SUMMARY
Technological improvements enable single-cell epigenetic analyses of organ development. We reasoned that high-resolution single-cell chromatin accessibility mapping would provide needed insight into the epigenetic reprogramming and transcriptional regulators involved in normal mammary gland development. Here, we provide a single-cell resource of chromatin accessibility for murine mammary development from the peak of fetal mammary stem cell (fMaSC) functional activity in late embryogenesis to the differentiation of adult basal and luminal cells. We find that the chromatin landscape within individual cells predicts both gene accessibility and transcription factor activity. The ability of single-cell chromatin profiling to separate E18 fetal mammary cells into clusters exhibiting basal-like and luminal-like chromatin features is noteworthy. Such distinctions were not evident in analyses of droplet-based single-cell transcriptomic data. We present a web application as a scientific resource for facilitating future analyses of the gene regulatory networks involved in mammary development.
Graphical Abstract
In Brief
The ability to deconstruct complex tissues into their constituent cell states and identify molecular mechanisms involved in cell differentiation is enabling deeper understanding of normal development and disease. Chung et al. use snATAC-seq to agnostically determine the chromatin states correlated with cell-state changes during embryonic and postnatal mammary development.
INTRODUCTION
The specialized functions of tissues require the coordinated activities of diverse differentiated cell types derived from stem or progenitor antecedents (Donati and Watt, 2015). The epigenetic programming of stem cells enables them to either retain their multi-potentiality or differentiate into the specific cell types. In some cases, epigenetic reprogramming allows cells to gain developmental plasticity to repair tissue injury (Ge and Fuchs, 2018). Determining the epigenetic and molecular programs that generate unique cell identities or developmental plasticity is critical for understanding the mechanisms for generating cell-type heterogeneity during normal tissue homeostasis and for enabling repair after injury. Perturbation of these mechanisms by oncogene activation, tumor suppressor loss, and inflammatory stimuli likely contributes to the cell-state reprogramming increasingly observed during the progression of many cancers (Feinberg et al., 2016; Kawamura et al., 2009; Koren et al., 2015; Schwitalla et al., 2013; Van Keymeulen et al., 2015).
The mammary gland is an excellent system for studying mechanisms of cellular specification because of its accessibility; the dramatic changes it undergoes in embryogenesis and postnatal development in response to puberty, pregnancy, and involution; and the substantial knowledge gained about factors involved in these cell-state transitions (Inman et al., 2015; Makarem et al., 2013; Veltmaat et al., 2003). However, there is also considerable debate on the nature of the mammary stem cells that generate and sustain the gland and on the mechanisms for establishing the basal and luminal cell lineages (Visvader and Stingl, 2014). One model proposes that bipotent mammary stem cells arise during embryogenesis (herein called fetal mammary stem cells [fMaSCs]) and that they generate basal, luminal progenitor (LP), and mature luminal (ML) populations that are postnatally maintained by lineage-restricted progenitors (Davis et al., 2016; Giraddi et al., 2015; Van Keymeulen et al., 2011; Wuidart et al., 2016). But the precise time and mechanisms by which fMaSC bipotency becomes luminally or basally restricted remains unknown. Based on recent lineage-tracing studies, it has been proposed that basal and luminal lineage specifications occur before birth (Elias et al., 2017; Lilja et al., 2018; Wuidart et al., 2018) but epigenetic and molecular profiling evidence for the existence of embryonic cell populations poised to adopt these lineages has not been presented.
One way of determining when primitive, undifferentiated embryonic cells acquire characteristics of lineage-committed cells is to use agnostic single-cell molecular profiling. Analysis of large cell populations isolated from different developmental stages using single-cell RNA sequencing (scRNA-seq) combined with bioinformatic analyses to generate lineage relationships and pseudotime developmental trajectories has been used for this purpose. One recent scRNA-seq study analyzed hundreds of embryonic day (E) 18 mammary cells by both droplet-based and C1 sequencing strategies. These analyses showed that these cells, which have the highest in vitro and in vivo fMaSC activity, comprise a single diffuse transcriptomic cluster, with most cells sharing characteristics of both basal and luminal cells, as might be expected of undifferentiated bipotent cells (Giraddi et al., 2018). An independent study using a limited number of E14 cells for RNA-seq came to a similar conclusion about the mixed-lineage nature of the bipotent cells and showed that the E14 cells could be traced into adult luminal and basal cells (Wuidart et al., 2018). Pseudotime analyses produced a trajectory in which the E18 cluster generated a basal subset and a LP subset shortly after birth. The LP was then inferred to generate a ML component when analyzed in the pre-pubertal adult (Giraddi et al., 2018). This study was consistent with an independent analysis that focused on postnatal and adult cells (Bach et al., 2017), but it differed from the results of another study (Pal et al., 2017), which concluded that a uniform, basally oriented cell cluster was present after birth and that this basal cluster generated the luminal lineages. However, the latter results are not consistent with the luminal-specific lineage-tracing studies that show the lack of basal contributions to the luminal cell populations postnatally (Elias et al., 2017; Lilja et al., 2018; Van Keymeulen et al., 2011; Wuidart et al., 2018, 2016).
The inability of scRNA-seq to reveal evidence of cells with basal or luminal characteristics is not an argument against their existence; rather, it may reflect technical or computational limitations such as low numbers of reads per cell and potential for regulatory factors expressed at low transcript numbers to fall below the detection threshold (dropouts). We therefore sought another strategy to bypass these limitations. We performed single-nucleus assay for transposase-accessible chromatin (ATAC) sequencing (snATAC-seq) for efficient and high-quality profiling of chromatin accessibility at single-cell resolution. Because chromatin accessibility is less transient than transcript expression, and because it indicates regulatory potential rather than the current cell state as implicated by the transcriptome, we reasoned that analyzing chromatin-state profiles may better separate cell types than gene expression alone (Corces et al., 2016; Shema et al., 2019). We therefore wanted to determine whether snATAC-seq enables greater resolution of cell-state heterogeneity in mammary tissues. The large sample size of single-cell studies also allows for the application of machine-learning techniques that can identify cell-type-specific genes and regulatory mechanisms that are involved in establishing mammary cell-state heterogeneity (Pott and Lieb, 2015). Here we agnostically profile mammary cells at different developmental stages using snATAC-seq to determine whether this approach clarifies the timing, extent, and underlying molecular mechanisms associated with lineage specification in mammary development. The data and strategies described provide a resource for future epigenetic studies of mammary cell regulation, a catalog of upstream control elements containing binding sites for cell-state-determining transcription factors, computational approaches that provide finer distinction of mammary cell states, and a pseudotime progression of mammary differentiation.
RESULTS
Profiling the Chromatin Accessibility Landscape at Single-Cell Resolution during Mammary Development
We used a combinatorial indexing-assisted single-cell ATAC sequencing (ATAC-seq) strategy to interrogate chromatin accessibility in mammary tissue at single-cell resolution spanning the interval from late embryogenesis to the adult (single-nucleus ATAC-seq [snATAC-seq]) (Figure 1A) (Cusanovich et al., 2015; Preissl et al., 2018). This approach allows scalable profiling of thousands of single nuclei while maintaining sufficient read depth to obtain high-quality chromatin accessibility data. All cell populations were first purified using fluorescence-activated cell sorting (FACS) to obtain EpCAM+, Lin− cells, which enabled removal of non-epithelial stromal and blood cells (Figure S1A). The aggregated fetal and adult single-nucleus profiles revealed nucleosomal fragmentation patterns, high correlation between biological replicates, and good signal-to-noise levels (Figures S1B–S1D; Table S1). Importantly, the snATAC-seq data showed enrichment of accessible regions that were previously identified by our bulk ATAC-seq profiling of FACS-enriched mammary cells analyzed at the same fetal and adult stages (Dravis et al., 2018) (Figure S1E).
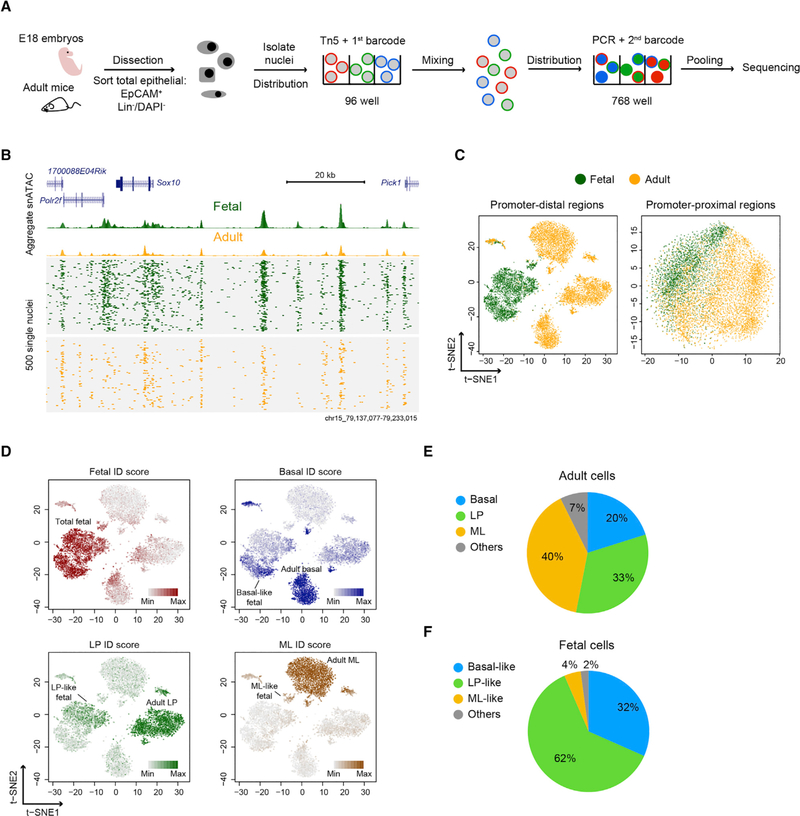
Figure 1. snATAC-Seq in Fetal and Adult Mammary Epithelial Cells.
(A) Overview of snATAC-seq experimental strategy.
(B) Aggregate ATAC-seq profile (top) and single-nucleus ATAC-seq profile (bottom) of mammary cells. Reads from 500 randomly selected cells are plotted to represent the single-nucleus profile.
(C) t-SNE representation of the snATAC-seq profile generated from either distal regions (>±1.5 kb transcription start site [TSS]) or promoter regions (<±1.5 kb TSS). The plots shown were derived by combining two biological replicates.
(D) Single-cell ID score of major mammary cell types overlaid on the snATAC-seq profile. Cell cluster annotations are shown.
(E) Cellular composition of regions adult cell population derived from snATAC-seq data.
(F) Cellular composition of regions adult-like cell types in the fetal cell population.
See also Figures S1–S3 and Table S1.
We employed a computational framework that has previously been shown to be able to identify cell clusters in diverse tissues to analyze snATAC-seq data (Cusanovich et al., 2015; Preissl et al., 2018). We first filtered out low-quality nuclei with stringent selection criteria, including read depth per cell (>2,000) and percentage of reads in peaks (>20%) (Figure S1F). This resulted in 7,846 high-quality single nuclei derived from 2,577 fetal and 5,269 adult cells (Figure 1B). For all replicates, the median reads per nucleus ranged from 4,420 to 7,488, and the median reads in peaks ranged from 69% to 74%. We then used the aggregate snATAC-seq profile from each replicate to determine open chromatin regions. This revealed 21,179 promoter-proximal regions (<±1.5 kb transcriptional start site [TSS]) and 145,453 distal regions (R±1.5 kb TSS). A 1.5 kb cutoff distance from TSS was chosen according to the mean read coverage around TSS (Figure S1G). Within both promoter-proximal and promoter-distal regions, we constructed a single-cell binary matrix of chromatin accessibility and then performed latent semantic indexing (LSI) and dimension reduction for signal normalization and processing (Figure S2A) (see STAR Methods). Because previous single-nuclei analyses demonstrated that promoter-distal regions more effectively resolve distinct clusters of cell types than promoter-proximal regions (Preissl et al., 2018), we also used t-Distributed Stochastic Neighbor Embedding (t-SNE) to ascertain patterns of relatedness of snATAC-seq data derived from promoter-distal regions (Figure 1C) (van der Maaten and Hinton, 2008). The two biological replicates analyzed reproducibly resulted in one fetal and three adult major cell clusters (Figure S2B). Such cluster structures were not observed using only promoter-proximal snATAC-seq data or using control shuffled nuclei profiles (Figures 1C and S2C). E18 cells were also better grouped into subclusters using promoter-distal regions versus analyses combining both promoter-distal and promoter-proximal regions (Figures 1C and S2D). These results show that the cell clusters observed are highly specific and are consistent with previous reports showing that accessibility at promoter-distal elements is more specific to cell type than accessibility at promoter-proximal regions (Corces et al., 2016; Shlyueva et al., 2014; Wu et al., 2016).
Identification of Major Mammary Cell Types from the snATAC-Seq Profile
Density peak clustering on cells after t-SNE identified 14 clusters, some of which represent subclusters within major cell types (Figures S2E and S2F). We excluded clusters 10 and 14, because their unusually high read count distributions suggest they may result from barcode collision events (Figure S2G). To annotate the remaining 12 cell clusters, we examined the chromatin accessibility at cell-type-specific open and closed regions that we had previously identified and verified using FACS-purified mammary cell populations and bulk ATAC-seq data (Dravis et al., 2018). We used these data to calculate a single-cell ID score for fetal, adult basal, LP, and ML cells. Higher ID scores represent greater similarity to the reference cell type (see STAR Methods). We determined which cell clusters correspond to fetal, adult basal, LP, and ML populations by overlaying the four ID scores onto the t-SNE (Figures 1D and S2H). We inferred from this approach that adult basal, LP, and ML cells represent 20%, 33%, and 40% of the total adult mammary population, respectively (Figure 1E). These estimates are consistent with previous FACS-based studies (Shackleton et al., 2006; Stingl et al., 2006).
Interestingly, and in contrast to other cell types that possess relatively tight cluster structures, the described approach previously separated fetal cells into three subclusters (two large and one small) (Figure 1D). Although each of the three clusters possesses fetal characteristics, they can be distinguished by separate enrichment with adult basal, LP, and ML ID scores. The basal-, LP-, and ML-like fetal cells represent 32%, 62%, and 4% of the total fetal population, respectively (Figure 1F). This separation of fetal cells into adult-like subclusters is also evident when the data are visualized using uniform manifold approximation and projection (UMAP) (Figure S3) (McInnes et al., 2018), another dimensionality reduction method that preserves lineage pairwise distances of embedding better than t-SNE (Becht et al., 2018). Altogether, these results show that the snATAC-seq data are of high quality, that they can reveal cell types based on chromatin accessibility, and that they provide finer distinctions among fetal cells than previously possible using scRNA-seq.
E18 Fetal Mammary Cells Show Features Consistent with Partial Lineage Specification
Our use of cell ID score (Figure 1D) indicates that fetal cells can be subdivided into distinct clusters with some adult-like features. To validate this observation and investigate lineage relatedness, we calculated a basal-to-luminal score by combinatorial analysis of the top basal and luminal accessible genes (see STAR Methods). As expected, our analysis shows strong enrichment of a basal score in basal cells and a luminal score in ML cells (Figure 2A). LP cells mostly have an intermediate-to-luminal status, suggesting that these are luminal cells that possess some basal characteristics, which is consistent with our previous scRNA-seq analyses (Giraddi et al., 2018). Interestingly, the three subclusters of fetal cells show clear indications of acquiring basal-, LP-, or ML-like gene accessibility (Figure 2A).
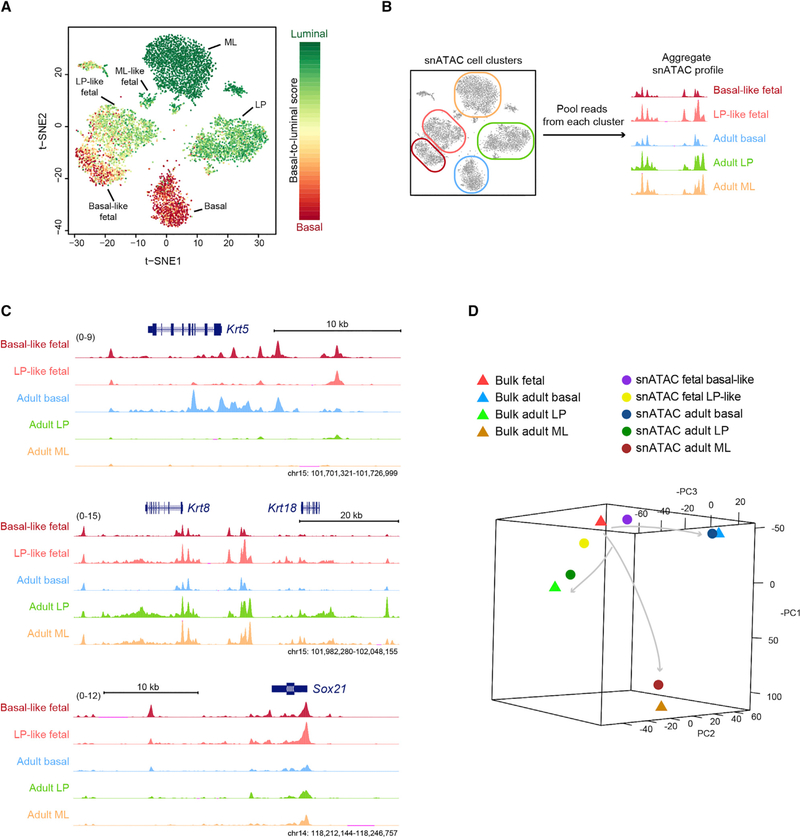
Figure 2. Fetal Mammary Cells at E18 Show Epigenetic Features of Partial Lineage Specification.
(A) Single-cell basal-to-luminal score overlaid on the snATAC t-SNE plot. Cell cluster identities are shown.
(B) Approach to generating an aggregate snATAC profile based on single-cell clustering.
(C) Signal tracks of the aggregate snATAC profile. Signal ranges are shown in the parentheses.
(D) PCA comparing the aggregate snATAC profile with bulk ATAC-seq of sorted mammary populations. Arrows indicate putative mammary differentiation paths.
See also Figure S4.
We also separated reads based on the single-cell clustering and aggregated them to compare the cell cluster profiles as aggregated bulk (ML-like fetal cells were not analyzed, because the cell number is too low to obtain an accurate aggregate signal) (Figure 2B). We found that the basal-like fetal cells are more accessible at basal markers, including Krt5 and Acta2, while LP-like fetal cells are more accessible at LP and pan-luminal markers, such as Krt8, Krt18, and Kit (Figures 2C and S4). Both fetal subclusters are accessible at fetal-enriched genes, such as Sox21 and Sox10. We next compared our aggregate snATAC-seq profile with our previously published bulk ATAC-seq data using principal-component analysis (PCA) (Dravis et al., 2018). We found strong concordance between the aggregate snATAC-seq profiles and our previous bulk ATAC-seq profiling (Figure 2D). Interestingly, these data also indicate that LP-like and basal-like fetal cells have indeed moved toward the adult LP and basal chromatin state, respectively, although they still remain close to bulk fetal cells (Figure 2D). This suggests that fetal cells at this stage of mammary development are starting to acquire adult-like chromatin accessibility, but they still largely possess their fetal-specific features. These results indicate that the E18 fetal cell epigenetic landscape is partially specified into states similar to the three major adult cell types. This type of poised landscape would position the cells to differentiate rapidly into the corresponding cell types after birth, as has been observed using RNA-seq and immune fluorescence analyses (Giraddi et al., 2018).
Single-Cell Transcription Factor Dynamics of Mammary Development
Transcription factors and the programs they control mediate many cell specification events. We therefore wanted to use the snATAC-seq data to infer potential transcriptional regulators of cell-state control during mammary development. We used chromVar, a package designed for analyzing sparse snATAC-seq data by inferring transcription actor (TF) activity using variability of TF DNA binding motif enrichment at accessible chromatin regions (Schep et al., 2017). The chromVar package calculates a TF Z score, which infers for each single cell the TFs that are binding to open chromatin regions based on the TF motifs present within these regions. Our analysis reveals that TFs previously known to function in regulating mammary cell state are highly enriched in their corresponding cell clusters. For example, Nfix and Sox10 are enriched in fetal cells, while Elf5, Foxa1, and P63 are enriched in LP, ML, and basal cells, respectively (Figure 3A). The TF Z scores of these major factors also highly correlate with their mRNA expression (Figure 3B), suggesting their relevance as transcriptional regulators. In addition, TF Z scores for members of the Smarcc, Fos/Jun, Fox, P63, Nf1, nuclear factor κB (NF-κB), and Sox family TFs are significantly variable for the single mammary cells when compared with the permutated background (Figure S5A; Table S2), suggesting that these factors may contribute to cellular differentiation programs.
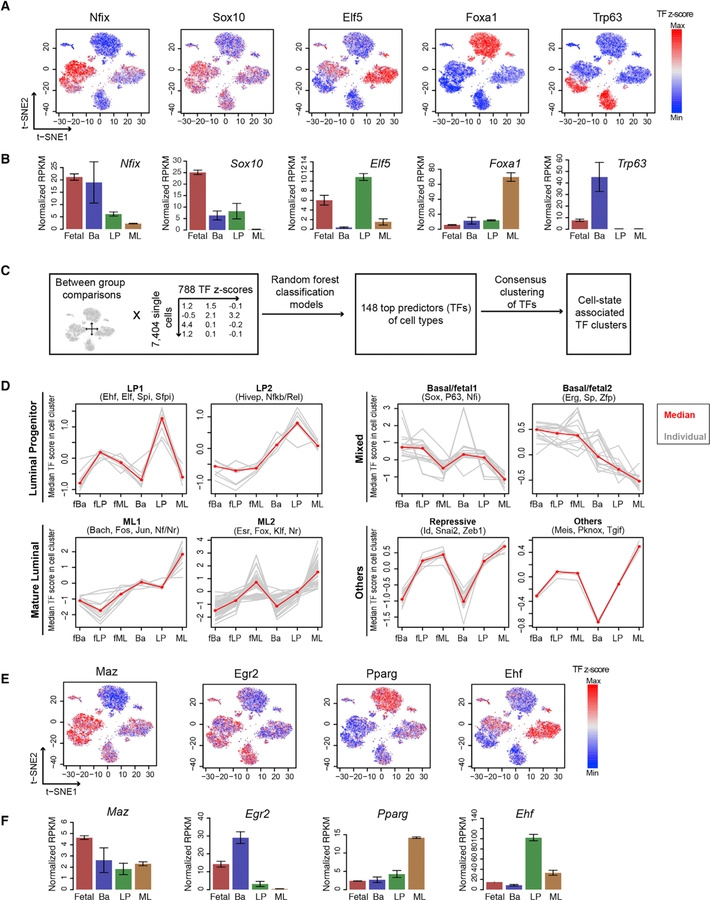
Figure 3. Single-Cell Transcription Factor Dynamics during Mammary Development.
(A) TF Z score calculated from chromVar overlaid on the snATAC t-SNE plot.
(B) RNA-seq expression of TFs from (A). Mean ± SEM (n = 2).
(C) Computational framework to identify cell-state-predictive TFs.
(D) TF Z score profile in cell types for the 8 TF clusters, grouped into luminal progenitor, mature luminal, mixed basal/fetal, and other TFs. Individual (gray) and median (red) TF Z scores in cell type are shown. Examples of a TF family in a cluster are shown on the top.
(E and F) TF Z score (E) and RNA-seq expression (F) of identified mammary cell-state factors that are less known. Mean ± SEM (n = 2).
See also Figures S5 and S6 and Table S2.
The preceding cell ID score and aggregate snATAC-seq analyses indicate that E18 fetal cells can be separated into clusters with basal-, LP-, and ML-like features. Supporting this observation, orthogonal analysis from the TF Z score shows that each of the three fetal cell subclusters is enriched with TFs associated with adult LP cells (Elf5), basal cells (P63), or ML cells (Foxa1), despite all of these cells retaining significant fetal-like representation of embryonic factors such as Nfix (Figure 3A).
To systematically identify TFs that are associated with each mammary cell state, we employed a bioinformatic framework that leverages the large sample size of single-cell datasets by combining machine learning and data clustering (see STAR Methods) (Figure 3C). We used random forest, a high-accuracy and low-predictor bias machine-learning approach (Geurts et al., 2009), to extract TFs that are important in predicting whether a cell belongs to the fetal, basal, LP, or ML cluster. To improve the accuracy of the classification, cells corresponding to the stromal/mesenchymal cluster (cluster 6, Figure S2F) or that likely result from barcode collisions (clusters 10 and 14, Figure S2F) were removed before applying random forest. The random forest model was tuned to ensure optimal accuracy (Figures S5B–S5D). This resulted in 148 TFs as strong predictors of cell state (Table S2). We then performed consensus clustering on these TFs based on their TF Z scores across all single cells. Using unsupervised cluster number selection, we identified 8 major TF clusters (Figures S5E and S5F) that can largely be separated into five categories: LP, ML, basal/fetal mix, repressive, and other (Figure 3D; Table S2). Many factors were co-enriched in fetal and basal cells, suggesting the similarity of the transcriptional regulatory landscape of these two cell types. Consistent with previous findings (Dravis et al., 2018, 2015), members of the Nf1 and Sox families are enriched in basal/fetal clusters, while Elf and Fox, Fos/Jun-related factors are enriched in LP and ML clusters, respectively (Figures 3A and 3D).
We arranged these TFs into single-cell correlation networks to help visualize the similarities among the TFs, their corresponding clusters, and the cell-type annotations (Figure S6A). Specifically, ML- and basal/fetal-associated factors are at opposite ends of the network, indicating they are highly dissimilar. LP-specific factors are centrally positioned, suggesting an intermediate level of relatedness along the luminal-basal differentiation axis. Of note, although some transcriptional repressors, such as Snai2, Id3/4, and Zeb1, cluster with other luminal factors, these TFs likely function as basal factors by serving as repressors of luminal characteristics, as indicated by their chromatin accessibility profiles and previous reports (Phillips et al., 2014). Interestingly, in addition to the more well-known factors, we identified TFs that have not typically been associated with mammary cell state. Examples of these include Maz, Sp2, and Zfp148 in fetal cells; Egr2/4 and Cebpb in basal cells; Pparg and Meis1 in ML cells; Ehf in LP cells; Zfp105 in all luminal cells; and NF-αB1 and Rela in all adult cells (Figures 3E and S6B). In support of the inferences made by the preceding computational approach, we found that the mRNA expression patterns of some of these factors correlate well with their TF Z scores (Figure 3F). The TF mRNA expression data also suggest that these TFs may be subject to cell-type-specific regulation and may contribute to cell-type determination. The combination of TF transcriptome data (Dravis et al., 2018; Giraddi et al., 2018), single-cell TF profiles, and chromatin accessibility data presented here provides a valuable resource for future characterization of candidate mammary cell-state regulators. However, we note that members within the same TF family may have similar DNA binding motif profiles, and thus may not always be distinguishable, and that functional validation will be required for more precise assignment of regulatory roles.
Mammary Differentiation Trajectory Inferred by Pseudotime Ordering of Single-Cell TF Profile
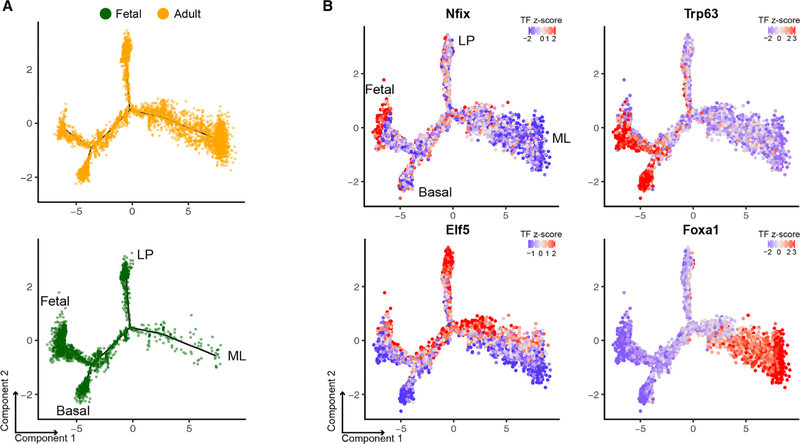
A local dimensionality reduction method such as t-SNE enables cluster identification in single-cell data. However, t-SNE does not reveal cell-state developmental trajectories that occur during tissue development. We thus used the Monocle 2 algorithm to organize single mammary cells into a pseudotime trajectory based on their TF profiles (see STAR Methods) (Qiu et al., 2017). Monocle 2 does not assume branch number and learns a single-cell trajectory in a fully unsupervised manner. We again used our cell ID scores to identify the likely state of each branch and endpoint (Figure S7A). Our analysis reveals a continuum of developmental states between E18 fetal cells and postnatal luminal and basal cells, in which the LP state branches off halfway between the fetal and the ML state. The closest cell state to fetal is basal, followed by LP and ML (Figure S7B). This is consistent with previous reports that LP and basal cells share transcriptional and epigenetic similarities with fetal cells (Dravis et al., 2018). Cells of the intermediate state are also observed spanning end states, demonstrating the levels of heterogeneity and potential plasticity within the mammary gland. Interestingly, we observed that some fetal cells are already entering the basal, intermediate, or LP state but rarely the ML state (Figure 4A), which is concordant with the cell ID score and TF Z score analyses. Superimposing the TF Z score onto the pseudotime trajectory, we observed enrichment of state-specific factors in their associated cell type (Figure 4B). We also applied Monocle 2 pseudotime ordering to our previously generated mammary gland scRNA-seq data (E16, E18, postnatal day [P] 4, and adult stages) (Giraddi et al., 2018). This resulted in a similar pseudotime trajectory (Figures S7C–S7E). Altogether, these observations validate our approach and show that the differentiation trajectory projected by this algorithm is consistent across different analytical approaches.
Figure 4. Pseudotime Ordering of Single-Cell TF Profile Infers the Mammary Differentiation Trajectory.
(A) Fetal (green) and adult (yellow) cells along the DDRTree pseudotime trajectory. The cell state associated with each branch is indicated.
(B) Representative TF Z score profile overlaid on the pseudotime trajectory plot.
See also Figure S7.
Predicting cis-Regulatory Chromatin Interactions in Mammary Cells
Chromatin accessibility changes at distal enhancers are generally more strongly correlated with cell type than changes at gene promoters. Such changes can illuminate regulatory enhancers, suggest factors involved in altering gene expression to elicit phenotype, and indicate target genes of such regulation. Deciphering which of the many putative upstream elements comprise regulatory enhancers and which of the potential targets they control is crucial for gaining a more precise understanding of the epigenetic and transcriptomic underpinnings of cell-state regulation. However, the nearest-gene approach of mapping enhancer-promoter interactions is suboptimal, because enhancers can be separated by significant distances from genes (Corces et al., 2018). Moreover, critical enhancer-promoter interactions may involve bypassing adjacent genes to reach the intended target.
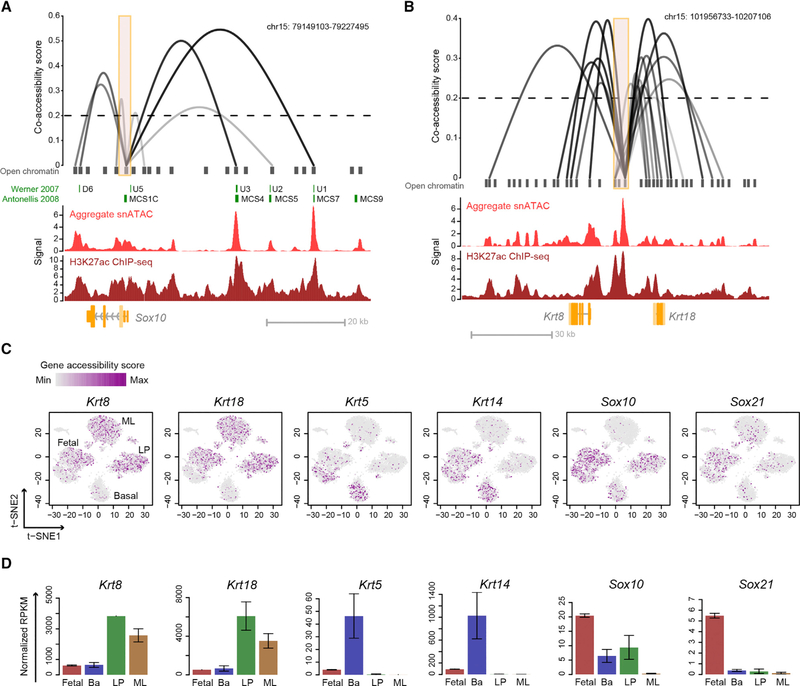
We used the Cicero algorithm to identify putative enhancergene interactions relevant to cell-state determination (Pliner et al., 2018). Cicero uses single-cell chromatin co-accessibility to predict a genome-wide cis-regulatory map that is concordant with chromatin interaction data. Cicero identified 111,604 putative sites above the co-accessibility threshold of 0.2 in our snATAC-seq database. To validate the predicted co-accessible sites, we focused on the Sox10 locus, because Sox10 enhancers have been functionally characterized in the developing mouse embryo and in transgenic zebrafish (Antonellis et al., 2008, p. 10; Betancur et al., 2010). We found that many enhancers that have strong putative co-accessibility with the Sox10 promoter were functionally verified as the main drivers of Sox10 expression (Figure 5A). These strong enhancers also overlap with the histone activation mark histone H3 lysine 27 acetylation (H3K27ac) from published E18 epithelial cell chromatin immunoprecipitation sequencing (ChIP-seq) data (Dravis et al., 2018) (Figure 5A), indicating that the Cicero-predicted sites correlate with sites of known regulatory importance. In addition to the Sox10 locus, we identified putative co-accessible sites at important mammary cell-state indicator genes. For example, we found a large enhancer cluster enriched with the activating H3K27ac mark connected to both Krt8 and Krt18 genes (Figure 5B). This observation suggests the potential importance of this enhancer cluster in regulating luminal cell state. We also found putative distal sites that may be important in regulating genes associated with LP (Kit) and basal (Krt5 and Krt14) cell states (Figure S8). Collectively, these data indicate how chromatin accessibility mapping can serve as a valuable resource to predict factors relevant to mammary cell-state determination.
Figure 5. Putative cis-Regulatory Interactions and Gene Accessibility Derived from snATAC-Seq Data.
(A and B) Linkage plots of Cicero-predicted cis-regulatory interactions at Sox10 (A) and Krt8 and Krt18 (B) loci (orange boxes indicate Sox10 promoter region or Krt8 and Krt18 putative enhancer region). Previously characterized Sox10 enhancers are shown in green. Signal tracks from aggregate snATAC-seq and H3K27ac ChIP-seq of fetal cells are shown. The height and opaqueness of the loop corresponds to the co-accessibility score between two linked elements. Dotted lines indicate the co-accessibility cutoff.
(C) Single-cell gene accessibility score of cell markers overlaid on the snATAC t-SNE plot. Cell cluster identities are annotated.
(D) RNA-seq expression of genes from (C) in mammary cell types. Mean ± SEM (n = 2).
See also Figure S8.
Quantifying Genome-wide Gene Accessibility at Single-Cell Resolution
We used the inferred chromatin interaction map to profile the overall accessibility of each gene by quantifying the accessibility of all enhancers and their linked gene. This approach yielded a gene accessibility score for 18,938 individual genes across 7,846 single cells. We observed that expressed genes are generally associated with higher gene accessibility scores, whereas most genes that are not expressed in mammary cells (reads per kilobase of transcript per million mapped reads [RPKM] < 3; see STAR Methods) are associated with near-zero gene accessibility scores (Figure S9A). We filtered out genes that are not expressed in mammary cells based on RNA-seq, resulting in accessibility profiles for 11,327 genes (see STAR Methods). Superimposing the gene accessibility score on the single-cell t-SNE plot shows enrichment of luminal markers Krt8 and Krt18 in both LP and ML cells, while basal markers Krt5 and Krt14 are enriched in basal cells (Figure 5C). In addition, the fetal-associated genes Sox10 and Sox21 are most accessible in fetal cells (Figure 5C). In each of these cases, the chromatin accessibility score correlates well with mRNA expression (Figure 5D). These data validate the utility and specificity of the gene accessibility score as a resource to examine genome-wide gene accessibility. Importantly, we observed inverse enrichment of luminal (Krt8 and Krt18) and basal (Krt5 and 14) markers in the fetal subclusters that show characteristics of adult lineages (Figure 5C).
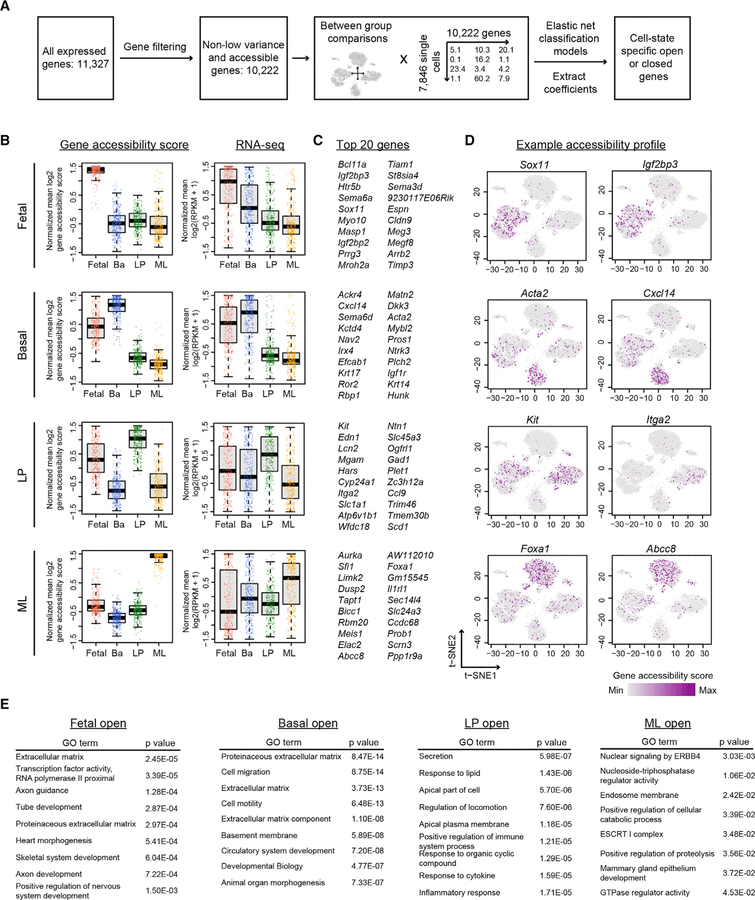
Identification of Mammary Cell-Type-Specific Accessible Genes
To identify genes that are specifically open or closed in each mammary cell type, we employed a machine-learning strategy to find genes whose accessibility can predict cluster identity (see STAR Methods). We used elastic net (Zou and Hastie, 2005), a penalized model that is suitable for high-dimensional data learning but is more computationally efficient than random forest in processing the large number of genes presented here (Figures 6A and S9B). Our approach identified top genes that are specifically open or closed in fetal, adult basal, LP, and ML cells (Figures 6C and S9D; Table S3). The accessibility of these genes also correlates well with gene expression (Figures 6B and S9C), suggesting a connection between the chromatin accessibility and the cell-type-specific gene expression pattern. Interestingly, many basal- or LP-specific genes also have increased levels of accessibility in fetal cells, again indicating the partial basal and LP specification of fetal cells and the epigenetic relatedness of these cell types. Cluster 6 (Figure S2F), identified earlier, was unusual in that it includes both fetal and adult cells but is distinct from the known epithelial cell types. We found that genes involved in extracellular matrix (ECM) interactions and collagen formation are enriched in this cluster (Figure S10; Table S3). Given that this cluster also generally lacks accessibility at epithelial cytokeratins (Figure 5C), we infer that it represents stromal cell contamination or an unknown mesenchymal-like population. Validation of this analytical method came from our identification of previously known cell-type-specific genes, such as Sox11 in fetal cells, Acta2 in basal cells, Kit in LP cells, and Foxa1 in ML cells (Figure 6D, left). We found distinct cell-type-correlated genes such as Igf2bp3 in fetal cells, Cxcl14 in basal cells, Itga2 in LP cells, and Abcc8 in ML cells (Figures 6C and 6D; Table S3). Gene Ontology (GO) analyses of these gene groups show that fetal cells are open with genes related to ECM, transcriptional activity, and tissue development; basal cells are open with genes associated with ECM, migration, and motility; LP cells exhibit accessibility in genes related to secretion, lipid, and stimulus responses; and ML cells have open chromatin in genes linked to signaling processes and mammary gland development and closed chromatin in genes involved in cell motility, lipid response, and ECM, all of which are terms associated with basal and LP identity (Figures 6E and S9E; Table S3).
Figure 6. Identification and Analysis of Mammary Cell-Type-Specific Accessible Genes.
(A) Computational framework to identify cell-type-specific open or closed genes.
(B) Boxplot of the gene accessibility score and RNA-seq expression of the top 300 accessible genes in fetal, basal, LP, and ML cells. Each dot is one gene, the thick horizontal middle line is the median, the height of the box is the interquartile range (IQR), and the dotted vertical line is 1.5 × IQR.
(C and D) Top 20 genes (C) and their representative accessibility profile (D) from (B).
(E) Gene Ontology (GO) analysis of the top 300 accessible genes from each cell type. The p values are Bonferroni corrected for multiple testing.
See also Figures S9–S12 and Table S3.
To identify differentially accessible genes between basal-like and LP-like fetal cells, we used random forest instead of elastic net because of the relatively smaller number of cells in these two fetal clusters. Using a stringent cutoff based on checking the ranked gene accessibility profile on the t-SNE plot, we selected the top 65 most important genes as differentially accessible signature genes for basal-like and LP-like fetal cells (Table S3). Our approach successfully identified previously known basal lineage-associated genes, such as Trp63, Krt5, and Acta2, and luminal lineage-associated genes, such as Ehf, Krt8, Krt18, and Krt19. By overlaying the enrichment score of the basal-like fetal signatures and LP-like fetal signatures on our previously generated 103 scRNA-seq data of E16, E18, P4, and adult mammary cells, we found that although E18 cells remain in one tight cluster, they start to show signs of basal and luminal lineage bifurcation (Figures S11A and S11B). In contrast, fetal E16 cells remain in a single population with no significant enrichment of these basal-like and LP-like signatures. Similar signature enrichment analysis of the top open or closed gene signatures of fetal E18, adult basal, adult LP, and adult ML cells confirmed that the gene accessibility scores could accurately predict major mammary epithelial types and identify cell-state-associated genes (Figure S11C; Table S3). This validates our snATAC-seq analytical approaches and, more importantly, indicates the ability of snATAC-seq to better resolve cell states during development. In addition, we found cell surface genes that exhibited differential accessibility between basal-like and LP-like fetal cells (Figure S11D) and were differentially transcribed between basal-like and LP-like fetal E18 cells (Figure S11E). They may serve as candidate markers for separating the fetal subpopulations for future functional studies.
Integration of snATAC-Seq and scRNA-Seq Data Using Cicero Gene Accessibility Scores
snATAC-seq and scRNA-seq reveal two distinct aspects of gene regulation. snATAC-seq profiles chromatin accessibility, which provides a leading indicator of the potential to express a set of genes. scRNA-seq profiles transcript abundance, which reflects the current or past state of gene activity. Therefore, it is reasonable to expect that snATAC-seq and scRNA-seq may reveal distinct profiles, especially during periods of rapid developmental change. To further evaluate the overall correlation of snATAC-seq and scRNA-seq in terms of predicting cell states, we used the recently published Seurat3 package (Stuart et al., 2019) in an attempt to develop a unified dataset based on similarities between single-nucleus ATAC (snATAC) gene accessibility scores and scRNA-seq expression profiles (see STAR Methods). Dimension reduction by UMAP on the unified dataset revealed high concordance between the methods in terms of predicting major mammary cell types (Figure 7). To confirm the concordant cell clustering based on snATAC-seq and scRNA-seq, we compared the cell-type annotations derived from previous independent snATAC-seq and scRNA-seq analyses in a split view (Figure 7B). Cells corresponding to the same cell type were grouped into the same major cell clusters in the unified dataset. Importantly, E18 cells from the snATAC-seq still fall into three separate subclusters, and E18 cells identified using scRNA-seq overlapped with all three snATAC E18 subclusters. This indicates that integrating snATAC-seq data with 10x Genomics-derived scRNA-seq data improves the ability of the latter to identify cell states.
Figure 7. Integration of snATAC-Seq and scRNA-Seq Data.
(A) UMAP representation of the co-embedded snATAC and single-cell RNA (scRNA) dataset. Cells from two assays were labeled with two colors as indicated in the plot.
(B) Split view of the UMAP representation of the co-embedded snATAC and scRNA dataset by assay techniques. Cell-type annotations obtained from previous independent snATAC-seq and scRNA-seq analysis were superimposed onto each cell. Each major cell type was represented in a different color.
An Online Resource for Visualization of Mammary snATAC-Seq Data
We developed and launched a web application for our snATAC-seq data to serve as a scientific resource for further investigation of genes and regulatory mechanisms involved in mammary differentiation and to facilitate data visualization (https://wahl-lab-salk.shinyapps.io/Mammary_snATAC/). Features such as cell ID score, TF Z score, and gene accessibility score (11,327 expressed genes) across single mammary cells can be plotted with the t-SNE, UMAP, or pseudotime plots seen throughout this report. We have included the published bulk RNA-seq data for easy comparison between chromatin accessibility and gene expression. To facilitate the identification of putative enhancers of a gene of interest, we also included the Cicero co-accessible connections in our shiny app website. This plot is integrated with aggregated snATAC-seq signals derived from major cell-state-associated clusters and previously published H3K27ac ChIP-seq profiles derived from FACS-purified major mammary epithelial cell types. This resource enables users to select a gene in which they are interested to visualize Cicero-inferred putative enhancer-promoter connections and to infer tissue specificity of putative enhancers and gene activities. We expect this to be a valuable hypothesis-generating resource for the mammary gland community.
DISCUSSION
Our snATAC-seq computational framework can be scaled for larger single-cell epigenetic studies, but here we focused on two stages of mammary gland development (comprising late embryogenesis and the adult gland) and developed a reference epigenome database based on thousands of embryonic and adult virgin cell types. Analysis of the E18 embryonic cells enabled us to ascertain whether pre-natal mammary cells retain bipotent features, as evident in previous lineage-tracing studies (Van Keymeulen et al., 2011), or are poised for lineage specification before birth, as suggested by recent lineage-tracing reports (Elias et al., 2017; Lilja et al., 2018; Wuidart et al., 2018). Interestingly, our snATAC-seq analysis reveals that the E18 fMaSC population is composed of cells with luminal and basal-oriented chromatin features, with the fetal-like LP cells and fetal-like basal cells equidistant from the bulk fetal ATAC principal-component coordinate. Together with the RNA-seq data (Giraddi et al., 2018), we interpret this finding to suggest that most cells at this stage are weakly committed and biased toward either a luminal or a basal fate. This likely positions these cells to differentiate rapidly into the respective cell type in response to appropriate microenvironmental cues after birth.
Monocle 2 analysis of the snATAC-seq data is consistent with the proposal that cells exhibiting characteristics of basal and luminal cells are present before birth. The pseudotime trajectory shows two clear branches: a basal branch that generates adult basal cells and a luminal branch that generates an intermediate that coincides with the LP. ML cells then descend from the LP. This pseudotime analysis is not consistent with a prior suggestion of a basal intermediate for LPs (Pal et al., 2017), but it is consistent with our and other scRNA-seq analyses (Bach et al., 2017; Giraddi et al., 2018). Pseudotime trajectories from the adult human mammary gland suggests that adult luminal and myoepithelial cells may originate from a less differentiated bipotent MaSC population within the basal compartment (Nguyen et al., 2018). This is consistent with our observations in the Monocle 2 pseudotime trajectory based on chromVar-inferred TF activity profiles (Figure S7A), although our pseudotime data indicate that the bipotent progenitor arises in fetal development, a period that was not analyzed in the human study. It will be important to use snATAC-seq to assess earlier embryonic stages to determine when the first evidence of luminal and basal orientations occur and to deduce the cues leading to such bifurcation. Our database provides a reference resource for future analyses of the epigenomes of cells undergoing pubertal expansion (P21–P35), hormone-induced changes in the adult during estrus cycling, pregnancy-associated changes such as development of the lactation-competent gland and involution, and subtypes of breast cancer. Such studies should improve our understanding of how cellular and epigenetic heterogeneity correlate and identify the putative underlying cell-state regulators for subsequent functional validation.
The snATAC-seq pipeline we used enabled epigenetic profiling of thousands of adult mammary cells, which produced an agnostic assessment of the heterogeneity of the basal and luminal cell lineages. Studies have consistently shown that the basal mammary cells transplant to form full and functional outgrowths with an efficiency of ~1%–2%. Luminal cells largely lack this capacity. One interpretation of this observation is that basal cells are heterogeneous and contain a minority subset correlating to a bipotential progenitor that is not revealed by lineage tracing. We therefore interrogated our large snATAC-seq database of adult basal cells to try to identify either a 1%–2% subpopulation that may possess these characteristics or the predicted stem-enriched populations obtained by sorting for markers such as Lgr5, Procr, and Tspan8 (Fu et al., 2017; Plaks et al., 2013; Wang et al., 2015). However, our chromatin profiling data show that basal cells comprise one major cluster. Although there is heterogeneous expression of these markers within the basal cell fraction, our data suggest that such expression does not correspond to significant differences in net chromatin accessibility or the existence within the basal population of a subset with the characteristics of the highly plastic fMaSC population. One caveat to these conclusions is that because EpCAM selection was necessary to obtain sufficient numbers of mammary cells for our analyses, if EpCAM-negative mammary cell populations exist within fetal or adult mammary tissues, they would be missed in our analyses.
Our research group and others have previously performed single-cell transcriptomic profiling on mammary gland cells at similar stages of embryonic and adult development (Bach et al., 2017; Giraddi et al., 2018; Pal et al., 2017; Wuidart et al., 2018). In agreement with the chromatin profiling data described here, these analyses found clear distinctions among the three separate basal, LP, and ML lineages of the adult mammary gland. Interestingly, our scRNA-seq analysis of embryonic mammary cells revealed that despite significant transcriptional heterogeneity, E18 mammary cells presented as a single net population (Giraddi et al., 2018). By evaluating the enrichment of fetal-, basal-, and LP-like signature genes inferred from the Cicero gene accessibility scores, we found that although E18 cells remain in one tight cluster, they start to show signs of basal and luminal lineage bifurcation (Figure S11B). By contrast, E16 mammary cells remain in a single population with no enrichment for E18 differentially accessible basal- and LP-like signatures. It is possible that the epigenetic changes occurring at E18 have yet to be fully reflected at the transcriptomic level because of the lag time between generating an epigenetic change and consequent change in transcription. Other factors that may complicate deduction of cell state inferred by RNA include the stability of historically produced mRNAs and the production of key regulatory transcription factors below the level of detection. Consistent with the lag between change in chromatin and transcriptional readout, we note that mammary cells from P4 mice show clear separation of basal and luminal populations (Figure S11B, and see previous immune fluorescence and RNA-SCOPE analyses; Giraddi et al., 2018). Altogether, these results indicate the power of snATAC-seq to reveal developmental cell states and differentiation programs.
In our previous analyses of embryonic and adult mammary cells using bulk transcriptomic and epigenetic profiling (Dravis et al., 2018), we found that chromatin profiling indicated that LP was the adult cell type that most closely resembled the chromatin features associated with fMaSCs. This was of interest because the LP cell type has consistently been suggested as the cell of origin for the aggressive basal-like breast cancer subtype, which we have previously shown possesses transcriptomic similarities to fMaSCs (Lim et al., 2009; Molyneux et al., 2010; Spike et al., 2012). The close relationship between fMaSCs and LP cells is consistent with the single-cell chromatin analyses presented here, because we found that the adult LP profile is closer to LP-like fMaSCs and bulk fMaSCs than adult basal cells by PCA. We also found that most E18 cells have adopted LP-associated chromatin features. Interestingly, snATAC-seq revealed that LP cells uniquely exhibit several features associated with a basal cell identity, and such features are not apparent in the ML population. Thus, it is possible that LP cells may inherently possess fMaSC-like and mixed-lineage characteristics associated with cell-state plasticity. This may explain why the LP cell type appears to be the preferred cell of origin of basal-like breast cancers in several mouse models and its ability to acquire expanded cell-state plasticity in response to oncogene activation (Koren et al., 2015; Van Keymeulen et al., 2015).
The single-cell epigenetic data presented constitute a technical resource for deconstructing the cell-state heterogeneity of a developing tissue and for identifying putative regulatory elements and transcription factors involved in cell-type specification. Our chromatin profiling of individual mammary cells at embryonic and adult developmental stages, and accompanying analyses that predict transcription factor activity and gene accessibility in relation to distinct mammary cell states, provide valuable resources to discover and validate cell-state regulators. The links between mammary development and breast cancer suggest that this resource, which we have made available as a web-based app, will have significant utility in target discovery for breast cancers.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Geoffrey M. Wahl (wahl@salk.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
CD1 mice were obtained from Charles River and housed in Salk Institute animal facilities. All mice used in these studies were female. All animals were handled in accordance with Salk Institute IACUC and AAALAC approved protocols and other ethics guidelines.
METHOD DETAILS
Mammary cell isolation
Normal mammary epithelial cells were isolated based on previously described procedures. All cells were isolated from CD1 mice. E18 mammary rudiments (all rudiments), and 8-week old adult mammary glands (#4 gland only) were dissected and pooled from multiple animals into dissection media (Epicult-B Basal medium (Stem Cell Technologies #05610) supplemented with 5% FBS, pen/strep, fungizone, hydrocortisone, and collagenase/hyaluronidase (1500/500 U, Stem Cell Technologies)), and agitated with shaking for 1.5 h for the fetal tissue, and 3 h for the adult tissue at 37C. Erthyrocytes were removed with ammonium chloride exposure for 4 min on ice, followed by cell trituration with dispase (5 U) and DNase (100 ug). Final suspensions were passed through a 40 um filter to remove cell aggregates, and stored in Hank’s Balanced Salt Solution with 2% FBS for immunostaining and flow cytometry sorting. Cells were then stained with EpCAM-AF647 (BioLegend #AB_1134101) and lineage markers (Biotin-Ter-119 (BD #553672), Biotin-CD45 (BD #553078) and Biotin-CD31 (BD #553371)), with mouse Fc Block (BD #553141) on ice for 15 min, followed with lineage markers conjugated with SA-APC-Cy7 (BioLegend #405208). For cell sorting, DAPI+ and Lin+ cells were excluded, and total mammary epithelial populations were flow sorted as EpCAM+, using a BD InFlux cell sorter. Immediately after sorting, cells were resuspended in 10% DMSO, 20% FBS in DMEM and frozen at −80. To obtain sufficient cells (> 200k per sample) for the snATAC-seq protocol, each adult biological replicate was pooled from sorted cells of three adult mice, while each fetal biological replicate was pooled from sorted cells of approximately 80 fetuses.
Combinatorial barcoding assisted single-nucleus ATAC-seq
Combinatorial ATAC-seq was performed as described previously with modifications (Cusanovich et al., 2015; Preissl et al., 2018). For the fetal samples, mammary epithelial cells from roughly 65 E18 embryos were pooled for each biological replicate. For the adult samples, cells from 3 8-week old mice were pooled per biological replicate. For each sample two biological replicates were processed. Cells were thawed and pelleted in a swinging bucket centrifuge (500 × g, 5 min, 4°C; 5920R, Eppendorf). Cell pellets were resuspended in 250 μl nuclei permeabilization buffer (10 mM Tris-HCl pH 7.4 (Sigma), 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL-CA630 (Sigma), 0.1% Tween-20, and 0.01% Digitonin (Promega, G9441), cOmplete (Roche) in water) (Corces et al., 2017), incubated for 5 min at 4°C with rotation and pelleted again (500 × g, 5 min, 4°C; 5920R, Eppendorf). Nuclei were resuspended in 500 μL high salt tagmentation buffer (36.3 mM Tris-acetate (pH = 7.8), 72.6 mM potassium-acetate, 11 mM Mg-acetate, 17.6% DMF) and counted using a hemocytometer. Concentration was adjusted to 2000 nuclei/9 ml, and 2,000 nuclei were dispensed into each well of a 96-well plate - 32 wells for each of the adult replicates and 16 wells for the fetal replicates, respectively. All downstream pipetting steps were performed on a Biomek i7 Automated Workstation (Beckman Coulter). For tagmentation, 1 μL barcoded Tn5 was added (Picelli et al., 2014), mixed and incubated for 60 min at 37°C with shaking (500 rpm). To inhibit the Tn5 reaction, 10 μL of 40-mM EDTA were added to each well and the plate was incubated at 37°C for 15 min with shaking (500 rpm). Next, 5 μL 5 × sort buffer (5% BSA, 5 mM EDTA in PBS) were added. All wells were combined and filtered using a 30-μm CellTric (Sysmex) into a FACS tube and stained with 3 μM Draq7 (Cell Signaling). Using a SH800 (Sony), 20 nuclei were sorted per well into eight 96-well plates (total of 768 wells) containing 10.5 μL EB (25 pmol primer i7, 25 pmol primer i5, 200 ng BSA (Sigma)). After addition of 1 μL 0.2% SDS, samples were incubated at 55°C for 7 min with shaking (500 rpm). We added 1 μL 12.5% Triton-X to each well to quench the SDS and 12.5 μL NEBNext High-Fidelity 2 × PCR Master Mix (NEB). Samples were PCR-amplified for 12 cycles (72°C 5 min, 98°C 30 s, (98°C 10 s, 63°C 30 s, 72°C 60 s) × 11, held at 72°C). After PCR, all wells were combined. Libraries were purified according to the MinElute PCR Purification Kit manual (QIAGEN) and size selection was performed with SPRI Beads (Beckman Coulter, 0.55x and 1.5x). Libraries were quantified using a Qubit fluorimeter (Life technologies) and the nucleosomal pattern was verified using a Tapestation (High Sensitivity D1000, Agilent). The library was sequenced on a HiSeq2500 sequencer (Illumina) using custom sequencing primers, 25% spike-in library and following read lengths: 50 + 43 + 37 + 50 (Read1 + Index1 + Index2 + Read2). The library was sequenced twice to obtain enough reads depth.
Single-nucleus ATAC-seq data processing and cluster analysis
All bioinformatic analyses were performed with bash script and R. Fastq files from the two sequencing runs were merged. Pair-end sequencing reads were trimmed with sickle (https://github.com/najoshi/sickle) to remove low quality base pairs, and mapped to mm10 reference genome with bowtie2 with the following parameters: bowtie2 -p 16 -t -X 2000–no-mixed–no-discordant (Langmead et al., 2009). Data processing, which includes low quality reads filtering (MAPQ < 30), removal of duplicates and mitochondrial reads, adjusting for Tn5 insertion and separation of single-cell reads, were performed with the snATAC-seq pipeline as previously described (Preissl et al., 2018) with the following parameters: snATAC pre -t 16 -m 30 -f 2000 -e 75. Open chromatin regions were determined by calling peaks for each replicate using macs2 with these parameters: macs2 callpeak–nolambda–nomodel–shift −100–extsize 200–keep-dup all–bdg–SPMR -q 5e-2 (Zhang et al., 2008). Peaks from each replicate were merged with bedtools into non-overlapping regions (Quinlan and Hall, 2010), resized to 1 kb and separated into promoter proximal (< ± 1.5 kb TSS) and distal (> = ± 1.5 kb TSS) regions. For cell selection, we filtered out cells that did not pass our quality threshold: reads per cell > = 2000 and reads in peak ratio > = 0.2. Data statistics for each biological replicate is shown in Table S1. The aggregate snATAC profile was plotted with UCSC genome browser (https://genome.ucsc.edu), and the single-cell reads profile was plotted with the R package Sushi. Average signal profile was plotted with deepTools (Ramírez et al., 2014).
To reveal single-cell clusters with proper data normalization, we adapted a previously described workflow to process the snATAC-seq data (Cusanovich et al., 2015). First, we calculated a binary matrix of cell versus open chromatin regions using the snATAC pipeline tool snATAC bmat, with 1 indicating > = 1 reads and 0 indicating < 1 reads within a specific region. We next performed latent semantic indexing and singular value decomposition, keeping only the top 50 dimensions. Of note, since we did not observe strong correlation between PC1 and read depth as described previously (Figure S2A), we kept all 50 dimensions for further analysis. Next, we performed dimension reduction with t-distributed stochastic neighbor embedding (t-SNE) (Rtsne package in R; parameters: dims = 2, perplexity = 30, max_iter = 1000, theta = 0.5, pca = FALSE, exaggeration_factor = 12; the optimal t-SNE was selected based on lowest Kullback–Leibler divergence value among 9 random runs), and uniform manifold approximation and projection (UMAP) (umap package in R; parameters: n_components = 2, n_neighbors = 15, random_state = 524). Unbiased cluster identification was performed with density peak clustering using the R package densityClust, with rho = 50 and delta = 3.5 (Rodriguez and Laio, 2014).
Our snATAC-seq procedure may generate barcode collision (Preissl et al., 2018), meaning that reads from two different cells possess the same barcode. As library collision may confound cell type identification, we excluded cell clusters whose library complexity (defined as numbers of unique reads per cell) are significantly higher than expected, as has been previously described (Cusanovich et al., 2018). To do so, we sampled the population with cell number corresponding to each cluster for 10,000 times, and plotted this background library complexity profile with 99% confidence intervals against the actual profile from each cell cluster. Clusters that have significantly higher library complexity distribution than the background were considered as barcode collision cells, and were not further analyzed.
To annotate cluster identity, we calculated single-cell normalized accessibility at our previously defined cell-type specific regions: uniquely accessible regions (UARs) and uniquely repressed regions (URRs) (Dravis et al., 2018). We first resized the UARs/URRs to 1 kb based on peak center so that downstream calculation will not be biased by peak size. We next generated a binary matrix of cells versus UARs/URRs using snATAC pipeline described above, and calculated the read depth normalized ‘cell ID score’ defined as: (sum of UAR counts / reads per cell) - (sum of URR counts / reads per cell). This cell ID score was calculated for each single-cell and for each cell type UARs/URRs, including those of fetal, basal, LP and ML cells. To reduce outlier effect and better reveal the intermediate range, the cell ID score was capped at upper and lower 10% quantile and plotted.
Transcription factor dynamics, clustering, and network analysis
TF dynamics were inferred with chromVar package as previously described (Schep et al., 2017). Open chromatin regions called from aggregate profile described above was used to calculate read counts at open chromatin. mm10 reference genome was used for correcting GC bias, and chromVar curated motif V2 (mouse_pwms_v2) was used for generating TF z-score. A total of 788 TFs were used for downstream analysis. TF variability was calculated as the variance of TF z-score across all the single cells, and it is compared against a ‘expected variability’ calculated by the mean of the same TF z-score profile permutated for 1000 times. TF z-score score was capped at upper and lower 10% quantile when plotted to reduce outlier effect and better reveal the dynamic range.
We employed a random forest classification framework to find TFs whose TF z-scores predict major cell types. We used random forest because it is highly accurate, robust to noise and is suitable for high dimension data modeling. When applied with permutation importance (rather than Gini importance), random forest variable analysis is relatively resistant to bias caused by predictor co-linearity (https://explained.ai/rf-importance/index.html#6.1). We used the R package Caret (http://topepo.github.io/caret/index.html) to individually tune our models and extract important variables, with these parameters: mtry = 5–200, ntree = 500, importance = TRUE, metric = ROC. The predictors are the 788 TFs, with read depth per cell as a covariate. The class comparisons for the model are: fetal versus all adults, basal versus all other adults, LP versus all other adults, ML versus all other adults, and basal only versus ML only. To improve accuracy, cells corresponding to the stromal contamination (cluster 6 in Figure S3B) and barcode collision doublets (cluster 10 and 14 in Figure S3B) were excluded from random forest analysis. We excluded fetal cells in the adult classification model as fetal cells show characteristics of adult cell types, and thus may confound our model. The top TFs from each model were further checked by visualization to ensure the effectiveness of the variable analysis.
To cluster TFs based on their single-cell profile, we pooled the top TFs from the random forest variable analysis to generate a list of cell-type predictive TFs, and then performed clustering on these key TFs. The pooling was done with the top 50 TFs from each classification model, except for the ‘basal only versus ML only’ model, where we selected the top 100 TFs. The pooling resulted in 148 TFs. We next performed K-medoid consensus clustering with Pearson correlation on these 148 TFs based on their chromVar TF z-score, using the R package ConsensusClusterPlus (Wilkerson and Hayes, 2010), with these parameters: maxK = 15, reps = 1000, pItem = 0.8 (80% of TFs sampled), pFeature = 0.5 (50% of cells sampled), distance = pearson, clusterAlg = pam, seed = 2018. We used cophenetic coefficient to determine the optimal cluster number. The median TF z-scores for each TF cluster in fetal-Basal-like, fetal-LP-like, fetal-ML like, basal, LP and ML cells were plotted to check the enrichment profile.
We constructed a weighted pearson correlation network with the TF z-score across the single cells, using the R package igraph (Csárdi and Nepusz, 2006). We removed weak edges that have correlation < 0.2, and used the Fruchterman Reingold algorithm to construct the network.
scRNA-seq data processing
10x scRNA-seq data were download from SRA with accession number GSE111113. Fastq files were processed with cellranger v3.0.1 using default parameters. Then the filtered feature barcode-count matrix of each stage was read into Seurat object for cell quality check and filtering. Individual Seurat object of each stage were combined into a single Seurat object by using the MergeSeurat (Butler et al., 2018) function with do.normalize = FALSE. Cell selection in Seurat2 was performed as described previously by selecting the percent of mitochondria (low.thresholds = −Inf, high.thresholds = 0.07), number of UMI per cell (low.thresholds = 3000, high.thresholds = 45000), and number of genes detected (low.thresholds = 500, high.thresholds = 7000). This resulted in 647 E16, 1174 E18, 1320 P4, and 1941 adult high quality single cells. Then the expression matrix was read depth normalized and log transformed with the NormalizeData function of Seurat package (Butler et al., 2018). Dimension reduction and visualization was done in R using the UMAP package with default parameters. Top 1000 most variable genes across all the single cells were used. Enrichment of snATAC gene accessibility derived fetal Ba-like and LP-like signature genes in each individual cell was evaluated by using the AUCell R package (Aibar et al., 2017) with the default settings. Then the fetal Ba-like and LP-like signature enrichment scores were overlaid on the UMAP dimension 1 and dimension 2 plot.
Pseudotime analysis
We used Monocle2 with the TF z-score profile for pseudotime ordering of single cells as described previously (Qiu et al., 2017). The top 200 most variable TFs across all the single cells were used for ordering cells based on the ranked TF z-score variability (Figure S6A). Dimension reduction and trajectory learning were performed with these parameters: max_components = 2, method = DDRTree, norm_method = none; the cell ordering was performed with the default settings. For scRNA-seq pseudotime trajectory, the filtered gene count matrix was imported from the processed expression matrix in the Seurat object with the importCDS function of monocle2 package (Qiu et al., 2017). The top 1000 most differentially expressed genes across 12 clusters were selected for ordering cells in pseudotime. Cluster numbers were selected based on the tSNE plots. Cells were reduced into 3 DDRTree dimensions (norm_method = “log,” reduction_method = “DDRTree,” max_components = 3) and then ordered in pseudotime with default parameters. Branch of E16 cells was set as the root.
Prediction of cis-regulatory interaction and gene accessibility analysis
Cicero analysis was performed as described previously (Pliner et al., 2018). We input Cicero with binarized chromatin accessibility at all promoter and distal regions as defined above. We used the t-SNE coordinates calculated above for dimension reduction and nearest neighbor calculation, with mouse mm10 as the reference genome. In total, we revealed 3,622,246 chromatin interactions with Cicero co-accessibility score > 0. The R package Gviz was used for plotting cis-regulatory interaction maps and genomic tracks (Hahne and Ivanek, 2016).
To calculate genome-wide gene accessibility score, we used the Cicero function ‘build_gene_activity_matrix’, with co-accessibility cutoff = 0.2. Instead of only using promoter regions, we used UTRs and exons to annotate genes and then calculated gene activity scores for each single cell. Compared to only using promoters, we found that including UTRs and exons improves the results in the following ways: 1) genes transcribed from alternative promoters are captured; 2) there is better signal coverage across single cells while not compromising cluster-specificity (examples are shown in Figure S12A); 3) more genes are covered (11327 versus 11063) and gene accessibility score of these additional genes are largely concordant with RNA-seq data (examples from the top 20 open genes are shown in Figures S12B and S12C). We filtered out non-expressed genes based on published bulk RNA-seq data (Dravis et al., 2018), resulting in 11,327 genes with accessibility scores. Non-expressed genes were defined as those that have a RPKM value < 3 in all normal and tumorigenic mammary cells. For visualization, gene accessibility scores were capped at upper and lower 5% quantiles to reduce outlier effects and better reveal those in the intermediate range.
To identify cell-type specific open or closed genes, we applied an elastic net classification framework on the gene accessibility score. We used elastic net instead of random forest as described above, because the latter is inefficient with large numbers of predictors. Although elastic net is generally less accurate and flexible than random forest, the penalization function of this algorithm allows effective variable importance analysis even when co-linearity is present. We first filtered out low variance and low accessibility genes of which their accessibility scores variance and sum across the single cells are at the bottom 5%, which results in 10,222 genes being kept. We next used the R package Caret to perform elastic net tuning with these parameters: method = glmnet, preProc = c(“center,” “scale”), tuneLength = 3, metric = ROC. The predictors are the 10,222 genes, with 7846 single cells as samples. The class comparisons for the model are: fetal versus all adults, basal versus all other adults, LP versus all other adults, ML versus all other adults, and basal only versus ML only. The top open and closed genes in each cell type are defined as those having the highest and lowest elastic net coefficient, respectively. The top genes were checked by visualization to ensure the effectiveness of the variable analysis. Gene ontology analysis was performed with ClueGO network analysis under Cytoscape (Bindea et al., 2009; Shannon et al., 2003), with these term libraries: GO molecular function, GO cellular component, GO biological process, KEGG, Reactome and Wiki pathway.
To derive the fetal Ba-like and LP-like accessible signature genes, we used random forest instead of elastic net due to the smaller number of fetal cells. To avoid potential outlier effects on data centering and scaling, we capped the Cicero gene accessibility scores at top and bottom 1% before running random forest. We used the R package Caret to tune the model and extract important variables, with these parameters: mtry = 5–200, ntree = 500, importance = TRUE, metric = ROC. Tuned mtry = 200, AUC = 0.9151 with 95%CI: 0.9037–0.926 (DeLong). Accessibility profiles of top variables were visually checked on t-SNE plot and the top 65 genes were selected as the most important signature genes to distinguish fetal Ba-like versus LP-like cells. Stringent cutoff was applied to reduce false positives. Each of these 65 genes was assigned to Ba- or LP-like class according to its mean expression in these two clusters.
To calculate the ‘basal-to-luminal score’, the top 300 basal and ML specific genes from the ‘basal only versus ML only’ model were extracted. For each cell, the score is calculated as: (sum of accessibility score at basal specific genes) / (sum of accessibility score at luminal specific genes). For visualization on the t-SNE plot, the scores were capped at upper and lower 10% quantiles.
To perform principle comportment analysis (PCA) with bulk ATAC-seq and aggregate snATAC-seq profile, we used deepTools to calculate ATAC signal enrichment of each sample/cell type at all the UARs and URRs. For normalization, the sample-wise signals were centered and scaled, while the region-wise signals were centered, and then subjected to PCA with the R function ‘svd’. The 3D PCA plot was plotted with the R package Rgl (https://cran.r-project.org/web/packages/rgl/vignettes/rgl.html).
Integration of snATAC-seq and scRNA-seq data
We used the Seurat 3 package to integrate snATAC-seq and 10x scRNA-seq data obtained from E18 and adult mammary cells (Stuart et al., 2019). Integration was performed by following the official snATAC and scRNA integration vignette (https://satijalab.org/seurat/v3.0/atacseq_integration_vignette.html). Briefly, to process the 10x scRNA-seq datasets, normalized gene expression matrix was obtained from the Seurat 2 processed scRNA-seq data above and updated into Seurat 3 object format. Only E18 and adult cells were subset out. To prepare snATAC dataset, Cicero gene accessibility score matrix after filtering out non-expressed genes (see STAR Methods above) was first stored in an “assay” of a Seurat 3 object and was subsequently processed by running FindVariableFeatures(), NormalizeData(), and ScaleData() with default parameters. QC-filtered binary peak count matrix at promoter-distal regions was processed by RunLSI(n = 50, scale.matrix = NULL) and RunUMAP(reduction = “lsi,” dims = 1:50). To identify the “anchors” between cells from the two different assays, scRNA-seq dataset was used as reference and snATAC gene accessibility score was used as query. Features parameter was defined through running VariableFeatures() on the scRNA-seq data. Reduction method was “cca.” To co-embed snATAC and scRNA datasets, we used the anchors obtained by comparing the snATAC gene accessibility scores to scRNA expression profiles. The anchors were used to impute RNA-seq values for the snATAC-seq cells (see details in Stuart et al., 2019). We then merged the measured and imputed scRNA-seq data and run a standard UMAP analysis to visualize all the cells together. To confirm the concordant clustering of the same cell types, cell-type annotations derived from previous independent snATAC and scRNA analysis were superimposed onto corresponding cells in an UMAP split view of the co-embed dataset.
Mammary snATAC-seq web application
The snATAC-seq web application is written with R Shiny and served on the Shiny server (https://www.rstudio.com/products/shiny/). Cicero predicted putative connections were filtered to only retain connections with co-accessibility scores above 0.1 to improve computing efficiency.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses, data processing, and heatmap plotting were performed in R (https://www.r-project.org/), unless otherwise noted. Details of the statistical tests used in this manuscript and the number of replicates (n) are presented in the figures and figure legends, are reiterated in the text, and described in the detail in the Method Details section above. They are also summarized below:
Background level of library complexity were determined by sampling the population with cell number corresponding to each cluster for 10,000 times. This background library complexity profile with 99% confidence intervals was compared against the actual mean profile from each cell cluster.
Expected background variability of chromVAR TF z-scores was calculated by the mean of the same TF-score profile across cells randomly permutated for 1000 times.
The single-cell correlation of chromVAR TF z-scores was calculated using weighted pearson correlation network in R package igraph. We removed edges that have correlation < 0.2, and used the Fruchterman Reingoldalgorithm to construct the network.
Random forests models were tuned using the ROC metric in R package Caret.
K-medoid consensus clustering was performed with Pearson correlation on the 148 TF z-scores in R package ConsensusClusterPlus.
Elastic net models were tuned using the ROC metric and the glmnet package in R package Caret.
DATA AND CODE AVAILABILITY
Raw and processed data have been deposited to NCBI Gene Expression Omnibus with the accession number GSE125523. Previous published 10x scRNA-seq data were download from NCBI Gene Expression Omnibus with accession number GSE111113. The basic scripts for snATAC-seq analysis can be accessed here: https://github.com/jaychung10010/Mammary_snATAC-seq
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor® 647 anti-mouse CD326 (Ep-CAM) | Biolegend | RRID:AB_1134101 |
| Biotin Rat Anti-Mouse TER-119/Erythroid Cells | BD Biosciences | Cat #: 553672; RRID:AB_394985 |
| Biotin Rat Anti-Mouse CD31 | BD Biosciences | Cat #: 553371; RRID:AB_394817 |
| Biotin Rat Anti-Mouse CD45 | BD Biosciences | Cat #: 553078; RRID:AB_394608 |
| Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block) | BD Biosciences | Cat #: 553142; RRID:AB_394657 |
| APC Cy7 Streptavidin | Biolegend | Cat #: 405208 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase/Hyaluronidase | Stem Cell Technologies | Cat #: 07912 |
| Dispase | Stem Cell Technologies | Cat #: 07913 |
| DAPI | Thermo Fisher Scientific | Cat #: 62248 |
| IGEPAL-630 | Sigma | Cat #: I8896 |
| Digitonin | Promega | Cat #: G9441 |
| Tn5 | Picelli et al., 2014 | N/A |
| DRAQ7 | Cell Signaling Technologies | Cat #: 7406 |
| Ammonium Chloride | Stem Cell Technologies | Cat #:07800 |
| Critical Commercial Assays | ||
| EpiCult-B Mouse Medium Kit | Stem Cell Technologies | Cat #: 05610 |
| Fetal Bovine Serum | Biowest | Cat #: S1620 |
| NEB Next High Fidelity 2× PCR Master Mix | New England BioLabs | Cat #: M0541 |
| MinElute PCR Purification Kit | QIAGEN | Cat #: 28004 |
| SPRI Beads | Beckman Coulter | Cat #: B23317 |
| Deposited Data | ||
| snATAC-seq FastQ files | This paper | GEO: GSE125523 |
| 10× scRNA-seq FastQ files | Giraddi et al., 2018 | GEO:GSE111113 |
| Bulk RNA-seq, ATAC-seq, and ChlP-seq sequencing files (bed, bigwig, and FastQ files) | Dravis et al., 2018 | GEO: GSE116386 |
| Experimental Models: Organisms/Strains | ||
| CD1 mice | Charles River | Strain code: 022 |
| Software and Algorithms | ||
| snATAC bioinformatics pipeline | Preissl et al., 2018 | https://github.com/r3fang/snATAC |
| Sickle 1.33 | Joshi and Fass, 2011 | https://github.com/najoshi/sickle |
| Bowtie2 | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| MACS2 | Zhang et al., 2008 | https://github.com/taoliu/MACS |
| Samtools | Li et al., 2009 | http://www.htslib.org |
| Bedtools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/ |
| UCSC genome browser | Kent et al., 2002 | https://genome.ucsc.edu |
| Sushi (R) | Phanstiel et al., 2014 | http://bioconductor.org/packages/release/bioc/html/Sushi.html |
| Deeptools | Ramírez et al., 2014 | http://deeptools.ie-freiburg.mpg.de/ |
| Rtsne v0.13 (R) | Krijthe, 2015 | https://github.com/jkrijthe/Rtsne |
| umap v0.2.0.0 (R) | McInnes et al., 2018 | https://cran.r-project.org/web/packages/umap/index.html |
| densityClust (R) | Rodriguez and Laio, 2014 | https://cran.r-project.org/web/packages/densityClust/index.html |
| Rgl (R) | Adler and Murdoch, 2019 | https://cran.r-project.org/web/packages/rgl/index.html |
| Barcode collisions identification | Cusanovich et al., 2018 | http://doi.org/10.1038/nature25981 |
| chromVAR | Schep et al., 2017 | https://github.com/GreenleafLab/chromVAR |
| Caret (R) | Kuhn, 2008 | http://topepo.github.io/caret/index.html |
| ConsensusClusterPlus (R) | Wilkerson and Hayes, 2010 | https://bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html |
| Monocle 2 (R) | Qiu et al., 2017 | http://cole-trapnell-lab.github.io/monocle-release |
| igraph (R) | Csárdi and Nepusz, 2006 | http://igraph.org |
| Cellranger v3.0.1 | 10x Genomics | https://support.10xgenomics.com/developers/software/downloads/latest |
| Seurat v2.3 (R) | Satija et al., 2015 | https://satijalab.org/seurat |
| Seurat v3.0.2 (R) | Stuart et al., 2019 | https://satijalab.org/seurat |
| AUCell | Aibar et al., 2017 | https://github.com/aertslab/AUCell |
| Cicero (R) | Pliner et al., 2018 | https://cole-trapnell-lab.github.io/cicero-release |
| Gviz (R) | Hahne and Ivanek, 2016 | https://bioconductor.org/packages/release/bioc/html/Gviz.html |
| ClueGO | Bindea et al., 2009 | http://www.ici.upmc.fr/cluego/cluegoDownload.shtml |
| Cytoscape v3.7.1 | Shannon et al., 2003 | https://cytoscape.org |
| shiny v1.3.2 (R) | Chang, 2019 | https://cran.r-project.org/web/packages/shiny/index.html |
| shinydashboard v.0.7.1 (R) | Chang, 2018 | https://cran.r-project.org/web/packages/shinydashboard/index.html |
| R v.3.5 (Mac OS X and windows 10) | The R Project for Statistical Computing | https://www.r-project.org |
| Basic snATAC-seq analysis scripts | This paper | https://github.com/jaychung10010/Mammary_snATAC-seq |
| Other | ||
| Resource website for the snATAC-seq data | This paper | https://wahl-lab-salk.shinyapps.io/Mammary_snATAC/ |
Highlights.
Performed single-nucleus (sn)ATAC-seq profiling of fetal and adult mammary cells
snATAC-seq reveals chromatin changes correlating with basal or luminal cell states
Cells with luminal- or basal-oriented chromatin features areevident before birth
A web resource for single-cell profile of embryonic and postnatal mammary development
ACKNOWLEDGMENTS
We thank Cynthia Ramos and Luke Wang for laboratory management, Grace Lee and Annie Odelson for laboratory assistance, and Conor Fitzpatrick and Caz O’Connor at the Salk Flow Cytometry Core, Max Shokirev at the Salk Bioinformatics Core, Nasun Hah at the Salk Next Generation Sequencing Core, and Nikki Lytle and Eugene Ke for critical evaluation of the manuscript. Work in the laboratory of G.M.W. was supported, in part, by a Cancer Center Core Grant (5 P30CA014195); NIH/National Cancer Institute (R35 CA197687); the Susan G. Komen Foundation (SAC110036); the Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002); the Breast Cancer Research Foundation (BCRF); the Freeberg Foundation; the Copley Foundation; and the William H. Isacoff MD Research Foundation for Gastrointestinal Cancer. C.D. was supported by CA174430. B.R. was supported by the Ludwig Institute for Cancer Research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.089.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adler D, and Murdoch D (2019). 3D Visualization Using OpenGL. R package version 0.100.30.
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J-C, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Huynh JL, Lee-Lin S-Q, Vinton RM, Renaud G, Loftus SK, Elliot G, Wolfsberg TG, Green ED, McCallion AS, and Pavan WJ (2008). Identification of neural crest and glial enhancers at the mouse Sox10 locus through transgenesis in zebrafish. PLoS Genet. 4, e1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach K, Pensa S, Grzelak M, Hadfield J, Adams DJ, Marioni JC, and Khaled WT (2017). Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun 8, 2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, and Sauka-Spengler T (2010). Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. USA 107, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, and Galon J (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W (2018). shinydashboard: Create Dashboards with “Shiny.”.
- Chang W (2019). shiny: Web Application Framework for R.
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. ; Cancer Genome Atlas Analysis Network (2018). The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csárdi G, and Nepusz T (2006). The igraph Software Package for Complex Network Research (InterJournal Complex Systems). [Google Scholar]
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, and Shendure J (2015). Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R, Pliner HA, Christiansen L, Qiu X, Steemers FJ, et al. (2018). The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FM, Lloyd-Lewis B, Harris OB, Kozar S, Winton DJ, Muresan L, and Watson CJ (2016). Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun 7, 13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, and Watt FM (2015). Stem cell heterogeneity and plasticity in epithelia. Cell Stem Cell 16, 465–476. [DOI] [PubMed] [Google Scholar]
- Dravis C, Spike BT, Harrell JC, Johns C, Trejo CL, Southard-Smith EM, Perou CM, and Wahl GM (2015). Sox10 Regulates Stem/Progenitor and Mesenchymal Cell States in Mammary Epithelial Cells. Cell Rep 12, 2035–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravis C, Chung C-Y, Lytle NK, Herrera-Valdez J, Luna G, Trejo CL, Reya T, and Wahl GM (2018). Epigenetic and Transcriptomic Profiling of Mammary Gland Development and Tumor Models Disclose Regulators of Cell State Plasticity. Cancer Cell 34, 466–482.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias S, Morgan MA, Bikoff EK, and Robertson EJ (2017). Long-lived unipotent Blimp1-positive luminal stem cells drive mammary gland organogenesis throughout adult life. Nat. Commun 8, 1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Koldobskiy MA, and Göndör A (2016). Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet 17, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu NY, Rios AC, Pal B, Law CW, Jamieson P, Liu R, Vaillant F, Jackling F, Liu KH, Smyth GK, et al. (2017). Identification of quiescent and spatially restricted mammary stem cells that are hormone responsive. Nat. Cell Biol 19, 164–176. [DOI] [PubMed] [Google Scholar]
- Ge Y, and Fuchs E (2018). Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet 19, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts P, Irrthum A, and Wehenkel L (2009). Supervised learning with decision tree-based methods in computational and systems biology. Mol. Biosyst 5, 1593–1605. [DOI] [PubMed] [Google Scholar]
- Giraddi RR, Shehata M, Gallardo M, Blasco MA, Simons BD, and Stingl J (2015). Stem and progenitor cell division kinetics during postnatal mouse mammary gland development. Nat. Commun 6, 8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraddi RR, Chung C-Y, Heinz RE, Balcioglu O, Novotny M, Trejo CL, Dravis C, Hagos BM, Mehrabad EM, Rodewald LW, et al. (2018). Single-Cell Transcriptomes Distinguish Stem Cell State Changes and Lineage Specification Programs in Early Mammary Gland Development. Cell Rep 24, 1653–1666.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F, and Ivanek R (2016). Visualizing Genomic Data Using Gviz and Bioconductor. Methods Mol. Biol 1418, 335–351. [DOI] [PubMed] [Google Scholar]
- Inman JL, Robertson C, Mott JD, and Bissell MJ (2015). Mammary gland development: cell fate specification, stem cells and the microenvironment. Development 142, 1028–1042. [DOI] [PubMed] [Google Scholar]
- Joshi N, and Fass J (2011). Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files version 1.33.
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, and Izpisúa Belmonte JC (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O, et al. (2015). PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 525, 114–118. [DOI] [PubMed] [Google Scholar]
- Krijthe J (2015). Rtsne: T-Distributed Stochastic Neighbor Embedding using a Barnes-Hut Implementation.
- Kuhn M (2008). The caret Package.
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja AM, Rodilla V, Huyghe M, Hannezo E, Landragin C, Renaud O, Leroy O, Rulands S, Simons BD, and Fre S (2018). Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cell Biol 20, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. ; kConFab (2009). Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med 15, 907–913. [DOI] [PubMed] [Google Scholar]
- Makarem M, Spike BT, Dravis C, Kannan N, Wahl GM, and Eaves CJ (2013). Stem cells and the developing mammary gland. J. Mammary Gland Biol. Neoplasia 18, 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, and Melville J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv, arXiv:1802.03426. https://arxiv.org/abs/1802.03426. [Google Scholar]
- Molyneux G, Geyer FC, Magnay F-A, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. (2010). BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417. [DOI] [PubMed] [Google Scholar]
- Nguyen QH, Pervolarakis N, Blake K, Ma D, Davis RT, James N, Phung AT, Willey E, Kumar R, Jabart E, et al. (2018). Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun 9, 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, Wilcox S, Fu N, Liu KH, Jackling FC, et al. (2017). Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun 8, 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Araya CL, and Snyder MP (2014). Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley SH, Perou CM, Gill G, et al. (2014). Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports 2, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Björklund AK, Reinius B, Sagasser S, Winberg G, and Sandberg R (2014). Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24, 2033–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, and Werb Z (2013). Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3, 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. (2018). Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell 71, 858–871.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott S, and Lieb JD (2015). Single-cell ATAC-seq: strength in numbers. Genome Biol 16, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. (2018). Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci 21, 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, and Manke T (2014). deep-Tools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, and Laio A (2014). Machine learning. Clustering by fast search and find of density peaks. Science 344, 1492–1496. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat M-L, Wu L, Lindeman GJ, and Visvader JE (2006). Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Bernstein BE, and Buenrostro JD (2019). Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet 51, 19–25. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, and Stark A (2014). Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet 15, 272–286. [DOI] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, and Wahl GM (2012). A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 10, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, and Eaves CJ (2006). Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, and Hinton G (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, and Blanpain C (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, et al. (2015). Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525, 119–123. [DOI] [PubMed] [Google Scholar]
- Veltmaat JM, Mailleux AA, Thiery JP, and Bellusci S (2003). Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation 71, 1–17. [DOI] [PubMed] [Google Scholar]
- Visvader JE, and Stingl J (2014). Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev 28, 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Cai C, Dong X, Yu QC, Zhang X-O, Yang L, and Zeng YA (2015). Identification of multipotent mammary stem cells by protein C receptor expression. Nature 517, 81–84. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, and Hayes DN (2010). ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. (2016). The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657. [DOI] [PubMed] [Google Scholar]
- Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, and Blanpain C (2016). Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev 30, 1261–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuidart A, Sifrim A, Fioramonti M, Matsumura S, Brisebarre A, Brown D, Centonze A, Dannau A, Dubois C, Van Keymeulen A, et al. (2018). Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol 20, 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, and Hastie T (2005). Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B. Stat. Methodol 67, 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data have been deposited to NCBI Gene Expression Omnibus with the accession number GSE125523. Previous published 10x scRNA-seq data were download from NCBI Gene Expression Omnibus with accession number GSE111113. The basic scripts for snATAC-seq analysis can be accessed here: https://github.com/jaychung10010/Mammary_snATAC-seq