agonist through a mechanism involving agonist-promoted down-regulation of receptor expression [10,19]. Recent reports demonstrate that other synonymous SNPs in this gene can affect RNA stability and thus alter the amount of protein [20]. Accordingly, the variants rs1042717 (Leu84Leu), rs1042718 (Arg175Arg), and rs1042719 (Gly351Gly) have been associated with malaria susceptibility, hypertension, longevity, and asthma [20–23].
Although ADRB2 gene variants play an important role in disease susceptibility and drug responses, they have been scarcely studied among MEZs [24,25] and there are no previous reports of the geographic distribution of these variants in the MA population. In the present study, we aimed to investigate the distribution of five coding SNPs in ADRB2 within the MA population, as well as their contribution to the ethnic structure of the MEZ population.
Methods
Study population
This study included 2,011 unrelated MAs, belonging to 62 different ethnic groups distributed throughout the Mexican territory, from the Metabolic Analysis in an Indigenous Sample (MAIS) cohort study [3]. The participants identified themselves as indigenous, spoke the same native language as their parents and grandparents, and were born in the same region as their parents and grandparents. Our study also included 1,980 unrelated MEZ adults whose parents and grandparents were born in Mexico. This study was conducted in accordance with the Declaration of Helsinki, and was approved by the ethics and human research committees of the National Institute of Genomic Medicine in Mexico City, Mexico. All participants provided written informed consent, and their confidentiality was preserved at all times.
Since admixture of the MEZ population has generated great genetic diversity throughout the Mexican territory, we also investigated the Amerindian influence on the regional admixture of the MEZs based on the frequency of ADRB2 polymorphisms. For this analysis, we compared the genotypic and allelic frequencies of the five studied SNPs between 1,851 Amerindian individuals (representing 31 Amerindian groups, each including at least 10 individuals) and 1,980 MEZ individuals matched by geographic region. Both MAs and MEZs were sorted into five geographic regions: North, Central East, Central West, South, and South East [3,4].
Genotyping
Genomic DNA was extracted from whole blood using the QIAmp DNA Blood Maxi kit (Qiagen Systems, Inc., Valencia CA), following the manufacturer’s protocol. All subjects were genotyped for five SNPs localized within the coding region of ADRB2: the non–synonymous SNPs rs1042713 (G/A, Gly16Arg) and rs1042714 (C/G, Gln27Glu), and the synonymous SNPs rs1042717 (G/A, Leu84Leu), rs1042718 (C/A, Arg175Arg), and rs1042719 (G/C, Gly351Gly). Genotyping was performed using the TaqMan Allelic Discrimination assay on an ABI PRISM 7900 thermocycler (Applied Biosystems, Foster City, CA, USA). The genotyping call rate was over 96% in all tested SNPs, and no discordant genotypes were found in samples run in duplicate (15%). The TaqMan results were validated by direct sequencing of random samples from each genotype (10%) using an automated ABI PRISM 310 Genetic Analyzer (Applied Biosystems Foster City, CA, USA) with 100% reproducibility. The MA population cohort had an average Amerindian ancestry of 95 ± 5%, as previously described [3].
Statistical analysis
Allele frequency comparisons were performed by using a chi-square test with the PLINK v1.07 program [26]. A P value of <0.05 after Bonferroni correction was considered significant. To measure the level of population differentiation, individual allelic and genotypic data were used to calculate the Wright’s fixation index (FST) using GENEPOP software version 1.2 [27]. Linkage disequilibrium (LD) and haplotype structure were analyzed using Haploview software version 4.2 (http://www.broad.mit.edu/mpg/). All maps were constructed with QGIS software version 2.14, and were modified from the National Commission of Knowledge and Use of Biodiversity (CONABIO) [28]. For the MA population, we estimated the correlation coefficient between the allele frequencies of each variant and various geographic coordinates of the ethnic groups (including altitude, latitude, and longitude), and the significance was evaluated by the Pearson’s test, using R version 3.4.4 statistical software [29].
Results
Distribution of ADRB2 polymorphisms and haplotype analysis in MA and MEZ populations
The allele and genotype distributions of the five presently analyzed ADRB2 SNPs were in Hardy–Weinberg equilibrium among both MAs and MEZs. We further found that the allelic and genotypic frequencies of rs1042713A (Arg16) were similar between these two populations (P > 0.05). In contrast, the frequency of the Glu27 (G) allele of rs1042714 was significantly lower in MAs than in MEZs (P = 1×10−8), and the GG homozygous genotype was not observed in any Amerindian ethnic group. On the other hand, the allelic and genotypic frequencies of rs1042717A, rs1042718A, and rs1042719C were significantly higher in MAs than MEZs (P < 0.001; Tables 1 and S1).
Table 1. Comparison of allele frequencies of ADRB2 SNPs in MAs, MEZs and other continental population.
| Allele Frequencies | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP (Allele) | Present study | 1000 genomes (phase 3) | ||||||
| MAs (n = 2011) | MEZs (n = 1980) |
MXL (n = 64) |
CEU (n = 99) |
IBS (n = 107) |
YRI (n = 108) |
JPT (n = 104) |
CHB (n = 103) |
|
| rs1042713 (A) | 0.47 | 0.44 | 0.48 | 0.35b,c | 0.38a | 0.53c | 0.44 | 0.55a,c |
| rs1042714 (G) | 0.03 | 0.15b | 0.14b | 0.47b,d | 0.43b,d | 0.12b | 0.06a,d | 0.11b |
| rs1042717 (A) | 0.51 | 0.43b | 0.38a | 0.19b,d | 0.19b,d | 0.35b,c | 0.46 | 0.34b,c |
| rs1042718 (A) | 0.50 | 0.42b | 0.38a | 0.17b,d | 0.15b,d | 0.34b,c | 0.47 | 0.35b,c |
| rs1042719 (C) | 0.53 | 0.47b | 0.49 | 0.26b,d | 0.28b,d | 0.35b,d | 0.54 | 0.44a |
a MAs vs other populations P < 0.05.
b MAs vs other populations P < 0.0001 (Range,10−4 to 10−8).
c MEZ vs other populations P < 0.05.
d MEZ vs other populations P < 0.0001 (Range,10−4 to 10−8).
To obtain a global perspective regarding the behavior of these variants, we compared the presently observed allele frequencies with those reported for CPs in the 1000 Genomes database. Compared populations included Utah Residents (CEPH) with Northern and Western European Ancestry (CEU); Yoruba in Ibadan, Nigeria (YRI); Han Chinese in Beijing, China (CHB); Japanese in Tokyo, Japan (JPT); Mexican Ancestry from Los Angeles USA (MXL); and Iberian Population in Spain (IBS). This last population was included because the European contribution to Mexican genetic admixture is mainly from Spain [5,30].
These comparisons indicated that the frequency of the rs1042713A (Arg16) allele among MAs was similar to the rates reported for MXL, YRI, and JPT (P > 0.05), but significantly different from CEU, IBS, and CHB (P < 0.05). Remarkably, MAs exhibited the lowest frequency of the rs1042714G (Glu27) allele (0.03) compared to all CPs [MXL, 0.14; CEU, 0.47; IBS, 0.43; YRI, 0.12; and CHB, 0.11 (P < 0.001); JPT, 0.06 (P = 0.02); and MEZs, 0.15 (P < 0.001)]. On the other hand, the frequencies of the synonymous alleles rs1042717A, rs1042718A, and rs1042719C were higher among MAs than in the other populations (P < 0.05) with the exception of JPT (Table 1). Importantly, the allele frequencies of rs1042717A and rs1042718A were significantly higher among MAs than in MXL or MEZs. In contrast, the frequency of the rs1042719C allele was similar between the MA and MXL groups, although it significantly differed between MAs and MEZs (Table 1).
The frequencies of all ADRB2 alleles in the MEZ group were significantly different from those in the CEU and IBS populations, with the exception of rs1042713A in IBS. As expected, none of the variants frequencies significantly differed between MEZs and MXL. Similar to the findings in MAs, all of the variants (except rs1042714) exhibited very similar behavior in MEZs and JPT, but not in YRI and CHB (Table 1). Notably, the frequencies of all of the analyzed variants in the MEZ population exhibited an intermediate relationship to those observed in their ancestral populations, with IBS on one side and MAs on the other.
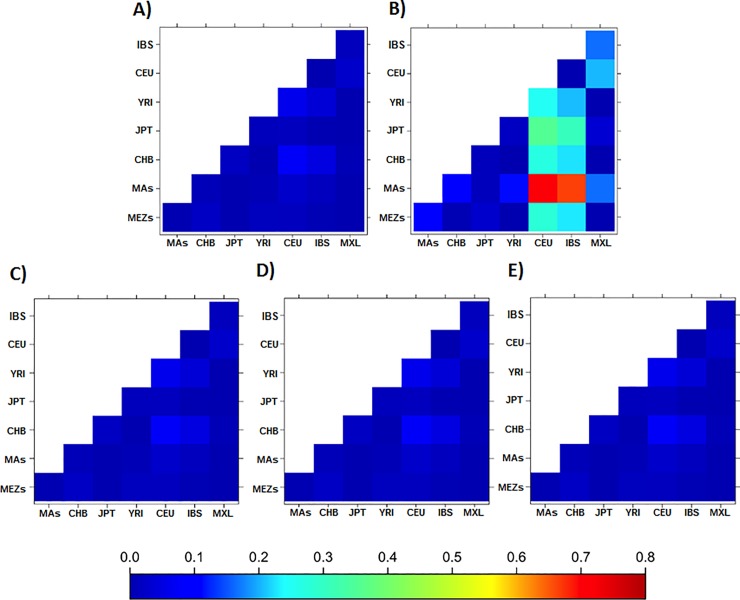
We also investigated the level of differentiation (FST) of the five ADRB2 polymorphisms between MAs, MEZs, and the CPs. The rs1042713 variant exhibited the lowest level of differentiation among all of the analyzed populations, whereas the rs1042714 variant exhibited the most extreme level of differentiation, particularly between MAs and CEU (0.709), and between MAs and IBS (0.665) (Fig 1 and S2 Table). With regards to rs1042717, rs1042718, and rs1042719, the highest levels of population differentiation were found between MAs and CEU (0.172, 0.185, and 0.125, respectively), and the lowest FST values were found between MAs and JPT (0.002, 0.000, and 0.000, respectively; Fig 1 and S2 Table).
Fig 1. Level of genetic differentiation (FST) for the five ADRB2 variants among MAs, MEZs and other continental populations (see also S2 Table).
Populations from the 1000 Genomes database were used. A) rs1042713A; B) rs1042714G; C) rs1042717A; D) rs1042718A; E) rs1042719C. The darkest blue indicates the lowest level of differentiation, whereas red indicates the highest FST value.
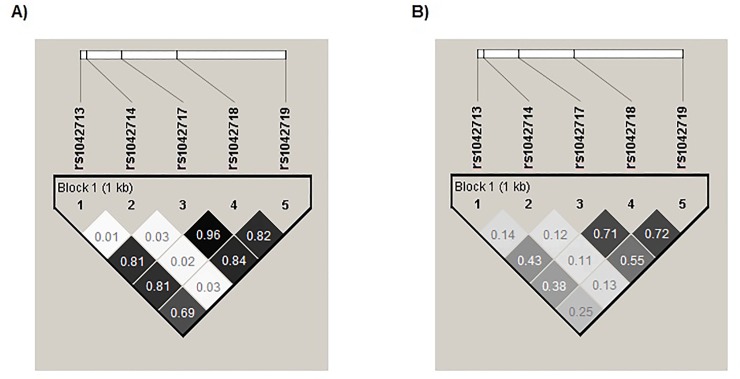
The synonymous variants rs1042717, rs1042718, and rs1042719 exhibited high LD in both the MA and MEZ populations, with higher values among MAs (r2 = 0.96, 0.84, and 0.82) than MEZs (r2 = 0.71, 0.55, and 0.72; Fig 2). In contrast, the non-synonymous SNP rs1042714 exhibited no evidence of LD with any other SNPs in either population (r2 = 0.01 to 0.03 among MAs, and r2 = 0.11 to 0.14 among MEZs; Fig 2). This analysis revealed five haplotypes with frequencies greater than 1%, of which four were shared by MEZs and MAs, as well as one haplotype with a low frequency (0.02) found only in MEZ individuals (Table 2). Interestingly, the frequencies of the four shared haplotypes significantly differed between the two groups (P < 0.001; Table 2).
Fig 2.
Linkage disequilibrium (LD) structure of ADRB2 SNPs in A) MAs and B) MEZs. The r2 value was calculated using Haploview software 4.2.
Table 2. Haplotype frequencies of ADRB2 SNPs in MA and MEZ populations.
| Haplotypes | Frequencies | P value | ||
|---|---|---|---|---|
| MEZs | MAs | |||
| 1 | GCAAC | 0.36 | 0.48 | 1x10-8 |
| 2 | ACGCG | 0.35 | 0.43 | 1x10-8 |
| 3 | GGGCG | 0.12 | 0.03 | 1x10-8 |
| 4 | ACGCC | 0.05 | 0.03 | 8x10-5 |
| 5 | GCACC | 0.02 | NF | - |
NF, not found
Since the three synonymous variants rs1042717, rs1042718, and rs1042719 showed a high LD in both MEZs and MAs, we only show the rs1042717 variant data from our further analyses.
Allele frequencies of ADRB2 variants among MA ethnic groups
We also compared the distribution of ADRB2 variants between MAs belonging to different ethnic groups. This analysis included only Amerindian groups represented by at least 10 individuals which totaled 31 of the 62 ethnic groups (n = 1,851 individuals). We found that the ADRB2 variants exhibited high heterogeneity among all of the ethnic groups. For example, the allele frequency of rs1042713A ranged from 0.23 in the Nahuatl group from Morelos to 0.72 in the Seri group, whereas that of rs1042717A ranged from 0.31 in Seris to 0.77 in Nahuatls from Morelos (Table 3). In contrast, the rs1042714G (Glu27) allele exhibited very low frequencies in all ethnic groups, with frequencies of <0.05 in 24 of the 31 analyzed groups, and frequencies of >0.10 among only the Purepechas and Mayos (0.107 and 0.125, respectively). This SNP was monomorphic in the Chuj, Huasteco, Huave, Kanjobal, Mocho, Tojolabal, and Nahuatl from Morelos ethnic groups (Table 3).
Table 3. Distribution of allele frequencies of ADRB2 SNPs in MAs.
| SNP | AF | SNP | AF | SNP | AF |
|---|---|---|---|---|---|
| rs1042713(G/A) | A | rs1042714(C/G) | G | Tag SNP rs1042717(G/A) | A |
| Nahuatl Mora | 0.23 | Nahuatl Mora | 0.000 | Seria | 0.31 |
| Purepecha | 0.29 | Kanjobala | 0.000 | Chontal Oaxa | 0.32 |
| Kanjobala | 0.36 | Huasteco | 0.000 | Pame | 0.35 |
| Zapoteco | 0.40 | Chuj | 0.000 | Jakalteko | 0.36 |
| Nahuatl Edo Mex | 0.40 | Mocho | 0.000 | Mam | 0.38 |
| Popoluca | 0.41 | Tojolabal | 0.000 | Mocho | 0.40 |
| Otomi | 0.42 | Huave | 0.000 | Nahuatl Pue | 0.45 |
| Nahuatl CDMX | 0.43 | Zapoteco | 0.008 | Mazateco | 0.46 |
| Nahuatl SLP | 0.43 | Mixteco | 0.008 | Mixteco | 0.46 |
| Maya | 0.44 | Nahuatl SLP | 0.011 | Chuj | 0.47 |
| Huasteco | 0.45 | Mam | 0.012 | Mayo | 0.47 |
| Mazahua | 0.45 | Kaqchikel | 0.014 | Yaqui | 0.47 |
| Kaqchikel | 0.46 | Chinanteco | 0.019 | Chinanteco | 0.48 |
| Totonaco | 0.47 | Tarahumara | 0.022 | Tarahumara | 0.49 |
| Yaqui | 0.47 | Totonaco | 0.022 | Mixe | 0.50 |
| Mixe | 0.48 | Mixe | 0.024 | Mazahua | 0.50 |
| Mayo | 0.48 | Nahuatl Edo Mex | 0.024 | Tojolabal | 0.51 |
| Chinanteco | 0.49 | Mazateco | 0.025 | Nahuatl CDMX | 0.52 |
| Huave | 0.50 | Otomi | 0.026 | Maya | 0.53 |
| Tojolabal | 0.50 | Serib | 0.026 | Otomi | 0.53 |
| Mazateco | 0.53 | Yaqui | 0.029 | Totonaco | 0.53 |
| Mixteco | 0.53 | Nahuatl Pue | 0.039 | Popoluca | 0.54 |
| Nahuatl Pue | 0.53 | Jakalteko | 0.040 | Huave | 0.55 |
| Tarahumara | 0.56 | Maya | 0.045 | Nahuatl SLP | 0.56 |
| Mocho | 0.57 | Mazahua | 0.050 | Purepecha | 0.57 |
| Chuj | 0.58 | Pame | 0.050 | Nahuatl Edo Mex | 0.57 |
| Pame | 0.60 | Nahuatl CDMX | 0.050 | Huasteco | 0.58 |
| Mam | 0.62 | Popoluca | 0.071 | Kaqchikel | 0.60 |
| Chontal Oaxb | 0.62 | Chontal Oaxb | 0.091 | Zapoteco | 0.60 |
| Jakalteko | 0.65 | Purepecha | 0.107 | Kanjobalb | 0.64 |
| Serib | 0.72 | Mayo | 0.125 | Nahuatl Morb | 0.77 |
a Ethnicities that showed the lowest frequency in all SNPs analyzed.
b Ethnicities that showed the highest frequency in all SNPs analyzed.
AF = allele frequencies
We determined the FST of the five ADRB2 variants among 31 Amerindian groups, and found the highest level of differentiation in the Seri, Pame, Nahuatl from Morelos, Chontal, and Kanjobal groups (Fig 3). Notably, the Nahuatls from Morelos exhibited high population differentiation compared to most of the other groups (24 with FST > 0.10), but mainly compared to the Chontal and Pame (FST = 0.30 and 0.28, respectively). The Seri, Chontal, and Pame groups did not show any degree of differentiation between each other (FST = 0.00, 0.00, and 0.00, respectively; Fig 3, S3 Table).
Fig 3. Pairwise FST values among the 31 MA ethnicities ordered geographically (see also S3 Table).
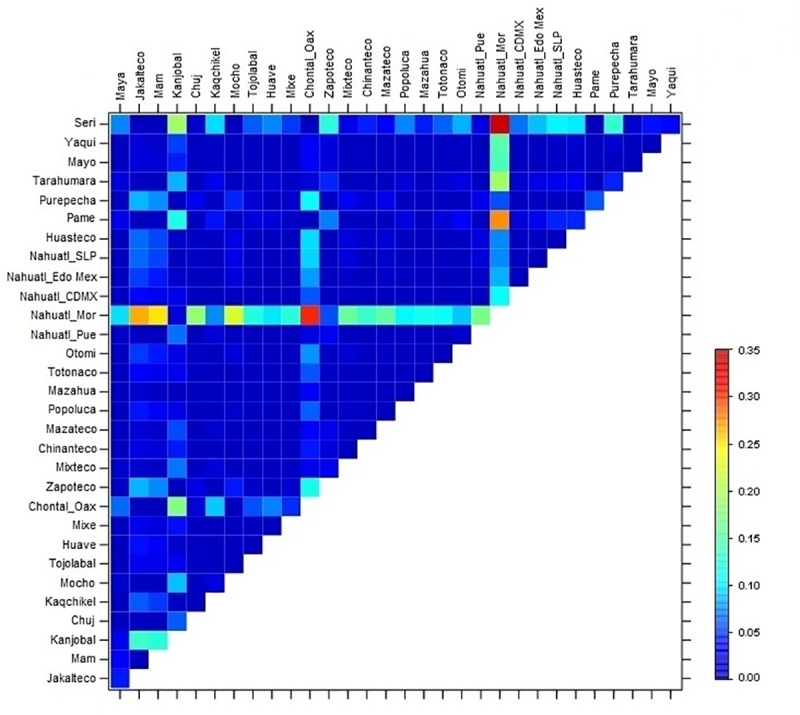
Calculations were performed with the five ADRB2 variants using GENEPOP software version 1.2. The darkest blue indicates the lowest level of differentiation, whereas red color indicates the highest FST value.
Geographic distribution of ADRB2 variants among MA and MEZ individuals
We sorted MA and MEZ individuals into five geographic regions, and found that the Seris in the North, Pames and Nahuatls from Morelos in the Central East, Chontals from the South, and Kanjobals from the South East exhibited extreme frequencies compared to other geographically close groups. Therefore, we removed these ethnic groups from the geographic analysis (Table 4). The geographic distribution of the rs1042713 variant did not significantly differ between MEZ and MA individuals (Fig 4A). In contrast, the Glu27 (G) allele of rs1042714 exhibited a significantly lower frequency among MAs than MEZs in all regions except the Central West region (Fig 4B). Interestingly, within the MEZ group, the frequency of rs1042714G decreased from 22% in the North to 11% in the South; whereas MAs exhibited a similar distribution of this allele in all regions except the Central West (Fig 4B). Similarly, the geographic distribution of the rs1042717 SNP did not significantly differ between the MEZ and MA populations, except for in the Central East region (Fig 4C).
Table 4. Geographic distribution and comparison of allele frequencies of ADRB2 SNPs in 31 MA ethnic groups and the MEZ population.
| Geographic Region | Ethnic Group (n) | rs1042713 A | rs1042714 G | rs1042717 A | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AF |
P value |
P value* |
AF | P value |
P value* |
AF |
P value |
P value* |
||
| North | Mayo (29) | 0.483 | 0.38 | NS | 0.125 | 3x10-3b | 0.01 | 0.466 | 0.87 | NS |
| Seria (19) | 0.722 | 0.026 | 0.306 | |||||||
| Tarahumara (93) | 0.557 | 0.022 | 0.439 | |||||||
| Yaqui (37) | 0.472 | 0.029 | 0.472 | |||||||
| MAs (159) | 0.520 | 0.04b | NS | 0.040 | 1x10-8b | 1x10-8b | 0.450 | 0.26 | NS | |
| MEZs (122) | 0.430 | 0.220c | 0.400 | |||||||
| Central West | MAs [Purepecha (14)] | 0.286 | 0.19 | NS | 0.107 | 0.32 | NS | 0.571 | 0.12 | NS |
| MEZs (180) | 0.410 | 0.180c | 0.420 | |||||||
| Central East | Huasteco (79) | 0.447 | 0.79 | NS | 0.000 | 0.09 | NS | 0.576 | 0.8 | NS |
| Mazahua (10) | 0.450 | 0.050 | 0.500 | |||||||
| Nahuatl CDMX (53) | 0.431 | 0.050 | 0.520 | |||||||
| Nahuatl Edo Mex (22) | 0.405 | 0.024 | 0.568 | |||||||
| Nahuatl Mora (45) | 0.227 | 0.000 | 0.773 | |||||||
| Nahuatl Pue (52) | 0.529 | 0.039 | 0.451 | |||||||
| Nahuatl SLP (44) | 0.430 | 0.011 | 0.558 | |||||||
| Otomi (223) | 0.424 | 0.026 | 0.533 | |||||||
| Pamea(10) | 0.600 | 0.050 | 0.350 | |||||||
| Popoluca (36) | 0.412 | 0.071 | 0.543 | |||||||
| Totonaco (97) | 0.468 | 0.022 | 0.527 | |||||||
| MAs (616) | 0.440 | 0.72 | NS | 0.027 | 1x10-8b | 1x10-8b | 0.530 | 1x10-8b | 1x10-8b | |
| MEZs (1435) | 0.440 | 0.140c | 0.430 | |||||||
| South | Chinanteco (83) | 0.494 | 0.31 | NS | 0.019 | 0.57 | NS | 0.475 | 0.15 | NS |
| Chontal Oaxa (44) | 0.622 | 0.091 | 0.429 | |||||||
| Huave (26) | 0.500 | 0.000 | 0.477 | |||||||
| Mazateco (61) | 0.526 | 0.025 | 0.458 | |||||||
| Mixe (90) | 0.481 | 0.030 | 0.500 | |||||||
| Mixteco (137) | 0.534 | 0.008 | 0.459 | |||||||
| Zapoteco (66) | 0.396 | 0.008 | 0.600 | |||||||
| MAs (463) | 0.500 | 0.50 | NS | 0.015 | 1x10-8b | 1x10-8b | 0.490 | 0.14 | NS | |
| MEZs (180) | 0.480 | 0.110c | 0.440 | |||||||
| South East | Chuj (17) | 0.577 | 6x10-3b | NS | 0.000 | 0.13 | NS | 0.469 | 0.01b | NS |
| Jakalteko (40) | 0.647 | 0.039 | 0.363 | |||||||
| Kanjobala (29) | 0.362 | 0.000 | 0.643 | |||||||
| Kaqchikel (37) | 0.457 | 0.014 | 0.600 | |||||||
| Mam (45) | 0.615 | 0.012 | 0.377 | |||||||
| Maya (252) | 0.437 | 0.045 | 0.526 | |||||||
| Mocho (15) | 0.571 | 0.000 | 0.400 | |||||||
| Tojolabal (46) | 0.500 | 0.000 | 0.511 | |||||||
| MAs (452) | 0.490 | 0.62 | NS | 0.030 | 1x10-8b | 1x10-8b | 0.490 | 0.05 | NS | |
| MEZs (63) | 0.470 | 0.160 | 0.400 | |||||||
| Total | MAs (1704) | 0.480 | 2x10-3b | 0.01 | 0.030 | 1x10-8b | 1x10-8b | 0.500 | 1x10-8b | 1x10-8b |
| MEZs (1980) | 0.440 | 0.150 | 0.430 | |||||||
a Ethnicities with divergent frequencies; not considered in the comparison.
b P values show the regions having significant differences.
c Decreased frequency from the North to South.
*P values (<0.05) after Bonferroni correction.
AF = Allele frequency
Fig 4. Geographic distribution of ADRB2 alleles in five regions among MAs and MEZs.
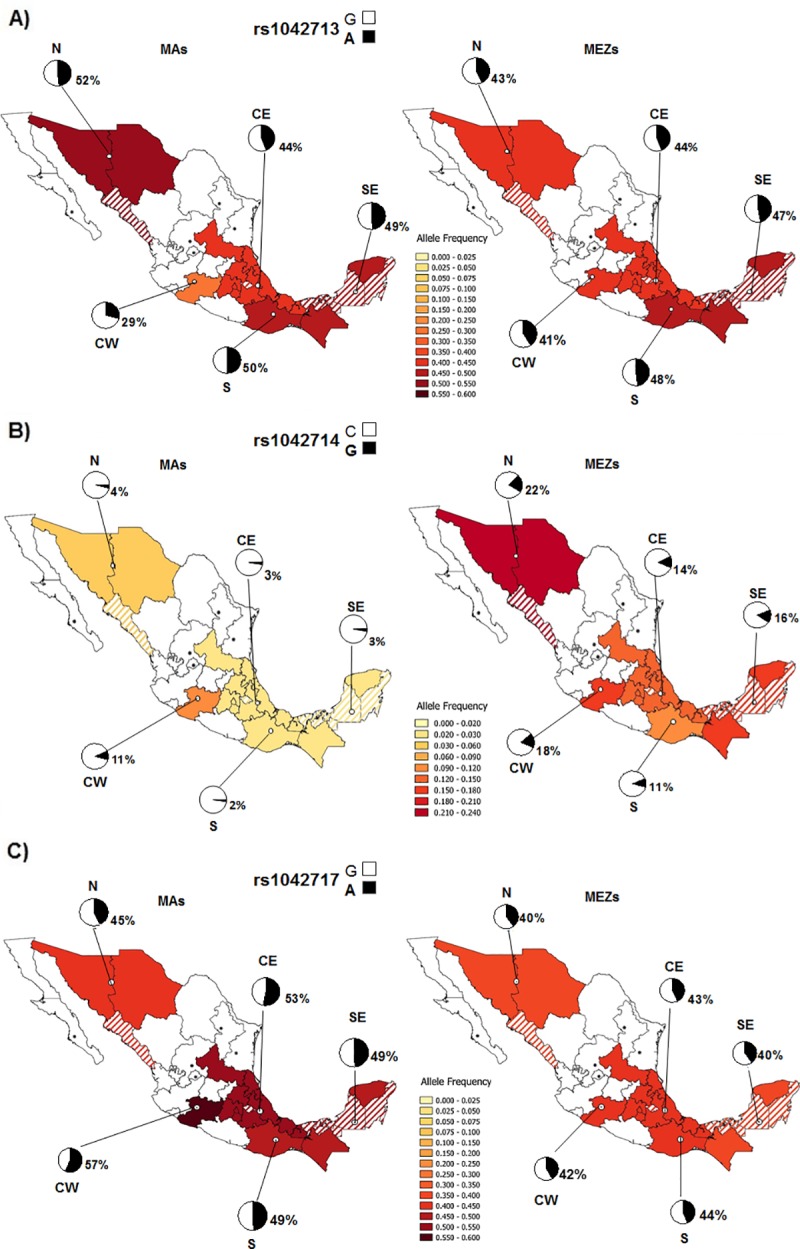
A) rs1042713A; B) rs1042714G; and C) tag SNP rs1042717A; North (N), Central East (CE), Central West (CW), South (S) and South East (SE). Striped States were not sampled because they are inhabited by neighboring indigenous included in this study. *States without indigenous population.
We investigated whether the geographic parameters of latitude, longitude and altitude might influence the distribution of the variants analyzed in this study. Our results indicated that only the frequency of the Glu27 (G) allele of rs1042714 showed a tendency of a significant negative correlation with longitude (P = 0.05), exhibiting a decreasing frequency from West to East.
Discussion
After the initial out-of-Africa expansion, the combination of human long-range migration, genetic adaptations to changing environments, and admixture have led to great differences in the genetic structures of human groups with different ancestries [16,31–33]. Several studies demonstrate how these genetic differences influence people’s susceptibility to developing a diversity of chronic diseases, generating potential group-specific genetic risk factors [34]. It has been proposed that ancestral variants that conferred selection advantages during the early development of human populations may become maladaptive under current environmental conditions [35]. Thus, human geneticists are performing detailed investigations of the geographic distribution of genetic variations, enabling reevaluation of current models of peopling through the world, and of the importance of natural selection in determining the geographic distribution of phenotypes [33].
In the present study, we investigated frequency distributions of the alleles and genotypes of five risk-associated SNPs located in the coding region of ADRB2 within both MEZ and MA groups. Our analyses revealed great diversity in the frequency distributions of the individual variants, not only between MEZs and MAs, but also among the different studied MA groups.
Among the five analyzed variants, the Glu27 (G) allele of rs1042714G exhibited the greatest differences in frequency between MAs and other populations worldwide. MA individuals showed the lowest frequency of this G allele, reported to date. In fact, this allele was absent in most of the analyzed MA groups, with frequencies of >0.05 in only four ethnic groups (Popoluca, Chontal Oax, Purepecha, and Mayo). These frequencies were still significantly lower than those observed among MEZs (0.15) or IBS (0.43), an ancestral population of MEZ individuals. These findings are relevant because this variant, which has a Gln substituted for Glu at position 27 in the protein, shows strong association with a variety of chronic degenerative diseases, including asthma, obesity, coronary artery disease, myocardial infarction, type 2 diabetes and, more recently, with longevity and acclimatization [10,14,36–38].
Notably, the Gln27 (C) allele of rs1042714 has been considered an energy-expense allele, which may protect humans from extreme temperature changes [18]. Thus, the high frequency of Gln27 in MA individuals (97%) may have resulted from selection pressures due to extreme low temperatures during the glacial period before humans emigrated from Beringia, which worked against the ancestral Glu27 allele and favored selection of the derived Gln27 allele. The almost exclusive presence of the energy-expense Gln27 allele in the MA population similar to observations in the Japanese and Han Chinese populations (94.2% and 95.2%, respectively) supports the notion that MAs may be descendants from groups that came from East Asia, which were subjected to extreme low temperatures during the glacial period. It has been hypothesized that human dispersion in northeast Asia immediately before and after the Last Glacial Maximum most likely led to the settlement of Beringia, and ultimately of the Americas [39,40]. Similar observations have been reported with the variants -217A, 825T, and -246G in the AGT, GNB3, and ENaCα genes, respectively, which are associated with hypertension. This differential susceptibility may be due to exposure to selection pressures during human adaptation to climate change [16].
We also found that the Nahuatl from Morelos exhibited the most divergent frequencies of the five ADRB2 markers, followed by the Seris. Previous reports have described the high level of differentiation within the Seri group [3, 4], but not the high differentiation in the Nahuatl from Morelos group. Importantly, we found a high degree of population differentiation between the Nahuatl from Morelos in the Central East region and Seris from the North (0.341). Using these five SNPs, differences were higher than that previously observed between the CEU and CHB populations from the 1000 Genomes (0.108). These findings support the previously reported by Moreno et al., who by using a genome wide scan technology, also observed a higher differentiation between some Mexican Amerindian groups (Seris and Lacandon: 0.136) than that found between CEU and CHB populations (0.11) [4,41].
On limitation of this study is that we analyzed only five markers in a single gene. However the high differentiation observed among the different Amerindian ethnic groups may still be interpreted as indicating several possible events: 1) the settlement of new colonies by founder effects; 2) the presence of strong “bottlenecks”; 3) positive selection for alleles that were appropriate in the new environments; and 4) increased allele frequency due to allele surfing, a process in which a small subset of individuals expands and multiplies into an unsettled territory. Despite the highly diverse frequencies of the analyzed SNPs, we identified the same types of haplotypes among MAs and MEZs, with the exception of one low-frequency haplotype that was found only among MEZ individuals. This behavior was most likely due to the high LD observed between the three synonymous SNPs.
Of the five analyzed variants, rs1042714 was the only one to show a geographic gradient across the Mexican territory, with a decreasing frequency from North to South, among MEZ individuals but not among MAs. It is well known that MA individuals have contributed along with Caucasian and, to a lesser extent, African individual towards the generation of the current MEZ population in Mexico, with a gradient of Caucasian ancestry decreasing from North to South [3,4]. Many reports of European, African, and Asian populations suggest that ADRB2 has been subjected to either balancing selection or a selective sweep [17,18]. However, the Glu27 (G) allele of rs1042714, which is almost absent among MAs but carried at a high frequency among Europeans, may have been enriched in our population at the time that Spanish people colonized Mexico. Notably, Gorlov et al. [42] tested the hypothesis that SNPs that influence disease risk undergo positive or negative selection more frequently than the average SNP in the human genome. They suggested that diseases play a central role in human evolution, directly or indirectly influencing the population frequencies of genetic variants via hitchhiking or bottleneck effects.
In conclusion, the Gln27Glu variant in ADRB2, which is associated with a broad range of phenotypes, is an excellent paragon supporting Gorlov’s hypothesis that risk alleles may be susceptible to different selection pressures. The extreme low temperatures during the glacial period could have worked against the ancestral Glu27 allele, which suffered negative selection in groups that came from East Asia and settled the Americas, while Gln27, an energy-expense allele that may protect humans from extreme temperature changes, experienced positive selection. Moreover, our results showed that the majority of Glu27 alleles in the MEZ population seemed to be an exclusively Caucasian contribution. Consequently, in the MEZ population, the phenotypes associated with this variant could have a Caucasian heritage, whereas the traits associated with Gln27 may have a predominantly Amerindian contribution. Similar to ADRB2, other disease susceptibility genes may also undergo selection pressure. This kind of study is critical for understanding the importance of assessing the population structure and analyzing the behaviors of the genetic components of populations that harbor great diversity, such as MAs, which may contribute and influence biomedical traits in the MEZ population. The present in depth analysis of ADRB2 variants and haplotypes among MAs and MEZs improves our understanding of ethnic and individual differences in the contribution of ADRB2 to disease susceptibility within the Mexican population.
Supporting information
Geographic distribution of genotype frequencies of ADRB2 SNPs among 31 Mexican Amerindians (MAs) Ethnic Groups and Mexican Mestizos (MEZs).
(DOC)
FST values among Mexican Mestizos (MEZs), Mexican Amerindians (MAs), and five continental populations (CPs) for the ADRB2 variants analyzed in this study.
(DOC)
FST values for the ADRB2 variants; rs1042713A, rs1042714G rs1042717A, rs1042718A and rs1042719C among the Mexican Amerindian groups sorted by geographic region.
(DOC)
Acknowledgments
The authors are grateful to Olaf Iván Corro Labra and José Luis de Jesús García Ruíz from the “Comisión Nacional para el Desarrollo de los Pueblos Indígenas”. This study was submitted is part fulfillment of the PhD requirements for María Guadalupe Salas Martínez at Posgrade in Genomics Sciences Program, Universidad Autónoma de la Ciudad de México, CDMX, México.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Consejo Nacional de Ciencia y Tecnología Mexico, (CONACyT) grant numbers: S008-2014-1-233970. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript
References
- 1.Johnson NA, Coram MA, Shriver MD, Romieu I, Barsh GS, London SJ, et al. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 2011;7(12): e1002410 10.1371/journal.pgen.1002410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Instituto Nacional de Estadística y Geografía (INEGI), 2010. México. http://www.censo2010.org.mx.
- 3.Contreras-Cubas C, Sánchez-Hernández BE, García-Ortiz H, Martínez-Hernández A, Barajas-Olmos F, Cid M, et al. Heterogenous Distribution of MTHFR Gene Variants among Mestizos and Diverse Amerindian Groups from Mexico. PLoS One. 2016;11(9): e0163248 10.1371/journal.pone.0163248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Estrada A, Gignoux CR, Fernández-López JC, Zakharia F, Sikora M, Contreras AV, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344(6189): 1280–1285.28. 10.1126/science.1251688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422): 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myles S, Davison D, Barrett J, Stoneking M, Timpson N. Worldwide population differentiation at disease-associated SNPs. BMC Med Genomics. 2008;1:22 10.1186/1755-8794-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol. 2014;133(1): 16–26. 10.1016/j.jaci.2013.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wechsler ME, Castro M, Lehman E, Chinchilli VM, Sutherland ER, Denlinger L, et al. NHLBI Asthma Clinical Research Network. Impact of race on asthma treatment failures in the asthma clinical research network. Am J Respir Crit Care Med. 2011;184(11): 1247–1253. 10.1164/rccm.201103-0514OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21(11): 596–601. 10.1016/j.tig.2005.08.007 [DOI] [PubMed] [Google Scholar]
- 10.Liang SQ, Chen XL, Deng JM, Wei X, Gong C, Chen ZR, et al. Beta-2 adrenergic receptor (ADRB2) gene polymorphisms and the risk of asthma: a meta-analysis ofcase-control studies. PLoS One. 2014;9(8): e104488 10.1371/journal.pone.0104488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litonjua AA, Gong L, Duan QL, Shin J, Moore MJ, Weiss ST, et al. Very important pharmacogene summary ADRB2. Pharmacogenet Genomics. 2010;20(1): 64–69. 10.1097/FPC.0b013e328333dae6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anwar MS, Iskandar MZ, Parry HM, Doney AS, Palmer CN, Lang CC. The future of pharmacogenetics in the treatment of heart failure. Pharmacogenomics. 2015;16(16): 1817–1827. 10.2217/pgs.15.120 [DOI] [PubMed] [Google Scholar]
- 13.Masuo K. Roles of beta2- and beta3-adrenoceptor polymorphisms in hypertension and metabolic syndrome. Int J Hypertens. 2010; 832821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Wu J, Yu L. Association of Gln27Glu and Arg16Gly polymorphisms in Beta2-adrenergic receptor gene with obesity susceptibility: a meta-analysis. PloS One. 2014;9(6): e100489 10.1371/journal.pone.0100489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia K, Ding R, Zhang Z, Li W, Shang X, Yang X, et al. The association of eight potentially functional polymorphisms in five adrenergic receptor-encoding genes with myocardial infarction risk in Han Chinese. Gene. 2017;624: 43–49. 10.1016/j.gene.2017.04.045 [DOI] [PubMed] [Google Scholar]
- 16.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, Levine MA, et al. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1(6): e82 10.1371/journal.pgen.0010082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cagliani R, Fumagalli M, Pozzoli U, Riva S, Comi GP, Torri F, et al. Diverse evolutionary histories for beta-adrenoreceptor genes in humans. Am J Hum Genet. 2009;85(1): 64–75. 10.1016/j.ajhg.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takenaka A, Nakamura S, Mitsunaga F, Inoue-Murayama M, Udono T, Suryobroto B. Human-specific SNP in obesity genes, adrenergic receptor beta2 (ADRB2), Beta3 (ADRB3), and PPAR γ2 (PPARG), during primate evolution. PLoS One. 2012;7(8): e43461 10.1371/journal.pone.0043461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larocca N, Moreno D, Garmendia JV, Velasquez O, Martin-Rojo J, Talamo C, et al. Beta 2 adrenergic receptor polymorphisms, at codons 16 and 27, and bronchodilator responses in adult Venezuelan asthmatic patients. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2013;57(4): 374–378. [DOI] [PubMed] [Google Scholar]
- 20.Valencia DM, Naranjo CA, Parra MV, Caro MA, Valencia AV, Jaramillo CJ, et al. Association and interaction of AGT, AGTR1, ACE, ADRB2, DRD1, ADD1, ADD2, ATP2B1, TBXA2R and PTGS2 genes on the risk of hypertension in Antioquian population. Biomedica, 2013;33(4): 598–614. 10.7705/biomedica.v33i4.1489 [DOI] [PubMed] [Google Scholar]
- 21.Zhao L, Yang F, Xu K, Cao H, Zheng GY, Zhang Y, et al. Common genetic variants of the β2-adrenergic receptor affect its translational efficiency and are associated with human longevity. Aging Cell. 2012;11(6): 1094–1101. 10.1111/acel.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saadi AV, Gupta H, Angural A, Dhanya SK, Mony S, Oberoi D, et al. Single nucleotide polymorphisms of ADRB2 gene and their association with susceptibility for Plasmodium falciparum malaria and asthma in an Indian population. Infect Genet Evol. 2013; 20: 140–147. 10.1016/j.meegid.2013.08.026 [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Aguilar NE, Del Río-Navarro BE, Navarro-Olivos E, García-Ortíz H, Orozco L, Jiménez-Morales S. SPINK5 and ADRB2 haplotypes are risk factors for asthma in Mexican pediatric patients. J Asthma. 2015; 52(3): 232–239. 10.3109/02770903.2014.966913 [DOI] [PubMed] [Google Scholar]
- 24.Vargas-Alarcón G, Fragoso JM, Cruz-Robles D, Vargas A, Martinez A, Lao-Villadóniga JI, et al. Association of adrenergic receptor gene polymorphisms with different fibromyalgia syndrome domains. Arthritis Rheum. 2009;60(7): 2169–2173. 10.1002/art.24655 [DOI] [PubMed] [Google Scholar]
- 25.Messina Baas O, Pacheco Cuellar G, Toral-López J, Lara Huerta SF, Gonzalez-Huerta LM, Urueta-Cuellar H, et al. ADRB1 and ADBR2 gene polymorphisms and the ocular hypotensive response to topical betaxolol in healthy Mexican subjects. Curr Eye Res. 2014;39(11): 1076–1080. 10.3109/02713683.2014.900807 [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3): 559±575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rousset F. genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8(1): 103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 28.Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO), cartographer División política estatal 1:250000.2005. Available from: http://www.conabio.gob.mx/informacion/metadata/gis/destdv250k_2gw.xml?_httpcache=yes&_xsl=/db/metadata/xsl/fgdc_html.xsl&_indent=no.
- 29.R Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: (2013). URL: http://www.R-project.org/. [Google Scholar]
- 30.Ordoñez G, Romero S, Orozco L, Pineda B, Jiménez-Morales S, Nieto A, et al. Genomewide admixture study in Mexican Mestizos with multiple sclerosis. Clin Neurol Neurosurg. 2015;130: 55–60. 10.1016/j.clineuro.2014.11.026 [DOI] [PubMed] [Google Scholar]
- 31.Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, Tamiya G, et al. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet. 2004;74(5): 898–916. 10.1086/420793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75(6): 1059–1069. 10.1086/426406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30(9): 377–389. 10.1016/j.tig.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimentidis YC, Abrams M, Wang J, Fernandez JR, Allison DB. Natural selection at genomic regions associated with obesity and type-2 diabetes: East Asians and sub-Saharan Africans exhibit high levels of differentiation at type-2 diabetes regions. Hum Genet. 2011;129(4): 407–418. 10.1007/s00439-010-0935-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansley MK, Ivy JR, Bailey MA. ISN Forefronts Symposium 2015: The Evolution of Hypertension-Old Genes, New Concepts. Kidney Int Rep. 2016;1(3):197–203. 10.1016/j.ekir.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang DW, Liu M, Wang P, Zhan X, Liu YQ, Zhao LS. ADRB2 polymorphisms predict the risk of myocardial infarction and coronary artery disease. Genet Mol Biol. 2015; 38(4): 433–443. 10.1590/S1415-475738420140234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomar A, Malhotra S, Sarkar S. Polymorphism profiling of nine high altitude relevant candidate gene loci in acclimatized sojourners and adapted natives. BMC Genet. 2015;16: 112 10.1186/s12863-015-0268-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulminski AM, Culminskaya I, Ukraintseva SV, Arbeev KG, Land KC, Yashin AI. Beta2-adrenergic receptor gene polymorphisms as systemic determinants of healthy aging in an evolutionary context. Mech Ageing Dev. 2010;131(5): 338–345. 10.1016/j.mad.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidd JR, Friedlaender F, Pakstis AJ, Furtado M, Fang R, Wang X, et al. Single nucleotide polymorphisms and haplotypes in Native Americanpopulations. Am J Phys Anthropol. 2011;146(4): 495–502. 10.1002/ajpa.21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505(7481): 87–91. 10.1038/nature12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International HapMap 3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311): 52–58. 10.1038/nature09298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorlov IP, Gorlova OY, Amos CI. Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases. PLoS Genet. 2015;11(7): e1005371 10.1371/journal.pgen.1005371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic distribution of genotype frequencies of ADRB2 SNPs among 31 Mexican Amerindians (MAs) Ethnic Groups and Mexican Mestizos (MEZs).
(DOC)
FST values among Mexican Mestizos (MEZs), Mexican Amerindians (MAs), and five continental populations (CPs) for the ADRB2 variants analyzed in this study.
(DOC)
FST values for the ADRB2 variants; rs1042713A, rs1042714G rs1042717A, rs1042718A and rs1042719C among the Mexican Amerindian groups sorted by geographic region.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.