Abstract
Somatic mutations can result in the formation of neoantigens, immunogenic peptides that are presented on the tumor cell surface via HLA molecules. These mutations are expected to be under negative selection pressure, but the extent of the resulting neoantigen depletion remains unclear. Based on HLA affinity predictions, we annotated the human genome for its translatability to HLA binding peptides and screened for reduced single nucleotide substitution rates in large genomic datasets from untreated cancers. Apparent neoantigen depletion signals became negligible when considering trinucleotide-based mutational signatures, either due to lack of power or efficient immune evasion mechanisms active early during tumor evolution.
Cancer is caused by somatic mutations in driver genes. These genomic alterations result in a selective growth advantage and positive selection of the affected cells1. With the rise of next-generation sequencing technologies, increasing insights into the cancer genome have led to a comprehensive characterization of the frequencies and patterns of somatic mutations across different cancers2,3. For a tumor to evolve, it also needs to develop ways to avoid immune destruction, a process referred to as immunoediting and one of the more recent hallmarks of cancer4,5. Mouse studies have shown that T lymphocyte recognition of tumor-specific antigens is crucial for immunoediting to occur6. The accumulation of somatic mutations in the tumor genome results in the formation of neoantigens, small peptides presented on the cell surface that can stimulate cytotoxic (CD8+) T lymphocytes (CTLs). To attenuate these CTL responses, a cancer cell can upregulate ligands for checkpoint receptors7. Therapeutically blocking these checkpoint pathways has been shown effective in several cancers such as metastatic melanoma and non-small cell lung cancer7–9. However, responses to immune checkpoint blockade (ICB) therapy are still largely unpredictable, and it is not completely understood why some tumors do not respond or develop resistance to therapy.
Several genomic alterations (e.g. CASP8 mutations, B2M mutations, HLA loss) have been discovered that can partially explain this ICB therapy unresponsiveness10–16. Furthermore, as stimulation of CTLs is critically dependent on the formation and presentation of neoantigens, it is not surprising that one of the main determinants of therapy responsiveness is mutation burden17–19. Indeed, the higher the mutation burden, the higher the number of potential neoantigens and hence ways to stimulate the immune system. On the other hand, negative (or purifying) selection is expected to act on neoantigen-forming mutations. This should result in a depletion of such mutations and escape from immune-induced cancer cell death. The presence of neoantigen depletion has been suggested in several cancers such as colorectal cancer, metastatic melanoma, esophageal, bladder, cervical and lung cancer10,13,20,21. As the main determinant of CTL immunogenicity is a peptide’s capacity to bind to the cell’s human leukocyte antigens (HLA) from the type I major histocompatibility complex (MHC-I), the conclusions of these studies are mostly based on lower-than-expected numbers of non-synonymous somatic mutations in predicted HLA-binding peptides, using the number of synonymous mutations as a reference.
Somatic mutations are caused by different mutational processes that are active during tumor evolution. A widely used method for characterizing the properties of mutational processes are trinucleotide-based mutational signatures, which describe frequencies for all single nucleotide substitutions in all possible sequence contexts in terms of adjacent upstream and downstream nucleotides, resulting in a total of 96 substitution types3. This implies that the mutation probability at any genomic position is dependent on the immediate sequence context in combination with the active mutational processes. It has now been clearly demonstrated that mutational signatures need to be accounted for in any model aiming at finding signals of selection in cancer22–24. However, it is currently not clear whether and how mutational signatures and their sequence context preferences influence signals of neoantigen depletion.
Here we show that, when mutational signatures are considered, putative signals of neoantigen depletion become weak to absent in cancer genomics data from treatment-naïve tumor samples. Our results are in line with the overall weak signals of negative selection in cancer and challenge the idea that neoantigen depletion signals are detectable based on HLA affinity predictions in large-scale cancer mutation datasets.
Results
Annotation of HLA-binding regions in the human genome
Somatic mutations are expected to result in neoantigen formation when (i) the resulting peptides are presented via MHC-I and (ii) they are recognized by CTLs through specific T-cell receptor (TCR) binding, which only occurs when there is no immune tolerance, i.e. when presented peptides are new to the immune system. Given sufficient co-stimulatory signals, this will result in CTL-mediated killing of neoantigen-presenting cancer cells, enforcing a negative selection pressure during tumor evolution (Fig. 1a). We hypothesized that this specific form of negative selection and hence neoantigen depletion should be detectable as reduced mutation rates in genomic regions that can be translated to HLA-binding peptides. Therefore, our first aim was to define these regions, thereby generating an HLA-binding genomic annotation.
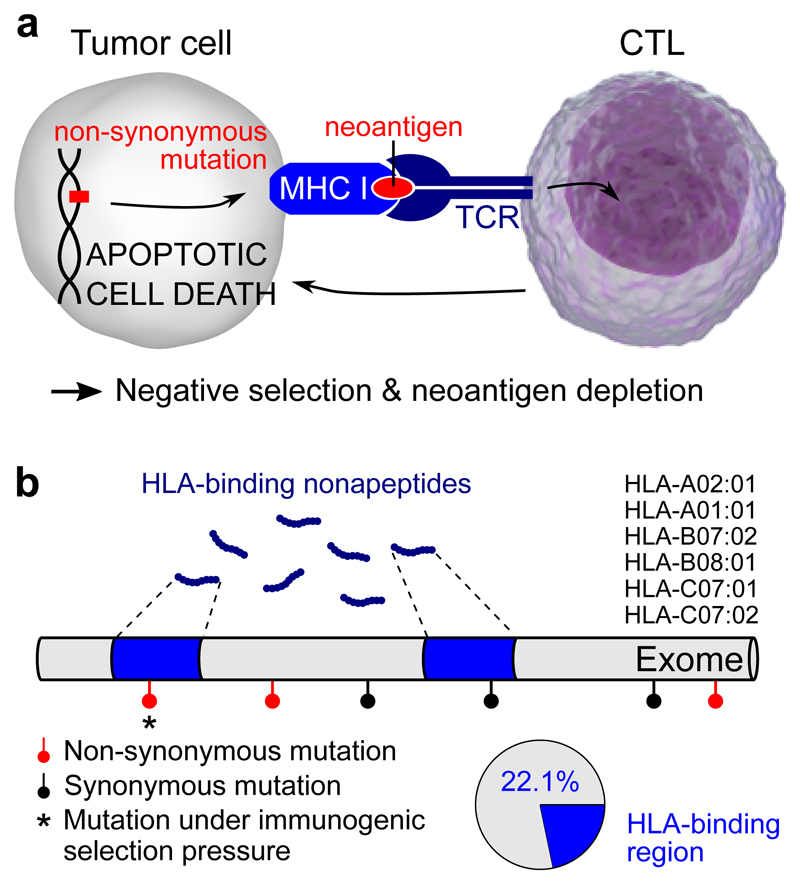
Figure 1. Development of an HLA-binding genomic annotation to detect somatic mutations under immunogenic selective pressure.
a, Neoantigen formation is expected when a non-synonymous mutation leads to a structural change in the CTL (CD8+ cytotoxic T lymphocyte) contact residues of an HLA-binding nonapeptide. This can result in CTL-mediated apoptotic cell death and hence negative selection of the underlying somatic mutation. TCR, T cell receptor; MHC-I, type I major histocompatibility complex. b, Binding affinities of all possible nonapeptides were determined for the six most common HLA alleles as indicated. Peptides were considered HLA-binding when their aggregated Kd over the six alleles was below 500 nM (see Methods); HLA-binding peptides mapped to 22.1% of the exome as indicated.
HLA-binding affinities are determined by both the amino acid sequence and by patient-specific HLA genotypes, composed of a combination of two HLA-A, two HLA-B and two HLA-C alleles. We initially considered a single prototypical HLA genotype consisting of the two most common HLA-alleles (HLA-A01:01, HLA-A02:01 HLA-B07:02, HLA-B08:01, HLA-C07:01 and HLA-C07:02; Supplementary Fig. 1), enabling us to define a single HLA-binding genome annotation to use throughout the analyses. For these six HLA alleles, the affinities were predicted for all possible nonapeptides (9-mers) translated from the coding genome and were aggregated in a single affinity, a similar approach to what has been described recently25 (see Methods and Supplementary Fig. 1). By considering a nonapeptide HLA-binding when the aggregated Kd was lower than 500 nM26, we found that the complete pool of HLA-binding nonapeptides mapped to 22.1% of the exome (Fig. 1b).
Apparent negative selection signals in HLA-binding regions
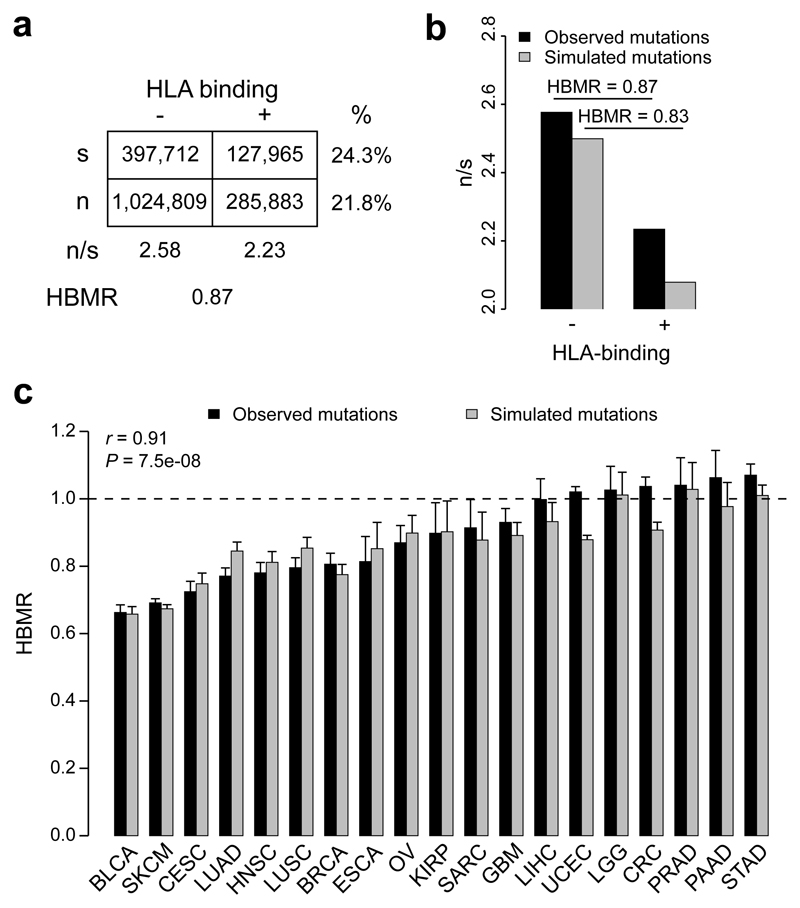
Having annotated the human exome for the HLA-binding properties of its translated peptides, we next aimed to search for signals of immune-induced negative selection in the cancer genome. All available synonymous and non-synonymous (i.e. missense) somatic mutation data were downloaded from The Cancer Genome Atlas (TCGA), encompassing 1,836,369 mutations from 8,683 different samples and spanning 32 cancer types (Supplementary Table 1). As only non-synonymous mutations in HLA-binding regions are expected to be under immunogenic selection pressure, we used the number of synonymous mutations as a background reference and determined the ratio between the observed numbers of non-synonymous and synonymous mutations (n/s) in HLA-binding as well as non-binding regions. We found that n/s was lower in HLA-binding regions on a pan-cancer level (n/s 2.23 in HLA-binding vs. 2.58 in non-binding regions, P = 3.24 × 10-298, Fisher’s exact test; Fig. 2a,b). To quantify the extent of this putative neoantigen depletion signal, we defined an HLA-binding mutation ratio (HBMR) as the ratio of n/s in HLA-binding to non-binding peptides. This way, negative immunogenic selection of somatic mutations is expected to result in HBMR values lower than 1 (or higher than 1 if these mutations have been influenced by positive selection). For the pan-cancer analysis this implied an HBMR of 0.87, suggesting the overall loss of 13% of non-synonymous mutations due to negative selection (Fig. 2a,b).
Figure 2. Analysis of somatic mutation rates in HLA-binding annotated genomic regions.
a, Contingency table showing the total number of synonymous (s) and non-synonymous (n) mutations in the HLA-binding and non-binding exome. The HLA-binding mutation ratio (HBMR) indicates the ratio of n/s in HLA-binding to non-binding regions. b, Bar plot comparing the n/s ratios of observed and simulated mutations. c, HBMR calculated for observed and simulated mutations from 19 cancer types containing at least 10,000 somatic mutations per cancer type. Error bars indicate 95% confidence intervals, calculated using two-sided Fisher’s exact test. Pearson correlation coefficient (r) and P value indicated on top left. See Supplementary Table 1 for cancer type abbreviations and sample sizes.
We next aimed to determine how these signals differed between cancer types focusing on the 19 cancer types with at least 10,000 mutations in the TCGA dataset. Given the observed mutation burdens, we estimate sufficient power (0.8 at P < 0.05) to detect negative selection operating on between 2% (UCEC) and 13% (KIRP) of the predicted neoantigens (Supplementary Fig. 2). We observed HBMR values that were significantly below 1 for 12 out of 19 analyzed cancer types, including bladder cancer (BLCA, HBMR = 0.66, P = 1.5 × 10-127), metastatic melanoma (SKCM, HBMR = 0.69, P = 0), cervical cancer (CESC, HBMR = 0.72, P = 1.3 × 10-51), lung adenocarcinoma (LUAD, HBMR = 0.77, P = 2.3 × 10-60), head and neck cancer (HNSC, HBMR = 0.78, P = 6.6 × 10-36) and squamous cell lung cancer (LUSC, HBMR = 0.80, P = 1.4 × 10-34) (Fig. 2c and Supplementary Table 2).
Reduced non-synonymous mutations in HLA-binding regions are not caused by selection processes
To be able to determine whether and to what extent selection processes and hence neoantigen depletion are indeed responsible for the observed reduction in non-synonymous mutations in HLA-binding regions, we determined the expected mutation rates in the absence of any selection pressure. For every observed somatic mutation, we simulated one mutation by randomly sampling from all possible point mutations with the same trinucleotide substitution type (e.g. TCC>TTC), resulting in a simulated mutation dataset with a similar size as the observed data. As expected, all signals of positive selection in driver genes disappeared in the simulated mutation data (Supplementary Fig. 3). Using this simulated mutation database, we recalculated the mutation rates and HBMR values. Strikingly, a strong signal of apparent negative selection and hence neoantigen depletion, similar to the real mutation data, was still present (HBMR = 0.83, P = 0; Fig. 2b). This similarity was also present for the individual cancer types (Pearson’s r = 0.91, P = 7.5 × 10-8), with the strongest signals again observed for bladder cancer and metastatic melanoma (Fig. 2c). The fact that a set of randomly generated mutations, upon which selection cannot have acted, gave results that closely mimicked those from actual mutation data casts doubt on the apparent neoantigen depletion signals. As the simulated and real mutations were only matched with respect to trinucleotide substitution types, this analysis suggests that sequence differences between HLA-binding and non-binding regions, combined with specific sequence preferences of relevant mutagenic exposures, introduce biases in n/s ratios, leading to apparent signals of neoantigen depletion.
We noted that these findings were robust to the way HLA-binding capacity was determined. Determining HLA affinities using patient-specific HLA genotypes (rather than the six most frequent alleles), focusing on the best binding allele only and using a more stringent Kd cut-off of 50 nM or a percentile-based cut-off of 1%, did not substantially alter the observed reduction in HBMR values (Supplementary Fig. 4). Similar results were also obtained when the analysis was restricted to genomic regions encoding known epitopes from IEDB (immune epitope database; Supplementary Fig. 5).
While the exclusion of non-expressed or cancer driver genes did not change the observed differences between tumor types either (Supplementary Fig. 4), we observed a lower overall percentage of somatic mutations in HLA-binding regions for expressed compared to non-expressed genes (21.7% vs. 28.3% respectively for the pan-cancer dataset, Fisher’s exact test P = 0), and an opposite effect for driver compared to non-driver genes (18.8% vs. 22.8% respectively, P = 7.6 × 10-238; Supplementary Fig. 6). Similar differences were observed for both non-synonymous and synonymous mutations, again raising doubts about a putative interpretation as immunogenic selection signals. These findings also imply that mutations in non-expressed transcripts should not be used as background reference when studying immunogenic selection pressures in cancer genomics data.
Different trinucleotide substitution probabilities explain lower non-synonymous mutation rates in HLA-binding regions
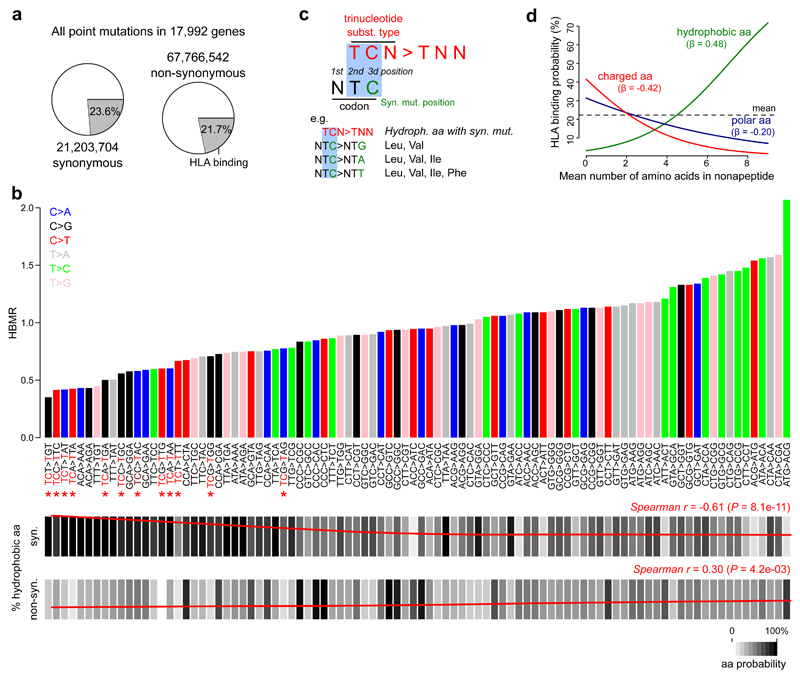
To better understand the association between trinucleotide substitution types and HLA-binding regions, we simulated all possible point mutations in 17,992 genes (21,203,704 synonymous and 67,766,542 non-synonymous mutations; Fig. 3a) and used the HBMR metric to quantify the difference between expected mutation rates in HLA-binding and non-binding regions for each trinucleotide substitution type. There was a notable variability between the trinucleotide substitution types, with HBMR values ranging from 0.35 for TCT>TGT substitutions to 2.07 for ATG>ACG substitutions (Fig. 3b). The trinucleotide substitution types with the lowest HBMR values were the most abundant in the cancer types with low overall HBMRs (e.g. 23.9% of all malignant melanoma mutations are TCC>TTC, the trinucleotide substitution type with the second to lowest HBMR; Supplementary Fig. 7). Remarkably, many of the substitution types with the lowest HBMR values were TCN>TNN (Fig. 3b), and a strong negative correlation was indeed observed between a cancer type’s HBMR value and its proportion of TCN>TNN mutations (Pearson’s r = -0.81, P = 2.4 × 10-5; Supplementary Fig. 7). Mutational signatures 2 and 3 (APOBEC-related) and the UV-induced signature 7, which are both related to these patterns, consequently had the lowest HBMR values (Supplementary Fig. 7).
Figure 3. Association between trinucleotide substitution types and HLA-binding properties.
a, All possible synonymous and non-synonymous mutations were determined in 17,992 genes. Pie charts indicate the proportions of mutations that are located in HLA-binding regions. b, Bar plot on top indicates the expected HBMR value for each trinucleotide substitution type, determined from all possible mutations from a given type in the complete exome (numbers shown in a). Main substitution types are colored as indicated by the legend on top left. Note that HBMR values are not derivable for four trinucleotide substitution types (ATT>AAT, ATT>AGT, ACT>AGT and ACT>AAT) due to the absence of synonymous mutations resulting from these substitution types (e.g. an ATT>AAT substitution can never be synonymous). TCN>TNN substitutions are indicated by red asterisks. Below the bar plot, the frequency of synonymous and non-synonymous mutations hitting hydrophobic amino acids is indicated for each substitution type (scale indicated on bottom right). Loess regression line in red with Spearman correlation coefficient (r) and P value indicated on top right (correlation between HBMR and mutation frequency for 92 different substitution types). c, Illustration of TCN>TNN mutations mainly resulting in synonymous mutations in hydrophobic amino acid codons. d, Logistic regression line indicating the correlation between a nonapeptide’s mean number of hydrophobic/charged/polar amino acids (0 to 9) and the HLA-binding probability. Regression coefficients (β) are given for each amino acid class. The mean number of amino acids for each class was determined for 1 million random exome locations (9 nonapeptides per position) to make the analysis comparable to the other analyses. A similar analysis on individual nonapeptides is shown in Supplementary Figure 10.
High synonymous mutation probabilities in hydrophobic amino acid codons correlate to lower perceived mutation rates in HLA-binding regions
We next aimed to explain the association between trinucleotide substitution types and HLA-binding properties. Because different sequence contexts imply different amino acid codon probabilities on the one hand, while different physicochemical properties of amino acids influence binding to HLA on the other hand, we investigated the relationships between trinucleotide substitution types, the amino acid content of peptides, and their expected HMBR values.
We first focused on the correlation between HBMR values and amino acid classes (hydrophobic, polar or charged) in our annotated genome. For synonymous mutations, a strong negative correlation was observed between a trinucleotide substitution type’s HBMR value and the frequency of hydrophobic amino acid codons (Spearman’s r = -0.61, P = 8.1 × 10-11; Fig. 3b), while an opposite, weaker and positive correlation was noted for non-synonymous mutations (Spearman’s r = 0.30, P = 4.2 × 10-3; Fig. 3b). This effect was mainly related to Leu, Val and Iso (Supplementary Fig. 8); hydrophobic amino acids encoded by codons with a thymine on the second codon position (Supplementary Fig. 9). Combined with the observation that most of the corresponding trinucleotide substitution types conform to the pattern TCN>TNN, this association can be explained by the upstream T of the substitution type matching with the T at the second codon position and the substituted nucleotide matching with the third codon position (Fig. 3c). Indeed, when a codon with a T at the second position is hydrophobic, any C mutation at the third position of a Leu or Val codon always results in a synonymous mutation. This is also the case for most mutations that affect the same position in Ile and for some mutations at the Phe codon as exemplified in Figure 3c.
Secondly, as hydrophobic amino acids are known to influence HLA-binding affinities27, we determined the correlation between the number of amino acids from a certain class in a nonapeptide and its HLA-binding capacity. By randomly sampling from 1 million coding regions and determining the translated peptides’ HLA-binding affinity, we observed a positive association between the number of hydrophobic amino acids in a peptide and its HLA-binding capacity (logistic regression coefficient β = 0.48, Fig. 3d and Supplementary Fig. 10).
These results demonstrate that certain trinucleotide substitution types, like TCN>TNN, which occur frequently in metastatic melanoma, bladder cancer and cervical cancer, are likely to lead to synonymous mutations in Leu, Val and Ile codons. Because these amino acids are more frequent in HLA-binding peptides, this leads to lower perceived non-synonymous mutation rates when synonymous mutations are used as a background reference. The earlier described difference in apparent neoantigen depletion in expressed vs. non-expressed genes is also related to hydrophobic amino acid content, as a gene enrichment analysis of non-expressed genes showed a strong membrane protein enrichment (e.g. olfactory receptors; Supplementary Fig. 6).
Weak to absent neoantigen depletion signals after correcting for trinucleotide substitution effects
Our study shows that differential mutation rates between HLA binding and non-binding peptides mainly result from differences in sequence composition. We next aimed to determine whether any remaining signal of neoantigen depletion would be detectable after correcting for these trinucleotide substitution effects.
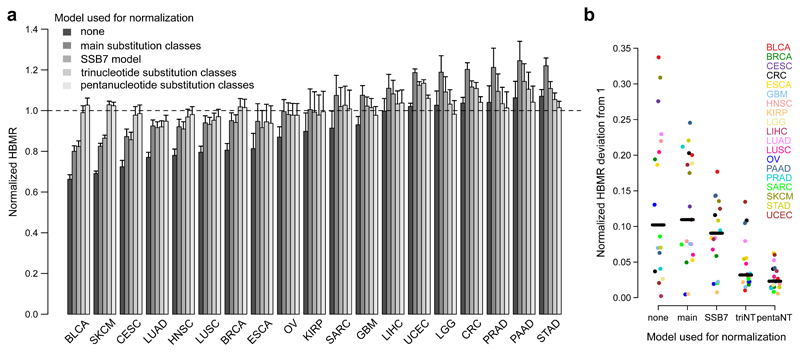
As a first approach, we normalized the observed HBMR value to its expected value for each cancer, under a trinucleotide substitution model and considering the HLA-binding annotation developed in this study (see Methods). We reanalyzed all cancers and observed a disappearance of neoantigen depletion signals, except for a limited signal in lung cancer (Fig. 4a and Supplementary Table 2). In line with our earlier findings (Supplementary Fig. 4), results did not substantially change when different criteria were used to calculate HLA binding capacity or when mutations were called using the more recent MC3 mutation caller28 (Supplementary Fig. 11). Similarly, dN/dS values did not suggest any signal of negative selection after correcting for differing trinucleotide sequence contexts in HLA binding vs. non-binding regions (Supplementary Fig. 12).
Figure 4. Weak to absent neoantigen depletion signals after correcting for trinucleotide-based mutational signature effects.
a, Bar plot showing normalized HBMR values for 19 different cancer types. HBMR values were obtained by normalization of the observed HBMR values to the expected tumor-type specific values. The latter were calculated using mutation probabilities derived from different models as indicated on top left. Error bars indicate 95% confidence intervals, calculated using two-sided Fisher’s exact test. See Supplementary Table 1 for cancer type abbreviations and sample sizes and Supplementary Table 2 for detailed results. b, Comparison of HBMR deviations from 1 after normalization using different substitution models as indicated. Each dot represents a cancer type. Median values are indicated by horizontal lines.
Notably, correcting using mutation probabilities derived from the SSB7 or other models that do not consider the complete adjacent sequence context resulted in corrected signals falsely suggestive of neoantigen depletion in e.g. melanoma and bladder cancer (Fig. 4a and Supplementary Fig. 12). Conversely, normalization using an extended sequence context (pentanucleotide substitution model) further decreased the apparent selection signals, with loss of significance in lung squamous cell carcinoma (Fig. 4a,b).
The previous results were all derived for a prototypical HLA genotype and for the reference genome (i.e. wild-type peptides). While this approach was useful in gathering new insights into associations between substitution types and HLA affinities, there is a risk of missing selection signals that are HLA genotype-specific and/or only act on mutations that result in new HLA binders (i.e. hit the HLA-binding residues of a nonapeptide, rather than the CTL contact residues). We thus searched for neoantigen depletion signals in mutated HLA-binding peptides, where binding affinities were predicted for sample-specific genotypes. We noted that only 1.88% of all non-synonymous mutations resulted in a non-binding peptide gaining HLA-binding properties (Supplementary Fig. 13). Similar numbers (1.92%) were found using our simulated mutation database, thus again providing no convincing support for selection acting on these specific mutations.
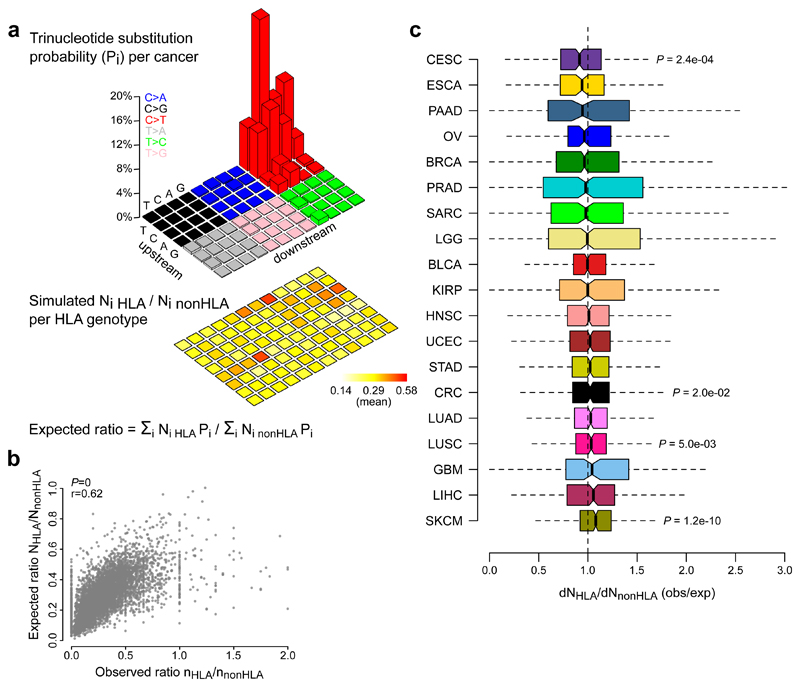
Finally, given that we have shown that synonymous mutation counts are particularly vulnerable to the effects of mutational signatures, we considered a selection metric (dNHLA/dNnonHLA) that was independent of synonymous mutations. This metric compares the observed ratio between the number of non-synonymous mutations in HLA-binding and non-binding peptides with the corresponding expected ratio. The latter was determined for each HLA genotype from all TCGA samples, using mutated peptides from 960,000 randomly simulated mutations (10,000 for each trinucleotide substitution type) and considering the aggregated mutational signature from each cancer type (Fig. 5a,b). By normalizing the observed to the expected ratios for each sample, all tumor types were reanalyzed for putative selection signals. This analysis generally confirmed the absence of detectable neoantigen depletion, except for a signal in cervical cancer (median dNHLA/dNnonHLA = 0.91, one-sample Wilcoxon signed-rank test P = 2.4 × 10-4; Fig. 5c and Supplementary Table 2). Further, dNHLA/dNnonHLA did not correlate with immune cytolytic activity (Supplementary Fig. 14). Notably, 3 out of 19 tumor types had values significantly above 1. These signals were comparable in effect size to cervical cancer, most pronounced in melanoma (median dNHLA/dNnonHLA = 1.08, P = 1.2 × 10-10), and remained when using a pentanucleotide rather than trinucleotide model (Supplementary Fig. 14). As these positive signals are unlikely to indicate true positive selection, they may rather reflect limitations of the dNHLA/dNnonHLA model, which does not consider synonymous mutation rates. Finally, neoantigen depletion signals were absent when the number of non-synonymous mutations in HLA-binding peptides was normalized to an expected number that was estimated directly from the pan-cancer dataset, as suggested previously10 (Supplementary Fig. 14). Notably, we observed that the neoantigen depletion signals in colorectal and kidney cancer, as reported by Rooney et al.10, disappeared after excluding samples with miscalled HLA genotypes from the original dataset (results obtained using authors’ source code; Supplementary Fig. 14).
Figure 5. An HLA genotype-specific analyses of mutated peptides confirms the absence of neoantigen depletion signals in most tumor types.
a, Methodological approach. For each trinucleotide substitution type (i), 10,000 mutations were randomly simulated (960,000 mutations in total). The expected number of non-synonymous mutations in HLA-binding and non-binding peptides were derived for each substitution type considering the mutated peptides’ HLA affinities for the sample-specific HLA genotype (heatmap on bottom). From these numbers, the expected ratio between non-synonymous mutations in HLA-binding and non-binding peptides was calculated using the substitution probabilities of the corresponding cancer type (legoplot on top). b, Scatter plot shows the correlation between observed and expected ratios, with Pearson correlation coefficients (r) and P values indicated on top left. c, dNHLA/dNnonHLA values were calculated for each TCGA sample and grouped by tumor types. Boxplots indicate median values and lower/upper quartiles with whiskers extending to 1.5x the interquartile range. Two-sided Wilcoxon signed-rank test was used to test deviation from 1. P values are given for cancers with q values below 0.1. Mutations in cancer driver genes or non-expressed genes were excluded. See Supplementary Table 1 for cancer type abbreviations and sample sizes and Supplementary Table 2 for detailed results.
Taken together, these results point to a general absence of detectable neoantigen depletion signals in large-scale mutation data from untreated tumors and emphasize the importance of using accurate background mutation models to correct for sequence biases introduced by relevant mutational processes.
Discussion
In this study, we initially observed an apparent reduction of somatic point mutations in genomic regions encoding HLA-binding nonapeptides. Rather than being an effect of negative selection acting on immunogenic mutations, we demonstrated correlative relationships between the probability of mutagenesis in different nucleotide sequences and predicted HLA affinities for corresponding peptides. In particular, the number of hydrophobic amino acids are a major determinant of HLA binding capacity for a peptide while simultaneously being a strong determinant of mutation rate, depending on the mutational processes at play. When correcting for these correlations, detectable negative selection signals were weak to absent. Our results demonstrate that mutation rate differences between peptides with variable HLA affinities should be interpreted with care and have broad relevance for other studies that derive selection signals from HLA affinity predictions.
To detect immunogenic selection signals, we initially annotated the human exome with respect to HLA-binding capacity by determining which segments are translatable to HLA-binding peptides, for simplicity assuming a single prototypical HLA genotype for all samples. This implies a focus on wild-type peptides under the hypothesis that mutations in CTL contact residues are subject to negative selection pressures. Using this annotation, the approach can be easily reproduced on any mutation dataset, without the need for complex and time-consuming HLA-typing or HLA affinity predictions. The theoretical drawback is that this does not capture neoantigenic mutations that lead to new HLA-binding peptides (i.e. increase the HLA affinities) and/or effects that are HLA genotype-specific. However, additional analyses addressing patient-specific HLA genotypes as well as de novo HLA-binding peptides likewise failed to produce strong support for neoantigen depletion signals.
Synonymous mutations are often used as a background mutation reference when analyzing non-synonymous substitutions with respect to selection, resulting in metrics such as dN/dS. Recent studies have shown that these metrics get confounded when not considering the adjacent sequence context22,23. A key finding of our study is that simplistic substitution models will lead to biased immunogenic selection signals, due to HLA affinity predictions also being sequence dependent. An important advantage of any metric that considers synonymous mutations as a background reference (like HBMR) is that any unexpected property that equally effects synonymous and non-synonymous mutation rates will be cancelled out (such as differential mutation burdens in expressed and non-expressed genes). However, given that we observed strong dependencies specifically between synonymous mutation probabilities and HLA-binding properties of corresponding encoded peptides, leaving synonymous mutations out of the equation may also have advantages. We did this by considering the ratio between the observed number of non-synonymous mutations in HLA-binding and non-binding regions and normalizing this ratio to an expected ratio, estimated under a trinucleotide substitution model for each individual HLA genotype. Calculation of the resulting dNHLA/dNnonHLA metric for each sample did not provide clear evidence of neoantigen depletion, similar to our initial analysis taking synonymous mutations into account. We could only detect a weak signal in cervical cancer and demonstrated that the previously reported neoantigen depletion signal in colorectal adenocarcinoma10 was due to HLA genotyping problems in samples that were later removed from TCGA. Notably, the dNHLA/dNnonHLA approach also indicated positive signals in some cancers, at effect sizes comparable to the depletion in cervical cancer. Since positive selection in HLA-binding regions is improbable, this likely reflects limitations in the accuracy of the expectation model, casting doubt on the observed negative signal in cervical cancer as well. While this may reflect exclusion of synonymous mutations in this metric, it can also be noted that mutational signatures were here determined at the tumor type level, and it is possible that consideration of patient-specific mutational signatures from whole genome sequencing datasets may potentiate more refined analyses in the future.
In addition to point mutations, which have been the main focus of studies of neoantigen depletion, future studies should also address frameshifting indels in this context. This is a different challenge, as single indels may generate large numbers of unnatural peptides through introduction of novel open reading frames, which may or may not be subject to nonsense-mediated decay29. Consistently, indels have been described as more strongly associated with response to immunotherapy30, and it can be noted that microsatellite unstable colon cancers, which harbor larger numbers of indels, appear responsive to checkpoint inhibitors while normal colon carcinomas are not31.
In summary, our results indicate that signals of neoantigen depletion, detected using HLA affinity predictions, are overall weak to absent in the untreated cancer genome. While we cannot exclude that this is related to poor accuracy to predict neoantigen formation (Supplementary Fig. 2), it is noteworthy that signals of negative selection in general are weak in cancer mutation data22,23,32,33. Therefore, either only a very small fraction of predicted neoantigenic sites are immunogenic, or the lack of negative selection signals suggests that developing tumors possess or evolve efficient immune evasion mechanisms (e.g. HLA loss or PDL1 amplification). If this is indeed the case, detectable signals of neoantigen depletion are only expected in the absence of these escape mechanisms, such as after ICB therapy21.
Methods
TCGA mutation and expression data
MuTect2-called whole exome sequencing (WES) mutation annotation format (maf) files from all 33 available cancer types from The Cancer Genome Atlas (TCGA) were downloaded from the Genomic Data Commons (GDC) Data Portal (data release v7). Colon and rectal adenocarcinoma were considered as a single cancer type for the analysis. All mutation data were fused in a single mutation database and were converted from hg38 to hg19 using UCSC’s liftOver34. Variants were reannotated using ANNOVAR35. For each mutation, the main substitution type (i.e. C>A, C>G, C>T, T>A, T>C and T>G) was derived by converting each purine substitution to its complementary base substitution. To determine the trinucleotide substitution type, additional information was added regarding the identity of the upstream and downstream base. Sequence information was derived from UCSC hg1934.
TCGA Level 3 RNASeqV2 (RSEM normalized) mRNA expression data were downloaded from the Broad Institute TCGA Genome Data Analysis Center (2016): Firehose stddata__2016_01_28 run (Broad Institute of MIT and Harvard; doi:10.7908/C11G0KM9). Expression data were fused in a single gene x sample matrix. Each mutation’s gene expression value was added to the mutation database.
HLA typing
HLA typing of all TCGA samples was performed using Polysolver11. WES normal bam files from all available TCGA samples were accessed using FireCloud36, the HLA regions from the main HLA-alleles (HLA-A, HLA-B and HLA-C) in chromosome 6 (coordinates 6:29909037-29913661; 6:31321649-31324964; 6:31236526-31239869) were extracted and the resulting bam files were downloaded. Polysolver was run on these bam files using default settings and without setting prior population probabilities, resulting in the successful genotyping of 8,968 TCGA samples. The resulting output was converted in a sample x HLA allele matrix. To validate this HLA typing, the derived frequencies for each HLA allele were compared with the allele frequencies from a healthy US blood donor population, downloaded from Allele frequency net37 (Supplementary Fig. 1).
HLA affinity predictions and annotation of the HLA-binding genome
Using the R GenomicRanges package38 and UCSC hg19 genome sequence information, a GPos object was created containing information about the complete exome. For each coding DNA sequence (CDS) position, the amino acid sequences of the nine possible translated 9-mers (nonapeptides) were determined using Ensembl 75. Genes with unavailable or ambiguous protein information in Ensembl were discarded, resulting in a GPos object containing nonapeptide information of 17,992 genes. HLA affinities of these nonapeptides were predicted for the most frequent HLA alleles (A02:01, A01:01, B07:02, B08:01, C07:01, C07:02; a combination referred to as the prototypical genotype) using netMHCPan3.039. For each CDS position, the best binding peptide (peptide with the lowest predicted Kd value) was determined for each of the six HLA alleles. Finally, one aggregated Kd value was calculated using the harmonic mean value of the Kd values of the six different peptides (one from each allele) and all genomic regions with aggregated Kd values below 500 nM were considered as HLA-binding regions. The same methodology was used to predict HLA affinities in TCGA somatic mutation data. These TCGA predictions were done for both the prototypical and the sample-specific HLA genotype (specific combination of two HLA-A, two HLA-B and two HLA-C alleles) and for wild-type as well as mutated peptides.
Simulation of somatic mutations
All possible point mutations were determined for 17,992 genes by considering for each CDS position the three possible substitutions (any nucleotide can be substituted in three different nucleotides). ANNOVAR35 was used to annotate the variants and determine the reference and alternative amino acids for each mutation. This information was added to the higher described GPos object.
To determine the expected somatic mutation rates in the absence of any selection pressure, a simulated mutation database was created, with a similar size as the TCGA mutation database. To match this simulation database for differences in trinucleotide substitution probabilities, we randomly sampled the observed number of mutations from each corresponding substitution type from the GPos object. Like for the observed TCGA mutations, HLA affinities were predicted for the wild-type and the mutated nonapeptides and for both the prototypical and the sample-specific genotype. The later was determined by scrambling the columns from the sample x HLA allele matrix. This way, completely random HLA genotypes were generated, with the same allele frequency and mutation frequency per type as in the real data.
Amino acid analysis
To derive the probability of any substitution type to hit a certain amino acid or class of amino acids, we used the GPos object containing all possible mutations and determined the amino acid frequency for each substitution type and separately for synonymous and non-synonymous mutations. Amino acids were grouped in three classes: hydrophobic (Gly, Ala, Pro, Val, Leu, Iso, Met, Trp and Phe), polar (Ser, Thr, Tyr, Asn, Gln and Cys) and charged (Lys, Arg, His, Asp and Glu).
Calculation of the HLA-binding mutation ratio (HBMR) and related metrics
To quantify putative signals of immunogenic selection, we defined an HLA-binding mutation ratio (HBMR):
where n+ and n- are the total number of non-synonymous mutations located in HLA-binding and non-binding regions, respectively. Similarly, s+ and s- are the number of synonymous mutations in- and outside HLA-binding genomic regions. A similar metric, called the epitope mutation ratio (EMR) was calculated for the analysis of the IEDB epitopes. Here, + and – refer to the location inside and outside of epitope mapped regions. HBMR P values and 95% confidence intervals were calculated using a two-sided Fisher’s exact test.
dN/dS was calculated considering differences in specific trinucleotide substitution probabilities between cancer types22:
where Ni and Si are the number of (non-)synonymous sites with class i substitutions and Pi is the probability of substitution class i. The normalized HBMR was calculated as follows:
where Ni+ and Si+ are the number of (non-)synonymous sites with class i substitutions in HLA-binding regions, Ni- and Si- are the number of (non-)synonymous sites with class i substitutions in non-HLA-binding regions respectively and Pi is the probability of substitution class i.
The dNHLA/dNnonHLA ratio was calculated for each TCGA sample as follows:
with variables as defined above, but with HLA affinities determined for mutated peptides from individual genotypes. The number of HLA-binding and non-binding sites was determined for each individual TCGA genotype, under a trinucleotide substitution model. To achieve this, 960,000 substitutions were randomly sampled from the complete exome (10,000 for each substitution type) and HLA affinities were predicted for all the mutations, considering the cancer-type-specific mutational signature.
The ratio R of observed to expected neoantigens as described by Rooney et al.10 was calculated for each TCGA sample as follows:
where is the expected number of non-synonymous mutations per synonymous site and refers to the expected number of HLA-binders per non-synonymous site, both for substitution type i and estimated empirically from the pan-cancer dataset. Note that these variables are similar to the originally defined variables and respectively. Similarly, n+ and N+ were originally called Bobs and Bpred, while n and N were originally referred to as Nobs and Npred. They were defined here as such to be consistent with the rest of the methodology.
Calculation of these metrics was always based on a trinucleotide substitution model as indicated (i index). The normalized HBMR, dN/dS and dNHLA/dNnonHLA were also calculated using alternative substitution models, either based on the six main substitution classes, pentanucleotide substitution classes or using the SSB7 model. The latter is based on the six main substitution classes but considers CpG mutations as a separate class20.
Neoantigen depletion simulation and power analysis
All metrics developed in this study were evaluated using an in silico analysis of neoantigen depletion by removing increasing amounts of non-synonymous mutations hitting HLA-binding regions from the mutation dataset.
Statistical power of the HBMR metric was evaluated using the R exact2x2 package (Fisher’s exact test at significance level 0.05) for different amounts of neoantigen depletion, numbers of mutations and neoantigen prediction accuracies. For this analysis, the non-synonymous mutation proportion (71%) and HLA-binding proportion (22.1%) were fixed to values derived from the pan-cancer dataset and the HLA-binding annotation respectively.
For the power analysis of the dNHLA/dNnonHLA ratio, the ratios obtained from the simulated mutation database (containing no selection signals) were log-transformed to obtain a normal distribution. After resampling 1,000 times a predefined amount of values from this normal distribution and adding an in silico amount of neoantigen depletion, power was determined based on the number of significant deviations from 0 (corresponding to 1 in non-logtransformed data) using Wilcoxon signed-rank test at P < 0.05. This analysis was performed again for different amounts of neoantigen depletion, sample numbers and neoantigen prediction accuracies.
Human epitope mapping
Data from 66,698 known human IEDB (Immune Epitope Database) epitopes were downloaded from synapse at https://www.synapse.org/ (id syn11935058)20. These epitopes were mapped to the human genome (hg19) using the proteinToGenome function from the ensembldb R package and the EnsDb.Hsapiens.v75 R library. Mapping was successful for 66,536 (99.8%) epitopes.
Statistical analysis
The R statistical package was used for all data processing and statistical analysis. Details on statistical tests used are reported in the respective sections. Further information on research design is available in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
The results published here are in whole or part based upon data generated by TCGA. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov. We are most grateful to the patients, investigators, clinicians, technical personnel, and funding bodies who contributed to TCGA, thereby making this study possible.
This work was supported by grants from Wenner-Gren Foundation (SSv2017-005 to E.L.), the Swedish Cancer Society (CAN 2015/541 to E.L.), Knut and Alice Wallenberg Foundation (KAW 2014.0057 and KAW 2015.0144 to E.L.), Swedish Medical Research Council (2018-02852 to E.L.), Swedish Foundation for Strategic Research (RB13-0204 to E.L.), EMBO (STF7729 to J.V.d.E.) and Cancer Research UK (C14303/A17197 and A21141 to M.L.M.).
Footnotes
Data Availability
This study is based on public data (open or controlled access) from The Cancer Genome Atlas Network. Downstream data and source code are available at zenodo (https://doi.org/10.5281/zenodo.2621365 and https://doi.org/10.5281/zenodo.3461642, respectively).
Author Contributions
J.V.d.E., E.L. and M.L.M. designed and conceptualized the study. A.J.-S. provided input on study design and contributed to the interpretation of the results. J.V.d.E. was responsible for data analysis and drafted the manuscript. All authors discussed the results, edited and finalized the manuscript.
Competing Interests Statement
The authors declare they have no conflicts of interest.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research. N et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 6.DuPage M, Mazumdar C, Schmidt LM, Cheung AF, Jacks T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature. 2012;482:405–409. doi: 10.1038/nature10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–6. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGranahan N, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355 doi: 10.1126/science.aaf8399. eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutledge WC, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. 2013;19:4951–4960. doi: 10.1158/1078-0432.CCR-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown SD, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–485. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi NA et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zapata L, et al. Negative selection in tumor genome evolution acts on essential cellular functions and the immunopeptidome. Genome Biol. 2018;19:67. doi: 10.1186/s13059-018-1434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riaz N, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e15. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van den Eynden J, Larsson E. Mutational signatures are critical for proper estimation of purifying selection pressures in cancer somatic mutation data when using the dN/dS metric. Front Genet. 2017;8:74. doi: 10.3389/fgene.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martincorena I, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marty R, et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell. 2017;17:1272–1283.e15. doi: 10.1016/j.cell.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul S, et al. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chowell D, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci USA. 2015;112:E1754–E1762. doi: 10.1073/pnas.1500973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellrott K, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turajlic S, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18:1009–1021. doi: 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 30.Mandal R, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364:485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein A, Folprecht G. Immunotherapy of colon cancer. Oncol Res Treat. 2018;41:282–285. doi: 10.1159/000488918. [DOI] [PubMed] [Google Scholar]
- 32.Van den Eynden J, Basu S, Larsson E. Somatic mutation patterns in hemizygous genomic regions unveil purifying selection during tumor evolution. PLoS Genet. 2016;12:e1006506. doi: 10.1371/journal.pgen.1006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weghorn D, Sunyaev S. Bayesian inference of negative and positive selection in human cancers. Nat Genet. 2017;49:1785–1788. doi: 10.1038/ng.3987. [DOI] [PubMed] [Google Scholar]
- 34.Rosenbloom KR, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 2014;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birger C, et al. FireCloud, a scalable cloud-based platform for collaborative genome analysis: Strategies for reducing and controlling costs. bioRxiv. 2017 doi: 10.1101/209494. 209494. [DOI] [Google Scholar]
- 37.González-Galarza FF, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 2015;43:D784–D788. doi: 10.1093/nar/gku1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen M, Andreatta M. NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 2016;8:33. doi: 10.1186/s13073-016-0288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.