Abstract
Background:
Numerous researches have demonstrated that miR-142-5p plays significant roles in several cancers, although the functional characteristic of miR-142-5p in breast cancer has not been determined. This study is designed to explore the biological significance of miR-142-5p in breast cancer clinical implication and mechanism of action.
Methods:
The differential expression patterns of miR-142-5p and Sorbin and SH3 domain-containing protein 1 and correlations between them and clinical significances were analyzed based on data from database. The expression levels of miR-142-5p in breast cancer cells were detected using quantitative real-time polymerase chain reaction. Cell counting kit-8, transwell, and wound healing assays were used to explore the potential functions of miR-142-5p in breast cancer cells. In addition, bioinformatics prediction analysis and luciferase reporter assay were utilized to predict and identify the potential target gene of miR-142-5p. A rescue experiment was conducted by transfecting miR-142-5p inhibitors and si-Sorbin and SH3 domain-containing protein 1 into cells to explore miR-142-5p/Sorbin and SH3 domain-containing protein 1 pairs on breast cancer cells behaviors.
Results:
The analysis results showed that miR-142-5p was highly expressed in patients with breast cancer, while Sorbin and SH3 domain-containing protein 1 presented a trend of low expression. The clinical significances analysis suggested that the overexpression of miR-142-5p is closely correlated with metastasis, while low expression of Sorbin and SH3 domain-containing protein 1 is correlated with clinicopathological characteristics and poor overall survival in patients with breast cancer. In vitro exploration, the expression of miR-142-5p was upregulated in breast cancer cells and inhibition of miR-142-5p expression significantly reduced the proliferation, invasion, and migration of breast cancer cells. Through rescue experiments, breast cancer cells proliferation, invasion, and migration reduction induced by silencing of miR-142-5p were reversed via knockdown Sorbin and SH3 domain-containing protein 1.
Conclusion:
Our findings insinuate that miR-142-5p functions as a positive regulator of promoting breast cancer cells biological behaviors and clinical metastasis, possibly regulated by targeting Sorbin and SH3 domain-containing protein 1, thus providing valuable information in the development of preventive or even therapeutic strategies for utilizing miR-142-5p as a promising target.
Keywords: miR-142-5p, invasion, migration, breast cancer, SORBS1
Introduction
Breast cancer is ranked as the most common cancer affecting women and the most frequent cause of cancer death worldwide.1 The etiology of breast cancer is not utterly clear, and the early stage of breast cancer does not have typical symptoms and signs, thus making the diagnosis and treatment of breast cancer difficult.2 At present, surgery is the main treatment for early-stage breast cancer.3 Meanwhile, the use of chemotherapy, radiotherapy, and hormone therapy significantly improved survival rate.4 In recent years, our understanding of metastatic breast cancer has made great progress; however, metastasis is still the main cause of breast cancer morbidity and mortality,5 and there is no satisfactory treatment for metastatic breast cancer. Therefore, defining metastasis-specific regulators makes it possible to classify, inform, and improve treatment options of breast cancer.6 Recently, many studies found that the discovery of molecular mechanisms has played a positive role in the treatment of breast cancer, although it has limitations. Meanwhile, published research indicated that the expression of aberrant microRNA (miRNA) is associated with many cancers, including breast cancer.7 Therefore, it is necessary to study the efficacy of miRNA in the treatment of breast cancer.
MicroRNA is a class of small noncoding RNA with a length of 21 to 25, which plays a role in gene regulation by inhibiting degradation and translation of target RNA.8 MicroRNA is thought to be involved in regulation of diverse biological processes, including cell differentiation, proliferation, and apoptosis.9 Numerous miRNAs, such as tumor suppressors or oncogenes, have been reported to play specific roles in cancer initiation, tumor growth, and metastasis.10 MicroRNAs are expected to be used in diagnosis, treatment, and prognosis of clinical cancer.11 In previous research, miR-142-5p is considered a key regulator of cell survival12 and can promote development of colorectal cancer and facilitate generation of aerobic glycolysis,13 accelerate cell growth and migration in renal cell carcinoma,14 regulate apoptosis in ovarian cancer cells,15 control tumor cell programmed death ligand 1 expression and enhance antitumor immunity,16 suppress tumorigenesis in non-small cell lung cancer,17 and induce cancer stem cell–like properties of cutaneous squamous cell carcinoma.18 Notably, downregulation of miR-142-5p promotes tumor metastasis in gastric cancer.19 Whereas the implication of miR-142-5p in the development and metastasis of breast cancer has not been fully elucidated and the mechanism remains unclear until recently. Therefore, we conducted a detailed study on the role of miR-142-5p in breast cancer progression.
In this study, we identified the expression of miR-142-5p in human breast cancer tissues and cells and its clinical significance; detected the function of miR-142-5p on breast cancer cells proliferation, migration, and invasion; and determined the correlations between them and clinical implications. Then, the direct interaction between miR-142-5p and Sorbin and SH3 domain-containing protein 1 (SORBS1) was determined. Also, we found that SORBS1 was downregulated and led to poor overall survival in patients with breast cancer. Rescue assay indicated that SORBS1 took part in the migration and invasion of breast cancer cells and reversed the promotion effect of miR-142-5p on breast cancer cells. All results in this study reveal that miR-142-5p regulated SORBS1 functions as a significant modulator in breast cancer progression, thus providing potential diagnostic and therapeutic targets for the treatment of breast cancer.
Materials and Methods
Data Acquisition and Analysis
The differential expression of miR-142-5p and clinical parameters was obtained from OncomiR (https://www.oncomir.org), the expression pattern of SORBS1 and clinical characteristics was got from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). The prognosis of SORBS1 in 869 patients with breast cancer was analyzed using Kaplan-Meier plotter (http://kmplot.com/analysis/) and resulted 434 with low SORBS1 expression and 435 with high SORBS1 expression. And in the survival curve analysis, low and high expression of SORBS1 in patients with breast cancer were defined according to the median from TCGA database: Those above the median were high expression and those under the median were low expression. A P < .05 was considered statistically significant.
Cell Culture
The Shanghai Cell Bank of the Chinese Academy of Medical Sciences (Shanghai, China) provided all the cell lines, including human breast cancer cell lines MCF7, MDA-MB-231, MDA-MB-451, MDA-MB-453, and HCC1937, and normal cell line HS578Bst. The cells were incubated with RPMI-1640 at 37°C, 5% CO2, 10% serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin.
Transfection
Cells were transfected with 20 nmol/L of miR-142-5p mimics, miR-142-5p inhibitors or negative control (NC) duplex (GenePharma, Shanghai, China), and the SORBS1 small interfering RNA (siRNA) and NC siRNA (Sangon, Shanghai, China) using Lipofectamine 2000 transfection kit (Invitrogen, Carlsbad, California), according to the manufacturer’s protocol. The siRNA sequences were as follows: si-SORBS1: 5′-GCTTGGAGAATGAGAGCCAAA-3′ and si-con: 5′-AATTCTCCGAACGTGTCACGT-3′.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
RNA extraction kit (Invitrogen) was used to extract total RNA from the cells. Messenger RNA (mRNA) was reverse transcribed into complementary DNA (cDNA) by SuperScript III reverse transcriptase (Invitrogen) and performed quantitative real-time polymerase chain reaction (qRT-PCR) by SYBR Green Master Mix (Applied Biosystems, Foster City, California). MiScript reverse transcription kit (Qiagen, Germany) was utilized to convert miRNA into cDNA and MiScript SYBR-Green PCR kit (Qiagen, Germany) was applied to carry out the experiment of qRT-PCR. Expression level of mRNA was normalized to actin and miRNA was normalized to U6. The process was performed as follows: 40 cycles consisting of 95°C for 5 minutes, 95°C for 30 seconds, an extension step at 60°C for 45 seconds, and 72°C for 30 minutes. 2−ΔΔCT method was used to calculate the relative expression levels of mRNA and miRNA. The primers were used as follows:
miR-142-5p: F: 5′-CATAAAGTAGAAAGCACTAC-3′
R: 5′-GAACATGTCTGCGTATCTC-3′
U6: F: 5′-AGATTAGCATGGCCCCTGC-3′
R: 5′-GCAGGGGCCATGCTAATCT-3′
SORBS1: F: 5′-TATCAGCCTGGCAAGTCTTCCG-3′
R: 5′-CCCGTCTGATTCCCTCTTCACT-3′
Actin: F: 5′-CACCATTGGCAATGAGCGGTTC-3′
R: 5′-AGGTCTTTGCGGATGTCCACGT-3′
Cell Counting Kit-8 Assay
A 100 μL of cell suspension was added into 96-well plates at a density of 1 × 103 cells/well after transfected for 24 hours and the cells were incubated in carbon dioxide incubator. Cell counting kit-8 (CCK-8) kit was used to analyze the cells activity every other 24 hours, 10 μL of CCK-8 agent was added into the plates before detection and cultured for 1.5 hours in incubator. The optical density value was detected by a microplate reader (Bio-Rad, Hercules, California), and the proliferation curve was obtained using GraphPad Prism 5.0.
Transwell Assay
Transwell chamber (Corning, Lowell, Massachusetts) was used to detect the invasion and migration of the cells. Matrigel (BD Biosciences) should be precoated into the upper chamber of the transwell chamber, put 100 μL of cell suspension at a density of 1 × 105 cells/chamber into it, and added 500 μL of complete culture solution into the lower chamber for invasion. After overnight, the upper chamber was wiped off with cotton swab and the invasion cells were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.1% crystal violet for 20 minutes; then 5 visual fields were randomly captured with microscope for observation and counting. Unlike the invasion experiment, the transwell chamber of migration does not need to be coated with matrigel.
Wound Healing Assay
The cells were seeded in the 6-well plates to form a monolayer of fused cells after transfected with miR-142-5p inhibitors and si-SORBS1. The wounds were formed by 100 μL tips in the monolayer of fused cells. The width of the wound was captured using a phase-contrast microscope at 0 and 24 hours after wound scratching.
Luciferase Reporter Assay
Bioinformatics prediction softwares such as miRanda, miRDB, miRWalk, and TargetScan were used to predict the target genes of miR-142-5p and select candidate target genes. Dual-luciferase reporter assay was used to further confirm the prediction results of bioinformatics softwares and Dual Luciferase reporting system was used in Dual luciferase reporting kit. Wild-type or mutant-type SORBS1 was subcloned into the pMIR-Report Luciferase (Ambion, Waltham, MA, USA ) and then cotransfected with miR-142-5p mimic and miR-142-5p mimic NC into cells. The transfected cells were planted on 96-well plates at a density of 1 × 104 cells/hole and incubated for 48 hours, and the lysate was added to each well. Finally, the relative luciferase activity was measured by firefly luciferin to Marine luciferin intensity using a Dual Luciferase reporting system to reflect the effect of miR-142-5p on SORBS1 level.
Statistical Analysis
SPSS version 22.0 software and GraphPad Prism 5.0 were used for statistical analysis. The measurement data with normal distribution were expressed as mean ± standard deviation, Student t test was used for comparison between the 2 groups, and comparison among multiple groups was analyzed using a 1-way analysis of variance, followed Dunnett post hoc test (compared all groups vs control group) and Bofferornie post hoc test (compared all pairs of groups). The χ2 test was used for comparison between gene expression and clinicopathologic characteristics. A P < .05 was considered statistically significant.
Results
Highly Expression of MiR-142-5p in Breast Cancer Results in Distant Metastasis
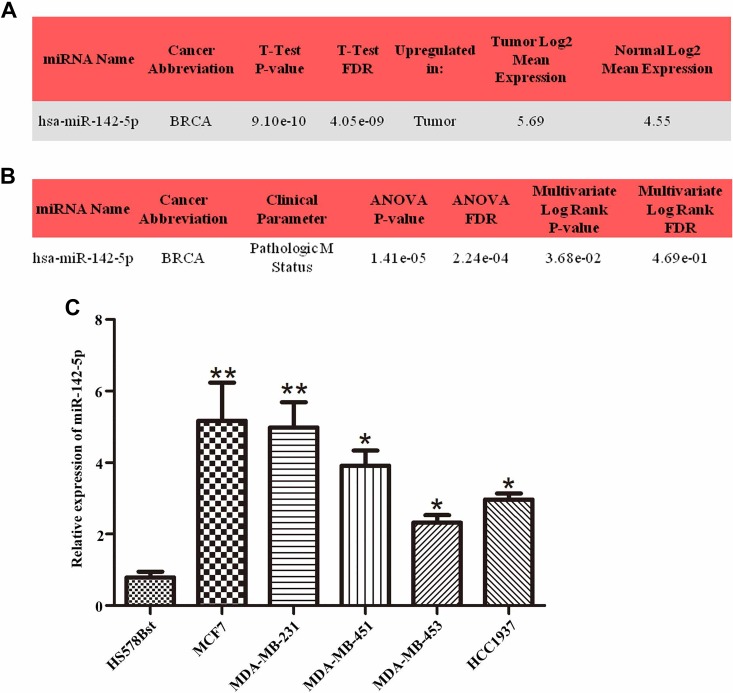
According to the analysis results of OncomiR library, miR-142-5p was upregulated in the breast cancer tissues (Figure 1A) and strongly correlated with distant metastasis (Figure 1B). In vivo experiment, we detected miR-142-5p was highly expressed in breast cancer cell lines, including MCF7, MDA-MB-231, MDA-MB-451, MDA-MB-453, and HCC1937, compared to normal cell line HS578Bst by qRT-PCR, especially highly expressed in MCF7 and MDA-MB-231 (Figure 1C, *P < .05, **P < .01). Therefore, MCF7 and MDA-MB-231 cells were selected in the following experiments. The findings indicated that miR-142-5p was highly expressed in breast cancer samples, resulting in poor prognosis.
Figure 1.
High expression of miR-142-5p in breast cancer closely associated with distant metastasis. A, The data from OncomiR website indicated that miR-142-5p was highly expressed in breast tumor tissues compared with normal (P < .0001). B, Clinical parameters analysis revealed that the expression level of miR-142-5p was associated with distant metastasis in patients with breast cancer (P < .0001). C, The results from quantitative real-time polymerase chain reaction investigated that miR-142-5p was promoted at different degrees in breast cancer cell lines compared with human normal cell line HS578Bst (*P < .05, **P < .01).
MiR-142-5p Promotes Breast Cancer Cells Proliferation, Migration, and Invasion
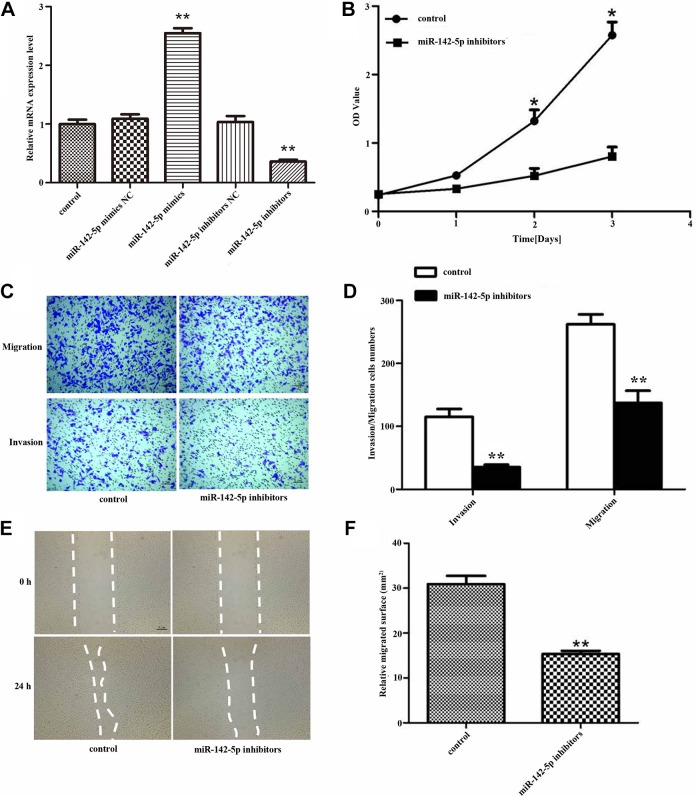
In order to explore the effect of miR-142-5p on breast cancer cells, first, we attempted to up-/downregulated miR-142-5p in breast cancer cells with mimics/inhibitors. Quantitative RT-PCR was used to detect the relative expression of miR-142-5p after the cells were transfected with miR-142-5p mimics/inhibitors. The results showed that after transfected with miR-142-5p mimics, the expression of miR-142-5p could be promoted, while downregulation of miR-142-5p had the opposite effect (Figure 2A, **P < .01). The CCK-8 assay showed that the cells proliferation capacity was reduced after transfected with miR-142-5p inhibitors compared to the untreated cells (Figure 2B, *P < .05). To further investigate the effect of miR-142-5p on breast cancer cells migration and invasion, we transfected the cells with miR-142-5p inhibitors and then evaluated migration and invasion abilities. As shown in Figure 2C and D, the number of migrated and invaded cells that were treated with the miR-142-5p inhibitors was distinctively reduced (**P < .01) compared with the control. And the wound healing assay showed the same results, compared with the control, the relative migrated surface was reduced after transfected with miR-142-5p inhibitors (Figure 2E and F, **P < .01). These observations suggested that miR-142-5p holds an important role in accelerating the proliferation, migration, and invasion abilities of breast cancer in vitro.
Figure 2.
Deletion of miR-142-5p blocked breast cancer cells proliferation, migration, and invasion. A, The relative expression of Sorbin and SH3 domain-containing protein 1 (SORBS1) after MCF7 cells transfected with miR-142-5p mimics/inhibitors (**P < .01). B, The proliferation of MCF7 cells measured by cell counting kit-8 (CCK-8) after transfected with miR-142-5p inhibitors (*P < .05). C, The migration and invasion (**P < .01) of MCF7 cells detected by transwell after transfected with miR-142-5p inhibitors and (D) is quantification. E, The relative migrated surface was decreased after the MCF7 cells transfected with miR-142-5p inhibitors and (F) is quantification (**P < .01).
Sorbin and SH3 Domain-Containing Protein 1 Is a Direct Target of MiR-142-5p and Negatively Regulated by MiR-142-5p
To the best of our knowledge, miRNAs executed their function through suppressing the expression of their target genes. In order to better understand the molecular mechanism of miR-142-5p, we used the miRNA target analysis tools such as TargetScan, MiRanda, miRDB, and miRWalk to predict possible miRNAs binding sites to SORBS1. According to the analysis, the program predicated a good binding sequence between miR-142-5p and SORBS1. The bioinformatics analysis showed that there was a potential relationship between SORBS1 and miR-142-5p.
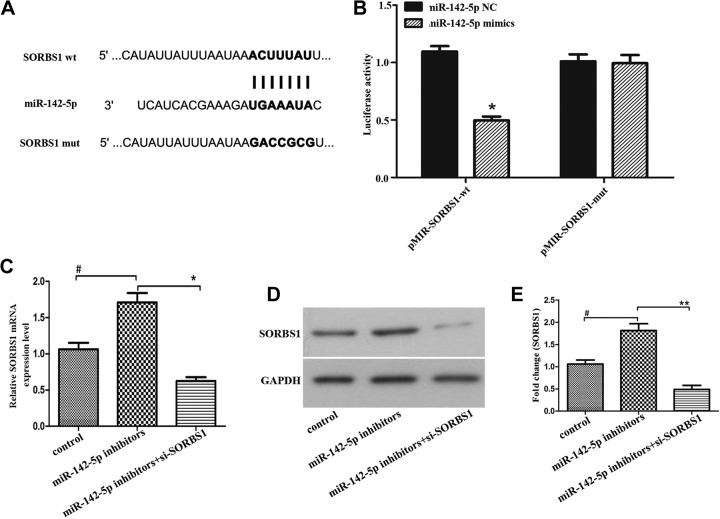
To further explore the interactive action between SORBS1 and miR-142-5p, we performed a luciferase reporter assay in breast cancer cells. The wild-type SORBS1 sequence containing the miR-142-5p binding site or a mutated SORBS1 sequence was cloned into luciferase reporter vectors (Figure 3A) and then transfected into cells with the miR-142-5p mimics and NC. As shown in Figure 3B, the overexpression of miR-142-5p significantly reduced the luciferase reporter activity in wild-type group but had little effect on luciferase reporter activity in mutant-type group (*P < .05). Subsequently, we transfected cells with miR-142-5p inhibitors or miR-142-5p inhibitors+si-SORBS1 to detect the effect of miR-142-5p on SORBS1. The data from Figure 3C to E indicated that inhibition of miR-142-5p upregulated the RNA and protein expression levels of SORBS1, indicating that SORBS1 was negatively modulated by miR-142-5p. The data also stated that transfected with si-SORBS1 reversed the promoting effect of miR-142-5p inhibitor on SORBS1 expression in both mRNA and protein levels. The results provided overwhelming evidence, supporting that SORBS1 was a particular target of miR-142-5p and negatively regulated by miR-142-5p in breast cancer cells.
Figure 3.
Sorbin and SH3 domain-containing protein 1 (SORBS1) was a target of miR-142-5p and negatively regulated by miR-142-5p. A, The predicted miR-142-5p binding site of SORBS1 and a mutated version. B, Luciferase reporter assay showed that the luciferase activity in wide-type and mutant-type SORBS1 groups after transfected with miR-142-5p mimics and negative control (*P < .05). The messenger RNA (C) and protein (D) expression of SORBS1 after transfected with miR-142-5p inhibitors, or miR-142-5p inhibitors+si-SORBS1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) regarded as a control, and (E) is quantification of (D), (*P < .05, **P < .01).
Patients With Low Expression of SORBS1 Tended to Poor Overall Survival
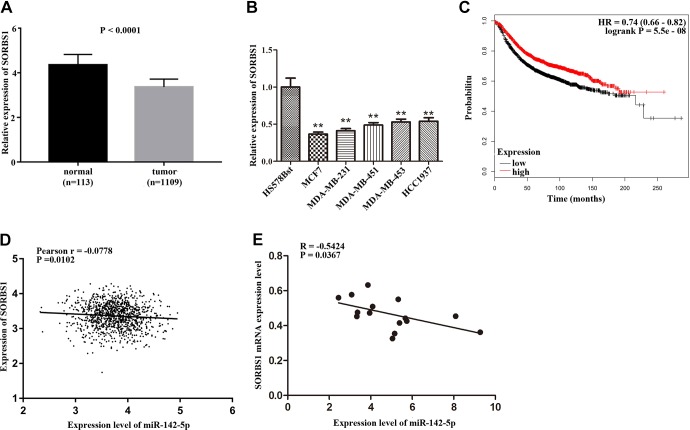
Based on the above research, we conducted a comprehensive analysis of SORBS1; according to the data from TCGA and Kaplan-Meier plotter websites, we found that SORBS1 was downregulated in patients with breast cancer (Figure 4A, P < .0001). The expression of SORBS1 in several breast cancer cell lines was analyzed. As shown in Figure 4B, SORBS1 was significantly downregulated in MCF7, MDA-MB-231, MDA-MB-451, MDA-MB-453, and HCC1937 compared with normal cell line HS578Bst (P < .01). Moreover, patients with breast cancer had poor overall survival with low expression of SORBS1 compared with the high expression of SORBS1 (Figure 4C, P = .009). A correlation between miR-142-5p and SORBS1 in patients with breast cancer (Figure 4D, P = .0102) and breast cancer cells (Figure 4E, P = .0367) was analyzed. The results indicated that miR-142-5p was negatively correlated with SORBS1 expression in both breast cancer cells and TCGA BRCA patients. The clinical parameters from the TCGA database showed that low expression of SORBS1 was associated with pathological staging (P = .03), death (P = .007), and the status of estrogen receptor (P = .0003), progesterone receptor (P = .00004), and HER2 (P = .000005) in patients with breast cancer (Table 1). The results suggested that downregulation of SORBS1 may promote the development of breast cancer and lead to poor prognosis and emphasized that miR-142-5p and SORBS1 were negatively correlated in breast cancer.
Figure 4.
Downregulation of Sorbin and SH3 domain-containing protein 1 (SORBS1) communicated with poor prognosis. A, The expression of SORBS1 was downregulated in breast cancer tissues compared with normal tissues (P < .0001). B, SORBS1 was downregulated in several breast cancer cell lines compared with HS578Bst cell line (**P < .01). C, Patients in breast cancer with low expression of SORBS1 had poor overall survival (P = .009). D, The correlation between miR-142-5p and SORBS1 in patients with breast cancer (P = .0102). E, The correlation between miR-142-5p and SORBS1 in breast cancer cells (P = .0367).
Table 1.
Relationship Between SORBS1 Expression and Clinicopathologic Features of Patients With Breast Cancer.
| Characteristics | Expression of SORBS1 | P Value | |
|---|---|---|---|
| Low | High | ||
| Age | .077 | ||
| <60 | 227 | 254 | |
| ≥60 | 206 | 181 | |
| Stage | .030a | ||
| I + II | 318 | 346 | |
| III + IV | 116 | 89 | |
| Tumor stage | .554 | ||
| T1 + T2 | 368 | 375 | |
| T3 + T4 | 66 | 60 | |
| Metastasis | .311 | ||
| M0 | 424 | 429 | |
| M1 | 10 | 6 | |
| Lymph node metastasis | .072 | ||
| N0 | 199 | 226 | |
| N1 + N2 + N3 | 235 | 209 | |
| Death | .007a | ||
| No | 355 | 384 | |
| Yes | 79 | 51 | |
| ER status | |||
| Positive | 375 | 426 | .0003a |
| Negative | 143 | 94 | |
| PR status | |||
| Positive | 316 | 376 | .00004a |
| Negative | 203 | 140 | |
| HER2 status | |||
| Positive | 103 | 61 | .000005a |
| Negative | 236 | 323 | |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; SORBS1, Sorbin and SH3 domain-containing protein 1.
a P < .05.
Aberrant Expression of SORBS1 Reversed the Promotion Effect of MiR-142-5p on Breast Cancer Cells
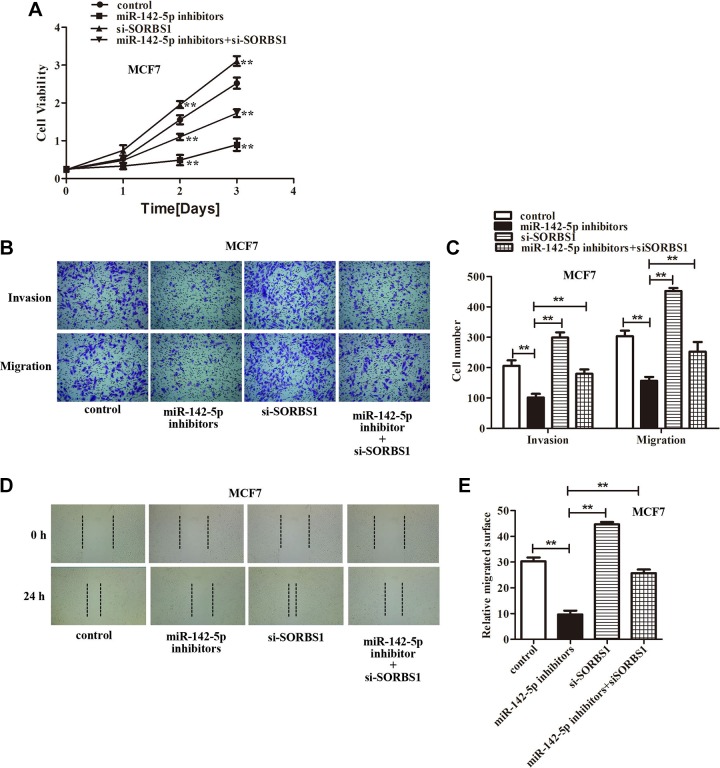
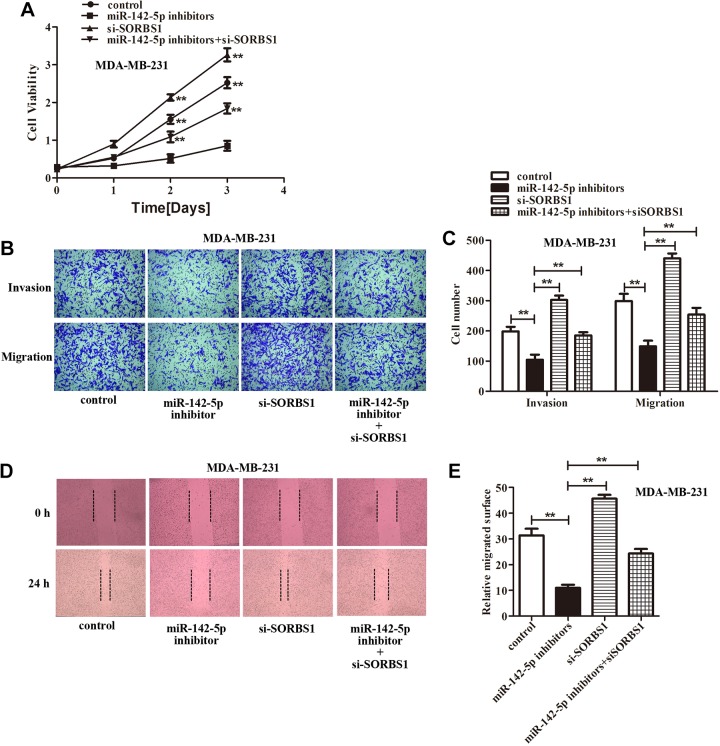
In light of the abovementioned findings, we speculated that miR-142-5p might promote breast cancer cells proliferation, invasion, and migration by downregulation SORBS1 expression. Accordingly, we cotransfected the miR-142-5p inhibitors and si-SORBS1 into the MCF7 and MDA-MB-231 cells to assess the miR-142-5p/SORBS1 on breast cancer progression. The CCK-8 assay showed that inhibition of miR-142-5p suppressed breast cancer cells growth compared with the control, silencing SORBS1 increased the breast cancer cells growth compared with miR-142-5p inhibitors group. However, downregulation of SORBS1 significantly relieved the inhibitory effect of miR-142-5p inhibitors on the growth suppression of breast cancer cells after transfected with miR-142-5p inhibitors+si-SORBS1 compared with miR-142-5p inhibitors group (Figures 5A and 6A). Transwell assay demonstrated that knockdown miR-142-5p inhibited breast cancer cells invasion and migration abilities compared with the control, and attenuating SORBS1 promoted breast cancer cells movement compared with miR-142-5p inhibitors group. Although downregulation of SORBS1 reversed the inhibitory effect of miR-142-5p inhibitors on breast cancer cells invasion and migration abilities after transfected with miR-142-5p inhibitors+si-SORBS1 compared with miR-142-5p inhibitors group (Figures 5B-C and 6B-C). Wound healing assay also found exhausting of miR-142-5p reduced the breast cancer cells migrated surface compared with the control, and depletion of SORBS1 increased the migrated surface of breast cancer cells compared with miR-142-5p inhibitors group. But downregulation of SORBS1 reversed the inhibitory effect of miR-142-5p inhibitors on breast cancer cells–migrated surface after transfected with miR-142-5p inhibitors+si-SORBS1 compared with miR-142-5p inhibitors group (Figures 5D and E and 6D and E). In summary, we found that depletion of SORBS1 reversed the inhibition of breast cancer cells proliferation, invasion, and migration abilities caused by miR-142-5p inhibitors proved that miR-142-5p acted as a promoting role in breast cancer by targeting SORBS1 (**P <.01 represented vs miR-142-5p inhibitors group).
Figure 5.
Knockdown Sorbin and SH3 domain-containing protein 1 (SORBS1) reversed the inhibiting effect of miR-142-5p inhibitors on MCF7 cells proliferation, migration, and invasion abilities. A, The cell counting kit-8 (CCK-8) assay indicated that silenced SORBS1 rescued the MCF7 cells proliferation after downregulation of miR-142-5p expression (**P < .01). B, Transwell assays demonstrated that downregulation of SORBS1 expression reversed the inhibitory effect of miR-142-5p inhibitors on MCF7 cells invasion and migration, and (C) is quantification of (B), (**P < .01). D, The wound healing assay confirmed that silenced SORBS1 obviously reversed the inhibitory effect of miR-142-5p inhibitors on MCF7 cells migration. E, Quantification of (D), (**P < .01 represented miR-142-5p inhibitors group).
Figure 6.
Depletion of Sorbin and SH3 domain-containing protein 1 (SORBS1) rescued the negative influence of miR-142-5p inhibitors on MDA-MB-231 cells proliferation, migration, and invasion abilities. A, The cell counting kit-8 (CCK-8) assay showed that knockdown SORBS1 reversed the inhibition effect on MDA-MB-231 cells proliferation caused by miR-142-5p inhibitors (**P < .01). B, The data from transwell assay indicated that silencing SORBS1 rescued the inhibition effect on MDA-MB-231 cells aggressiveness ability induced by miR-142-5p inhibitors. C, Quantification of (B), (**P < .01). D, The wound healing assay exhibited that exhausting SORBS1 alleviated the inhibition effect on MDA-MB-231 cells migrated surface caused by miR-142-5p inhibitors. E, Quantification of (D), (**P < .01 represented miR-142-5p inhibitors group).
Discussion
In this study, we found that miR-142-5p was frequently upregulated in breast cancer and significantly correlated with lymph node metastasis by bioinformatics analysis. Meanwhile, in vitro, we also investigated that miR-142-5p could promote breast cancer cells proliferation, invasion, and migration. With prediction softwares and dual-luciferase reporter assay, we also verified that SORBS1 was a direct functional target of miR-142-5p. Through analyzing TCGA data, SORBS1 was downregulated in breast cancer tissues, associated with the pathological staging and death, and its low-expression level led to worse prognosis in patients with breast cancer. The data from OncomiR indicated that the expression levels of miR-142-5p were correlated with metastasis. However, the data from TCGA database showed that the expression levels of SORBS1 were not correlated with metastasis. But the results from our study exhibited that miR-142-5p promoted breast cancer cells invasion and migration through targeting SORBS1, suggesting that SORBS1 was involved in the metastasis of breast cancer cells. Some research reported that SORBS1 could suppress tumor metastasis in breast cancer,20 which was consistent with our results. The clinical characteristics of SORBS1 were obtained from the TCGA database, including 868 cases of patients with breast cancer with complete clinical data. However, it is not clear whether these patients with breast cancer have metastasized. Therefore, there is a certain deviation in the clinical characteristics analysis. From rescue study, increased SORBS1 expression was stimulated by miR-142-5p inhibitor; we also observed that SORBS1 participated in the migration and invasion abilities of breast cancer cells and reversed the promotion effect of miR-142-5p on breast cancer cells. In summary, our findings suggested that upregulated miR-142-5p expression contributed to breast cancer progression by promoting inhibition of SORBS1 expression.
MiR-142-5p has been repeatedly reported to be involved in the development of various cancers. For instance, miR-142-5p suppresses tumorigenesis in non-small cell lung cancer by targeting PIK3CA,17 promotes development of colorectal cancer through targeting SDHB,13 adjusts cell growth and migration in renal cell carcinoma by targeting BTG3,14 regulates apoptosis in ovarian cancer by targeting MCL1,15 and so on. However, the effect of miR-142-5p on breast cancer has rarely been reported. In this study, we found that miR-142-5p was highly expressed in both breast cancer tissues and cell lines. And the data from OncomiR indicated that the overexpression of miR-142-5p led to distant metastasis of breast cancer, combined with reducing the expression of miRNA-142-5p can inhibit the proliferation, invasion, and migration of breast cancer cells, demonstrated the significance of miR-142-5p on breast cancer metastasis. These results indicated that miR-142-5p plays potently facilitated role in breast cancer progression.
The regulation of miR-142-5p by downstream target genes played a crucial role in revealing its cancer-promoting mechanism. Therefore, we analyzed the downstream genes of miR-142-5p with multiple bioinformatics prediction softwares and verified it with dual-luciferase reporter assay. It was found that SORBS1 was an important target gene of miR-142-5p downstream. Previous research had found that SORBS1, as an adaptor protein, also known as CAP, involved in much biological functions, such as control glucose transport,21 transcriptional activity,22 and insulin signaling.23 Meanwhile, SORBS1 can regulate cell adhesion, motility and spreading,24 and cytoskeleton organization25 by interacting with cytoskeleton regulators. Recently, some researches demonstrated that SORBS1 restrains tumor metastasis and improves the sensitivity of cancer to chemotherapy drug.20 In the present study, we found that SORBS1 was downregulated in tumor tissues and led to poor prognosis, clinical parameters analysis demonstrated that SORBS1 associated with pathological stage and death in breast cancer. These results suggested that miR-142-5p/SORBS1, as a collaborator, may act as a significant role in breast cancer progression, and rescue assay found that silenced SORBS1 could relieve the inhibition of miR-142-5p inhibitors on breast cancer cells biological behaviors, thus promoting breast cancer cells proliferation, invasion, and migration. Therefore, together with the previous study, we speculated that miR-142-5p was significantly associated with breast cancer progression and worse clinical outcomes and could inhibit the migration and invasion of breast cancer cells via directly targeting SORBS1.
In general, our outcomes demonstrate that miR-142-5p enhanced breast cancer progression by silencing SORBS1. The findings investigated that miR-142-5p was overexpressed in breast cancer tissues and cell lines and resulted in metastasis, while the expression of SORBS1 was lower in breast cancer than that in the control and caused worse overall survival. Downregulation of miR-142-5p impeded the abilities of proliferation, invasion, and migration on breast cancer cells. The potential oncogene function of miR-142-5p was mediated by reducing its target gene SORBS1. These results indicate that miR-142-5p/SORBS1 may be a hopeful target for the development of new therapies for breast cancer.
Abbreviations
- CCK-8
cell counting kit-8
- cDNA
complementary DNA
- miRNA
microRNA
- NC
negative control
- mRNA
messenger RNA
- qRT-PCR
quantitative real-time polymerase chain reaction
- siRNA
small interfering RNA
- SORBS1
Sorbin and SH3 domain-containing protein 1
- TCGA
The Cancer Genome Atlas
Footnotes
Authors’ Note: Our study did not involve any human or animal. Neither did we involve patients nor the data were obtained from OncomiR website.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the National Natural Science Foundation of China (project no: 81702783) and the Natural Science Foundation of Guangdong Province (project no: 2017A030310574).
ORCID iDs: Weixuan Yu  https://orcid.org/0000-0001-9388-6178
https://orcid.org/0000-0001-9388-6178
Yunda Zhang  https://orcid.org/0000-0003-1220-8987
https://orcid.org/0000-0003-1220-8987
References
- 1. Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69(3):313–317. [DOI] [PubMed] [Google Scholar]
- 2. Bhatelia K, Singh K, Singh R. TLRs: linking inflammation and breast cancer. Cell Signal. 2014;26(11):2350–2357. [DOI] [PubMed] [Google Scholar]
- 3. Bertozzi N, Pesce M, Santi PL, Raposio E. Oncoplastic breast surgery: comprehensive review. Eur Rev Med Pharmacol Sci. 2017;21(11):2572–2585. [PubMed] [Google Scholar]
- 4. Fyles AW, Mccready DR, Manchul LA, et al. Tamoxifen with or without breast irradiation in women 50 years of age or older with early breast cancer. N Engl J Med. 2004;16(1):67–69. [DOI] [PubMed] [Google Scholar]
- 5. Scully OJ, Bay BH, Yip G, Yu Y. Breast cancer metastasis. Cancer Genomics Proteomics. 2012;9(5):311–320. [PubMed] [Google Scholar]
- 6. Petrovic N, Davidovic R, Bajic V, Obradovic M, Isenovic RE. MicroRNA in breast cancer: the association with BRCA1/2. Cancer Biomark. 2017;19(2):119–128. [DOI] [PubMed] [Google Scholar]
- 7. McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015;34(1):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. [DOI] [PubMed] [Google Scholar]
- 10. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. [DOI] [PubMed] [Google Scholar]
- 11. Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300(1):10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhan L, Lei S, Li W, et al. Suppression of microRNA-142-5p attenuates hypoxia-induced apoptosis through targeting SIRT7. Biomed Pharmacother. 2017;94:394–401. [DOI] [PubMed] [Google Scholar]
- 13. Liu S, Xiao Z, Ai F, et al. miR-142-5p promotes development of colorectal cancer through targeting SDHB and facilitating generation of aerobic glycolysis. Biomed Pharmacother. 2017;92:1119–1127. [DOI] [PubMed] [Google Scholar]
- 14. Liu L, Liu S, Duan Q, et al. MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am J Transl Res. 2017;9(5):2394–2402. [PMC free article] [PubMed] [Google Scholar]
- 15. Su J, Ruan S, Dai S, Mi J, Chen W, Jiang S. NF1 regulates apoptosis in ovarian cancer cells by targeting MCL1 via miR-142-5p. Pharmacogenomics. 2018;20(3):155–165. [DOI] [PubMed] [Google Scholar]
- 16. Jia L, Xi Q, Wang H, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun. 2017;488(2):425–431. [DOI] [PubMed] [Google Scholar]
- 17. Wang Z, Liu Z, Fang X, Yan H. MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in non-small cell lung cancer. Cell Physiol Biochem. 2017;43(6):2505–2515. [DOI] [PubMed] [Google Scholar]
- 18. Bai X, Zhou Y, Chen P, Yang M, Xu J. MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J Cell Biochem. 2018;119(2):2179–2188. [DOI] [PubMed] [Google Scholar]
- 19. Yan J, Yang B, Lin S, Xing R, Lu Y. Downregulation of miR-142-5p promotes tumor metastasis through directly regulating CYR61 expression in gastric cancer. Gastric Cancer. 2018; 22(2):302–313. [DOI] [PubMed] [Google Scholar]
- 20. Song L, Chang R, Dai C, et al. SORBS1 suppresses tumor metastasis and improves the sensitivity of cancer to chemotherapy drug. Oncotarget. 2016;8(6):9108–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiang SH, Baumann CA, Kanzaki M, et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature. 2001;410(6831):944–948. [DOI] [PubMed] [Google Scholar]
- 22. Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27(1):1–7. [DOI] [PubMed] [Google Scholar]
- 23. Lin WH, Huang CJ, Liu MW, et al. Cloning, mapping, and characterization of the human sorbin and SH3 domain containing 1 (SORBS1) gene: a protein associated with c-Abl during insulin signaling in the hepatoma cell line Hep3B. Genomics. 2001;74(1):12–20. [DOI] [PubMed] [Google Scholar]
- 24. Mei Z, Jun L, Alan C, et al. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2014;25(22):5284–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langhorst MF, Jaeger FA, Mueller S, Sven Hartmann L, Luxenhofer G, Stuermer CA. Reggies/flotillins regulate cytoskeletal remodeling during neuronal differentiation via CAP/ponsin and Rho GTPases. Eur J Cell Biol. 2008;87(12):921–931. [DOI] [PubMed] [Google Scholar]