Abstract
Subtype-specific leukemia oncogenes drive aberrant gene expression profiles that converge on common essential mediators to ensure leukemia self-renewal and inhibition of differentiation. The transcription factor c-MYB functions as one such mediator in a diverse range of leukemias. Here we show for the first time that transcriptional repression of myeloid differentiation associated c-MYB target genes in AML is enforced by the AAA+ ATPase RUVBL2. Silencing RUVBL2 expression resulted in increased binding of c-MYB to these loci and their transcriptional activation. RUVBL2 inhibition resulted in AML cell apoptosis and severely impaired disease progression of established AML in engrafted mice. In contrast, such inhibition had little impact on normal hematopoietic progenitor differentiation. These data demonstrate that RUVBL2 is essential for the oncogenic function of c-MYB in AML by governing inhibition of myeloid differentiation. They also indicate that targeting the control of c-MYB function by RUVBL2 is a promising approach to developing future anti-AML therapies.
Introduction
Acute Myeloid Leukemia (AML) is a heterogeneous disease affecting both children and adults. The use of intensive chemotherapy, risk stratification and hematopoietic stem cell transplantation have improved outcomes. However, cure rates for paediatric (60-70%), young (40-45%) and older adults (10-20%) remain poor [1, 2]. Although primary oncogenic drivers represent attractive targets for novel therapies, a complimentary approach has focussed on inhibiting the expression and activity of transcription factors, such as c-MYB, that are required to integrate oncogenic programs downstream of driving oncogenes across a broad spectrum of cancers [3, 4]. Thus, anti-AML activity has been demonstrated for small molecules and peptidomimetics that inhibit the interaction between c-MYB and the CBP/p300 transcriptional co-activator complexes [5, 6], and for drugs that target c-MYB for proteasomal degradation [7].
The AAA+ (ATPases associated with diverse cellular activities) ATPases, RUVBL1 and RUVBL2, were originally isolated as components of transcriptional complexes and shown to function in a number of different cellular processes, including transcriptional regulation, chromatin remodelling and DNA damage responses [8]. They were also found to be essential for the oncogenic activity of c-MYC [9] and for survival and progression of multiple different cancer types [8]. Recently, the RUVBL1/RUVBL2 complex was implicated in hepatocellular carcinogenesis, through amplifying the transcriptional response of E2F factors [10]. We previously showed that in MLL-rearranged AML cells, MLL-fusions are responsible for maintenance of RUVBL2 expression, mediated via transcriptional activation of the RUVBL2 gene by c-MYB [11]. Silencing expression of endogenous RUVBL2 or over-expression of a mutant RUVBL2(DN) molecule (capable of ATP binding but not its hydrolysis) in human and mouse AML cells, induced differentiation and apoptosis, and inhibited colony formation. Dependence on RUVBL2 expression was a general feature of AML cells and not limited to the MLL-rearranged subtype [11]. This is consistent with data from recent CRISPR essentiality screens in AML [12, 13].
Here, we report that inhibition of RUVBL2 expression or function impairs progression of established leukemia. In contrast, RUVBL2 function is dispensable for differentiation of normal hematopoietic progenitor cells. We demonstrate that RUVBL2 binds c-MYB and ensures that its transcriptional activity is compatible with differentiation arrest and self-renewal of AML cells by enforcing repression of a subset of c-MYB target genes. These data suggest that therapeutic targeting of RUVBL2 represents an opportunity to disrupt the c-MYB oncogenic program in AML, while sparing normal hematopoiesis.
Materials and methods
Mice
Mice were maintained in the UCL GOS ICH animal facilities and experiments were performed according to and approved by the United Kingdom Home Office regulations and followed UCL GOS ICH institutional guidelines.
Western blot and co-immunoprecipitation (Co-IP) analysis
Western blot analysis was performed as previously described [7, 14]. Antibody clone names are available in Supplementary Methods. For Co-IP analyses, THP1 and HA-RUVBL2 expressing THP1 cells were washed in cold PBS, proteins cross-linked for 10 minutes with 0.1 mM disuccinimidyl glutarate (DSG) and quenched for 10 minutes with 1 mM Tris pH7.4 on ice. Cells were washed three times with cold PBS, lysed and proteins immunoprecipitated using the Pierce Classic Magnetic IP/Co-IP Kit (ThermoFisher Scientific).
Flow cytometry and apoptosis assays
Flow cytometry was performed as previously described [11]. The antibodies and kits used are available in Supplementary Methods.
Colony formation assays
CD117+/lineage- mouse hematopoietic progenitor cells (HPC) were purified from C57BL/6-CD45.1 mouse bone marrow by magnetic-activated cell sorting, using the Lineage Cell Depletion Kit (Miltenyi Biotec) followed by positive selection using anti-CD117-PE (Biolegend) and anti-PE microbeads (Miltenyi Biotec). Colony assays were performed in M3434 (StemCell Technologies) methylcellulose.
Lentiviral and retroviral vector cloning and transduction
For inducible expression, shRNA and cDNA were cloned into the pTRIPZ (Open Biosystems/Thermo Scientific) lentiviral vector. Induction was achieved by treatment of cells every 48 hours with 1 μg/ml doxycycline (Clontech-Takara Bio). Constitutive shRNA expression was performed using MISSION pLKO.1 shRNA constructs (Sigma Aldrich), and cDNA expression using the pMSCV-hCD2T [15] and pCSGW-PIG [7] vectors. The RUVBL2(DN) [11] cDNA was described previously. The HA-RUVBL2 cDNA from pCDNA-3xHA-Reptin, a gift from Steven Artandi (Addgene plasmid # 51636) [16], was cloned into pCSGW [17]. The pCSGW-LUC2 vector was previously described [7]. Lentiviral and retroviral transductions were performed as previously described [11, 18, 19].
In vivo transplantation
Inducible shSCR and shRUVBL2 THP1 clones were transduced with a luciferase expressing lentiviral vector as previously described [7]. Cells were transplanted into NOD-SCID-γ-/- mice (NSG; The Jackson Laboratory) and imaging performed as described previously [7]. For transplantation of normal transduced mouse HPC, lethally γ-irradiated (split dose: 5Gy & 4Gy) C57BL/6 mice were injected intravenously with 2×105 C57BL/6-CD45.1 transduced HPC. Peripheral blood was analysed 27 days after transplantation and recipients sacrificed 4 months after transplantation. For mouse MLL-ENL cell transplantation, sublethally γ-irradiated (6Gy) C57BL/6 or C57BL/6J-CD45.1 mice were intravenously injected with 0.5-1×106 leukemic cells. In vivo induction was initiated with 3 intraperitoneal injections (1mg of doxycycline in 100µl of PBS, Clontech-Takara Bio) every other day, followed by continual treatment with doxycycline in the diet (625mg/kg, Harlan). For inducible RUVBL2(DN) MLL-ENL transplanted mice, doxycycline was administered in the drinking water (200µg/ml doxycycline and 5% sucrose).
Quantitative RT-PCR (qRT-PCR) analysis
Quantitative RT-PCR (qRT-PCR) was performed on isolated mRNA using TaqMan probe based chemistry and an ABI Prism 7900HT fast Sequence Detection System (Life Technologies), as previously described [7]. All primer/probe sets were from Applied Biosystems, Life Technologies.
RNA sequencing (RNA-seq) and Gene set enrichment analysis (GSEA)
Total cellular RNA was purified from control and doxycycline treated samples from three independent experiments for each time-point and submitted to UCL Genomics for RNA-sequencing, as detailed in Supplementary Methods. GSEA (https://software.broadinstitute.org/gsea/) was used to examine enrichment of gene sets for c-MYC activated target genes [20], AML LSC,[21] c-MYB target genes [7], gene expression changes following shRNA [22], CRISPR-mediated [23] and peptidomimetic [6] c-MYB targeting, PMA-induced myeloid differentiation [24, 25] and monocyte terminal differentiation [26] in gene expression changes following RUVBL2 silencing. Enrichment of the 36 genes with increased expression and c-MYB binding following RUVBL2 silencing was examined in gene expression changes following siRNA [25], CRISPR-mediated [23] and peptidomimetic [6] c-MYB targeting, and following 24 hours exposure of THP1 cells to PMA [25]. RNA-seq data is available on the GEO repository, GSE117106.
Chromatin immunoprecipitation and sequencing (ChIP-seq)
Chromatin immunoprecipitation using commercially available antibodies and sequencing of isolated DNA was performed, as detailed in Supplementary Methods. Pre-processed data were then aligned to the genome (UCSC hg19) with BWA14 and deduplicated. Peak calling was conducted using MACS1.3.3 [27] at a P-value cut-off of 10-6. Bigwig files were generated using bam2bw. Tags within a given region were counted and adjusted to represent the number of tags within a 1 kb region. Subsequently the percentage of these tags as a measure of the total number of sequenced tags of the sample was calculated and displayed as heat map or boxplot as before [28, 29]. ChIP-seq data is available on the GEO repository, GSE117224.
Statistics
Statistical significance was determined using Prism (GraphPad) software. Statistical analysis of survival curves was performed using the logrank (Mantel-Cox) test. Statistical analysis of means was performed using the one sample t test or unpaired Student’s t test, two-tailed P values < 0.05 being considered statistically significant. Variance was similar between groups. For RNA-seq analysis, statistically significant changes in gene expression were P < 0.05 using the Wald test. For analysis of H3K27ac changes on dynamic c-MYB peaks, P values were calculated using the Mann–Whitney U test.
Results
RUVBL2 inhibition impairs AML progression
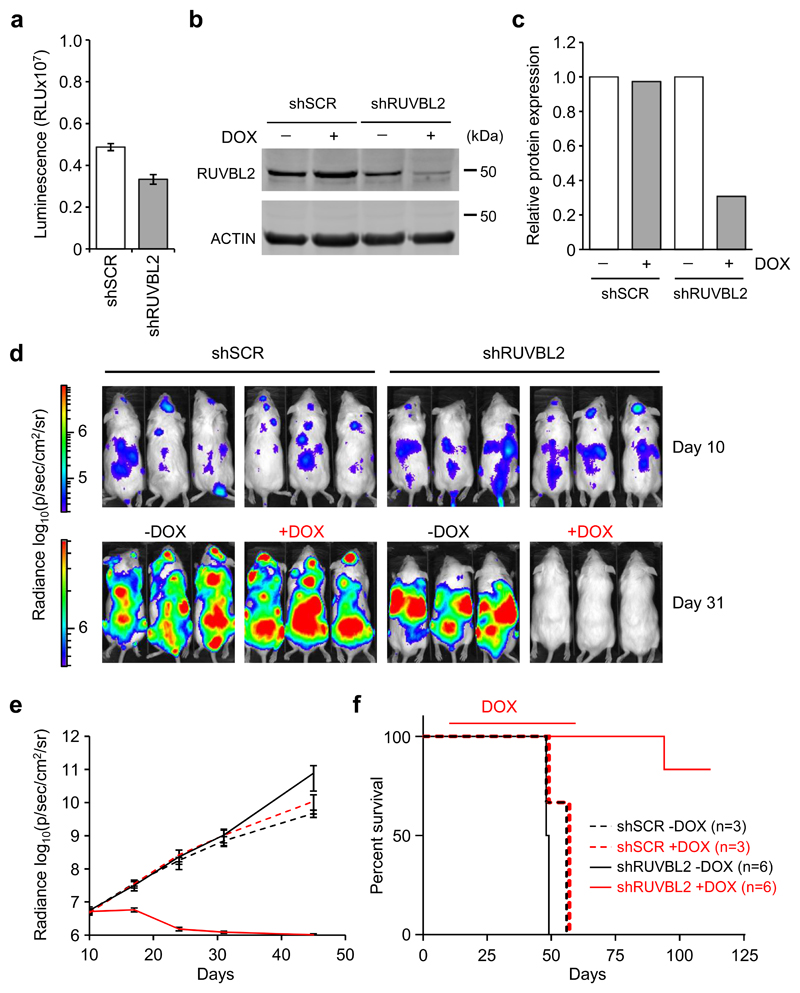
Previously, we demonstrated that silencing RUVBL2 expression with two independent shRNA resulted in AML cell differentiation and apoptosis [11]. Here, we examined whether this would result in impairment of leukemia progression in vivo. The most effective shRNA was cloned into the TRIPZ inducible expression vector and used to transduce THP1 cells. Clones were then generated from the inducible control shSCR and shRUVBL2 THP1 cells and further transduced with a luciferase-expressing lentiviral vector (Fig. 1a-c). NSG mice were transplanted with control and shRUVBL2 THP1 clones and disease progression measured by bioluminescence imaging. Ten days after transplantation, when bioluminescence signal was detected in all recipient mice confirming AML engraftment (Fig. 1d,e), doxycycline treatment of the experimental groups was initiated and maintained until day 59 post-transplantation. Whereas bioluminescence increased steadily in untreated groups and in doxycycline-treated control shSCR mice, the signal declined to background levels in shRUVBL2 mice following treatment with doxycycline (Fig. 1d,e). Disease latency was similar for the untreated groups and the doxycycline-treated shSCR mice, all mice succumbing to leukemia within 8 days of each other (Fig. 1f). In contrast, most of the doxycycline-treated shRUVBL2 mice survived through to the end of the experiment, a striking result in this aggressive disease model (Fig. 1f). Leukemia was undetectable in 4 out of the 5 surviving mice at the end of the experiment, 112 days after transplantation, with localised disease progression evident in the remaining mouse (Supplementary Figure 1).
Fig. 1. Elimination of established AML following RUVBL2 silencing.
a Luminescence signal in shSCR and shRUVBL2 THP1-LUC2 clones, prior to transplantation. b Western blot analysis of RUVBL2 protein in shSCR and shRUVBL2 clones treated with doxycycline for 3 days, or left untreated. c Quantification of RUVBL2 expression in b. Data are normalised to Actin loading control and to untreated shSCR and shRUVBL2 clones. d Bioluminescence imaging examples of NSG recipient mice 10 days after injection with shSCR or shRUVBL2 THP1-LUC2 clones, and before doxycycline treatment, (day 10, top), and 21 days later, after treatment with doxycycline (+DOX) or not (-DOX) (day 31, bottom). Bars for luminescence signal represent photons/s/cm2/steradian. e Graph depicting AML progression, measured by bioluminescence imaging, in untreated shSCR (black dashed lines, n = 3) and shRUVBL2 (black solid lines, n = 6) recipient mice or following doxycycline treatment in shSCR (red dashed lines, n = 3) and shRUVBL2 (red solid lines, n = 6) mice. There was no significant difference in bioluminescence signal between groups subjected to doxycycline treatment and those left untreated, at day 10 after transplantation, and before doxycycline treatment started (P = 0.37 for shSCR and P = 0.58 for shRUVBL2 groups), unpaired Student’s t test. f Survival curve for recipient mice in e. Red line above curves indicates length of doxycycline treatment. Survival curves were significantly different for doxycycline treated and untreated groups for shRUVBL2 mice (P = 0.0009), but not shSCR mice (P = 0.23), logrank (Mantel-Cox) test.
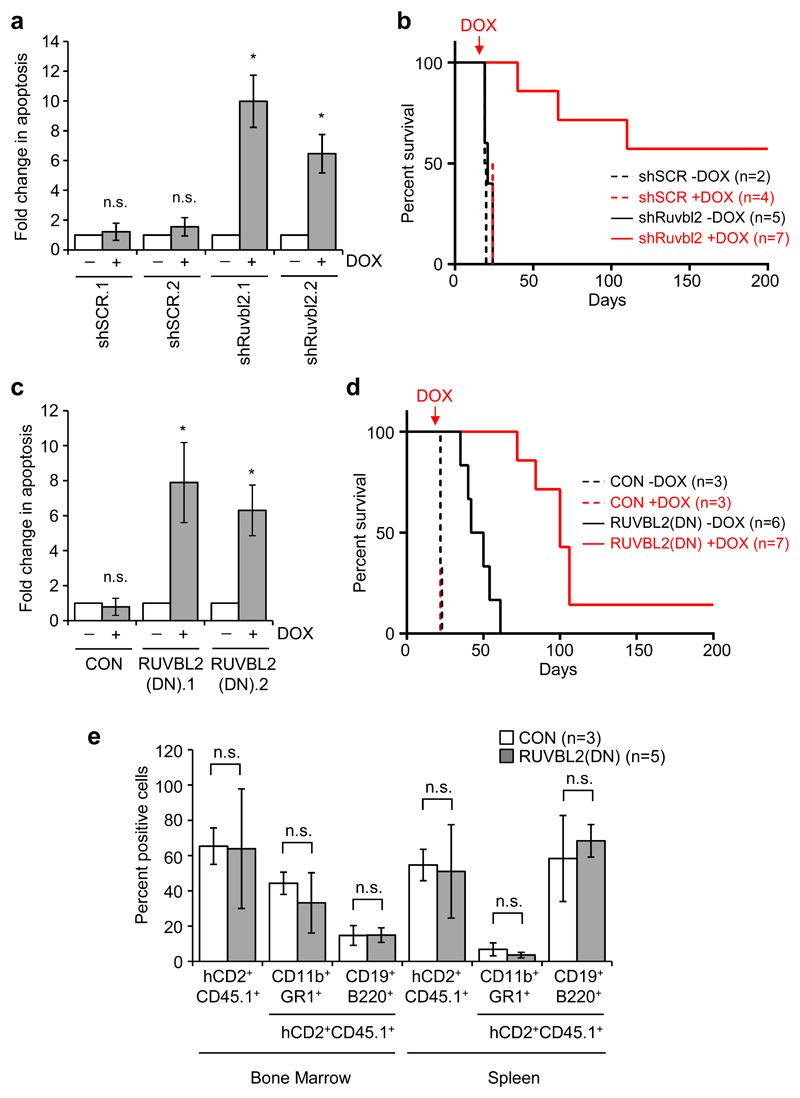
Next, we examined the impact of RUVBL2 inhibition on in vivo progression of AML using two different mouse models. In the first model, we generated MLL-ENL mouse myeloid leukemia cells, as previously described [18, 19], and transduced them with inducible TRIPZ vectors containing control shRNA or shRNA targeting mouse Ruvbl2. Treatment of shRuvbl2 MLL-ENL clones, derived from these cells, with doxycycline in vitro resulted in significant apoptosis in comparison to untreated cells or to control cells (Fig. 2a and Supplementary Figure 2a). MLL-ENL clones were also transplanted into sub-lethally irradiated mice and experimental groups were exposed to doxycycline treatment 10 days later. Disease latency was significantly impaired in shRuvbl2 transplanted mice treated with doxycycline, with more than half of the group surviving the course of the experiment (Fig. 2b).
Fig. 2. RUVBL2 inhibition impairs AML progression but not normal hematopoiesis.
a Apoptosis induction in mouse MLL-ENL clones, inducibly expressing control (shSCR) or mouse Ruvbl2-specific (shRuvbl2) shRNA 6 days after doxycycline treatment. Bars and error bars represent means and SD of fold changes in apoptosis (annexin V positive cells) from three independent experiments. *P < 0.05; n.s. not significant (relative to untreated cells), one sample t test. b Survival curves for recipient mice transplanted with shSCR.1 (dashed lines) and shRuvbl2.2 (solid lines) clones, untreated (black lines, n = 2 for shSCR.1 and n = 5 for shRuvbl2.2) or treated with doxycycline (red lines, n = 4 for shSCR.1 and n = 7 for shRuvbl2.2). Red arrow indicates point at which doxycycline treatment started. P = 0.0002 for doxycycline treated versus untreated shRuvbl2 mice, logrank (Mantel-Cox) test. c Apoptosis induction in inducible control (CON) and human RUVBL2(DN) mouse MLL-ENL clones 48 hours after doxycycline treatment. Bars and error bars represent means and SD of fold changes in apoptosis from three independent experiments. *P < 0.05; n.s. not significant (relative to untreated cells), one sample t test. d Survival curves for recipient mice transplanted with CON (dashed lines) and RUVBL2(DN).1 (solid lines) clones, untreated [black lines, n = 3 for CON and n = 6 for RUVBL2(DN).1] or treated with doxycycline [red lines, n = 3 for CON and n = 7 for RUVBL2(DN).1]. Red arrow indicates point at which doxycycline treatment started. P = 0.0005 for doxycycline treated versus untreated RUVBL2(DN) mice, logrank (Mantel-Cox) test. e Percentages of total (hCD2+CD45.1+), myeloid (hCD2+CD45.1+CD11b+GR1+) and B lymphoid (hCD2+CD45.1+CD19+B220+) transduced donor cells in the bone marrow and spleen of recipient mice, four months after reconstitution with mouse HPC, transduced with control (CON) or RUVBL2(DN) expressing lentiviral vectors. Bars and error bars are means and SD of percentages from three control and five RUVBL2(DN) mice. n.s. = not significant, unpaired Student’s t test.
Since the capacity of RUVBL2 to bind and hydrolyse ATP is central to many of its diverse cellular functions [8], we next examined the effect of targeting the RUVBL2 ATPase activity on in vivo disease progression. MLL-ENL cells were transduced with the inducible TRIPZ vector, containing RUVBL2(DN), or empty vector control, and clones derived. The D299N point mutation in RUVBL2(DN) abrogates the ability of RUVBL2 to hydrolyse bound ATP. We demonstrated previously that over-expression of this mutant acts in a dominant negative manner over normal RUVBL2 function in AML cells in vitro [11]. Exposure of inducible RUVBL2(DN) cells to doxycycline in vitro resulted in rapid induction of apoptosis (Fig. 2c and Supplementary Figure 2b). Inducible clones were transplanted into sub-lethally irradiated recipient mice and after 14 days treatment of mice with doxycycline was initiated. Although doxycycline treatment made no impact on AML latency in mice transplanted with empty vector control MLL-ENL cells, consistent with previous results [19], disease progression of RUVBL2(DN) was significantly impaired by doxycycline treatment (Fig. 2d). Importantly, analysis of shRuvbl2 and RUVBL2(DN) leukaemia cells harvested from doxycycline treated mice that did succumb to disease had lost their ability to induce shRNA or RUVBL2(DN) expression (Supplementary Figure 2c, d).
In contrast to the deleterious effects of RUVBL2(DN) expression in AML cells, normal HPC expressing RUVBL2(DN) exhibited robust myeloid colony forming activity in vitro, similar to empty vector transduced HPC (Supplementary Figure 3a,b). Furthermore, neither short-term nor long-term hematopoietic reconstitution were significantly altered by expression of the RUVBL2(DN) mutant, recipient mice exhibiting equivalent reconstitution of both myeloid and B cell compartments of bone marrow and spleen to those transduced with empty vector transduced control HPC (Supplementary Figure 4a,b and Fig. 2e). This is consistent with our previous demonstration that RUVBL2 silencing has significantly less impact on the proliferation of normal human HPC than AML cells [11]. Collectively, these data indicate that AML cells are more sensitive than normal HPC to inhibition of RUVBL2 function, and that a therapeutic window exists for its therapeutic targeting.
RUVBL2 regulates expression of c-MYB target genes in AML cells
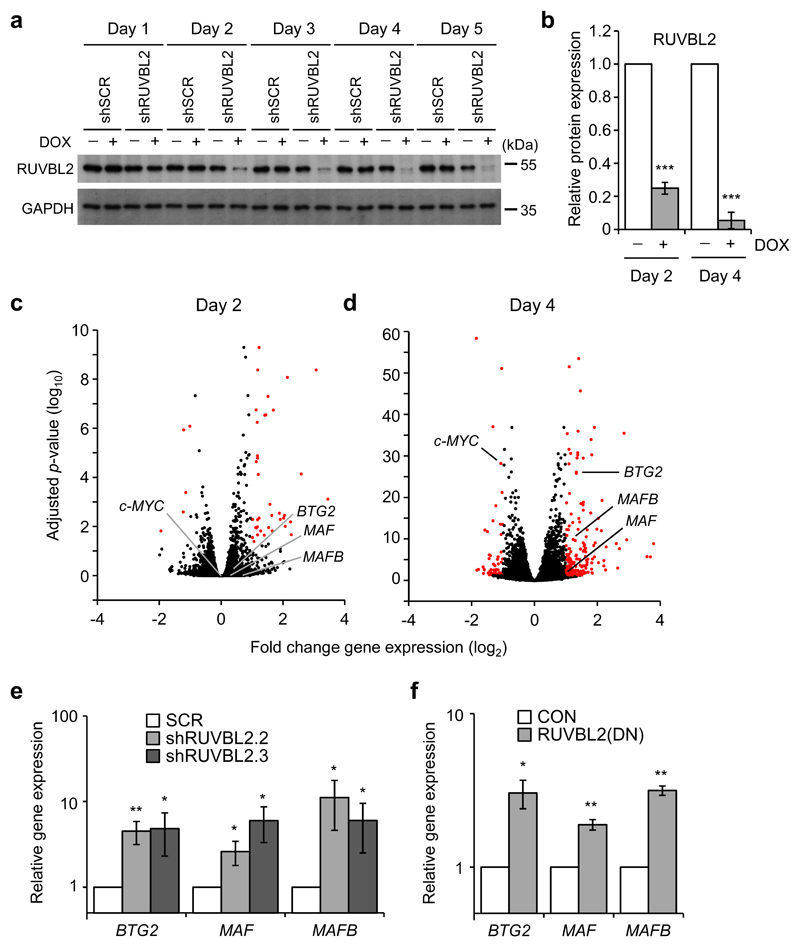
To determine the impact of RUVBL2 on global gene expression in AML cells we transduced bulk THP1 cells with the TRIPZ inducible shRUVBL2 and shSCR lentiviral vectors. Significant decreases in RUVBL2 protein expression were first detected at day 2 after induction of RUVBL2-specific, but not control, shRNA, and decreased further by day 4 (Fig. 3a,b). As expected, loss of RUVBL2 expression eventually resulted in apoptosis of THP1 cells, 8 days after doxycycline treatment (Supplementary Figure 5a,b). We then analysed changes in transcriptome profiles of THP1 cells by RNA sequencing (RNA-seq), at 2 days and 4 days following induction of RUVBL2-specific shRNA. RUVBL2 silencing resulted in 194 and 2,878 significant gene expression changes after 2 days and 4 days doxycycline treatment, respectively (Fig. 3c,d). Of these, the expression of 52 genes changed more than 2-fold at day 2 (6 down and 46 up), and 219 at day 4 (55 down and 164 up). These gene expression changes were validated in independent experiments by analysing the expression of a selected gene panel by qRT-PCR (Supplementary Figure 6). The RNA-seq data indicate that the predominant changes were increases in gene expression, consistent with the reported function of RUVBL2 as a co-factor in transcriptional repression, although there were some notable decreases in gene expression, such as c-MYC [8]. Among these were genes encoding the transcription factors BTG2, MAF and MAFB, known to promote myelomonocytic differentiation and growth arrest (Fig. 3d,e) [30–34]. Inhibition of RUVBL2 function by transduction of THP1 cells with RUVBL2(DN) also resulted in increased BTG2, MAF and MAFB expression (Fig. 3f). This function of RUVBL2 was not limited to THP1 cells, since increased BTG2 and MAFB expression following RUVBL2 silencing was also evident in a panel of AML cells lines (Supplementary Figure 7). These data indicate that RUVBL2 functions to repress expression of transcription factors that promote AML cell differentiation.
Fig. 3. Changes in gene expression profiles following RUVBL2 silencing in AML cells.
a Western blot analysis of RUVBL2 protein expression (upper panel) following doxycycline (DOX) treatment of THP1 cells transduced with inducible RUVBL2-specific shRNA (shRUVBL2) or control shRNA (shSCR). GAPDH (lower panel) was used as a loading control. b Quantification of RUVBL2 protein expression at days 2 and 4 after doxycycline treatment of shRUVBL2 THP1 cells. Bars and error bars are means and SD of three (day 2) and five (day 4) independent experiments. Data are normalised to GAPDH loading control and to untreated shRUVBL2 THP1 cells. ***P < 0.001, one sample t test. c, d Volcano plots of fold gene expression changes in shRUVBL2 THP1 cells following treatment with doxycycline for c 2 and d 4 days. Expression changes greater than 2-fold and P < 0.05 are shown in red, Wald test. e, f qRT-PCR validation of changes in BTG2, MAF and MAFB expression in THP1 cells following RUVBL2 inhibition by e two independent shRNA or f RUVBL2(DN) over-expression. Gene expression 24 hours after the end of puromycin selection is shown, normalised to e shSCR (SCR) or f empty vector (CON) transduced cells. Bars and error bars are means and SD of e five, and f three, independent experiments. *P < 0.05; **P < 0.01 (relative to controls), one sample t test.
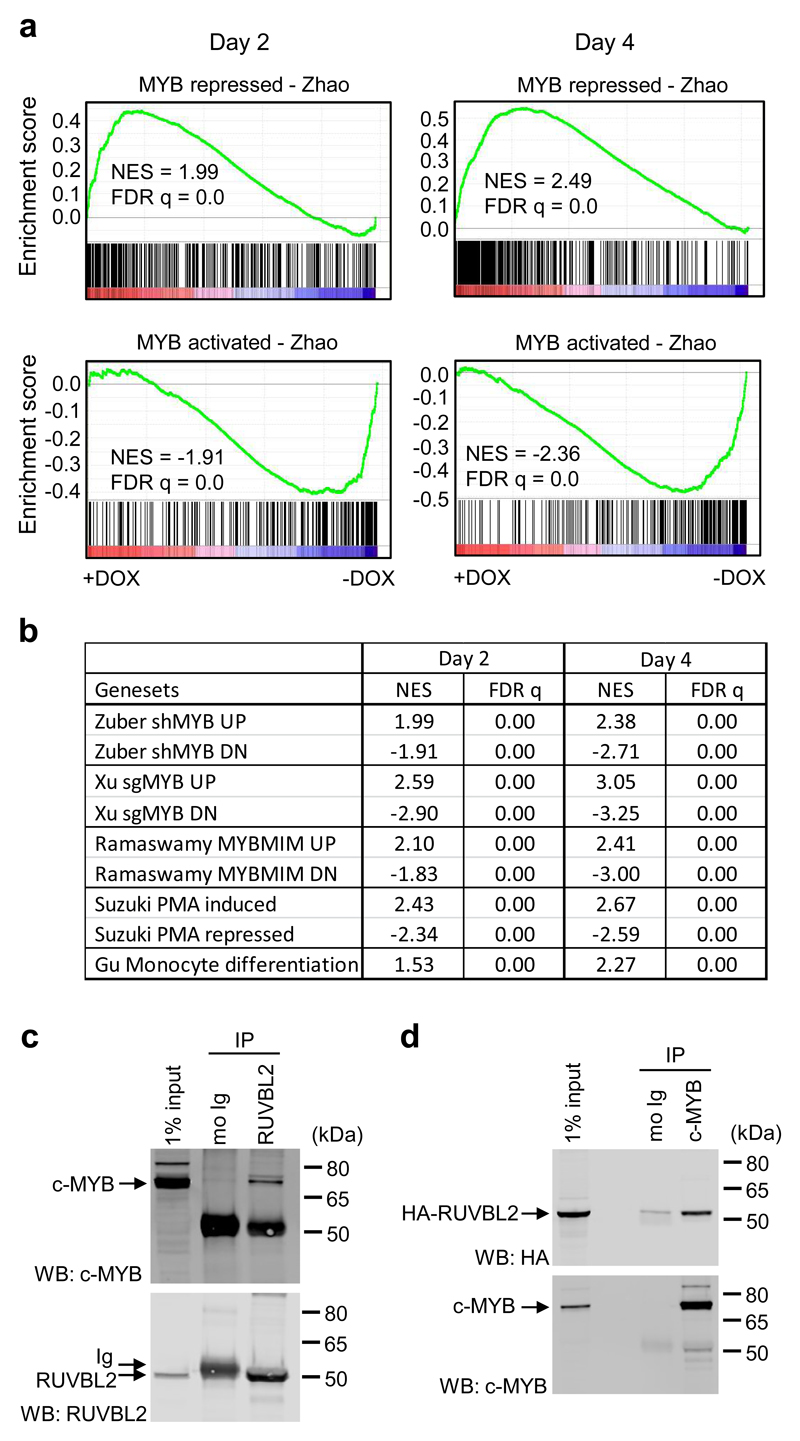
Since c-MYC expression is known to be regulated by the transcription factor c-MYB in AML cells, the expression of which did not change at the RNA or protein levels after RUVBL2 silencing (Supplementary Figure 8a-c), we next examined whether c-MYB function was impaired by loss of RUVBL2. Indeed, gene expression changes at both day 2 and day 4 following RUVBL2 silencing were found to be significantly enriched in direct c-MYB target genes (Fig. 4a), previously defined by integrating gene expression data [25] with target gene occupancy [35]. c-MYB gene sets derived from other studies, generated from shRNA targeting of c-MYB expression [22], CRISPR-based targeting of the c-MYB DNA-binding domain [23] and peptidomimetic inhibition of the interaction between c-MYB and the CBP-P300 co-activators [6], were all similarly enriched in gene expression changes at both time-points (Fig. 4b). A previous study reported that gene expression changes induced by c-MYB silencing, including increased expression of BTG2, MAF and MAFB, overlapped significantly with those following induction of THP1 cell differentiation by phorbol myristate acetate (PMA) treatment [25]. Indeed, genes affected by RUVBL2 silencing also showed enrichment for PMA-induced and monocyte terminal differentiation gene sets (Fig. 4b), as previously defined [24, 26]. This suggests that RUVBL2 arrests AML cell differentiation, a prerequisite for leukemia progression, by regulating the transcriptional activity of c-MYB. Consistent with these data, RUVBL2 silencing was associated with a significant down-regulation of genes linked to the AML leukemic stem cell signature (Supplementary Figure 8d) [21].
Fig. 4. RUVBL2 binds c-MYB and maintains its transcriptional program in AML.
a GSEA of c-MYB repressed (top) and activated (bottom) gene sets, as previously defined [7], in gene expression changes in shRUVBL2 THP1 cells following 2 (left panels) and 4 (right panels) days doxycycline treatment. b Table summarizing GSEA of c-MYB gene sets derived from shRNA [22], CRISPR-mediated [23] and peptidomimetic [6] c-MYB targeting in AML cells, and of a myeloid differentiation gene set [24] derived from global gene expression changes following PMA treatment of THP1 cells [25] and a terminal monocyte differentiation program [26]. c Western blot analysis of mouse IgG and anti-RUVBL2 immunoprecipitates from THP1 cells, following DSG cross-linking, stained with anti-c-MYB (top) and anti-RUVBL2 (bottom). d Western blot analysis of mouse IgG and anti-c-MYB immunoprecipitates from HA-RUVBL2 expressing THP1 cells, following DSG cross-linking, stained with anti-HA (top) and anti-c-MYB (bottom). Representative data c, d from one of three independent experiments.
RUVBL2 has been reported to interact with and regulate the activity of several different transcription factors in different cancer types [8, 36], including most recently E2F1 in hepatocellular carcinoma [10]. To determine whether RUVBL2 interacted with c-MYB in AML cells, we performed co-immunoprecipitation experiments in THP1 cells. Following in vivo protein crosslinking with disuccinimidyl glutarate (DSG), c-MYB was clearly found to co-immunoprecipitate with endogenous RUVBL2 (Fig. 4c). In the reciprocal experiment, c-MYB was immunoprecipitated from HA-RUVBL2 expressing THP1 cells, and shown to pull down the HA-tagged protein (Fig. 4d). These data suggest that RUVBL2 interacts with c-MYB in AML cells.
RUVBL2 is necessary to enforce transcriptional repression by c-MYB
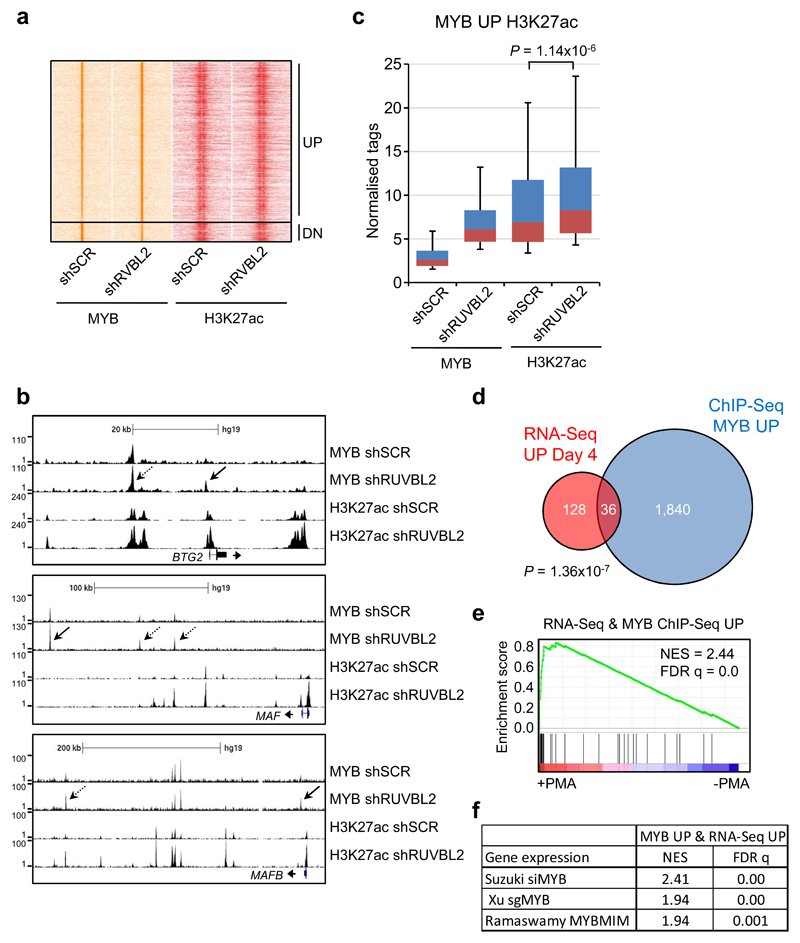
To examine the consequences of RUVBL2 silencing on genome-wide target gene occupancy by c-MYB, we performed chromatin immunoprecipitation combined with DNA sequencing (ChIP-seq) in THP1 cells, following transduction with constitutive RUVBL2-specific or control shRNA. A total of 17,254 c-MYB DNA-binding peaks were detected in control and shRUVBL2 cells, corresponding to 7,457 genes. We previously demonstrated that c-MYB controls RUVBL2 gene expression [11], and consistent with this finding, a c-MYB binding peak was evident at +462 bp relative to the RUVBL2 transcriptional start site (Supplementary Figure 9a). RUVBL2 silencing resulted in more than 2-fold increased binding of c-MYB at 2,355 peaks (MYB UP) and more than 2-fold decreased binding at 275 peaks (MYB DN), corresponding to 1,876 and 267 genes, respectively (Fig. 5a-b and Supplementary Figure 9b). We then performed further ChIP-seq analysis to determine the extent of co-localization of c-MYB binding peaks with regions of histone 3 lysine 27 acetylation (H3K27ac), a mark of transcriptional activity. Significant levels of H3K27ac were found to be associated with 568 MYB UP peaks and 109 MYB DN peaks. Furthermore, the H3K27ac signal increased significantly at the 568 MYB UP peaks upon RUVBL2 silencing (Fig. 5c), whereas it was not found to change significantly at the MYB DN peaks (Supplementary Figure 9c). We then compared the list of genes that were associated with increased c-MYB binding (1,876 genes) with those whose expression increased more than 2-fold (164 genes) following RUVBL2 silencing. There was a significant overlap of 36 genes between these two lists (Fig. 5d), which included BTG2, MAF and MAFB (Fig. 5b). This indicates that increased binding of c-MYB to these target genes correlated with increases in their expression. These genes were found to be positively enriched in the previously published PMA-induced gene expression changes (Fig. 5e) [25], suggesting that RUVBL2 is responsible for maintaining repression of c-MYB target genes involved in myeloid differentiation. Surprisingly, the 36 genes were also found to be positively enriched in gene expression data from studies targeting c-MYB expression and function (Fig. 5f) [6, 23, 25]. This analysis suggests that at these loci, loss of RUVBL2 expression results in conversion c-MYB from a transcriptional repressor into a transcriptional activator, correlating with increased binding of c-MYB and H3K27ac signal.
Fig. 5. Loss of RUVBL2 results in increased binding of c-MYB to repressed target genes and relieves their repression.
a Heatmap showing ChIP signal for c-MYB and histone H3K27ac for the dynamic c-MYB peaks (more than 2-fold changed) following shRNA mediated RUVBL2 silencing in THP1 cells. b Exemplar ChIP-Seq tracks for c-MYB peaks showing more than 1.5-fold (dashed arrows) and 2-fold (solid arrows) increased c-MYB binding (MYB UP) following RUVBL2 silencing. c Box plots showing ChIP-Seq signal for c-MYB and histone H3K27ac for MYB UP peaks (>2-fold) with a significant H3K27ac signal (MYB UP H3K27ac). H3K27ac signal increases significantly upon RUVBL2 silencing, Mann–Whitney U test. d Venn diagram showing overlap between genes whose expression increased (>2-fold, P < 0.05) in shRUVBL2 THP1 cells (RNA-Seq UP Day 4), following 4 days doxycycline treatment, and genes with increased MYB binding following RUVBL2 silencing (>2-fold, ChIP-Seq MYB UP). P-value obtained by hypergeometric test. e GSEA of the 36 genes, with increased expression and MYB binding following RUVBL2 silencing, in previously reported gene expression data from THP1 cells treated with PMA for 24 hours [25]. f Table summarising GSEA of the 36 genes in previously reported gene expression data following siRNA knockdown [25], CRISPR-mediated [23] and peptidomimetic [6] targeting of c-MYB in AML cells.
Discussion
In this study, we demonstrate that RUVBL2 is essential for the oncogenic activity of c-MYB in AML, ensuring transcriptional repression of myeloid differentiation-associated target genes. Myeloid differentiation arrest is a hallmark of AML, resulting in accumulation of aberrant immature myeloid progenitors. Reversing this differentiation block has long been a goal of novel anti-AML therapeutic strategies [37–39]. We present evidence for a molecular mechanism responsible for enforcing this block in AML differentiation. Our data indicate that RUVBL2 loss converts c-MYB from a repressor to a transcriptional activator of myeloid pro-differentiation genes. RUVBL2 inhibition results in increased c-MYB binding of these genes, associated with elevated H3K27 acetylation of c-MYB binding regions and activation of target gene expression. This is consistent with data from our previous study, which demonstrated that RUVBL2 inhibition led to growth inhibition, differentiation and eventual apoptosis of AML cells [11]. The present study demonstrates the importance of RUVBL2 in maintaining aberrant AML-associated transcriptional networks, highlighted by the ablation of established AML in vivo following RUVBL2 inhibition.
Previous analysis of myeloid transcription factor networks in AML suggested that c-MYB acts in part as an anti-differentiation transcriptional repressor [40]. Indeed, as well as activating its targets, c-MYB was found to repress half of its direct target genes, several of which are recognized positive regulators of myeloid differentiation [35]. Although the CBP/P300 co-factors are largely defined as transcriptional co-activators, they were also shown to be necessary for repression of target genes by c-MYB [35, 41] and peptidomimetic inhibition of c-MYB:CBP/P300 interaction was shown to result in increased expression of repressed c-MYB target genes, as well as decreased expression of activated genes [6]. This suggests that direct repression of positive regulators of myeloid differentiation and growth arrest by c-MYB is an essential component of its transforming activity. Indeed, our RNA-seq analysis demonstrate that in AML cells, RUVBL2 is required for c-MYB-dependent transcriptional repression of a pro-differentiation myeloid gene expression signature, including BTG2, MAF and MAFB genes. These genes are all expressed during normal myeloid differentiation. BTG2 is an anti-proliferative tumour suppressor and plays a role during differentiation of diverse tissues [42]. Much of its activity is linked to interaction with the PRMT1 arginine methyltransferase and methylation of histone and non-histone substrates. This complex has been shown to regulate gene expression as directly affecting post-transcriptional processes such as mRNA stability and cell cycle machinery dynamics [42]. Of particular interest in the context of our study, the BTG2-PRMT1 complex was reported to enhance myeloid differentiation of both AML cells and normal CD34+ HPC in response to retinoic acid (RA). This effect was found to be dependent on basal methylation of histone H4 at RA-responsive promoters by this complex, leading to more efficient histone H4 acetylation upon RA stimulation and consequent increases in target gene expression [32]. The myelomonocytic transcription factors MAF and MAFB have both been reported promote monocytic differentiation in AML cell lines and transformed myeloid cells, respectively [30, 31]. Moreover, MAFB was also shown to promote RA induced myeloid differentiation of THP1 cells, enhancing expression and histone H4 acetylation of RA target genes [43].
c-MYB is required for definitive hematopoiesis and plays an important role in differentiation of multiple hematopoietic lineages [44]. Although rarely mutated in leukemia, it has long been associated with hematopoietic malignancies [44, 45]. Indeed a number of studies demonstrated that c-MYB is an essential mediator of MLL-fusion activity in AML [46, 47], maintaining an aberrant self-renewal program downstream of the driving oncogenes [21, 22]. Furthermore, we recently demonstrated that c-MYB silencing impaired self-renewal of both MLL-rearranged and non-rearranged AML cells [7]. These properties of c-MYB in AML are consistent with cancer-associated master regulator activity [4], indicative of potential efficacy as a therapeutic target [3]. Our data demonstrate that RUVBL2 inhibition blocks the oncogenic activity of c-MYB without compromising normal haematopoiesis. This can be explained by the testable hypothesis that RUVBL2 interacts with c-MYB to repress pro-differentiation target genes, such as BTG2, MAF and MAFB, that would otherwise be transactivated by the increased levels of c-MYB associated with myeloid transformation.
RUVBL2 consists of three domains, the ATP binding pocket being formed by intramolecular interactions between domain I (containing the Walker motifs) and domain III [48]. The Walker A domain is required for binding of ATP, while the B domain is necessary for its hydrolysis. The RUVBL2(DN) Walker B mutant exerts a dominant negative effect over the transcriptional co-repressor function of endogenous RUVBL2 in our experiments. This indicates that the ATPase activity of RUVBL2 is essential for the oncogenic activity of c-MYB. In this respect, it is important to note that normal hematopoiesis exhibits no such dependence on RUVBL2 ATPase function.
Increased expression of RUVBL2 has been reported in a number of cancer types [8], and we previously found that its expression increased upon transformation of normal human hematopoietic progenitors and remained elevated in AML cells [11]. Interestingly, high RUVBL2 expression is found in a subset of t(4;11) infant acute lymphoblastic leukemia expressing high levels of IRX1/2 and low levels of HOXA cluster genes [49]. This subset has a significantly increased risk of relapse in comparison to the subset expressing high levels of HOXA genes [49–51]. Since t(4;11) infant ALL can often switch lineages to AML, it would be important to examine whether high RUVBL2 expression was linked to such lineage switches. RUVBL2 has been shown to regulate the transcriptional activity of numerous transcription factors implicated in oncogenesis, including c-MYC, β-catenin and E2F1 [8, 10, 36]. The relative importance of these interactions to maintenance of cancer cell transcriptional dystasis is likely to vary according to cancer tissue type. The central role of c-MYB in AML pathogenesis suggests that targeting its interaction with RUVBL2 represents a novel and promising approach to AML therapy.
Supplementary Material
Acknowledgments
The authors thank Ayad Eddaoudi and Stephanie Canning, UCL GOS ICH Flow Cytometry Facility, for providing assistance with flow cytometry, all the staff of the UCL GOS ICH Western Laboratories for excellent animal husbandry. E.A.M. was supported by a PhD studentship from Bloodwise (12068), L.Z. by a PhD studentship from the Medical Research Council (MRC DTG), and L.G. by a project grant from Children with Cancer UK (14-169). M.R.M. was supported by a Bennett Fellowship from Bloodwise and National Institute for Health Research University College London BRC Senior Clinical Research Fellowship, and J.d.B. by a fellowship from the Alternative Hair Charitable Foundation and Great Ormond Street Hospital Children’s Charity. Research in the laboratory of J.H.A.M. was supported by KIKA, and O.W. (V1305, V2617) was supported by grants from the Great Ormond Street Hospital Children’s Charity. This research was supported by the NIHR Great Ormond Street Hospital Biomedical Research Centre.
Footnotes
Author contributions E.A.M., L.Z., L.G. and O.W. performed the experiments and data analysis. T.B. and K.P. performed the RNA-sequencing and ChIP-sequencing experiments, and J.H.A.M. analysed ChIP-sequencing data. O.W., J.d.B. and M.R.M. provided project leadership and supervised the research. E.A.M. and O.W. wrote the paper. All authors read, provided critical comments and approved the manuscript.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bose P, Vachhani P, Cortes JE. Treatment of Relapsed/Refractory Acute Myeloid Leukemia. Curr Treat Options Oncol. 2017;18:17. doi: 10.1007/s11864-017-0456-2. [DOI] [PubMed] [Google Scholar]
- 3.Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat Rev Cancer. 2015;15:686–694. doi: 10.1038/nrc4018. [DOI] [PubMed] [Google Scholar]
- 4.Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer. 2017;17:116–130. doi: 10.1038/nrc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uttarkar S, Dasse E, Coulibaly A, Steinmann S, Jakobs A, Schomburg C, et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood. 2016;127:1173–1182. doi: 10.1182/blood-2015-09-668632. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy K, Forbes L, Minuesa G, Gindin T, Brown F, Kharas MG, et al. Peptidomimetic blockade of MYB in acute myeloid leukemia. Nat Commun. 2018;9:110. doi: 10.1038/s41467-017-02618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walf-Vorderwulbecke V, Pearce K, Brooks T, Hubank M, van den Heuvel-Eibrink MM, Zwaan CM, et al. Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia. 2018;32:882–889. doi: 10.1038/leu.2017.317. [DOI] [PubMed] [Google Scholar]
- 8.Grigoletto A, Lestienne P, Rosenbaum J. The multifaceted proteins Reptin and Pontin as major players in cancer. Biochim Biophys Acta. 2011;1815:147–157. doi: 10.1016/j.bbcan.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 10.Tarangelo A, Lo N, Teng R, Kim E, Le L, Watson D, et al. Recruitment of Pontin/Reptin by E2f1 amplifies E2f transcriptional response during cancer progression. Nat Commun. 2015;6 doi: 10.1038/ncomms10028. 10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osaki H, Walf-Vorderwulbecke V, Mangolini M, Zhao L, Horton SJ, Morrone G, et al. The AAA+ ATPase RUVBL2 is a critical mediator of MLL-AF9 oncogenesis. Leukemia. 2013;27:1461–1468. doi: 10.1038/leu.2013.42. [DOI] [PubMed] [Google Scholar]
- 12.Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017;168:890–903 e815. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangolini M, de Boer J, Walf-Vorderwulbecke V, Pieters R, den Boer ML, Williams O. STAT3 mediates oncogenic addiction to TEL-AML1 in t(12;21) acute lymphoblastic leukemia. Blood. 2013;122:542–549. doi: 10.1182/blood-2012-11-465252. [DOI] [PubMed] [Google Scholar]
- 15.Woodward MJ, de Boer J, Heidorn S, Hubank M, Kioussis D, Williams O, et al. Tnfaip8 is an essential gene for the regulation of glucocorticoid-mediated apoptosis of thymocytes. Cell Death Differ. 2010;17:316–323. doi: 10.1038/cdd.2009.125. [DOI] [PubMed] [Google Scholar]
- 16.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 18.Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, et al. Continuous MLL-ENL expression is necessary to establish a "Hox Code" and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- 19.Horton SJ, Walf-Vorderwulbecke V, Chatters SJ, Sebire NJ, de Boer J, Williams O. Acute myeloid leukemia induced by MLL-ENL is cured by oncogene ablation despite acquisition of complex genetic abnormalities. Blood. 2009;113:4922–4929. doi: 10.1182/blood-2008-07-170480. [DOI] [PubMed] [Google Scholar]
- 20.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Milazzo JP, Somerville TDD, Tarumoto Y, Huang YH, Ostrander EL, et al. A TFIID-SAGA Perturbation that Targets MYB and Suppresses Acute Myeloid Leukemia. Cancer Cell. 2018;33:13–28 e18. doi: 10.1016/j.ccell.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiques-Diaz A, Spencer GJ, Lynch JT, Ciceri F, Williams EL, Amaral FMR, et al. Enhancer Activation by Pharmacologic Displacement of LSD1 from GFI1 Induces Differentiation in Acute Myeloid Leukemia. Cell Rep. 2018;22:3641–3659. doi: 10.1016/j.celrep.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X, Ebrahem Q, Mahfouz RZ, Hasipek M, Enane F, Radivoyevitch T, et al. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub regulating granulo-monocytic fates. J Clin Invest. 2018 doi: 10.1172/JCI97117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandoli A, Singh AA, Prange KHM, Tijchon E, Oerlemans M, Dirks R, et al. The Hematopoietic Transcription Factors RUNX1 and ERG Prevent AML1-ETO Oncogene Overexpression and Onset of the Apoptosis Program in t(8;21) AMLs. Cell Rep. 2016;17:2087–2100. doi: 10.1016/j.celrep.2016.08.082. [DOI] [PubMed] [Google Scholar]
- 29.Prange KHM, Mandoli A, Kuznetsova T, Wang SY, Sotoca AM, Marneth AE, et al. MLL-AF9 and MLL-AF4 oncofusion proteins bind a distinct enhancer repertoire and target the RUNX1 program in 11q23 acute myeloid leukemia. Oncogene. 2017;36:3346–3356. doi: 10.1038/onc.2016.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hegde SP, Zhao J, Ashmun RA, Shapiro LH. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood. 1999;94:1578–1589. [PubMed] [Google Scholar]
- 31.Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, et al. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26:5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho BO, Jeong YW, Kim SH, Park K, Lee JH, Kweon GR, et al. Up-regulation of the BTG2 gene in TPA- or RA-treated HL-60 cell lines. Oncol Rep. 2008;19:633–637. [PubMed] [Google Scholar]
- 34.Ryu MS, Lee MS, Hong JW, Hahn TR, Moon E, Lim IK. TIS21/BTG2/PC3 is expressed through PKC-delta pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Exp Cell Res. 2004;299:159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Glazov EA, Pattabiraman DR, Al-Owaidi F, Zhang P, Brown MA, et al. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res. 2011;39:4664–4679. doi: 10.1093/nar/gkr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao YQ, Houry WA. The Role of Pontin and Reptin in Cellular Physiology and Cancer Etiology. Front Mol Biosci. 2017;4:58. doi: 10.3389/fmolb.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasine JP, Schiller GJ. Emerging strategies for high-risk and relapsed/refractory acute myeloid leukemia: novel agents and approaches currently in clinical trials. Blood Rev. 2015;29:1–9. doi: 10.1016/j.blre.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Greenblatt SM, Nimer SD. Chromatin modifiers and the promise of epigenetic therapy in acute leukemia. Leukemia. 2014;28:1396–1406. doi: 10.1038/leu.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, et al. Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell. 2016;167:171–186 e115. doi: 10.1016/j.cell.2016.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomaru Y, Simon C, Forrest AR, Miura H, Kubosaki A, Hayashizaki Y, et al. Regulatory interdependence of myeloid transcription factors revealed by Matrix RNAi analysis. Genome Biol. 2009;10:R121. doi: 10.1186/gb-2009-10-11-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kasper LH, Fukuyama T, Lerach S, Chang Y, Xu W, Wu S, et al. Genetic interaction between mutations in c-Myb and the KIX domains of CBP and p300 affects multiple blood cell lineages and influences both gene activation and repression. PLoS One. 2013;8:e82684. doi: 10.1371/journal.pone.0082684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN. Tumor suppressors BTG1 and BTG2: Beyond growth control. J Cell Physiol. 2019;234:5379–5389. doi: 10.1002/jcp.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Chen Q, Ross AC. Retinoic acid and tumor necrosis factor-alpha induced monocytic cell gene expression is regulated in part by induction of transcription factor MafB. Exp Cell Res. 2012;318:2407–2416. doi: 10.1016/j.yexcr.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 45.Pattabiraman DR, Gonda TJ. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–277. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 46.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin S, Zhao H, Yi Y, Nakata Y, Kalota A, Gewirtz AM. c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis. J Clin Invest. 2010;120:593–606. doi: 10.1172/JCI38030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–38929. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- 49.Trentin L, Giordan M, Dingermann T, Basso G, Te Kronnie G, Marschalek R. Two independent gene signatures in pediatric t(4;11) acute lymphoblastic leukemia patients. Eur J Haematol. 2009;83:406–419. doi: 10.1111/j.1600-0609.2009.01305.x. [DOI] [PubMed] [Google Scholar]
- 50.Stam RW, Schneider P, Hagelstein JA, van der Linden MH, Stumpel DJ, de Menezes RX, et al. Gene expression profiling-based dissection of MLL translocated and MLL germline acute lymphoblastic leukemia in infants. Blood. 2010;115:2835–2844. doi: 10.1182/blood-2009-07-233049. [DOI] [PubMed] [Google Scholar]
- 51.Kang H, Wilson CS, Harvey RC, Chen IM, Murphy MH, Atlas SR, et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2012;119:1872–1881. doi: 10.1182/blood-2011-10-382861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.