Abstract
Adenosine triphosphate–binding cassette subfamily C member 2 (ABCC2/Abcc2) is critically important to biliary excretion of many endobiotic and xenobiotic compounds, and is a major driving force for bile acid–independent bile flow. Abcc2 expression is reduced at the messenger RNA (mRNA) and protein levels in various forms of experimental cholestasis. In a microRNA (miRNA) screen of mouse liver after biliary obstruction, we found that miRNA let7a‐5p was significantly up‐regulated approximately 4‐fold. Similarly, ABCC2 mRNA was depleted and miRNA let7a‐5p was elevated over 4‐fold in livers of children with biliary atresia compared with normal livers. In silico analysis predicted that let7a‐5p would target the 3′ untranslated region (3′ UTR) of ABCC2/Abcc2 RNA. The objective of this study was to determine whether let7a‐5p contributes to the depletion of ABCC2/Abcc2 in cholestasis. To demonstrate the functional importance of miRNA let7a‐5p in regulating the expression of ABCC2, co‐transfection of a let7a‐5p mimic and an ABCC2‐3′ UTR luciferase construct into Huh‐7 cells led to a marked inhibition of luciferase activity by about 60%‐70% compared with controls, which was reversed by a let7a‐5p mimic inhibitor. Expression of this mimic led to a significant decrease in endogenous ABCC2 mRNA and protein levels in a Huh‐7 liver cell line, which could be blocked by expression of a let7a‐5p mimic inhibitor. Injection of a lentivirus let7a‐5p inhibitor into normal mouse liver or into mouse liver after common bile duct ligation led to a significant increase in endogenous Abcc2 mRNA and protein levels and a depletion of let7a‐5p mRNA levels compared with untreated, saline‐injected livers or livers treated with an inactive lentivirus control. Conclusion: These studies demonstrate that miR‐let7a‐5p is involved in regulating ABCC2/Abcc2 expression, and is aberrantly up‐regulated in obstructive cholestasis.
ABCC2 is critically important to biliary excretion of many endobiotic and xenobiotic compounds, and is a major driving force for bile acid–independent bile flow. ABCC2 expression is reduced at the mRNA and protein levels in various forms of experimental cholestasis, but the underlying mechanisms for this change have not been defined. Our studies demonstrate that miR‐let7a‐5p is involved in regulating ACCC2/Abcc2 expression, and is aberrantly up‐regulated in obstructive cholestasis.
![]()
Abbreviations
- ABCC2
adenosine triphosphate–binding cassette subfamily C member 2
- BSEP
bile salt export pump
- CBDL
common bile duct ligation
- cDNA
complementary DNA
- CT
threshold cycle
- FXR
farnesoid X receptor
- miRNA
microRNA
- mRNA
messenger RNA
- MRP2
multidrug resistance‐associated protein 2
- NC
negative control
- NTCP
Na/taurocholate co‐transporting polypeptide
- pCBDL
partial CBDL
- RPMI
Roswell Park Memorial Institute
- RT‐PCR
reverse transcription polymerase chain reaction
- 3′ UTR
3′ untranslated region
The adenosine triphosphate (ATP)‐binding cassette subfamily C member 2 (ABCC2, also known as the multidrug resistance protein 2, MRP2) is a multispecific, ATP‐dependent efflux transporter that is encoded by the ABCC2 gene and located on the liver canalicular membrane.1 ABCC2 transports a large number of amphipathic, usually multivalent organic anion conjugates from the liver into bile including bilirubin glucuronides, glutathione conjugates, leukotrienes, some heavy metals, and sulfated and glucuronidated substrates.2, 3 ABCC2 is also a major driving force for bile acid–independent bile flow through the secretion of reduced glutathione.4 Glucuronide‐conjugated and sulfate‐conjugated bile acids are also transported by ABCC2.4 ABCC2 plays a central role in the biliary excretion of conjugated bilirubin.1 Patients with Dubin‐Johnson Syndrome, who harbor polymorphisms in the ABCC2 gene, exhibit impaired secretion of conjugated bilirubin and other organic anions into bile.1 Abcc2 function is known to be impaired in various models of intrahepatic and obstructive cholestasis, resulting in conjugated hyperbilirubinemia.5 Both Abcc2 messenger RNA (mRNA) and protein levels are depleted in experimental cholestasis, but the mechanisms underlying these changes have not been well defined.5
The expression and function of ABCC2 are likely subject to complex regulatory mechanisms in health and disease. MicroRNAs (miRNAs) have emerged as important regulators of gene expression.6, 7 MiRNAs are a family of small noncoding RNA molecules (containing approximately 22 nucleotides) that post‐transcriptionally repress gene expression by base pairing to sequence motifs in the 3′ untranslated region (3′ UTR) of target mRNAs, leading to translational repression or mRNA degradation.6, 7 Over 1,800 miRNAs have been identified in the human, and more than 45,000 miRNA target sites are predicted to reside within 3′ UTRs of human RNAs.6, 8 MicroRNAs are important in normal cellular processes, but dysregulation of miRNA expression has been associated with disease.9, 10, 11 In a miRNA screen of mouse liver after biliary obstruction, we found that miRNA let7a‐5p was significantly up‐regulated about 4‐fold, and was predicted to target the 3′ UTR of ABCC2/Abcc2 RNA. In limited, published studies, let7a‐5p has not been well studied in various forms of experimental liver disease, including biliary obstruction when miRNAs were measured in plasma or liver tissue, and has no known role in regulating the expression of ABCC2/Abcc2. The goal of this study was to determine the functional importance of let7a‐5p in regulating ABCC2/Abcc2 through in vitro and in vivo studies.
Materials and Methods
Animals
Common bile duct ligation (CBDL) was performed in 8‐week‐old male C57Bl/6 mice as described previously, using a protocol approved by the Institutional Animal Care and Use Committee of the University of Colorado, Denver, Colorado.12 Mice were fed standard chow and were subjected to surgeries in the morning after an overnight fast. Briefly, laparotomy was performed on mice in which the common bile duct was ligated proximally and distally and severed in the middle. Total serum bile acids were estimated by a kit from Trinity Biotech (Ireland) to ensure that successful cholestasis was achieved (data not shown). Sham surgery was performed on control mice in which laparotomy and manipulation of the common bile duct was performed, but the bile duct was not ligated. Livers from sham‐operated and bile duct–ligated mice were collected at 3 days following ligation. In limited studies, partial bile duct ligation was done by looping a ligature around the common bile duct and an adjacent 7.0 surgical needle.13, 14 When the ligature is tied carefully and the needle is removed, a reproducibly narrowed but defined lumen remained that results in cholestasis. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health.
Human Samples
Liver biopsy samples from 3 infants with biliary atresia at the time of Kasai portoenterostomy and from three normal livers were obtained from a pediatric liver biobank at the University of Colorado. In this internal review board–approved protocol, consent was obtained from parents for use of residual tissue from clinically indicated liver biopsies and from unused pediatric donor tissue from reduced‐size liver transplants.
Cells and Cell Culture
The human hepatoma cell line Huh‐7 was cultured in Roswell Park Memorial Institute (RPMI) 1640 medium with fetal bovine serum (FBS) and antibiotics, as previously described.15 All cells were grown in 5% CO2 in a humidified incubator maintained at 37°C. The cell lines were obtained from the American Tissue Culture Collection (Manassas, VA).
Materials
Antibodies to MRP2/ABCC2 (ab3373) were obtained from Abcam (Cambridge, MA), anti‐FXR (farnesoid X receptor)/NRIH4 (252165) antibodies from Abbiotec (San Diego, CA), the anti‐BSEP (bile salt export pump)/ABCB11 (PAB4697) antibody from Abnova (Taipei, Taiwan), the NTCP (Na/taurocholate co‐transporting polypeptide)/SLC10A1 (GTX17430) antibody from Gene Tex (Irvine, CA), and the anti‐β‐actin (A5316) antibody from Sigma‐Aldrich (St. Louis, MO). Cell culture media, FBS, and Lipofectamine 2000 were obtained from Invitrogen (Carlsbad, CA). MicroRNA mimics, inhibitors, and negative controls were obtained from Exiqon/Qiagen (Boston, MA). Mission synthetic mmu‐let‐7a‐5p lenti‐miRNA inhibitors (MLTUD0195) were obtained from Sigma‐Aldrich. Ath‐miR416, a negative control (HLTUD001C) sequence from Arabidopsis thaliana with no homology to human and mouse gene sequences, was obtained from Sigma‐Aldrich. All other chemicals were from Sigma‐Aldrich or Fisher Scientific unless otherwise stated.
Plasmid Constructs
The 3′ UTR luciferase plasmids for the human pMIR/hABCC2 were a generous gift from Dr. M. Ananthanarayanan (Yale University, New Haven, CT). All of the positive clones containing 3′ UTR inserts were verified by restriction enzyme mapping and sequenced using the ABI 377 DNA sequencer model (SeqGen, Inc., Torrance, CA).
Bioinformatics
An in silico search for possible miRNA‐binding sites in the 3′ UTR of the ABCB11/Abcb11 (BSEP), ABCC2/Abcc2 (MRP2), SLC10A1/Slc10a1 (NTCP), and NR1H4/Nr1h4 (FXR) gene was performed using miRANDA (Memorial Sloan‐Kettering Cancer Center, New York, NY), DIANA‐microT‐CDS (Alexander Fleming Biomedical Science Research Center, Athens, Greece), and miRBase (Faculty of Life Science, University of Manchester, Manchester, United Kingdom).16, 17
MicroRNA Polymerase Chain Reaction Array
An miRNA polymerase chain reaction (PCR) array used a kit and protocol from Qiagen.18 Briefly, the miScript PCR System (Qiagen) consists of the miScript II RT Kit, miScript miRNA PCR Array, and miScript SYBR Green PCR Kit. MicroRNA was extracted using the miRNeasy kit (Qiagen). Contaminating DNA was removed using the RNase‐Free DNase Set (Qiagen) on‐column digestion protocol as per manufacturer’s instructions. The single complementary DNA (cDNA) was prepared by the miScript II RT kit. MiScript miRNA PCR Arrays are mature miRNA‐specific forward primers (miScript Primer Assays) that are arrayed in biologically relevant pathway‐focused and whole miRNome panels. These PCR arrays are provided in ready‐to‐use 96‐well plates. Each assay in a miScript miRNA PCR Array was verified to ensure sensitive and specific detection of mature miRNA by real‐time PCR. A web‐based miScript miRNA PCR Array data analysis tool (Qiagen) was used for the analysis of real‐time PCR data. After the raw threshold cycle (CT) data were uploaded, the tool automatically performed all fold‐change calculations using the ΔΔCT method of relative quantification.
Transient Transfection With miR‐199a‐5p, miR223‐3p, and let‐7a‐5p
To investigate the effect of let‐7a‐5p, miR‐199a‐5p, and miR‐223‐3p on ABCC2 mRNA and protein expression, Huh‐7 cells were seeded in 12‐well plates (8 × 104 cells/mL). miR‐199a‐5p and miR‐223‐3p were also elevated 4‐5‐fold after CBDL (Supporting Table S1), but were not predicted to target the 3′ UTR of ABCC2/Abcc2. After 24 hours, cells were transfected with 100 nM miRNA mimics or anti‐miRNA inhibitors and the corresponding negative controls (Exiqon/Qiagen) by using Lipofectamine RNAiMax diluted in Opti‐MEM I (both purchased from Invitrogen) at a final concentration of 3 mM. After 8 hours of incubation, the transfection medium was replaced with fresh complete growth medium. At 48 hours and 72 hours after transfection, the total mRNA and protein were isolated by using TRIzol reagent (Life Technologies, Carlsbad, CA) and M‐PER (Mammalian Protein Extraction Reagent, Pierce Biotechnology, Waltham, MA) containing proteolytic and phosphatase inhibitor mixture (Sigma‐Aldrich). Messenger RNA and protein were quantified in three independently performed experiments.
Because the expression of SLC10A1 (NTCP) is very low in Huh‐7 cells, Huh‐7 cells were transfected with an NTCP expression plasmid 24 hours before transfection of miRNA mimics and inhibitors.
Western Blotting After Mimic, Inhibitor, or Negative Control Transfections
To examine the effect of let7a‐5p, miR‐199a‐5p, and miR‐223‐3p overexpression, Huh‐7 cells were plated in 6‐well plates at a density of 1 × 105 cells/well 24 hours before transfection. The following day, the cells were transfected with mimics, inhibitors, or negative controls at a concentrations of 50, 75, and 100 nm after complexing with Lipofectamine RNAiMax at a ratio of 1:3 (μL/μL) in Opti‐MEM medium. The cells were fed with RPMI 1640 medium after 24 hours. Forty‐eight and 72 hours after transfection, protein extracts of Huh‐7 cells transiently transfected with control, mimics, and inhibitors were lysed for 15 minutes on ice in 150 μL of M‐PER (Pierce Biotechnology) containing protease and phosphatase inhibitor mixtures. Cell debris was removed by centrifugation at 16,000 rpm (21,130g) for 15 minutes at 4°C in an Eppendorf microcentrifuge. Protein concentration in the supernatant was estimated using bovine serum albumin as standard (Bio‐Rad Laboratories, Hercules, CA). A total of 50‐75 μg of protein in 1 × Laemmli buffer was loaded per lane of 4%‐20% Mini‐Protean TGX precast gels (Bio‐Rad) and run at 200 volts for 40 minutes. Prestained Precision Plus Protein Dual Color Standards (Bio‐Rad) were also run on the same gels to estimate the protein molecular size. The fractionated proteins were blotted using a Bio‐Rad Semi‐dry blotter (Trans‐Blot SD semi‐dry blotter cell) to precut the PVDF membranes (Immunoblot PVDF membrane, 7.0 × 8.5 cm) in Tris glycine/methanol (20%) buffer at 20 V for 90 minutes. Following protein transfer, the blots were blocked with 5% Nonfat Dry Blot Omniblok (American BioAnalytical, Natick, MA) in 1 × blocking buffer (diluted from 10 × Tris‐buffered saline with 0.5% Tween 20 [Kirkegaard & Perry Lab Inc., Gaithersburg, MD]). Following blocking, incubation with primary antibodies (to BSEP, MRP2, NTCP, FXR, and β‐actin) with secondary peroxidase‐conjugated antibodies and the washing steps was done as previously described.15 Signals developed with Clarity western ECL substrate (Bio‐Rad) were quantitated using the Bio‐Rad ChemiDoc System.
Total RNA Isolation and Real‐Time PCR Assays
Total RNA was isolated from Huh‐7 cells using the RNAeasy Mini Kit (Qiagen, Gaithersburg, MD) according to the manufacturer’s instructions.19 Quantitation of RNA was done using Nanodrop 2000. Complementary DNA synthesis was carried out using the Affinity Script Multi Temperature cDNA Synthesis Kit (Agilent Biotechnologies, Santa Clara, CA) on 2 μg of total RNA according to the manufacturer’s directions. The cDNA was diluted 10‐fold, and 5 μL of the diluted cDNA (100 ng) per well was used for real‐time quantitative PCR using SYBR Green as the detection method with Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA) gene expression. Oligonucleotide primers used in the SYBR Green real‐time quantitative PCR assays were provided on request. In SYBR Green assays, normalization was achieved using 36B4 as a housekeeping control, and β‐actin level assays. All real‐time quantitative PCR were done using an ABI QuantStudio 7 Flex Real‐Time PCR System machine (SeqGen). Relative expression was calculated using the comparative CT method (ΔΔCT method) according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA).
Luciferase Reporter Gene Assays
A 440–base pair DNA amplicon of the 3′ UTR region of the human ABCC2 mRNA with let7a ‐5p target site was obtained by reverse‐transcription PCR (RT‐PCR) using specific oligonucleotides forward (CAGTGCTCCCTGGCTAGCGGACT) and reverse (AGACAGGGTCTCTTCCTGATCTG). Point mutations were introduced into human ABCC2 3′ UTR using a QuikChange II XL Site‐Directed Mutagenesis Kit (Agilent, Santa Clara, CA) and appropriate mutant oligos. Mutants were confirmed by nucleic acid sequencing, and those plasmids were used for the transfection.
Wild type:
hABCC2wt‐3′UTR‐5′tatgaatacagcacaaTGTATCAGTTTTAATATTggggatcattagcat‐3′
Mutant:
hABCC2mu‐3′UTR‐5′tatgaatacagcacaaTGGCTCGTTGCTAGCATTggggatcattagcat‐3′
The fragments were subcloned into the pMIR‐ REPORT luciferase vector (Applied Biosystems), resulting in a cytomegalovirus‐driven expression construct Luc‐ABCC2‐3′ UTR. Huh‐7 cells were plated in 24‐well plates at a density of 1 × 105 cells/well 24 hours before transfection. MicroRNA mimics and inhibitors and negative controls 1 and 2 (Exiqon) were transfected at a final concentration of 50 nm in combination with Luc‐ABCC2‐3′ UTR (0.25 μg/well) in Opti‐MEM complexed to Lipofectamine 2000 at a ratio of 1:3 (μg:μL) according to the manufacturer’s instructions. LNA‐modified mimics and inhibitors (Exiqon) were used because the unmodified reagents showed nonspecific effects. Twenty‐four hours later, the medium was changed to RPMI 1640 medium. Forty‐eight hours after transfection, cell lysates were prepared in 250 μL of 1 × Passive Lysis Buffer (Promega, Madison, WI). Firefly and Renilla luciferase activities were assayed in 50 μL of the lysate using the Dual Luciferase Kit (Promega) according to the manufacturer’s instructions using a luminometer (Promega). Relative luciferase activities were reported after normalization of the individual values to Renilla luciferase.20
Mission Synthetic miRNA Inhibitor–Lentiviral Constructs
Each miRNA inhibitor construct was cloned and the sequence verified to ensure a match to the target miRNA. The lentiviral transduction particles were produced from sequence‐verified lentiviral plasmid vectors. The lentiviral miRNA inhibitors were cloned into the TRC2‐pLKO‐puro vector (Sigma‐Aldrich). The manufacturer’s instructions were followed to produce more virus. Huh‐7 cells were transduced with lentiviral particles containing the miRNA inhibitors. MicroRNA targets were validated by quantitating target protein and/or mRNA levels in response to miRNA down‐regulation.
Lentiviral constructs were injected directly into the left lobe of mouse liver immediately after CBDL or sham surgery, as done by others with injection into tissue.21, 22 A total of 100 µL of the concentrated lentivirus solution (1.9 × 107 TU/mL) was withdrawn by a 0.5‐cc insulin syringe with a 28.5G needle. The left lateral lobe was slowly infused with the viral construct to a total volume of 100 μL. The slow infusion allowed for effective spreading of the virus into the tissue without back flow. Livers from sham‐operated and bile duct–ligated mice were collected at 3 days after ligation (fourth day).
Statistical Analysis
Data are expressed as mean ± SEM. When two groups were compared, the two‐tailed paired Student t test was used. For comparison of multiple groups, an ordinary one‐way analysis of variance followed by Tukey’s post hoc multiple comparison test were done using Prism 8 software. A P value of less than 0.05 was considered statistically significant. All experiments using cultured cells or mouse livers were repeated at least 3 times. A minimum of 3 animals were used in each experiment.
Results
Abcc2 Is a Direct Target of miRNA let7a‐5p
A Mouse Liver miFinder miScript miRNA PCR Array from Qiagen was used to study the expression of miRNAs abundantly expressed or best characterized in liver tissue after biliary obstruction. Among the numerous miRNAs that were significantly up‐regulated or down‐regulated in mouse livers (n = 3 in each group) after CBDL compared with sham‐operated mice, miRNA let7a‐5p was up‐regulated about 4‐fold (Supporting Table S1). A 4‐fold increase in let7a‐5p miRNA levels was confirmed by RT‐PCR analysis (Fig. 1). In silico analysis using miRANDA, DIANA‐microT‐CDS, and miRBase microRNA target prediction algorithms all identified a potential miRNA let7a‐5p binding site in the 3′ UTR of Abcc2 and ABCC2 mRNAs. The alignments of let7a‐5p with mouse and human 3′ UTRs are shown in Fig. 2.
Figure 1.
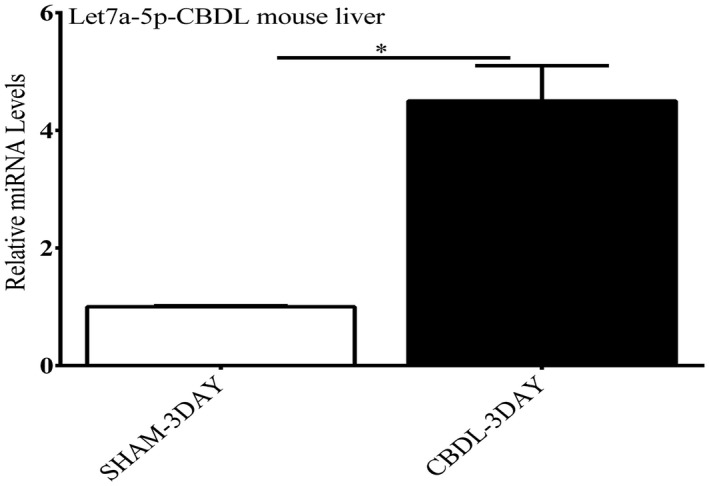
The effect of CBDL on expression of let7a‐5p. Three days of CBDL ligation led to an over 4‐fold increase in let7a‐5p miRNA levels compared with livers from sham‐operated mice on RT‐PCR analysis. *P < 0.01.
Figure 2.
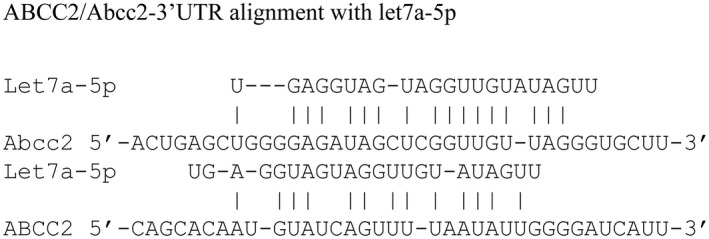
Predicted binding sites for the seed sequence of let7a‐5p in the 3′ UTR of Abcc2 (mouse) and ABCC2 (human) mRNAs using the bioinformatic tools.
As previously reported, Abcc2 mRNA and protein levels were significantly depleted after 3 days of CBDL in the mouse (Fig. 3A,B).5 Similar results were found in a less severe cholestatic model of partial CBDL (pCBDL) with depletion of Abcc2 mRNA and protein levels and up‐regulation of let7a‐5p miRNA (Supporting Fig. S1A‐D).
Figure 3.
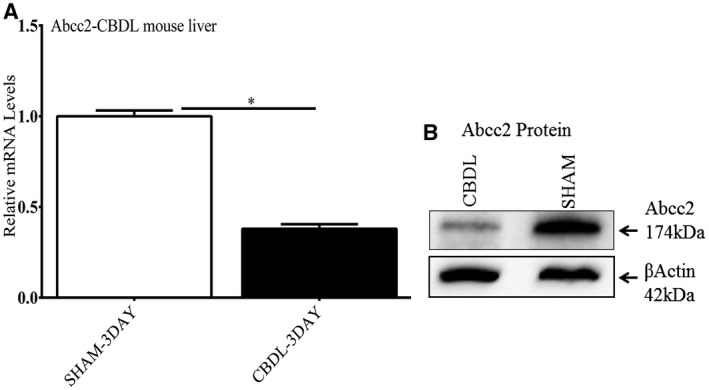
Effect of 3 days of CBDL in the mouse on Abcc2 mRNA and protein expression. There was significant depletion of Abcc2 mRNA (Fig. 1A) and protein levels (Fig. 1B) on RT‐PCR and western blot analysis, respectively, in CBDL versus sham‐operated mice. Experiments with CBDL mice were repeated 3 times. *P < 0.01.
ABCC2 mRNA was also significantly depleted and miRNA Let7a‐5p elevated over 4‐fold in livers of infants with biliary atresia compared with normal livers (Fig. 4A,B).
Figure 4.
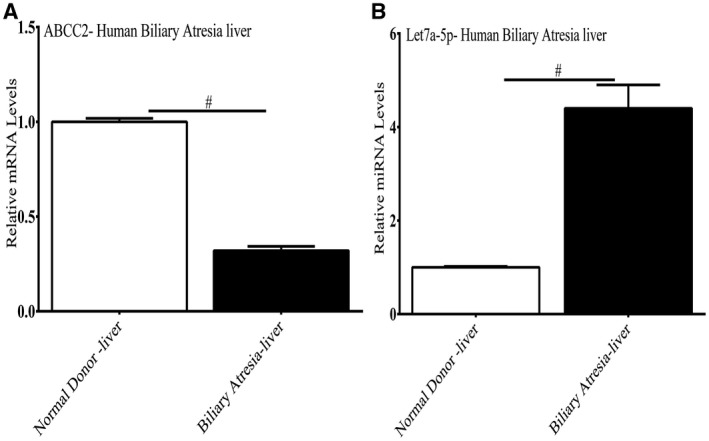
Expression of ABCC2 mRNA and let7a‐5p in infants with biliary atresia. Decreased expression of ABCC2 mRNA (Fig. 4A) and up‐regulation of let7a‐5p (Fig. 4B) was found in livers of 3 children with biliary atresia versus livers of 3 healthy children on RT‐PCR analysis. *P < 0.01.
Abcc2 Has a Functional let7a‐5p Responsive Element in the 3′ UTR
To demonstrate the functional importance of miRNA let7a‐5p in regulating the expression of ABCC2, we cloned the 3′ UTR of ABCC2 mRNA immediately downstream of a luciferase reporter. Co‐transfection of a let7a‐5p mimic and the ABCC2‐3′ UTR‐luciferase construct into Huh‐7 cells led to a marked inhibition of luciferase activity by about 60%‐70% compared with controls (Fig. 5A), which was reversed by a let7a‐5p mimic inhibitor. Site‐directed mutagenesis of the putative let7a‐5 binding site in the ABCC2‐3′ UTR prevented the inhibitory effect of the let7a‐5p mimic on luciferase expression (Fig. 5B). miR‐223‐5p and miR199b‐3p were also up‐regulated about 4‐fold after CBDL (Supporting Table S1), but overexpression of mimics for these miRNAs had no effect on the expression of the ABCC‐3′ UTR‐luciferase construct, which lacks a binding site for these miRNAs (Fig. 5C,D).
Figure 5.
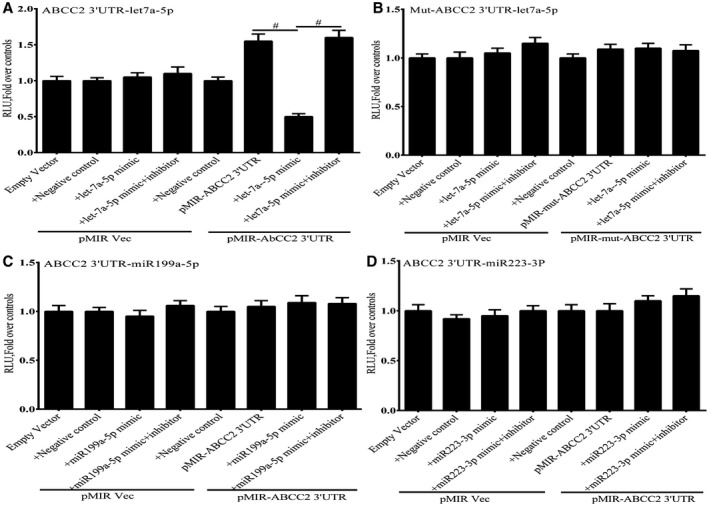
Effect of a let7a‐5p mimic and a mimic inhibitor on expression of an ABCC2‐3′ UTR‐luciferase construct expressed in Huh7 cells 48 hours after transfection. (A) Expression of a let7a‐5p mimic significantly repressed luciferase activity. The activity of the mimic was completely reversed by a mimic inhibitor. (B) The effect of the let7a‐5p mimic on luciferase activity was completely abrogated by mutation of the putative let7a‐5p binding site in the ABCC2‐3′ UTR. The miR199a‐5p (C) and miR223‐3p (D) mimics had no effect on the expression of an ABCC2‐3′ UTR‐luciferase construct expressed in Huh7 cells. Each panel represents the mean + SEM of three separate experiments. # P < 0.01.
Expression of let7a‐5p in Huh‐7 Cells Leads to Depletion of ABCC2 mRNA and Protein
To investigate the effects of let7a‐5p on endogenous ABCC2 expression, Huh‐7 cells were transfected with a let7a‐5p mimic. Overexpression of the let7a‐5p mimic but not the miR 223‐3p and miR199b‐3p mimics led to a significant decrease in ABCC2 mRNA levels (Fig. 6A). Overexpression of a let7a‐5p mimic inhibitor blocked the effect of the let7a‐5p mimic, and led to an increase in endogenous ABCC2 mRNA levels (Fig. 6B). In contrast, expression of the miR 223‐3p and miR199b‐3p mimics and mimic inhibitors had no effect on ABCC2 mRNA levels. Expression of let7a‐5p also markedly suppressed ABCC2 protein levels in Huh7 cells but had no effect on the amounts of BSEP (ABCB11), NTCP (SLC10A1), or FXR (NR1H4) proteins (Fig. 7A,B.). This effect was blocked by co‐transfection of let7a‐5p mimic inhibitor.
Figure 6.
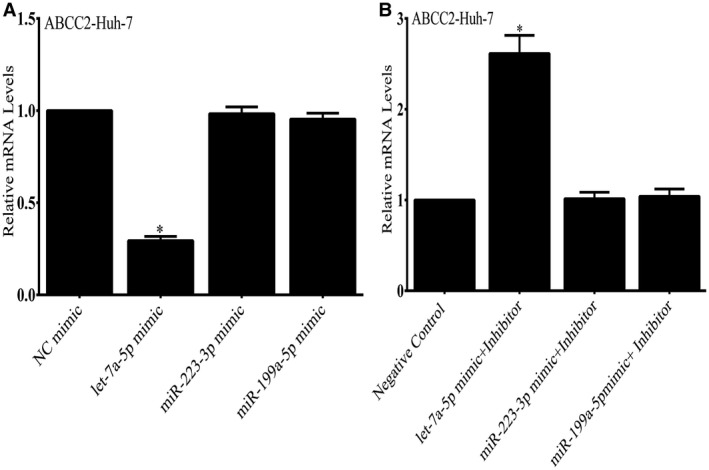
Effect of a let7a‐5p mimic on endogenous ABCC2 mRNA in Huh‐7 cells. Overexpression of the let7a‐5p mimic but not the miR 223‐3p and miR199b‐3p mimics led to a significant decrease in ABCC2 mRNA levels. In contrast, expression of the miR 223‐3p and miR199b‐3p mimics had no effect on ABCC2 levels. Expression of a let7a‐5p mimic and mimic inhibitor led to an increase in endogenous ABCC2 mRNA levels (Fig. 6B), whereas expression of the miR 223‐3p and miR199b‐3p mimics and inhibitors had no effect on ABCC2 mRNA levels. *P < 0.01.
Figure 7.
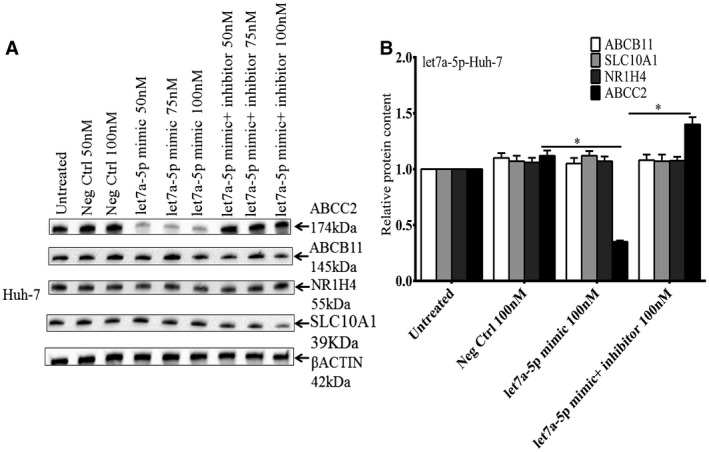
Western blot showing the effect of a let7a‐5p mimic on endogenous ABCC2 protein levels in Huh‐7 cells (Fig. 7A). Transfection of a let7a‐5p mimic into Huh‐7 cells led to a significant decrease in ABCC2 protein levels which could be blocked by expression of a let7a‐5p mimic inhibitor. Expression of let7a‐5p had no effect on the amounts of BSEP, NTCP, or FXR proteins in Huh‐7 cells (Fig. 7B). Quantification of blots using the Bio‐Rad ChemiDoc System. *Mean + SEM of three separate experiments. *P < 0.01.
Silencing of let7a‐5p by Lentivirus let‐7a‐5p Inhibitor Construct Increases Expression of Hepatic Abcc2
Before the in vivo experiments, we tested whether a lentivirus let‐7a‐5p inhibitor construct could affect let7a‐5p and ABCC2 expression in Huh‐7 cells. This construct led to a significant decrease in let7a‐5p mRNA (Fig. 8A) and a marked increase in ABCC2 mRNA (Fig 8B) and protein levels (Fig. 8C,D) in these cells compared with controls.
Figure 8.
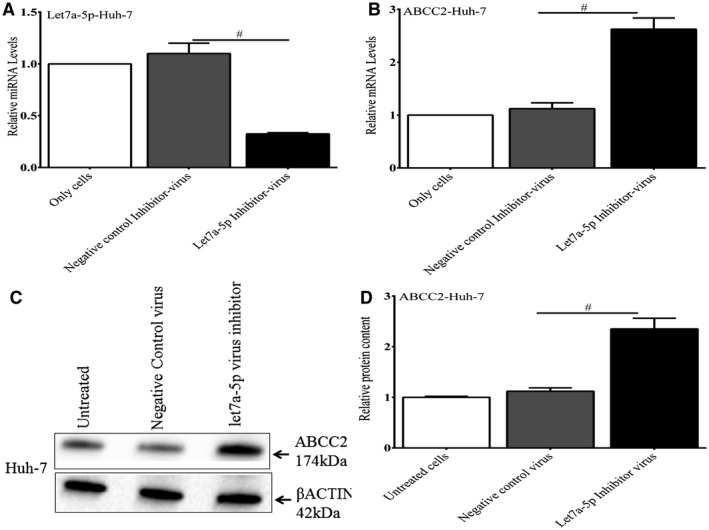
The effect of a lentivirus let7a‐5p inhibitor on endogenous let7a‐5p and ABCC2 expression in Huh‐7 cells. Cells were infected with a lentivirus let‐7a‐5p inhibitor construct and analyzed after 48 hours. This construct led to a significant decrease in let7a‐5p mRNA (A) and a marked increase in ABCC2 mRNA (B) and protein levels (C,D) in these cells compared with controls. *P < 0.001.
In an effort to understand the physiological role of let7a‐5p, we injected normal mouse liver or mouse liver after BDL with saline, a lentivirus‐let7a‐5p inhibitor, or an inactive lentivirus control. The objective was to competitively bind the inhibitor to let7a‐5p and prevent it from regulating its endogenous target. In normal mouse liver, injection of the lentivirus‐let7a‐5p inhibitor construct led to a significant depletion of let7a‐5p mRNA levels and an increase in endogenous Abcc2 mRNA and protein levels, compared with untreated mouse livers or livers treated with an inactive lentivirus control (Fig. 9A‐D). Similar to the in vitro data and experiments in normal mouse liver, expression of the lentivirus‐let7a‐5p inhibitor construct in livers after CBDL significantly decreased the let7a‐5p miRNA (Fig. 10A) levels and led to an increase in Abcc2 mRNA and protein levels (Fig. 10B‐D). Similar results were also observed in mice after pCBDL (Supporting Fig. S2A‐D). As expected, the lentivirus‐let7a‐5p inhibitor construct also increased the Abcc2 protein levels in sham‐operated mice above the untreated and control virus‐treated livers (Fig. 10D; Supporting Fig. S2D).
Figure 9.
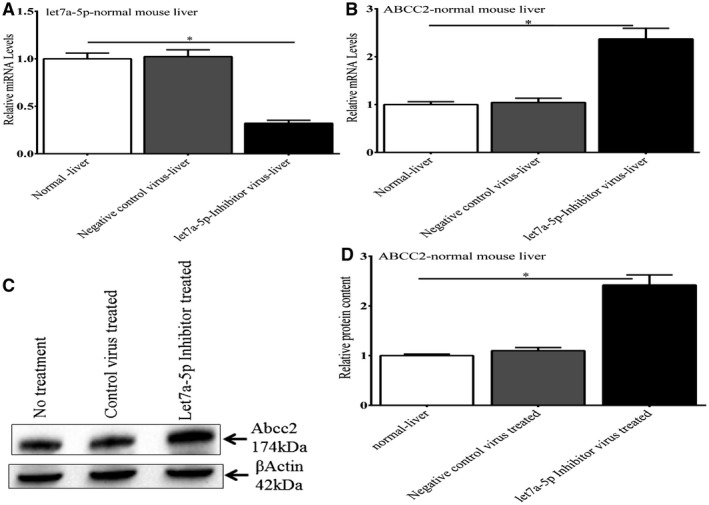
Effect of a lentivirus let7a‐5p inhibitor on endogenous let7a‐5p and Abcc2 expression in normal mouse liver. In normal mouse livers, injection of the lentivirus‐let7a‐5p inhibitor construct led to a significant depletion of let7a‐5p mRNA levels and an increase in endogenous Abcc2 mRNA and protein levels compared with untreated mouse liver or liver treated with an inactive lentivirus control. *P < 0.01.
Figure 10.
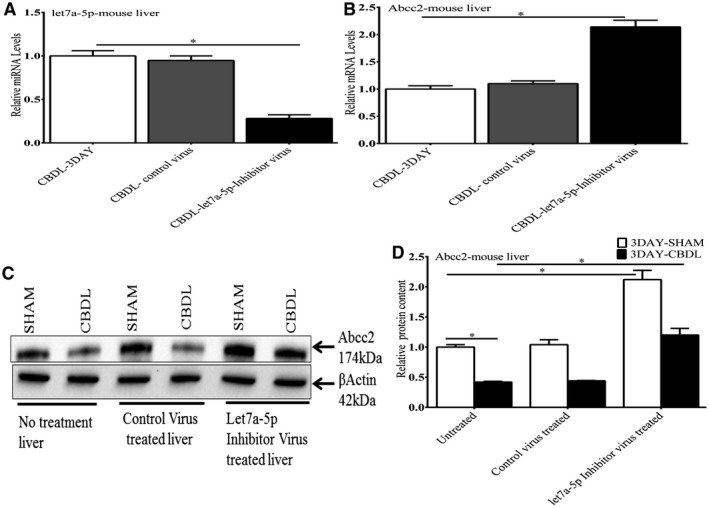
Effect of a lentivirus let7a‐5p inhibitor on let7a‐5p and Abcc2 expression in mouse liver after CBDL. After CBDL, injection of a lentivirus‐let7a‐5p inhibitor led to a significant decrease in let7a‐5p mRNA levels (10A) and an increase in endogenous Abcc2 mRNA (10B) and protein levels (10C and 10D) compared with untreated mouse liver or liver treated with an inactive lentivirus control. *P < 0.01.
Discussion
The excretion of amphipathic, multivalent organic anion conjugates across the canalicular membrane into bile is mediated by an adenosine triphosphate–dependent conjugate export pump, which has been designated as MRP2 or ABCC2. ABCC2 primarily functions to mediate the elimination of bile acid conjugates, glutathione conjugates, and conjugated metabolites of numerous drugs and other xenobiotics.1 ABCC2 is primarily an organic anion transporter, but weakly basic drugs may be co‐transported with glutathione.
Conjugated hyperbilirubinemia is a cardinal feature of obstructive cholestasis, and is closely linked to the activity of ABCC2.23 Abcc2 function is impaired in various experimental models of intrahepatic and obstructive cholestasis, including treatment by endotoxin, ethinylestradiol, or CBDL. In a study by Trauner et al., cholestasis in these rat models resulted in a marked decrease in Abcc2 protein and its tissue localization on the canalicular membrane.5 Abcc2 mRNA levels diminished profoundly after endotoxin and CBDL, but did not change after ethinylestradiol treatment. The underlying molecular mechanisms for these changes were unclear. The decreased expression of ABCC2 in complete biliary obstruction may be beneficial, as continued transport into an obstructed bile duct would be futile. However, the same depletion of Abcc2 also occurs in other forms of cholestasis, including partial biliary obstruction, which is demonstrated in Supporting Fig. S2. As a compensatory mechanism, the expression of other transporters such as basolateral MRP3 (ABCC3) is up‐regulated in cholestasis, allowing transport of anionic conjugates into sinusoidal blood.1
There is increasing recognition that the expression of numerous miRNAs is altered in almost all acute and chronic liver diseases.7, 9 MicroRNAs have well‐defined roles in cell proliferation, differentiation, cell death, organogenesis, and maintenance of organ function. MicroRNAs act by imperfect binding to complementary sequences in the 3′ UTR of target mRNA, leading to either cleavage of the mRNA or repression of protein translation.6 Let7a‐5p has not been previously associated with regulation of ABCC2/Abcc2 or other liver transporters that contribute to bile formation, and its behavior in cholestatic liver disease has not been well studied. In a study focusing primarily on cholangiocytes, Glaser et al. observed an over 15‐fold elevation of let‐7 miRNA in hepatocytes isolated from mice after CBDL.24 The major finding in our study was that microRNA let7a‐5p regulates the expression of Abcc2 in normal and cholestatic mouse liver. Let7a‐5p was found to be up‐regulated about 4‐fold after CBDL in the mouse and in children with biliary atresia, and in silico analysis predicted that it would target Abcc2. In vitro experiments then demonstrated that expression of a let7a‐5p mimic depressed Abcc2 mRNA and protein levels as well as the activity of an ABCC2‐3′UTR‐luciferase construct in Huh‐7 cells. Mutation of the putative binding site in the ABCC2‐3′UTR abrogated the inhibitory effect of the let7a‐5p mimic. Transfection of mimics for other miRNAs up‐regulated in obstructive cholestasis had no effect on ABCC2 expression. Moreover, injection of a lentivirus‐let7a‐5p inhibitor construct into normal mouse liver or mouse liver after CBDL led to a significant increase in endogenous Abcc2 mRNA and protein levels and a depletion of let7a‐5p mRNA levels compared with livers injected with saline or an inactive lentivirus construct.
Let‐7 miRNA was originally discovered in the nematode with a high level of evolutionary conservation from the worm to the human.25 The let‐7 miRNA family consists of 11 closely related genes.26 The gene for let‐7a is located on the chromosome 22 q13.31 associated with CpG islands. Its known functions include inhibition of cell migration, invasion, as well as epithelial‐mesenchymal transition by targeting several oncogenes. However, the biological functions of this and other miRNAs in health and disease are complex, and likely are tissue, cell, and context‐specific. Over 329 target genes of let‐7a‐5p have been identified in low‐throughput or high‐throughput miRNA targeting studies from the MiRTarBase miRNA Targets data set, but did not include ABCC2.25, 27, 28 ABCC5 and ABCC10 are apparent targets but the localization and function of these transporters are not well understood in liver.29 Because a gene can be regulated by many different miRNAs depending on the cellular context, it is not surprising that ABCC2 expression in HepG2 cells and in human peripheral blood monocytic cells is regulated by miRNA‐379.29, 30 Moreover, miR‐297 modulates multidrug resistance in human colorectal carcinoma by down‐regulating ABCC2.31
ABCC2 is regulated at multiple levels in health and disease, including membrane retrieval and reinsertion, translation, and transcription.1 Our studies indicate that miRNA let7a‐5p regulates Abcc2 in normal liver, and is dysregulated after CBDL. Although we have provided evidence that let7a‐5p has a role in regulating ABCC2/Abcc2, there are other epigenetic and transcriptional mechanisms that also affect ABCC2/Abcc2 expression in obstructive cholestasis. We have previously shown that mixed lineage leukemia 3, a histone H3 lysine 4 lysine methyltransferase, is depressed after CBDL in mice, and this epigenetic mark is essential to activation of Abcc2 by the nuclear receptor FXR.32 We have also reported that transcription factor nuclear factor kappa B (NF‐κB) suppresses the transactivation of ABCC2 by FXR in vitro, and that NF‐κB activation in the context of obstructive cholestasis in mice establishes a repressive chromatin environment at the promoters of several FXR‐target genes, including the promoter of Abcc2 through recruitment of HDACs and corepressors.15 It is likely that these and other mediators act in concert with let7a‐5p to repress ABCC2 expression in cholestasis, but the relative importance of each pathway during the progression of liver disease has not been defined. However, our finding that injection of a lentivirus‐let7a‐5p inhibitor construct into mouse liver after CBDL restored Abcc2 mRNA and protein levels to levels measured in control livers would suggest that this miRNA is particularly important in regulating Abcc2 expression. It is interesting that the effects of cholestasis on Abcc2 expression are organ‐specific in that renal Abcc2 expression is preserved, allowing urinary excretion of toxic organic anions and xenobiotics under conditions in which biliary excretion is impaired.33
Supporting information
Potential conflict of interest: Dr. Sokol advises Alexion. He consults for Retrophin, Mirum, and Albireo.
References
Author names in bold designate shared co‐first authorship.
- 1. Keppler D. Progress in the molecular characterization of hepatobiliary transporters. Dig Dis 2017;35:197‐202. [DOI] [PubMed] [Google Scholar]
- 2. Keppler D, Konig J. Hepatic canalicular membrane 5: expression and localization of the conjugate export pump encoded by the MRP2 (cMRP/cMOAT) gene in liver. FASEB J 1997;11:509‐516. [DOI] [PubMed] [Google Scholar]
- 3. Keppler D, Konig J, Buchler M. The canalicular multidrug resistance protein, cMRP/MRP2, a novel conjugate export pump expressed in the apical membrane of hepatocytes. Adv Enzyme Regul 1997;37:321‐333. [DOI] [PubMed] [Google Scholar]
- 4. Boyer JL. Bile formation and secretion. Compr Physiol 2013;3:1035‐1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trauner M, Arrese M, Soroka CJ, Ananthanarayanan M, Koeppel TA, Schlosser SF, et al. The rat canalicular conjugate export pump (Mrp2) is down‐regulated in intrahepatic and obstructive cholestasis. Gastroenterology 1997;113:255‐264. [DOI] [PubMed] [Google Scholar]
- 6. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis 2015;35:3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 8. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schueller F, Roy S, Vucur M, Trautwein C, Luedde T, Roderburg C. The role of miRNAs in the pathophysiology of liver diseases and toxicity. Int J Mol Sci 2018;19:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Otsuka M, Kishikawa T, Yoshikawa T, Yamagami M, Ohno M, Takata A, et al. MicroRNAs and liver disease. J Hum Genet 2017;62:75‐80. [DOI] [PubMed] [Google Scholar]
- 11. Banales JM, Saez E, Uriz M, Sarvide S, Urribarri AD, Splinter P, et al. Up‐regulation of microRNA 506 leads to decreased Cl‐/HCO3‐ anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology 2012;56:687‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balasubramaniyan N, Luo Y, Sun AQ, Suchy FJ. SUMOylation of the farnesoid X receptor (FXR) regulates the expression of FXR target genes. J Biol Chem 2013;288:13850‐13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heinrich S, Georgiev P, Weber A, Vergopoulos A, Graf R, Clavien PA. Partial bile duct ligation in mice: a novel model of acute cholestasis. Surgery 2011;149:445‐451. [DOI] [PubMed] [Google Scholar]
- 14. Yokota S, Ono Y, Nakao T, Zhang P, Michalopoulos GK, Khan Z. Partial bile duct ligation in the mouse: a controlled model of localized obstructive cholestasis. J Vis Exp 2018;133:56930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balasubramaniyan N, Ananthanarayanan M, Suchy FJ. Nuclear factor‐kappaB regulates the expression of multiple genes encoding liver transport proteins. Am J Physiol Gastrointest Liver Physiol 2016;310:G618‐G628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffiths‐Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36:D154‐D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oulas A, Karathanasis N, Louloupi A, Pavlopoulos GA, Poirazi P, Kalantidis K, et al. Prediction of miRNA targets. Methods Mol Biol 2015;1269:207‐229. [DOI] [PubMed] [Google Scholar]
- 18. Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem 2009;394:1117‐1124. [DOI] [PubMed] [Google Scholar]
- 19. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 4th ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2012. [Google Scholar]
- 20. Gould SJ, Subramani S. Firefly luciferase as a tool in molecular and cell biology. Anal Biochem 1988;175:5‐13. [DOI] [PubMed] [Google Scholar]
- 21. Balkow A, Hoffmann LS, Klepac K, Glöde A, Gnad T, Zimmermann K, et al. Direct lentivirus injection for fast and efficient gene transfer into brown and beige adipose tissue. J Biol Methods 2016;3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han X, Wang C, Li Y, Jin Z, Zhang B, Dong Y. miR‐29b in regulating blood pressure and cardiac function in the rat model of hypertension. Exp Ther Med 2019;17:3361‐3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zollner G, Trauner M. Mechanisms of cholestasis. Clin Liver Dis 2008;12:1‐26, vii. [DOI] [PubMed] [Google Scholar]
- 24. Glaser S, Meng F, Han Y, Onori P, Chow BK, Francis H, et al. Secretin stimulates biliary cell proliferation by regulating expression of microRNA 125b and microRNA let7a in mice. Gastroenterology 2014;146:1795‐1808.e1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let‐7 miRNAs and their functional implications. Protein Cell 2016;7:100‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016; 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA‐target interactions. Nucleic Acids Res 2011;39:D163‐D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, et al. miRTarBase update 2018: a resource for experimentally validated microRNA‐target interactions. Nucleic Acids Res 2018;46:D296‐D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haenisch S, Werk AN, Cascorbi I. MicroRNAs and their relevance to ABC transporters. Br J Clin Pharmacol 2014;77:587‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haenisch S, Laechelt S, Bruckmueller H, Werk A, Noack A, Bruhn O, et al. Down‐regulation of ATP‐binding cassette C2 protein expression in HepG2 cells after rifampicin treatment is mediated by microRNA‐379. Mol Pharmacol 2011;80:314‐320. [DOI] [PubMed] [Google Scholar]
- 31. Xu K, Liang X, Shen K, Cui D, Zheng Y, Xu J, et al. miR‐297 modulates multidrug resistance in human colorectal carcinoma by down‐regulating MRP‐2. Biochem J 2012;446:291‐300. [DOI] [PubMed] [Google Scholar]
- 32. Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, et al. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 2011;300:G771‐G781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Denson LA, Bohan A, Held MA, Boyer JL. Organ‐specific alterations in RAR alpha:RXR alpha abundance regulate rat Mrp2 (Abcc2) expression in obstructive cholestasis. Gastroenterology 2002;123:599‐607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


