Abstract
In the past few years, the use of specific cell types derived from induced pluripotent stem cells (iPSCs) has developed into a powerful approach to investigate the cellular pathophysiology of numerous diseases. Despite advances in therapy, heart disease continues to be one of the leading causes of death in the developed world. A major difficulty in unravelling the underlying cellular processes of heart disease is the extremely limited availability of viable human cardiac cells reflecting the pathological phenotype of the disease at various stages. Thus, the development of methods for directed differentiation of iPSCs to cardiomyocytes (iPSC-CMs) has provided an intriguing option for the generation of patient-specific cardiac cells. In this review, a comprehensive overview of the currently published iPSC-CM models for hereditary heart disease is compiled and analysed. Besides the major findings of individual studies, detailed methodological information on iPSC generation, iPSC-CM differentiation, characterization, and maturation is included. Both, current advances in the field and challenges yet to overcome emphasize the potential of using patient-derived cell models to mimic genetic cardiac diseases.
Keywords: Disease modelling , Induced pluripotent stem cells , Cardiomyocytes , Inherited heart disease , Cardiac differentiation
1. Introduction
Advances in the field of molecular biology and genomics have induced a shift in medicine towards a more precise, personalized, and causal approach. In the field of cardiology, this approach is thought to be especially beneficial for patients with inherited cardiac disease,1 currently often lacking sufficient standard of care. Unravelling the underlying mechanisms of cardiac disease and developing new treatment options have been hampered by limitations of available models. While established transgenic small animal models of inherited cardiac disease do provide some insights into the pathogenic mechanisms of these diseases,2,3 substantial differences between the physiology of cardiomyocytes from small animal models and humans limit extrapolation of results, leading to a failure to translate findings from small animals to humans.4 Likewise, transgenic non-cardiac human cell lines, e.g. hERG-overexpressing HEK-293 cells, were shown to model cardiac diseases insufficiently since they do not recapitulate the complex cardiac phenotype, e.g. sarcomere organization, calcium handling, metabolism, and (electro)physiology.5
The development of induced pluripotent stem cells (iPSCs) by Takahashi and Yamanaka6 has helped to overcome these challenges by enabling the generation of patient-specific iPSC-derived cardiomyocytes (iPSC-CMs), carrying disease-specific mutations. With these cells, researchers are able to recapitulate the disease phenotype in vitro. With refined protocols for iPSC generation and differentiation into somatic cell types, including cardiomyocytes, a sustainable source of non-transgenic human cardiac cells has become available for in vitro disease modelling. Cardiomyocytes generated from iPSCs have several advantages over human embryonic stem cells or organ derived stem cell models, as iPSCs can be generated from a variety of easily accessible cell sources, including cells from the skin, urine, and blood.7–9 Furthermore, the generated cardiomyocytes are donor (patient) specific, enabling genotype-phenotype associations, and offering a personalized drug-screening platform for individualized patient therapy.
Since the first study in 2010,10 much progress has been made using patient specific-iPSC models to characterize cardiac diseases and study their molecular pathogenesis. Over 90 studies using iPSC-CM models are now available, including long QT syndromes (LQTSs), catecholaminergic polymorphic ventricular tachycardia (CPVT), arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVC), familial dilated cardiomyopathy (DCM), familial hypertrophic cardiomyopathy (HCM), and many more. In this review, we provide an in-depth overview of the current iPSC-CM models of inherited cardiac diseases. Methods for differentiation and characterization of iPSC-CMs, including functional parameters like cellular electrophysiology, calcium handling, and contraction kinetics are evaluated. Finally, challenges, limitations, and future perspectives of iPSC-CM models of inherited cardiac disease will be discussed.
2. Generation of iPSC-CM models
2.1 iPSC generation
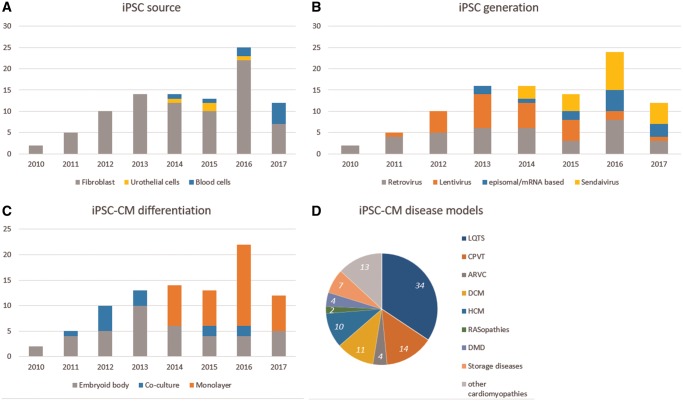
Patient-specific iPSC-CM models rely on the generation of iPSC lines from a tissue sample. To minimize the invasiveness of this procedure, a trend from using dermal fibroblasts from skin biopsies towards blood or urine cells can be observed8,9 (Figure 1A). Similarly, introduction of reprogramming factors6 into patient’s cells is moving from integrating vectors, e.g. retro- and lentiviruses, towards non-integrating vectors, e.g. Sendai virus11 or virus-free methods like episomal transfection,12 or mRNA delivery,13 avoiding insertional mutagenesis and potential transgene reactivation (Figure 1B). Details on methods used for generation and characterization of 192 iPSC lines (covering 152 different mutations; published in 91 papers) are provided in Supplementary material online, Table S1.
Figure 1.
Methods used to generate iPSC-CM models. (A) Cell source. (B) Reprogramming method. (C) Differentiation method. (D) Proportion of iPSC cell lines generated per disease. Data were retrieved from Supplementary material online, Table S1.
2.2 iPSC-CM differentiation
The directed differentiation of cardiomyocytes from pluripotent stem cells is currently performed by either co-culture with visceral endoderm-like (END-2) cells,14 embryoid body (EB) formation in suspension,15 or monolayer cell culture with supplementation of specific differentiation factors.16 Although details, efficiencies, and yields vary considerably between individual studies, the chemically defined monolayer differentiation protocol is increasingly used, likely due to its high efficiency and ease of use (Figure 1C and Supplementary material online, Table S1).17,18
2.3 iPSC-CM characterization
No consensus has been reached on the definition of a bona fide in vitro human cardiomyocyte sufficiently recapitulating it’s in vivo counterpart. iPSC-CMs have frequently been described displaying an immature, foetal-like phenotype, e.g. lacking mature sarcomeric organization,19 low ratios of multinucleation,20 underdeveloped t-tubule networks,21 and altered Ca2+ handling.22 Cardiac maturation involves changes in gene expression levels, structural reorganization (e.g. myofibrils), and importantly, functional changes (Ca2+ handling, contractility, and action potential characteristics) rather than the mere expression of certain markers.23 Thus, analyses of electrophysiological properties, contraction and contractile force, cell–cell coupling, metabolism, mitochondrial content and morphology, cell size and morphology, and sarcomere density and organization need to be considered. These differences between iPSC-CMs and adult cardiomyocytes have to be taken into account when establishing disease-in-a-dish models and interpreting results.
Studies within the scope of this review characterized iPSC-CMs to some degree, including immunofluorescence imaging, electron microscopy, fluorescence-activated cell sorting, and qRT-PCR. Most studies (53 of 91) report at which day of the differentiation protocols spontaneously contracting cells were first observed (Day 6–22 days, mean: Day 11). Characterization of the iPSC-CMs was carried out on average on Day 30 (between Day 1 and Day 150) after start of the differentiation protocol. A substantial fraction of studies (26 of 91) included functional measurements, e.g. multi-electrode arrays, to assess electrophysiological maturation of the iPSC-CMs. Details on characterization and assessment of maturation of patient specific iPSC-CMs are listed in Supplementary material online, Table S1.
Differences in protocols for generation of patient specific iPSC lines, differentiation to cardiomyocytes, and characterization of iPSC-CMs make studies on disease modelling difficult to compare and reproduce. However, the following studies highlight the evidence that iPSC-CMs exhibit clinically relevant phenotypes and that the use of these disease models in pathophysiological and molecular research can result in the discovery of new therapies for inherited cardiac diseases.
3. iPSC-CM models of cardiac disease
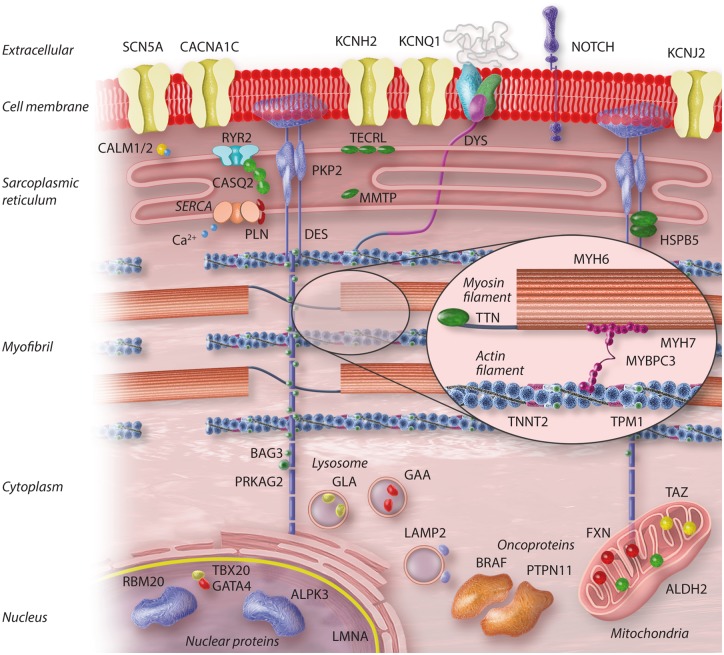
To provide an in-depth overview of the current iPSC-CM models of inherited cardiac diseases, Supplementary material online, Table S1 provides detailed information on all studied patient iPSC-CM lines, with a quantitative outline in Figure 1D. Supplementary material online, Table S2 lists the cardiac diseases, prevalence, and known associated genes, both with and without iPSC-CMs studies. The major results are discussed in the text and Figure 2 shows the subcellular localization of all mutated genes.
Figure 2.
Subcellular localization of cardiac disease-associated proteins studied using iPSC-CM models.
3.1 Long QT syndrome
The LQTS is an autosomal dominant cardiac disease, affecting up to 1 in 1000 live births. It is associated with over 500 different mutations in at least 15 genes24 encoding ion channel (interacting) proteins. Patients may only show a prolonged repolarization phase (the QT phase) on ECG measurements, but this can predispose to potentially life-threatening ventricular arrhythmias, so-called Torsades de pointe, and sudden cardiac arrest. In the last few years, patient-specific iPSC-CM research has mainly focused on LQTS1, LQTS2, and LQTS3, but Andersen-Tawil syndrome (LQTS7), Timothy syndrome (TS, LQTS8), and calmodulinopathies (LQTS14/15) have also been investigated.
Mutations in the genes KCNQ1 and KCNH2 (Figure 2) are most common and cause LQTS1 and LQTS2, respectively. The KCNQ1 gene encodes for the α-subunit of the voltage-gated K+ channel mediating the slow delayed rectifier K+ current (IKS current). The KCNH2 or hERG channel is a K+ channel needed for the rapid delayed rectifier current IKr. Both the IKs and the IKr are outward currents. A total of seven different mutations in the KCNQ1 gene have been studied.25–34 Multiple LQTS1 iPSC-CM models showed a dominant negative effect of a KCNQ1 mutation leading to a diminished IKs current due to a sarcolemmal deficiency of KCNQ1 channels.25,26,29 In addition, electrophysiological abnormalities, including a prolonged corrected field potential duration were recorded in LQTS1 iPSC-CMs.26 Cross-talk between the KCNQ1 mutation and Ca2+ handling abnormalities was reported as well, including the observation that Ca2+ antagonists could rescue the electrophysiological phenotype.28 Other studies reported protective effects of β-adrenergic antagonists25 or ML277, a selective IKs activator.29
KCNQ1 is also affected in Jervell and Lange-Nielsen syndrome (JLNS), which is characterized by life-threatening arrhythmias like ventricular tachycardia and bilateral deafness. JLNS is caused by a homozygous or compound heterozygous mutation in KCNQ1 or KCNE1,35 disrupting the function of the voltage-gated K+ channel needed for the IKs current. One study using engineered and patient-derived JLNS iPSC-CMs showed that both mutations led to the electrophysiological JLNS phenotype, with increased action potential duration (APD) and sensitivity to proarrhythmic drugs.36
Similarly to LQTS1 iPSC-CM models, LQTS2 iPSC-CM models showed APD prolongation.37,38 In total, eight different KCNH2 mutations were modelled.28,31,37–45 In general, the LQTS2 clinical phenotype was mimicked in vitro by a diminished IKr current and arrhythmia, caused by a decrease in hERG function due to the KCNH2 mutation. A study on the KCNH2 N996I mutation reported only a mild increase in APD without early-after depolarizations (EADs),42 agreeing well with the mild KCNH2 N996I clinical phenotype.46 Concerning the molecular mechanisms, trafficking defects were found to occur in the LQTS2 models40,44 possibly due to a defect in the glycosylation of KCNH2, which could be alleviated by blocking Calpain proteins,44 consequently rescuing the electrophysiological phenotype. As shown for LQTS1, LQTS2 also showed Ca2+ handling disturbances which could be rescued with Ca2+ antagonists.39 One study showed that RNAi-mediated knockdown of the mutant allele could rescue the electrophysiological phenotype, suggesting an at least partially dominant negative effect.44
LQTS3 is caused by gain-of-function mutations in the gene SCN5A. In total, nine LQTS3 associated mutations in SCN5A have been studied using iPSC-CMs.39,47–54 This gene encodes the main cardiac Na+ voltage-gated channel NaV1.5, essential for the Na+ current that determines the fast upstroke of the cardiac action potential. LQTS3 iPSC-CM studies have shown prolonged APD, possibly due to a slower inactivation of the sodium channel49 or a faster channel recovery from inactivation.47 While one study reported an arrhythmogenic effect of mexiletine due to hERG interaction,47 this was not observed in other studies, possibly due to a dosage effect.50 Detection of EADs varied with the mutation studied, with one study not recording any EADs or delayed after-depolarizations (DADs),48 which may be the result of significant cardiomyocyte subtype variability.48,49 Next to LQTS3, mutations in the SCN5A gene can cause Brugada syndrome (BrS), conduction disease and atrial standstill.55 However, mixed phenotypes are often seen, known as ‘overlap syndrome’. In an iPSC-CM model of overlap syndrome, electrophysiological abnormalities were found, including a significantly diminished INa current density due to the loss-of-function mutation.51 BrS was studied in three papers using iPSC-CMs.56–58 In one of those, three iPSC-CM models were made from patients with BrS where no BrS-associated mutations were found.56 While a BrS-associated SCN5A-1795insD mutation line did show effects due to a reduced sodium channel function, no abnormalities were seen in the aforementioned three iPSC-CM models, indicating that SCN5A dysfunction may not be a prerequisite for BrS. The fact that BrS often only presents in adulthood and is thought to be associated with ventricular fibrosis59 raises the question whether an iPSC-CM model can successfully model such a multifactorial disease at this stage.
Andersen-Tawil syndrome (ATS) or LQTS7 is a rare autosomal dominant genetic disorder linked to mutations in KCNJ2 (ATS1). Patients with mutations in KCNJ2 account for approximately 70% of cases, while the genetic cause of the remaining 30%, labelled as Type 2 (ATS2), remains unknown.60KCNJ2 encodes the voltage gated, inward-rectifying potassium channel (Kir2.1, Figure 2) that contributes to the inward-rectifier potassium current (IK1). So far three ATS1 mutations have been studied using an iPSC-CM model to elucidate the pathogenesis and find potential drug candidates.61 ATS iPSC-CMs recapitulated the abnormal electrophysiological phenotype of ATS, showing strong arrhythmic events and irregular Ca2+ release, which could be suppressed by the antiarrhythmic agent, flecainide, through modulation of the NCX current.
TS (LQTS8) is characterized by cardiac arrhythmias in combination with, among others, dysmorphic facial features and syndactyly. Up to 2005, only 18 cases were reported.62 TS is caused by a mutation in the gene CACNA1C, encoding for the sarcolemmal voltage-gated Ca2+ channel (Figure 2), CaV1.2, the main cardiac L-type calcium channel, which is indispensable for the generation of the cardiac action potential and excitation-contraction coupling.63 To date, one TS iPSC-CM model has been studied, which showed DADs and a slow intrinsic beating rate in ventricular iPSC-CMs, agreeing well with bradycardia found in TS patients.64
LQTS-associated calmodulinopathies (LQTS14 and 15) arise due to mutations in one of the three Calmodulin genes, affecting the Ca2+ binding properties of this protein.65 Clinically, the patients present with life-threatening arrhythmias that are often resistant to conventional therapies. LQT14-iPSC-CMs showed prolonged repolarization with altered rate-dependency, which resulted in augmented inward current during the plateau phase and failure to adapt to high pacing rates.66 Importantly, this study used addition of simulated IK1 by dynamic-clamp, to overcome the extremely low IK1/Kir2.1 expression in iPSC-CMs and mimic a mature action potential profile. Two studies using iPSC-CM models of LQT15 were able to show a dominant negative effect of single heterozygous CALM2 mutations on the suppression of L-type Ca2+ channel inactivation.67,68 Consistent with clinical phenotypes,69 LQT15-iPSC-CMs exhibited significantly lower beating rates and prolonged APD, compared with control cells, including allele-specific knockout cells.
3.2 Catecholaminergic polymorphic ventricular tachycardia
Affecting around 1 in 10 000 people, CPVT is characterized by stress-induced ventricular arrhythmia, potentially causing sudden cardiac death in young individuals.70 While one subtype of CPVT (CPVT1) is caused by mutations in the ryanodine receptor Type 2 gene (RYR2), a second subtype is caused by a mutated calsquestrin-2 gene (CASQ2). Both RYR2 and CASQ2 are needed for correct Ca2+ handling in the cardiomyocyte: while the RYR2 channel allows for Ca2+ flow out of the sarcoplasmic reticulum in the case of depolarization, CASQ2 is an abundant Ca2+-binding protein in the sarcoplasmic reticulum. Next to RYR2 and CASQ2, mutations in triadin (TRDN), CALM1/2, KCNJ2, and TECRL are also known (Figure 2 and Supplementary material online, Tables S1 and S2).
Altogether, 16 different mutations causing CPVT have been investigated.71–86 Induction of the arrhythmic phenotype by stressors including adrenergic agonists and pacing was observed in all CPVT iPSC-CM models. It was postulated that the deviant effects of adrenergic stimulation in CPVT are caused by altered CaMKII signalling.76 Pacing also induced a negative inotropic effect as well as DADs in both CPVT171 and CPVT2 models.75 Importantly, a more immature phenotype of CPVT iPSC-CMs was observed compared with control iPSC-CMs,75,79,80 raising the question whether this comparison is adequate. However, the validity of the CPVT iPSC-CM model has been shown,78 as responses to dantrolene agrees well with the responses in patients, depending on their respective mutations.
3.3 Arrhythmogenic right ventricular cardiomyopathy/dysplasia
ARVC is an inherited primary cardiomyopathy associated with increased risk of ventricular arrhythmia and sudden cardiac death. One in 5000 people are thought to be affected worldwide.87 Forty to fifty percent of ARVC patients harbour mutations in desmosomal proteins, including plakoglobin, plakophilin, desmoplakin, desmoglein 2, and desmocollin 2.88 The desmosome is needed for cell-to-cell attachment, connecting the cytoskeleton of neighbouring cells (Figure 2). So far, six different mutations have been studied using iPSC-CM models.89–94 In these studies, on baseline (before treatment with adipogenic medium), desmosome abnormalities were observed, and in two studies darker89 or clustered90 lipid droplets were seen. Signalling pathways including the canonical Wnt pathway and the PPARγ pathway were found to be up-regulated, which became more pronounced after treatment with so-called 3 Factor or 5 Factor adipogenic medium.90,91 The latter caused metabolic changes that could not be reversed by introducing a wild-type plakoglobin transcript.91 The same study showed that, by enriching for Isl+ cardiac progenitor cells and thus the amount of right ventricular (RV) cardiomyocytes, Isl1+ cells confer the dominant RV pathology seen in ARVC patients, such as lipogenesis. Additionally, the reduction in desmosomal protein levels in ARVC iPSC-CM models is in close agreement with histopathological data from ARVC patients.95 Nonetheless, it remains questionable whether iPSC-CMs treated with the 3 Factor and/or 5 Factor method, protocols using different small-molecule compounds and hormones to switch metabolism and accelerate pathogenesis, truly reflect the physiological environment in vivo, although PPARγ upregulation might reflect a more mature iPSC-CMs model.19 In addition, the clinical ARVC phenotype may be provoked by exercise, possibly causing more mechanical stress on the cardiomyocytes.96 It is challenging to model all the external influences in vitro in an iPSC-CM model, but this will likely help to elicit a more accurate disease phenotype.
3.4 Dilated cardiomyopathy
DCM is the most common cardiomyopathy, with a prevalence of 1 in 250. The aetiology is divers and can be metabolic, valvar, neuromuscular, or genetic. The clinical phenotype of DCM includes left ventricular dilation and impaired systolic function, ultimately leading to heart failure. The inherited form of DCM is associated with mutations in more than 20 different genes, coding for proteins in the cytoskeleton, nuclear lamina, and sarcomere.97 Lamin A/C (LMNA) DCM is among the most malignant subtypes, its clinical phenotype is characterized by early-onset atrial fibrillation and conduction delay as well as sudden cardiac death.98 Mutations in the desmin (DES) gene, in turn, can cause HCM, DCM,99 or ARVC.100 Of all DCM cases, 30% arise due to mutations in the titin (TTN) gene.101
Recent studies using iPSC-CM disease modelling have aimed to elucidate the molecular pathogenesis of mutations in lamin A/C (LMNA),102 desmin (DES),103 cardiac muscle troponin T (TNNT2),27,104,105 phospholamban (PLN),106 RNA-binding motif protein 20 (RBM20),107,108TTN,109,110 and BAG3111 (Figure 2 and Supplementary material online, Table S1). A total of 13 mutations have been studied, in some cases both homozygous and heterozygous forms. A significant amount of these studies describe disrupted sarcomeres, decreased contractile force and Ca2+ handling impairment, which is in agreement with what is seen in heart failure patients.112 In addition, as seen in vivo, β-adrenergic blockers and Ca2+ antagonists were proven to attenuate the phenotype,105,107 including sarcomeric disarray and increased apoptosis. Importantly, by using targeted gene correction, two iPSC-CM studies on PLN R14Del could fully revert the in vitro disease phenotype.106,113
3.5 Hypertrophic cardiomyopathy
HCM is estimated to be one of the most prevalent hereditary heart diseases in the world,114 affecting up to 1 in 500 individuals worldwide. It is characterized by left ventricle hypertrophy in the absence of a causative haemodynamic burden. Genetically, it is associated with mutations in sarcomeric protein-coding genes, most frequently myosin heavy chain-β (MYH7) and cardiac myosin binding protein C (MYBPC3),115–117 but also TPM1118,119 (Figure 2). To date, more than 1400 different mutations are known to cause HCM.118
Ten HCM-causing mutations in MYH7, MYBPC3, TPM1, ALPK3, and PRKAG2 (HCM-like) were studied using iPSC-CM models.27,115,117–124 These in vitro models recapitulate some of the disease phenotypes by reporting Ca2+ handling irregularities, cardiomyocyte hypertrophy, sarcomere disarray, arrhythmias, and hypercontractility.118,120,121,123 Several pathways were shown to be affected by the mutations, including Endothelin-1 signalling,119 the canonical Wnt-pathway,121 and most prominently, calcineurin/NFATc4 signalling.119–121 Both NFATc4 nuclear localization and disrupted Ca2+ handling were found to be causal factors in the development of the pathognomonic cellular hypertrophic phenotype. Some conflicting results were also found: while one study observed decreased RyR2 and SERCA2a expression levels,121 another reported the opposite.118 Additionally, the type of arrhythmogenic events differs significantly; for some mutations, DADs were prominent arrhythmic events,120 while in others, they were accompanied by EADs118 or no DADs were seen.121 The exact cause of these heterogeneities is not clear but could be attributed to the maturity of the cells, as the time point of measurement differs significantly between the studies.
3.6 Left ventricular non-compaction cardiomyopathy
Left ventricular non-compaction cardiomyopathy (LVNC) is characterized by a prominent LV trabecular meshwork, deep intertrabecular recesses in the ventricular wall and a thin-compacted layer.125 Major clinical manifestations include systolic and diastolic dysfunction, systemic embolism, and arrhythmias. LVNC has similar traits as, and shares many of its associated genes with DCM and HCM.126 However, mutations in MIB1 have been shown to cause LVNC without additional phenotypes.127 As an isolated myocardial trait, prevalence is very low, but as a morphologic trait shared by different cardiomyopathies, it is more common. So far, one iPSC-CM LVNC disease model has been developed.128 In this study, it was shown that iPSC-CMs generated from LVNC patients with a mutation in the cardiac transcription factor TBX20 revealed reduced baseline proliferative capacity by approximately 50%, which recapitulates the cell cycle defects thought to play a role in LVNC pathogenesis. Additionally, the LVNC iPSC-CM model was associated with perturbed transforming growth factor beta signalling through PRDM16, a gene in which specific mutations are known to cause LVNC.129
3.7 Hypoplastic left heart syndrome
Three iPSC-CM models of hypoplastic left heart syndrome (HLHS)130–132 have been established. HLHS is a rare congenital heart defect (CHD) that is characterized by a severely underdeveloped left heart. HLHS iPSC-CMs showed an immature cellular phenotype and a more foetal-like gene expression profile, consistent with observations in vivo.133 In one study mutations in the MYH6 gene were correlated to defective heart development, as they documented defective cardiomyogenesis in heterozygous MYH6 mutated iPSC lines,131 whereas a more recent study found several NOTCH mutations and showed re-activation of NOTCH signalling to partly rescue the in vitro disease phenotype.132
3.8 GATA4 congenital heart disease
CHDs are the most prevalent of all human developmental malformations with an incidence of 6–8 per 1000 live births.134 Atrial and ventricular septum defects are the most common type of CHD, present in 50% of all cases of CHD. Mutations in GATA4, a cardiac transcription factor, have been shown to cause cardiac septal defects and cardiomyopathy.135 Mechanistically, the GATA4 G296S mutation results in a reduced affinity for its binding element, decreased transcriptional activity, and disrupted interaction with Tbx5. Using iPSC-CMs, this mutation was shown to impair contractility, calcium handling, and metabolic activity, through disruption of transcriptional cooperativity.136 Namely, the mutation disrupted (i) TBX5 recruitment, associated with dysregulation of genes related to cardiac septation, (ii) GATA4 and TBX5-mediated repression at non-cardiac genes, and (iii) enhanced open chromatin states at endothelial/endocardial promoters.
3.9 Monogenic diseases with cardiac traits
3.9.1 RASopathies: cardiofaciocutaneous syndrome and Noonan syndrome with multiple lentigines
The genes that are mutated in cardiofaciocutaneous syndrome (CFCS) and Noonan syndrome with multiple lentigines (NSML) (formerly known as LEOPARD syndrome) encode proteins that function in the RAS/MAP kinase pathway. CFCS is characterized by the simultaneous occurrence of multiple congenital abnormalities and mental retardation. Along with e.g. macrocephaly and hyperkeratosis, most patients present with CHDs like pulmonic stenosis or HCM.137 To date, around 60 cases of CFCS have been published,138 and two BRAF mutations were characterized using iPSC-CMs, where an upregulation of a foetal gene programme, including ANP, was shown as well as arrhythmogenicity.139,140 The model also showed the involvement of fibroblasts in developing the pathognomonic cardiomyocyte phenotype.139 NSML is another very rare inherited disorder, with approximately 85% of patients carrying a mutation in the PTPN11 gene and commonly developing HCM, in addition to many other disease features. The HCM phenotype was recapitulated in an NSML iPSC-CM model where cardiomyocyte enlargement and preferential nuclear localization of NFATc4 was shown.10
3.9.2 Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is a severe X-linked neuromuscular disorder that affects approximately 1 in 3500 new-born males. DMD is caused by mutations in dystrophin, which encodes a rod-shaped cytoplasmic protein that anchors the sarcomere to the extracellular matrix. DMD usually presents in early childhood with progressive muscle weakness. Because of therapeutic improvements for DMD, patients have a longer lifespan, but resulting in cardiomyopathy to become a prevalent cause of mortality.141 The cardiac phenotype observed in DMD patients includes arrhythmias, structural alterations and haemodynamic abnormalities, and patients still die young.
Nine different mutations in the dystrophin gene, affecting different isoforms of the protein, have been studied using iPSC-CM models.142–146 Part of the initial work done on DMD iPSC-CMs models focused on gene expression and its rescue.142,143 CM differentiation was observed to be less efficient in the DMD cell lines,143 and iPSC-CMs showed increased cellular damage, apoptosis, and altered Ca2+ handling.144,145 The use of 3D-engineered cardiac tissues has shed some light on the impaired response to external cues in DMD iPSC-CMs.146 Guan et al.144 found elevated cardiac injury markers in vitro with DMD iPSC-CMs, which is in agreement with clinical data.147
3.9.3 Other myopathies
Myotonic Dystrophy Type 1 (DM1) is a genetic multisystem disorder with a minimum prevalence of 8–10 in 100 000.148 DM1 can occur from birth to old age, and symptoms can vary significantly between patients, from minor muscle pain to severe muscle weakness, respiratory issues, myotonia, and cardiac conduction defects. DM1 results from an unstable trinucleotide (CTG) repeat expansion within the 3’ end of the dystrophia myotonica protein kinase (DMPK) gene.149 As there is no effective therapy, a recent study aimed to develop genome therapy in DM1 iPSCs to reverse the phenotype for developing autologous stem cell therapy for DM1.150 While DM1 iPSC-CMs showed presence of nuclear RNA foci and aberrant splicing of MBNL1/2, insulin receptor, and cardiac troponin T, removal of mutant transcripts by genome-treatment in DM1 iPSC-CMs led to reversal of the disease phenotype.
Another gene related to multisystem disorders featuring cardiomyopathy and skeletal myopathy is HSPB5, encoding a small heat shock protein.151 The 343delT mutation has been shown to cause early-onset skeletal myopathy presenting with limb weakness and hypotonia in early childhood. In an iPSC-CM model of early-onset skeletal myopathy, aggregation of insoluble mutated protein and induction of a cellular stress response was observed.152In vitro refolding of 343delT in the presence of wild-type rescued its solubility, concordant with the recessive inheritance of the disease.
3.9.4 Friedreich’s ataxia
Friedreich’s ataxia (FRDA) is an autosomal recessive disorder causing neurodegeneration and cardiomyopathy. It is estimated to occur in up to 1 in 20 000 Caucasians, and is caused by an excessively expanded GAA repeat within the first intron of the frataxin gene, leading to epigenetic silencing of the gene. Frataxin is a small mitochondrial protein involved in iron sulphur cluster biosynthesis. FRDA usually presents with progressive ataxia between 10 and 15 years of age, causing dysarthria, muscle weakness and spasticity. Around two-thirds of all FRDA patients also suffer from cardiomyopathy.153
In FRDA iPSC-CMs, fibrillary disarray, mitochondrial damage, and calcium handling deficiency were observed under standard culture conditions.154,155 However, challenging the iPSC-CMs with increasing concentrations of iron led to hypertrophy, mitochondrial damage, and Ca2+ handling disruptions.156,157 Deferiprone, a drug counteracting iron overload in β-thalassemia, was reported to relieve stress-stimulation in FRDA iPSC-CMs,157 consistent with clinical studies reporting a reduction in heart hypertrophy in FRDA patients.158 However, the GAA repeat length reportedly changed during iPSCs generation,154 which has to be taken into account when directly comparing the in vitro findings to the clinical phenotype.
3.9.5 Barth syndrome
Barth syndrome (BTHS) is caused by a mutation in the TAZ1 gene coding for tafazzin, a CoA-independent phospholipid acyltransferase. This enzyme is necessary for the maturation of cardiolipin, a component of the inner mitochondrial membrane. The mutation results in a multidimensional phenotype: DCM, skeletal myopathy, neutropenia, growth retardation, and increased urinary excretion of 3-methylglutaconic acid in early childhood.159 So far, one BTHS iPSC-CM model exists.160 Using modified RNA to rescue TAZ1 expression and CRISPR/Cas9-based scarrless genome editing to correct the TAZ1 mutation, this study showed that the TAZ1 mutation was causal for the biochemical phenotype including cardiolipin levels, mitochondrial and ATP deficits, as well as of the functional cellular phenotype, including sarcomeric disarray and diminished contractility. Normalizing mitochondrial reactive oxygen species (ROS) production could attenuate this phenotype, and the therapeutic agent bromoenol lactone increased cellular cardiolipin levels, thereby improving sarcomere organization as well as twitch strength.
3.9.6 Storage diseases: Pompe, Fabry, and Danon disease
In the US, around 1 in 40 000 new-borns have Pompe disease,161 also called Glycogen Storage disease Type II. It is caused by an autosomal recessive mutation in the acid alpha-glucosidase (GAA) gene, encoding for a lysosomal glycogen-degrading enzyme. Depending on the residual function in the mutated GAA gene, there are early and late onset forms. The classic clinical presentation of infantile-onset Pompe disease includes generalized muscle weakness as well as cardiomegaly and HCM.162 Enzyme replacement therapy is the golden standard, using recombinant human GAA (rhGAA). The first successful iPSC generation from an early onset Pompe disease patient was not possible without the introduction of an inducible GAA transgene,163 whereas two other studies were able to generate early and late onset Pompe iPSCs without the addition of GAA.164,165 The early onset Pompe disease iPSC-CMs were shown to have the typical cellular Pompe phenotype, including large glycogen-containing vacuoles, multiple large lysosomes, and autophagosomes as well as deteriorating mitochondria, in close agreement with previous histological findings.166 RhGAA was able to partially rescue this phenotype in Pompe iPSC-CMs. Research on a late-onset Pompe cell line recapitulated part of the aforementioned findings and reported a successful attenuation of the cellular phenotype using a lentiviral GAA rescue.164 A more hypertrophic phenotype, as expected with Pompe disease cardiomyocytes, could not be observed, which could partly be due to the immaturity of the examined iPSC-CMs. Notably, Raval et al.165 were able to detect protein hypoglycosylation in Pompe iPSC-CMs, which is associated with altered Ca2+ handling167 that, in turn, may cause cardiac hypertrophy.120
Fabry disease, another storage disease, is caused by an X-linked alpha-galactosidase A (GLA) deficiency, leading to progressive build-up of globotriaosylceramide (GL-3) in lysosomes. Clinically, the deficiency results in renal failure, left ventricular hypertrophy, and possibly strokes.168 Similar to Pompe disease, enzyme replacement therapy is the current treatment option. In the first Fabry iPSC-CM model,169 low-GLA activity and the accumulation of GL-3 was shown to cause abnormal sarcomeric structures, consistent with observations in endomyocardial biopsies.170 Glucosylceramide synthase inhibition and recombinant GLA were shown to both prevent and reverse the cellular phenotype. Subsequent studies of Fabry iPSC-CMs showed cellular hypertrophy, impaired contractility, decreased metabolism,171 and increased IL-18 levels in both iPSC-CMs and patient sera, in parallel with LV hypertrophy progression.172 Importantly, IL-18 neutralization reduced progression of hypertrophy in vitro.
Danon disease (DD) is an X-linked lysosomal and glycogen storage disorder characterized by impaired autophagy caused by mutations in the LAMP2 gene, encoding the lysosomal-associated membrane protein Type 2. Patients present with severe cardiac and skeletal muscle abnormalities resulting in HCM, heart failure and sudden cardiac death.173 In an iPSC-CM model of DD, the impairment of autophagic flux, mitochondrial damage, and increased apoptosis were observed.174 Additionally, DD iPSC-CMs were significantly larger and exhibited increased calcium decay times compared with healthy iPSC-CMs, thereby recapitulating the hypertrophy and decrease of contractile function seen in DD patients. While DD causes early-onset fatal cardiomyopathy in male patients, female patients show a later onset and less severe clinical phenotype, which has been attributed to random inactivation of the LAMP2 gene on the X chromosome.175 Indeed, a recent study using DD iPSC-CMs provided evidence that this random inactivation is responsible for the phenotype in female patients.176
3.9.7 Other diseases with cardiac traits
iPSC-CMs have also been used to model familial transthyretin amyloidosis (ATTR), an autosomal-dominant protein-folding disorder leading to multi-organ failure and death.177 Hepatocytes of ATTR patients with mutations in the transthyretin (TTR) gene produce mutant TTR that forms aggregates and fibrils in target organs, mainly the heart and peripheral nervous system. Patient specific iPSCs were differentiated into hepatic, neuronal, and cardiac lineages thereby modelling the three major tissue types involved in this disease. The demonstration that hepatic secreted mutant TTR decreased iPSC-CM cell survival, supports the use of patient-specific iPSC as models for studying disorders affecting multiple organs.
An iPSC model of abetalipoproteinemia (ABL) has also been used to illustrate a disease involving multiple organ.178 ABL or Bassen-Kornzweig syndrome, is a rare autosomal recessive disorder of lipoprotein metabolism, resulting from mutations in the gene encoding the microsomal triglyceride transfer protein (MTTP).179 In addition to expression in the liver and intestine, MTTP is also expressed in cardiomyocytes, a finding that may explain why several ABL patients exhibit cardiac arrhythmias and heart failure.180 ABL iPSC-CMs failed to secrete apoB and show elevated lipid storage defects, associated with ABL disease, and are hypersensitive to metabolic stress.178
iPSC-CM models may also be useful to potentially improve risk management of coronary artery disease (CAD), as shown by a study using iPSCs with an aldehyde dehydrogenase 2 (ALDH2) mutation (ALDH2*2), which is linked to an increased risk of CAD and more severe outcomes, and occurs in 8% of the human population.181,182 It was shown that the ALDH2*2 mutation resulted in elevated ROS and toxic aldehydes, inducing cell cycle arrest and apoptosis, especially during ischaemia.
3.10 Patient-independent applications of iPSC-CMs
Next to monogenetic diseases affecting the heart, several studies have established iPSC-CM models for acquired cardiac diseases as well. Diabetic cardiomyopathy was modelled by switching fatty acid-adapted wild-type iPSC-CMs to a glucose-rich medium containing endothelin 1 and cortisol, resulting in induction of a diabetic stress phenotype, showing hypertrophy, lipid accumulation, and peroxidation.183 An iPSC-CM model of Takotsubo cardiomyopathy has also been established, which showed that enhanced β-adrenergic signalling and a higher sensitivity to catecholamine-induced toxicity are associated with the disease phenotype.184 In addition, iPSC-CM disease modelling has been successfully extended towards infectious diseases affecting the heart, namely coxsackievirus-induced viral myocarditis,185,186 and endotoxin-induced inflammation,187 showing that iPSC-CMs possess the reaction system involved in both viral and endotoxin-induced inflammation. In the viral myocarditis models, multiple drugs, including Interferon β1, but not Interferon α, were shown to attenuate the infection by abrogating virus proliferation in vitro.
iPSC-CM models have been used successfully to recapitulate cardiotoxic drug effects, as reviewed in greater detail elsewhere.188 A deeper insight into the individual susceptibility of breast cancer patients to doxorubicin-induced cardiotoxicity (DIC) was provided using patient specific iPSC-CMs.189 However, the use of this model for the discovery of novel DIC cardioprotectants might be limited, as one of two tested cardioprotectants increased toxicity, possibly due to the use of immature iPSC-CMs. In another study, the effect of the drug combination Sofosbuvir and Amiodarone, known to cause severe bradycardia in vivo, was studied using iPSC-CMs. Indeed, this combination, among others, was shown to cause impaired Ca2+ handling in vitro.190 iPSC-CMs have also been used to screen for cardiovascular toxicities associated with anticancer tyrosine kinase inhibitors.191 Importantly, the results correlated with clinical phenotypes and this study led to the finding that toxicity can be alleviated via cardioprotective insulin/IGF signalling. Additionally, efforts to establish alcoholic cardiomyopathy and peripartum cardiomyopathy iPSC-CM models are ongoing.
4. Discussion
Using human iPSC-CMs for disease modelling has certainly made progress in recent years with more than 25 iPSC lines carrying new disease specific mutations published every year since 2013 (Supplementary material online, Table S1). Currently, as shown in our review, over 150 cardiac disease specific iPSC lines have been described, some available from repositories and cell banks. The relevance of using these iPSC-CMs as disease models is well established as they reflect pathophysiological characteristics of a broad range of inherited cardiac diseases.
iPSC-CM models of LQTS, CPVT, and ARVC mimicked the electrophysiological abnormalities and drug responses seen in these inherited arrhythmic diseases which emphasizes the potential use of these models to develop patient-specific clinical regimens and as drug screening platforms. A striking example of the predictive value is the susceptibility to arrhythmogenic effects of cisapride seen in LQTS (and HCM) iPSC-CMs, which was previously observed in patients and the reason to withdraw this drug from the market.27 In another study, a panel of genetically diverse iPSC-CMs reproduced the patients susceptibility to develop a cardiotoxic response to sotalol, a QT-prolonging drug.192 Moreover, an iPSC-CM model of LQTS3 could be treated with a combined pacing and pharmacological approach similar to the treatment regimen used in these patients.47
iPSC-CM models of DCM and HCM also agreed well with in vivo characteristics, showing impaired Ca2+ handling, decreased contractile force, and sarcomeric disarray in DCM, and Ca2+ handling irregularities, cardiomyocyte hypertrophy, and arrhythmias in HCM. Additionally, iPSC-CM models of LQTS, DCM, HCM, LVNC, CHD, Pompe disease, NSML, and Takotsubo cardiomyopathy enabled researchers to gain new insights on the molecular pathogenesis leading to the disease phenotype observed in these models and patients, providing potential new leads for individualized therapies.10,44,104,119–121,136,165,184,193 As one example, a DCM TNNT2 R137W iPSC-CM model showed epigenetic activation of phosphodiesterase (PDE) expression, a newly discovered pathological mechanism in this DCM subtype, presenting the potential of using PDE inhibitors in these patients.104
Genome editing has particularly helped to progress iPSC-CM disease modelling. In several disease models, genome editing was used to reverse mutated, disease causing genes to the healthy wild type,28,106,110,113,150 validating the pathogenicity of the mutation by showing a reversal of the disease phenotype, and providing an ideal diseased cell to healthy cell comparison to gain novel insight on the involved signalling pathways. Moreover, these studies lay the groundwork for developing targeted gene therapy for patients with hereditary cardiac disease.
In addition to the successes of inherited cardiac disease models, iPSC-CM models of anti-cancer related drug toxicity produced novel insight on how to combat drug toxicity, provided a cardiac safety index for tyrosine kinase inhibitors,191 and showed patient-specific vulnerability to doxorubicin to potentially guide decision making of using doxorubicin.189
To summarize the progress, the presented studies have shown that iPSC-CM disease models reflect important clinical phenotypes and can be used to elucidate the molecular mechanisms and cellular phenotypes of a large variety of cardiac diseases. Moreover, iPSC-CM models can be used to predict patient responses to novel or existing drugs.
However, despite the progress, pitfalls are still present and several limitations and challenges need to be considered when using iPSC-CMs for cardiac disease modelling.
Firstly, patient characteristics and the choice of cell type for reprogramming will influence individual iPSC line characteristics and hence the quality as a disease model system. It has been postulated that while the pathogenic mutation is the ‘first hit’, age, gender, ethnicity, comorbidities, and environmental factors, or so-called ‘second hits’ may play an important role in developing a clinical phenotype.194 For example, ethnicity can modulate the clinical presentation of HCM,195 as well as show differences in basic muscle metabolism as measured by CK levels.196 Most papers do not include detailed patient characteristics, and the ‘second hit’ hypothesis has yet to be tested in iPSC-CM models.
The cell type used for generation of the iPSC line can also influence characteristics of iPSC-CM modelling of cardiac disease, since the original cell type-specific epigenetic pattern can persist in reprogrammed iPSC lines, and differentiation efficiency is affected.197,198 In the studies covered by our review, iPSCs were generated either from skin biopsies, urine or blood samples, but none of the studies assessed potential differences between iPSCs, and derivative iPSC-CMs, generated from different cell types of the same donor. Furthermore, the X-inactivation status in female iPSCs, which can either be retained or erased during the generation of iPSCs, depending on the protocol used,199 has not been assessed.
Secondly, the choice of controls is another critical aspect when assessing and comparing results from studies using iPSC-CM models. Some papers have used asymptomatic carriers as controls, which offer some insight in the importance of second hits. Most often the cellular phenotype of iPSC-CM models has been compared with healthy control iPSC-CMs, sometimes including (gender-matched) family members, to ensure a similar genomic background. Isogenic control iPSC lines, generated by genome editing, so far are the ideal controls as they have the same genomic background of the original disease model iPSC line and provide a relevant and true ‘healthy control’.31,42
Thirdly, there is considerable variability in protocols for differentiation and characterization of iPSC-CMs (Supplementary material online, Table S1). This may lead to observed variances in differentiation efficacy, defined as the ratio of cardiomyocyte marker positive cells at a specific time point of the differentiation protocol, and kinetics, defined as the time point in the differentiation protocol for the emergence of a specific cardiomyocyte marker, e.g. spontaneous beating. iPSC-CM models also differ frequently by the cardiomyocyte subtype generated, with varying ratios of ventricular, atrial, and nodal cells, affecting comparability of essential read-outs, like electrophysiology and contractile force. Depending on the disease studied, culture conditions for iPSC-CMs can affect outcomes notably. The arrhythmic phenotype of CPVT is present only when stimulated with adrenergic agonists, comparable to the clinical phenotype, whereas the ARVC iPSC-CM phenotype is not as overt at baseline as after stimulation with adipogenic medium.
Finally, we have compared assays used to assess cardiomyocyte maturation in the different studies. Only few studies compare iPSC-CM gene expression levels to adult cardiac tissue and assess sarcomeric and mitochondrial organization or other morphological features (Supplementary material online, Table S1). Although electrophysiological properties of spontaneously beating iPSC-CMs have been measured to some extent, spontaneous beating is not standard in adult cardiomyocytes. Preferably, iPSC-CMs should be paced to analyse potential arrhythmogenicity of their respective mutations. Furthermore, with several specific cardiac diseases manifesting exclusively during adulthood, the relatively immature iPSC-CMs might only partly recapitulate the disease phenotypes. Therefore, assessment of maturation of iPSC-CMs used for cardiac disease modelling is advisable, and we have listed key characteristics and related recommended measurements in Table 1.
Table 1.
Key characteristics of iPSC-CM maturation and related recommended measurements to asses these characteristics
| Characteristics | Recommended measurement |
|---|---|
| Cardiomyocyte-specific gene expression | Transcriptome analysis |
| Methylation patterns | |
| Structural features | Sarcomere number, density, and organization |
| Length, shape, and % binucleated | |
| Mitochondrial volume and morphology | |
| Electrophysiological phenotypes | Action potential profile |
| Ion current densities and gating properties | |
| Cardiomyocyte subtype determination | |
| Calcium handling and contractility | Ability to contract; spontaneous vs. stimulation |
| Calcium transients, sparks, and waves | |
| Contractile force | |
| Metabolic profile | Lipid oxidation vs. glycolysis |
| Oxygen consumption and maximum respiration | |
| Response to sympathetic stimulation | Positive inotropic, chronotropic, and lusitropic effects |
| Electrical coupling | Gap junction localization, density, and composition Conduction velocity |
As outlined so far, significant progress has been made with iPSC-CM disease models, while several pitfalls still need attention. Several suggestions to improve and exploit the full potential are currently discussed200 and several research groups have started to implement them.
In order to make results reproducible and inter-comparable between research groups, standardization of protocols for the generation of iPSC lines and iPSC-CMs is essential. Standardization is needed on various methodological levels, from selecting patients (as cell donors), cell types and reprogramming protocols, in order to minimize confounding effects on the iPSC lines and thus potential differences in derivative iPSC-CMs, to protocols for directed differentiation and characterization of iPSC-CMs. Only if studies use iPSC-CM populations of comparable purity (CMs vs. non-CMs), subtype (atrial, ventricular, nodal), and maturity, robust and eventually more clinically relevant results can be acquired.
Additionally, to increase rigidity of cardiac disease modelling studies, not only multiple patients as donors of cells to generate iPSC lines, but also multiple cell lines from each reprogramming201 should be included to account for variability between individual patients with different genetic background and comorbidities, and between different clonal lines with potentially different cellular characteristics arising from stochastic reprogramming events, especially regarding epigenetics.202
For modelling more complex diseases involving multiple cell types and specific spatial organization, like ARVC, which is characterized by fibro-fatty depositions inside the myocardial tissue and crosstalk between different cell types,89 an increase in model complexity from a homogeneous 2D cell layer to 3D structures with multiple cell types will be needed.203,204 These can be potentially generated by the same iPSC line, leading to patient specific myocardial tissue engineering, but also could use a mixture of different backgrounds, e.g. diseased and healthy. Recently, the necessity of complex models was shown for a specific type of DCM linked to mutated titin, with engineered heart tissues, composed as 3D structures of iPSC-CMs and stromal cells in a collagen Type I/fibrinogen matrix, showing the typical contractile deficient phenotype, whereas the phenotype was absent in the 2D iPSC-CM model.110
Finally, especially if standardization issues have been addressed, iPSC lines and iPSC-CMs from various disease specific backgrounds should be collected, validated, stored and made accessible to researchers205,206 to build a comprehensive and cohesive body of knowledge on inherited cardiac disease. This will lead to improved disease models and generation of a critical amount of reproducible and robust data to draw clinically relevant conclusions, which would otherwise rely on animal and first-in-man studies, without having assessed the fundamental disease mechanisms and potential new therapies in a valid human, cardiac disease specific modelling system, as represented by iPSC-CMs.
5. Conclusion
Patient-specific iPSC-CM models have demonstrated the ability to facilitate the study of many aspects of the clinical disease phenotype and are particularly useful in elucidating the molecular mechanisms of certain cardiac diseases. However, major differences between the complex in vivo architecture and pathophysiology and simplified in vitro conditions can result in limitations for recapitulating functional mechanisms. It is evident that the use of iPSC-CMs for disease modelling requires a set of criteria for thorough characterization, which we have listed in Table 1. Additionally, future optimization of iPSC-CM maturation, functional tissue engineering and culture conditions should lead to the establishment of more predictive iPSC-CM disease models that more closely mimic the disease. Finally, a consensual multi-dimensional approach to standardize generation of iPSC, differentiation to iPSC-CM, functional characterization of iPSC-CM and to define valid controls is necessary to generate more robust and reproducible iPSC-CM disease models.
Conflict of interest: none declared.
Funding
This work was supported by Horizon2020 European Research Council 2016 Consolidator Grant EVICARE (725229), Technobeat (668724), the Project SMARTCARE-II of the BioMedicalMaterials institute, co-funded by the ZonMw-TAS programme (#116002016) (J.P.G.S.); the Dutch Ministry of Economic Affairs, Agriculture and Innovation and the Netherlands CardioVascular Research Initiative (CVON): the Dutch Heart Foundation, Dutch Federations of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Sciences (P.A.D.); the NIH Director’s Pioneer Award (DP1 LM012179-04), the American Heart Association Established Investigator Award, and the Endowed Faculty Scholar Award of the Lucile Packard Foundation for Children and Child Health Research Institute at Stanford (S.M.W.); and UCL Hospitals NIHR Biomedical Research Centre (F.W.A.).
Supplementary Material
References
- 1. MacRae CA, Vasan RS.. The future of genetics and genomics. Circulation 2016;133:2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Papadatos GA, Wallerstein PMR, Head CEG, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AEO, Huang CL-H, Vandenberg JI, Colledge WH, Grace AA.. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci U S A 2002;99:6210–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yutzey KE, Robbins J.. Principles of genetic murine models for cardiac disease. Circulation 2007;115:792–799. [DOI] [PubMed] [Google Scholar]
- 4. Hackam DG, Redelmeier DA.. Translation of research evidence from animals to humans. JAMA 2006;296:1727–1732. [DOI] [PubMed] [Google Scholar]
- 5. Sallam K, Li Y, Sager PT, Houser SR, Wu JC.. Finding the rhythm of sudden cardiac death: new opportunities using induced pluripotent stem cell-derived cardiomyocytes. Circ Res 2015;116:1989–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takahashi K, Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 7. Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Belmonte JCI.. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol 2008;26:1276–1284. [DOI] [PubMed] [Google Scholar]
- 8. Seki T, Yuasa S, Fukuda K.. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc 2012;7:718–728. [DOI] [PubMed] [Google Scholar]
- 9. Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, Hao L, Chan Y-C, Ng K-M, Ho JC, Wieser M, Wu J, Redl H, Tse H-F, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA.. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol 2011;22:1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang Y-S, Schaniel C, Lee D-F, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR.. Patient-specific induced pluripotent stem cell derived models of LEOPARD syndrome. Nature 2010;465:808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M.. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 2009;85:348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu J, Vodyanik MA, Smuga-Otto K, Ntosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA.. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 13. Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ.. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010;7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, Brink S, van den Hassink R, Heyden M, van der Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L.. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation 2003;107:2733–2740. [DOI] [PubMed] [Google Scholar]
- 15. Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ.. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res 2009;104:e30–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O'Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE.. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 2007;25:1015–1024. [DOI] [PubMed] [Google Scholar]
- 17. Burridge PW, Keller G, Gold JD, Wu JC.. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 2012;10:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batalov I, Feinberg AW.. Differentiation of cardiomyocytes from human pluripotent stem cells using monolayer culture. Biomark Insights 2015;10:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C.. Transcriptional landscape of cardiomyocyte maturation. Cell Rep 2015;13:1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Remedios C, Dos Malm T, Andrä M, Jashari R, Nyengaard JR, Possnert G, Jovinge S, Druid H, Frisén J.. Dynamics of cell generation and turnover in the human heart. Cell 2015;161:1566–1575. [DOI] [PubMed] [Google Scholar]
- 21. Lieu DK, Liu J, Siu C-W, McNerney GP, Tse H-F, Abu-Khalil A, Huser T, Li RA.. Absence of transverse tubules contributes to non-uniform Ca2+ wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev 2009;18:1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hwang HS, Kryshtal DO, Feaster TK, Sánchez-Freire V, Zhang J, Kamp TJ, Hong CC, Wu JC, Knollmann BC.. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. J Mol Cell Cardiol 2015;85:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veerman CC, Kosmidis G, Mummery CL, Casini S, Verkerk AO, Bellin M.. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev 2015;24:1035–1052. [DOI] [PubMed] [Google Scholar]
- 24. Nakano Y, Shimizu W.. Genetics of long-QT syndrome. J Hum Genet 2016;61:51–55. [DOI] [PubMed] [Google Scholar]
- 25. Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker DD, Schömig A, Laugwitz K-L.. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 2010;363:1397–1409. [DOI] [PubMed] [Google Scholar]
- 26. Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T, Kuroda Y, Yae K, Hashimoto H, Tanaka T, Hattori F, Sato T, Miyoshi S, Takatsuki S, Murata M, Kurokawa J, Furukawa T, Makita N, Aiba T, Shimizu W, Horie M, Kamiya K, Kodama I, Ogawa S, Fukuda K.. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res 2012;95:419–429. [DOI] [PubMed] [Google Scholar]
- 27. Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Wang PJ, Nguyen PK, Bers DM, Robbins RC, Wu JC.. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013;127:1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, Hu S, Kay MA, Urnov FD, Shinnawi R, Gold JD, Gepstein L, Wu JC.. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol 2014;64:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma D, Wei H, Lu J, Huang D, Liu Z, Loh LJ, Islam O, Liew R, Shim W, Cook SA.. Characterization of a novel KCNQ1 mutation for type 1 long QT syndrome and assessment of the therapeutic potential of a novel IKs activator using patient-specific induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther 2015;6:39.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kiviaho AL, Ahola A, Larsson K, Penttinen K, Swan H, Pekkanen-Mattila M, Venäläinen H, Paavola K, Hyttinen J, Aalto-Setälä K.. Distinct electrophysiological and mechanical beating phenotypes of long QT syndrome type 1-specific cardiomyocytes carrying different mutations. Int J Cardiol Heart Vasc 2015;8:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sala L, Yu Z, Ward-van Oostwaard D, Veldhoven JPD, van Moretti A, Laugwitz K, Mummery CL, IJzerman AP, Bellin M.. A new hERG allosteric modulator rescues genetic and drug—induced long-QT syndrome phenotypes in cardiomyocytes from isogenic pairs of patient induced pluripotent stem cells. EMBO Mol Med 2016;8:1065–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuusela J, Kujala VJ, Kiviaho A, Ojala M, Swan H, Kontula K, Aalto-Setälä K.. Effects of cardioactive drugs on human induced pluripotent stem cell derived long QT syndrome cardiomyocytes. Springerplus 2016;5:234.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuusela J, Kim J, Räsänen E, Aalto-Setälä K.. The effects of pharmacological compounds on beat rate variations in human long QT-syndrome cardiomyocytes. Stem Cell Rev Rep 2016;12:698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuusela J, Larsson K, Shah D, Prajapati C, Aalto-Setälä K.. Low extracellular potassium prolongs repolarization and evokes early afterdepolarization in human induced pluripotent stem cell-derived cardiomyocytes. Biol Open 2017;6:777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwartz PJ, Spazzolini C, Crotti L, Bathen J, Amlie JP, Timothy K, Shkolnikova M, Berul CI, Bitner-Glindzicz M, Toivonen L, Horie M, Schulze-Bahr E, Denjoy I.. The Jervell and Lange-Nielsen syndrome: natural history, molecular basis, and clinical outcome. Circulation 2006;113:783–790. [DOI] [PubMed] [Google Scholar]
- 36. Zhang M, D’Aniello C, Verkerk AO, Wrobel E, Frank S, Ward-van Oostwaard D, Piccini I, Freund C, Rao J, Seebohm G, Atsma DE, Schulze-Bahr E, Mummery CL, Greber B, Bellin M.. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc Natl Acad Sci USA 2014;111:E5383–E5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L.. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 2011;471:225–229. [DOI] [PubMed] [Google Scholar]
- 38. Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C.. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J 2011;32:952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spencer CI, Baba S, Nakamura K, Hua EA, Sears MAF, Fu CC, Zhang J, Balijepalli S, Tomoda K, Hayashi Y, Lizarraga P, Wojciak J, Scheinman MM, Aalto-Setälä K, Makielski JC, January CT, Healy KE, Kamp TJ, Yamanaka S, Conklin BR.. Calcium transients closely reflect prolonged action potentials in iPSC models of inherited cardiac arrhythmia. Stem Cell Rep 2014;3:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jouni M, Si-Tayeb K, Es-Salah-Lamoureux Z, Latypova X, Champon B, Caillaud A, Rungoat A, Charpentier F, Loussouarn G, Baró I, Zibara K, Lemarchand P, Gaborit N.. Toward personalized medicine: using cardiomyocytes differentiated from urine-derived pluripotent stem cells to recapitulate electrophysiological characteristics of type 2 long QT syndrome. J Am Heart Assoc 2015;4:e002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lahti AL, Kujala VJ, Chapman H, Koivisto A-P, Pekkanen-Mattila M, Kerkela E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setala K.. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech 2012;5:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bellin M, Casini S, Davis RP, D'Aniello C, Haas J, Ward-van Oostwaard D, Tertoolen LGJ, Jung CB, Elliott DA, Welling A, Laugwitz K-L, Moretti A, Mummery CL.. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J 2013;32:3161–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, Kemp PJ, Staniforth A, Mellor I, Denning C.. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J 2014;35:1078–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehta A, Sequiera GL, Ramachandra CJA, Sudibyo Y, Chung Y, Sheng J, Wong KY, Tan TH, Wong P, Liew R, Shim W.. Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc Res 2014;102:497–506. [DOI] [PubMed] [Google Scholar]
- 45. Mura M, Mehta A, Ramachandra CJ, Zappatore R, Pisano F, Ciuffreda MC, Barbaccia V, Crotti L, Schwartz PJ, Shim W, Gnecchi M.. The KCNH2-IVS9-28A/G mutation causes aberrant isoform expression and hERG trafficking defect in cardiomyocytes derived from patients affected by long QT syndrome type 2. Int J Cardiol 2017;240:367–371. [DOI] [PubMed] [Google Scholar]
- 46. Moss AJ, Zareba W, Kaufman ES, Gartman E, Peterson DR, Benhorin J, Towbin JA, Keating MT, Priori SG, Schwartz PJ, Vincent GM, Robinson JL, Andrews ML, Feng C, Hall WJ, Medina A, Zhang L, Wang Z.. Increased risk of arrhythmic events in long-QT syndrome with mutations in the pore region of the human ether-a-go-go-related gene potassium channel. Circulation 2002;105:794–799. [DOI] [PubMed] [Google Scholar]
- 47. Terrenoire C, Wang K, Chan Tung KW, Chung WK, Pass RH, Lu JT, Jean JC, Omari A, Sampson KJ, Kotton DN, Keller G, Kass RS.. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol 2013;141:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NBE, Tan TH, Wong KY, Shim W, Wong P, Cook SA, Liew R.. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol 2013;168:5277–5286. [DOI] [PubMed] [Google Scholar]
- 49. Fatima A, Kaifeng S, Dittmann S, Xu G, Gupta MK, Linke M, Zechner U, Nguemo F, Milting H, Farr M, Hescheler J, Saric T.. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS One 2013;8:e83005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malan D, Zhang M, Stallmeyer B, Müller J, Fleischmann BK, Schulze-Bahr E, Sasse P, Greber B.. Human iPS cell model of type 3 long QT syndrome recapitulates drug-based phenotype correction. Basic Res Cardiol 2016;111:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis RP, Casini S, Berg CW, Van Den Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW, Van Wilde AAM, Bezzina CR, Verkerk AO, Freund C, Mummery CL.. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 2012;125:3079–3091. [DOI] [PubMed] [Google Scholar]
- 52. Veerman CC, Mengarelli I, Lodder EM, Kosmidis G, Bellin M, Zhang M, Dittmann S, Guan K, Wilde AAM, Schulze-Bahr E, Greber B, Bezzina CR, Verkerk AO.. Switch from fetal to adult SCN5A isoform in human induced pluripotent stem cell–derived cardiomyocytes unmasks the cellular phenotype of a conduction disease–causing mutation. J Am Heart Assoc 2017;6:e005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Okata S, Yuasa S, Suzuki T, Ito S, Makita N, Yoshida T, Li M, Kurokawa J, Seki T, Egashira T, Aizawa Y, Kodaira M, Motoda C, Yozu G, Shimojima M, Hayashiji N, Hashimoto H, Kuroda Y, Tanaka A, Murata M, Aiba T, Shimizu W, Horie M, Kamiya K, Furukawa T, Fukuda K.. Embryonic type Na+ channel β-subunit, SCN3B masks the disease phenotype of Brugada syndrome. Sci Rep 2016;6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Portero V, Casini S, Hoekstra M, Verkerk AO, Mengarelli I, Belardinelli L, Rajamani S, Wilde AAM, Bezzina CR, Veldkamp MW, Remme CA.. Anti-arrhythmic potential of the late sodium current inhibitor GS-458967 in murine Scn5a-1798insD+/- and human SCN5A-1795insD+/- iPSC-derived cardiomyocytes. Cardiovasc Res 2017;113:829–838. [DOI] [PubMed] [Google Scholar]
- 55. Remme CA, Wilde AAM, Bezzina CR.. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med 2008;18:78–87. [DOI] [PubMed] [Google Scholar]
- 56. Veerman CC, Mengarelli I, Guan K, Stauske M, Barc J, Tan HL, Wilde AAM, Verkerk AO, Bezzina CR.. hiPSC-derived cardiomyocytes from Brugada syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci Rep 2016;6:30967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kosmidis G, Veerman CC, Casini S, Verkerk AO, Pas S, Van De Bellin M, Wilde AAM, Mummery CL, Bezzina CR.. Readthrough-promoting drugs gentamicin and PTC124 Fail to Rescue Na v 1.5 function of human-induced pluripotent stem cell-derived cardiomyocytes carrying nonsense mutations in the sodium channel gene SCN5A. Circ Arrhythm Electrophysiol 2016;9:e004227.. [DOI] [PubMed] [Google Scholar]
- 58. Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC.. Patient-specific and genome-edited induced pluripotent stem cell–derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol 2016;68:2086–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nademanee K, Raju H, Noronha SV, De Papadakis M, Robinson L, Rothery S, Makita N, Kowase S, Boonmee N, Vitayakritsirikul V, Ratanarapee S, Sharma S, Wal AC, Van Der Christiansen M, Tan HL, Wilde AA, Nogami A, Sheppard MN, Veerakul G, Behr ER.. Fibrosis, connexin-43, and conduction abnormalities in the Brugada syndrome. J Am Coll Cardiol 2015;66:1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nguyen H-L, Pieper GH, Wilders R.. Andersen–Tawil syndrome: clinical and molecular aspects. Int J Cardiol 2013;170:1–16. [DOI] [PubMed] [Google Scholar]
- 61. Kuroda Y, Yuasa S, Watanabe Y, Ito S, Egashira T, Seki T, Hattori T, Ohno S, Kodaira M, Suzuki T, Hashimoto H, Okata S, Tanaka A, Aizawa Y, Murata M, Aiba T, Makita N, Furukawa T, Shimizu W, Kodama I, Ogawa S, Kokubun N, Horigome H, Horie M, Kamiya K, Fukuda K.. Flecainide ameliorates arrhythmogenicity through NCX flux in Andersen-Tawil syndrome-iPS cell-derived cardiomyocytes. Biochem Biophys Rep 2017;9:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gillis J, Burashnikov E, Antzelevitch C, Blaser S, Gross G, Turner L, Babul-Hirji R, Chitayat D.. Long QT, syndactyly, joint contractures, stroke and novel CACNA1C mutation: expanding the spectrum of Timothy syndrome. Am J Med Genet A 2012;158A:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gez LS, Hagalili Y, Shainberg A, Atlas D.. Voltage-driven Ca2+ binding at the L-type Ca2+ channel triggers cardiac excitation-contraction coupling prior to Ca2+ influx. Biochemistry 2012;51:9658–9666. [DOI] [PubMed] [Google Scholar]
- 64. Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE.. Using iPS cells to investigate cardiac phenotypes in patients with Timothy syndrome. Nature 2011;471:230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Crotti L, Johnson CN, Graf E, Ferrari GM, De Cuneo BF, Ovadia M, Papagiannis J, Feldkamp MD, Rathi SG, Kunic JD, Pedrazzini M, Wieland T, Lichtner P, Beckmann BM, Clark T, Shaffer C, Benson DW, Kääb S, Meitinger T, Strom TM, Chazin WJ, Schwartz PJ, George AL.. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 2013;127:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rocchetti M, Sala L, Dreizehnter L, Crotti L, Sinnecker D, Mura M, Pane LS, Altomare C, Torre E, Mostacciuolo G, Severi S, Porta A, Ferrari GM, De George AL, Schwartz PJ, Gnecchi M, Moretti A, Zaza A.. Elucidating arrhythmogenic mechanisms of long-QT syndrome CALM1-F142L mutation in patient-specific induced pluripotent stem cell-derived cardiomyocytes. Cardiovasc Res 2017;113:531–541. [DOI] [PubMed] [Google Scholar]
- 67. Limpitikul WB, Dick IE, Tester DJ, Boczek NJ, Limphong P, Yang W, Choi MH, Babich J, Disilvestre D, Kanter RJ, Tomaselli GF, Ackerman MJ, Yue DT.. A precision medicine approach to the rescue of function on malignant calmodulinopathic long-QT syndrome. Circ Res 2017;120:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamamoto Y, Makiyama T, Harita T, Sasaki K, Wuriyanghai Y, Hayano M, Nishiuchi S, Kohjitani H, Hirose S, Chen J, Yokoi F, Ishikawa T, Ohno S, Chonabayashi K, Motomura H, Yoshida Y, Horie M, Makita N, Kimura T.. Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum Mol Genet 2017;26:1670–1677. [DOI] [PubMed] [Google Scholar]
- 69. Makita N, Yagihara N, Crotti L, Johnson CN, Beckmann B-M, Roh MS, Shigemizu D, Lichtner P, Ishikawa T, Aiba T, Homfray T, Behr ER, Klug D, Denjoy I, Mastantuono E, Theisen D, Tsunoda T, Satake W, Toda T, Nakagawa H, Tsuji Y, Tsuchiya T, Yamamoto H, Miyamoto Y, Endo N, Kimura A, Ozaki K, Motomura H, Suda K, Tanaka T, Schwartz PJ, Meitinger T, Kääb S, Guicheney P, Shimizu W, Bhuiyan ZA, Watanabe H, Chazin WJ, George AL.. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ Cardiovasc Genet 2014;7:466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Leenhardt A, Denjoy I, Guicheney P.. Catecholaminergic polymorphic ventricular tachycardia. Circ Arrhythm Electrophysiol 2012;5:1044–1052. [DOI] [PubMed] [Google Scholar]
- 71. Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, Rosa AO, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U, Beyer V, Christian H, Rosenkranz S, Klauke B, Abdul S, Haverkamp W, Pfitzer G, Farr M, Morad M, Milting H, Hescheler J, Šaric T, Nguemo F, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U.. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem 2011;28:579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, Lahtinen AM, Toivonen L, Kontula K, Swan H, Laine M, Silvennoinen O, Aalto-Setälä K.. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. PLoS One 2012;7:e44660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz KL.. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med 2012;4:180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L.. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol 2012;60:990–1000. [DOI] [PubMed] [Google Scholar]
- 75. Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O.. Cardiomyocytes generated from CPVT D307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med 2012;16:468–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, Nakahama H, Portararo P, Bloise R, Napolitano C, Condorelli G, Priori SG.. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis 2013;4:e843.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang X-H, Morad M.. Calcium signaling in human stem cell-derived cardiomyocytes: evidence from normal subjects and CPVT afflicted patients. Cell Calcium 2016;59:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Penttinen K, Swan H, Vanninen S, Paavola J, Lahtinen AM, Kontula K, Aalto-Setälä K.. Antiarrhythmic effects of dantrolene in patients with catecholaminergic polymorphic ventricular tachycardia and replication of the responses using iPSC models. PLoS One 2015;10:e0134746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Novak A, Barad L, Lorber A, Gherghiceanu M, Reiter I, Eisen B, Eldor L, Itskovitz-Eldor J, Eldar M, Arad M, Binah O.. Functional abnormalities in iPSC-derived cardiomyocytes generated from CPVT1 and CPVT2 patients carrying ryanodine or calsequestrin mutations. J Cell Mol Med 2015;19:2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang X-H, Haviland S, Wei H, Sarić T, Fatima A, Hescheler J, Cleemann L, Morad M.. Ca2+ signaling in human induced pluripotent stem cell-derived cardiomyocytes (iPS-CM) from normal and catecholaminergic polymorphic ventricular tachycardia (CPVT)-afflicted subjects. Cell Calcium 2013;54:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Paavola J, Väänänen H, Larsson K, Penttinen K, Toivonen L, Kontula K, Laine M, Aalto-Setälä K, Swan H, Viitasalo M.. Slowed depolarization and irregular repolarization in catecholaminergic polymorphic ventricular tachycardia: a study from cellular Ca2+ transients and action potentials to clinical monophasic action potentials and electrocardiography. Europace 2016;18:1599–1607. [DOI] [PubMed] [Google Scholar]
- 82. Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, Dalal A, Mceachin ZT, Rossoll W, Hales CM, Fischbach PS, Wagner MB, Xu C.. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis Model Mech 2016;9:927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sasaki K, Makiyama T, Yoshida Y, Wuriyanghai Y, Kamakura T, Nishiuchi S, Hayano M, Harita T, Yamamoto Y, Kohjitani H, Hirose S, Chen J, Kawamura M, Ohno S, Itoh H, Takeuchi A, Matsuoka S, Miura M, Sumitomo N, Horie M, Yamanaka S, Kimura T.. Patient-specific human induced pluripotent stem cell model assessed with electrical pacing validates S107 as a potential therapeutic agent for catecholaminergic polymorphic ventricular tachycardia. PLoS One 2016;11:e0164795.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lodola F, Morone D, Denegri M, Bongianino R, Nakahama H, Rutigliano L, Gosetti R, Rizzo G, Vollero A, Buonocore M, Napolitano C, Condorelli G, Priori SG, Di Pasquale E.. Adeno-associated virus-mediated CASQ2 delivery rescues phenotypic alterations in a patient-specific model of recessive catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis 2016;7:e2393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Maizels L, Huber I, Arbel G, Tijsen AJ, Gepstein A, Khoury A, Gepstein L.. Patient-specific drug screening using a human induced pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia type 2. Circ Arrhythm Electrophysiol 2017;10:e004725. [DOI] [PubMed] [Google Scholar]
- 86. Devalla HD, Gélinas R, Aburawi EH, Beqqali A, Goyette P, Freund C, Chaix M, Tadros R, Jiang H, Béchec A, Le Monshouwer-Kloots JJ, Zwetsloot T, Kosmidis G, Latour F, Alikashani A, Hoekstra M, Schlaepfer J, Mummery CL, Stevenson B, Kutalik Z, Vries AA, de Rivard L, Wilde AA, Talajic M, Verkerk AO, Al-Gazali L, Rioux JD, Bhuiyan ZA, Passier R.. TECRL, a new life-threatening inherited arrhythmia gene associated with overlapping clinical features of both LQTS and CPVT. EMBO Mol Med 2016;8:1390–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Norman MW, McKenna WJ.. Arrhythmogenic right ventricular cardiomyopathy: perspectives on disease. Z Kardiol 1999;88:550–554. [DOI] [PubMed] [Google Scholar]
- 88. Finsterer J, Stöllberger C.. Arrhythmogenic right ventricular dysplasia in neuromuscular disorders. Clin Med Insights Cardiol 2016;10:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, Wong P, Liew R.. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J 2013;34:1122–1133. [DOI] [PubMed] [Google Scholar]
- 90. Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L.. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet 2013;6:557–568. [DOI] [PubMed] [Google Scholar]
- 91. Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen H-SV.. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013;494:105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP, Rothenberg E, Chen HSV, Napolitano C, Priori SG, Delmar M.. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a brugada syndrome phenotype. Circulation 2014;129:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Te Riele ASJM, Agullo-Pascual E, James CA, Leo-Macias A, Cerrone M, Zhang M, Lin X, Lin B, Sobreira NL, Amat-Alarcon N, Marsman RF, Murray B, Tichnell C, van der Heijden JF, Dooijes D, van Veen TAB, Tandri H, Fowler SJ, Hauer RNW, Tomaselli G, van den Berg MP, Taylor MRG, Brun F, Sinagra G, Wilde AAM, Mestroni L, Bezzina CR, Calkins H, Peter van Tintelen J, Bu L, Delmar M, Judge DP.. Multilevel analyses of SCN5A mutations in arrhythmogenic right ventricular dysplasia/cardiomyopathy suggest non-canonical mechanisms for disease pathogenesis. Cardiovasc Res 2017;113:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Akdis D, Saguner AM, Shah K, Wei C, Medeiros-Domingo A, Eckardstein A, von Lüscher TF, Brunckhorst C, Chen HSV, Duru F.. Sex hormones affect outcome in arrhythmogenic right ventricular cardiomyopathy/dysplasia: from a stem cell derived cardiomyocyte-based model to clinical biomarkers of disease outcome. Eur Heart J 2017;38:1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Asimaki A, Tandri H, Huang H, Halushka MK, Gautam S, Basso C, Thiene G, Tsatsopoulou A, Protonotarios N, McKenna WJ, Calkins H, Saffitz JE.. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N Engl J Med 2009;360:1075–1084. [DOI] [PubMed] [Google Scholar]
- 96. Kirchhof P, Fabritz L, Zwiener M, Witt H, Schafers M, Zellerhoff S, Paul M, Athai T, Hiller K-H, Baba HA, Breithardt G, Ruiz P, Wichter T, Levkau B.. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006;114:1799–1806. [DOI] [PubMed] [Google Scholar]
- 97. Burkett EL, Hershberger RE.. Clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol 2005;45:969–981. [DOI] [PubMed] [Google Scholar]
- 98. Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Spudich S, Girolami U, De Seidman JG, Muntoni F, Müehle G, Johnson W, McDonough B, Seidman CE.. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 1999;341:1715–1724. [DOI] [PubMed] [Google Scholar]
- 99. Kostera-Pruszczyk A, Pruszczyk P, Kamińska A, Lee HS, Goldfarb LG.. Diversity of cardiomyopathy phenotypes caused by mutations in desmin. Int J Cardiol 2008;131:146–147. [Google Scholar]
- 100. Hedberg C, Melberg A, Kuhl A, Jenne D, Oldfors A.. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy 7 is caused by a DES mutation. Eur J Hum Genet 2012;20:984–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Herman DS, Lam L, Taylor MRG, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Lenarda A, Di Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJR, Cook SA, Mestroni L, Seidman JG, Seidman CE.. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Siu CW, Lee YK, Ho JCY, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J, Lay KW, Colman A, Tse HF.. Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging 2012;4:803–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tse HF, Ho JCY, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC, Au KW, Zhang J, Lay KWJ, Esteban MA, Nicholls JM, Colman A, Sham PC.. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet 2013;22:1395–1403. [DOI] [PubMed] [Google Scholar]
- 104. Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC.. Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an ipsc model of dilated cardiomyopathy. Cell Stem Cell 2015;17:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC.. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 2012;4:130ra47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D, Kolokathis F, Kremastinos D, Katoulis A, Ren L, Cohen N, Gho JM, Tsiapras D, Vink A, Wu JC, Asselbergs FW, Li RA, Hulot JS, Kranias EG, Hajjar RJ.. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun 2015;6:6955.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wyles SP, Hrstka SC, Reyes S, Terzic A, Olson TM, Nelson TJ.. Pharmacological modulation of calcium homeostasis in familial dilated cardiomyopathy: an in vitro analysis from an RBM20 patient-derived iPSC model. Clin Transl Sci 2016;9:158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wyles SP, Li X, Hrstka SC, Reyes S, Oommen S, Beraldi R, Edwards J, Terzic A, Olson TM, Nelson TJ.. Modeling structural and functional deficiencies of RBM20 familial dilated cardiomyopathy using human induced pluripotent stem cells. Hum Mol Genet 2016;25:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xiang X, Werner G, Bohrmann B, Liesz A, Mazaheri F, Capell A, Feederle R, Knuesel I, Kleinberger G, Haass C.. TREM2 deficiency reduces the efficacy of immunotherapeutic amyloid clearance. EMBO Mol Med 2016;8:992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE.. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 2015;349:982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Judge LM, Perez-Bermejo JA, Truong A, Ribeiro AJS, Yoo JC, Jensen CL, Mandegar MA, Huebsch N, Kaake RM, So P-L, Srivastava D, Pruitt BL, Krogan NJ, Conklin BR.. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight 2017;2:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yano M, Ikeda Y, Matsuzaki M.. Altered intracellular Ca2+ handling in heart failure. J Clin Invest 2005;115:556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stillitano F, Turnbull IC, Karakikes I, Nonnenmacher M, Backeris P, Hulot JS, Kranias EG, Hajjar RJ, Costa KD.. Genomic correction of familial cardiomyopathy in human engineered cardiac tissues. Eur Heart J 2016;37:3282–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE.. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Circulation 1995;92:785–789. [DOI] [PubMed] [Google Scholar]
- 115. Dambrot C, Braam SR, Tertoolen LGJ, Birket M, Atsma DE, Mummery CL.. Serum supplemented culture medium masks hypertrophic phenotypes in human pluripotent stem cell derived cardiomyocytes. J Cell Mol Med 2014;18:1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M.. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 2003;107:2227–2232. [DOI] [PubMed] [Google Scholar]
- 117. Birket MJ, Ribeiro MC, Kosmidis G, Ward D, Leitoguinho AR, Pol V, van de Dambrot C, Devalla HD, Davis RP, Mastroberardino PG, Atsma DE, Passier R, Mummery CL.. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep 2015;13:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ojala M, Prajapati C, Pölönen RP, Rajala K, Pekkanen-Mattila M, Rasku J, Larsson K, Aalto-Setälä K.. Mutation-specific phenotypes in hiPSC-derived cardiomyocytes carrying either myosin-binding protein C or α-tropomyosin mutation for hypertrophic cardiomyopathy. Stem Cells Int 2016;2016:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tanaka A, Yuasa S, Mearini G, Egashira T, Seki T, Kodaira M, Kusumoto D, Kuroda Y, Okata S, Suzuki T, Inohara T, Arimura T, Makino S, Kimura K, Kimura A, Furukawa T, Carrier L, Node K, Fukuda K.. Endothelin-1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell-derived cardiomyocytes. J Am Heart Assoc 2014;3:e001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC.. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013;12:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GCL, Tobita K, Rasmusson RL, Yang L.. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res 2014;104:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pioner JM, Racca AW, Klaiman JM, Yang K-C, Guan X, Pabon L, Muskheli V, Zaunbrecher R, Macadangdang J, Jeong MY, Mack DL, Childers MK, Kim D-H, Tesi C, Poggesi C, Murry CE, Regnier M.. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rep 2016;6:885–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Phelan DG, Anderson DJ, Howden SE, Wong RCB, Hickey PF, Pope K, Wilson GR, Pébay A, Davis AM, Petrou S, Elefanty AG, Stanley EG, James PA, Macciocca I, Bahlo M, Cheung MM, Amor DJ, Elliott DA, Lockhart PJ.. ALPK3-deficient cardiomyocytes generated from patient-derived induced pluripotent stem cells and mutant human embryonic stem cells display abnormal calcium handling and establish that ALPK3 deficiency underlies familial cardiomyopathy. Eur Heart J 2016;37:2586–2590. [DOI] [PubMed] [Google Scholar]
- 124. Hinson JT, Chopra A, Lowe A, Sheng CC, Gupta RM, Kuppusamy R, O’Sullivan J, Rowe G, Wakimoto H, Gorham J, Burke MA, Zhang K, Musunuru K, Gerszten RE, Wu SM, Chen CS, Seidman JG, Seidman CE.. Integrative analysis of PRKAG2 cardiomyopathy iPS and microtissue models identifies AMPK as a regulator of metabolism, survival, and fibrosis. Cell Rep 2017;19:2410.. [DOI] [PubMed] [Google Scholar]
- 125. Weiford BC, Subbarao VD, Mulhern KM.. Noncompaction of the ventricular myocardium. Circulation 2004;109:2965–2971. [DOI] [PubMed] [Google Scholar]
- 126. Arbustini E, Weidemann F, Hall JL.. Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol 2014;64:1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Luxán G, Casanova JC, Martínez-Poveda B, Prados B, D'Amato G, MacGrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, Izquierdo-García JL, Fernández-Friera L, Sabater-Molina M, Kong Y-Y, Pizarro G, Ibañez B, Medrano C, García-Pavía P, Gimeno JR, Monserrat L, Jiménez-Borreguero LJ, de la Pompa JL.. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med 2013;19:193–201. [DOI] [PubMed] [Google Scholar]
- 128. Kodo K, Ong S-G, Jahanbani F, Termglinchan V, Hirono K, InanlooRahatloo K, Ebert AD, Shukla P, Abilez OJ, Churko JM, Karakikes I, Jung G, Ichida F, Wu SM, Snyder MP, Bernstein D, Wu JC.. iPSC-derived cardiomyocytes reveal abnormal TGFβ signaling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol 2016;18:1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Arndt AK, Schafer S, Drenckhahn JD, Sabeh MK, Plovie ER, Caliebe A, Klopocki E, Musso G, Werdich AA, Kalwa H, Heinig M, Padera RF, Wassilew K, Bluhm J, Harnack C, Martitz J, Barton PJ, Greutmann M, Berger F, Hubner N, Siebert R, Kramer HH, Cook SA, Macrae CA, Klaassen S.. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am J Hum Genet 2013;93:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jiang Y, Habibollah S, Tilgner K, Collin J, Barta T, Al-Aama JY, Tesarov L, Hussain R, Trafford AW, Kirkwood G, Sernagor E, Eleftheriou CG, Przyborski S, Stojković M, Lako M, Keavney B, Armstrong L.. An induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes. Stem Cells Transl Med 2014;3:416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Tomita-Mitchell A, Stamm KD, Mahnke DK, Kim M-S, Hidestrand PM, Liang HL, Goetsch MA, Hidestrand M, Simpson PM, Pelech AN, Tweddell JS, Benson DW, Lough JW, Mitchell ME. The impact of MYH6 variants in hypoplastic left heart syndrome. Physiol Genomics 2016;48:912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Yang C, Xu Y, Yu M, Lee D, Alharti S, Hellen N, Ahmad Shaik N, Banaganapalli B, Sheikh Ali Mohamoud H, Elango R, Przyborski S, Tenin G, Williams S, O’Sullivan J, Al-Radi OO, Atta J, Harding SE, Keavney B, Lako M, Armstrong L.. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Hum Mol Genet 2017;26:3031–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Bohlmeyer TJ, Helmke S, Ge S, Lynch J, Brodsky G, Sederberg JH, Robertson AD, Minobe W, Bristow MR, Perryman MB.. Hypoplastic left heart syndrome myocytes are differentiated but possess a unique phenotype. Cardiovasc Pathol 2003;12:23–31. [DOI] [PubMed] [Google Scholar]
- 134. Hoffman JI, Kaplan S.. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890–1900. [DOI] [PubMed] [Google Scholar]
- 135. Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D.. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003;424:443–447. [DOI] [PubMed] [Google Scholar]
- 136. Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D.. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 2016;167:1734–1749.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Roberts A, Allanson J, Jadico SK, Kavamura MI, Noonan J, Opitz JM, Young T, Neri G.. The cardiofaciocutaneous syndrome. J Med Genet 2006;43:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Weiss G, Confino Y, Shemer A, Trau H.. Cutaneous manifestations in the cardiofaciocutaneous syndrome, a variant of the classical Noonan syndrome. Report of a case and review of the literature. J Eur Acad Dermatol Venerol 2004;18:324–327. [DOI] [PubMed] [Google Scholar]
- 139. Josowitz R, Mulero-Navarro S, Rodriguez NA, Falce C, Cohen N, Ullian EM, Weiss LA, Rauen KA, Sobie EA, Gelb BD.. Autonomous and non-autonomous defects underlie hypertrophic cardiomyopathy in BRAF-mutant hiPSC-derived cardiomyocytes. Stem Cell Rep 2016;7:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Cashman TJ, Josowitz R, Johnson BV, Gelb BD, Costa KD.. Human engineered cardiac tissues created using induced pluripotent stem cells reveal functional characteristics of BRAF-mediated hypertrophic cardiomyopathy. PLoS One 2016;11:e0146697.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Spurney CF. Cardiomyopathy of Duchenne muscular dystrophy: current understanding and future directions. Muscle Nerve 2011;44:8–19. [DOI] [PubMed] [Google Scholar]
- 142. Dick E, Kalra S, Anderson D, George V, Ritso M, Laval SH, Barresi R, Aartsma-Rus A, Lochmueller H, Denning C, Lochmüller H, Denning C.. Exon skipping and gene transfer restore dystrophin expression in human induced pluripotent stem cells-cardiomyocytes harboring DMD mutations. Stem Cells Dev 2013;22:2714–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zatti S, Martewicz S, Serena E, Uno N, Giobbe G, Kazuki Y, Oshimura M, Elvassore N.. Complete restoration of multiple dystrophin isoforms in genetically corrected Duchenne muscular dystrophy patient-derived cardiomyocytes. Mol Ther Methods Clin Dev 2014;1:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Guan X, Mack DL, Moreno CM, Strande JL, Mathieu J, Shi Y, Markert CD, Wang Z, Liu G, Lawlor MW, Moorefield EC, Jones TN, Fugate JA, Furth ME, Murry CE, Ruohola-Baker H, Zhang Y, Santana LF, Childers MK.. Dystrophin-deficient cardiomyocytes derived from human urine: new biologic reagents for drug discovery. Stem Cell Res 2014;12:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GCL, Rasmusson RL, Denning C, Yang L.. Modeling and studying mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from Duchenne Muscular Dystrophy patients. Dis Model Mech 2015;8:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Macadangdang J, Guan X, Smith AST, Lucero R, Czerniecki S, Childers MK, Mack DL, Kim DH.. Nanopatterned human iPSC-based model of a dystrophin-null cardiomyopathic phenotype. Cel Mol Bioeng 2015;8:320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Ramaciotti C, Iannaccone ST, Scott WA.. Myocardial cell damage in Duchenne muscular dystrophy. Pediatr Cardiol 2003;24:503–506. [DOI] [PubMed] [Google Scholar]
- 148. Romeo V. Myotonic dystrophy type 1 or Steinert’s disease. Adv Exp Med Biol 2012;724:239–257. [DOI] [PubMed] [Google Scholar]
- 149. Fu YH, Pizzuti A, Fenwick RG, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, De Jong P, Wieringa B, Korneluk R, Perryman MB, Epstein HF, Caskey CT. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 1992;255:1256–1258. Fu YH, Pizzuti A, Fenwick RG, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, De Jong P, [DOI] [PubMed] [Google Scholar]
- 150. Gao Y, Guo X, Santostefano K, Wang Y, Reid T, Zeng D, Terada N, Ashizawa T, Xia G.. Genome therapy of myotonic dystrophy type 1 iPS cells for development of autologous stem cell therapy. Mol Ther 2016;24:1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Christians ES, Ishiwata T, Benjamin IJ.. Small heat shock proteins in redox metabolism: implications for cardiovascular diseases. Int J Biochem Cell Biol 2012;44:1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Mitzelfelt KA, Limphong P, Choi MJ, Kondrat FDL, Lai S, Kolander KD, Kwok W-M, Dai Q, Grzybowski MN, Zhang H, Taylor GM, Lui Q, Thao MT, Hudson JA, Barresi R, Bushby K, Jungbluth H, Wraige E, Geurts AM, Benesch JLP, Riedel M, Christians ES, Minella AC, Benjamin IJ.. The human 343delT HSPB5 Chaperone associated with early-onset skeletal myopathy causes defects in protein solubility. J Biol Chem 2016;291:14939–14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bidichandani SI, Delatycki MB, Friedreich Ataxia, Pagon R, Adam M, Ardinger H, Wallace S, Amemiya A, Bean L, Bird T, Ledbetter N, Mefford H, Smith R, Stephens K (eds). GeneReviews. Seattle, WA: University of Washington, Seattle, 2011. pp. 1–18. [Google Scholar]
- 154. Hick A, Wattenhofer-Donzé M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, André C, Reutenauer L, Rai M, Teletin M, Messaddeq N, Schiffmann SN, Viville S, Pearson CE, Pandolfo M, Puccio H.. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich’s ataxia. Dis Model Mech 2013;6:608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Crombie DE, Curl CL, Raaijmakers AJA, Sivakumaran P, Kulkarni T, Wong RCB, Minami I, Evans-Galea MV, Lim SY, Delbridge L, Corben LA, Dottori M, Nakatsuji N, Trounce IA, Hewitt AW, Delatycki MB, Pera MF, Pébay A.. Friedreich’s ataxia induced pluripotent stem cell-derived cardiomyocytes display electrophysiological abnormalities and calcium handling deficiency. Aging (Albany NY) 2017;9:1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Lee YK, Ho PWL, Schick R, Lau YM, Lai WH, Zhou T, Li Y, Ng KM, Ho SL, Esteban MA, Binah O, Tse HF, Siu CW.. Modeling of Friedreich ataxia-related iron overloading cardiomyopathy using patient-specific-induced pluripotent stem cells. Pflugers Arch Eur J Physiol 2014;466:1831–1844. [DOI] [PubMed] [Google Scholar]
- 157. Lee YK, Lau YM, Ng KM, Lai WH, Ho SL, Tse HF, Siu CW, Ho PWL.. Efficient attenuation of Friedreich’s ataxia (FRDA) cardiomyopathy by modulation of iron homeostasis-human induced pluripotent stem cell (hiPSC) as a drug screening platform for FRDA. Int J Cardiol 2016;203:964–971. [DOI] [PubMed] [Google Scholar]
- 158. Pandolfo M, Hausmann L.. Deferiprone for the treatment of Friedreich’s ataxia. J Neurochem 2013;126:142–146. [DOI] [PubMed] [Google Scholar]
- 159. Barth PG, Scholte HR, Berden JA, Van Der Klei-Van Moorsel JM, Luyt-Houwen IEM, Van'T Veer-Korthof ET, Van Der Harten JJ, Sobotka-Plojhar MA.. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 1983;62:327–355. [DOI] [PubMed] [Google Scholar]
- 160. Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang D-Z, Li K, Wang J, Wanders RJ. A, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT.. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014;20:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Martiniuk F, Chen A, Mack A, Arvanitopoulos E, Chen Y, Rom WN, Codd WJ, Hanna B, Alcabes P, Raben N, Plotz P. Carrier frequency for glycogen storage disease type II in New York and estimates of affected individuals born with the disease. Am J Med Genet 1998;79:69–72. [DOI] [PubMed] [Google Scholar]
- 162. Nishigaki T. Glycogen storage disease type II Ryoikibetsu Shokogun Shirizu 1998;19:349–352. [PubMed] [Google Scholar]
- 163. Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, Chien CL, Li LT, Chiang SC, Chen HF, Ho HN, Chen CH, Kuo HC.. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet 2011;20:4851–4864. [DOI] [PubMed] [Google Scholar]
- 164. Sato Y, Kobayashi H, Higuchi T, Shimada Y, Era T, Kimura S, Eto Y, Ida H, Ohashi T.. Disease modeling and lentiviral gene transfer in patient-specific induced pluripotent stem cells from late-onset Pompe disease patient. Mol Ther Methods Clin Dev 2015;2:15023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Raval KK, Tao R, White BE, Lange WJ, De Koonce CH, Yu J, Kishnani PS, Thomson JA, Mosher DF, Ralphe JC, Kamp TJ.. Pompe disease results in a Golgi-based glycosylation deficit in human induced pluripotent stem cell-derived cardiomyocytes. J Biol Chem 2015;290:3121–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Thurberg BL, Lynch Maloney C, Vaccaro C, Afonso K, Tsai AC-H, Bossen E, Kishnani PS, O'Callaghan M.. Characterization of pre-and post-treatment pathology after enzyme replacement therapy for pompe disease. Lab Invest 2006;86:1208–1220. [DOI] [PubMed] [Google Scholar]
- 167. Fermini B, Nathan RD.. Removal of sialic acid alters both T- and L-type calcium currents in cardiac myocytes. Am J Physiol 1991;260:H735–H743. [DOI] [PubMed] [Google Scholar]
- 168. Jakubowska E, Ryba M, Hruby Z.. Fabry disease Przegl Lek 2006;63:218–219. [PubMed] [Google Scholar]
- 169. Itier JM, Ret G, Viale S, Sweet L, Bangari D, Caron A, Le-Gall F, Bènichou B, Leonard J, Deleuze JF, Orsini C.. Effective clearance of GL-3 in a human iPSC-derived cardiomyocyte model of Fabry disease. J Inherit Metab Dis 2014;37:1013–1022. [DOI] [PubMed] [Google Scholar]
- 170. Chimenti C, Hamdani N, Boontje NM, DeCobelli F, Esposito A, Bronzwaer JGF, Stienen GJM, Russo MA, Paulus WJ, Frustaci A, Velden van der J.. Myofilament degradation and dysfunction of human cardiomyocytes in Fabry disease. Am J Pathol 2008;172:1482–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Chou S-J, Yu W-C, Chang Y-L, Chen W-Y, Chang W-C, Chien Y, Yen J-C, Liu Y-Y, Chen S-J, Wang C-Y, Chen Y-H, Niu D-M, Lin S-J, Chen J-W, Chiou S-H, Leu H-B.. Energy utilization of induced pluripotent stem cell-derived cardiomyocyte in Fabry disease. Int J Cardiol 2017;232:255–263. [DOI] [PubMed] [Google Scholar]
- 172. Chien Y, Chien C-S, Chiang H-C, Huang W-L, Chou S-J, Chang W-C, Chang Y-L, Leu H-B, Chen K-H, Wang K-L, Lai Y-H, Liu Y-Y, Lu K-H, Li H-Y, Sung Y-J, Jong Y-J, Chen Y-J, Chen C-H, Yu W-C.. Interleukin-18 deteriorates Fabry cardiomyopathy and contributes to the development of left ventricular hypertrophy in Fabry patients with GLA IVS4 + 919 G>A mutation. Oncotarget 2016;7:87161–87179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, Sue CM, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M.. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature 2000;406:906–910. [DOI] [PubMed] [Google Scholar]
- 174. Hashem S, Perry C, Bauer M, Han S, Clegg S, Ouyang K, Deacon D, Spinharney M, Panopoulos A, Izpisua Belmonte JC, Frazer K, Chen J, Gong Q, Zhou Z, Chi N, Adler E.. Brief report: oxidative stress mediates cardiomyocyte apoptosis in a human model of danon disease and heart failure. Stem Cells 2015;33:2343–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Majer F, Vlaskova H, Krol L, Kalina T, Kubanek M, Stolnaya L, Dvorakova L, Elleder M, Sikora J.. Danon disease: a focus on processing of the novel LAMP2 mutation and comments on the beneficial use of peripheral white blood cells in the diagnosis of LAMP2 deficiency. Gene 2012;498:183–195. [DOI] [PubMed] [Google Scholar]
- 176. Ng KM, Mok PY, Butler AW, Ho JCY, Choi SW, Lee YK, Lai WH, Au KW, Lau YM, Wong LY, Esteban MA, Siu CW, Sham PC, Colman A, Tse HF.. Amelioration of X-linked related autophagy failure in danon disease with DNA methylation inhibitor. Circulation 2016;134:1373–1389. [DOI] [PubMed] [Google Scholar]
- 177. Leung A, Nah SK, Reid W, Ebata A, Koch CM, Monti S, Genereux JC, Wiseman RL, Wolozin B, Connors LH, Berk JL, Seldin DC, Mostoslavsky G, Kotton DN, Murphy GJ.. Induced pluripotent stem cell modeling of multisystemic, hereditary transthyretin amyloidosis. Stem Cell Rep 2013;1:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Liu Y, Conlon DM, Bi X, Slovik KJ, Shi J, Edelstein HI, Millar JS, Javaheri A, Cuchel M, Pashos EE, Iqbal J, Hussain MM, Hegele RA, Yang W, Duncan SA, Rader DJ, Morrisey EE.. Lack of MTTP activity in pluripotent stem cell-derived hepatocytes and cardiomyocytes abolishes apoB secretion and increases cell stress. Cell Rep 2017;19:1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wetterau JR, Aggerbeck LP, Bouma ME, Eisenberg C, Munck A, Hermier M, Schmitz J, Gay G, Rader DJ, Gregg RE.. Absence of microsomal triglyceride transfer protein in individuals with abetalipoproteinemia. Science 1992;258:999–1001. [DOI] [PubMed] [Google Scholar]
- 180. Zamel R, Khan R, Pollex RL, Hegele RA.. Abetalipoproteinemia: two case reports and literature review. Orphanet J Rare Dis 2008;3:19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, De Almeida P, Lan F, Diecke S, Burridge PW, Gold JD, Mochly-Rosen D, Wu JC.. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med 2014;6:255ra130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Zhang H, Gong DX, Zhang YJ, Li SJ, Hu S.. Effect of mitochondrial aldehyde dehydrogenase-2 genotype on cardioprotection in patients with congenital heart disease. Eur Heart J 2012;33:1606–1614. [DOI] [PubMed] [Google Scholar]
- 183. Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, Gérard R, Badi L, Kam-Thong T, Bu L, Jiang X, Hoflack JC, Kiialainen A, Jeworutzki E, Aoyama N, Carlson C, Burcin M, Gromo G, Boehringer M, Stahlberg H, Hall BJ, Magnone MC, Kolaja K, Chien KR, Bailly J, Iacone R.. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep 2014;9:810–820. [DOI] [PubMed] [Google Scholar]
- 184. Borchert T, Hübscher D, Guessoum CI, Lam TDD, Ghadri JR, Schellinger IN, Tiburcy M, Liaw NY, Li Y, Haas J, Sossalla S, Huber MA, Cyganek L, Jacobshagen C, Dressel R, Raaz U, Nikolaev VO, Guan K, Thiele H, Meder B, Wollnik B, Zimmermann WH, Lüscher TF, Hasenfuss G, Templin C, Streckfuss-Bömeke K.. Catecholamine-dependent β-adrenergic signaling in a pluripotent stem cell model of takotsubo cardiomyopathy. J Am Coll Cardiol 2017;70:975–991. [DOI] [PubMed] [Google Scholar]
- 185. Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC, Che Y, Ebert A, Diecke S, Liang P, Red-Horse K, Carette JE, Wu SM, Wu JC.. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res 2014;115:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Belkaya S, Kontorovich AR, Byun M, Mulero-Navarro S, Bajolle F, Cobat A, Josowitz R, Itan Y, Quint R, Lorenzo L, Boucherit S, Stoven C, Di Filippo S, Abel L, Zhang SY, Bonnet D, Gelb BD, Casanova JL.. Autosomal recessive cardiomyopathy presenting as acute myocarditis. J Am Coll Cardiol 2017;69:1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Yücel G, Zhao Z, El-Battrawy I, Lan H, Lang S, Li X, Buljubasic F, Zimmermann W-H, Cyganek L, Utikal J, Ravens U, Wieland T, Borggrefe M, Zhou X-B, Akin I.. Lipopolysaccharides induced inflammatory responses and electrophysiological dysfunctions in human-induced pluripotent stem cell derived cardiomyocytes. Sci Rep 2017;7:2935.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Stack JP, Moslehi J, Sayed N, Wu JC.. Cancer therapy-induced cardiomyopathy: can human induced pluripotent stem cell modelling help prevent it? Eur Heart J 2018;0:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Burridge PW, Li YF, Matsa E, Wu H, Ong S-G, Sharma A, Holmström A, Chang AC, Coronado MJ, Ebert AD, Knowles JW, Telli ML, Witteles RM, Blau HM, Bernstein D, Altman RB, Wu JC.. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016;22:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Millard DC, Strock CJ, Carlson CB, Aoyama N, Juhasz K, Goetze T. A, Stoelzle-Feix S, Becker N, Fertig N, January CT, Anson BD, Ross JD.. Identification of drug-drug interactions in vitro: a case study evaluating the effects of sofosbuvir and amiodarone on hiPSC-derived cardiomyocytes. Toxicol Sci 2016;154:174–182. [DOI] [PubMed] [Google Scholar]
- 191. Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmström A, Matsa E, Zhang Y, Kumar A, Fan AC, Del Álamo JC, Wu SM, Moslehi JJ, Mercola M, Wu JC.. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med 2017;9:eaaf2584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Stillitano F, Hansen J, Kong CW, Karakikes I, Funck-Brentano C, Geng L, Scott S, Reynier S, Wu M, Valogne Y, Desseaux C, Salem JE, Jeziorowska D, Zahr N, Li R, Iyengar R, Hajjar RJ, Hulot JS.. Modeling susceptibility to drug-induced long QT with a panel of subject-specific induced pluripotent stem cells. 2017;6:e19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Caballero R, Utrilla RG, Amorós I, Matamoros M, Pérez-Hernández M, Tinaquero D, Alfayate S, Nieto-Marín P, Guerrero-Serna G, Liu Q-H, Ramos-Mondragón R, Ponce-Balbuena D, Herron T, Campbell KF, Filgueiras-Rama D, Peinado R, López-Sendón JL, Jalife J, Delpón E, Tamargo J.. Tbx20 controls the expression of the KCNH2 gene and of hERG channels. Proc Natl Acad Sci U S A 2017;114:E416–E425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Roden DM. Long QT syndrome: reduced repolarization reserve and the genetic link. J Intern Med 2006;259:59–69. [DOI] [PubMed] [Google Scholar]
- 195. Sheikh N, Papadakis M, Chandra N, Raju H, Zaidi A, Ghani S, Muggenthaler M, Gati S, Sharma S.. 170 Ethnic differences in phenotypic expression of hypertrophic cardiomyopathy. Heart 2011;97:A95. [Google Scholar]
- 196. Neal RC, Ferdinand KC, Yčas J, Miller E.. Relationship of ethnic origin, gender, and age to blood creatine kinase levels. Am J Med 2009;122:73–78. [DOI] [PubMed] [Google Scholar]
- 197. Kim K, Zhao R, Doi. A, Ng K, Unternaehrer J, Cahan P, Hongguang H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ.. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 2011;29:1117–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Sanchez-Freire V, Lee AS, Hu S, Abilez OJ, Liang P, Lan F, Huber BC, Ong SG, Hong WX, Huang M, Wu JC.. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J Am Coll Cardiol 2014;64:436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Tomoda K, Takahashi K, Leung K, Okada A, Narita M, Yamada NA, Eilertson KE, Tsang P, Baba S, White MP, Sami S, Srivastava D, Conklin BR, Panning B, Yamanaka S.. Derivation conditions impact x-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 2012;11:91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC.. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the American Heart Association. Circ Genom Precis Med 2018;11:e000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Vitale AM, Matigian NA, Ravishankar S, Bellette B, Wood SA, Wolvetang EJ, Mackay-Sim A.. Variability in the generation of induced pluripotent stem cells: importance for disease modeling. Stem Cells Transl Med 2012;1:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Saha K, Jaenisch R.. Technical challenges in using human induced pluripotent stem cells to model disease. Cell Stem Cell 2009;5:584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Vunjak Novakovic G, Eschenhagen T, Mummery C.. Myocardial tissue engineering: in vitro models. Cold Spring Harb Perspect Med 2014;4:a014076.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Mills RJ, Voges HK, Porrello ER, Hudson JE.. Disease modeling and functional screening using engineered heart tissue. Curr Opin Physiol 2018;1:80–88. [Google Scholar]
- 205. Stacey GN, Crook JM, Hei D, Ludwig T.. Banking human induced pluripotent stem cells: lessons learned from embryonic stem cells? Cell Stem Cell 2013;13:385–388. [DOI] [PubMed] [Google Scholar]
- 206. De Sousa PA, Steeg R, Wachter E, Bruce K, King J, Hoeve M, Khadun S, McConnachie G, Holder J, Kurtz A, Seltmann S, Dewender J, Reimann S, Stacey G, O'Shea O, Chapman C, Healy L, Zimmermann H, Bolton B, Rawat T, Atkin I, Veiga A, Kuebler B, Serano BM, Saric T, Hescheler J, Brüstle O, Peitz M, Thiele C, Geijsen N, Holst B, Clausen C, Lako M, Armstrong L, Gupta SK, Kvist AJ, Hicks R, Jonebring A, Brolén G, Ebneth A, Cabrera-Socorro A, Foerch P, Geraerts M, Stummann TC, Harmon S, George C, Streeter I, Clarke L, Parkinson H, Harrison PW, Faulconbridge A, Cherubin L, Burdett T, Trigueros C, Patel MJ, Lucas C, Hardy B, Predan R, Dokler J, Brajnik M, Keminer O, Pless O, Gribbon P, Claussen C, Ringwald A, Kreisel B, Courtney A, Allsopp TE.. Rapid establishment of the European Bank for induced Pluripotent Stem Cells (EBiSC)—the hot start experience. Stem Cell Res 2017;20:105–114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.