Abstract
Senescence is a cellular stress response that results in the stable arrest of old, damaged or preneoplastic cells. Oncogene-induced senescence is tumor suppressive but can also exacerbate tumorigenesis through the secretion of pro-inflammatory factors from senescent cells. Drugs that selectively kill senescent cells, termed senolytics, have proved beneficial in animal models of many age-associated diseases. Here, we show that the cardiac glycoside, ouabain, is a senolytic agent with broad activity. Senescent cells are sensitized to ouabain-induced apoptosis, a process mediated in part by induction of the pro-apoptotic Bcl2-family protein NOXA. We show that cardiac glycosides synergize with anti-cancer drugs to kill tumor cells and eliminate senescent cells that accumulate after irradiation or in old mice. Ouabain also eliminates senescent preneoplastic cells. Our findings suggest that cardiac glycosides may be effective anti-cancer drugs by acting through multiple mechanism. Given the broad range of senescent cells targeted by cardiac glycosides their use against age-related diseases warrants further exploration.
Introduction
Senescence is a protective stress response that limits the replication of damaged, preneoplastic or aged cells 1. Senescence can be induced by stresses including replicative exhaustion, oncogenic activation or exposure to genotoxic agents. Upon senescence induction, cells enter a stable cell cycle arrest, a process mediated by the upregulation of the cyclin dependent kinase inhibitor p16INK4a 2. Senescent cells also reorganize their chromatin, reprogram their metabolism, undergo changes in gene expression 1,3 and secrete a complex combination of factors collectively referred to as the senescence-associated secretory phenotype (SASP) 4. The SASP has many roles 5,6 and it is thought to mediate many of the pathophysiological consequences associated to senescence 7.
Senescent cells are present in pre-neoplastic and fibrotic lesions, they accumulate in old tissues and are associated with an increasing list of pathologies 8. Despite the fact that senescence protects against cancer and limits most types of fibrosis, the aberrant accumulation of senescent cells during ageing and disease is largely detrimental 9. This negative role of senescent cells in ageing was first demonstrated with the help of genetic models that allow for the selective ablation of senescent cells 10,11. The use of these mouse models has shown that clearing senescent cells from progeroid or naturally aging mice improves healthspan 10, increases lifespan 12 and benefits an array of pathologies that include atherosclerosis 13, osteoarthritis 14 and neurodegenerative diseases 15,16.
These successful genetic studies prompted a search for drugs that can selectively kill senescent cells, termed senolytics. Several senolytic compounds have been identified, including dasatinib and quercetin 17, piperlongumine 18, HSP90 inhibitors 19 or Bcl2 family inhibitors such as ABT-263 (also known as navitoclax) and ABT-737 20–22. Currently Bcl2 family inhibitors are the most widely used senolytics, having been shown effective at killing a range of senescent cells in vivo and reproducing the effects observed in transgenic mice modelling senescence ablation 23. Bcl2 inhibitors were initially developed as therapies for lymphoma. ABT-737 is a small molecule inhibitor of BCL-2, BCL-XL, and BCL-W but has low solubility and oral bioavailability. ABT-263 inhibits the same molecules and is better suited for use in vivo, but it causes significant thrombocytopenia. Due to the important side-effects associated to known senolytic compounds, there is a need to identify further compounds with senolytic properties. Despite the many roles that senescence plays in cancer initiation, progression and treatment, how senolytics can affect these processes is poorly defined. Here, we describe drug screens aimed at finding novel senolytics that can target cells undergoing oncogene-induced and therapy-induced senescence. As a result, we identified cardiac glycosides (CGs) as a novel class of broad-spectrum senolytic compounds.
Results
A drug screen identifies ouabain as a broad-spectrum senolytic
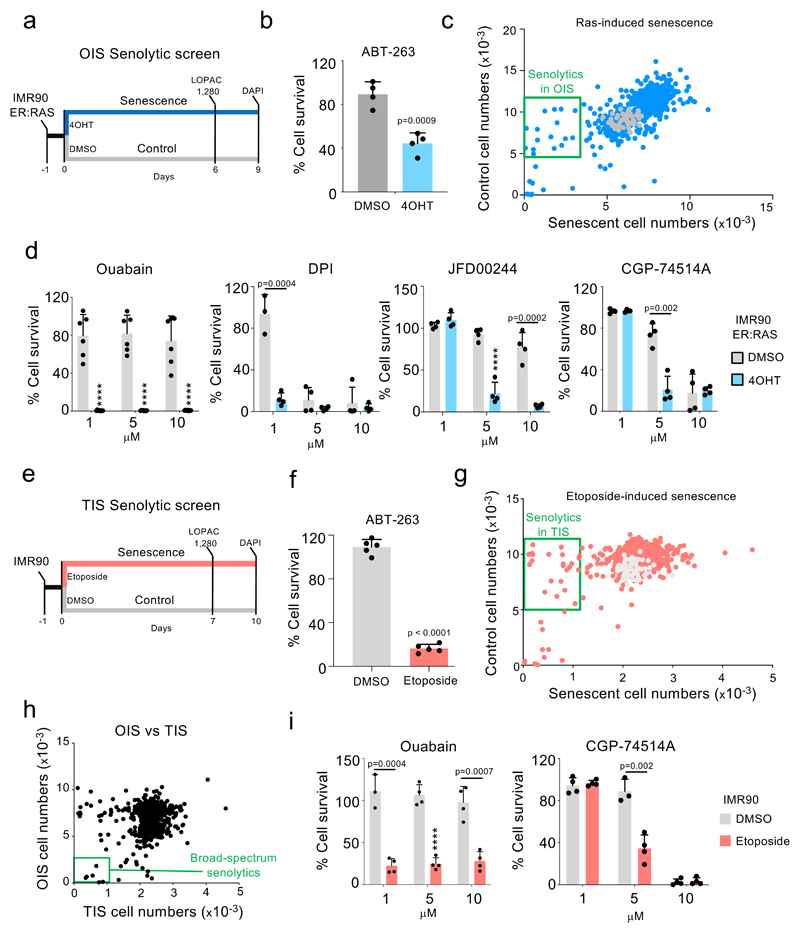
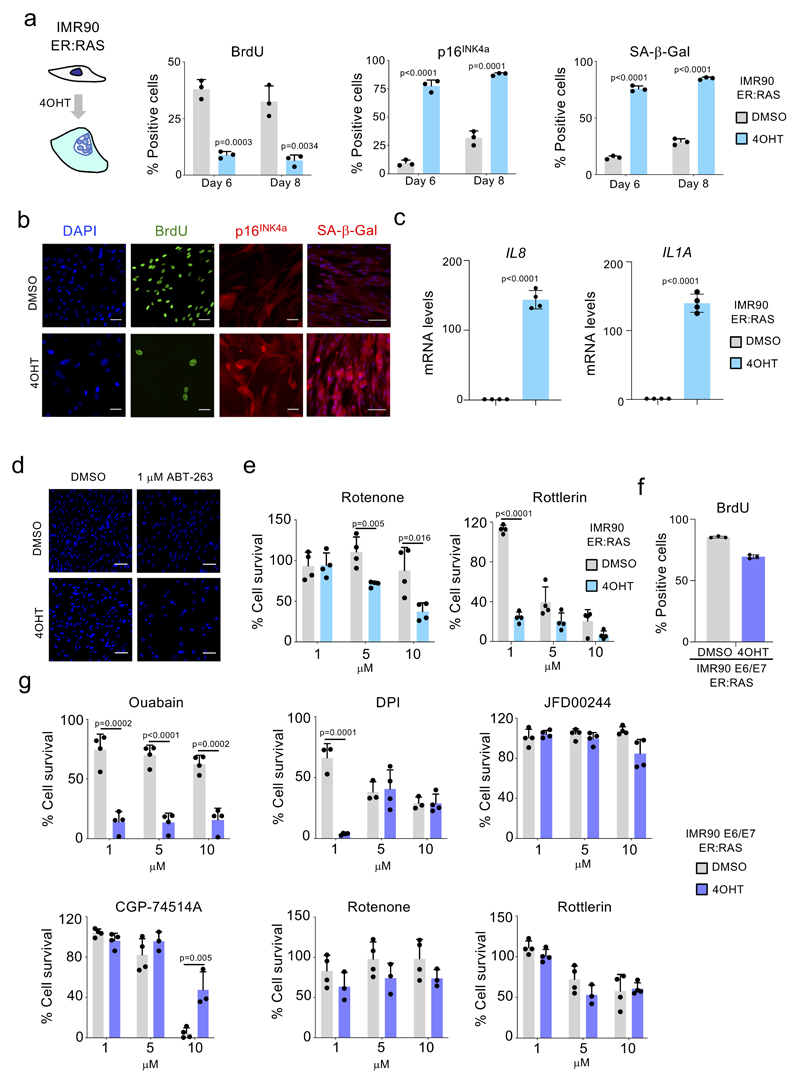
In an effort to identify compounds modulating senescence, we carried out unbiased drug screens. First, we used IMR90 ER:RAS cells as a model of oncogene-induced senescence (OIS) 24. In these cells, activation of oncogenic RAS with 4-hydroxy-tamoxifen (4OHT) induces senescence (Extended data Fig 1a-c). We set up a screen to compare the effects that compounds have on the viability of control and senescent cells (Fig 1a). We confirmed that treatment with 1 µM of the known senolytic preferentially eliminated cells undergoing OIS (Fig 1b and Extended data Fig 1d). By screening the LOPAC 1,280 library, comprised of 1,280 pharmacologically active compounds, we identified several drugs which preferentially killed cells undergoing Ras-induced senescence (Fig 1c-d and Extended data Fig 1e).
Figure 1. Drug screens identify ouabain as a broad-spectrum senolytic.
a, Experimental design for the senolytic screen on oncogene-induced senescence. b, Quantification of cell survival of senescent and control IMR90 ER:RAS cells after treatment with 1 μM ABT-263 for 3 days (n = 4). c, Screen results. LOPAC 1,280 library compounds were assessed at 10 μM for 3 days. Hits were selected based on their ability to specifically kill senescent cells. Blue dots represent library drugs, grey dots represent DMSO controls. Each dot is the mean of three replicates. d, Senolytic activity of the indicated drugs in the context of oncogene-induced senescence in IMR90 ER:RAS cells (n = 4; n = 6 for ouabain; DMSO versus 4OHT, ****P < 0.0001). e, Experimental design for the senolytic screen on therapy-induced senescence. f, Quantification of cell survival of senescent and control IMR90 cells after treatment with 1 μM ABT-263 for 3 days (n = 5). g, Screen results. LOPAC 1,280 library compounds were assessed at 10 μM for 3 days. Hits were selected based on their ability to specifically kill senescent cells. Red dots represent library drugs, grey dots represent DMSO controls. Each dot is the mean of three replicates. h, Comparison of senolytic activity for the LOPAC 1,280 library compounds in the context of OIS vs therapy-induced senescence. i, Senolytic activity of the indicated drugs in the context of therapy-induced senescence in IMR90 (n = 4; DMSO versus etoposide, ****P < 0.0001). All error bars represent mean ± s.d; n represents independent experiments. All statistical significances were calculated using unpaired two-tailed Student’s t-tests and Holm-Sidak method for multiple comparisons test.
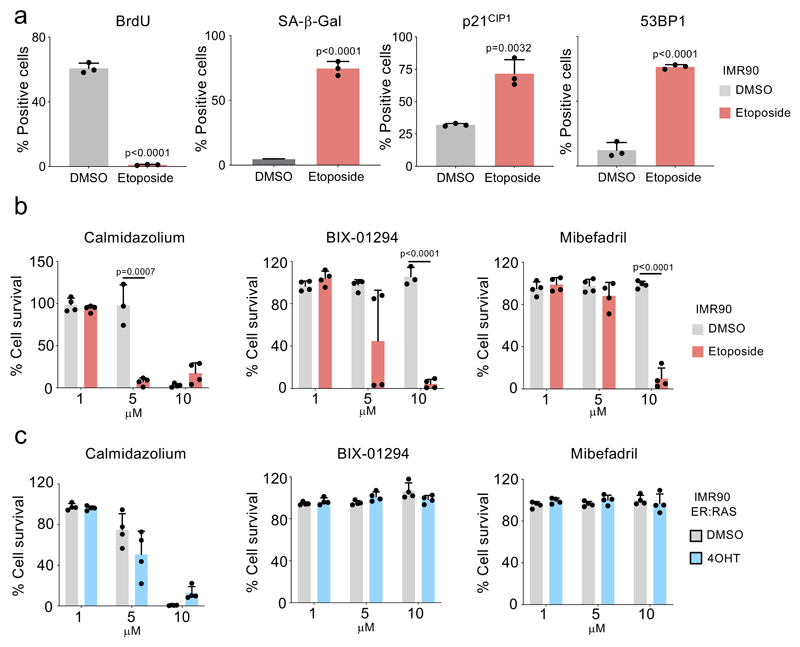
To understand whether the selective killing observed is specifically associated with activated RAS signalling or dependent on OIS, we used IMR90 E6/E7 ER:RAS cells. Papillomavirus E6 and E7 proteins interfere with the p53 and Rb pathways respectively, and activation of RAS in these cells does not cause a senescence-associated growth arrest (Extended data Fig 1f). While most of the compounds tested (4/6, JFD00244, CGP-74514A, rotenone and rottlerin) did not selectively kill IMR90 E6/E7 ER:RAS cells, the other 2 (ouabain and diphenyleneiodonium, DPI) did (Extended data Fig 1g).
Senescence can be triggered by a variety of insults that range from oncogenic activation, replicative stress or exposure to chemotherapeutic drugs 1,2. To identify senolytics targeting different types of senescence, we screened a model of therapy-induced senescence (TIS, Fig 1e). Treatment of IMR90 cells with etoposide caused senescence (Extended data Fig 2a), and sensitised cells to death with 1 µM ABT-263 (Fig 1f). A screen using the LOPAC 1,280 library identified compounds that behaved as senolytics in TIS (Fig 1g). By excluding toxic compounds and combining the data from both screens (Fig 1h), we observed that while a subset of the identified drugs only killed cells undergoing either TIS or OIS (Extended data Fig 2b-c), others (indicated by the green box in Fig 1h) selectively killed both types of senescent cells over their respective controls. Upon retesting, we confirmed that CGP-74514A (a CDK1 inhibitor) and ouabain share this ability to selectively kill cells undergoing both OIS and TIS (Fig 1i).
Cardiac glycosides induce apoptosis of senescent cells
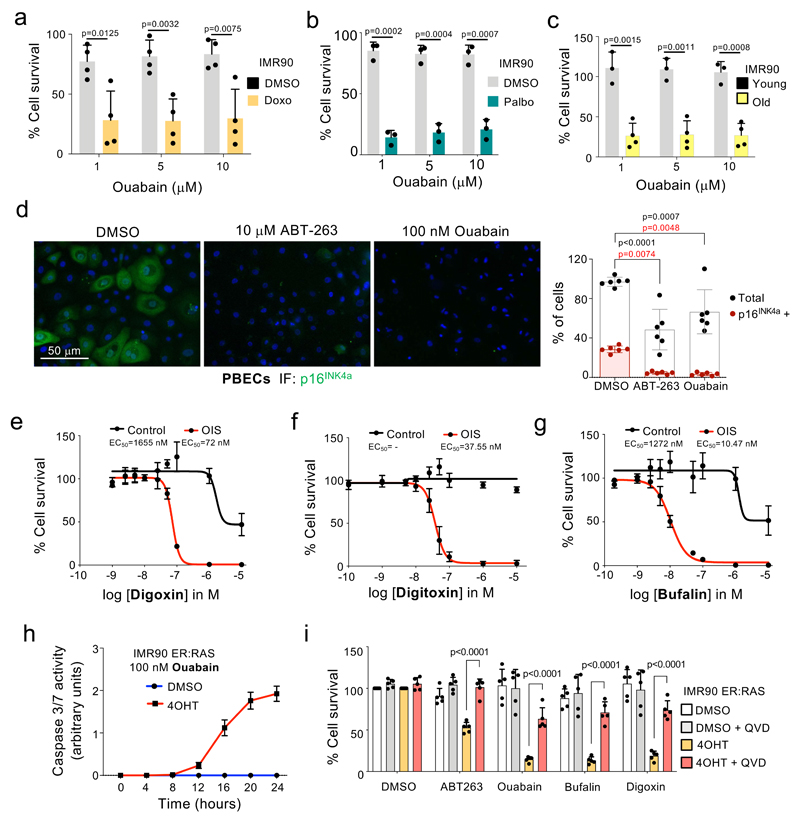
Ouabain is a natural compound belonging to the cardiac glycoside (CG) family first described as an specific inhibitor of the Na+, K+-ATPase 25, although CGs are notoriously pleiotropic and can inhibit other targets 26. Ouabain treatment not only preferentially killed cells undergoing senescence triggered by oncogenic RAS (Fig 1d), or etoposide (Fig 1i), but also those exposed to doxorubicin (Fig 2a), the CDK4/6 inhibitor palbociclib (Fig 2b) or undergoing replicative senescence (Fig 2c). Mid-passage cultures of primary bronchial epithelial cells (PBEC), contain a mixture of senescent (p16INK4a-positive) and normal (p16INK4a-negative) cells. Treatment with ouabain or ABT-263 resulted in the selective elimination of p16INK4a-positive-cells (Fig 2d). Taken together these results further demonstrate that ouabain behaves as a broad-spectrum senolytic agent.
Figure 2. Cardiac glycosides induce apoptosis of senescent cells.
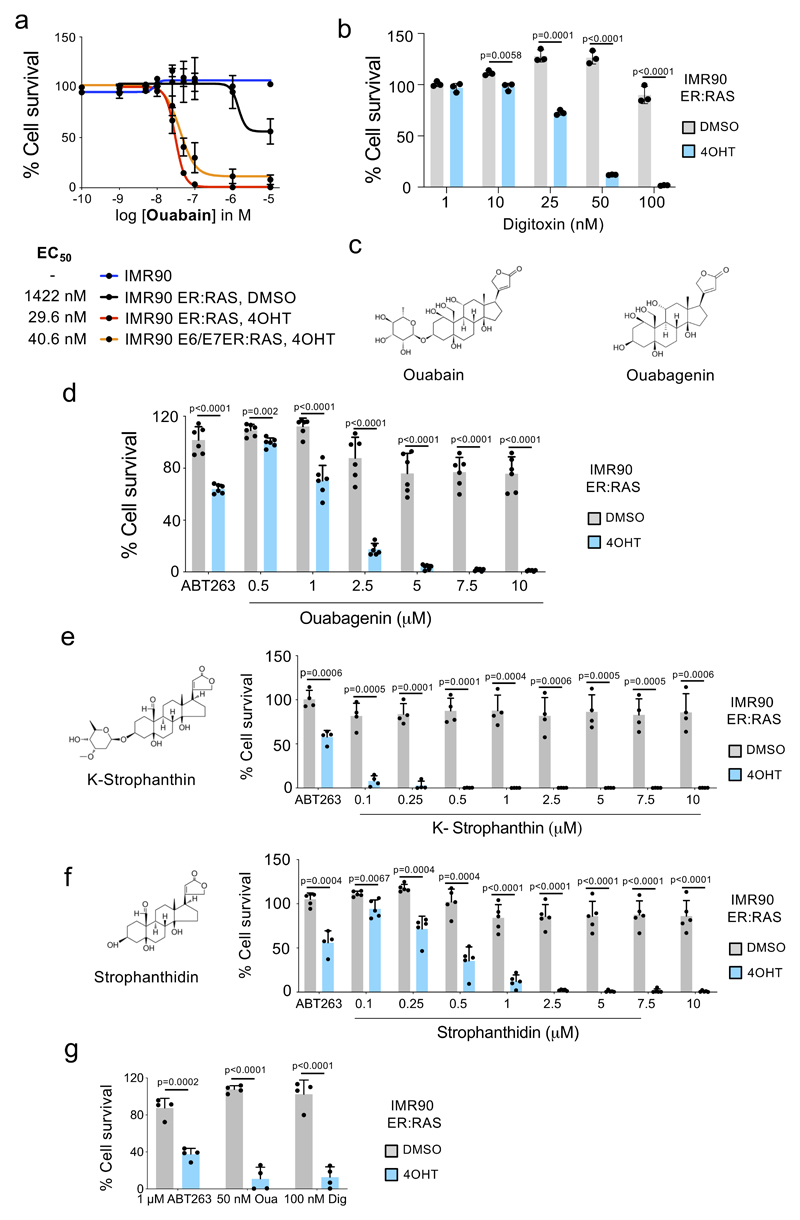
a-c, Senolytic activity of Ouabain in the context of therapy-induced senescence (doxorubicin, n = 4; palbociclib, n =3) and replicative senescence (n = 4). Statistical significance was calculated using unpaired two-tailed Student’s t-tests. d, Representative pictures (left) of immunofluorescence (IF) staining for p16INK4a in PBECs after treatment with ABT-263, Ouabain or vehicle (DMSO). p16INK4a is stained green. Scale bar, 50 μm. Quantification of total and p16INK4a-positive PBECs (right, n =6). Statistical significance was calculated using two-way ANOVA (Dunnett’s multiple comparisons test). Red stars refer to comparison of p16INK4a-positive PBECs, black stars refer to comparison of total PBEC numbers. e-g, Dose response analysis of senolytic activity of digoxin (e), digitoxin (f) and bufalin (g) in IMR90 ER:RAS cells (n = 4). h, Ouabain treatment of senescent cells induce caspase-3/7 activity. IMR90 ER:RAS were treated with 4-OHT or vehicle (DMSO) for 6 days to induce senescence. 100 nM ouabain was then added together with NucLight Rapid Red reagent for cell labelling and Caspase-3/7 reagent for apoptosis (IncuCyte). Caspase 3/7 activity was measured at 4h intervals (n = 3). i, Pan-caspase inhibition (20 μM Q-VD-OPh) rescues senolytic activity of cardiac glycosides (50 nM ouabain, 50 nM bufalin, 100 nM digoxin) and 1 μM ABT-263 on IMR90 ER:RAS cells (n = 5). Statistical significance was calculated using two-way ANOVA (Tukey’s test). All error bars represent mean ± s.d; n represents independent experiments.
To better understand the specificity of ouabain as a senolytic agent, we calculated its EC50 and observed that this was around 50-fold lower on senescent cells than on normal cells and also confirmed that ouabain killed non-senescent cells with increased RAS signalling (Extended data Fig 3a). Distinct cardiac glycosides such as digoxin and digitoxin, which are currently used for treating heart failure and atrial fibrillation 27, also behaved as senolytics (Fig 2e-f). Importantly, digitoxin behave as a senolytic at concentrations close to those observed in the plasma of cardiac patients treated with this drug (20–33nM) 28 (Fig 2f and Extended data Fig 3b). These data suggest that clinical benefits for human patients may be achievable at within the safe therapeutic dosing window for CGs.
Senescent cells have an increased lysosome mass and display higher activity of lysosomal enzymes including glycosidases such as ß-Galactosidase or α-fucosidase 29,30. To assess whether the glycoside chain of CGs could account for their differential effects on senescent cells, we tested bufalin, a cardiac steroid that shares a similar structure to CGs but lacks a glycoside chain. Bufalin also displayed senolytic properties (Fig 2g) as did ouabagenin, the aglycone of ouabain (Extended data Fig 3c-d). Furthermore, we tested k-stropanthin and strophanthidin, another CG and its correspondent aglycone, which shared a similar senolytic profile (Extended data Fig 3e-f), arguing against an underlying role for the glycoside chain in the specificity of CGs towards senescent cells. CGs can trigger different types of cell death depending on the context. Treatment with CGs resulted in cleaved caspase 3 and induced caspase 3/7 activity in senescent cells (Fig 2h and Supplementary Fig 1a). Moreover, the killing of senescent cells by CGs was prevented by pan-caspase inhibitors but not by inhibitors of ferroptosis, pyroptosis or necroptosis (Fig 2i and Supplementary Fig 1b). The above results suggest that CGs behave as senolytic agents by preferentially triggering apoptosis in senescent cells.
Mechanism explaining the senolytic properties of cardiac glycosides
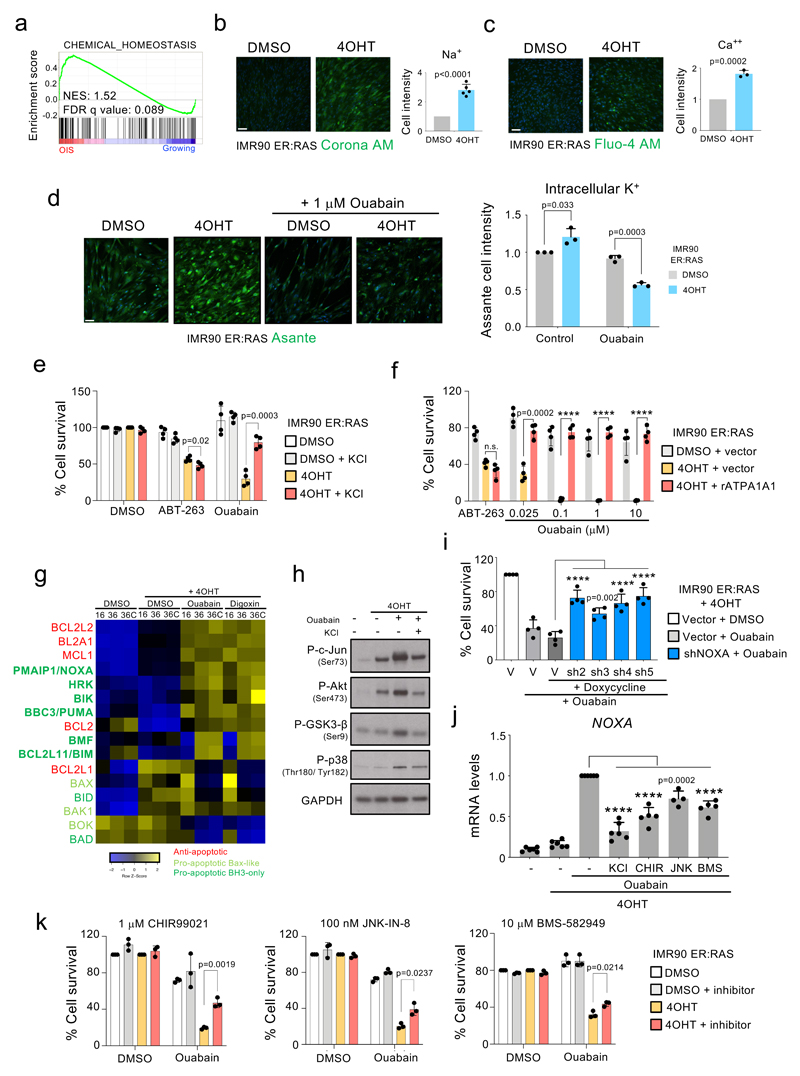
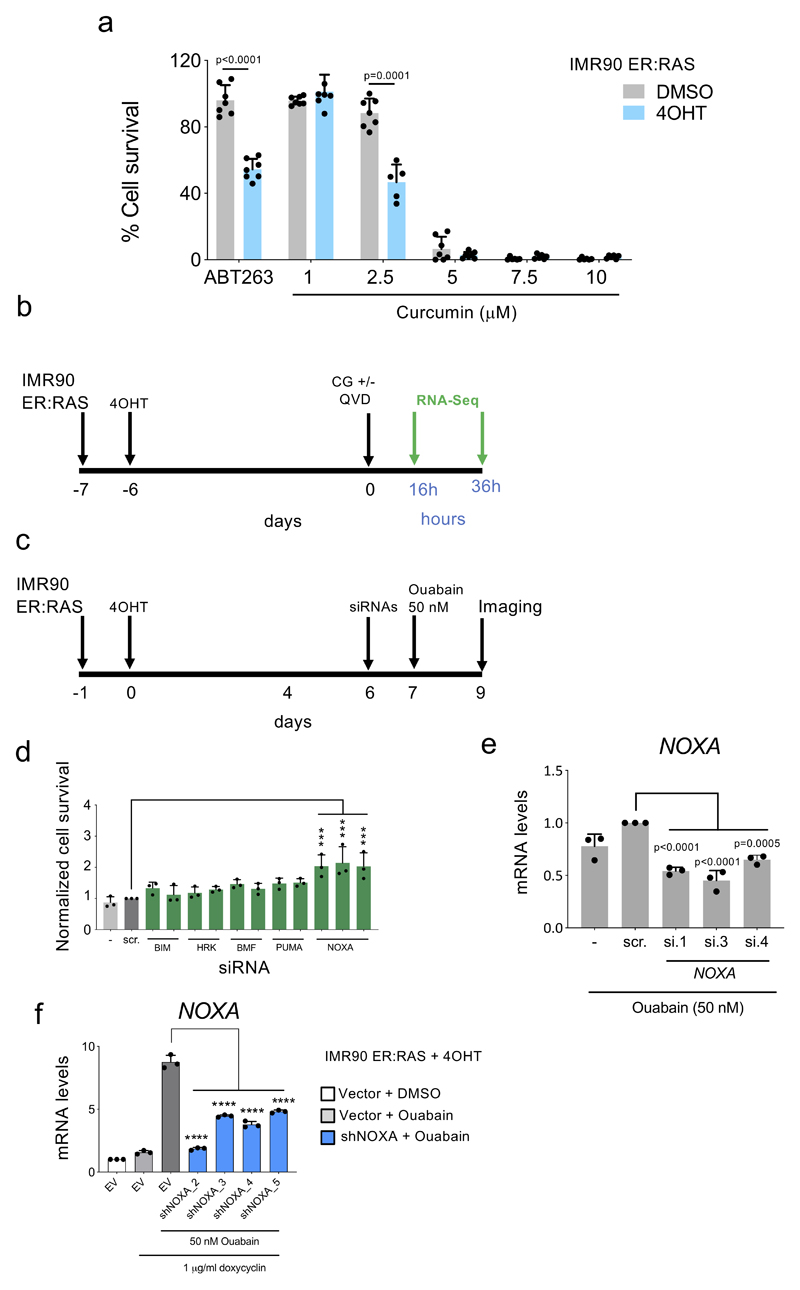
Cardiac glycosides inhibit the Na+, K+-ATPase, a member of the P-type family of cation pumps that import 2 K+ in exchange for 3 Na+ molecules 25. Gene set enrichment analysis (GSEA) suggested that cells undergoing OIS are subjected to alterations in cellular chemical homeostasis (Fig 3a). To better assess this, we compared the intracellular concentrations of Na+ (Fig 3b), Ca++ (Fig 3c) and K+ (Fig 3d) using fluorescent probes. Senescent cells displayed increased intracellular concentrations of all these cations. Interestingly, treatment with ouabain caused a more marked decrease of the intracellular levels of K+ in senescent than in normal cells, while supplementation with KCl prevented ouabain-induced (but not ABT-263-induced) death of senescent cells (Fig 3e). Curcumin, a general inhibitor of P-type pumps 31, also displayed senolytic activity (Extended data Fig 4a) suggesting that the senolytic properties of CGs could be related to the inhibition of their canonical target, Na+, K+-ATPase, although we cannot formally exclude that CGs induce senolysis via other cellular mechanisms. The rat Na+,K+-ATPase α1 subunit (rATP1A1) harbours two point mutations at Gln111 and Asn122 (analogous to those present in mouse Atp1a1) that reduce affinity to ouabain 32. Overexpression of rATP1A1 prevented the senolytic effects of ouabain, but not those of ABT-263 (Fig 3f).
Figure 3. Mechanism explaining the senolytic properties of cardiac glycosides.
a, Chemical homeostasis GSEA signature in cells undergoing OIS compared to growing cells. b-c, Intracellular levels of Na+ (n = 5) b) and Ca2+ (n = 3) c) in IMR90 ER:RAS senescent cells compared to the corresponding controls. Representative pictures (left) and quantification (right) are shown. d, Intracellular levels of K+ in IMR90 ER:RAS senescent cells compared to the corresponding controls treated with 1 μM ouabain for 18 hours or with vehicle (DMSO) (n = 3). Representative pictures (left) and quantification (right) are shown. e, Senolytic activity of 1 μM ABT-263 and 50 nM ouabain assessed after supplementing the media with 10 mM KCl (n = 4). f, Senolytic activity of ABT-263 and ouabain assessed in IMR90 ER:RAS overexpressing the ouabain-insensitive rat ATP1A1 or an empty vector (n = 4; 4OHT + vector versus 4OHT + rAtp1a1, ****P < 0.0001). Statistical significance was calculated using unpaired two-tailed Student’s t-test (b – f). g, Heatmap showing the differential expression of BCL-2 family genes in senescent compared to non-senescent cells after treatment with ouabain, digoxin or vehicle (DMSO). The timeline of the experiment is shown in Supplementary Figure 5b. 36C: cells analyzed 36h post-CG treatment on the presence of caspase inhibitor (Q-VD-OPh). h, IMR90 ER:RAS were treated with 4OHT or vehicle (DMSO) for 6 days to induce senescence. Cells were then treated with 1 μM ouabain for 6 hours, in the presence of 50 mM KCl or vehicle. Protein extracts were prepared. Immunoblots are a representative experiment out of 3. i, IMR90 ER:RAS were infected with four different doxycycline-inducible shRNAs targeting NOXA (n = 4; vector versus each different shRNA, ****P < 0.0001). Cells were subsequently treated with 4OHT or vehicle (DMSO) for 6 days to induce senescence, and with 1 μg/μl doxycycline or vehicle (DMSO) to induce NOXA knockdown. Cells were then treated with 50 nM ouabain for 48 hours. j, IMR90 ER:RAS were treated with 4OHT or vehicle (DMSO) for 6 days to induce senescence. Cells were then treated with 50 nM Ouabain for 20 hours, in the presence of 50 mM KCl, GSK3-β inhibitor, JNK inhibitor, p38MAPK inhibitor, or vehicle (DMSO). The effect of ouabain and all the different inhibitors on NOXA expression was determined by qRT-PCR (n = 5; ouabain versus ouabain + inhibitors, ****P < 0.0001). Statistical significance was calculated using one-way ANOVA (Dunnett’s test) (i – j). k, IMR90 ER:RAS were treated with 4OHT or vehicle (DMSO) for 6 days to induce senescence. Cells were then treated with 50 nM ouabain for 48 hours, in the presence of GSK3-β inhibitor, JNK inhibitor, p38MAPK inhibitor or vehicle (DMSO) (n = 3). Quantification of cell survival was performed as described in methods. Statistical significance was calculated using two-way ANOVA (Tukey’s test). All error bars represent mean ± s.d; n represents independent experiments.
To better understand the selectivity of CGs towards senescence, we analyzed the transcriptome of senescent cells treated with CGs (Extended data Figure 4b). We noted that both ouabain and digoxin induced a subset of pro-apoptotic Bcl2 family proteins (Fig 3g). We also observed an ouabain-dependent activation of JNK, GSK3-ß and p38 in senescent cells (Fig 3h). Knock down experiments suggested that several Bcl2 family proteins, most notably NOXA, mediated the senolytic actions of CGs (Fig 3i and Extended data Fig 4d and Extended data Fig 4e-f). Treatment with inhibitors of JNK, GSK3-ß or p38 signalling, confirmed the involvement of these pathways on the induction of NOXA (Fig 3j) and apoptosis in senescent cells treated with CGs (Fig 3k). In summary, the above experiments suggest that senescent cells are more sensitive to apoptosis induced by CGs in a way partially dependent on NOXA.
Ouabain eliminates preneoplastic senescent cells
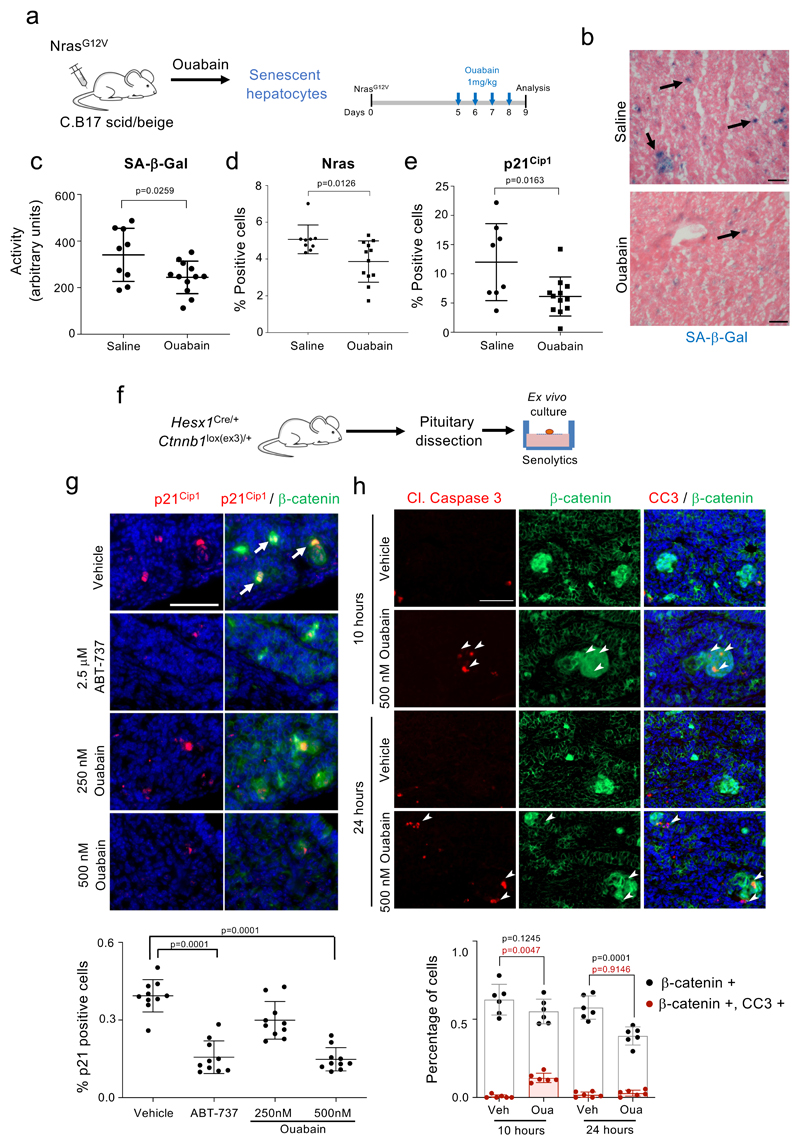
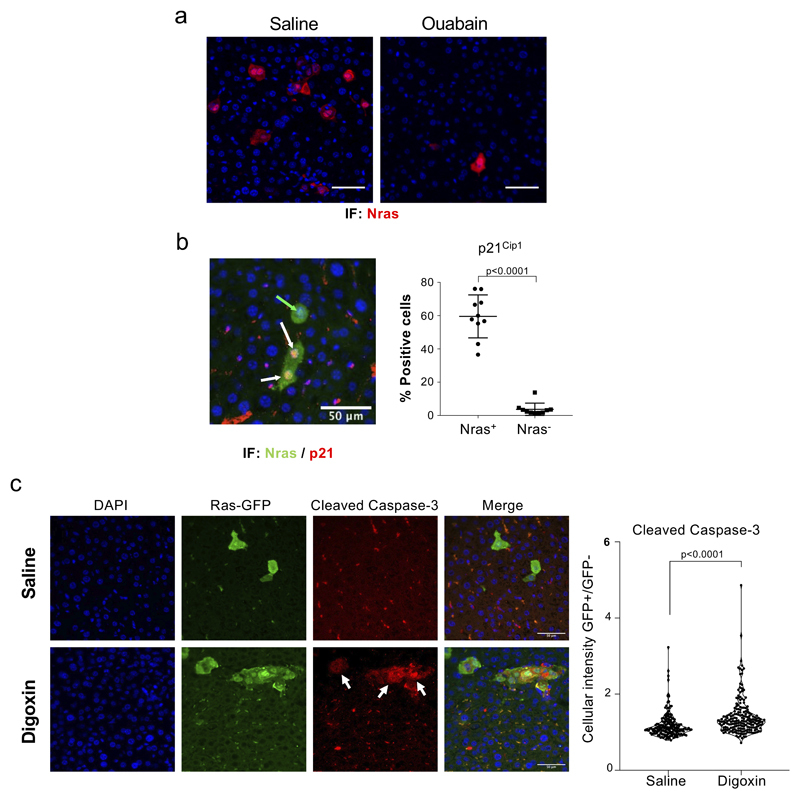
Murine cells are known to be more resistant to ouabain than human cells due to point mutations in the mouse Na+,K+-ATPase α1 subunit (mATP1A1) 32. While senescent MEFs were not killed by high doses of ouabain, ouabain behaved as a senolytic in BNL CL.2 murine liver cells (Supplementary Fig 2), suggesting that mouse models can be used to assess the senolytic effects of ouabain in vivo. Cancer-associated senescent cells modulate tumor development and therapy response at multiple levels 33. Oncogene-induced senescence protects against tumor initiation 34–37. These preneoplastic senescent cells are the subject of active immune surveillance and failure of this process results in increased tumor risk 5. To understand if ouabain could potentiate the elimination of cells undergoing OIS, we took advantage of a model of tumor initiation in the liver in which senescence is induced in hepatocytes by transposon-mediated transfer of oncogenic NRAS (NRASG12V) 5. We expressed NRASG12V in immunosuppressed mice (C.B17 scid/beige) and treated a cohort with ouabain (Fig 4a). Mice treated with CGs displayed reduced numbers of Nras-positive senescent hepatocytes due to increased apoptosis (Fig 4b-e and Extended data Fig 5), what validates the senolytic effects of CGs in vivo.
Figure 4. Ouabain selectively eliminates cells undergoing oncogene-induced senescence.
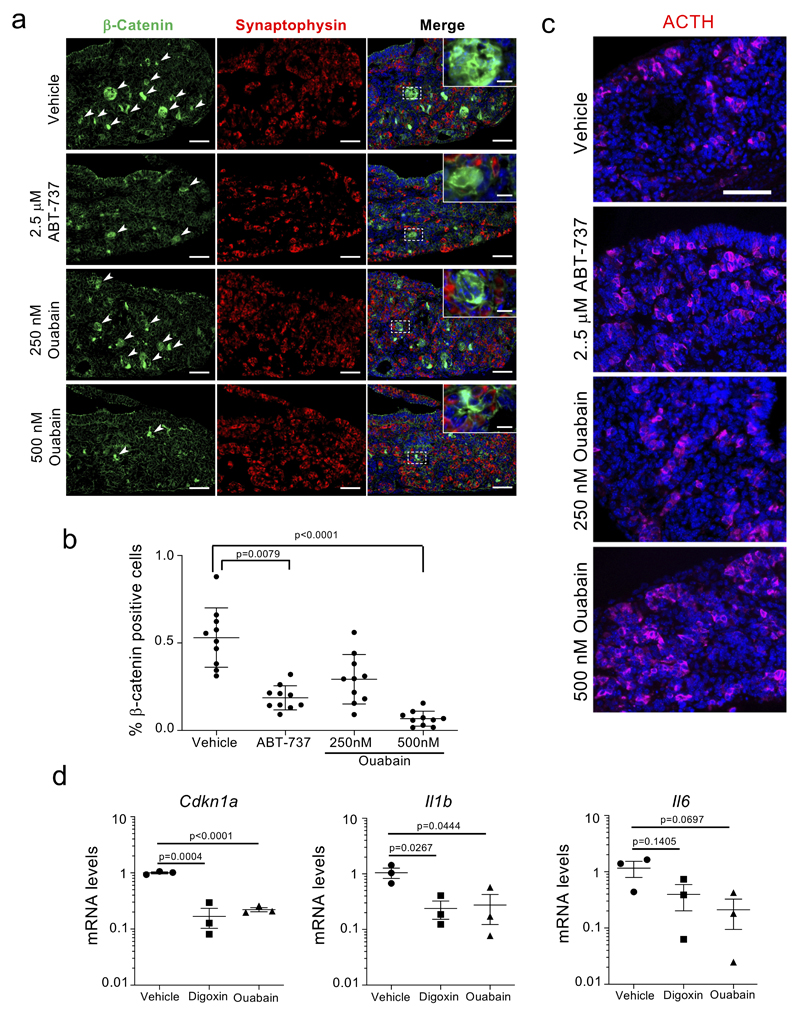
a, Experimental design for the oncogene-induced senescence experiment in the liver. b, Representative images of SA-β-Galactosidase staining in the liver. Arrows indicate examples of SA-β-Galactosidase-positive cells. Scale bar, 50 μm. c-e, Quantification of SA-β-galactosidase activity (c), Nras-positive cells (d), and p21Cip1-positive cells (e) in mice treated with vehicle (n = 9) or Ouabain (n = 12). Statistical significance was calculated using unpaired two-tailed Student's t-test. f, Experimental design for the senolytic experiment in an ACP mouse model. g, Tumoral pituitaries from 18.5dpc Hesx1Cre/+;Ctnnb1lox(ex3)/+ mice were cultured in the presence of either ABT-737 (2.5 μM), Ouabain (250 nM and 500 nM) or vehicle (DMSO) and processed for analysis after 72 hours (n = 10 mice per group). Immunofluorescence staining against β-catenin (green) and p21Cip1 (red). Arrows indicate examples of double positive cells. Scale bar, 50μm. Quantitative analysis of the immunofluorescence demonstrates that ABT-737 and ouabain significantly reduce the number of β-catenin-positive and of p21-positive cells. Statistical significance was calculated using Kruskal-Wallis and Dunn’s multiple comparisons test. h, Immunofluorescence staining against β-catenin (green) and Cleaved-caspase 3 (red) and quantitative analysis (n = 6 mice per group). Arrows indicate examples of double positive cells. Scale bar, 50 μm. Statistical significance was calculated using two-way ANOVA (Tukey’s multiple comparisons test). Red refers to comparison of double positive cells, black refers to comparison of β-catenin-positive cells. Data represent mean ± s.d; n represents number of mice; ns, not significant; **P < 0.01; ***P < 0.001.
OIS is mostly considered as a tumor suppressive mechanism but senescent cells present in the tumor microenvironment can drive tumor progression. In this regard, we recently showed that in adamantinomatous craniopharyngioma (ACP), a clinically-relevant pituitary paediatric tumor, clusters of β-catenin + preneoplastic senescent cells positively influence tumor progression in a paracrine manner 38. We used a mouse model driving the expression of oncogenic β-catenin in Hesx1+ embryonic precursors 39, and confirmed, as previously shown 38, that preneoplastic clusters of β-catenin positive cells were senescent: they were arrested (ki67 -) and enriched for markers of senescent such as p21CIP1 or Glb1 (the lysosomal β-galactosidase responsible of SA-β-Gal activity, Supplementary Fig 3). To understand if ouabain could also eliminate these pro-tumorigenic senescent lesions, embryonic pituitaries (18.5 dpc) were dissected and cultured ex vivo with or without senolytics (Fig 4f). Treatment with ABT-737 or ouabain specifically eliminated the β-catenin-positive senescent cells (Fig 4g and Extended data Fig 6a-b) without affecting other cell types in the pituitary such as synaptophysin + or ACTH + cells (Extended data Fig 6a, c). Moreover, treatment with ouabain or digoxin reduced the levels of Cdkn1a (encoding for p21Cip1), Il1b and Il6 transcripts (Extended data Fig 6d), suggesting that ouabain treatment resulted on reduced senescence and decreased SASP. Finally, co-staining with antibodies recognizing cleaved caspase 3 showed that ouabain selectively induced apoptosis of β-catenin + senescent cells (Fig 4h). The above results imply that CGs could be used as a prophylactic anti-cancer strategy to eliminate incipient preneoplastic cells.
Dual benefit of ouabain on therapy-induced senescence
Therapy-induced senescence plays an important role in determining the outcome of anti-cancer treatment 40, whilst also being responsible for some of its side effects 41. Irradiation, chemotherapy and some targeted anti-cancer drugs, including aurora kinase inhibitors, all induce senescence 40,42. It has been proposed that a two-step combination of senescence-inducing anti-cancer agents followed by senolytics could be an improved strategy to treat cancer 43.
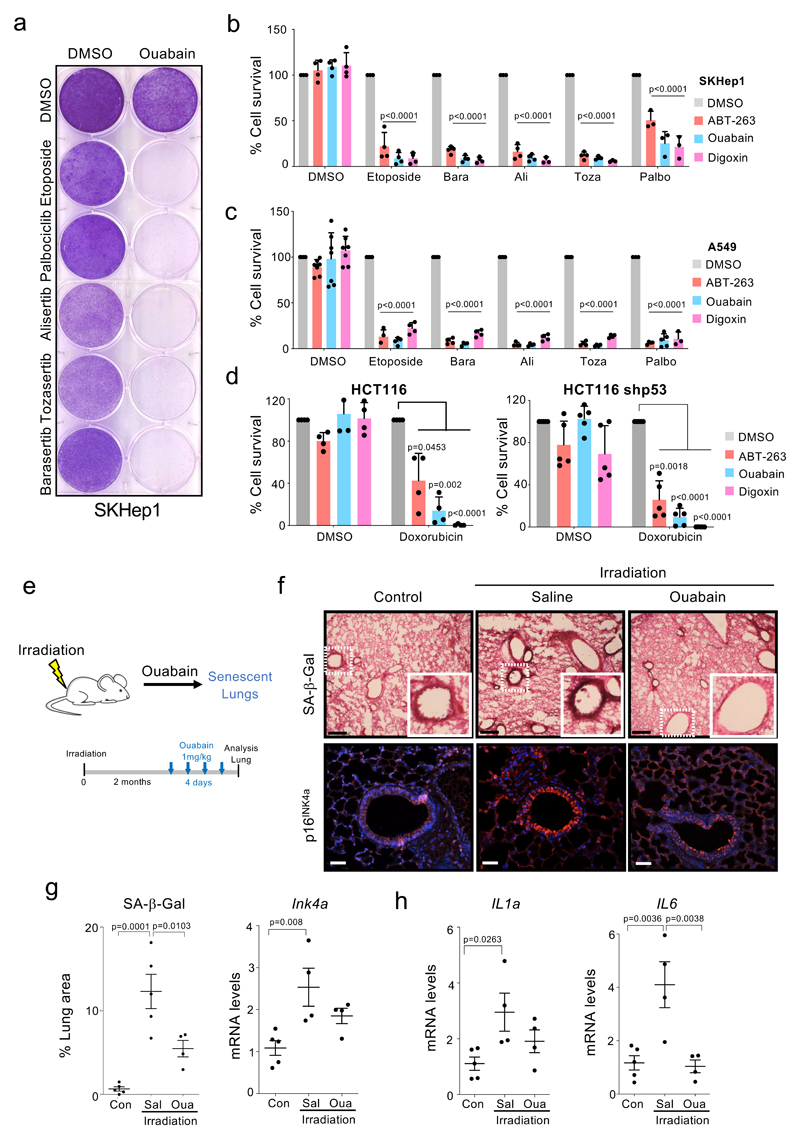
To investigate how CGs might behave in such a therapeutic protocol, we treated SKHep1 liver cancer cells or A549 lung cancer cells with senescence-inducing anti-cancer drugs. Treatment with etoposide (a Topoisomerase II inhibitor), palbociclib (that inhibits CDK4/6), or the aurora kinase inhibitors barasertib, alisertib and tozasertib, triggered senescence in SKHep1 and A549 cells (Supplementary Figure 4, 5). Following the initial drug treatment, sequential use of ouabain, digoxin, or ABT-263, a known senolytic 21 (Supplementary Figure 4a), resulted in the efficient killing of senescent cancer cells (Fig 5a-c and Supplementary Fig 5b). Similar results were obtained when two other liver cancer cells, HuH7 and HLF, were treated with alisertib followed by CGs (Extended data Fig 7a-b). The sequential use of senescence-inducing drugs and CGs also killed melanoma (SK-Mel-5, Extended data Fig 7c), breast cancer (MCF7 Extended data Fig 7d) and colon cancer (HCT116, Fig 5d) cells. Moreover, the use of syngenic lines displaying reduced p53 expression (Extended data Fig 7e-f) suggested that the effects were p53 independent (Fig 5d and Extended data Fig 7d). Finally, to assess whether the synergistic effects observed in the sequential treatments depended of senescent induction, we took advantage of the multikinase inhibitor sorafenib. Treatment with sorafenib induced growth arrest, but not senescence in SK-Hep1 and A549 cells (Supplementary Fig 4c and 5a, c-d), consistent with previous observations 44. Interestingly, we noted that sequential treatment with ouabain did not result in enhanced killing of these cancer cells (Supplementary Fig 4c and 5e-f)
Figure 5. Dual benefits of treatment with ouabain on therapy-induced senescence.
a, Crystal-violet stained 6-well dishes of SKHep1 cells that underwent therapy-induced senescence (etoposide, palbociclib or aurora kinase inhibitors) and were subsequently treated with ouabain or vehicle (DMSO). Timeline of the experiment is shown in Supplementary Fig 4a. Images are a representative experiment out of 3. b-d, Quantification of cell survival of SKHep1 cells (b), A549 cells (c) or HCT-116 cells (d) after treatment with the indicated drug combinations. Timeline of the experiment is shown in Supplementary Fig 4a (n = 4). Statistical significance was calculated using two-way ANOVA (Tukey’s test). e, Experimental design of the whole-body irradiation-induced senescence experiments. f, Representative immunofluorescence images of p16Ink4a (bottom) and SA-β-Galactosidase staining (top) in the lungs. Scale bar, 50 μm. g, Quantification of area positive for SA-β-Galactosidase staining and mRNA expression levels of p16Ink4a (n = 5 mock-irradiated control mice; n = 4 irradiated mice treated with ouabain; n = 4 irradiated mice treated with vehicle). h, Expression levels of ll1α and ll6 in lungs of non–irradiated mice (n = 5), irradiated mice treated with vehicle (n = 4) or irradiated mice treated with Ouabain (n = 4). Statistical significance was calculated using unpaired two-tailed Student's t-test. Data represent mean ± s.d; n represents independent experiments in b-d and number of mice in g-h.
Although senescence plays an important role in the outcome of anti-cancer therapies, off-target induction of senescence by irradiation or chemotherapy can also partially explain their side effects 41. To understand whether ouabain could also eliminate bystander senescent cells, we irradiated mice with 6 Gy (Fig 5e). We analysed the lung, because it is the organ where the accumulation of senescent cells after irradiation is noted first 45. Treatment with ouabain resulted in a reduced presence of senescent cells and lower expression of cytokines such as Il1a or Il6 (Fig 5f-h). A similar effect of ouabain on eliminating bystander senescent cells was observed in mice treated with doxorubicin (data not shown). The above results suggest that CGs can be combined with anti-cancer therapies to both enhance the elimination of cancer cells and avoid the accumulation of bystander senescent cells responsible of some of the side effects of these treatments.
Ouabain treatment resets immune infiltration in old mice
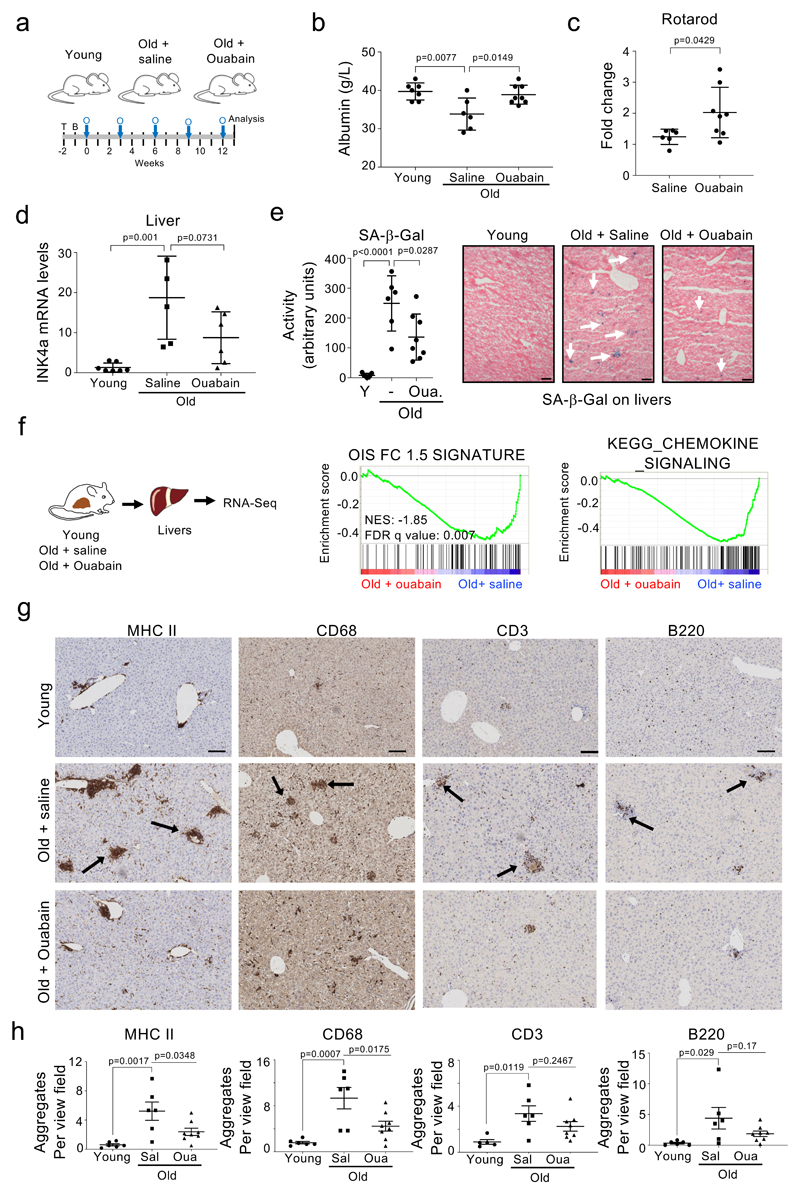
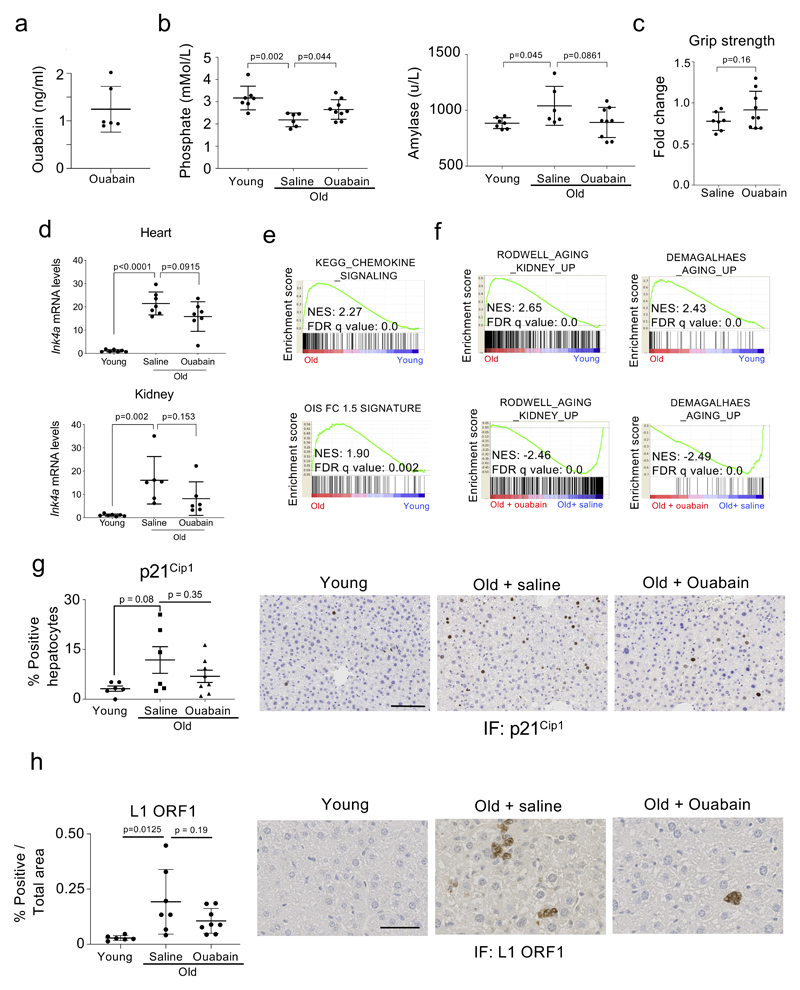
The accumulation of senescent cells in ageing organisms contributes to many diseases, including cancer 46. Since CGs are broad-spectrum senolytics, we investigated the effect that they have in the elimination of senescent cells in old mice. To this end, we subjected 24-month old female mice to a regime of intermittent ouabain treatment (Fig 6a). This regime was well tolerated in mice, with levels of ouabain in plasma reaching 1.24+/-0.48 ng/ml 24h after a round of treatment (Extended data Fig 8a). Consistent with its known pharmacological properties 47, levels of ouabain in blood dropped to undetectable levels at the end of the ‘holiday’ period. After 5 rounds of treatment, we measured different metabolic parameters in blood in a cohort of young mice and in old mice treated with ouabain or saline as a control. We observed several age-associated changes and a general trend to the reversal of such changes in mice treated with ouabain (Fig 6b and Extended data Fig 8b). For example, blood albumin levels, which are a general biomarker of well-being and fall during ageing and in a wide range of disease states 48, were decreased in old mice, a change that was significantly reversed upon treatment with ouabain (Fig 6b). Similarly, blood phosphate levels were lower in old mice and this decrease was significantly reversed upon ouabain treatment (Extended data Fig 8b). We also observed trends in the ouabain-treated cohorts to reverse the increase in amylase observed in old mice (Extended data Fig 8b). Furthermore, rotarod activity, a marker of motor performance and coordination that declines with age, was improved upon ouabain treatment (Fig 6c). There was also a non-significant improvement in the grip strength in the treated group (Extended data Fig 8c). To evaluate how ouabain treatment influences the presence of senescent cells in old mice, we assessed the expression of the senescent marker p16INK4a in different tissues. We observed reduced p16INK4a levels when comparing ouabain-treated old mice with their age-matched counterparts in several tissues, including liver, heart and kidney (Fig 6d and Extended data Fig 8d). In liver, there was also a significant reduction in SA-ß-Gal activity, suggesting that ouabain treatment diminished the number of senescent cells present in old mice (Fig 6e). Consistent with this we observed a non-significant decrease of other markers of senescence such as p21Cip1and expression of the ORF1 protein of the LINE1 transposon (Extended data Fig 8g-h). Interestingly, whereas expression of the CG-resistant Atp1a1 isoform is downregulated in the liver of old mice, the expression of a CG-sensitive isoform of mouse Na+,K+-ATPase α1 subunit, Atp1a3, increases (Supplementary Fig 6).
Figure 6. Ouabain resets immune infiltration in old mice.
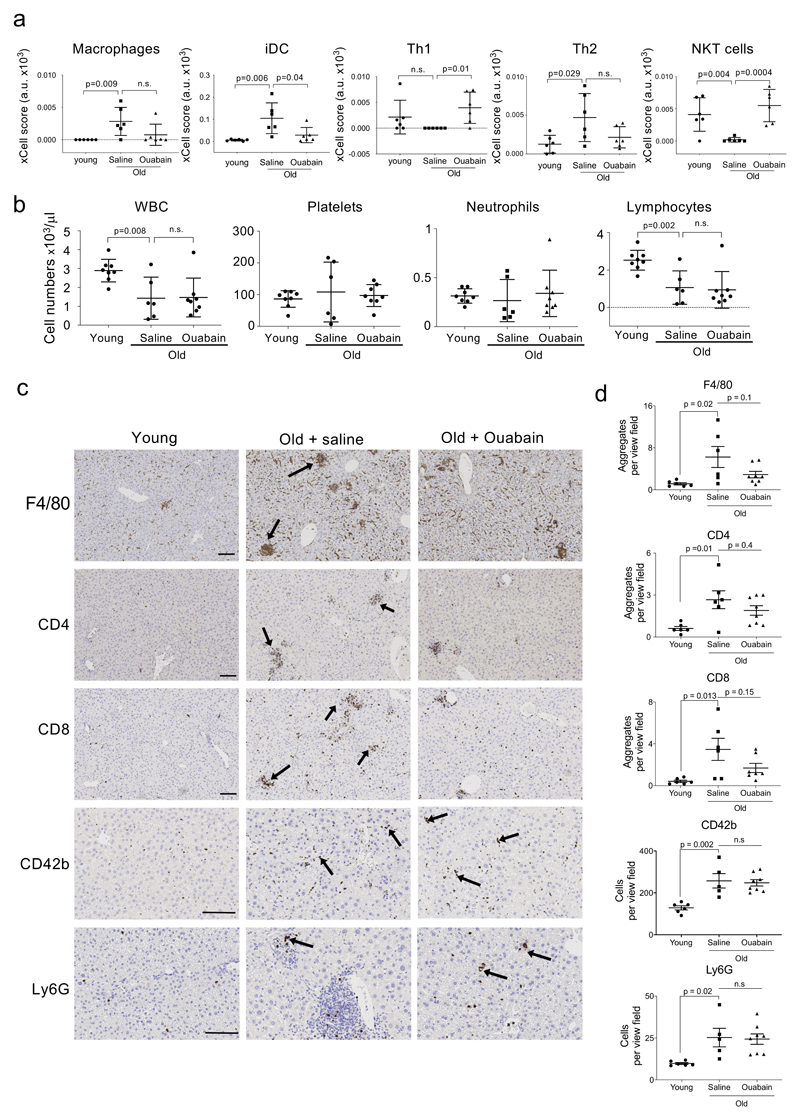
a, Experimental design for the ouabain treatment in old mice. T, time of enrolment. B, basal measures. O, ouabain treatment (3 consecutive days). b, Albumin levels of young (n = 7) and old mice, either treated with vehicle (n = 6) or ouabain (n = 8), were determined in whole-blood samples at the endpoint of the experiment. Data represent mean ± s.d; unpaired two-tailed Student's t-test. c, Motor coordination, balance and strength of old mice treated with vehicle (n = 6) or ouabain (n = 8) was assessed by performing the Rotarod test at 13 weeks after start of the experiment. Represented is the fold change versus the basal test. Data represent mean ± s.d; unpaired two-tailed Student's t-test. d, Expression levels of p16Ink4a in liver was determined by qRT-PCR following treatment with or vehicle (n = 5) or ouabain (n = 6). mRNA expression levels in young mice (n = 7) were used as reference. Data represent mean ± s.d; unpaired two-tailed Student's t-test. e, Left, quantification of SA-β-Galactosidase activity in the liver of young (n = 7) and old mice, either treated with vehicle (n = 6) or Ouabain (n = 8). Right, representative images of SA-β-Galactosidase activity in liver. Arrows indicate examples of SA-β-Galactosidase-positive cells. Scale bar, 50 μm. Data represent mean ± s.d; unpaired two-tailed Student's t-test. f, Left, experimental design for transcriptional profiling of livers from young or aged mice treated with ouabain or vehicle. Right, GSEA signature for oncogene-induced senescence and chemokines. g-h, Representative IHC images (g) and quantification (h) of the indicated immune cell markers in the liver of young (n = 6) and old mice, either treated with vehicle (n = 6) or ouabain (n = 8). Scale bar: 100µm. Data represent mean ± SEM; one-way ANOVA with Tukey’s post hoc comparison.
To further analyse the impact of ouabain during ageing, we conducted RNA-Seq experiments on these livers (Fig 6f). Using GSEA analysis, we observed the upregulation of gene signatures associated with senescence, inflammation and aging in old mice, a trend that significantly reversed upon ouabain treatment (Fig 6f and Extended data Fig 8e-f). We further analysed the transcriptome data using the xCell software, that predicts the cellular composition of the tissue 49. Using xCell, we observed that the immune infiltration in the liver changed in old mice. Interestingly, many of these changes were significantly reversed in ouabain-treated old mice (Extended data Fig 9a). Some senolytic agents, such as Bcl2 family inhibitors, affect the viability of specific immune cell populations, resulting in neutropenia and thrombocytopenia 50. To investigate whether changes in immune infiltration associated with ouabain treatment could be due to the selective killing of immune cell subsets, we conducted blood analysis at the end of the experiment. These studies did not reveal any significant alterations on the immune composition in blood upon ouabain treatment (Extended data Fig 9b), suggesting local effects on immune infiltration. This is in contrast with ABT-263 treatment that caused a drop of platelets in blood and decreased platelet infiltration in livers of mice (Supplementary Fig 7), which is consistent with what has been widely reported in human patients 50. To validate the predicted changes in immune infiltration on the livers of old mice treated with ouabain, we carried out IHC staining of liver sections using different immune cell markers. Significant increases in the formation of myeloid (MHCII+, CD68+ and F4/80+) and lymphoid (CD3+, CD4+, CD8+ and B220+) infiltrates were observed in livers of old mice compared to young mice. This trend was reversed with ouabain treatment (Fig 6g-h and Extended data Fig 9c-d). In contrast, treatment with ouabain did not affect the increased infiltration of platelets (CD42b +) and granulocytes (Ly6G +) observed in old mice (Extended data Fig 9c-d), therefore suggesting specificity. In conclusion, the above results suggest that ouabain can be used as a senolytic beyond cancer and that besides eliminating senescent cells, ouabain reduced the levels of local inflammation and immune infiltration, which could have important effects on a wide-range of pathologies.
Discussion
Senescent cells are associated with cancer, accumulate with age and are present in many pathologies 9. Genetic and pharmacological strategies have demonstrated the benefits of eliminating senescent cells in the context of aging and disease 10,12,51. Despite the multiple roles that senescence holds during cancer initiation, progression and treatment, how senolytics impact on these processes is understudied. Senescence can be triggered by a variety of insults ranging from oncogenes to genotoxic stresses, inflammation or replicative exhaustion 2. In this work, we carried out screens for drugs preferentially killing cells undergoing oncogene-induced or therapy-induced senescence. A subset of the identified compounds behave as broad-spectrum senolytics while others are just selective against a specific type of senescent cells. These differences in specificity open up possibilities for the choice of senolytic depending on their intended clinical use. One of the broad-specificity senolytics identified in the screen was ouabain, a natural compound belonging to the cardiac glycoside family. Ouabain also eliminates IMR90 E6/E7 ER:RAS cells that display increased Ras-signalling but do not undergo a senescence-associated growth arrest. This result suggests that ouabain is not just a senolytic compound but it also has a synthetic lethal interaction with RAS, which potentially makes it a cancer therapy with a useful dual mechanism of action. The results presented here are consistent with those of the Collado group, that also identified CGs as senolytic in the context of bleomycin-induced senescence of cancer cells (crossreference). Similar to ouabain, other CGs such as digoxin or digitoxin, drugs currently used in clinical practice, selectively induce apoptosis of senescent cells. Importantly, digitoxin behave as a senolytic at concentrations close to those observed in the plasma of cardiac patients treated with this drug (20–33nM) 28, suggesting the potential use of CGs as senolytics in the clinic.
The broad-spectrum senolytic action of CGs suggest that they might target a common vulnerability induced by senescence. The canonical activity of CGs is to inhibit the Na+, K+-ATPase 25 but other activities have been suggested to explain their pleiotropic effects 26. Although our data suggest that the senolytic effects of CGs are due to on-target inhibition of the Na+/K+-ATPase, it needs to be clarified whether this inhibition (fully) explains the senolytic properties of CGs. Eventually, treatment with ouabain or digoxin results in elevated levels of several proapoptotic Bcl2 family proteins. Amongst them, induction of NOXA explains in part how CGs trigger apoptosis in senescent cells.
Mice have been described more resistant than human to the canonical effects of CGs due to differences in the Atp1a1 protein 32. Therefore, a conundrum arises as to why CGs behave as senolytics in mice. One possible explanation is that CGs act via a non-canonical activity. Alternatively, we observed that the expression of the catalytic components of the Na+, K+-ATPase varies during aging. While CG-resistant Atp1a1 is downregulated in the liver of old mice, the expression of CG-sensitive Atp1a3 increases (Supplementary Fig 6). Eventually, how CGs exert their senolytic effects and how much of the benefits observed upon ouabain treatment of old mice are caused specifically by senolysis or via other effects will need to be clarified. In addition, whether the senolytic activity of CGs leads to improvements in other clinically relevant disease models associated with the accumulation of senescent cells, including cancer models, and whether CGs have senolytic activity in humans, remains to be established.Senescence occurs at different stages of cancer evolution and treatment. OIS is a powerful tumor suppressor mechanism 34–37. Therapy-induced senescence determines the response to chemo- and radiotherapy 40 whilst also contributing to their side effects 41. Our results suggest that treatment with CGs and by extension, with other senolytics can have multi-stage beneficial effects on the context of cancer therapies.
Given that chemotherapy, radiotherapy and some targeted anti-cancer drugs rely on senescence induction, a follow-up senolytic treatment has been proposed as a way to improve cancer therapy 42. Our data suggest that broad-spectrum senolytics such as CGs, will also eliminate other subsets of senescent cells such as preneoplastic cells, bystander senescent cells induced by anti-cancer therapy and age-associated senescent cells which contribute to chronic inflammation. This broad eradication of senescent cells should result in additional benefits besides those derived from the direct effect on cancer cells. In particular, the fact that CGs will eliminate incipient preneoplastic cells, should reduce the rate of cancer initiation and could suggest a prophylactic use to diminish cancer incidence. CGs have been used in the clinic for a long time. Digoxin and digitoxin are used for the treatment of heart failure and atrial arrhythmia. Interestingly, it has been suggested that CGs could be used as anti-cancer agents 52. In agreement with our results, a retrospective study showed that administration of digoxin during chemotherapy had a positive impact on overall survival in several cancer types 53. It was reasoned that the effect was due to the ability of CGs to induce immunogenic cell death. Our results provide an additional explanation for those observations: CGs synergize with chemotherapy by killing senescent cells. Moreover, the broad specificity of CGs as senolytics, and the causal role of senescence in age-related disease suggest that CGs might have benefits that extend beyond their anti-cancer properties. In this regard, studies in mice have shown that CGs reduced atherosclerosis 54 and ameliorates bleomycin induced pulmonary fibrosis 55, which is consistent with senolytic activity. Given the pleiotropic effects of CGs, we cannot discard alternative or complementary explanations beyond their senolytic effects, including direct killing of specific immune infiltrates.
We showed that intermittent ouabain treatment reduced the number of senescent cells in old mice. In that context, ouabain not only targeted senescent cells, but also had a broader impact that was reflected in improved metabolic parameters and physical fitness. Moreover, the ouabain-induced reduction of senescence diminished chronic inflammation and reversed changes in immune infiltration observed in old mice. Importantly, those effects seem to be specific as ouabain treatment did not impact into granulocytes (Ly6G +) or platelets (CD42b +). This data constitutes the first evidence of tissue remodelling happening in response to the elimination of senescent cells.
Despite the long-standing suggestions for novel clinical indications of CGs beyond heart disease, a factor limiting their use has been the potential for side effects. However, their senolytic action may require intermittent rather than continuous usage, and depending on the indication, administration could be local rather than systemic. The present study therefore suggests that the use of CGs as senolytics could be safer than initially thought. Given that digoxin has a long history of use in the clinic and its side effects can be monitored and managed 56, our pre-clinical data suggest that cardiac glycosides should be trialled as senolytics in the context of cancer therapies and beyond.
Methods
Drugs
The LOPAC 1,280 library was acquired from Sigma-Aldrich (LO1280). The following compounds were used in this study: ABT-263 (Selleckchem, S1001), Ouabain octahydrate (Sigma-Aldrich, O3125), Diphenyleneiodonium chloride (Sigma-Aldrich, D2926), JFD00244 (Sigma-Aldrich, J4829), CGP-74514A hydrochloride (Sigma-Aldrich, C3353), Etoposide (Sigma-Aldrich, E1383), Palbociclib HCl (Selleckchem, S1116), Digoxin (Sigma-Aldrich, D6003), Digitoxin (MedChemExpress HY-B1357), Bufalin (Sigma-Aldrich, B0261), Q-VD-OPh hydrate (Sigma-Aldrich, SML0063), KCl (BioVision, 2115-100), Doxycycline hydrate (Sigma-Aldrich, D9891), 4-Hydroxytamoxifen (Sigma-Aldrich, H7904), CHIR-99021 (Selleckchem, S2924), JNK-IN-8 (Selleckchem, S4901), BMS-582949 (Selleckchem, S8124), ABT-737 (Selleckchem, S1002), Alisertib (Selleckchem, S1133), Barasertib (Selleckchem, S1147), Tozasertib (Selleckchem, S1048), Doxorubicin hydrochloride (Cayman chemical, 15007), Rotenone (Sigma-Aldrich, R8875), Rottlerin (Sigma-Aldrich, R5648), Calmidazolium chloride (Tocris, 2561), BIX-01294 (Selleckchem, S8006), Mibefadril (Sigma-Aldrich, M5441), Ouabagenin (Santa Cruz, sc-295983), Strophanthidin (Sigma-Aldrich, S6626), Strophanthin K (Sigma-Aldrich, S355445), Liproxstatin-1 (Selleckchem S7699), Necrostatin-1 (Selleckchem S8037), Belnacasan (VX-765; Selleckchem S2228), Curcumin (Sigma-Aldrich, 08511), Sorafenib (Selleckchem S7397).
Antibodies
The following primary antibodies were used in this study: mouse monoclonal anti-BrdU (3D4; BD Biosciences, 555627) 1:2000, rabbit polyclonal anti-GAPDH (Abcam, ab22555) 1:2500, mouse monoclonal anti-p16INK4a (JC-8; from CRUK) 1:1000, rabbit polyclonal anti-p21 (M-19; Santa Cruz, sc-471) 1:200, rabbit monoclonal anti-phospho-c-Jun (Ser73; D47G9, Cell Signaling Technology, 3270) 1:1500, rabbit monoclonal anti-phospho-GSK-3β (Ser9; D85E12, Cell Signaling Technology, 5558) 1:2000, rabbit polyclonal anti-phospho-Akt (Ser473; Cell Signaling Technology, 9271) 1:2000, rabbit polyclonal anti-phospho-p38 MAPK (Thr180/Tyr182; Cell Signaling Technology, 9211) 1:2000, rabbit polyclonal anti-β-Catenin (Thermo, RB-9035-P1) 1:500, mouse monoclonal anti-ACTH (Fitzgerald, N/A) 1:1000, mouse polyclonal anti-p21 (BD Biosciences, 556431) 1:200, mouse monoclonal anti-Synaptophysin (27G12; Leica, NCL-L-SYNAP-299) 1:250, rat anti-MHCII (M5/114.15.2; Novus Biologicals, NBP1-43312) 1:500, rabbit polyclonal anti-CD68 (Abcam, ab125212) 1:100, rabbit monoclonal anti-CD3 (SP7; Zytomed, RBK024) 1:250, rat monoclonal anti-B220 (BD Biosciences, 553084) 1:3000, rat monoclonal anti-F4/80 (Linaris, T2006) 1:120, rat monoclonal anti-CD4 (eBioscience, 14-9766) 1:1000, mouse monoclonal anti-N-Ras (F155, Santa Cruz, sc-31), rabbit monoclonal anti-cleaved caspase 3 (Asp175; 5A1E; Cell Signaling Technology, 9664) 1:400, rabbit monoclonal anti-LINE-1 ORF1p (EPR21844-108; Abcam, ab216324) 1:500, rat monoclonal anti-CD8alpha (Invitrogen, 14-0808-82) 1:200, rabbit monoclonal anti-CD42b (Abcam, ab183345) 1:200, rat Ly-6g (BD Biosciences, 551459) 1:800, rabbit polyclonal anti-GLB1 (Proteintech, 15518-1AP) 1:100, rabbit monoclonal anti- Ki67 (Abcam, ab16667) 1:100, rat monoclonal anti-GFP (3H9, Chromotek) 1:200.
We used the following secondary antibodies: goat anti-mouse IgG (H+L, AlexaFluor 488 conjugated, Thermo Fischer Scientific, A11029), goat anti-mouse IgG (H+L), AlexaFluor 594 conjugated, Thermo Fischer Scientific, A11032), goat anti-rabbit IgG (H+L, AlexaFluor 594 conjugated, Thermo Fischer Scientific, A11037) and goat anti-rabbit IgG-HRP (Santa Cruz, sc-2004).
Cell lines
IMR-90 (ATCC® CCL-186™), HEK 293T (ATCC® CRL-11268™), SK-HEP-1 (ATCC® HTB-52™), SK-MEL-5 (ATCC® HTB-70™), HCT 116 (ATCC® CCL-247™), Primary Bronchial/Tracheal Epithelial cells (PBEC; ATCC® PCS-300-010™), MEF (ATCC® SCRC-1008™), MCF7 (ATCC® HTB-22), BNL CL.2 (ATCC® TIB-73), and A549 (ATCC® CCL-185™) were obtained from ATCC. HLF (JCRB0405) and HuH-7 (JCRB0403) were obtained from JCRB Cell Bank. IMR90 ER:RAS and IMR90 ER:RAS cells expressing E6 and E7 proteins of HPV16 were generated by retroviral infection of IMR90 cells and have been described elsewhere 57,58. IMR90, SK-Hep-1, HCT116, MCF-7, BNL CL.2 and MEFs were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Sigma) and 1% antibiotic-antimycotic solution (Gibco). A549 were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin-streptomycin (Gibco). Sk-Mel-5 were cultured in RPMI Medium 1640 (Gibco) supplemented with 10% fetal bovine serum (Sigma) and 1% antibiotic-antimycotic solution (Gibco). PBEC were cultured in Airway Epithelial Cell Basal Medium (ATCC, PCS-300-030) supplemented with Bronchial Epithelial Cell Growth Kit (ATCC, PCS-300-040). Huh7 and HLF were cultured in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin (Gibco), 1% non-essential aminoacids (Gibco), and 1% Na-Pyruvate (Gibco). To induce OIS, IMR90 ER:RAS were treated with 100 nM 4-hydroxytamoxifen (4OHT, Sigma) reconstituted in DMSO. To induce chemotherapy-induced senescence IMR90 cells were treated with 0.5 μM Doxorubicin (Sigma), for 24 hours; with 50 μM Etoposide (Sigma), for 48 hours; or with aurora kinase inhibitors (0.2 μM tozasertib, 1 μM alisertib, 1 μM barasertib), for 7 days as described elsewhere 42. To induce senescence by ionizing radiation, MEFs were γ-irradiated (10 Gy) and analysed at the indicated times.
Vector construction
The rat Na+,K+-ATPase α1 subunit (rAlpha1) construct was a gift from Mauro Giacca (ICGEB, Italy) and has been described before 59. To generate a rAlpha1 retroviral expressing vector, first the cDNA was PCR amplified using primers FwratATP1A1SnaB1 (5’-CGTACGTAGCCATGGGGAAGGGGGTT-3’) and RvratATP1A1SalI (5’-CGGTCGACGCCCTAGTAGTAGGTTTCCTT-3’), and the Platinum® PCR SuperMix High Fidelity (Invitrogen™) according to manufacturer’s instructions. Then, rAlpha1 cDNA was sub-cloned into a pBabe empty vector using standard techniques.
For de novo generation of miRE-based inducible shRNAs against NOXA, the human 97-mer oligonucleotides (IDT Ultramers) coding for the respective shRNAs (shNOXA.2, PMAIP1_5366_1221; shNOXA.3, PMAIP1_5366_1235; shNOXA.4, PMAIP1_5366_574; shNOXA.5, PMAIP1_982) were PCR amplified using the primers miRE-Xho-fw (5’-TGAACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3’) and miRE-EcoOligo-rev (5’-TCTCGAATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGC-3’), and the AccuPrime™ Pfx DNA Polymerase (Invitrogen™) according to manufacter’s instructions as described 60. Amplification products were subsequently cloned into the pRRL-TetON-Puro-Amp lentiviral backbone.
Retroviral and lentiviral infection
To generate IMR90 ER:RAS expressing rAlpha1 or inducible shRNAs against NOXA, HEK293T cells were transfected with retroviral/lentiviral and packaging vectors using PEI (PEI 2500, Polysciences). Two days after transfection, HEK293T lentiviral supernatants were collected, filtered (0.45 μM), diluted 1/4, supplemented with 4μg/ml polybrene and added to IMR90 ER:RAS cells plated the day before at a density of 1 million cells per 10 cm dish. Four hours later, lentivirus-containing media was replaced with fresh media. Two days after transfection, HEK293T undiluted retoviral supernatants were collected, filtered (0.45 μM) and supplemented with 4μg/ml polybrene and added to IMR90 ER:RAS cells plated the day before at a density of 1 million cells per 10 cm dish. Three and six hours later, fresh retrovirus-containing media was added to IMR90 ER:RAS. Three days after infection, all cells were passaged and cultured for three days in the presence of 1μg/ml puromycin (InvivoGen) to select for infected cells.
Growth assays
For BrdU incorporation assays, the cells were incubated with 10 μM BrdU for 16-18 hours and then fixed with 4% PFA (w/v). BrdU incorporation was assessed by Immunofluorescence and High Content Analysis microscopy. For crystal violet staining, the cells were seeded at low density on 6-well dishes and fixed at the end of the treatment with 0.5% glutaraldehyde (w/v). The plates were then stained with 0.2% crystal violet (w/v).
Immunofluorescence staining of cells
Cells were grown in 96-well plates, fixed with 4% PFA (w/v), permeabilised in 0.2% Triton® X-100 (v/v) diluted in PBS for 10 min, and blocked with 1% BSA (w/v) and 0.4% fish gelatin (v/v) (Sigma) for 30 min. Cells were then incubated with a primary antibody for 45 min, followed by the corresponding fluorescence-labelled secondary antibody (Alexa Fluor®) for 30 min and 1 μg/ml DAPI for 15 min. Antibodies were diluted in blocking solution. After every step, cells were washed with PBS three times.
Cytochemical SA-β-Galactosidase assay
Cells were grown on 6-well plates, fixed with 0.5% glutaraldehyde (w/v) (Sigma) in PBS for 10-15 min, washed with 1mM MgCl2/PBS (pH 6.0) and then incubated with X-Gal staining solution (1 mg/ml X-Gal, Thermo Scientific, 5 mM K3[Fe(CN)6] and 5 mM K4[Fe(CN)6] for 8 hr at 37°C. Bright field images of cells were taken using the DP20 digital camera attached to the Olympus CKX41 inverted light microscope. The percentage of SA-β-Gal positive cells was estimated by counting at least 100 cells per replicate sample facilitated by the “point picker” tool of ImageJ software (NIH).
For SA-β-Galactosidase staining in tissues, frozen sections (6 μm) were fixed in ice-cold 0.5% glutaraldehyde (w/v) solution for 15 min, washed with 1mM MgCl2/PBS (pH 6.0) for 5 min and then incubated with X-Gal staining solution for 16-18 hr at 37°C. After the staining, the slides were counterstained with eosin, dehydrated, mounted and analysed by phase-contrast microscopy. SA-β-Gal tissue staining was quantified using ImageJ software (NIH) by measuring the percentage of stained area in each section and multiplying it by its mean intensity value as described before 61. To exclude the luminal spaces in the lung sections, the percentage of SA-β-Gal positive area was divided by the total lung area, as determined by eosin-positive area using ImageJ (NIH).
Fluorescence SA-β-Galactosidase assay
Cells were grown on 96-well plates and incubated with DDAOG (D-6488, Life Technologies) for 2 hours. Cells were then fixed with 4% PFA for 15 min and nuclei was stained with 1 μg/ml DAPI. Images were taken using high-throughput fluorescent microscope IN Cell Analyzer 2000 (GE Healthcare) with a 20x objective. The percentage of SA-β-Gal positive cells was estimated using the INCell Investigator 2.7.3 software based on differences on cell intensity over an arbitrary threshold.
Determining senolytic activity
For oncogene-induced senescence experiments, IMR90 ER:RAS cells were plated in 96-well dishes and induced to undergo senescence by treating them with 100 nM 4OHT for 6 days. At that point, 10 µM of the indicated drugs were added, unless otherwise stated. In parallel, the same treatments were carried out in IMR90 ER:RAS cells treated with DMSO (-4OHT). These cells do not undergo senescence. Cells were fixed at day 9 after 4OHT induction and stained with 1 μg/ml DAPI for 15 min to assess cell numbers using automated microscopy. Different models of senescence were used to test the senolytic compound activity in cell culture in a similar fashion. Briefly, for therapy-induced senescence IMR90 cells were treated with 50 μM etoposide (48h), 0.5 μM doxorubicin (24h) or left untreated, and then kept in drug-free complete media until day 7, when the senolytics were added. Cells were fixed at day 10 after senescence induction. In all senescence types tested, 3-day course of senolytics was applied. The percentage of cell survival was calculated dividing the number of cells after drug treatment by the number of cells treated with vehicle.
For Huh7 and HLF, cells were plated in 6-well dishes and induced to undergo senescence by treating them with 500 nM Alisertib for 4 days, removing the compound and allowing for senescence establishment for another 4 days. For senolysis, compounds were subsequently added for 3 days at concentration of 1 µM or 5 µM. Quantification of cell viability was done by collecting all cells and determining the number of dead vs. viable cells by trypan blue exclusion.
High Content Analysis (HCA)
IF imaging was carried out using the automated high-throughput fluorescent microscope IN Cell Analyzer 2000 (GE Healthcare) with a 20x objective with the exception of DNA damage foci analysis which required a 40x objective. Fluorescent images were acquired for each of the fluorophores using built-in wavelength settings (‘DAPI’ for DAPI, ‘FITC’ for AlexaFluor® 488 FITC, ‘Texas Red’ for AlexaFluor® 594 and ‘Cy5’ for DDAOG). Multiple fields within a well were acquired in order to include a minimum of 1,000 cells per sample-well. HCA of the images were processed using the INCell Investigator 2.7.3 software as described previously 62. Briefly, DAPI served as a nuclear mask hence allowed for segmentation of cells with a Top-Hat method. To detect cytoplasmic staining in cultured cells, a collar of 7-9 μm around DAPI was applied. To detect cytoplasmic staining in tissue sections, a multiscale top-hat parameter was set on the reference wavelength (typically NRAS staining). Nuclear IF in the reference wavelength, i.e. all the other wavelengths apart from DAPI, was quantitated as an average of pixel intensity (grey scale) within the specified nuclear area. Cytoplasmic IF in the reference wavelength was quantitated as a coefficient of variance (CV) of the pixel intensities within the collar area. Nuclear foci IF in the reference wavelength was quantified as n number of foci per nucleus. In samples of cultured cells, a threshold for positive cells was assigned above the average intensity of unstained or negative control sample unless otherwise specified. In tissue sections, a threshold for positive cells was assigned above background staining using the built-in ‘cell to background ratio’ measurement. Immunohistochemistry imaging and quantification was also automated.
Gene expression analysis
Total RNA was extracted using Trizol reagent (Invitrogen) and the RNeasy isolation kit (Qiagen). cDNA was generated using random hexamers and SuperScript II reverse transcriptase (Invitrogen). Quantitative real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems) in a CFX96 real-time PCR detection system (Bio-Rad). GAPDH or RPS14 expression were used for normalization. Human primer pairs are:
PMAIP1: ACCAAGCCGGATTTGCGATT, ACTTGCACTTGTTCCTCGTGG.
RPS14: CTGCGAGTGCTGTCAGAGG, TCACCGCCCTACACATCAAACT.
Mouse primer pairs are:
Gapdh: AACTTTGGCATTGTGGAAGG, ACACATTGGGGGTAGGAACA
and ATGACATCAAGAAGGTGGTG, CATACCAGGAAATGAGCTTG.
Il1α: CGCTTGAGTCGGCAAAGAAAT, TGGCAGAACTGTAGTCTTCGT.
Il6: TGATTGTATGAACAACGATGATGC, GGACTCTGGCTTTGTCTTTCTTGT.
Ink4A: CCCAACGCCCCGAACT, GCAGAAGAGCTGCTACGTGAA.
Cxcl1: CTGGGATTCACCTCAAGAACATC, CAGGGTCAAGGCAAGCCTC.
Rps14: GACCAAGACCCCTGGACCT, CCCCTTTTCTTCGAGTGCTA.
Cdkn1a: CAGATCCACAGCGATATCCA, ACGGGACCGAAGAGACAAC.
Il1β: TGCCACCTTTTGACAGTGATG, TGATGTGCTGCTGCGAGATT.
Il6: CAAGAAAGACAAAGCCAGAGTC, GAAATTGGGGTAGGAAGGAC.
Atp1a1: GGGGTTGGACGAGACAAGTAT, CGGCTCAAATCTGTTCCGTAT.
Atp1a2: TGAGCTGGGCCGAAAATACC, GGGTCCATCTCTAGCCAGAAT.
Atp1a3: TCAGGGACCTCTTTCGACAAG, GCATCAGCTTTACGGAACCC.
Immunoblot
Cells were lysed in RIPA buffer (80 mM Tris pH 8.0, 150 mM NaCl, 1% Triton® X-100, 0.5% Na-Doc, 0.1% SDS, 1mM EDTA) supplemented with 1 tablet of phosphatase and 1 tablet of protease inhibitors (Roche) per 10 ml RIPA. Lysis was performed on ice for 20 min with occasional vortex followed by centrifugation at 13,000 rpm for 15 min at 4°C to collect supernatant containing the protein extracts. Immunobloting was carried out using standard technique. Briefly, proteins were separated by size using precast gels (Bio-Rad) and then transferred to nitrocellulose membranes. The membranes were blocked for 1 hour in PBS supplemented with 0.1% Tween® 20 (v/v) and 5% BSA (w/v) (Sigma) and incubated with primary antibodies at 4°C overnight. After three washes with PBS supplemented with 0.1% Tween® 20 (v/v), the membranes were incubated with the corresponding secondary antibody conjugated with horseradish peroxidase and visualized using ECL system (GE Healthcare) for chemiluminescent detection of the protein bands.
Transfection of siRNAs
IMR90 ER:RAS cells in suspension (100 μl) were reverse-transfected with siRNAs (Dharmacon) on a well of a 96-well plate. The suspension media was DMEM supplemented with 10% FBS only. The transfection mix for each sample contained 0.2 μl of DharmaFECT™ 1 (Dharmacon) in 17.4 μl plain DMEM mixed with 3.6 μl siRNA 30 min before to cell seeding. 18 hours after transfection, the media was replaced in fresh complete media. 24 hours after transfection, 50 nM ouabain or vehicle (DMSO) were added. The cells were fixed 72 hours after transfection with 4% PFA (w/v) and stained with DAPI (1 μg/ml) for 15 min as previously described. The following siRNAs were acquired from Dharmacon and used in this study: scrambled siRNAs (D-001210-01 and D-001210-02). siRNAs targeting the following human genes: PMAIP1 (D-005275-07 and D-005275-08), BBC3 (D-004380-05 and D-004380-06), BMF (D-004393-25 and D-004393-26), HRK (D-008216-04 and D-008216-05) and BCL2L11 (D-004383-17 and D-004383-18).
IncuCyte analysis
IMR90 ER:RAS cells were plated in 96-well dishes and induced to undergo senescence as previously described. Different concentrations of ouabain and digoxin were added as normally. Cell culture media was supplemented with IncuCyte NucLight Rapid Red reagent for cell labelling (Essen Bioscience) and IncuCyte Caspase-3/7 reagent for apoptosis (Essen Bioscience). Four images per well were collected every 2 hr for 3 days using a 10x objective.
Measurement of intracellular ions
Intracellular K+, Na+and Ca2+ were measured using Asante Potassium Green-2 AM (Abcam, 2 μM), CoroNa Green, AM (Invitrogen, 5 μM) and the Fluo-4 AM (Invitrogen, 2 μM) probes, respectively. Cells were incubated with the probes for 30 min (CoroNa Green and Fluo-4) or 60 min (Asante Potassium Green-2 AM). Probes were diluted in HBSS (Hank’s Balanced Salt Solution, Sigma). After treatment, cells were washed and fixed 4% PFA (w/v) as normally for Asante Potassium Green-2 AM and CoroNa Green. For cells incubated with Fluo-4 AM, cells were incubated with Hoechst (1 μg/ml) for the last 10 min of the incubation and were imaged alive.
Mouse models and drug treatments
All mice were purchased from Charles River UK. For induction of senescence, C57BL/6J mice at of 8–12 weeks of age were exposed to a sublethal dose (6 Gy) of total body irradiation. 8 weeks after, mice were injected with 1 mg/kg ouabain (i.p.) or vehicle for 4 consecutive days. Mice were killed 24 h after the last injection. Mice lungs were harvested for RNA extraction, paraffin embedded for immunohistology, or frozen in OCT/Sucrose 15% (1:1) solution for cryosectioning and SA-β-gal stains. The mice used for all experiments were randomly assigned to control or treatment groups. Both sexes were used throughout the study.
For in vivo treatment, ABT-263 was prepared in ethanol:polyethylene glycol 400:Phosal 50 PG at 10:30:60 as previously described 45. Mice were gavaged with vehicle (ethanol:polyethylene glycol 400:Phosal 50 PG) or ABT-263 (50mg/kg). Peripheral whole blood and liver samples were collected 6 hours after dosing.
Female C57BL/6J mice aged 98 to 103 weeks at the start of the experiment (they were not littermates) were treated with 1 mg/kg ouabain (i.p.) (n = 9) or vehicle (n = 7) for 13 weeks. Mice were injected 3 times per week every other day (day 1, 3 and 5). Every week of treatment was followed by two weeks of rest (except for the experiment endpoint). A group of young female C57BL/6J mice (10 weeks old) was used as reference. Mice were trained twice for all the physical tests before starting treatment. Tests were always performed the week after ouabain treatment.
Female C.B-17 SCID/beige (CB17.Cg-PrkdcscidLystbg-J/Crl) mice were injected at 5–8 weeks of age. Vectors for hydrodynamic injection were prepared with the Sigma-Aldrich GenElute HP Endotoxin-Free Plasmid Maxiprep Kit. Transposon-mediated gene transfer was previously described 5; briefly 20 μg of the indicated vector containing NrasG12V and 5 μg of SB13 transposase-expressing plasmid were diluted in sterile-filtered phosphate-buffered saline to a total volume of 10% of the body weight of the animal before being injected into the lateral tail vein in under 10 s. 5 days after HTVI, mice were treated with 1 mg/kg ouabain (i.p.) or vehicle for 4 consecutive days. The mice were randomly assigned to control or treatment group. 24 h after the last injection of the drug (9 days after HTVI) mice were culled and livers collected. All mouse procedures were performed under licence, following UK Home Office Animals (Scientific Procedures) Act 1986 and local institutional guidelines (UCL or Imperial College ethical review committees).
Physical tests
The rotarod apparatus (Ugo Basile, Italy) was used to measure fore- and hind-limb motor coordination, balance and strength. Mice received three trials per day with an inter-trial interval of 1 h for 3 consecutive days. The rod accelerated from 5 to 60 rpm over a period of 570 s and the latency to fall was recorded. For the subsequent analysis the mean latency was taken for the 9 trials and plotted as a value for the week.
A grip strength meter was used to measure forelimb strength. To measure grip strength the mouse is swung gently by the tail so that its forelimbs contact the bar. The mouse instinctively grips the bar and is pulled horizontally backwards, exerting a tension. When the tension becomes too great, the mouse releases the bars. The maximum load is recorded by the grip strength meter, which is then returned to zero before the next test. The mouse is placed in its home cage for a minute to rest before the next test. Each mouse performs five consecutive tests, and the three best scores are used for statistical analysis. This protocol is preferred because lower scores are often due to the mouse failing to grip the bar effectively, rather than reflecting muscular strength.
Vetscan VS2 for analysis of blood parameters
Whole blood was collected into lithium-heparin coated tubes (Abaxis, UK) from the carotid artery following cervical dislocation of mice. 120-140 μL of whole blood was then added into either the comprehensive diagnostic profile rotor for geriatric testing (Abaxis, UK) and ran on the VetScan VS2 Chemistry Analyzer (Abaxis, UK).
Peripheral whole blood analysis of immune cell composition
Blood was collected using citrate or EDTA (Microvette® CB 300 K2E, Sarstedt) as anticoagulant from tail vein whole blood and diluted using saline to a volume of at least 200 ml. Complete blood counts were obtained using the Sysmex XE2100 automated cell counter (Sysmex Corporation, Japan).
IHC staining of liver sections
Livers were fixed overnight in 4% PFA prior to embedding and sectioning. Paraffin-embedded liver sections (2 μm) were processed for immunohistochemistry (IHC) thereafter. Automated staining was performed on BOND-MAX (Leica Biosystems). Antigen retrieval was carried out with Bond™citrate solution (AR9961, Leica), Bond™EDTA solution (AR9640, Leica) or Bond™ proteolytic enzyme kit (AR9551, Leica) after which sections were incubated with antibodies against antigens in Bond™ primary antibody diluent (AR9352, Leica Biosystems). Primary antibody exposure was followed by secondary antibody (Leica Biosystems) and staining using the Bond Polymer Refine Detection Kit (DS9800, Leica Biosystems). For quantification of stainings, slides were scanned using a SCN400 slide scanner (Leica Biosystems) at 20x magnification.
ELISA for ouabain
C57BL/6J mice were treated with 1 mg/kg ouabain (i.p.) for 4 consecutive days. 24 hours after the last injection, blood was collected from tail vein using lithium heparin coated Microvette tubes (Sarstedt CB300 LH). Samples were centrifuged at 2000g for 5min at room temperature and the supernatant was collected as plasma for subsequent ELISA analysis (Cloud-Clone Corp., CEV857Ge).
Ex vivo culture of mouse pituitaries
Neoplastic pituitaries were dissected from Hesx1Cre/+;Ctnnb1lox(ex3)/+ embryo at 18.5dpc. Wild type pituitaries were dissected from Hesx1+/+;Ctnnb1lox(ex3)/+ embryo at 18.5dpc. The sex is not determined. These embryos are obtained in the same litters by crossing Hesx1Cre/+ males 2 to 6 months old (MGI:3822473; 63; C57BL/C6 background) with Ctnnb1lox(ex3)/lox(ex3) females 2 to 6 months old (MGI:1858008; 64; C57BL/C6 background). Genotype is assessed by phenotypic discrimination. After dissection, pituitaries were placed on top of 0.2 μM Whatman filters (SLS) in 24 well plates containing 500μl of media (DMEM-F12, Gibco, 1% Pen/Strep, Sigma and 1% FBS, PAA) supplemented with either ABT-737 (2.5 μM), ouabain (250 nM and 500 nM) or vehicle (DMSO). Media was changed every 24h, pituitaries were processed for analysis after 72 hours. Immunofluorescence staining was performed as previously described 38. The proportion of β-catenin-accumulating and p21-positive cells was calculated as an index out of the total DAPI-stained nuclei. The proportion of β-catenin-accumulating, cleaved-caspase-3 and p21-positive cells was calculated as an index out of the total DAPI-stained nuclei. Over 300,000 DAPI nuclei were counted from ten histological sections per sample, in a total of twelve neoplastic pituitaries. For gene expression analysis, after 72h of ex vivo culture, pituitaries were passed through a Qiashredder column (Qiagen) and processed for total RNA extraction using the RNeasy Micro kit (Qiagen). 150ng of total RNA was reverse transcribed to cDNA using the Transcriptor First Strand cDNA Synthesis Kit and random hexamers (Roche).
RNA-Seq analysis
Quality of the raw sequencing reads were assessed using FASTQC. Human RNA-Seq samples were aligned to Human genome hg19 by suing Tophat (v 2.0.11) with parameters " --library-type fr-firststrand" and using Ensembl version 72 gene annotation. Mouse RNA-Seq samples were aligned to Mouse genome mm9 by Tophat (v 2.0.11) with parameters " --library-type fr-firststrand" and using Ensembl version 67 gene annotation. The remaining downstream analysis steps are common to both human and mouse datasets.
Gene expression levels were quantified as number of uniquely aligned reads overlapping with gene coordinates using feature counts function form Rsubread R package. Differentially expressed genes between conditions were identified using DESeq2 R package and genes with Benjamini–Hochberg corrected p-values < 0.05 were defined as differentially expressed. For PCA and unsupervised clustering, the read counts were normalised using rlog from DESeq2 65. Heatmaps were generated using heatmap.2 function available in gplots R package.
For GSEA, genes were ranked using 'Wald statistics' from DESeq2 results and GSEA was performed using GSEA desktop application version 2.2.7 with Molecular Signature Database version 3.1.
We used xCell 49 to infer the various cell types enriched in our samples. xCell uses gene signatures derived from 64 immune and stroma cell types to determine the cell type composition in each sample. Normalised expression values (FPKM) were computed using R script and gene symbol with highest expression was selected for duplicate gene symbols. Human homologs for mouse genes were retrieved using MGI Human-Mouse homology report:
(http://www.informatics.jax.org/downloads/reports/HOM_MouseHumanSequence.rpt). Abundance scores were then computed using xCell online version (http://xcell.ucsf.edu/) and visualised using boxplots.
Statistical analysis
GraphPad Prism 8 was used for statistical analysis. Two-tailed unpaired Student’s t-tests were used to estimate statistically significant differences between two groups. Two-way ANOVA with Tukey’s post hoc comparison was used for multiple comparisons. Values are presented as mean ± s.d. unless otherwise indicated. Asterisks (*) always indicate significant differences as follows: ns = not significant, * = P < 0.5, ** = P < 0.01, *** = P < 0.001.
For in vivo studies, mice were randomly assigned to treatment groups. All replicates in this study represent different mice.
Extended Data
Extended Figure 1. IMR90 ER:RAS cells as a model of OIS.
a, Quantification of immunofluorescence staining for BrdU, p16INK4a, and SA-β-Galactosidase of IMR90 ER:RAS cells 6 or 8 days after treatment with 4-OHT or vehicle (DMSO) (n = 3). b, Representative immunofluorescence images. BrdU incorporation, which indicates proliferation, is stained green; p16INK4a is stained red. Scale bar, 50 μm. SA-b-Galactosidase is stained red. Scale bar, 100 μm. c, Expression levels for IL8 and IL1A of senescent and control IMR90 ER:RAS cells 6 days after 4-OHT or vehicle (DMSO) (n = 4). d, DAPI staining of senescent and control IMR90 ER:RAS cells after 1 μM ABT-263 treatment for 3 days showing reduced numbers of senescent cells after ABT-263 treatment. Scale bar, 100 μM. e, Senolytic activity of the indicated drugs in the context of oncogene-induced senescence in IMR90 ER:RAS cells (n = 4). f, Quantification of immunofluorescence staining for BrdU in IMR90 ER:RAS cells expressing E6 and E7 proteins of HPV16 (n = 3). g, Senolytic activity of the indicated drugs in IMR90 ER:RAS cells expressing E6 and E7 proteins of HPV16. All error bars represent mean ± s.d; n represents independent experiments. All statistical significances were calculated using unpaired two-tailed Student’s t-tests.
Extended Figure 2. Senolytic drug screen in therapy-induced senescence.
a, Quantification of immunofluorescence staining for BrdU, SA-b-Galactosidase activity, p21CIP1 and 53BP1 in IMR90 cells treated with 50 μM etoposide (n = 3). b-c, Senolytic activity of the indicated drugs in the context of therapy-induced senescence in IMR90 (n = 4, b) and oncogene-induced senescence in IMR90 ER:RAS cells (n = 4, c). All error bars represent mean ± s.d; n represents independent experiments. All statistical significances were calculated using unpaired two-tailed Student’s t-tests.
Extended Figure 3. The glycoside chain in CGs is dispensable for their senolytic activity.
a, Dose response analysis of senolytic activity of ouabain in IMR90, control IMR90 ER:RAS cells (DMSO), senescent IMR90 ER:RAS cells (4-OHT) and IMR90 ER:RAS cells expressing E6 and E7 proteins of HPV16 (n = 3). b, Dose response analysis of senolytic activity of digitoxin in the context of oncogene-induced senescence in IMR90 ER:RAS cells (n = 3). c, Chemical structure of ouabain and its aglycone version, ouabagenin. d, Quantification of cell survival in senescent and control IMR90 ER:RAS cells after treatment with ouabagenin, the aglycone version of ouabain (n = 6). e-f, Quantification of cell survival of IMR90 ER:RAS cells undergoing OIS and the corresponding controls after treatment with the CG KStrophanthin (e) (n = 4) or its aglycone version Strophantidin (f) (n = 5). g, Quantification of cell survival in senescent and control IMR90 ER:RAS 3 days after 1 μM ABT-263 or CG treatment (50 nM ouabain, 100 nM digoxin). Senolytic drugs were added 8 days after 4-OHT or vehicle (DMSO) (n = 4). All error bars represent mean ± s.d; n represents independent experiments. All statistical significances were calculated using unpaired two-tailed Student’s t-tests.
Extended Figure 4. Senescent cells are more sensitive to CGs due to their altered osmotic balance.
a, Quantification of cell survival of senescent and control IMR90 ER:RAS cells after treatment with curcumin (n = 6). Statistical significance was calculated using unpaired two-tailed, Student’s t-test. b, Experimental design for the transcriptional profiling of senescent and control IMR90 ER:RAS cells after treatment with cardiac glycosides (CG). QVD indicates treatment with a general caspase inhibitor (Q-VD-OPh). c, IMR90 ER:RAS cells were transfected with 2 independent siRNAs targeting BCL-2 family genes at day 6 after senescence induction as indicated in the scheme. d, IMR90 ER:RAS were transfected with at least two independent siRNAs targeting BCL-2 family genes at day 6 after senescence induction (n = 3; scrambled siRNA versus three different siRNAs against NOXA, ***P < 0.001). The timeline of the experiment is shown in (c). Statistical significance was calculated using one-way ANOVA (Dunnett’s test). e, Expression levels of NOXA after knock down with three independent siRNAs (n = 3). Statistical significance was calculated using one-way ANOVA (Dunnett’s test). f, Expression levels of NOXA after knock down with four independent shRNAs (n = 3; vector versus different shRNAs against NOXA, ****P < 0.0001). Statistical significance was calculated using one-way ANOVA (Dunnett’s test). All error bars represent mean ± s.d; n represents independent experiments.
Extended Figure 5. Ouabain eliminates liver preneoplastic senescent cells.
a, Representative images of immunofluorescence staining of Nras. Mice were treated with vehicle (n = 9) or ouabain (n = 12) as explained in Fig 4a. Nras is stained in red. Scale bar, 70 μm. b, Immunofluorescence staining and quantification of p21Cip1 in Nras-positive senescent hepatocytes vs Nras-negative normal hepatocytes. Nras is stained in green, p21Cip1 is stained in red. White arrows indicate Nras-positive, p21Cip1-positive cells; green arrow indicates a Nras-positive, p21Cip1-negative cells (n = 10 per group). c, SCID/beige mice were treated with saline or Digoxin (1mg/kg) on two consecutive days, 5 days after hydrodynamic transduction of Nras-GFP. Mice were culled 6 hours after the second treatment. Representative images of immunofluorescence staining of GFP and cleaved caspase-3 and quantification of intensity levels in 1/2 independent experiments (n = 200 cells). Scale bar, 50 μm. Statistical significance was calculated using unpaired two-tailed Student's t-test. Data represent mean ± s.d; n represents number of mice.
Extended Figure 6. Ouabain eliminates preneoplastic senescent cells.
a, Representative images of immunofluorescence staining of β-catenin (green) and synaptophysin (red) in tumoral pituitaries from 18.5dpc Hesx1Cre/+;Ctnnb1lox(ex3)/+ mice that were cultured in the presence of either ABT-737 (2.5 μM), ouabain (250 nM and 500 nM) or vehicle (DMSO) (n = 10 per group). Scale bar, 50 μm. b, Quantitative analysis of the immunofluorescence in (a) demonstrates that ABT-737 and ouabain significantly reduce the number of β-catenin-positive cells. Statistical significance was calculated using Kruskal-Wallis and Dunn’s multiple comparisons test. c, Representative images of immunofluorescence staining of ACTH (adrenocorticotropic hormone; magenta). Scale bar: 50μm. d, qRT-PCR analysis revealing that the senescent marker Cdkn1a (encoding for p21Cip1) and the SASP components Il1b and Il6 are reduced in neoplastic pituitaries treated with 100 nM ouabain and 100 nM digoxin relative to vehicle controls (n = 3 per group). Statistical significance was calculated using unpaired two-tailed Student's t-test; data represent mean ± s.d; n represents number of mice.
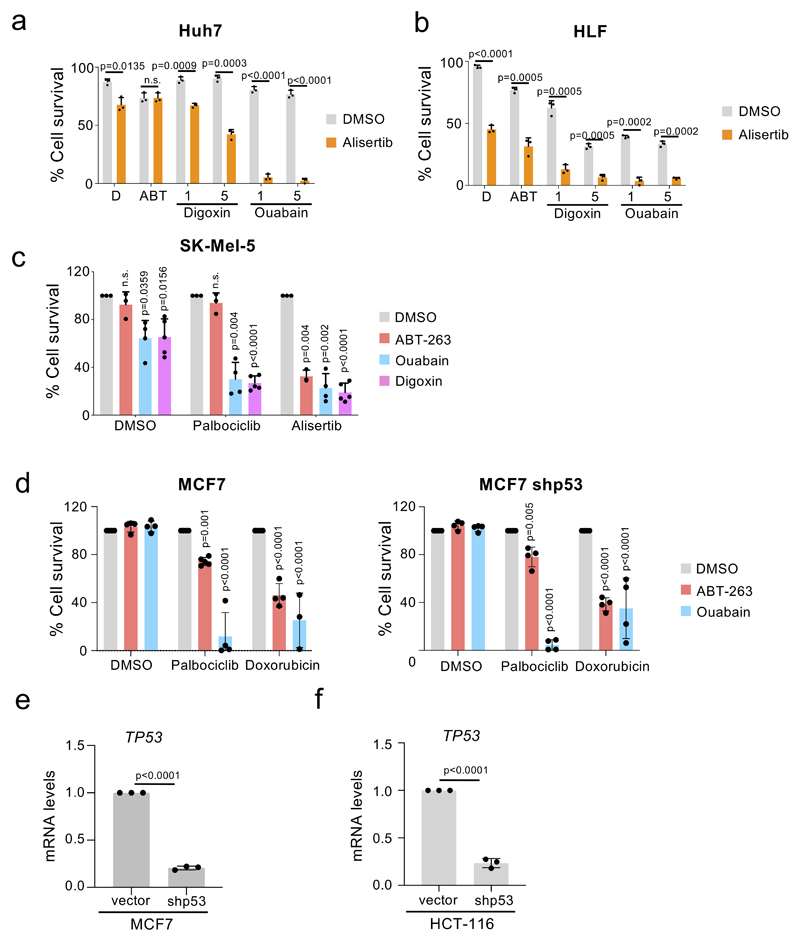
Extended Figure 7. Anti-cancer effect of cardiac glycosides across different human cancer cell lines.
a-b, Quantification of cell survival by trypan blue staining of Huh7 cells (a) and HLF cells (b) after treatment with the indicated drug combinations (n = 3). Timeline of the experiment is shown in Supplementary Fig 4a. Statistical significance was calculated using unpaired two-tailed Student's t-test. c, Quantification of cell survival of senescent (alisertib, palbociclib) and control (DMSO) SK-Mel-5 melanoma cells (n = 4). Statistical significance was calculated using two-way ANOVA (Dunnett’s test). d, Quantification of cell survival of senescent (doxorubicin, palbociclib) and control (DMSO) MCF-7 or MCF-7 breast cancer cells infected with a shRNA against TP53 (n = 4). Statistical significance was calculated using twoway ANOVA (Dunnett’s test). e-f, mRNA expression levels of TP53 in MCF-7 cells (e) and HCT-116 cells (n = 3). Statistical significance was calculated using unpaired two-tailed, Student’s t-test. Data represent mean ± s.d; n represents independent experiments; ns, not significant.
Extended Figure 8. Ouabain treatment reverses age-associated changes in old mice.
a, Ouabain levels in plasma were assessed by ELISA 24 hours after finishing a 4-day course of daily 1mg/kg ouabain i.p. injections. (n = 6). b, Phosphate and amylase levels of young (n = 7) and old mice, either treated with vehicle (n = 6) or ouabain (n = 9), were determined in whole-blood samples at the endpoint of the experiment. Statistical significance was calculated using unpaired two-tailed Student's t-test. c, Grip strength assessment in old mice treated with vehicle (n = 7) or ouabain (n = 9) 10 weeks after the start of the experiment, referred to the basal test. Statistical significance was calculated using unpaired two-tailed Student's t-test. d, Expression levels of p16Ink4a in heart and kidney were determined by qRT-PCR following treatment with vehicle (n = 6) or ouabain (n = 7). mRNA expression levels in young mice (n = 7) were used as reference. Statistical significance was calculated using unpaired two-tailed Student's t-test. e-f, GSEA signature for chemokines, oncogene-induced senescence (e) and ageing (f). g, Quantitative analysis (left) and representative IHC pictures (right) of p21Cip1 positive hepatocytes in the liver of young (n = 6) and old mice treated with ouabain (n = 8) or vehicle (saline) (n = 6). Scale bar, 100 μm. Data represent mean ± s.e.m. Statistical significance was calculated using one-way ANOVA with Tukey’s post hoc comparison. h, Quantitative analysis (left) and representative IHC pictures (right) of LINE-1 ORF in the liver of young (n = 6) and old mice treated with ouabain (n = 8) or vehicle (saline) (n = 7). Statistical significance was calculated using one-way ANOVA with Tukey’s post hoc comparison. Scale bar, 50 μm. Data represent mean ± s.d; n represents number of mice.
Extended Figure 9. Ouabain treatment resets immune infiltration in old mice.
a, xCell analysis of the transcriptome data predicts changes in immune infiltration in the liver of old mice that could be reverted with ouabain. RNA-Seq data from the livers of young (n = 6) and old mice, either treated with vehicle (n = 6) or ouabain (n = 6) was used. Statistical significance was calculated using unpaired two-tailed Student's t-test. b, Blood analysis at the end of the experiment show that ouabain treatment does not change immune composition. Blood from young (n = 8) and old mice, either treated with vehicle (n = 6) or ouabain (n = 8) was used. Statistical significance was calculated using unpaired two-tailed Student's t-test. c-d, Representative IHC images (c) and quantification (d) of the indicated immune cell markers in the liver of young (n = 6) and old mice, either treated with vehicle (n = 6) or ouabain (n = 8). Scale bar: 100μm. Statistical significance was calculated using one-way ANOVA with Tukey’s post hoc comparison. Data represent mean ± s.e.m.; n represents number of mice; ns, not significant.
Supplementary Material
Acknowledgements
We are grateful to members of J. Gil’s laboratory for reagents, comments and other contributions to this project. We thank R. Laberge and A. Bhushan for feedback and members of the Genomics LMS facility (L. Game, K. Rekopoulou and A. Ivan) for help with RNASeq. Core support from MRC (MC-A652-5PZ00 and MC_U120085810), and grants from LifeArc and Unity Biotechnology funded this research in J. Gil’s laboratory. D.J.W. was funded by a Wellcome Trust Strategic Award (098565) and core support from MRC (MC-A654-5QB40). L.Z. is supported by the German Research Foundation: DFG EXC 2180 – 390900677 [Image Guided and Functionally Instructed Tumour Therapies“ (iFIT)], FOR2314, SFB-TR209, Gottfried Wilhelm Leibniz Program], the German Ministry for Education and Research (BMBF): eMed/Multiscale HCC, the European Rresearch Council: Concolidartor grant ‘CholangioConcept’ and the German Center for Translational Cancer Research (DKTK). J.P.M.-B. was funded by the Brain Tumour Charity (SIGNAL and EVEREST), Great Ormond Street Hospital (GOSH) Children’s Charity and National Institute of Health Research Biomedical Research Centre at GOSH for Children NHS Foundation Trust and University College London. J.P.M.-B. is a GOSH for Children’s Charity Principal Investigator. S.M. is a PhD fellow funded by Boehringer Ingelheim Fonds.
Footnotes
Reporting Summary. Further information on research design is available in the Reporting Summary linked to this article.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request. The RNA-seq data generated in this study have been deposited in the GEO database under accession number GSE122081.
Author Contributions
A.G. and N.H. performed, designed, analyzed experiment sand wrote the manuscript.
B.S., V. W., A.J.I., J.Bi., J. H., A. O., S.G., R.G., K. W., J.P., E.E.I., D.H., Ju.G. and S.M. performed, designed and analyzed experiments.
G.D. carried out bioinformatics analysis.
J. Be., D. D., A.G.U., L.Z., S.V., J.P. M.-B. and D.J.W. designed experiments and secured funding.
Je.G. conceived and designed the project, secured funding and wrote the manuscript, with all authors providing feedback.
Competing Interests
J.G. owns equity and has acted as a consultant for Unity Biotechnology and Geras Bio. Unity Biotechnology funds research on senolytics in J.G.’s laboratory. J.G., A.G. and N.H. are named inventors in a MRC patent related to senolytic therapies (PCT/GB2018/051437).
References
- 1.Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128:1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang TW, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 6.Acosta JC, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tchkonia T, Zhu Y, van Deursen J, Campisi J, Kirkland JL. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest. 2013;123:966–972. doi: 10.1172/JCI64098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McHugh D, Gil J. Senescence and aging: Causes, consequences, and therapeutic avenues. J Cell Biol. 2018;217:65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 10.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Childs BG, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354:472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinta SJ, et al. Cellular Senescence Is Induced by the Environmental Neurotoxin Paraquat and Contributes to Neuropathology Linked to Parkinson's Disease. Cell Rep. 2018;22:930–940. doi: 10.1016/j.celrep.2017.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussian TJ, et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature. 2018;562:578–582. doi: 10.1038/s41586-018-0543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, et al. The Achilles' heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Discovery of piperlongumine as a potential novel lead for the development of senolytic agents. Aging (Albany NY) 2016;8:2915–2926. doi: 10.18632/aging.101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuhrmann-Stroissnigg H, et al. Identification of HSP90 inhibitors as a novel class of senolytics. Nat Commun. 2017;8:422. doi: 10.1038/s41467-017-00314-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, et al. ABT-263 induces apoptosis and synergizes with chemotherapy by targeting stemness pathways in esophageal cancer. Oncotarget. 2015;6:25883–25896. doi: 10.18632/oncotarget.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y, et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15:428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yosef R, et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat Commun. 2016;7 doi: 10.1038/ncomms11190. 11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ovadya Y, Krizhanovsky V. Strategies targeting cellular senescence. J Clin Invest. 2018;128:1247–1254. doi: 10.1172/JCI95149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgilis A, et al. PTBP1-Mediated Alternative Splicing Regulates the Inflammatory Secretome and the Pro-tumorigenic Effects of Senescent Cells. Cancer Cell. 2018;34:85–102 e109. doi: 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Therien AG, Blostein R. Mechanisms of sodium pump regulation. Am J Physiol Cell Physiol. 2000;279:C541–566. doi: 10.1152/ajpcell.2000.279.3.C541. [DOI] [PubMed] [Google Scholar]
- 26.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 27.Cheng JW, Rybak I. Use of digoxin for heart failure and atrial fibrillation in elderly patients. Am J Geriatr Pharmacother. 2010;8:419–427. doi: 10.1016/j.amjopharm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Lazaro M. Digitoxin as an anticancer agent with selectivity for cancer cells: possible mechanisms involved. Expert Opin Ther Targets. 2007;11:1043–1053. doi: 10.1517/14728222.11.8.1043. [DOI] [PubMed] [Google Scholar]
- 29.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(Pt 20):3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrand DG, et al. alpha-Fucosidase as a novel convenient biomarker for cellular senescence. Cell Cycle. 2013;12:1922–1927. doi: 10.4161/cc.24944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dao TT, et al. Demethoxycurcumin Is A Potent Inhibitor of P-Type ATPases from Diverse Kingdoms of Life. PLoS One. 2016;11:e0163260. doi: 10.1371/journal.pone.0163260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price EM, Lingrel JB. Structure-function relationships in the Na,K-ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry. 1988;27:8400–8408. doi: 10.1021/bi00422a016. [DOI] [PubMed] [Google Scholar]
- 33.Sieben CJ, Sturmlechner I, van de Sluis B, van Deursen JM. Two-Step Senescence-Focused Cancer Therapies. Trends Cell Biol. 2018;28:723–737. doi: 10.1016/j.tcb.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 35.Michaloglou C, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 36.Braig M, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 37.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Meljem JM, et al. Stem cell senescence drives age-attenuated induction of pituitary tumours in mouse models of paediatric craniopharyngioma. Nat Commun. 2017;8:1819. doi: 10.1038/s41467-017-01992-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaston-Massuet C, et al. Increased Wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc Natl Acad Sci U S A. 2011;108:11482–11487. doi: 10.1073/pnas.1101553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt CA, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 41.Demaria M, et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov. 2016 doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, et al. High-Throughput Functional Genetic and Compound Screens Identify Targets for Senescence Induction in Cancer. Cell Rep. 2017;21:773–783. doi: 10.1016/j.celrep.2017.09.085. [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Bernards R. Taking advantage of drug resistance, a new approach in the war on cancer. Front Med. 2018;12:490–495. doi: 10.1007/s11684-018-0647-7. [DOI] [PubMed] [Google Scholar]
- 44.Rudalska R, et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med. 2014;20:1138–1146. doi: 10.1038/nm.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selden R, Smith TW. Ouabain pharmacokinetics in dog and man. Determination by radioimmunoassay. Circulation. 1972;45:1176–1182. doi: 10.1161/01.cir.45.6.1176. [DOI] [PubMed] [Google Scholar]
- 48.Salive ME, et al. Serum albumin in older persons: relationship with age and health status. J Clin Epidemiol. 1992;45:213–221. doi: 10.1016/0895-4356(92)90081-w. [DOI] [PubMed] [Google Scholar]
- 49.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson WH, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Childs BG, et al. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov. 2017;16:718–735. doi: 10.1038/nrd.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mijatovic T, et al. Cardiotonic steroids on the road to anti-cancer therapy. Biochim Biophys Acta. 2007;1776:32–57. doi: 10.1016/j.bbcan.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Menger L, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med. 2012;4:143ra199. doi: 10.1126/scitranslmed.3003807. [DOI] [PubMed] [Google Scholar]
- 54.Shi H, et al. Digoxin reduces atherosclerosis in apolipoprotein E-deficient mice. Br J Pharmacol. 2016;173:1517–1528. doi: 10.1111/bph.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, et al. Ouabain ameliorates bleomycin induced pulmonary fibrosis by inhibiting proliferation and promoting apoptosis of lung fibroblasts. Am J Transl Res. 2018;10:2967–2974. [PMC free article] [PubMed] [Google Scholar]
- 56.Whayne TF., Jr Clinical Use of Digitalis: A State of the Art Review. Am J Cardiovasc Drugs. 2018 doi: 10.1007/s40256-018-0292-1. [DOI] [PubMed] [Google Scholar]
- 57.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agostini S, et al. Inhibition of Non Canonical HIV-1 Tat Secretion Through the Cellular Na(+),K(+)-ATPase Blocks HIV-1 Infection. EBioMedicine. 2017;21:170–181. doi: 10.1016/j.ebiom.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fellmann C, et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 61.Tordella L, et al. SWI/SNF regulates a transcriptional program that induces senescence to prevent liver cancer. Genes Dev. 2016;30:2187–2198. doi: 10.1101/gad.286112.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herranz N, et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat Cell Biol. 2015;17:1205–1217. doi: 10.1038/ncb3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andoniadou CL, et al. Lack of the murine homeobox gene Hesx1 leads to a posterior transformation of the anterior forebrain. Development. 2007;134:1499–1508. doi: 10.1242/dev.02829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.