Abstract
Molecular mechanisms that define patterns of neuropeptide expression are essential for the formation and rewiring of neural circuits. The prodynorphin gene (PDYN) gives rise to dynorphin opioid peptides mediating depression and substance dependence. We here demonstrated that PDYN is expressed in neurons in human dorsolateral prefrontal cortex (dlPFC), and identified neuronal differentially methylated region in PDYN locus framed by CCCTC-binding factor binding sites. A short, nucleosome size human-specific promoter CpG island (CGI), a core of this region may serve as a regulatory module, which is hypomethylated in neurons, enriched in 5-hydroxymethylcytosine, and targeted by USF2, a methylation-sensitive E-box transcription factor (TF). USF2 activates PDYN transcription in model systems, and binds to nonmethylated CGI in dlPFC. USF2 and PDYN expression is correlated, and USF2 and PDYN proteins are co-localized in dlPFC. Segregation of activatory TF and repressive CGI methylation may ensure contrasting PDYN expression in neurons and glia in human brain.
Keywords: cell type-specific expression, DNA methylation, human brain, neuropeptides, transcription
Introduction
Neuropeptides, the largest family of neuromodulators, exert specific and coherent effects on the formation and rewiring of brain neural circuits and, consequently, on behavior (Bargmann 2012; Marder 2012; Leinwand and Chalasani 2014). Most neurons express neuropeptides and multiple neuropeptides are expressed by a single neuron. As a rule, each neuropeptide is expressed in tiny neuronal subpopulations characterized by specific distribution patterns across and within central nervous system (CNS) areas.
Cell type-specific gene expression is controlled by complex interplay of epigenetic and transcription factors (TFs). The epigenetic-transcriptional codes dictate the generation and specification of distinct neuronal and glial populations (Ciernia and LaSalle 2016; Yao et al. 2016). Cytosine methylation and hydroxymethylation enriched in neurons are essential for the development and function of the mammalian brain, dynamically regulated by neuronal activity and altered in neurodegenerative and psychiatric disorders (Hahn et al. 2013; Lister et al. 2013; Yao et al. 2016). DNA differentially methylated regions (DMRs) between neurons and other cell types may regulate cell type-specific transcription. Neuronal DMRs are over-represented in CpG island (CGI) shores, but not in CGIs which often overlap with gene promoters, and harbor binding motifs for neuron-specific and activity-dependent TFs (Telese et al. 2013; Ciernia and LaSalle 2016; Yao et al. 2016). TF binding is strongly influenced by methylation of CpG sites within their recognition sequences, pointing to a cause-and-effect relationship between CpG methylation and gene repression.
Prodynorphin (PDYN) is a precursor protein to the dynorphin opioid peptides that elicit their effects through κ-opioid receptor (Schwarzer 2009; Chavkin 2013). PDYN is expressed in specific neural circuits in the basal ganglia, frontal lobes, hippocampus and other brain areas. Pharmacological, genetic and clinical data demonstrates a role for dynorphins in the processing of reward and mood control, and cognitive processes including impulsivity, learning, and memory (Schwarzer 2009; Tejeda et al. 2012; Chavkin 2013). Molecular dysregulation of dynorphins in cortical areas and striatum is implicated in depression, stress, and substance dependence (Butelman et al. 2012; Tejeda et al. 2012; Van’t Veer and Carlezon 2013; Koob and Volkow 2016). A fundamental role of these peptides in shaping and dissipation of neural circuits is emphasized by generalized pathological changes in the human brain caused by PDYN missense mutations that affect the core neuropeptide sequence (Bakalkin et al. 2010; Smeets et al. 2015).
Remarkable patterns of cell type- and neuron-specific neuropeptide expression may be controlled by orchestrated activities of epigenetic and transcriptional mechanisms, each of which permits or restricts, activates or inhibits gene expression depending on the cellular context. We address this hypothesis by analysis of the expression of the PDYN gene in the dorsolateral prefrontal cortex (dlPFC) of human brain. This region is responsible for executive functions including working memory, cognitive flexibility and planning, and emotional processing. Dysregulation of dlPFC contributes to major depressive and post-traumatic stress disorders, schizophrenia, and substance dependence, in which dynorphins may have a pathogenic role (Butelman et al. 2012; Tejeda et al. 2012; Van’t Veer and Carlezon 2013). We here undertook a focused, in-depth study of PDYN regulation that complement, specify, and further elaborate epigenome-wide analyses of human brain relevant for psychiatric disorders. We aimed to identify epigenetic and transcriptional mechanisms of cell type-specific PDYN expression in dlPFC, particularly to pinpoint regions in the PDYN locus which are differentially methylated in neurons, and to identify methylation-sensitive sequence-specific TF(s) that target these regions and may establish neuron-specific pattern of PDYN expression. Molecular processes in human brain were analyzed using postmortem human specimens and cellular models.
Materials and Methods
Quantitative analysis of DNA methylation using real-time polymerase chain reaction (PCR), hairpin-bisulfite PCR, DNA hydroxymethylation, electrophoretic mobility shift assay, and luciferase reporter assays, RNA purification and western blot analysis were performed according to standard protocols and are described in detail in Supplementary Material. Plasmid construction, antibodies used, and analysis of TF binding sites are also described there.
Human Subjects
Tissues were collected at the New South Wales Brain Tissue Resource Centre (BTRC), University of Sydney, Australia (http://sydney.edu.au/medicine/pathology/btrc/; see Supplementary Table S1). All subjects were males of European descend (n = 138) with the mean age of 56.3 years (range 34–83 years). Cases with a prolonged agonal life support or with a history of cerebral infarction, head injury, or neurodegenerative disease (e.g., Alzheimer’s disease) were excluded. Informed written consent for autopsy was obtained from the next-of-kin and collection was approved by the Human Research Ethics Committees of the Sydney Local Health District (X15-0199) and the University of Sydney. The study was approved by the Swedish Central Ethical Review Board.
DNA Methylation Analysis
Infinium HumanMethylation 450k BeadChip Array
DNA was purified from human brain samples or fluorescence-activated nuclear sorting (FANS)-sorted nuclei using the DNeasy Blood & Tissue kit (Qiagen) and bisulfite converted using EZ DNA methylation Gold kit (Zymo Research) according to manufacturers’ instructions. Methylation profiling was performed using Infinium HumanMethylation450 BeadChip assay (Illumina) by The SNP&SEQ Technology Platform at Science for Life Laboratory (Sweden). Data were analyzed using the R/Bioconductor 3.3 package minfi 1.18.2 (Fortin et al. 2014). Poor quality samples (mean detection P-value > 0.01) were discarded; probes with a detection P-value > 0.01 in at least 1 sample within a data set were also discarded, as were X and Y chromosome probes, probes with single nucleotide polymorphisms (SNPs) at the CpG or single base extension site and cross-reactive probes identified by (Chen et al. 2013). Neuronal proportions were estimated by CETS 3.0.3 package (Guintivano et al. 2013).
Pyrosequencing Analysis
Total, neuronal and glial DNA methylation status of 1.25 kb human PDYN promoter region (see Supplementary Fig. S2) was quantified by direct bisulfite pyrosequencing. DNA was bisulphite treated using EpiTect Bisulfite Kit (Qiagen) according to manufacturer’s protocol and PCR-amplified with HotStart-IT Taq DNA polymerase (USB Corporation). Primers for quantitative analysis of methylation were designed by Pyrosequencing Assay Design Software 1.0.6 (Biotage). Genomic coordinates of interrogated regions and primer sequences are shown in Supplementary Table S4. All products were confirmed to be single bands of expected size by agarose gel electrophoresis. The biotinylated antisense strands of the target regions were immobilized using Streptavidin-coated Sepharose HP beads (GE Healthcare) followed by purification using Vacuum Prep Tool (Biotage), according to standard procedures. The PCR products, with annealed sequencing primers, were sequenced on a PyroMark ID System (Biotage). The targeted CpGs for each assay were evaluated by converting the pyrograms to numerical values for peak heights using Pyro Q-CpG software (Biotage). Pyrosequencing was performed at varionostic GmbH (Germany). Assay validation and controls are described in Supplementary Material.
Gene Expression Analysis
SYBR Green-Based Assay
Primers were designed with “Vector NTI advance 11” software (Invitrogen). Primer sequences and PCR conditions are reported in Supplementary Table S3. qPCRs were carried out using iQ SYBR Green Supermix (Bio-Rad) with a CFX96 Real-Time Detection System (Bio-Rad). Melting curve analysis of the PCR products was performed to ensure primer specificity and lack of primer dimers. Amplicons were separated on agarose gel and sequenced to ensure correct amplification. Gene expression in SK-N-MC cells was normalized to expression of ACTB and GUSB reference genes.
TaqMan®-Based Assay
TaqMan assays for GFAP (Hs00909233_m1), PDYN (Hs00225770_m1), POLR2A (Hs00172187_m1), RBFOX3 (Hs01370653_m1), RPLP0 (Hs99999902_m1), and USF2 (Hs00231528_m1) (Applied Biosystems) were used. cDNAs were mixed with TaqMan assay and iTaq Universal Probes supermix (Applied Biosystems) for qPCR with a CFX96 Real-Time Detection System (Bio-Rad). Gene expression in dlPFC was calculated by relative quantification using POLR2A and RPLP0 reference genes (Johansson et al. 2007).
RNA purified form FANS-sorted nuclei was analyzed using Droplet Digital™ PCR (McDermott et al. 2013). cDNAs were mixed with TaqMan assay, ddPCR Supermix for Probes (Bio-Rad) and Droplet Generation Oil (Bio-Rad), partitioned into 14 000–17 000 droplets in QX200 Droplet Generator and used for PCR with T100 Thermal Cycler (Bio-Rad) according to manufacturer’s instructions. The fluorescence intensity of the droplets was measured using the QX200 Droplet Reader (Bio-Rad). The data analysis was performed with QuantaSoft droplet reader software (Bio-Rad). mRNA amount was calculated using the Poisson statistics (Hindson et al. 2013). The absolute transcript levels were expressed in RNA copies per ng of total RNA.
Fluorescence-Activated Nuclei Sorting
Tissue samples were Dounce homogenized in lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM Mg(CH3COO)2, 0.1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 0.1% Triton X-100, 1 mM DTT), gently suspended in sucrose solution (1.7 M sucrose, 3 mM Mg(CH3COO)2, 1 mM DTT, 10 mM Tris-HCl, pH 8.0), layered onto a sucrose cushion and centrifuged at 30 000 g for 2.5 h. Nuclei pellets were resuspended in PBS and filtered through a 40 μm Nitex mesh to remove remaining clumps. Nuclei were incubated with 1:600 anti-Neuronal Nuclei (NeuN) antibody conjugated with mouse IgG labeling reagent Alexa 488 (Molecular Probes) for 18 h at +4 °C in the dark. NeuN were sorted on a FACSAria III flow cytometer (BD BioSciences). To ensure sorting single but not aggregated nuclei the preparations were stained with a Hoesch dye, and a gate was set to isolate singlets only that were readily discerned from doublets, triplets, and higher-order aggregates based on their fluorescence intensity. The purity of NeuN was confirmed by re-analysis of the sorted preparations. For RNA preparation, nuclei were incubated with anti-NeuN antibody for 30 min and directly sorted in the RLT lysis buffer (Qiagen).
Chromatin Immunoprecipitation
Protocol developed by Zuccato et al. (2007) was modified and optimized for analysis of smaller amount of frozen human brain tissue samples. Protocol validation and controls are described in Supplementary Material. About 100 mg powdered human brain tissue were cross-linked with 1% formaldehyde for 10 min on ice. Crosslinking was quenched by adding glycine solution to 125 mM final concentration. Tissue pellet was precipitated by centrifugation at 400×g, washed 3 times in PBS and once in buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl), resuspended in buffer A supplemented with 0.1 mM benzamidine and 0.1 mM phenylmethylsulfonyl fluoride, incubated for 5 min on ice and homogenized with a Pestle A in a Dounce glass-glass homogenizer. After centrifugation at 5000×g, the supernatant was removed and Dounce buffer (10 mM Tris-HCl, pH 7.5, 4 mM MgCl2, 1 mM CaCl2) and 2.4U of micrococcal nuclease (MNAse) (Sigma) were added to the pelleted nuclear fraction. After incubation for 15 min at room temperature, EDTA (final 10 mM) was added to stop the MNAse digestion. Cell lysis buffer (final 50 mM Tris-HCl, pH 8.0, 0.1% SDS; 0.5% sodium deoxycholate) was added, and after brief sonication on Branson Sonifier B15, the samples were incubated for 5 min on ice and centrifuged at 10 000×g to remove debris. One hundred microliters of chromatin aliquots were diluted 5-fold with modified RIPA buffer (140 mM NaCl, 10 mM Tris, pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 1% TX-100, 0.01% SDS, 0.1% sodium deoxycholate) supplied with protease inhibitors. Four micrograms of rabbit polyclonal anti-USF2 or rabbit polyclonal anti-acetyl Histone H3 (K9/K14) antibodies, or pre-immune rabbit IgG were added, and the reaction mixture was incubated overnight with rotation at 4 °C. Twenty-five microliters of protein G paramagnetic beads (Invitrogen) were added to the sample followed by incubation for 4 h at 4°C with rotation. Beads were captured, supernatant removed and beads-antibody complexes were washed twice with Wash buffer I (20 mM Tris-HCl, pH 8.1, 50 mM NaCl, 2 mM EDTA, 1% TX-100, 0.1% SDS) for 2 min, once with Wash buffer II (10 mM Tris-HCl, pH 8.1, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 250 mM LiCl) and twice with TE buffer. Washed beads were resuspended in elution buffer (0.1 M NaHCO3, 1% SDS) and incubated to reverse crosslinks overnight at 65°C. Chromatin was then treated with Proteinase K (Qiagen) for 1.5 h at 37 °C. Following phenol/chloroform extraction, qPCR was performed with specific primers (see Supplementary Table S3). Amplicon positions are shown in Supplementary Figure S5D.
Mononucleosomal DNA Analysis using qPCR
Chromatin was prepared and de-crosslinked as described for Chromatin immunoprecipitation (ChIP). Briefly, tissue powder was cross-linked, homogenized and chromatin was digested with MNAse. Digested chromatin was incubated overnight at 65°C to reverse crosslinks and DNA was extracted using phenol/chloroform. MNAse-treated DNA was loaded onto 1.5% agarose gel and the fragment corresponding to mononucleosomal size was excised and purified using illustra GFX™ PCR DNA and Gel Band purifyication kit (GE Healthcare). Fragment enrichment was quantitated by qPCR with primers listed in Supplementary Table S3. Amplicon positions of are shown in Supplementary Figure S5D.
Immunofluorescence
Paraffin sections (4 μm) were dewaxed using the standard protocol in autostainer (Bond RX, Leica) followed by heat-induced epitope retrieval step in high pH EDTA buffer (HIER2, Leica) and incubated with rabbit anti-PDYN antibody (1:2000) in antibody diluent (0.1 M phosphate buffer containing 0.3% TX-100 and 0.1% sodium azide, pH 7.4) for 48 h at 4 °C (Kononenko et al. 2016). Sections washed in TBS containing 0.5% Tween 20 (TNT) were incubated in TBS containing 2.5% blocking reagent (TNB, Perkin Elmer), then with horseradish peroxidase conjugated secondary swine antibody against rabbit IgG (1:200; Dako), washed in TNT buffer, incubated in amplification diluent containing Cy5-conjugated tyramine (1:100, Perkin Elmer) and after washing in TNT probed with an antibody against glial fibrillary acidic protein (GFAP; 1:1000) raised in chicken and biotin-conjugated rabbit anti-NeuN (RBFOX3; 1:1000) in primary antibody diluent for 16 h at 4°C. Sections were then blocked in TNB and incubated with Alexa fluor 488 conjugated anti-chicken IgY raised in donkey (1:200, Jackson immunoresearch). NeuN immunoreactivity (ir) was visualized using peroxidase avidin/biotin (Vectastain Elite, Vector laboratories) followed by incubation in amplification diluent containing Cy3.5-conjugated tyramine (1:100, Perkin Elmer). Lipofuscin autofluorescence was blocked by incubating the sections in 70% EtOH containing 1% Sudan black. Slides were mounted using ProLong Gold antifade reagent (Invitrogen). Entire slides were scanned using a slide-scanning microscope (Vslide, Metasystems) equipped with a 10× objective and the appropriate excitation and emission filter sets for detection of used fluorophores. After scanning individual field of view images were stitched and background signal based on median pixel intensity was subtracted. Image segmentation based on thresholded NeuN and GFAP masks were used to calculate average PDYN-ir in both cell types and to create overview images color-coding astrocyte and neuronal PDYN-ir. All image processing was performed using the open source image analysis software ImageJ (NIH).
Immunolabeling and Light Microscopy
Formalin-fixed paraffin-embedded sections (7 μm) of human dlPFC were deparaffinaized in xylene and rehydrated in a graded ethanol series. Antigen retrieval was performed in sodium citrate buffer, pH 6.0 by microwave heating. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide (Sigma-Aldrich). Rabbit and goat serums (Dako) were used as a blocking reagent. Primary immunolabeling was performed at 4°C using polyclonal rabbit anti-human PDYN antibodies (1:1000) overnight and monoclonal mouse anti-USF2 antibody (1:7500) for 3 days. For double labeling sections were incubated with anti-USF2 antibody followed by incubation with anti-human PDYN antibody. Secondary antibody staining was performed using polyclonal rabbit anti-mouse IgG (1:300, Dako) and polyclonal goat anti-rabbit IgG (1:300, Dako) for 45 min at room temperature. Labeling by secondary IgG (Dako) or pre-immune rabbit serum IgG fraction (Nikoshkov et al. 2005) was used for negative control. Immunoreactivity visualization was performed using VECTASTAIN Elite ABC system (Vector Laboratories) followed by 3,3′-diaminobenzidine (DAB) plus nickel or DAB only treatment (Vector Laboratories). Single immunolabeled samples were counterstained by hematoxylin (for PDYN) and toluidine blue (for USF2) to visualize all cell nuclei.
For analysis of double immunolabeled samples 50–60 image fields (×20, minimum 50 cells per field) per section were taken uniformly across the layers II, III, and V and immunoreactivity of all cells within every image field was visually analyzed (ca. 2500–00 cells per every subject). Light microscopy analysis was carried out using Olympus BM-2 microscope equipped with Nikon DXM 1200 F digital camera and EclipseNet image software. All images were taken using the same exposure and RGB range.
Statistical Analysis
Statistical analysis was performed using Statistica version 12.0 (StatSoft Scandinavia, Uppsala, Sweden) and R version 3.3.0 (http://www.R-project.org/). Normality of data distribution was analyzed using Kolmogorov–Smirnov test. R/bumphunter package with b = 105 bootstrapping resamples (Jaffe et al. 2012) was used to identify neuronal DMRs given null hypothesis of no tissue effect for all measured CpGs. We formed DMRs only from CpGs detected by “bumphunter” at FWER < 10−6 both with running median smoothing (k = 5) and without smoothing. Statistical analysis was performed by 2-tailed Student’s t-test (Figs 1L, 4B, and 4C), or 1- (Figs 1K, 3I, and 4E) and 2-way (Figs 3G,H and 4A) ANOVAs followed by post hoc Tukey HSD-test on least squares means. z-transformed Pearson correlations of methylation beta-values were analyzed using 2-way ANCOVAs (Fig. 2C), for detailed description see Supplementary Material. PDYN expression (Figs 5A, B) was analyzed using linear regression model with the following cofactors: age, PMI, brain pH, RQI, RBFOX3, GFAP, USF2 mRNA levels, neuronal proportion, and alcoholism (DSM-IV). Backward stepwise linear regression by R/MASS package was used to refine the model in 2 independent cohorts. For analysis of effects of USF2 protein (Fig. 5C) initial linear model for PDYN expression was constructed with the following cofactors: age, PMI, brain pH, and RQI and USF2 protein level. Overly influential points with Cook’s distance ≥0.2 were removed when analyzing candidate models (Faraway 2002). A significance level of P < 0.05 was accepted as statistically significant and all tests were 2-tailed.
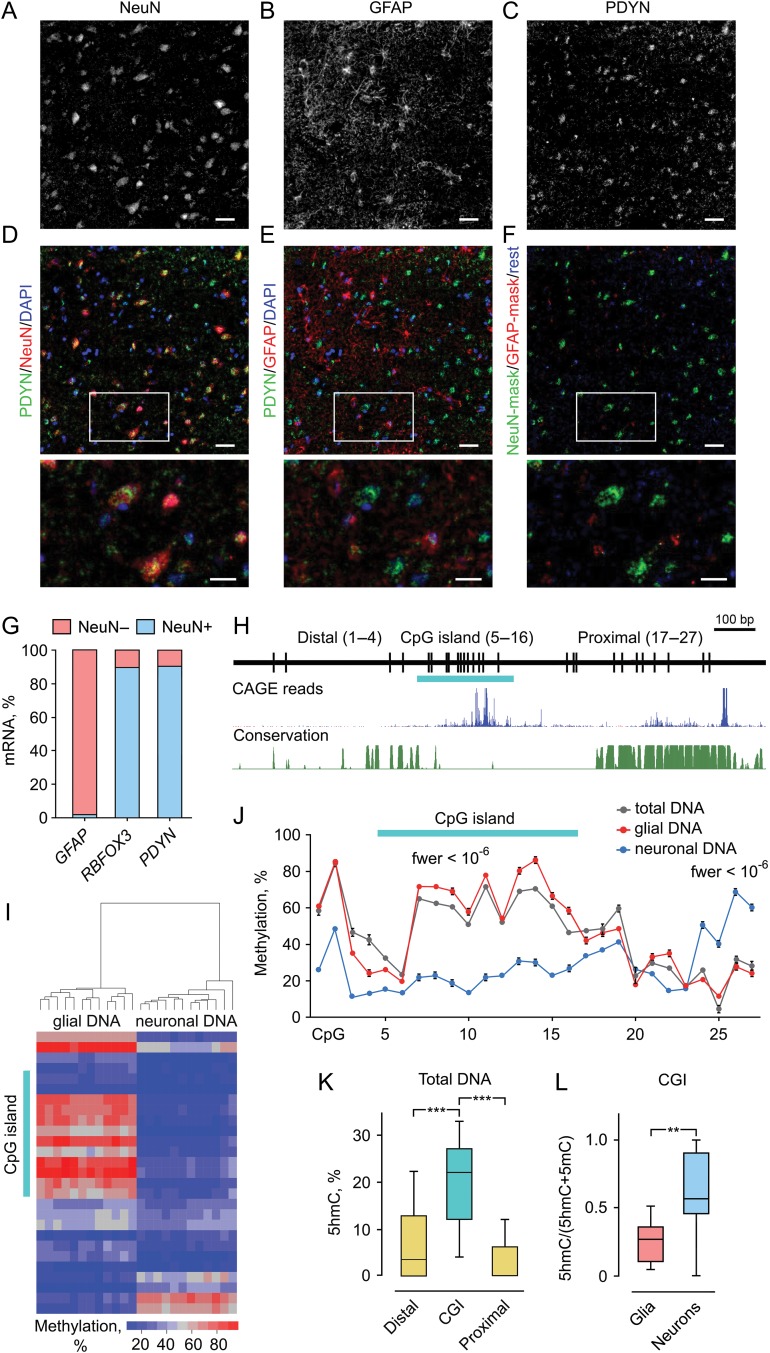
Figure 1.
Neuronal expression of PDYN in human dlPFC is associated with 2 DMRs and elevated 5hmC levels in the promoter CGI. (A–F) Immunofluorescence micrographs showing the distribution of PDYN immunoreactivity (ir) in NeuN-ir neurons (A and D) and GFAP-ir astrocytes (B and E) in the cortex. PDYN-ir located in the cytosol and proximal processes of NeuN-ir neurons (C) was mainly co-localized with NeuN-ir (D). Minor PDYN-ir was co-localized with GFAP-ir (E) located in astrocyte cell bodies and processes (B). Neuronal PDYN-ir is shown in green, astrocyte PDYN-ir in red, and PDYN-ir not co-existing with GFAP-ir or NeuN-ir in deep blue (F). Lower row shows magnified images of the area outlined in (D–F). The majority of PDYN-ir pixels especially those with high intensity are found in neurons. Scale bars, 50 μm (A–F); 25 μm (magnified images in the lower row). (G) Levels of RBFOX3 (neuronal marker) and PDYN mRNAs are high in NeuN while those of GFAP (astrocyte marker) in non-NeuN. Bar graphs show average mRNA amount in the NeuN+ and NeuN− fractions as % of total amount of these mRNAs. Nuclei were isolated by FANS from dlPFC of 3 subjects, mRNA analyzed using ddPCR and normalized to total RNA. (H) Diagram depicts CpG positions, short CpG island (CGI) (horizontal bar), and Distal and Proximal regions in 1.42 kb human PDYN promoter. Genome browser shot shows TSSs in human PDYN promoter (FANTOM5 UCSC track, GRCh37/hg19), followed by phastCons track showing conserved elements. (I) Heatmap depicting methylation levels measured by pyrosequencing across PDYN promoter. Clusters of neuronal and non-neuronal DNA are distinguished based on methylation levels. (J) Two neuronal DMRs identified by “bumphunter”. DMR1 (CpGs 1–19) is hypo- while DMR2 (CpGs 24–27) hypermethylated in neurons. The CGI CpGs demonstrate largest and most significant differences among those in DMR1. Average methylation level for 12 subjects and the errors of the means are shown. fwer values for DMRs computed using bootstrap method, as detailed in the “Materials and Methods” section, are indicated. (K) The 5hmC levels are significantly higher in the CGI compared to the Distal or Proximal promoter regions in total tissue DNA analyzed by 5hmC-qAMP assay (n = 21 subjects). (L) A proportion of 5hmC assessed as the (5hmC)/(5hmC + 5mC) ratio in the CGI is significantly higher in neurons compared to glial cells analyzed by 5hmC-qAMP and qAMP assays (n = 12 subjects). The bar graphs show averages and the errors of the means. In box plots, center line is the median, box spans the interquartile range (IQR), and whiskers are 1.5 × IQR from box limits. **P < 0.01, ***P < 0.001. See also Supplementary Figures S1–S3.
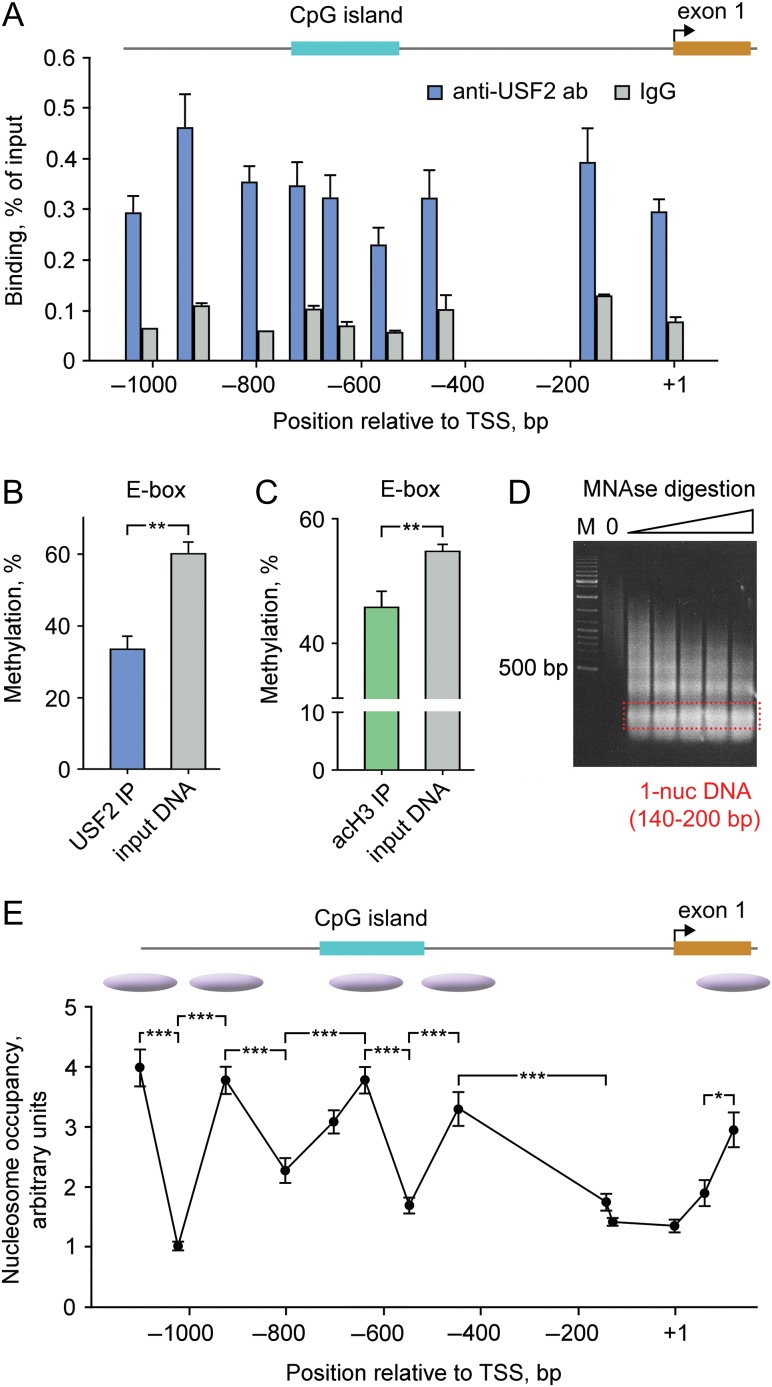
Figure 4.
USF2 binds to hypomethylated PDYN promoter in the human dlPFC. (A) Enrichment of USF2 across PDYN promoter. ChIP-qPCR results are shown as levels of DNA immunoprecipitated by anti-USF2 antibodies or control IgG for 9 amplicons. Six tissue samples all of each pooled from three subjects, altogether from 18 subjects were analyzed. Post hoc Tukey HSD-test showed significantly higher binding of USF2 comparing to control IgG (3.9-fold; P < 0.001) to the promoter. Bar graphs show mean ± SEM. (B and C) Methylation of the PDYN promoter E-box CpG 12 associated with USF2 (B) or acetylated histone 3 (H3K9/K14 acetylation, an activatory mark; (C)) was significantly lower compared to that of the total tissue DNA. Methylation was analyzed in DNA immunoprecipitated either with anti-USF2 antibodies or anti-H3-Ac antibodies by qAMP assay using PCR primer Set 7 (see Supplementary Table S3). Six tissue samples all of each pooled from 3 subjects, altogether from 18 subjects were analyzed. Bar graphs show mean ± SEM. **P < 0.01 (t-test). (D) Marker (M) and DNAs digested with increasing amounts of MNAse (1.25, 1.5, 1.75, 2.0, and 2.25 units) on an agarose gel. (E) qPCR analysis of MNAse-digested chromatin from dlPFC. Nucleosome occupancy and inferred positioning are shown. The bar graphs (A–C) and data points (E) show averages for 6 samples pooled from 18 subjects and the errors of the means. The lines connecting data points do not imply levels between points. Bent arrow, proximal TSS. *P < 0.05, **P < 0.01, ***P < 0.001. For experimental details, see Supplementary Figure S5.
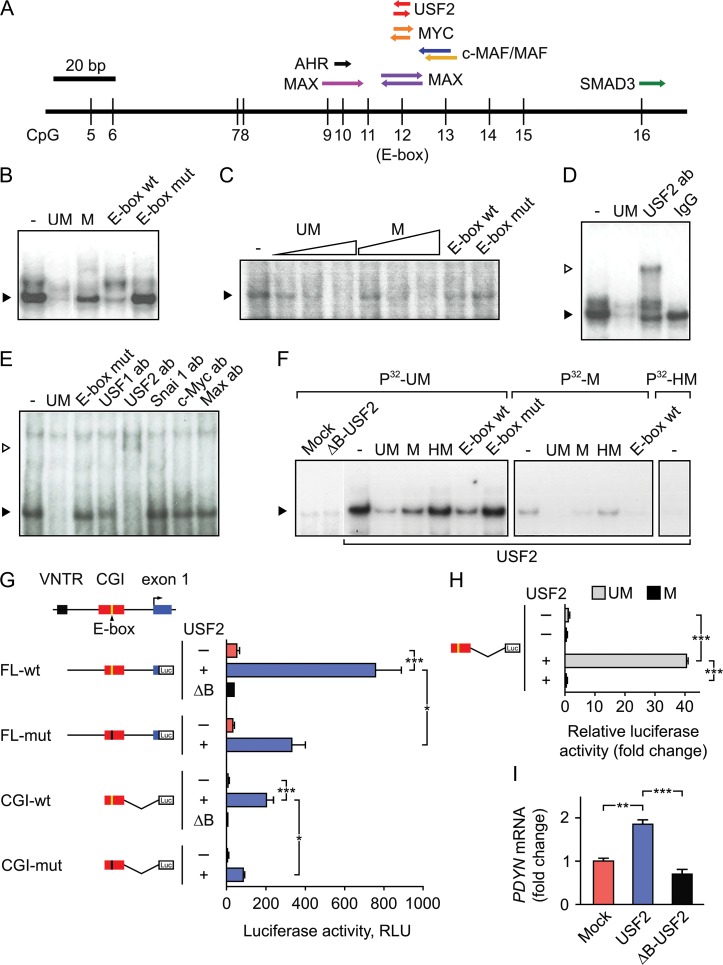
Figure 3.
USF2 binds to unmethylated E-box in the PDYN promoter CGI and activates gene transcription. (A) A putative CpG-containing TF binding sites (TF-BSs) in the PDYN promoter CGI identified by TRANSFAC. The CpG 12 is located in E-box sequence (CACGTG). (B and C) Methylation-sensitive DNA-binding factor interacting with the CGI E-box was identified by EMSA in nuclear extract of RFB (B) and human dlPFC (C). Labeled probe: unmethylated (UM) CGI E-box oligonucleotide. Competitors: UM- and methylated (M) CGI E-box oligonucleotides; and wild-type (E-box wt) and mutant (E-box mut) E-box oligonucleotides. Filled arrowhead shows the specific complex. (D and E) The CGI E-box-binding factor is identified as USF2 in RFB (D) and human dlPFC (E) in supershift and depletion experiment with antibodies against c-Myc, Max, Snai 1, USF1, and USF2. IgG, control IgG. Filled and open arrowheads show the specific and supershifted complex. (F) Binding of USF2 ectopically produced in SK-N-MC cells to the CGI E-box is inhibited by E-box methylation or hydroxymethylation. Labeled probes: E-box UM, M, or hydrohymethylated (HM) CGI oligonucleotides. ΔB-USF2, a dominant negative mutant lacking DNA-binding domain. (G and H) Luciferase reporter assays in SK-N-MC cells. Schematics of human PDYN promoter and luciferase reporter constructs are shown. Luc, luciferase coding sequence. Cells were transfected with USF2 (+) or ΔB-USF2 (ΔB) expressing vectors, or mock (−). RLU, relative light units normalized to “renilla” luciferase activity in (G) and to mock in (H). In (H), the unmethylated (UM) or methylated (M) CGI-wt inserted in pCpGL-basic, a CpG-free reporter vector, was transfected. (I) Expression of the endogenous PDYN gene is activated in SK-N-MC cells transfected with USF2- but not with ΔB-USF2-expressing vector, or mock transfected cells. The bar graphs show averages of 3 independent experiments and the errors of the means. *P < 0.05, **P < 0.01, ***P < 0.001.
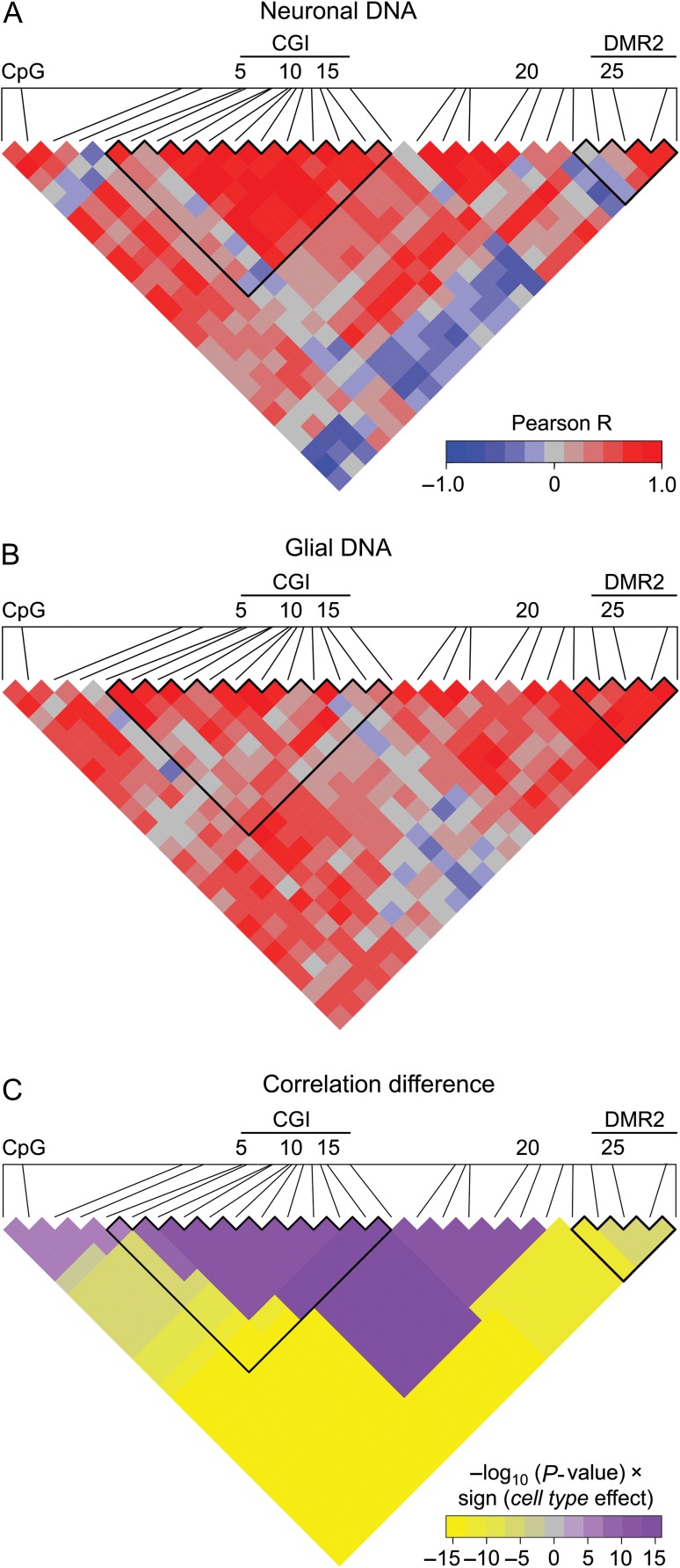
Figure 2.
Central promoter region including the most part of CGI demonstrates high coordination in methylation in neurons, whereas the rest of the promoter in glial cells. (A and B) Heatmap representation of correlations in methylation of 27 CpGs in PDYN promoter in neuronal (A) and glial DNA (B) from dlPFC (n = 12). Scale bar shows the color-coded pairwise Pearson correlation R-values with red and blue indicating high correlations and high anticorrelations, respectively. CGI and DMR2 boundaries are outlined. (C) Heatmap representation of direction and significance of differences between neurons and glia for pairwise Pearson correlations of CpG methylation levels. For each CpG pair in heatmap −log10(P-value) × sign (“cell type” effect) from ANCOVA with the most significant cell type effect (among all regions including this CpG pair, see “Materials and Methods” section) is shown. Purple and yellow colors indicate significantly higher correlations in neurons and glia, respectively. CGI and DMR2 boundaries are outlined.
Figure 5.
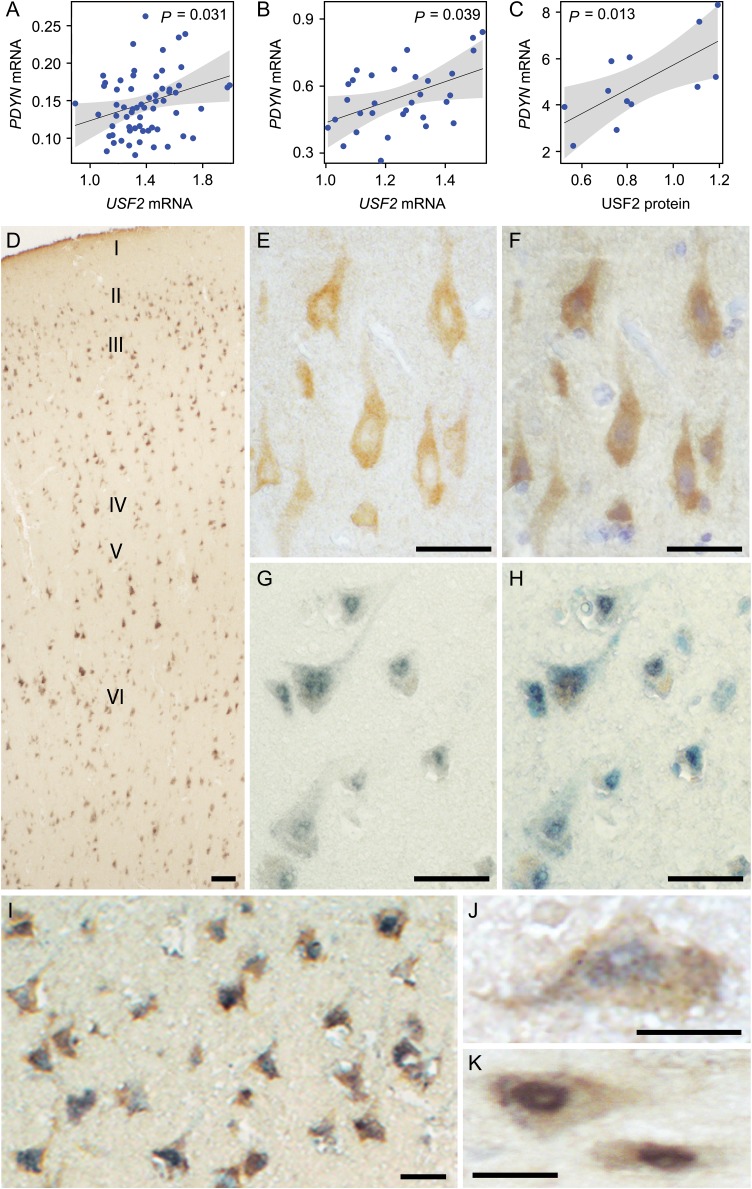
USF2 is positively associated with PDYN mRNA and is co-localized with PDYN in the human dlPFC. (A–C) Effect display for the main effect of USF2 mRNA (A, n = 64 subjects of the First cohort; B, n = 31 subjects of the Second cohort) and USF2 protein (C, n = 12 subjects) on PDYN mRNA. 95% confidence band is drawn around the estimated effect. P-values from ANOVAs are indicated. Potentially confounding factors such as age, postmortem interval, tissue pH, RBFOX3, GFAP levels, and alcoholism were included as covariates in the analysis but did not cause any significant effects. (D) Overview of dlPFC laminae immunolabeled for PDYN and USF2 proteins. Note the uniform distribution of the signal abundance and intensity across cortical laminae (shown with roman numerals). (E) A representative image showing PDYN immunoreactivity in neurons of the layer V. Note predominantly cytoplasmic localization of the signal. (F) Visualization of cell nuclei in (E) by hematoxylin staining. (G) A representative image showing USF2 signal in the layer V. Note predominantly nuclear localization of the signal. (H) Visualization of cell nuclei in (F) by toluidine blue staining. (I) Both PDYN and USF2 immunoreactivity are located in the same neurons in the layers III/IV. (J and K) High-magnification images of neurons in the layer V expressing both PDYN and USF2. Scale bars, 100 μm (D); 50 μm (E–I); and 25 μm (J and K).
Results
Neuronal Expression of PDYN is Associated with Differential and Coordinated Methylation Pattern and 5hmC Enrichment in PDYN Promoter in Human dlPFC
We examined cell type-specific PDYN expression in human dlPFC, first, using triple-labeling immunofluorescence for PDYN, NeuN, the neuronal marker transcribed from the RBFOX3 gene, and GFAP, the astrocyte marker (Fig. 1A–F); and second, analyzing PDYN, RBFOX3, and GFAP mRNAs by ddPCR in neuronal and non-neuronal (mostly glial) nuclei isolated by FANS (Fig. 1G). PDYN expression in the NeuN-expressing neurons and GFAP-expressing astrocytes was identified based on overlapping immunoreactity (ir). Anti-NeuN antibody labeled neuron-like perikarya (Fig. 1A,D) while anti-GFAP antibody astrocyte-like cells and processes (Fig. 1B,E). Anti-PDYN antibody revealed neuron-like and neuropil-like staining pattern (Fig. 1C–F). A most of the PDYN-ir co-existed with NeuN-ir (Fig. 1D,F) while only its small portion overlapped with GFAP-ir (Fig. 1E,F). Analysis of images segmented based on PDYN-ir, NeuN-ir, and GFAP-ir revealed a 5.5-fold higher level in neurons vs. astrocytes (Fig. 1F).
Consistently with immunofluorescence results, PDYN mRNA was highly enriched, approximately 10-fold in neuronal compared to non-NeuN (Fig. 1G). RBFOX3 and GFAP mRNAs were localized in neuronal and non-NeuN, respectively (Fig. 1G).
To identify PDYN DMR(s) compatible with its neuronal expression, we compared methylation profiles of the 42 kb PDYN locus framed by 2 strong CCCTC-binding factor (CTCF) peaks (see Supplementary Fig. S1A), between neurons and non-neuronal cells using Illumina Infinium HumanMethylation450 beadchip (450 K) data. CTCF is a DNA-binding factor that is involved in transcriptional regulation, insulator activity and regulation of the 3D structure of chromatin; it binds together strands of DNA, thus forming chromatin loops, anchors DNA to cellular structures, for example, the nuclear lamina and defines the boundaries between active and heterochromatic DNA (Phillips and Corces 2009). Analysis of methylation of 10 CpGs in this locus present on the 450 K array identified 2 adjacent promoter DMRs showing opposite methylation patterns. In neurons compared to glial cells CpGs in the DMR1 were hypomethylated (fwer < 10−15) while in DMR2 hypermethylated (fwer < 10−15) (see Supplementary Fig. S1B). The central segment of DMR1 represents a short, 247 bp CpG island (CGI) that overlaps with the distal cluster of transcription start sites (TSSs) (Fig. 1H) and is a unique human trait; it is conserved between humans and great apes but not other vertebrates (Fig. 1H). Bisulfite pyrosequencing of the 1.25 kb PDYN promoter fragment (nucleotide sequence is shown in Supplementary Fig. S2) validated this finding. Two DMRs were identified; DMR1 consisting of CpGs 1–19 and hypomethylated in neurons (fwer < 10−6), and DMR2 encompassing CpGs 24–27 and hypermethylated in neurons (fwer < 10−6) (Figs 1I,J). Total DNA methylation pattern was similar with that of glial DNA (Fig. 1J).
A high abundance of nonCpG methylation that is asymmetric in DNA molecules was found in the human brain (Lister et al. 2013; Varley et al. 2013). Frequencies of nonCpG and asymmetric cytosine methylation in the PDYN promoter CGI (CpGs 5–16) assessed using hairpin-bisulfite PCR assay were low, 6.4% and 1.2%, respectively. The assay also identified 2 methylation patterns of individual DNA molecules which were either virtually nonmethylated at the CGI, or densely methylated at this location, suggesting their origin from neurons and glia, respectively, (see Supplementary Fig. S3).
Active gene regulatory regions are enriched in 5-hydroxymethylcytosine (5hmC) (Feldmann et al. 2013; Lister et al. 2013; Wen et al. 2014). We examined whether 5hmC is present in PDYN promoter by quantitative analysis of 5hmC using real-time PCR (5hmC-qAMP) assay. The 5hmC levels were significantly higher in the CGI relative to the Distal (3.0-fold; P = 0.0001) and Proximal (5.3-fold; P = 0.0001) regions in total tissue DNA (Fig. 1K). Validation by Mirror-BS seq assay showed essentially the same 5 hmC levels in the CGI (data not shown). A proportion of 5hmC in total modified cytosine fraction in the CGI was significantly 2.4-fold higher in neurons compared to glia (P = 0.004) (Fig. 1L).
To examine whether GpG methylation within each of the 2 DMRs and between the DMRs is interdependent, we analyzed Pearson correlations between methylation levels for all pairs of 27 CpGs in the PDYN promoter for neurons and glia separately. A graphical representation of R-values revealed 2 regions with high positive intraregional correlations that were the DMR1/CGI in neurons (Fig. 2A) and DMR2 in glial cells (Fig. 2B). Interestingly, correlations between DMR1 and DMR2 were highly negative in neurons but not glia (Fig. 2A). The significance of differences in the pairwise correlations in methylation between neurons and glia, and the direction of these differences are visualized as the heatmap on Figure 2C. The central promoter region encompassing CpGs 7–22 demonstrated significantly higher correlations in neurons than in glia, whereas in the rest of the promoter methylation significantly higher correlated in glia compared to neurons. These patterns suggest coordinated regulation of CpG methylation of the central promoter segment including most of the CGI in neurons that may be a characteristic of active gene. In opposite, the coordinated methylation in the rest of the promoter may reflect the repressed gene state in non-neuronal cells.
In summary, PDYN expression is associated with opposing methylation patterns of the 2 DMRs, high coordination in CpG methylation within the CGI and enrichment of 5hmC in the CGI in neurons compared to glia. These findings suggested a local regulation of CGI methylation/hydroxymethylation in the context of PDYN promoter, and a CGI role in regulation of PDYN transcription.
USF2 Binds to E-box in the PDYN Promoter CGI and Activates Transcription
The CGI may serve as a regulatory module, a target for TF(s), which action is abrogated by CpG methylation. TRANSFAC analysis focused on canonical neuronal CpG-containing TF-binding sites (TF-BSs) identified target DNA elements for Upstream Stimulatory Factor 2 (USF2), MYC, MAX, c-MAF/MAF, AHR, and SMAD3 in the CGI (Fig. 3A), of which USF2, MYC, and MAX are the E-box TFs. Sequences aligned around CGI CpGs form a consensus E-box (CACGTG) (see Supplementary Table S5) while canonical E-box sequence is also present in the CGI (CpG 12).
A factor that binds to the nonmethylated CGI E-box oligonucleotide with 3.5- to 20-fold higher affinity compared to the E-box methylated oligonucleotide in both rat fetal brain (RFB, a rich source of TFs) and human dlPFC was identified using electromobility shift assay (EMSA) (Figs 3B,C; see Supplementary Table S6). The binding factor targeted E-box, as demonstrated with another canonical E-box (CAGGTG) oligonucleotide and its mutant. Antibodies against USF2, but not those against USF1, SNAI 1, c-MYC, or MAX, which are all E-box TFs, depleted and supershifted the complex formed by CGI E-box oligonucleotide (Figs 3D and E). USFs are basic helix-loop-helix E-box TFs that regulate activity-dependent transcription in neurons (Chen et al. 2003), recruit histone modification complexes and maintain a chromatin barrier (West et al. 2004; Huang et al. 2007; Gallagher et al. 2009). USF2 but not ΔB-USF2, a dominant negative mutant lacking DNA-binding domain (van Deursen et al. 2009) both ectopically expressed in SK-N-MC cells, produced the complex with the CGI E-box probe characterized by the same mobility and binding characteristics in EMSA as those formed by human dlPFC and RFB extracts (Fig. 3F). Methylation or hydroxymethylation impaired the ability of CGI E-box oligonucleotide to form the complex (Fig. 3F).
USF2 Activates Transcription from PDYN Promoter in Cellular Models
We next investigated whether USF2 activates transcription from PDYN promoter in reporter gene assay. The reporter expression was driven by the 1,3 kb PDYN promoter segment including 56 bp of exon 1 (FL-wt, full-length wt); FL with mutated E-box (FL-mut); a CGI (CpG 9–16) (CGI-wt); or CGI with mutated E-box (CGI-mut) (Fig. 3G). Expression was compared between SK-N-MC cells which were transfected with USF2– or ΔB-USF2-expressing plasmids or mock transfected. USF2 strongly, 14-fold and significantly (P = 0.0009) activated transcription from FL-wt construct, while E-box mutation reduced, 2.3-fold the induction (P = 0.018). Consistently, transcription from CGI-wt construct was 21-fold activated by USF2 (P = 0.0006) while E-box mutation 2.3-fold diminished this effect (P = 0.0103). ΔB-USF2 failed to activate transcription.
Effects of CGI methylation were studied using a construct with CGI (CpGs 9–16) inserted into pCpGL-basic, a CpG-free reporter vector to exclude influence of vector backbone DNA methylation. USF2 strongly activated, 40-fold transcription from nonmethylated construct (P = 0.0002) while CGI methylation virtually completely abolished this activation (P = 0.00023) (Fig. 3H).
Ectopic expression of USF2 increased transcription from the endogenous PDYN gene in SK-N-MC cells 1.9-fold relative to mock (P = 0.0015), and 2.6-fold relative to ΔB-USF2 (P = 0.0005) (Fig. 3I). Altogether, these results strongly suggest that USF2 is an activator of PDYN transcription that acts through the binding to PDYN CGI E-box, which is abrogated by CpG methylation/hydroxymethylation.
USF2 Occupies PDYN Promoter at the Hypomethylated E-box in Human dlPFC
USF2-PDYN promoter interactions in dlPFC were investigated by quantitative chromatin immunoprecipitation (ChIP-qPCR) assay adopted for analysis of human brain (see Supplementary Fig. S5). USF2 was found to be bound to DNA across the PDYN promoter at 3.9-fold higher average levels compared to those of IgG in dlPFC (P = 0.0001) (Fig. 4A). The CGI E-box CpG methylation analyzed by quantitative real-time PCR (qAMP) assay was significantly lower in DNA immunoprecipitated with anti-USF2 antibodies than those of input DNA (P = 0.002) (Fig. 4B). DNA methylation is generally coupled with histone acetylation (Cedar and Bergman 2009). Methylation of the E-box CpG in DNA fragments immunoprecipitated using anti-acetylated histone 3 (H3K9/K14 acetylation; the gene-activating mark) antibody was significantly (P = 0.0099) lower compared to that in the input chromatin fraction (Fig. 4C). These findings suggest that USF2 activates PDYN transcription in dlPFC through binding to nonmethylated promoter CGI that is associated with acetylated histones.
DNA methylation landscape may be shaped by nucleosome positioning and, conversely, may determine nucleosome occupancy (Allis and Jenuwein 2016). The length of the promoter CGI (247 bp) suggests that it may form a single nucleosome with the linker DNA. Analysis of nucleosome occupancy across the PDYN promoter in dlPFC by examination of promoter segments in mononucleosomal DNA fraction demonstrated that the PDYN CGI may be occupied by a well–positioned nucleosome (Fig. 4E). Two nucleosomes located upstream and two downstream of the CGI, including +1 nucleosome in exon 1, could be also predicted. This nucleosome positioning pattern was identified for total tissue chromatin and thereby may be a characteristic of the repressive chromatin state.
PDYN Expression is Associated with USF2 in Human dlPFC
Significant positive main effect of USF2 mRNA on PDYN mRNA in dlPFC was observed in two cohorts of human subjects (Figs 5A and B). The effect of USF2 protein on PDYN mRNA was also evident (Fig. 5C and see Supplementary Fig. S6A). These results support the notion on regulation of PDYN transcription by USF2 in dlPFC.
PDYN and USF2 Proteins are Co-Localized in Human dlPFC
USFs are ubiquitously expressed in animal tissues including brain (Sirito et al. 1998), and in inter-, pyramidal and trigeminal ganglion neurons, and in oligodendrocytes and astrocytes within the CNS (Zeisel et al. 2015). Analysis of USF2 transcripts by ddPCR in cell nuclei isolated by FANS identified similar USF2 mRNA levels in neuronal and non-neuronal cells in human dlPFC (see Supplementary Fig. S6B).
To examine whether this ubiquitous TF may direct neuron-specific PDYN expression, we analyzed localization patterns of USF2 and PDYN proteins in dlPFC. Cells immunopositive for PDYN and USF2 were located in most cortical layers showing uniform distribution across dlPFC (Fig. 5D). Immunoreactive PDYN was located in cytoplasm and axons (Figs 5E, J, and K) while USF2 predominantly in the cell nucleus (Figs 5F, J, and K). Counterstaining demonstrated that PDYN and USF2 immunopositive cells represented a fraction of cell population in the cortex (Figs 5F and H). Counting of single and double labeled cells demonstrated that PDYN-positive neurons were virtually all USF2-positive, and most USF2-immunoreactive cells were PDYN-positive (Figs 5D and I–K; see Supplementary Table S7). The co-localization pattern was evident in all cortical layers for neurons of different morphologies including pyramidal neurons. Together with molecular data (see Supplementary Fig. S6B), these results suggest that only neurons producing USF2 at high levels express PDYN in dlPFC.
Discussion
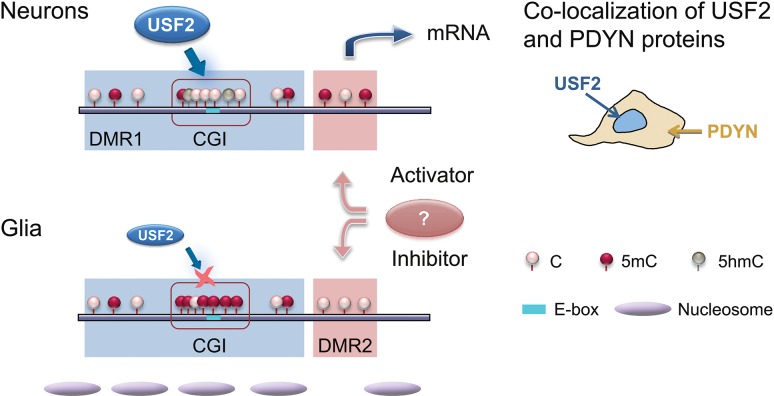
Our findings demonstrate that PDYN expression in neurons of human dlPFC is associated with differential methylation of its promoter. Two short adjacent PDYN promoter DMRs with methylation patterns, which are opposite (1) between the DMRs in each neurons and glia, and (2) in each DMR between neurons and glia, are located in the PDYN locus framed by strong CTCF binding sites. CTCF creates boundaries between topologically associating chromatin domains and forms chromatin loops binding distant DNA segments (Ong and Corces 2014). The presence of CTCF binding sites suggests that CTCF bridges flanks of the PDYN locus together forming a chromatin loop that creates a structural context for autonomic regulation of PDYN. USF2, a methylation-sensitive TF may target hypomethylated CGI in the DMR1 and activate PDYN transcription. Conversely, CGI hypermethylation in non-neuronal cells may hinder the USF2-mediated activation (Fig. 6). On average, neurons and non-neuronal cells express USF2 at similar levels. However, cell-to-cell differences in USF2 protein are substantial as demonstrated by immunohistochemistry. Only USF2-expressing cells synthesize PDYN. Thus, both the CGI hypomethylation and high USF2 expression are the conditions for cell type-specific PDYN transcription (Fig. 6).
Figure 6.
Model for integration of epigenetic and transcriptional mechanisms of cell type-specific PDYN expression in the human dlPFC. USF2, the ubiquitous TF expressed at high levels in subpopulation of neurons, activates PDYN transcription through binding to nonmethylated E-box in the short, nucleosome size human promoter-specific CGI in PDYN, which is hypomethylated in neurons. This results in co-localization of USF2 and PDYN proteins. In non-neuronal cells the CGI is hypermethylated that prevents activation of PDYN transcription by USF2, which is also present in these cells albeit at low levels. Methylation levels of DMR2 are opposite to those of the DMR1/CGI both in neurons and glia. This reciprocal pattern may contribute to differential PDYN transcription in neurons and non-neuronal cells; DMR2 may be targeted by a methylation-dependent transcriptional activator in neurons and/or methylation-sensitive transcriptional repressor in non-neuronal cells. Segregation of activating and restricting mechanisms could ensure contrasting PDYN expression in neurons and glia.
The current epigenome-wide view is that DMRs mostly overlap with CGI shores but not with CGIs (Doi et al. 2009; Ciernia and LaSalle 2016; Yao et al. 2016). The short PDYN promoter CGI may be an exception; it is differentially methylated between neurons and glia, and enriched in 5hmC suggesting high turnover rate of cytosine methylation, a mark of active regulatory regions. In neurons, methylation is highly coordinated within the DMR1/CGI but not between this domain and other PDYN promoter areas. In this feature, the PDYN CGI is similar to contiguous methylation clusters in human blood DNA and characterized by high between-CpG methylation correlations (Liu et al. 2014). Differences in the CGI methylation patterns may reflect differences in chromatin structure between neurons and glia. The CGI chromatin domain may form a nucleosome in glia and other cell types that may be a characteristic of the repressive chromatin state. This notion is supported by our analysis of positioning of nucleosome that occupies the PDYN CGI in glia. Altogether, our observations suggest that the PDYN CGI functions as a modular promoter element which methylation/hydroxymethylation is locally regulated, and which epigenetic DNA modifications and interactions with sequence-specific TFs may delineate patterns of PDYN expression.
In contrast to DMR1, DMR2 is hypermethylated in neurons. This opposite pattern along with negative correlations in methylation between 2 DMRs suggest autonomic for each DMR but coordinated between them regulation of methylation, and their complementary role in control of PDYN transcription (Fig. 6). Hypothetically, in neurons hypermethylated DMR2 may be targeted by a methylation-dependent DNA-binding factor (e.g., MeCP2) capable to activate gene transcription; while in non-neuronal cells PDYN may be repressed through binding of methylation-sensitive repressor to nonmethylated DMR2 (e.g., DREAM (Naranjo and Mellstrom 2012)). Thus, there may be one more mechanism for tight control of cell type-specific PDYN transcription in human dlPFC.
USFs are regulated by phosphorylation and mediate Ca2+- responsive transcription in neurons (Chen et al. 2003; Spohrer et al. 2016). These TFs are indispensable for normal embryonic development, and also play an essential role in adult mice exerting pleiotropic effects on brain function, fertility, and lifespan. Despite of ubiquitous expression and pleiotropic effects USFs may regulate cell lineage-specific transcription through control of TFs required for cell commitment and differentiation (Deng et al. 2013; Spohrer et al. 2016). In human brain, cellular segregation of USF2, which activates transcription, and the hypermethylated CGI, which restricts transcription, may be the mechanism that ensures the contrasting PDYN transcription in neurons and glia, and thereby determines PDYN cellular phenotype (Fig. 6). It is important to identify primary mechanism(s) that control(s) the elevated USF2 expression and PDYN CGI methylation in subpopulation of neurons versus other cells.
Compared to many other neuronal genes transcriptional regulation of human PDYN gene has not yet been well disentangled; and TFs and mechanisms of cell type- and brain area-specific and neuronal activity-dependent transcription have not been identified. The human PDYN promoter region is a unique human cis-regulatory trait that consistent with hypothesis on the recent positive selection of opioid cis-regulation in humans (Rockman et al. 2005). Early studies demonstrated the presence of promoter activity in 1 kb fragment of human PDYN promoter (Telkov et al. 1998) and identified a minimal inducible promoter fragment located −150 to +150 bp around the TSS (Carrion et al. 1998). Within this region a downstream response element functions as a transcriptional silencer which is targeted by the Ca2+-binding transcriptional regulator DREAM (Carrion et al. 1999; Ledo et al. 2000; Cheng et al. 2002; Campos et al. 2003). Several other TFs including Yin-Yang1 and NF-κB were implicated in PDYN regulation (Bakalkin et al. 1994, 1995, 1997). Target DNA elements for these TFs are located in the coding part of exon 4 where they overlap with the enkephalin-encoding sequences implying that short, repetitive and conservative neuropeptide-encoding sequences may function as target elements for TFs. Consistently, exon 4 PDYN fragment comprising these sequences demonstrates ability to activate a reporter gene in cellular experiments (Nikoshkov et al. 2005).
Structural genetic variants including variable number tandem repeat (VNTR) or SNPs may affect phenotype via changes in gene expression, possibly via creation or disruption of binding sites for TFs. The human PDYN gene harbors 1–5 copies of 68 bp VNTR located upstream of the TSS. The PDYN VNTR contains AP-1 (TRE)-like-binding site and weakly influences the inducibility of the gene (Zimprich et al. 2000). TPA stimulation elevated the expression of reporter gene from the promoter with 3 or 4 repeats compared to 1 or 2 repeats in cells (Rouault et al. 2011). In human striatum, a number of inducible repeats correlated with PDYN mRNA (Nikoshkov et al. 2008). Effects on transcription were assayed for 19 naturally occurring haplotypes of the PDYN 3 kb cis-regulatory region (Babbitt et al. 2010). The impact of these variants differed between cell types and across brain region with at least 6 different variants affected transcript abundance in vitro. In the human cortical areas, promoter SNP rs1997794 was strongly associated with expression level (Babbitt et al. 2010), while VNTR polymorphism did not affect PDYN expression (Cirulli and Goldstein 2007). Consistently, in the human hippocampus PDYN expression may also depend on the rs1997794 genotype (Taqi et al. 2011). The T, low risk allele of this SNP resides within noncanonical AP-1-binding element that may be targeted by JUND and FOSB proteins, the dominant AP-1 constituents in the human brain. The T to C transition abrogated AP-1 binding (Taqi et al. 2011). Thus, the impact of genetic variations on PDYN expression is complex and context dependent, partially due to the cis genotype-cell interactions, and epistatic interactions between nearby variants. Interestingly, rs1997794 resides in the PDYN promoter between the two DMRs that is strategical position for coordination of transcriptional events regulated by TFs targeting DMR1 that is USF2 and DMR2 that is DREAM.
Little is known about DNA methylation and its effects on PDYN gene transcription. DNA methylation of 2 short CpG islands located in the promoter and coding part of exon 4 of PDYN were analyzed in human brain tissues and peripheral blood mononuclear cells (PBMCs) (Yuferov et al. 2011). Methylation of the promoter CGI differed substantially between brain tissues and PBMC, and between caudate and anterior cingulate cortex. Thus, PDYN promoter DNA methylation profile was concluded to be typical for primary responsive genes with regulatory elements for both basal and tissue-specific transcription (Yuferov et al. 2011).
Rodent studies demonstrated that a cell lineage-specific Pdyn expression is controlled by Ptf1a, Pax2, Neurod1/2/6, and Bhlhb5 TFs in the dorsal spinal cord and Islet-1 in the striatum (Brohl et al. 2008; Huang et al. 2008; Wildner et al. 2013; Kardon et al. 2014; Lu et al. 2014). These TFs and USF2 are E-box-binding proteins (Ptf1a, Neurod1/2/6, and USF2), modulate E-box-dependent transcription, or are regulated by E-box TFs. Thus, E-box-mediated regulation may be a prerequisite for formation of cellular PDYN phenotype in rodent and human CNS.
As the limitation the absence of in vivo evidence on a role of USF2 in PDYN transcription that may be obtained in animal models should be mentioned. It is necessary to emphasize that PDYN promoter CGI is a unique human trait that is not a characteristic of rodent Pdyn promoter. Animal study would require a construction of humanized mice lines with a transgene in which human PDYN CGI would drive neuronal expression of a reporter gene. Three conditions should be met: (1) low CGI methylation and (2) the presence of high USF2 levels in expressing neurons. Furthermore, (3) effects of other E-box TFs (c-Myc, Max, Neuro D) that are widely expressed across the CNS should be excluded. Future studies aimed to generate a transgene, which fulfills these 3 conditions, are warranted.
Neuropeptides are special in their expression profiles restricted to specific neuronal subpopulations. Epigenetic and transcriptional mechanisms that define spatial, temporal, and adaptive neuropeptide expression patterns have not yet been identified. Our study is apparently the first that uncovers these mechanisms for human brain. Our findings suggest that the short, nucleosome size human gene-specific PDYN promoter CGI may regulate cell type-specific PDYN transcription as a chromatin element that integrates cellular signals through changes in methylation/demethylation and TF binding (Fig. 6). Interestingly, short CGIs in contrast to long islands mostly reside in intragenic regions and exhibit tissue-specific DNA methylation patterns that points to their specific functions (Zeng et al. 2014). Our findings emphasize the complexity of PDYN regulation that determines its cell type-specific expression in human brain through the dual epigenetic and transcriptional mechanism. Given the important role of dynorphins in addictive and psychiatric disorders, our findings provide an insight into fundamental molecular mechanisms that may be impaired in pathological human brain.
Supplementary Material
Notes
The authors thank Drs Q. Zhou, H. Geraschenko, A. Iatsyshyna, and W. Sun for assistance; D. Tan for sample processing and T. H. Chung for bioinformatics analysis for Mirror-BS seq, Dr M. Andersson for discussion; Prof. H. Druid and Dr K. Alkass for the help with FANS; Dr M. Rehli for luciferase reporter vector, Dr D. van Deursen for USF vectors, Prof I. Virtanen for anti-Snai 1 antibodies, Drs T. Axelsson and A. Haukkala for assistance with 450K assay. Conflict of interest: X.S. is an employee at Zymo Research.
Funding
The Swedish Science Research Council (K2014-62X-12190-19-5), Swedish Council for Working Life and Social Research (2009-1709, 259-2012-23) and Swedish Institute Visby to G.B., and National Institute of Health P30 GM103328 (G.R.). The content is solely the responsibility of the authors and does not necessarily represent the official National Institute of Health views. New South Wales BTRC was supported by The University of Sydney and US Government under the National Institutes of Health (NIAAA012725-15) to J.K.
References
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet. 17:487–500. [DOI] [PubMed] [Google Scholar]
- Babbitt CC, Silverman JS, Haygood R, Reininga JM, Rockman MV, Wray GA. 2010. Multiple functional variants in cis modulate PDYN expression. Mol Biol Evol. 27:465–479. [DOI] [PubMed] [Google Scholar]
- Bakalkin G, Telkov M, Yakovleva T, Terenius L. 1995. [Leu5]enkephalin-encoding sequences are targets for a specific DNA-binding factor. Proc Natl Acad Sci U S A. 92:9024–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalkin G, Watanabe H, Jezierska J, Depoorter C, Verschuuren-Bemelmans C, Bazov I, Artemenko KA, Yakovleva T, Dooijes D, Van de Warrenburg BPC, et al. 2010. Prodynorphin mutations cause the neurodegenerative disorder Spinocerebellar Ataxia Type 23 (vol 87, pg 593, 2010). Am J Human Genet. 87:736–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalkin G, Yakovleva T, Terenius L. 1994. Prodynorphin gene expression relates to NF-kappa B factors. Brain Res Mol Brain Res. 24:301–312. [DOI] [PubMed] [Google Scholar]
- Bakalkin G, Yakovleva T, Terenius L. 1997. The Leu-enkephalin-encoding sequence DNA-binding factor (LEF) is the transcription factor YY1. Biochem Biophys Res Commun. 231:135–139. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. 2012. Beyond the connectome: how neuromodulators shape neural circuits. BioEssays. 34:458–465. [DOI] [PubMed] [Google Scholar]
- Brohl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Muller T, Birchmeier C. 2008. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev Biol. 322:381–393. [DOI] [PubMed] [Google Scholar]
- Butelman ER, Yuferov V, Kreek MJ. 2012. kappa-opioid receptor/dynorphin system: genetic and pharmacotherapeutic implications for addiction. Trends Neurosci. 35:587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos D, Jimenez-Diaz L, Carrion AM. 2003. Ca(2+)-dependent prodynorphin transcriptional derepression in neuroblastoma cells is exerted through DREAM protein activity in a kinase-independent manner. Mol Cell Neurosci. 22:135–145. [DOI] [PubMed] [Google Scholar]
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. 1999. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 398:80–84. [DOI] [PubMed] [Google Scholar]
- Carrion AM, Mellstrom B, Naranjo JR. 1998. Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol Cell Biol. 18:6921–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 10:295–304. [DOI] [PubMed] [Google Scholar]
- Chavkin C. 2013. Dynorphin-still an extraordinarily potent opioid peptide. Mol Pharmacol. 83:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. 2003. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 23:2572–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-A, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, et al. 2002. DREAM is a critical transcriptional repressor for pain modulation. Cell. 108:31–43. [DOI] [PubMed] [Google Scholar]
- Ciernia AV, LaSalle J. 2016. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat Rev Neurosci. 17:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. 2007. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum Mol Genet. 16:1931–1939. [DOI] [PubMed] [Google Scholar]
- Deng C, Li Y, Liang S, Cui K, Salz T, Yang H, Tang Z, Gallagher PG, Qiu Y, Roeder R, et al. 2013. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet. 9:e1003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 41:1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraway JJ. 2002. Practical regression and ANOVA using R. https://cran.r-project.org/doc/contrib/Faraway-PRA.pdf. [Google Scholar]
- Feldmann A, Ivanek R, Murr R, Gaidatzis D, Burger L, Schubeler D. 2013. Transcription factor occupancy can mediate active turnover of DNA methylation at regulatory regions. PLoS Genet. 9:e1003994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD. 2014. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PG, Nilson DG, Steiner LA, Maksimova YD, Lin JY, Bodine DM. 2009. An insulator with barrier-element activity promotes alpha-spectrin gene expression in erythroid cells. Blood. 113:1547–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. 2013. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin S-G, Jiang Y, Pfeifer GP. 2013. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Reports. 3:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. 2013. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 10:1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L. 2008. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 322:394–405. [DOI] [PubMed] [Google Scholar]
- Huang S, Li X, Yusufzai TM, Qiu Y, Felsenfeld G. 2007. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol Cell Biol. 27:7991–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA. 2012. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 41:200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, Garrick T, Sheedy D, Hurd Y, Bakalkin G, Ekstrom TJ. 2007. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 1132:20–28. [DOI] [PubMed] [Google Scholar]
- Kardon AP, Polgar E, Hachisuka J, Snyder LM, Cameron D, Savage S, Cai X, Karnup S, Fan CR, Hemenway GM, et al. 2014. Dynorphin acts as a neuromodulator to inhibit itch in the dorsal horn of the spinal cord. Neuron. 82:573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononenko O, Bazov I, Watanabe H, Gerashchenko G, Dyachok O, Verbeek DS, Alkass K, Druid H, Andersson M, Mulder J, et al. 2016. Opioid precursor protein isoform is targeted to the cell nuclei in the human brain. Biochim Biophys Acta. 1861:246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. 2016. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry. 3:760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledo F, Carrion AM, Link WA, Mellstrom B, Naranjo JR. 2000. DREAM-alphaCREM interaction via leucine-charged domains derepresses downstream regulatory element-dependent transcription. Mol Cell Biol. 20:9120–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinwand SG, Chalasani SH. 2014. From genes to circuits and behaviors: neuropeptides expand the coding potential of the nervous system. Worm. 3:e27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science. 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Aryee MJ, Ekstrom TJ, Padyukov L, Klareskog L, Vandiver A, Moore AZ, Tanaka T, Ferrucci L, et al. 2014. GeMes, clusters of DNA methylation under genetic control, can inform genetic and epigenetic analysis of disease. Am J Hum Genet. 94:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KM, Evans SM, Hirano S, Liu FC. 2014. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc Natl Acad Sci U S A. 111:E168–E177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. 2012. Neuromodulation of neuronal circuits: back to the future. Neuron. 76:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott GP, Do D, Litterst CM, Maar D, Hindson CM, Steenblock ER, Legler TC, Jouvenot Y, Marrs SH, Bemis A, et al. 2013. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 85:11619–11627. [DOI] [PubMed] [Google Scholar]
- Naranjo JR, Mellstrom B. 2012. Ca2+-dependent transcriptional control of Ca2+ homeostasis. J Biol Chem. 287:31674–31680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoshkov A, Drakenberg K, Wang X, Horvath MC, Keller E, Hurd YL. 2008. Opioid neuropeptide genotypes in relation to heroin abuse: dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc Natl Acad Sci U S A. 105:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoshkov A, Hurd YL, Yakovleva T, Bazov I, Marinova Z, Cebers G, Pasikova N, Gharibyan A, Terenius L, Bakalkin G. 2005. Prodynorphin transcripts and proteins differentially expressed and regulated in the adult human brain. FASEB J. 19:1543–1545. [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. 2009. CTCF: master weaver of the genome. Cell. 137:1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Hahn MW, Soranzo N, Zimprich F, Goldstein DB, Wray GA. 2005. Ancient and recent positive selection transformed opioid cis-regulation in humans. PLoS Biol. 3:e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault M, Nielsen DA, Ho A, Kreek MJ, Yuferov V. 2011. Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity. Addict Biol. 16:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer C. 2009. 30 years of dynorphins—new insights on their functions in neuropsychiatric diseases. Pharmacol Ther. 123:353–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M. 1998. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci USA. 95:3758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets CJLM, Jezierska J, Watanabe H, Duarri A, Fokkens MR, Meijer M, Zhou Q, Yakovleva T, Boddeke E, den Dunnen W, et al. 2015. Elevated mutant dynorphin A causes Purkinje cell loss and motor dysfunction in spinocerebellar ataxia type 23. Brain. 138:2537–2552. [DOI] [PubMed] [Google Scholar]
- Spohrer S, Dimova EY, Kietzmann T, Montenarh M, Gotz C. 2016. The nuclear fraction of protein kinase CK2 binds to the upstream stimulatory factors (USFs) in the absence of DNA. Cell Signal. 28:23–31. [DOI] [PubMed] [Google Scholar]
- Taqi MM, Bazov I, Watanabe H, Nyberg F, Yakovleva T, Bakalkin G. 2011. Prodynorphin promoter SNP associated with alcohol dependence forms noncanonical AP-1 binding site that may influence gene expression in human brain. Brain Res. 1385:18–25. [DOI] [PubMed] [Google Scholar]
- Tejeda HA, Shippenberg TS, Henriksson R. 2012. The dynorphin/kappa-opioid receptor system and its role in psychiatric disorders. Cell Mol Life Sci. 69:857–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG. 2013. “Seq-ing” insights into the epigenetics of neuronal gene regulation. Neuron. 77:606–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telkov M, Geijer T, Terenius L. 1998. Human prodynorphin gene generates several tissue-specific transcripts. Brain Res. 804:284–295. [DOI] [PubMed] [Google Scholar]
- Van’t Veer A, Carlezon WA Jr. 2013. Role of kappa-opioid receptors in stress and anxiety-related behavior. Psychopharmacology (Berl). 229:435–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen D, van Leeuwen M, Vaulont S, Jansen H, Verhoeven AJ. 2009. Upstream Stimulatory Factors 1 and 2 activate the human hepatic lipase promoter via E-box dependent and independent mechanisms. Biochim Biophys Acta. 1791:229–237. [DOI] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. 2013. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, et al. 2014. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 15:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 16:453–463. [DOI] [PubMed] [Google Scholar]
- Wildner H, Das Gupta R, Brohl D, Heppenstall PA, Zeilhofer HU, Birchmeier C. 2013. Genome-wide expression analysis of Ptf1a- and Ascl1-deficient mice reveals new markers for distinct dorsal horn interneuron populations contributing to nociceptive reflex plasticity. J Neurosci. 33:7299–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Christian KM, He C, Jin P, Ming G-L, Song H. 2016. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 17:537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuferov V, Nielsen DA, Levran O, Randesi M, Hamon S, Ho A, Morgello S, Kreek MJ. 2011. Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs. Pharmacogenet Genom. 21:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- Zeng J, Nagrajan HK, Yi SV. 2014. Fundamental diversity of human CpG islands at multiple biological levels. Epigenetics. 9:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Kraus J, Woltje M, Mayer P, Rauch E, Hollt V. 2000. An allelic variation in the human prodynorphin gene promoter alters stimulus-induced expression. J Neurochem. 74:472–477. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, Buckley N, et al. 2007. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington’s disease. J Neurosci. 27:6972–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.