Abstract
Purpose
Cone rod-dystrophies (CRDs) are pigmentary retinopathies mainly involving cones. CRDs typically present with decreased visual acuity and loss of sensitivity in the central visual field, reflecting the primary dysfunction of cones associated with night blindness and concentric visual field loss due to rod dysfunction. We describe the phenotype, natural history, and molecular analysis results of an early onset form of CRD.
Methods
An otherwise healthy 25-year-old man from Sardinia, Italy, initially presented with subacute visual loss and central scotoma in both eyes. He underwent a complete ophthalmic examination, electrophysiologic testing, and genetic counseling. We first applied a candidate gene approach on ABCA4 to detect mutations; then, we performed exome sequencing (WES) on all family members to identify causative mutations.
Results
The ophthalmic examination was unremarkable except the fundus examination, which revealed a well-circumscribed ring-shaped area of choroidal and RPE atrophy surrounding the fovea in the left eye and small white patches of atrophy around the fovea in the right eye. The ocular features and medical history were consistent with a diagnosis of CRD. Twenty years later, he showed a marked impairment in visual function, secondary to severe atrophic maculopathy associated with sparse pigmentary deposits. Molecular analysis identified two novel frameshift mutations in C2orf71: c.3039dupC: p.Ser1014Leufs*93 and c.1804_1805delAG:p. His603Argfs*77.
Conclusions
The mutations in C2orf71 reported in this study comprise protein truncation mutations, which are likely to be involved in the pathogenesis of this severe form of early onset CRD.
Introduction
With a worldwide prevalence of 1 out of 4,000, typical retinitis pigmentosa, also called rod-cone dystrophies (RCDs), is the most common form of hereditary retinal degeneration. In RCDs, although rod and cone function is affected, the early manifestations are usually night blindness and progressive visual field constriction, which reflect the primary involvement of rods [1,2]. Conversely, cone-rod dystrophies (CRDs), with an estimated prevalence of 1 in 40,000, belong to the group of pigmentary retinopathies. CRDs primarily present with decreased visual acuity and loss of sensitivity in the central visual field due to cone dysfunction. The clinical course of CRDs is generally more severe and rapid than that of RCDs, leading to earlier legal blindness and disability [2].
Traditionally, diagnosis of CRDs is based on clinical history, multimodal imaging evaluation, and electrophysiological testing. Although it is true that retinal degenerations can overlap toward end-stage disease, advanced CRDs usually differ from RCDs especially in the residual visual fields topography, as well as in the levels of residual cone versus rod function. Therefore, genetic analysis is always advisable to confirm the clinical diagnosis. The four major causative genes involved in the pathogenesis of CRDs include ATP-binding cassette, subfamily A, member 4 (ABCA4; GeneID: 24; OMIM 601691; which causes Stargardt disease (STGD) and 30% to 60% of autosomal recessive CRDs), cone-rod homeobox-containing gene (CRX; GeneID: 1406; OMIM 602225) and guanylate cyclase 2D, membrane (GUCY2D; GeneID: 3000; OMIM 600179; which are responsible for many reported cases of autosomal dominant CRDs), and retinitis pigmentosa GTPase regulator (RPGR; GeneID: 6103; OMIM 312610; which causes about two thirds of X-linked RP and an undetermined percentage of X-linked CRDs) [1,3].
Recently, mutations in C2orf71, first described in 2010 in six patients with non-syndromic, autosomal recessive retinitis pigmentosa and two patients with early onset retinal dystrophy with nystagmus, have also been associated with other phenotypes, such as CRDs [4,5]. The current total number of C2orf71 mutations reported in the Human Gene Mutation Database (HGMD) is 35 (13 missense, 11 nonsense, nine small deletions, one insertion, and one insertion/deletion; accession date: 13 June 2019) [5].
In an individual with suspected CRD, it is important to investigate whether other retinal disease genes, apart from the five above, could lead to suspected STGD. In this report, we describe the clinical findings, natural history, and molecular analysis results of an early onset form of CRD.
Methods
Patients
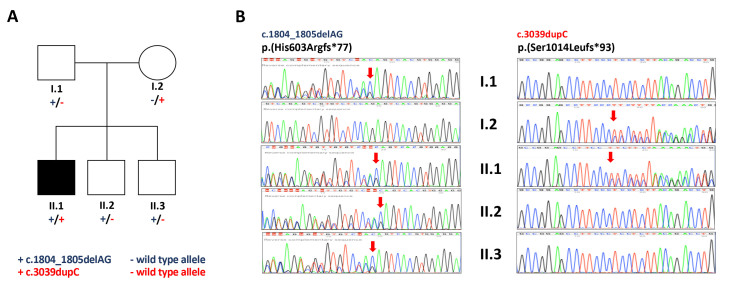
The analyzed family from Sardinia, Italy, included five members: Only one of them, a 45-year-old man, was affected by CRD with an unidentified molecular genetic diagnosis. The pedigree structure is shown in Figure 1. Taking into account the information derived from the pedigree, the disease was consistent with autosomal recessive inheritance or a single de novo mutation with dominant inheritance. Venous blood samples were collected from the affected patient, his parents, and unaffected siblings in EDTA tubes and stored at −20 °C, until DNA extraction. Genomic DNA extraction was carried out by salting out method according to Miller et al, 1988 [6].
Figure 1.
Genetic testing results. A: Pedigree of the family under study. All the members were recruited for whole exome analysis. Mutation status of C2orf71 is shown under the symbols for each subject (+/−, heterozygous carriers). B: Sanger sequencing confirmation: Chromatographs obtained with Sanger sequencing for all family members. The arrow indicates the frameshift start. Sanger sequencing that identified the mutation c.1804_1805delAG was performed using a reverse primer.
Approval from the local ethics committee/institutional review board was obtained, and the study was conducted in full accordance with the tenets of the Declaration of Helsinki and the ARVO statement on human subjects. Each participant received detailed information and provided written informed consent.
Clinical examination and molecular screening
The affected patient underwent a complete ophthalmic examination, including best-corrected visual acuity (BCVA) measured with Snellen charts, fundus autofluorescence (FAF), fluorescein angiography (FA; Heidelberg Retina Angiograph II; Heidelberg Engineering, Heidelberg, Germany), and spectral domain–optical coherence tomography (SD-OCT; Spectralis; Heidelberg Engineering), and electrophysiological testing. In most cases, CRDs are caused by mutations in ABCA4 inherited in an autosomal recessive model. Molecular analysis for the detection of point mutations in ABCA4 was performed for each of the 50 exons, using denaturing high-performance liquid chromatography (DHPLC) analysis and automatic sequencing of fragments with anomalous peaks according to Stenirri et al., 2004 [7], and automatic sequencing of fragments with anomalous peaks.
We performed exome sequencing (WES) on four family members (proband, father, mother, and healthy sibling). DNA libraries were prepared starting from 100 ng of genomic material according to the manufacturer’s instructions using the TruSeq™ Exome Enrichment Kit (Illumina Inc., San Diego, CA) that selects 62 Mb of genomic content, including exons, untranslated regions (UTRs), and miRNA. Each sample was sequenced by using the Illumina HiSeq2500 platform with paired-end reads of 101 bp, according to the manufacturer’s instructions.
Alignment, variant calling, and variant prioritization phase of the exome data were performed using an in-house pipeline based on Burrow-Wheeler Aligner (BWA), Genome Analysis ToolKit (GATK), Samtools, and Annovar [8-11]. We obtained about 30,000 variants per individual, which were filtered based on genotype quality, recessive pattern of inheritance, de novo events, gene feature, and an minimum allele frequency of less than 0.01 in reference databases (dbSNP, 1000 Genomes, ExAC, and gnomAD). SIFT, Polyphen2, and MutationTaster were used to predict the effect of amino-acid change on protein function).
A candidate gene approach was conducted by screening all the variants present in the genes causing retinal diseases in the RetNet database. Variants were evaluated for their phenotypic and biological function.
Segregation validation with Sanger sequencing
To verify the co-segregation of the C2orf71 variants, the forward and reverse primer sequences (GGA CAA AAC AGG ACC TCA GA and GAA GTT CGC CGC TTT GTG, GAT CAA GTT TGT CCC TGT GC and GCA CCC AGG GCA TAA AAT G, respectively for c.3039dupC and C.1804_1805delAG variants) used for PCR were designed using Primer3 software. One hundred nanogram of genomic DNA were amplified using the AmpliTaq Gold DNA Polymerase (Applied Biosystems Inc., Foster City, CA) and run at 56 ºC annealing temperature. The amplified PCR products were treated with ExoSAP-IT™ PCR Product Cleanup Reagent (ThermoFisher Scientific, Waltham, MA). Sequencing was run on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA) using the BigDye-Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sanger sequencing was performed using the DNA samples from the proband and all healthy available family members (Figure 1).
Results
Clinical findings
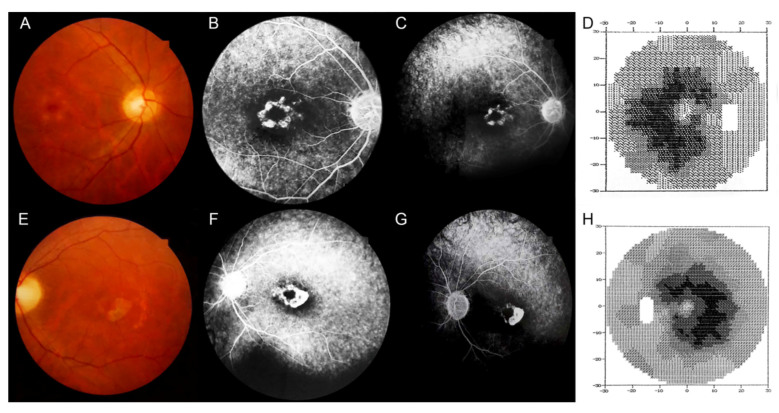
The patient was originally seen in 1998, when he was 25 years old. He initially complained of subacute central visual loss in both eyes. BCVA was 20/50 Snellen equivalent in his right eye (RE) and 20/80 Snellen equivalent in the left eye (LE). The ophthalmic examination was unremarkable except the fundus examination, which revealed a well-circumscribed ring-shaped area of choroidal and RPE atrophy surrounding the left fovea and small white patches of atrophy around the right fovea. In both eyes, a pale appearance of the optic disc was noted, but no pigment deposits were seen (Figure 2). A punctate retinal pigment epitheliopathy was observed bilaterally in the midperipheral retina, more evident on FA, which also revealed hyperfluorescent macular lesions suggestive of bull’s-eye maculopathy. No dark choroid was noted. In both eyes, visual field testing showed an absolute paracentral ring scotoma, surrounded by a relative annular scotoma extending 15° from the fovea bilaterally. Electroretinography (ERG) revealed early and predominant involvement of photopic over scotopic responses. In particular, rod and cone amplitudes were reduced, with delayed a- and b-wave single flash photopic response and implicit time shift at the 30 Hz flicker response. These results were consistent with a diagnosis of CRD.
Figure 2.
Multimodal imaging evaluation and visual fields, at baseline. A: Color fundus photography reveals small white patches of atrophy around the fovea in the right eye and a well-circumscribed ring-shaped area of choroidal and RPE atrophy surrounding the fovea in the left eye. A, E: In both eyes, there is a pale optic disc, but no pigment deposits are visible. A punctate retinal pigment epitheliopathy can be seen in the midperipheral retina bilaterally, more evident on fluorescein angiography, which also reveals hyperfluorescent macular lesions suggestive of bull’s-eye maculopathy (B, C, F, G). D, H: In both eyes, visual field testing showed an absolute paracentral ring scotoma, surrounded by a relative annular scotoma extending 15° from the fovea bilaterally.
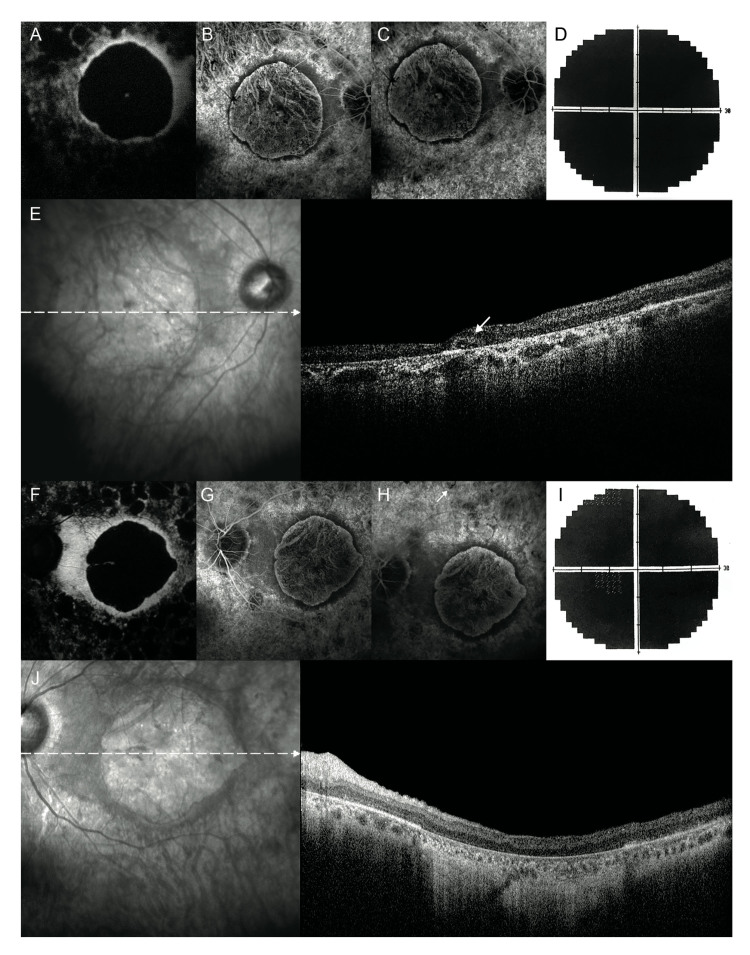
Twenty years later, in 2018, BCVA was bilateral light perception, with visual field extinction. FAF showed a central round area of decreased autofluorescence corresponding to the area of macular atrophy, surrounded by a ring of relatively increased autofluorescence. Furthermore, in the midperipheral retina, several roundish areas of reduced autofluorescence, suggestive of patchy atrophy, were noted. In both eyes, FA frames revealed severe macular atrophy surrounded by a ring of preserved RPE (Figure 3). Likewise, FA revealed the presence of sparse pigmentary deposits in the midperipheral retina of both eyes. SD-OCT showed severe bilateral retinal thinning with the disappearance of the external retinal layers. Additionally, SD-OCT scans of the RE revealed ovoid tubular structures with partially hyperreflective borders and hyperreflective material inside, suggestive of outer retina tubulations (ORTs), whereas in the LE, severe atrophy of the external retinal layers with backscattering was observed (Figure 3).
Figure 3.
Fundus autofluorescence, fluorescein angiography, and visual fields, 20 years later. A, F: In both eyes, fundus autofluorescence shows a central round area of decreased autofluorescence corresponding to the area of macular atrophy, surrounded by a ring of relatively increased autofluorescence. In the mid-peripheral retina, several roundish areas of reduced autofluorescence, suggestive of patchy atrophy, can be seen. B, C, G, H: In both eyes, fluorescein angiography frames reveal a severe macular atrophy surrounded by a ring of preserved retinal pigment epithelium. The mid-peripheral retina is extensively atrophic with some sparse pigmentary deposits (white arrow in H). D, I: Bilateral visual field extinction. E: In the right eye, simultaneous infrared and spectral domain optical coherence tomography show an ovoid tubular structure with a partial hyperreflective border and hyporreflective material inside, suggestive of an outer retina tabulation (white arrow) while (J) in the left eye reveal a severe retinal thinning secondary to the atrophy of the external retinal layers with backscattering.
Molecular findings
DHPLC molecular screening of the patient identified one heterozygous missense mutation in ABCA4 (NM_000350) in exon 30: c.4535C>G:p.P1512R, previously described in a heterozygous condition in a patient of Italian origin with STGD [7,12]. The other mutant allele remained undetected. The unidentified mutations may be located in parts of the gene that have not been screened, such as most of the intron sequences and the promoter region.
To improve the molecular diagnosis, we then performed exome sequencing on four family members. The average target coverage was about 80X, and in the target region, at least 10X sequence coverage was achieved for approximately 90% of the target bases for all samples. We confirmed the presence of the heterozygous mutation p.Pro1512ARG in ABCA4 in the proband and detected the same molecular condition in his healthy brother. We also identified five synonymous missenses in the coding region of ABCA4 and 10 intronic variants (see details in Appendix 1.) which have no effects on phenotype. After filtering of the variants using a candidate gene approach for the genes causing retinal diseases (see Appendix 2), and the quality assessment in the IGV browser, we identified two novel frameshift mutations in C2orf71 (NG_021427.1): c.3039dupC: p.Ser1014Leufs*93, maternally derived, and c.1804_1805delAG:p. His603Argfs*77, paternally derived. Sanger sequencing confirmed both mutations in the proband, the c.3039dupC variant in the mother, and the c.1804_1805delAG variant in the father and in the two healthy siblings (Figure 1). The parents, although they are carriers of mutations in C2orf71, do not have any of the abnormalities found in individuals with STGD.
Discussion
This case report describes the clinical presentation and long-term evolution of an early onset form of CRD in a patient with two novel mutations in C2orf71. Clinical features and medical history led us to suspect a recessive form of RP with a predominant involvement of cones. Our suspicion was confirmed by the ERG results, which were suggestive of a CRD.
Currently, more than 30 genes have been identified as causing non-syndromic RP with autosomal recessive inheritance (RetNet: Retinal Information Network). It is estimated that 50% of RP genes are still unknown [13,14]; however, mutations in ABCA4 represent the most common cause of autosomal recessive CRDs [15,16]. ABCA4 encodes a photoreceptor-specific ATP-binding cassette transporter [17] that is localized in cones and in the outer segment discs of rods [18-20].
This patient showed one heterozygous missense mutation in ABCA4-(c.4535C>G: p.P1512R). This mutation alone is not sufficient to cause a CRD: Subjects heterozygous for this mutation were found to have no clinical evidence of CRD or other eye diseases [12].
In an effort to identify the molecular cause of the retinal dystrophy in this patient, we performed WES, which detected two novel mutations in C2orf71. This gene consists of two exons encoding for 1,288 amino-acid residues forming a protein highly expressed in the retina and scarcely present in the other human tissues [4].
C2orf71 protein localization within the photoreceptor outer segments may explain their degeneration. Genes with the highest levels of retinal specificity have been reported to cause disorders of the retina in patients with CRD [5].
In patients with CRD with C2orf71 mutations, the disease commonly occurs in the fourth or fifth decade of life [21], presents with a ring scotoma around the fovea, and progresses to marked chorioretinal atrophy in the macular area. This condition is associated with pigmentary deposits in the peripheral retina during the late stages of the disease [21]. Interestingly, in this patient, the disease occurred earlier (i.e., in the third decade), presented with a bilateral paracentral scotoma without pigmentary deposits, and rapidly progressed to profound visual loss in both eyes.
The mutations in C2orf71 reported in this study comprise protein truncation mutations that played a causative role in the pathogenesis of this severe form of early onset CRD. Individuals heterozygous for some mutant alleles in ABCA4 are more susceptible to developing retinal degenerations [17,22,23]. Unfortunately, we do not have any evidence to say that the concomitant mutation in ABCA4 may have negatively affected the clinical presentation in this patient. Taking into account the high phenotypic variability of retinal dystrophies, many attempts have been made to characterize these disorders by using more precise diagnostic techniques, such as FAF, FA, and SD-OCT.
In the case presented here, at the last follow-up examination, the SD-OCT scans revealed the presence of round or ovoid hyporeflective spaces with hyperreflective borders located in the outer nuclear layer of the retina, suggestive of ORTs. ORTs were originally detected in patients with neovascular age-related macular degeneration (AMD) using SD-OCT, and since then, they have been described in a variety of advanced degenerative retinal disorders, including inflammatory diseases and retinal dystrophies [5,24-27].
Although the pathogenic mechanisms underlying the development of ORTs are far from clear, the microstructural loss of photoreceptor integrity and retinal layer atrophy with maintained laminar structure, even in the late stages of the disease, seem to point to a primary photoreceptor disorder. The development of ORTs could be the result of a long-lasting retinal degenerative process, originating from the rearrangement of photoreceptor and Müller cells in a reparative attempt [24,27]. Histological studies have shown that the presence of RPE and other cells within the ORTs’ lumen might provide trophic support to degenerative photoreceptor cells. ORTs have been suggested to promote the survival of degenerative photoreceptor cells, thus preserving visual function [28].
In summary, we described two novel frameshift mutations in C2orf71 in a patient with severe, early onset CRD. It is likely that these mutations may be somehow involved in the pathogenesis of this rare genetic retinal disorder. Further future studies are warranted to confirm these results.
Acknowledgments
The authors are sincerely grateful to the family described herein for their willing assistance and continuing cooperation in the study.
Appendix 1.
To access the data, click or select the words “Appendix 1.” List of all variants identified by exome sequencing in ABCA4 in the patient, his parents, and siblings. The chromosome and position of the variants are depicted according to the human genome assembly, GRCh37/hg19.
Appendix 2.
To access the data, click or select the words “Appendix 2.” Gene list of 262 causing retinal diseases present in RetNet database, used for candidate gene approach.
References
- 1.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokochi M, Li D, Horiguchi M, Kishi S. Inverse pattern of photoreceptor abnormalities in retinitis pigmentosa and cone-rod dystrophy. Doc Ophthalmol. 2012;125:211–8. doi: 10.1007/s10633-012-9348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–53. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 4.Collin RW, Safieh C, Littink KW, Shalev SA, Garzozi HJ, Rizel L, Abbasi AH, Cremers FP, den Hollander AI, Klevering BJ, Ben-Yosef T. Mutations in C2ORF71 Cause Autosomal-Recessive Retinitis Pigmentosa. Am J Hum Genet. 2010;86:783–8. doi: 10.1016/j.ajhg.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerth-Kahlert C, Tiwari A, Hanson JVM, Batmanabane V, Traboulsi E, Pennesi ME, Al-Qahtani AA, Lam BL, Heckenlively J, Zweifel SA, Vincent A, Fierz F, Barthelmes D, Branham K, Khan N, Bahr A, Baehr L, Magyar I, Koller S, Azzarello-Burri S, Niedrist D, Heon E, Berger W. C2orf71 Mutations as a Frequent Cause of Autosomal-Recessive Retinitis Pigmentosa: Clinical Analysis and Presentation of 8 Novel Mutations. Investig Opthalmology Vis Sci. 2017;58:3840. doi: 10.1167/iovs.17-21597. [DOI] [PubMed] [Google Scholar]
- 6.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenirri S, Fermo I, Battistella S, Galbiati S, Soriani N, Paroni R, Manitto MP, Martina E, Brancato R, Allikmets R, Ferrari M, Cremonesi L. Denaturing HPLC profiling of the ABCA4 gene for reliable detection of allelic variations. Clin Chem. 2004;50:1336–43. doi: 10.1373/clinchem.2004.033241. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli A, Ferrari M, Soriani N, Gessi A, Foglieni B, Martina E, Manitto MP, Brancato R, Dean M, Allikmets R, Cremonesi L. Mutational scanning of the ABCR gene with double-gradient denaturing-gradient gel electrophoresis (DG-DGGE) in Italian Stargardt disease patients. Hum Genet. 2001;109:326–38. doi: 10.1007/s004390100583. [DOI] [PubMed] [Google Scholar]
- 13.Maurizio BP, Pierluigi I, Stelios K, Stefano V, Marialucia C, Ilaria Z, Francesco B. Retro-mode imaging and fundus autofluorescence with scanning laser ophthalmoscope of retinal dystrophies. BMC Ophthalmol. 2012;12:8. doi: 10.1186/1471-2415-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manitto MP, Roosing S, Boon CJF, Souied EH, Bandello F, Querques G. Clinical Utility Gene Card for: autosomal recessive cone-rod dystrophy. Eur J Hum Genet. 2015;23:3–5. doi: 10.1038/ejhg.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacDonald IM, Tran M, Musarella MA. Ocular genetics: current understanding. Surv Ophthalmol. 2004;49:159–96. doi: 10.1016/j.survophthal.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet. 2000;67:960–6. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. Late-onset Stargardt disease is associated with missense mutations that map outside known functional regions of ABCR (ABCA4). Hum Genet. 2001;108:346–55. doi: 10.1007/s004390100493. [DOI] [PubMed] [Google Scholar]
- 18.Papermaster DS, Reilly P, Schneider BG. Cone lamellae and red and green rod outer segment disks contain a large intrinsic membrane protein on their margins: an ultrastructural immunocytochemical study of frog retinas. Vision Res. 1982;22:1417–28. doi: 10.1016/0042-6989(82)90204-8. [DOI] [PubMed] [Google Scholar]
- 19.Azarian SM, Travis GH. The photoreceptor rim protein is an ABC transporter encoded by the gene for recessive Stargardt’s disease (ABCR). FEBS Lett. 1997;409:247–52. doi: 10.1016/s0014-5793(97)00517-6. [DOI] [PubMed] [Google Scholar]
- 20.Mata NL, Weng J, Travis GH. Biosynthesis of a major lipofuscin fluorophore in mice and humans with ABCR-mediated retinal and macular degeneration. Proc Natl Acad Sci USA. 2000;97:7154–9. doi: 10.1073/pnas.130110497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, Hoyng CB, Roepman R, Klevering BJ. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157–86. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Lewis RA, Lupski JR. Macular degeneration: the emerging genetics. Hosp Pract. 1995;2000:41–50. [PubMed] [Google Scholar]
- 23.Allikmets R. Molecular genetics of age-related macular degeneration: current status. Eur J Ophthalmol. 1999;9:255–65. doi: 10.1177/112067219900900401. [DOI] [PubMed] [Google Scholar]
- 24.Zweifel SA, Engelbert M, Laud K, Margolis R, Spaide RF, Freund KB. Outer retinal tubulation: a novel optical coherence tomography finding. Arch Ophthalmol. 2009;127:1596–602. doi: 10.1001/archophthalmol.2009.326. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg NR, Greenberg JP, Laud K, Tsang S, Freund KB. Outer retinal tubulation in degenerative retinal disorders. Retina. 2013;33:1871–6. doi: 10.1097/IAE.0b013e318296b12f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heon E, Alabduljalil T, McGuigan DB, III, Cideciyan AV, Li S, Chen S, Jacobson SG. Visual Function and Central Retinal Structure in Choroideremia. Investig Opthalmology Vis Sci. 2016;57:377. doi: 10.1167/iovs.15-18421. [DOI] [PubMed] [Google Scholar]
- 27.Litts KM, Messinger JD, Dellatorre K, Yannuzzi LA, Freund KB, Curcio CA. Clinicopathological correlation of outer retinal tubulation in age-related macular degeneration. JAMA Ophthalmol. 2015;133:609–12. doi: 10.1001/jamaophthalmol.2015.126. [DOI] [PubMed] [Google Scholar]
- 28.Zweifel SA. Outer Retinal Tubulation. Arch Ophthalmol. 2009;127:1596. doi: 10.1001/archophthalmol.2009.326. [DOI] [PubMed] [Google Scholar]