Abstract
Liquid biopsy analysis, mainly based on circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), provides an extremely powerful tool for the molecular profiling of cancer patients in real time. In this study, we directly compared PIK3CA hotspot mutations (E545K, H1047R) in EpCAM‐positive CTCs and paired plasma‐ctDNA in breast cancer (BrCa). PIK3CA hotspot mutations in CTCs and ctDNA were analyzed using our previously developed highly sensitive (0.05%), specific, and validated assay in plasma‐ctDNA from 77 early and 73 metastatic BrCa patients and 40 healthy donors. We further analyzed and directly compared PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges (CTCs) and paired plasma‐ctDNA, in 56 cases of early and 27 cases of metastatic breast cancer, and 16 corresponding primary tumors. In plasma‐ctDNA,PIK3CA hotspot mutations were identified in 30/77(39.0%) early and 35/73(47.9%) metastatic BrCa cases; none (0/40, 0%) of the healthy donors’ plasma‐ctDNA samples were positive. Our direct comparison study in DNAs isolated from CellSearch® cartridges (CTCs) and paired plasma‐ctDNA from the same blood draws has shown a lack of concordance in early BrCa (27/56, 48.2%), while the concordance in the metastatic setting was higher (18/27, 66.6%). Our results were validated by ddPCR methodology, and the concordance between our assay and ddPCR for PIK3CA E545K hotspot mutation was 30/37 (81.1%). In many cases, PIK3CA hotspot mutations were detected in samples found to be negative for CTCs in CellSearch®. Our data demonstrated for the first time that (a) PIK3CA hotspot mutations are present at high frequencies in CTCs isolated from CellSearch® cartridges and paired plasma‐ctDNA both in early and metastatic BrCa, (b) the detection and concordance of PIK3CA hotspot mutations between plasma‐ctDNA and CTCs are higher in the metastatic setting, (c) PIK3CA mutational status significantly changes after therapeutic intervention, and (d) PIK3CA mutation detection in CTCs and plasma‐ctDNA provides complementary information.
Keywords: breast cancer, circulating tumor cells, circulating tumor DNA, liquid biopsy, mutation analysis, PIK3CA
Our direct comparison study on the presence of PIK3CA hotspot mutations (E545K, H1047R) in EpCAM‐positive CTCs and paired plasma‐ctDNA demonstrated for the first time that PIK3CA mutations are present at high frequencies in both CTCs and paired plasma‐ctDNA and that there is a high concordance in the metastatic setting. PIK3CA mutational status significantly changes after therapeutic intervention.
Abbreviations
- BrCa
breast cancer
- CTC
circulating tumor cells
- ctDNA
circulating tumor DNA
- ddPCR
droplet digital PCR
- EMT
epithelial–mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- NGS
next‐generation sequencing
- PB
peripheral blood
1. Introduction
Dynamic changes of tumor features over time, also known as tumor heterogeneity, are now recognized as one of the most significant issues in tumor biology (Bedard et al., 2014). A major limitation of the classic tissue biopsy approach is that genetically different subclones that are present at a minor frequency in the primary tumor and thus possibly not initially detected may be selected after treatment or differentially represented in the metastatic process, leading to treatment resistance (Bedard et al., 2014). The minimally invasive ‘liquid biopsy’ approach overcomes these limitations since by providing new ways to monitor tumor genetics and tumor dynamics in real time (Alix‐Panabières and Pantel, 2016).
Liquid biopsy has a high potential to significantly change the therapeutic strategy in cancer patients but there are still many challenging questions to be answered (Bardelli and Pantel, 2017). Detection, enumeration, and molecular characterization of circulating tumor cells (CTCs) and analysis of circulating tumor DNA (ctDNA) provide useful information regarding the individual molecular profile of each patient in real time, before and after treatment (Lianidou and Hoon, 2017). The field of liquid biopsy applications is growing exponentially, including molecular target identification, prognosis assessment, diagnosis of recurrence, monitoring of response to treatment, and monitoring of tumor genomic profiles over time (Lianidou and Hoon, 2017). Most importantly, blood‐based tests are very challenging and highly important in cases where tumor biopsies are not accessible (Ignatiadis et al., 2015; Lianidou et al., 2015; Wang et al., 2017). Recently, the US Food and Drug Administration (FDA) approved the first liquid biopsy‐based companion diagnostic test for the administration of first‐ to third‐generation TKI therapies in NSCLC patients, based on the presence of sensitive or resistant EGFR mutations in cell‐free DNA (cfDNA) (Malapelle et al., 2017).
Several studies so far have shown that both CTCs and ctDNA can be detected in peripheral blood of cancer patients not only in advanced but even at early stages (Bettegowda et al., 2014; Stathopoulou et al., 2002). ctDNA analysis in plasma has been suggested as an alternative to the classic biopsy approach since it essentially comprises a subtype of total cfDNA that is derived from tumor cells thus providing their genomic signature. However, there is an essential difference between CTCs and ctDNA; CTCs as viable cells circulating in peripheral blood can provide real‐time information on the metastatic spread and reveal active and possibly targetable signaling networks, while ctDNA can give specific information on the presence or absence of specific alterations deriving from the tumor, indicating therapy response or resistance (Bidard et al., 2018; Gold et al., 2015; Siravegna et al., 2017).
Circulating tumor cells enumeration using the CellSearch® system has been approved by the FDA more than a decade ago as a prognostic marker in metastatic breast, colorectal, and prostate cancer (Riethdorf et al., 2018). CTC detection and enumeration are associated with progression‐free survival (PFS) and overall survival (OS) in metastatic (Bidard et al., 2014; Cristofanilli et al., 2004) and early BrCa (Lucci et al., 2012; Rack et al., 2014; Stathopoulou et al., 2002). Beyond enumeration, CTC molecular characterization is very important, since it can provide significant information at the gene expression, DNA methylation, and DNA mutation level (Aktas et al., 2016; Chimonidou et al., 2011, 2013; Markou et al., 2011, 2014; Mastoraki et al., 2018; Mostert et al., 2015; Sieuwerts et al., 2018; Strati et al., 2011). However, CTCs are highly heterogeneous, and there is a lack of a unique marker for their isolation and identification, since there are not well‐defined universal surface targets among all malignant cell types (Lianidou and Hoon, 2017). This issue is becoming more complicated when surface CTC‐enrichment targets change during epithelial‐to‐mesenchymal transition (EMT) (Gorges et al., 2012). Based on that, a variety of EpCAM‐independent systems have been now developed to capture and identify CTCs (Lianidou and Hoon, 2017). CTC molecular characterization can offer very important information on patient prognosis and can also identify therapeutic targets or mechanisms of resistance, such as mutations in driver genes (Bingham et al., 2017; Kidess‐Sigal et al., 2016; Liu et al., 2017; Pestrin et al., 2015; Shaw et al., 2017).
Plasma‐ctDNA analysis is another important component of liquid biopsy. Increased concentrations of ctDNA fragments have been detected in plasma and serum of cancer patients with various tumor types, and their presence has been correlated with unfavorable outcome (Bettegowda et al., 2014; Dawson et al., 2013; Schwarzenbach et al., 2011; Wan et al., 2017). ctDNA analysis based on tumor‐derived genetic alternations is now increasingly used for treatment purposes (Malapelle et al., 2017), and the large dynamic range of plasma‐ctDNA presents a considerable correlation with changes in tumor burden (Bettegowda et al., 2014; Dawson et al., 2013).
The evaluation of mutational status of specific genes in liquid biopsy material is highly informative, but requires highly sensitive and robust methodologies. Recently, our group developed and validated an ultrasensitive and highly specific methodology for the detection of PIK3CA hotspot mutations (E545K, H1047R) in CTCs, based on the combination of allele‐specific priming, competitive probe blocking of wild‐type amplification, asymmetric PCR, and probe melting analysis (Markou et al., 2014). Using this highly sensitive assay (0.05%), we have shown that PIK3CA hotspot mutations are present at a relatively high frequency in the EpCAM‐positive CTC fraction not only in patients with metastatic but with early BrCa as well. Our study has also shown that PIK3CA mutational status can change during disease recurrence or progression in BrCa patients (Markou et al., 2014). The aim of the present study was to use this highly sensitive methodology to directly compare the mutational status of PIK3CA (E545K, H1047R) in CTC‐DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA from the same blood draws in patients with early and metastatic BrCa.
2. Materials and methods
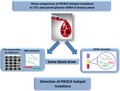
The outline of the workflow of our study is shown in Fig. 1.
Figure 1.

Experimental flowchart of the study.
2.1. Clinical samples
We initially analyzed PIK3CA hotspot mutations in 190 plasma‐ctDNA samples from (a) patients with early BrCa (n = 77), (b) patients with metastatic BrCa (n = 73), and (c) healthy donors (all female, n = 40). We further performed a direct comparison study of PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA in 56 samples of 43 patients with early BrCa and in 27 samples of 16 patients with metastatic breast cancer. Pretreatment and post‐treatment peripheral blood samples were available for 13 and 11 of these patients with early and metastatic BrCa, respectively, while corresponding formalin‐fixed paraffin‐embedded (FFPE) primary tumor samples were available for 16 of these patients. The study was conducted in accordance with the 1964 Declaration of Helsinki and was approved by the ethics and scientific committees of the participating Institutions. All participating patients signed an informed consent form to participate in the study, which was approved by the ethics and scientific committees of our institutions. The clinicopathological characteristics of the patients are shown in Table S1. We also included in our study 42 DNA samples isolated from CellSearch® cartridges from an independent group of 17 metastatic BrCa patients, before and after first cycle of treatment. For 8 of these patients, blood sampling at progression of disease was also available. The majority of these patients were ER+/PR+/HER2−.
2.2. Isolation of plasma‐ctDNA
Whole blood samples were collected into venous blood collection tubes using K2EDTA (BD Vacutainer, Plymouth) as anticoagulant. Samples were mixed thoroughly, and plasma was isolated within 2 h from sample collection by centrifugation at 530 g for 10 min at room temperature. Once isolated, plasma samples were further centrifuged twice at 2000 g for 10 min, before transferring into clean 2‐mL tubes and freezing at −70 °C until further processing. The QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) was used to isolate ctDNA from 2.00 mL of plasma according to manufacturer's instructions.
2.3. Isolation of gDNA from CellSearch® cartridges
CellSearch® analysis was performed according to the manufacturer's instructions; 7.5 mL of PB was used for each patient with metastatic disease, while in patients with early BrCa, three CellSave tubes were used for each patient (in total 22.5 mL PB). Following CTC analysis, CellSearch® cartridges were stored in a dark place at 4 °C until gDNA isolation. CTCs and WBCs (prestained with antibody to CD45, pan‐CK, and DAPI) were aspirated from the CellSearch® cartridge and underwent downstream gDNA extraction using the QIAamp DNA Micro Kit (Qiagen) according to manufacturer's instructions.
2.4. gDNA isolation from FFPEs
Formalin‐fixed paraffin‐embedded tissue sections of 10 mm containing > 80% of tumor cells were used for DNA extraction. gDNA was isolated with the QIAamp DNA FFPE Tissue Kit (Qiagen), according to the manufacturer's instructions.
2.5. Whole genome amplification
In the independent group of 42 clinical samples analyzed, the archival DNA material was not sufficient for downstream analysis. For this reason, a whole genome amplification (WGA) step for all these samples was included. Amplification with REPLI‐g kit (Qiagen) was performed according to the manufacturer's recommendations. A blank and DNA from human cell lines, MCF‐7 and T47D, were included for each experimental procedure as negative and positive controls, respectively. DNA concentration was determined using the Quant‐iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen™, Waltham, MA, USA), and DNA was diluted according to the manufacturers’ manual. To verify the specificity of our assay after WGA, 10 plasma samples from healthy donors were checked before and after WGA step.
2.6. Sample preparation
To avoid contamination, different rooms, dedicated labware, and dedicated areas were used for all procedures. All DNA preparation and handling steps took place in specific laminar‐flow hoods under DNase‐free conditions. DNA concentration in all cases was measured in a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) , calibrated with the recommended CF‐1 (concentrated aqueous potassium dichromate, K2Cr2O7) standard solution. The isolated DNA samples were stored at −70 °C until further use.
2.7. PIK3CA hotspot mutation analysis
The detection of PIK3CA hotspot mutations (E545K, H1047R) was based on our previously developed and validated ultrasensitive assay (analytical sensitivity 0.05%) (Markou et al., 2014).
2.8. Quality control
Synthetic gene fragments were synthesized as gBlocks by Integrated DNA Technologies (Coralville, IA, USA) and were used as positive controls for PIK3CA hotspot mutations (sequences are available upon request). Synthetic oligos for E545K and H1047R mutation sites were designed so that each sequence was represented by a unique gBlock that mutation position would be included. Lyophilized gBlocks were suspended in Tris/EDTA buffer to a stock concentration of 10 ng·mL−1 and were further diluted to working concentrations as needed. Also, two negative controls were included in each experimental procedure, one as negative control of PCR reaction (blank) and the second one as wild‐type control, which contained wild‐type as DNA template. All samples were checked for their DNA integrity prior to analysis, by amplifying a region of PIK3CA exon 20 that includes the hotspot mutation site.
2.9. Spiking experiments
To further verify the analytical sensitivity of the assay, we collected peripheral blood samples from two healthy donors in 2 different types of tubes: (a) K2EDTA (BD Vacutainer) and (b) CellSave (Menarini Silicon Biosystems). In both cases, the first 5 mL of blood was discarded to avoid skin epithelial cell contamination. In each tube, MCF‐7 cells (100 cells enumerated in a Malassez Hemocytometer) were spiked into peripheral blood (10 mL in EDTA tube and 7.5 mL in CellSave tube), and mixed immediately after spiking. In the EDTA tube, CTC isolation was performed by adding red cell lysis buffer and capture beads, coated with the monoclonal antibody BerEP4 against the human epithelial antigen EpCAM. In the CellSave tube, CellSearch® analysis was performed according to the manufacturer's instructions. Following CTC analysis, CellSearch® cartridges were used for downstream analysis of EpCAM‐positive CTCs. In both cases, DNA was isolated using QIAamp DNA Micro Kit and DNA concentration was measured in a NanoDrop 1000 spectrophotometer that was calibrated with the recommended CF‐1 standard solution.
2.10. Droplet digital PCR for the detection of PIK3CA E545K hotspot mutation
We analyzed 37 of these DNA samples using droplet digital PCR (ddPCR) technology for PIK3CA E545K hotspot mutation. All clinical samples were analyzed with the QX200 Droplet Digital PCR System (Bio‐Rad Laboratories, Inc., Hercules, CA, USA) using TaqMan hydrolysis probes for detecting and quantifying wild‐type PIK3CA, as well as PIK3CA E545K in exon 9. One probe was specific for the wild‐type sequence (wild‐type reference assay/HEX, Channel 2), and the other was specific for each PIK3CA E545K hotspot mutation (mutation assay/FAM, Channel 1). The PCR mix for one well was set up by mixing 2 μL DNA sample, 10 μL 2 × ddPCR Supermix for Probes (No dUTP) (Bio‐Rad Laboratories), 1 μL 20 × target primers/probe (FAM), 1 μL 20 × reference primers/probe (ΗΕΧ), in a reaction volume of 20 μL, adjusted with sterile water, according to the manufacturer's instructions. The cycling profile of the PIK3CA mutant detection assay was as follows: 95 °C for 10 min (1 cycle), 94 °C for 30 s, and 55 °C for 60 s (55 cycles), and infinite hold at 12 °C. Each experimental procedure included a negative control (nontemplate control) and a positive control (MCF‐7 cell line), which contained enough mutant and wild‐type DNA to create a double‐positive cluster of greater than 100 droplets. PCR products were loaded onto the QX200 Droplet Reader (Bio‐Rad Laboratories, Inc.), and the data were analyzed with QuantaSoft Software.
2.11. Statistical analysis
Statistical analysis was performed via SPSS version 24.0 (IBM® SPSS® Statistics, Armonk, NY, USA). A P‐value ≤ 0.05 was considered to be significant. Graphics were generated with MS Excel 2010 (Microsoft Corporation, Seattle, WA, USA). Agreement between the presence of PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA was assessed by using the chi‐square test and McNemar test.
3. Results
3.1. Detection of PIK3CA hotspot mutations in plasma‐ctDNA
We analyzed in total 190 plasma‐ctDNA samples for PIK3CA hotspot mutations from (a) early BrCa (n = 77), (b) metastatic BrCa (n = 73), and (c) healthy donors (all female, n = 40). We first evaluated the specificity of the PIK3CA hotspot mutations assay in plasma‐ctDNA samples from 40 female healthy donors. None of the plasma‐ctDNA (0/40, 0%) was found positive for both PIK3CA hotspot mutations (Fig. 2). Moreover, to verify further the specificity of this procedure, a WGA step was also included prior to analysis and 10 plasma samples from healthy donors were checked before and after WGA in order to ensure that no false‐positive results are detected. Indeed, both PIK3CA hotspot mutations (E545K, H1047R) were not detected before and after WGA using our highly sensitive assay. Concerning analytical sensitivity, our spiking experiment results have clearly shown that PIK3CA E545K hotspot mutation was detected by using our laboratory‐developed assay. PIK3CA hotspot mutations were identified in plasma‐ctDNA in 30/77(39.0%) and in 35/73(47.9%) cases of early BrCa and metastatic BrCa, respectively. In early BrCa, PIK3CA E545K hotspot mutation was identified in 30/77(39.0%) plasma‐ctDNA samples, and PIK3CA H1047R hotspot mutation was identified in 7/77(9.1%). In metastatic BrCa, 30/73(41.1%) ctDNA samples were found positive for PIK3CA E545K hotspot mutation and 15/73(20.5%) were found positive for PIK3CA H1047R hotspot mutation.
Figure 2.
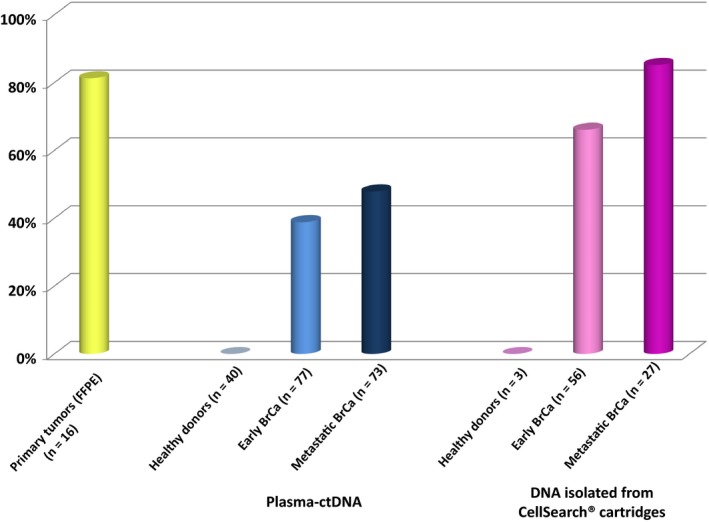
PIK3CA hotspot mutations in primary tumors, DNA isolated from plasma‐ctDNA and CellSearch cartridges.
3.2. Direct comparison of PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA
Circulating tumor cells enumeration using the CellSearch® and PIK3CA mutational analysis in plasma and DNAs isolated from CellSearch® cartridges were performed in a subgroup of these patients, using the same blood draw (same date and time), more specifically: (a) in 56 samples from 43 patients with early BrCa, (b) in 27 samples from 16 patients with metastatic BrCa, and (c) in 3 PB samples from healthy blood donors. None of DNA samples isolated, in exactly the same way, from CellSearch® cartridges from peripheral blood of these three healthy donors was found positive for PIK3CA hotspot mutations. In early BrCa, PIK3CA hotspot mutations were detected in 37/56(66.1%) DNA samples isolated from CellSearch® cartridges (Fig. 2). More specifically, PIK3CA E545K hotspot mutation was detected in 37/56(66.1%) DNA samples isolated from CellSearch® cartridges, whereas PIK3CA H1047R hotspot mutation was detected in 7/56(12.5%) (Fig. 3). CellSearch® detected at least one CTC in 13/56(23.2%) samples. PIK3CA hotspot mutations were identified in 9/13(69.2%) DNAs samples isolated from CellSearch® cartridges where at least one CTC was detected. Moreover, in 28/43(65.1%) cases, PIK3CA hotspot mutations were detected in CellSearch® cartridges that were found negative for CTCs (Fig. 3). In metastatic BrCa, PIK3CA hotspot mutations were detected in 23/27(85.2%) DNA samples isolated from CellSearch® cartridges (Fig. 2). PIK3CA E545K hotspot mutation was detected in 22/27(81.5%) DNAs isolated from CellSearch® cartridges, whereas PIK3CA H1047R hotspot mutation was detected in 8/27(29.6%) (Fig. 3). CellSearch® detected at least one CTC in 12/27(44.4%) samples, and PIK3CA hotspot mutations were identified in 10/12(83.3%) DNAs isolated from CellSearch® cartridges positive for CTCs. PIK3CA hotspot mutations were also detected in 13/15(86.7%) cases that were found negative for CTCs by CellSearch®. In two cases, where CellSearch® detected 122 and 23 CTCs, respectively, PIK3CA hotspot mutations were not detected (Fig. 3).
Figure 3.
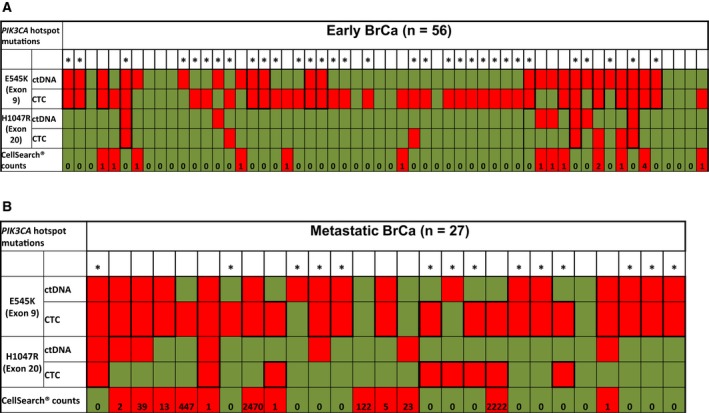
Direct comparison study of PIK3CA hotspot mutations in DNA isolated from CellSearch® cartridges and paired plasma‐ctDNA from (A) early and (B) metastatic BrCa patients.
Paired plasma‐ctDNA samples were analyzed for the same PIK3CA hotspot mutations. In early BrCa, PIK3CA hotspot mutations were detected in 24/56(42.9%) paired plasma‐ctDNA samples. More specifically, PIK3CA E545K hotspot mutation was detected in 24/56(42.9%), whereas PIK3CA H1047R hotspot mutation was detected in 7/56(12.5%) plasma‐ctDNA samples (Fig. 3). In metastatic BrCa, PIK3CA hotspot mutations were detected in 18/27(66.7%) paired plasma‐ctDNA samples. PIK3CA E545K hotspot mutation was detected in 17/27(63.0%) plasma‐ctDNA samples, whereas PIK3CA H1047R hotspot mutation was detected in 7/27(25.9%) (Fig. 3). Both PIK3CA hotspot mutations were identified in the same plasma samples in seven early and six metastatic BrCa cases.
In early BrCa, PIK3CA hotspot mutations were detected in 37/56(66.1%) DNA samples isolated from CellSearch® cartridges and in 24/56(42.8%) paired plasma‐ctDNA samples. In total, in these 56 samples, the concordance of PIK3CA hotspot mutations between DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA samples was 27/56 (48.2%) (Table 1). In metastatic BrCa, PIK3CA hotspot mutations were detected in 23/27(85.2%) DNA samples isolated from CellSearch® cartridges and in 18/27(66.7%) paired plasma‐ctDNA samples. In total, in these 27 samples, the concordance of PIK3CA hotspot mutations between DNA isolated from CellSearch® cartridges and paired plasma‐ctDNA samples was 18/27 (66.6%) (Table 1). The correlation of PIK3CA hotspot mutations and clinicopathological characteristics of early and metastatic BrCa patients used in the direct comparison study is shown in Table S2.
Table 1.
Direct comparison of PIK3CA hotspot mutations in ctDNA and CellSearch® cartridges isolated from early (n = 56) and metastatic (n = 27) BrCa patients
| PIK3CA hotspot mutations (early breast cancer) | ||||
|---|---|---|---|---|
| N = 56 | CellSearch® cartridges | |||
| ctDNA | Neg | Neg | Pos | Total |
| 11 | 21 | 32 | ||
| Pos | 8 | 16 | 24 | |
| Total | 19 | 37 | 56 | |
| Concordance: (27/56) 48.2% (P = 0.582, chi‐square test) | ||||
| PIK3CA hotspot mutations (metastatic breast cancer) | ||||
|---|---|---|---|---|
| N = 27 | CellSearch® cartridges | |||
| ctDNA | Neg | Neg | Pos | Total |
| 2 | 7 | 9 | ||
| Pos | 2 | 16 | 18 | |
| Total | 4 | 23 | 27 | |
| Concordance: (18/27) 66.6% (P = 0.407, chi‐square test) | ||||
3.3. PIK3CA mutational status in CTCs and paired plasma‐ctDNA changes before and after treatment
3.3.1. Early BrCa
In early BrCa, PIK3CA mutational status was evaluated in 13 cases where peripheral blood samples were available before and after treatment. Before treatment, PIK3CA hotspot mutations were detected in plasma‐ctDNA in 7/13(53.8%), and in corresponding DNAs isolated from CellSearch® cartridges in 8/13(61.5%) cases. After treatment, PIK3CA hotspot mutations were detected in plasma‐ctDNA in 9/13(69.2%), and in DNAs isolated from CellSearch® cartridges in 7/13(53.8%) cases (Fig. 4A). Before treatment, PIK3CA E545K hotspot mutation was detected in 8/13(61.5%) DNA samples isolated from CellSearch® cartridges and in 7/13(53.8%) paired plasma‐ctDNA samples. However, only one sample was positive for PIK3CA H1047R hotspot mutation in both DNA isolated from CellSearch® cartridge and the paired plasma‐ctDNA samples 1/13(7.7%) (Fig. 4B). After treatment, PIK3CA E545K hotspot mutation was detected in 7/13(53.8%) DNAs isolated from CellSearch® cartridges and in 9/13(69.2%) paired plasma‐ctDNA samples (Fig. 4B). PIK3CA H1047R hotspot mutation was detected in 4/13(30.8%) DNA isolated from CellSearch® cartridges and in 4/13(30.8%) paired plasma‐ctDNA samples. After treatment, five patients (#2, #4, #5, #8, and #13) retained their initial PIK3CA hotspot mutation status in plasma‐ctDNA and four patients (#4, #6, #9, and #13) in DNA isolated from CellSearch® cartridges (Fig. 4B). In five cases (#1, #3, #6, #7, and #9), PIK3CA hotspot mutations were detected only in post‐treatment plasma‐ctDNA samples, but not before whereas in three cases (#10, #11, and #12) PIK3CA hotspot mutations were detected in plasma‐ctDNA only before treatment. In four cases (#1, #5, #7, and #8), PIK3CA hotspot mutations were detected only in post‐treatment DNA samples isolated from CellSearch® cartridges, whereas their corresponding pretreatment samples were found negative. However, there were five cases (#2, #3, #10, #11, and #12) where PIK3CA hotspot mutations were detected in pretreatment but not in the corresponding post‐treatment samples. According to our results, after treatment, more plasma‐ctDNA samples but less DNA samples isolated from CellSearch® cartridges were found positive for PIK3CA hotspot mutations.
Figure 4.

(A) PIK3CA hotspot mutations in DNA isolated from CellSearch® cartridges and paired plasma‐ctDNA, before and after treatment from (i) early (n = 13) and (ii) metastatic (n = 11) BrCa patients. Heatmap for (B) early and (C) metastatic BrCa patients.
3.3.2. Metastatic BrCa
In metastatic BrCa, PIK3CA mutational status was evaluated in 11 cases where peripheral blood samples were available before and after completion of first‐line treatment. Before treatment, PIK3CA hotspot mutations were detected in plasma‐ctDNA in 8/11(72.7%), and in corresponding DNAs isolated from CellSearch® cartridges in 8/11(72.7%) cases (Fig. 4A). PIK3CA E545K hotspot mutation was detected in 8/11(72.2%) DNA isolated from CellSearch® cartridges and in 7/11(63.6%) paired plasma‐ctDNA samples (Fig. 4C). PIK3CA H1047R hotspot mutation was detected in 2/11(18.2%) DNA isolated from CellSearch® cartridges and in 4/11(36.6%) paired plasma‐ctDNA samples (Fig. 4C). After first‐line treatment, PIK3CA E545K hotspot mutation was detected in 9/11(81.8%) DNA isolated from CellSearch® cartridges and in 6/11(54.5%) paired plasma‐ctDNA samples. PIK3CA H1047R hotspot mutation was detected in 5/11(45.5%) DNA isolated from CellSearch® cartridges and in 1/11(9.1%) paired plasma‐ctDNA samples. After first‐line treatment, in five patients (#1, #2, #3, #6, and #10), the same PIK3CA hotspot mutations were detected in plasma‐ctDNA and in seven patients (#1, #2, #3, #4, #5, #6, and #10) the same PIK3CA hotspot mutations were detected in DNA isolated from CellSearch® cartridges (Fig. 4C). For two patients (#5 and #9), PIK3CA hotspot mutations were detected only in post‐treatment plasma‐ctDNA, but not before treatment. On the contrary, in four cases (#1, #4, #7, and #8), PIK3CA hotspot mutations were detected only in pretreatment plasma‐ctDNA samples, but not after treatment. In three cases (#6, #9, and #11), PIK3CA hotspot mutations were detected only in DNA isolated from CellSearch® cartridges post‐treatment, whereas the corresponding pretreatment samples were found negative. There was only one case (#8) where PIK3CA mutation was detected only before pretreatment sample, but not after treatment. According to our results, after treatment, more DNA samples isolated from CellSearch® cartridges but less plasma‐ctDNA samples were found positive for PIK3CA hotspot mutations.
3.4. Comparison of PIK3CA hotspot mutations in primary tumors, corresponding DNA isolated from CellSearch® cartridges and paired plasma‐ctDNA
For 16 cases, DNA isolated from corresponding primary tumors (FFPEs) was also available for comparison studies (Table 2). PIK3CA hotspot mutations were identified in 13/16(81.3%) of these FFPE samples (Fig. 2); PIK3CA E545K hotspot mutation was detected in 9/16(56.3%), and PIK3CA H1047R hotspot mutation was detected in 11/16(68.8%) (Table 2). In all cases where PIK3CA E545K hotspot mutation was detected in FFPEs, it was also detected in DNA isolated from CellSearch® cartridges. Moreover, there were six cases (#4, #6, #9, #10, #14, and #16) where PIK3CA E545K mutation was detected de novo in DNA isolated from CellSearch® cartridges, but was not detected in FFPEs. In three of these cases (#4, #14, and #16), E545K was also detected in paired plasma‐ctDNA samples. A high concordance for PIK3CA hotspot mutations between FFPE and DNAs isolated from CellSearch® cartridges was found (12/16, 75.0%), whereas this concordance was much lower between FFPE and plasma‐ctDNA samples (6/16, 37.5%) (Table 2).
Table 2.
PIK3CA hotspot mutations in primary tumors, DNA isolated from CellSearch® (CS) cartridges and plasma‐ctDNA from BrCa patients (n = 16)
| PIK3CA hotspot mutations | ||||||||
|---|---|---|---|---|---|---|---|---|
| # ID | E545K | H1047R | CellSearch® (CS) counts | Status | ||||
| Primary tumor | ctDNA | CTCs (CS) | Primary tumor | ctDNA | CTCs (CS) | |||
| 1 | + | − | + | + | − | − | 0 | Early |
| 2 | − | − | − | + | − | − | 0 | Early |
| 3 | + | − | + | + | − | − | 0 | Early |
| 4 | − | + | + | + | − | − | 0 | Early |
| 5 | + | − | + | + | − | − | 1 | Early |
| 6 | − | − | + | + | − | − | 0 | Early |
| 7 | + | + | + | + | + | − | 0 | Metastatic |
| 8 | + | + | + | + | − | − | NA | Early |
| 9 | − | − | + | − | − | + | 0 | Early |
| 10 | − | − | + | + | − | − | 0 | Early |
| 11 | + | + | + | + | + | − | 2 | Metastatic |
| 12 | + | − | + | + | − | − | 0 | Metastatic |
| 13 | + | − | + | − | − | − | 447 | Metastatic |
| 14 | − | + | + | − | + | + | 1 | Metastatic |
| 15 | + | + | + | − | + | − | 39 | Metastatic |
| 16 | − | + | + | − | + | + | 0 | Metastatic |
3.5. Comparison study between the combination of allele‐specific, asymmetric rapid PCR and melting analysis assay and ddPCR for the detection of PIK3CA E545K hotspot mutation
The results of the clinical sample analysis were based on the definition of threshold line for each subpopulation of mutant or wild‐type droplets. All positive droplets, those above the threshold line of 3.500 droplets, are scored as positive, whereas the negative droplets those above the threshold line of 3.500 are scored as negative (background). In case of clinical samples, at least one droplet of PIK3CA E545K DNA‐mutated sequence was considered as positive sample for PIK3CA E545K hotspot mutation by ddPCR.
For this study, we analyzed in total by ddPCR 37 samples: 17 genomic DNAs isolated from CellSearch® cartridges (EpCAM‐positive cells) and 20 plasma‐ctDNA samples for PIK3CA E545K mutation. More specifically, 14 and 3 DNAs isolated from CellSearch® cartridges (EpCAM‐positive cells) from early and metastatic patients, and 11 and 9 plasma‐ctDNA samples from early and metastatic patients, respectively, were used for this study.
In total, the concordance between our assay and ddPCR for PIK3CA E545K hotspot mutation was 30/37 (81.1%) (Table 3). More specifically, 21 samples were found positive by both methods, and 9 samples found negative by both methods. There were only 2 samples that were found positive by our method and negative by ddPCR, and 5 samples that were found positive by ddPCR and negative by our method. Our data support that our assay and ddPCR give comparable results (P = 0.001).
Table 3.
Comparison study between the combination of allele‐specific, asymmetric rapid PCR and melting analysis assay and droplet digital PCR for detecting PIK3CA E545K hotspot mutation in early and metastatic BrCa patients
| Combination of allele‐specific, asymmetric rapid PCR and melting analysis assay (Markou et al., 2014) | ddPCR | ||
|---|---|---|---|
| Neg | Neg | Pos | Total |
| 9 | 5 | 14 | |
| Pos | 2 | 21 | 23 |
| Total | 11 | 26 | 37 |
| Concordance: (30/37) 81.8% (P = 0.001, chi‐square test) | |||
3.6. PIK3CA hotspot mutations in CTCs from an independent group of DNA samples isolated from CellSearch® cartridges
In the independent patient group studied, PIK3CA hotspot mutations were detected in 25/42(59.5%) DNA samples isolated from CellSearch® cartridges most of which were positive for CTCs (Fig. S1). PIK3CA E545K hotspot mutation was detected in 12/42(28.6%) DNAs isolated from CellSearch® cartridges, whereas PIK3CA H1047R hotspot mutation was detected in 17/42(40.5%). CellSearch® detected at least one CTC in 37/42(88.1%) samples, and PIK3CA hotspot mutations were identified in 22/37(59.5%) DNAs isolated from CellSearch® cartridges positive for CTCs. Moreover, PIK3CA hotspot mutations were also detected in 3/5(60%) cases that were found negative for CTCs by CellSearch®. These data are in concordance with our previous results, indicating that PIK3CA mutations are also present in EpCAM‐positive cells where CellSearch® gave negative results for the presence of CTCs. In this independent patient group, we found in total 4/42(9.5%) samples where both PIK3CA mutations were detected. As can be seen in Fig. S1, in all these four cases, CTCs were detected in the CellSearch®. Moreover, in 15/42(35.7%) cases where CellSearch® was positive for CTCs, no PIK3CA mutations were detected.
4. Discussion
Liquid biopsy is now becoming a highly important tool for following up cancer patients in real time with a variety of solid tumors (Alix‐Panabières and Pantel, 2016; Bardelli and Pantel, 2017; Bedard et al., 2014; Bettegowda et al., 2014; Ignatiadis et al., 2015; Lianidou and Hoon, 2017; Lianidou et al., 2015; Malapelle et al., 2017; Stathopoulou et al., 2002; Wang et al., 2017). However, some questions still need to be answered (Bardelli and Pantel, 2017). One common question is whether CTCs and ctDNA give identical or complementary information, and which of the two is the best choice to follow up patients over time. PIK3CA mutational status has already been studied in CTCs and plasma‐ctDNA using different methodologies such as NGS and ddPCR; however, there are no studies till now where PIK3CA mutational status was evaluated in CTC and plasma‐ctDNA in identical blood draws using the same methodology. In the present study, we addressed this question for the first time by performing a direct comparison study of PIK3CA mutational status (E545K, H1047R) in DNA isolated from CellSearch® cartridges and paired plasma‐ctDNA from the same blood draws in early and metastatic BrCa, using our previously described highly sensitive real‐time PCR methodology (Markou et al., 2014).
PI3K is one among the most important downstream molecules in the pathway of tyrosine kinase growth factor receptors and one of the most promising targets for personalized medicine. PIK3CA mutations have been reported in 18%–40% (Barbareschi et al., 2007) of BrCa cases; three ‘hotspot’ mutations (E542K, E545K, and H1047R) comprise more than 80% (Campbell et al., 2004) of all PIK3CA mutations and are localized in exons 9 and 20. Moreover, early clinical studies have indicated that the presence of PIK3CA mutations is linked to the acquirement of enhanced sensitivity to PI3K pathway inhibitors (Janku et al., 2011). Although the presence of PIK3CA mutations in archival tissues has been evaluated in many clinical trials such as BOLERO‐2, FERGI, BELLE‐2, and BELLE‐3, the PFS benefit from PI3K or mTOR inhibitors seems to be largely maintained irrespectively of PIK3CA genotype (Hortobagyi et al., 2016); it is important to note that the detection limit of the next‐generation sequencing methodology used in this study for mutation allele frequency (MAF) was 5%.
However, after taking into consideration the advantages of noninvasive liquid biopsy analysis, we expect that identifying key mutations in CTCs and plasma‐ctDNA from peripheral blood samples may provide more actionable information on the molecular profile of metastatic tumors. Results of the BELLE‐2 and BELLE‐3 clinical studies, based on the combination of a pan‐PI3K inhibitor (buparlisib) with endocrine therapy, indicated a better PFS in the PIK3CA mutant subgroup identified through ctDNA analysis, but not for the PIK3CA mutant subgroup as identified in primary tumors (Baselga et al., 2017; Di Leo et al., 2018). However, the BOLERO‐2 study which examined the benefit of an inhibitor of the PI3K/AKT/mTOR pathway (everolimus), has shown a benefit for patients when everolimus was added to endocrine therapy regardless of the presence of a PIK3CA mutation (H1047R, E545K, and E542K) in either tumor tissues or ctDNA samples analyzed by droplet digital PCR (ddPCR), suggesting that PIK3CA mutation status is not a predictive determinant for everolimus benefit (Moynahan et al., 2017). mTOR, however, is downstream of PI3K, and studies have shown that AKT‐independent mechanisms of mTOR activation are involved in breast carcinogenesis (Tee et al., 2003). Recently, the mTOR inhibitors temsirolimus and everolimus and the PI3K inhibitors idelalisib and copanlisib have been approved by the FDA for clinical use in the treatment of a number of different cancers. Very recently, it was shown that early ctDNA dynamics in the commonly truncal mutations in PIK3CA may predict sensitivity to palbociclib, a CDK4/6 inhibitor (O'Leary et al., 2018a,2018b). Novel compounds with greater potency and selectivity, as well as improved therapeutic indices owing to reduced risks of toxicity, are clearly required. In addition, biomarkers that are predictive of a response, such as PIK3CA mutations for inhibitors of the PI3K catalytic subunit α isoform, must be identified and analytically and clinically validated (Janku et al., 2018).
PIK3CA mutational status has already been studied in CTCs (Bingham et al., 2017; Gasch et al., 2016; Markou et al., 2014; Schneck et al., 2013) and plasma‐ctDNA (Moynahan et al., 2017; Takeshita et al., 2017). When PIK3CA hotspot mutations were analyzed in CTCs isolated using a label‐free technology and corresponding ctDNA in 9 patients using 23 identical blood draws with colorectal cancer, mutational status of PIK3CA showed a match between CTCs and ctDNA in six blood draws and discordance in two, with 15 blood draws showing no mutations; however, in this study, PCR‐based Sanger sequencing (analytical sensitivity 7.5%) was used for mutation detection in CTCs and targeted panel NGS for cfDNA (Kidess‐Sigal et al., 2016). In another recent study, in a limited number of five metastatic breast cancer patients with more than 100 CTCs/7.5 mL PB (enumerated using the CellSearch), single EpCAM‐positive CTCs were isolated by DEPArray and compared with matched cfDNA and primary tumor tissue by targeted NGS for about 2,200 mutations in 50 cancer genes; according to this study, in these five patients, cfDNA profiles provided an accurate reflection of PIK3CA mutations seen in individual CTCs (Kidess‐Sigal et al., 2016; Shaw et al., 2017). According to these studies, the percentage of PIK3CA hotspot mutations in CTC and plasma‐ctDNA is around 30–40%. Our results indicate a much higher percentage of PIK3CA‐positive samples in both CTC and plasma‐ctDNA and this could possibly be explained by the higher analytical sensitivity of the molecular assay that we used (analytical sensitivity 0.05%) (Markou et al., 2014).
According to our results, in both early and metastatic BrCa, PIK3CA hotspot mutations were detected in the large majority of DNAs isolated from CellSearch® cartridges but in a lower number of paired plasma‐ctDNA samples. It is interesting to note that in most cases where CTCs were detected by CellSearch®, PIK3CA hotspot mutations were identified either in ctDNA or in CTCs or both, in the early and metastatic BrCa setting. There were two cases where CTCs were detected at high numbers, but PIK3CA mutations were not detected in CTCs. Moreover, there were many cases where CTC were not detected by CellSearch® but our downstream analysis in DNA material isolated from these cartridges was positive for PIK3CA mutations. This finding could be possibly explained by the fact that some EpCAM‐positive cells can be negative for CKs (8, 18, 19) due to EMT. These findings are in accordance with our previous results, where we identified PIK3CA hotspot mutations in EpCAM‐positive cells that were negative for CK‐19 mRNA expression (Lampignano et al., 2017; Markou et al., 2014). We have also shown in one of our previous studies (Markou et al., 2018) by using exactly the same blood draws that we can detect a lot of molecular alterations in EpCAM‐positive CTCs, in identical samples that were ‘officially’ negative for CTCs when using the CellSearch.
We realize that we report a high percentage of PIK3CA mutations in primary tumors; however, in all cases where PIK3CA E545K hotspot mutation was detected in FFPEs, it was also detected in DNA isolated from CellSearch® cartridges. However, we have to emphasize that the number of primary tumors that we tested was very low, since these are not randomly selected samples, but highly selected paired samples, where ctDNA and CTCs were also available, and our main aim was not to show the incidence of PIK3CA mutations in primary breast tumors but to compare the PIK3CA mutational status in primary tumors, CTCs and ctDNA. Our findings are in accordance with our previous paper (Markou et al., 2014), and we confirm again that there was no concordance in CTCs, ctDNA, and primary tumors.
Our aim was to evaluate the potential of our previously published assay for detecting PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA samples in early and metastatic breast cancer patients. The patients’ selection was performed independently from the receptor status of these patients. Our aim was clearly not to provide evidence regarding the potential utility of PIK3CA mutation status by plasma‐ctDNA or CTC in predicting response to specific therapy. This is not a clinical study, and the samples analyzed were not selected based on the clinical characteristics of the patients. On the contrary, the BELLE‐2 and BELLE‐3 clinical trial studies included a large number of clinical samples, and patients’ selection was based on specific inclusion and exclusion criteria, demonstrating the incidence of PIK3CA mutations in specific target populations.
Our direct comparison study revealed for the first time that in early breast cancer, PIK3CA hotspot mutations are present in CTCs at a higher percentage before than after treatment, when compared to paired plasma‐ctDNA samples. This finding may suggest that in early BrCa, standard treatment was more often successful in targeting a number of cancer cells, thus leading to the release of DNA derived from these dying cells into the systemic circulation. However, in metastatic BrCa, the percentage of PIK3CA hotspot mutations in DNAs isolated from CellSearch® cartridges was significantly higher in post‐treatment samples. This could be possibly explained by increased resistance to standard therapy which was not targeted therapy for PIK3CA hotspot mutations. Therefore, this nontargeted therapy seems to be ineffective to reduce the CTC/tumor burden in metastatic BrCa patients harboring PIK3CA hotspot mutations. In the same samples, the presence of PIK3CA hotspot mutations in plasma‐ctDNA was decreased after treatment, possibly indicating that the cells remained intact, hampering the release of ctDNA in peripheral blood. The concordance of PIK3CA mutations between CTCs and ctDNA was relatively poor in early breast cancer (48.2%) and higher in the metastatic setting (66.6%).
Since the high incidence of mutations found could raise questions, we performed a comparison study in the same samples using a commercially available highly sensitive methodology for PIK3CA mutations based on ddPCR, for ctDNA samples and CTCs that were available. Our results revealed a high concordance between our assay and ddPCR (81.1%, P = 0.001).
Our results in CTCs could only be compared with similar studies using highly sensitive methodologies, like ddPCR; however, till now so highly sensitive methodologies have not been applied in CTCs isolated using the CellSearch system. In the study of Schneck et al., the mutation rate reported for PIK3CA mutations in CTCs was only 15.8%. This percentage is much lower than the one that we report; however, this discrepancy can be clearly explained by the much lower analytical sensitivity of the SNaPshot methodology that is 5 to 10% (Hurst et al., 2009). Our results in ctDNA of metastatic breast cancer patients are in agreement with a very recent study by O'Leary et al. (2018b) where a percentage of 56.3% (9/16) is reported for PIK3CA mutations, in ctDNA samples, since we also report for metastatic breast cancer a similar percentage, even lower, more specifically 47.9% (35/73) in ctDNA samples. In the independent patient group, PIK3CA hotspot mutations were present at high frequencies (59.5%) in DNA samples isolated from CellSearch® cartridges. In most cases where CellSearch® detected at least one CTC, PIK3CA hotspot mutations were also identified. However, in 3 cases PIK3CA hotspot mutations were also detected in samples that were found negative for CTCs according to the CellSearch®. These data are in concordance with our previous results, indicating that PIK3CA mutations are also present in EpCAM‐positive cells where CellSearch gave negative results.
5. Conclusions
We performed for the first time a direct comparison study on the presence of PIK3CA hotspot mutations in CTCs and plasma‐ctDNA using identical blood draws and a highly sensitive and analytically validated methodology. Our results indicate that: (a) PIK3CA hotspot mutations are present at high frequencies in DNAs isolated from CellSearch® cartridges and paired plasma‐ctDNA both in early and metastatic BrCa, (b) the concordance between plasma‐ctDNA and CTCs is higher in the metastatic setting, (c) PIK3CA mutational status significantly changes after therapeutic intervention, and (d) PIK3CA mutation detection in CTCs and plasma‐ctDNA provides complementary information. Our data reflect significant differences in CTCs and ctDNA before and after treatment and point toward the necessity of following patients over time using an integrated liquid biopsy approach.
Conflict of interest
The authors declare no conflict of interest.
Author Contributions
ET and AM performed the experiments. EP performed the experiments in the CellSearch. ET, AM, and EL wrote the manuscript. ET, AM, AK, AP, DM, VG, and EL edited the manuscript. AK, AP, DM, and VG collected the clinical specimens. EL designed the study and supervised the experimental procedures.
Supporting information
Fig S1. Independent group: PIK3CA hotspot mutations in DNA isolated from CellSearch® cartridges, before and after treatment from metastatic BrCa patients (n = 17).
Table S1. Clinicopathological characteristics of early and metastatic BrCa patients used in the direct comparison study.
Table S2. Correlation of PIK3CA hotspot mutations and clinicopathological characteristics of early and metastatic BrCa patients used in the direct comparison study.
Acknowledgements
This study has been financially supported by the IMI contract no. 115749 CANCER‐ID: ‘Cancer treatment and monitoring through identification of circulating tumor cells and tumor related nucleic acids in blood’ (https://www.cancer-id.eu/). This study has been co‐financed by the European Union (European Regional Development Fund—ERDF) and Greek national funds through the Operational Program ‘Competitiveness and Entrepreneurship’ of the National Strategic Reference Framework (NSRF)—Research Funding Program: ‘Liquid biopsy: In vivo capturing and molecular characterization of circulating tumor cells as a novel tool for improving tertiary prevention in breast cancer’. This research project was also implemented under the IKY Scholarship which was funded by the Action ‘Strengthening Postdoctoral Researchers’ act from the resources of EP ‘Human Resources Development Program, Education and Lifelong Learning’ with priority axes 6, 8, and 9 and is co‐financed by the European Social Fund (ESF) and the Greek Government.
References
- Aktas B, Kasimir‐Bauer S, Müller V, Janni W, Fehm T, Wallwiener D, Pantel K, Tewes M, DETECT Study Group (2016) Comparison of the HER2, estrogen and progesterone receptor expression profile of primary tumor, metastases and circulating tumor cells in metastatic breast cancer patients. BMC Cancer 16, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix‐Panabières C and Pantel K (2016) Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6, 479–491. [DOI] [PubMed] [Google Scholar]
- Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, Ferro A, Dalla Palma P, Galligioni E and Marchetti A (2007) Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res 13, 6064–6069. [DOI] [PubMed] [Google Scholar]
- Bardelli A and Pantel K (2017) Liquid biopsies, what we do not know (Yet). Cancer Cell 31, 172–179. [DOI] [PubMed] [Google Scholar]
- Baselga J, Im S, Iwata H, Cortés J, De Laurentiis M, Jiang Z, Arteaga CL, Jonat W, Clemons M, Ito Y et al (2017) Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor‐positive, HER2‐negative, advanced breast cancer (BELLE‐2): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet Oncol 18, 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ and Siu LL (2014) Tumour heterogeneity in the clinic. Nature 501, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM et al (2014) Detection of circulating tumor DNA in early‐ and late‐stage human malignancies. Sci Transl Med 6, 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard FC, Michiels S, Riethdorf S, Mueller V, Esserman LJ, Lucci A, Naume B, Horiguchi J, Gisbert‐Criado R, Sleijfer S et al (2018) Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta‐analysis. J Natl Cancer Inst 110, 560–567. [DOI] [PubMed] [Google Scholar]
- Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert‐Criado R, Mavroudis D, Grisanti S, Generali D, Garcia‐Saenz JA, Stebbing J et al (2014) Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol 15, 406–414. [DOI] [PubMed] [Google Scholar]
- Bingham C, Fernandez SV, Fittipaldi P, Dempseym PW, Ruthm KJ, Cristofanilli M and Katherine Alpaugh R (2017) Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res Treat 163, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IG, Russell SE, Choong DYH, Montgomery KG, Ciavarella ML, Hooi CSF, Cristiano BE, Pearson RB and Phillips WA (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64, 7678–7681. [DOI] [PubMed] [Google Scholar]
- Chimonidou M, Strati A, Malamos N, Georgoulias V and Lianidou ES (2013) SOX17 promoter methylation in circulating tumor cells and matched cell‐free DNA isolated from plasma of patients with breast cancer. Clin Chem 59, 270–279. [DOI] [PubMed] [Google Scholar]
- Chimonidou M, Strati A, Tzitzira Α, Sotiropoulou G, Malamos N, Georgoulias V and Lianidou ES (2011) DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem 57, 1169–1177. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW et al (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351, 781–791. [DOI] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler‐Araujo B et al (2013) Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Di Leo A, Johnston S, Lee KS, Ciruelos E, Lønning PE, Janni W, O'Regan R, Mouret‐Reynier MA, Kalev D, Egle D et al (2018) Buparlisib plus fulvestrant in postmenopausal women with hormone‐receptor‐positive, HER2‐negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE‐3): a randomised, double‐blind, placebo‐controlled, phase 3 trial. Lancet Oncol 19, 87–100. [DOI] [PubMed] [Google Scholar]
- Gasch C, Oldopp T, Mauermann O, Gorges TM, Andreas A, Coith C, Müller V, Fehm T, Janni W, Pantel K et al (2016) Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2‐negative metastatic breast cancer. Mol Oncol 10, 1330–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Cankovic M, Furtado LV, Meier F and Gocke CD (2015) Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn 17, 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges TM, Tinhofer I, Drosch M, Röse L, Zollner TM, Krahn T and von Ahsen O (2012) Circulating tumour cells escape from EpCAM‐based detection due to epithelial‐to‐mesenchymal transition. BMC Cancer 12, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA, Pritchard KI, Campone M, Noguchi S, Perez AT, Deleu I et al (2016) Correlative analysis of genetic alterations and everolimus benefit in hormone receptor‐positive, human epidermal growth factor receptor 2‐negative advanced breast cancer: results from BOLERO‐2. J Clin Oncol 34, 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CD, Zuiverloon T, Hafner C, Zwarthoff EC and Knowles MA (2009) A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes 2, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis M, Lee M and Jeffrey SS (2015) Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res 21, 4786–4800. [DOI] [PubMed] [Google Scholar]
- Janku F, Tsimberidou AM, Garrido‐Laguna I, Wang X, Luthra R, Hong DS, Naing A, Falchook GS, Moroney JW, Piha‐Paul SA et al (2011) PIK3CA Mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther 10, 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janku F, Yap TA and Meric‐Bernstam F (2018) Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 15, 273–291. [DOI] [PubMed] [Google Scholar]
- Kidess‐Sigal E, Liu HE, Triboulet MM, Che J, Ramani VC, Visser BC, Poultsides GA, Longacre TA, Marziali A, Vysotskaia V et al (2016) Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label‐free platform: comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget 7, 85349–85364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampignano R, Yang L, Neumann MHD, Franken A, Fehm T, Niederacher D and Neubauer H (2017) A novel workflow to enrich and isolate patient‐matched EpCAMhigh and EpCAMlow/negative CTCs enables the comparative characterization of the PIK3CA status in metastatic breast cancer. Int J Mol Sci 18, 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianidou E, Hoon D (2017) Circulating tumor cells and circulating tumor DNA In Tietz Textbook of Clinical Chemistry and Molecular Diagnostics (Nader R, Horrath A, Wittwer C, eds). Sixth. pp. 1111–1144. Elsevier Ltd, Amsterdam, the Netherlands. [Google Scholar]
- Lianidou ΕS, Μarkou Α and Strati Α (2015) The role of CTCs as tumor biomarkers. Adv Exp Med Biol 867, 341–367. [DOI] [PubMed] [Google Scholar]
- Liu Y, Meucci S, Sheng L and Keilholz U (2017) Meta‐analysis of the mutational status of circulation tumor cells and paired primary tumor tissues from colorectal cancer patients. Oncotarget 8, 77928–77941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, Xiao L, Bedrosian I, Kuerer HM and Krishnamurthy S (2012) Circulating tumour cells in non‐metastatic breast cancer: a prospective study. Lancet Oncol 13, 688–695. [DOI] [PubMed] [Google Scholar]
- Malapelle U, Sirera R, Jantus‐Lewintre E, Reclusa P, Calabuig‐Fariñas S, Blasco A, Pisapia P, Rolfo C and Camps C (2017) Profile of the Roche cobas® EGFR mutation test v2 for non‐small cell lung cancer. Expert Rev Mol Diagn 17, 209–215. [DOI] [PubMed] [Google Scholar]
- Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, Georgoulias V and Lianidou E (2014) PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res 20, 5823–5834. [DOI] [PubMed] [Google Scholar]
- Markou A, Lazaridou M, Paraskevopoulos P, Chen S, Świerczewska M, Budna J, Kuske A, Gorges TM, Joosse SA, Kroneis T et al (2018) Multiplex gene expression profiling of in vivo isolated circulating tumor cells in high‐risk prostate cancer patients. Clin Chem 64, 297–306. [DOI] [PubMed] [Google Scholar]
- Markou A, Strati A, Malamos N, Georgoulias V and Lianidou ES (2011) Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin Chem 57, 421–430. [DOI] [PubMed] [Google Scholar]
- Mastoraki S, Strati A, Tzanikou E, Chimonidou M, Politaki E, Voutsina A, Psyrri A, Georgoulias V and Lianidou E (2018) ESR1 methylation: a liquid biopsy‐based epigenetic assay for the follow‐up of patients with metastatic breast cancer receiving endocrine treatment. Clin Cancer Res 24, 1500–1510. [DOI] [PubMed] [Google Scholar]
- Mostert B, Sieuwerts AM, Kraan J, Bolt‐de Vries J, Van der Spoel P, Van Galen A, Peeters DJ, Dirix LY, Seynaeve CM, Jager A et al (2015) Gene expression profiles in circulating tumor cells to predict prognosis in metastatic breast cancer patients. Ann Oncol 26, 510–516. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Ringeisen F, Hortobagyi GN et al (2017) Correlation between PIK3CA mutations in cell‐free DNA and everolimus efficacy in HR+, HER2‐ advanced breast cancer: results from BOLERO‐2. Br J Cancer 116, 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, André F, Loibl S, Loi S, Garcia‐Murillas I et al (2018b) The genetic landscape and clonal evolution of breast cancer resistance to Palbociclib plus Fulvestrant in the PALOMA‐3 trial. Cancer Discov 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, Liu Y, Bartlett CH, Koehler M, Cristofanilli M et al (2018a) Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun 9, 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, Pazzagli M, Di Leo A and Pinzani P (2015) Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol 9, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack B, Schindlbeck C, Jückstock J, Andergassen U, Hepp P, Zwingers T, Friedl TW, Lorenz R, Tesch H, Fasching PA et al (2014) Circulating tumor cells predict survival in early average‐to‐high risk breast cancer patients. J Natl Cancer Inst 106, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethdorf S, O'Flaherty L, Hille C and Pantel K (2018) Clinical applications of the Cell Search platform in cancer patients. Adv Drug Deliv Rev 125, 102–121. [DOI] [PubMed] [Google Scholar]
- Schneck H, Blassl C, Meier‐Stiegen F, Neves RP, Janni W, Fehm T and Neubauer H (2013) Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol Oncol 7, 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, Hoon DSB and Pantel K (2011) Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 11, 426–437. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Guttery DS, Hills A, Fernandez‐Garcia D, Page K, Rosales BM, Goddard KS, Hastings RK, Luo J, Ogle O et al (2017) Mutation analysis of cell‐free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res 23, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts AM, Mostert B, Van der Vlugt‐Daane M, Kraan J, Beaufort CM, Van M, Prager WJC, De Laere B, Beije N, Hamberg P et al (2018) An in‐depth evaluation of the validity and logistics surrounding the testing of AR‐V7 mRNA expression in circulating tumor cells. J Mol Diagnostics 20, 316–325. [DOI] [PubMed] [Google Scholar]
- Siravegna G, Marsoni S, Siena S and Bardelli A (2017) Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14, 531–548. [DOI] [PubMed] [Google Scholar]
- Stathopoulou A, Vlachonikolis I, Mavroudis D, Perraki M, Kouroussis C, Apostolaki S, Malamos N, Kakolyris S, Kotsakis A, Xenidis N et al (2002) Molecular detection of cytokeratin‐19‐positive cells in the peripheral blood of patients with operable breast cancer: evaluation of their prognostic significance. J Clin Oncol 20, 3404–3412. [DOI] [PubMed] [Google Scholar]
- Strati A, Markou A, Parisi C, Politaki E, Mavroudis D, Georgoulias V and Lianidou E (2011) Gene expression profile of circulating tumor cells in breast cancer by RT‐qPCR. BMC Cancer 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita T, Yamamoto Y, Yamamoto‐Ibusuki M, Tomiguchi M, Sueta A, Murakami K, Omoto Y and Iwase H (2017) Analysis of ESR1 and PIK3CA mutations in plasma cell‐free DNA from ER‐positive breast cancer patients. Oncotarget 8, 52142–52155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC and Blenis J (2003) Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase‐activating protein complex toward Rheb. Curr Biol 13, 1259–1268. [DOI] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia‐Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R and Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17, 223–238. [DOI] [PubMed] [Google Scholar]
- Wang J, Chang S, Li G and Sun Y (2017) Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med 11, 522–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Independent group: PIK3CA hotspot mutations in DNA isolated from CellSearch® cartridges, before and after treatment from metastatic BrCa patients (n = 17).
Table S1. Clinicopathological characteristics of early and metastatic BrCa patients used in the direct comparison study.
Table S2. Correlation of PIK3CA hotspot mutations and clinicopathological characteristics of early and metastatic BrCa patients used in the direct comparison study.


