ABSTRACT
Background
Early infant diet influences postnatal gut microbial development, which in turn can modulate the developing immune system.
Objectives
The aim of this study was to characterize diet-specific bioregional microbiota differences in piglets fed either human breast milk (HM) or infant formula.
Methods
Male piglets (White Dutch Landrace Duroc) were raised on HM or cow milk formula (MF) from postnatal day (PND) 2 to PND 21 and weaned to an ad libitum diet until PND 51. Piglets were euthanized on either PND 21 or PND 51, and the gastrointestinal contents were collected for 16s RNA sequencing. Data were analyzed using the Quantitative Insight into Microbial Ecology. Diversity measurements (Chao1 and Shannon) and the Wald test were used to determine relative abundance.
Results
At PND 21, the ileal luminal region of HM-fed piglets showed lower Chao1 operational taxonomic unit diversity, while Shannon diversity was lower in cecal, proximal colon (PC), and distal colon (DC) luminal regions, relative to MF-fed piglets. In addition, at PND 51, the HM-fed piglets had lower genera diversity within the jejunum, ileum, PC, and DC luminal regions, relative to MF-fed piglets. At PND 21, Turicibacter was 4- to 5-fold lower in the HM-fed piglets’ ileal, cecal, PC, and DC luminal regions, relative to the MF-fed piglets. Campylobacter is 3- to 6-fold higher in HM-fed piglets duodenal, ileal, cecal, PC, and DC luminal regions, in comparison to MF-fed piglets. Furthermore, the large intestine (cecum, PC, and rectum) luminal region of HM-fed piglets showed 4- to 7-fold higher genera that belong to class Bacteroidia, in comparison to MF-fed piglets at PND 21. In addition, at PND 51 distal colon lumen of HM-fed piglets showed 1.5-fold higher genera from class Bacteroidia than the MF-fed piglets.
Conclusions
In the large intestinal regions (cecum, PC, and rectum), MF diet alters microbiota composition, relative to HM diet, with sustained effects after weaning from the neonatal diet. These microbiota changes could impact immune system and health outcomes later in life.
Keywords: piglet, microbiota, human milk, infant formula, gastrointestinal bioregions
Introduction
Human breast milk (HM) contains bioactive components, some of which are highly conserved (such as lactoferrin, an iron-containing protein), and has evolved to nourish a developing infant. Epidemiologic evidence demonstrates that babies fed HM have improved short- and long-term immune health outcomes compared to their formula-fed counterparts (1–5). For example, preterm babies that receive HM are at a much lower risk of developing necrotizing enterocolitis, a potentially life-threatening disease (6). Bioactive components in HM are believed to sculpt the gut microbiota of neonates, which may infer immunologic protection (7–10).
HM- and cow milk formula (MF)-fed infants have been shown to have different microbiota profiles, measured in the stool (11, 12). There are many components of HM that may be driving this effect; however, human milk oligosaccharides (HMOs) may be among the most influential. HMOs are the third most abundant component of breast milk and are not utilized by the neonate, but instead are metabolized by specific bacteria in the gut (8). In clinical trials, a specific strain of Bifidobacterium, which is known to metabolize HMOs, has been shown to directly colonize the gut following co-exposure to HM (13). Additional components of HM, such as IgA (14), glycosylated proteins, and bacteria, may also modify the neonates’ gut microbiota.
Currently, limited data exist on whether HM components influence the microbiota in region-specific ways in the gastrointestinal (GI) tract. Considering the ethical constraints of obtaining multiple bioregional samples from human infants, researchers must turn to animal models to examine this question. The piglet is a good animal model for the study of GI tract development, because there are many similarities with humans in terms of the digestive and metabolic functions of the gut (15–18). Additionally, the establishment of microbiota in the pig is similar to infants with respect to being influenced by the mode of delivery (19) and early postnatal diet and weaning (20). We have previously established this model for the examination of HM feeding's effects on the humoral and cell-mediated immune response (19). The current study complements and extends our prior work by determining the bioregional microbiota in response to HM versus cow milk formula (MF). The objective was to determine the effects of HM and MF neonatal diets at postnatal day (PND) 21, as well as the impact after weaning (PND 51) on luminal microbiota.
Methods
Animal experiments
The piglet study design has been previously described (19). Animal maintenance and experimental treatments were conducted in accordance with the ethical guidelines for animal research established and approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. White Dutch Landrace Duroc sows were fed a soy-free diet and artificially inseminated in a commercial farm setting. All sows were multiparous and the resulting litter sizes were 8–11; male piglets were used. Piglets were not from the same mother; however, all the mothers and piglets were housed in the same open area, but in separate pens in the same farm. Male piglets were allowed to suckle for 48 h at the farm, then received an intramuscular injection of iron (Iron Hydrogenated Dextran, 1 mL at 100 mg/mL) at postnatal day (PND) 2, prior to transfer to the Arkansas Children's Nutrition Center vivarium. At the vivarium, piglets were individually housed, which allowed for monitoring of food intake and separation in the case of diarrhea. The temperature was maintained at 27°C for the first week and transitioned to 25°C in the second week. Piglets could see and hear each other but otherwise had no direct contact. They were fed an isocaloric diet of either HM, provided from the Mothers’ Milk Bank of North Texas, or a dairy-based MF (Similac Advance powder; Ross Products, Abbott Laboratories, Columbus, OH; n = 12/group) until PND 21. The feeding schedule was as follows: every 2 h in the first week of the study; every 4 h in the second week of study; and every 6 h in the third week of study. Each milk feeding was determined by a daily desired caloric intake (based on body weight), divided by the number of feeds. Piglets were trained to drink from nipples on a fixed schedule to provide 1.047 MJ/kg per day until PND 21. Dietary information has been published (19). At PND 14, a solid “starter pig food’ was introduced until PND 21, where full weaning to an ad libitum diet occurred (Teklad diet 140,608; Harlan). From PND 21 until PND 51, piglets were fed ad libitum in accordance with the nutrient recommendations of the NRC for growing piglets (21). Piglets were immunized on PND 21 and PND 35 with 100 μg of cholera toxin (C8052, Millipore Sigma) and 100 μg of cholera toxin subunit B (CTB; C9903, Millipore Sigma) by gavage (total volume 2 mL saline, with 0.2 M sodium bicarbonate). Piglets received omeprazole (20 mg, orally; Arkansas Children's Hospital pharmacy) and were deprived of food 8 h prior to immunization and 1 h postimmunization. We administered the DAPTACEL vaccine (0.5 mL; Arkansas Children's Hospital pharmacy) intramuscularly in the shank on the same days. Control piglets also received omeprazole and deprived of food similarly to the treatment group (19). Piglets were euthanized 6–8 h after the final feeding period by anesthetizing with isoflurane, followed by exsanguination. Upon collection, GI contents were separated into 7 sections: duodenum, jejunum, ileum, cecum, proximal colon (PC), distal colon (DC), and rectum. DC contents were collected 50 cm from the end of the colon, and PC contents were obtained 25 cm from the cecum. Ileal contents were collected 50 cm from the distal end of the small intestine. The jejunal contents were collected 229 cm from the distal end, while the duodenal contents were designated at 50 cm from the proximal end of the stomach. For the cecum contents, a 10-cm section from the middle was utilized, and the rectal content was collected right before necropsy. Luminal contents from each section were collected within a scintillation vial, immediately snap-frozen in liquid nitrogen, and stored at –80°C until use.
16s Ribosomal RNA amplicon sequencing
Bioregional GI contents were homogenized using the QIAamp Fast DNA stool mini kit (Germantown, MD; Cat#51,604) for DNA extraction. Amplification of variable region 4 of bacterial 16s ribosomal RNA (rRNA) genes was carried out using 515F/806R forward and reverse primers (22). The pool of amplicons was paired-end sequenced (2 × 250 bp) with ∼30% PhiX DNA using an Illumina Miseq. The data analysis was carried out by the Quantitative Insight into Microbial Ecology pipeline for operational taxonomic unit (OTU) clustering and taxonomic identification of amplicon sequences (23). We had a similarity threshold of 97% to cluster amplicon sequences for taxonomic annotation of OTUs using the Greengenes 16s rRNA database and a confidence threshold of 80%.
Statistical analysis
All tests utilized 16s rRNA gene sequencing data, and a P value of less than or equal to 0.05 was considered significant. All analyses were performed directly in R (version 3.5.2) or in the R-based HTML app, Dynamic Assessment of Microbial Ecology (24). Prior to the statistical analysis, sequencing counts were initially screened for sample depths <1000 to remove poorly sequenced samples. Following this screen, the mean sequencing depth for PND 21 and PND 51 samples were 24,822 and 14,958, respectively. We removed low abundant OTUs by filtering OTUs that had <10% of samples below 10 read counts. A 1-way ANOVA was used to identify diet and regional differences in Shannon and Chao1 measurements of alpha-diversity. Beta-diversity measurements were calculated using Bray–Curtis dissimilarities, and diet and regional differences were assessed by a permutational multivariate ANOVA (PERMANOVA). Dietary differences in individual taxa were assessed on shrunken log-fold differences by Wald's test for significance, as implemented in DESeq2 (25–27). Shrunken log2 fold estimates at the genera and phyla levels were calculated by DESeq2 with a false discovery rate. A P value <0.05 was considered significant. Standard log2 fold differences were also calculated and compared to the estimates derived from DESeq2. Taxa significantly altered by the diet with a mismatched DESeq2 log2 fold estimates and standard calculations were considered not significant. Immunization did not influence the measures of alpha- or beta-diversity; therefore, control and immunized animals were pooled in the analysis of the PND 51 piglet microbiota (Supplemental Tables 1 and 2). An analysis of the microbiota across time points had to be pulled from 2 miSeq runs. OTUs were summed across all samples to determine whether the total sums were different across the 2 data sets. No significant difference in total read counts was observed; thus, the data were rarefied to the lowest sample utilizing the Vegan package. The number of samples at PND 21 and PND 51 were 8 to 11 per group.
Results
Alpha-diversity is affected by diet in a region-specific manner
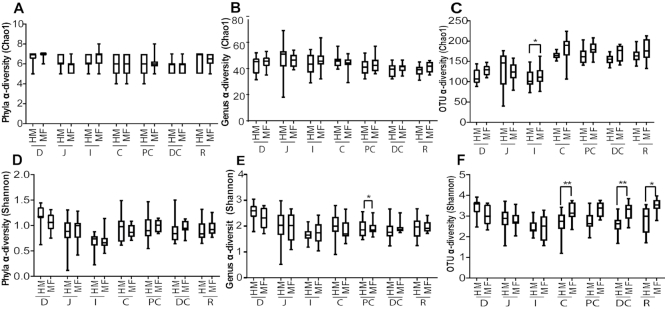
We compared PND 21 to PND 51 to determine microbial diversity (Supplemental Table 3). Statistical significance was noted for the interaction between time and tissue for Chao1 (P < 0.001). A 3-way interaction was observed for time, tissue, and diet for Shannon (P < 0.05; Supplemental Table 3) at the genera taxonomic level. Thus, we analyzed the 16s rRNA data for diet and region. At PND 21, HM- and MF-fed piglets showed similar diversity at phylum and genus taxonomic levels for Chao1 (Figures 1A and B). However, at the OTU level, the luminal region of the ileum of HM-fed piglets showed significantly lower diversity (Chao1 index) relative to MF-fed piglets (Figure 1C). For the Shannon index, no differences were noted at the phylum level between the diets across the luminal regions of the GI tract (Figure 1D). However, the HM group had a higher Shannon diversity index at the genera level in the PC lumen compared to MF-fed piglets (Figure 1E). Additionally, at the OTU level, HM-fed piglets had lower diversity (Shannon) in the luminal regions of the cecum, distal colon, and rectum compared to MF-fed piglets (Figure 1F).
FIGURE 1.
Dietary differences in alpha-diversity across luminal regions in piglets fed with HM or MF at PND 21. (A) Phyla Chao1, (B) genus Chao1, (C) OTU Chao1, (D) phyla Shannon, (E) genus Shannon, (F) OTU Shannon. Animal numbers were 9–11 for each diet group. Medians are shown with the 25th and 75th quartiles of Chao1 and Shannon diversity measurements, with significance between diets across the luminal regions.*P < 0.05, **P < 0.01. C, cecum; D, duodenum; DC, distal colon; HM, human milk; I, ileum; J, jejunum; MF, milk formula; PC, proximal colon; PND, postnatal day; OTU, operational taxonomic units; R, rectum.
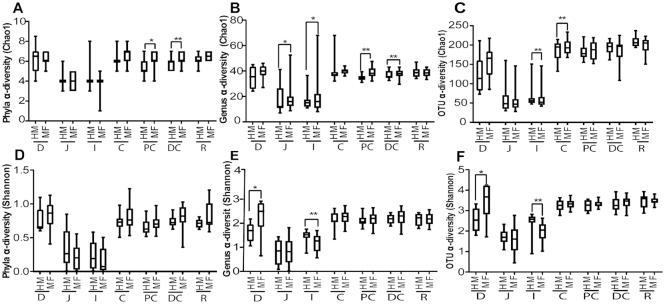
At PND 51 (weaning neonatal diet), the luminal regions of PC and DC of HM-weaned piglets showed significantly lower diversity (Chao1) relative to those weaned from the MF diet (Figure 2A). Interestingly, the HM weaned group also had lower diversity (Chao1) at the genus level within the jejunum, ileum, PC, and DC lumen, relative to those weaned from MF (Figure 2B). Similarly, at the OTU level, piglets weaned from the HM diet had lower Chao1 measures in the cecal lumen, but higher Chao1 in the ileal lumen, relative to piglets assigned to MF (Figure 2C). No dietary differences were seen at PND 51 at the phyla level using Shannon diversity measurements (Figure 2D). At the genera (Figure 2E) and OTU (Figure 2F) taxonomic levels, piglets weaned from HM diet showed significantly lower diversity (Shannon) in the duodenum lumen and higher diversity in the ileum lumen compared to those weaned from MF. Overall, HM-fed piglets had lower alpha-diversity measurements within all regions and at both feeding periods.
FIGURE 2.
Dietary differences in alpha-diversity across luminal regions in piglets fed with HM or MF at PND 51. (A) Phyla Chao1, (B) genus Chao1, (C) OTU Chao1, (D) phyla Shannon, (E) genus Shannon, (F) OTU Shannon. Animal numbers were 8–15 for each diet group. Medians are shown with the 25th and 75th quartiles of Chao1 and Shannon diversity measurements, with significance between diets across the luminal regions. *P < 0.05, **P < 0.01. C, cecum; D, duodenum; DC, distal colon; HM, human milk; I, ileum; J, jejunum; MF, milk formula; PC, proximal colon; PND, postnatal day; OTU, operational taxonomic units; R, rectum.
Region-specific beta-diversity shifts due to diet
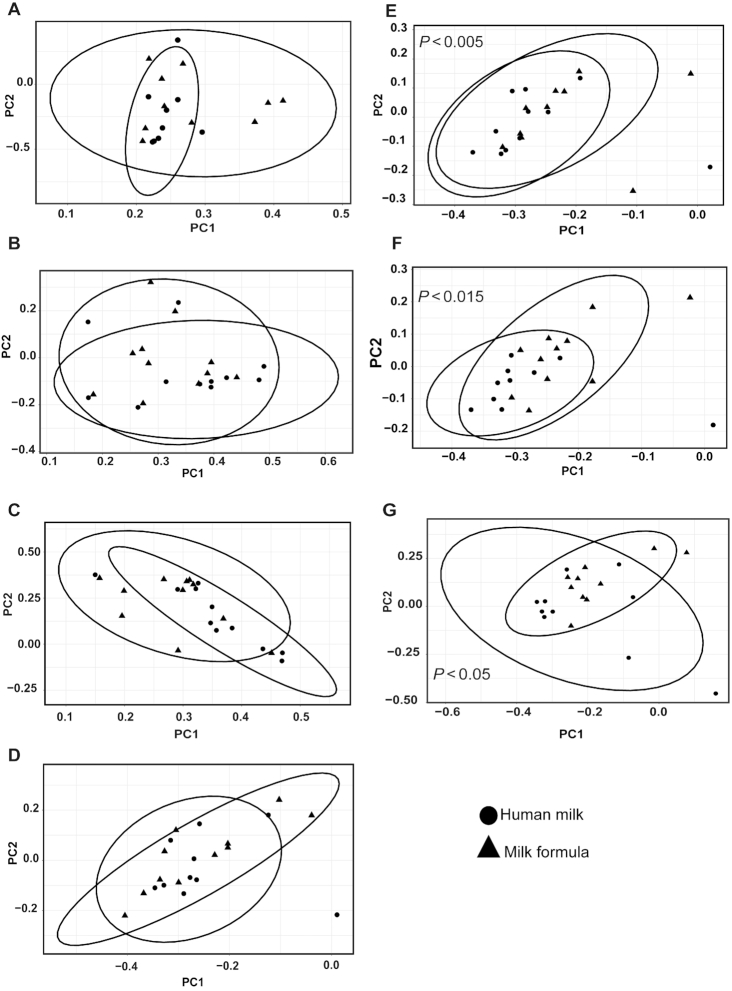
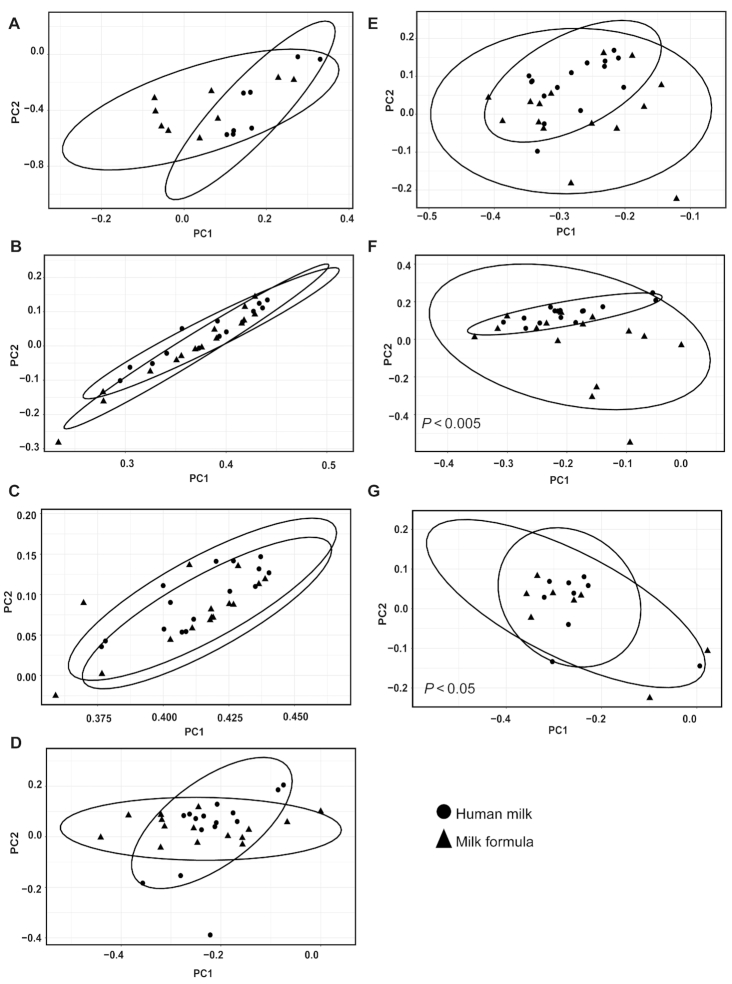
When we compared time differences from PND 21 to PND 51, the data demonstrated a clear separation of the microbiota by time at the genera taxonomic level (Supplemental Figure 1). Based on the PERMANOVA analysis, we observed an interaction between time and diet (P < 0.01) and time and tissue (P < 0.01), suggesting that differences by region and diet should be assessed within a given time (Supplemental Figure 1). Additionally, PERMANOVA at the phyla level shows a significant interaction between diet and tissue (P < 0.05). No interaction was seen at the OTU level; however, diet (P < 0.01) and tissue (P < 0.01) were independently significant (data are not shown). Figures 3 and 4 display the beta-diversity at PND 21 and PND 51 across regions at the genera taxonomic level. On PND 21, diet effect in beta-diversity was seen at the genera level in the PC (P < 0.01), DC (P < 0.01), and rectal luminal regions (P < 0.05; Figure 3). At the phyla and OTU levels, there were also significant effects of diet (phyla P < 0.01; OTU P < 0.01) and tissue (phyla P < 0.01; OTU P < 0.01; data are not shown). Moreover, significant diet-specific differences were observed in the cecum, PC, DC, and rectum luminal regions (data are not shown) at the OTU level. In the PND 51 piglets, HM-fed and MF-fed piglets showed a clear separation of the community structure in DC and rectal (Figure 4) luminal regions at the genera taxonomic level.
FIGURE 3.
Impact of neonatal diet on bacterial community across luminal regions in piglets fed with HM or MF at PND 21. Distances created with Bray–Curtis show genera taxonomic communities across luminal regions. (A) Duodenum, (B) jejunum, (C) ileum, (D) cecum, (E) proximal colon, (F) distal colon, and (G) rectum. P values represent group differences among PCoA scores along component 1. Animal numbers were 8–11 for each diet group. HM, human milk; MF, milk formula; PC, principal component; PCoA, principal coordinates analysis; PND, postnatal day.
FIGURE 4.
Impact of neonatal diet on bacterial community across luminal regions in piglets fed with HM or MF at PND 51. Distances created with Bray–Curtis show genera taxonomic communities across luminal regions. (A) Duodenum, (B) jejunum, (C) ileum, (D) cecum, (E) proximal colon, (F) distal colon, and (G) rectum. P values represent group differences among PCoA scores along component 1. Animal numbers were 8–15 for each diet group HM, human milk; MF, milk formula; PC, principal component; PCoA, principal coordinates analysis; PND, postnatal day.
Region-specific relative abundance shifts due to diet
Region-specific differences in relative abundance due to diet were seen across the GI tracts in both PND 21 (preweaned) and PND 51 (weaned) piglets. Table 1 describes significantly altered phyla due to diet across intestinal regions at PND 21. Tenericutes phylum was more abundant in the cecum, PC, and rectal luminal regions of HM-fed piglets, relative to the MF-fed group. Moreover, Fusobacteria phylum was lower in the HM-fed group in the cecum, PC, and rectum regions, in comparison to the MF group. Verrucomicrobia was more abundant only on the rectal region of the HM-fed group, relative to the MF-fed piglets. We further determined significantly altered genera at PND 21 across all regions, and only 8 genera were significantly different between the diet groups in the small intestinal regions (Table 2). In the duodenal region, genus Campylobacterwas more abundant in the HM group, while an unidentified genus from the Flavobacteriaceae family was lower in HM- compared to MF-fed piglets. In the jejunal region, Campylobacter was more abundant in HM-fed piglets, relative to the MF-fed group, while an unidentified genus from the Peptostreptococcaceae family was lower in the HM-fed group. In addition, Veillonella,Clostridium,Turicibacter,Sarcina, and an unidentified genus in the Clostridiaceae family were less abundant in the ileum region of the HM-fed group than in the MF-fed group. Campylobacter was more abundant in the HM-fed group than in the MF-fed group (Table 2). In contrast to the small intestine, 40 genera were altered across large intestinal regions between the HM- and MF-fed piglets (Table 2). In the cecal luminal region of HM-fed piglets, Dorea,Actinobacillus, Prevotella,Campylobacter,Haemophilus,Anaerovibrio, and an unassigned genus from the Prevotellaceae family, the S24-7 family, and the 2 classes Bacteroidia and Mollicutes were significantly more abundant, relative to MF-fed piglets. The genera Fusobacterium and rc4-4 were less abundant in the HM-fed group compared to MF fed-piglets. In the PC and DC regions of the large intestine, Dorea and Actinobacillus were significantly abundant in HM-fed piglets, relative to MF-fed piglets. In addition, Anaerovibrio and unassigned genera from families Prevotellaceae and S24-7 and class Bacteroidia were abundant in the PC luminal region of HM-fed piglets. In the DC region of HM-fed piglets, Prevotella and Haemophilus were more abundant, relative to MF-fed piglets. Additionally, Turicibacter was decreased in HM-fed piglets compared to MF-fed piglets, across all luminal regions of the large intestine. Interestingly, Campylobacter was found to be greater in the HM-fed group from the duodenum through the DC lumen compared to MF-fed piglets. Importantly, Campylobacter was most abundant in the duodenal lumen of the HM-fed group, at 2.11%, and decreased through the GI tract to the rectal lumen, with an abundance of 0.182%. Furthermore, in the rectal region, Dorea,Akkermansia, genera from class Bacteroidia, families S24-7 and Erysipelotrichaceae were more abundant in HM-fed piglets than in MF-fed piglets.
TABLE 1.
Relative abundance of phyla affected by human milk or milk formula in piglets at postnatal day 211
| Regions | Phylum | HM | MF | log2 FC | P adj |
|---|---|---|---|---|---|
| Cecum | Tenericutes | 3.137 ± 6.1 | 0.153 ± 0.4 | 5.2 | 0.004 |
| Cecum | Fusobacteria | 0.546 ± 1.4 | 3.935 ± 7.3 | −3.0 | 0.023 |
| Proximal colon | Tenericutes | 3.462 ± 6.5 | 0.178 ± 0.5 | 5.9 | 0.027 |
| Proximal colon | Fusobacteria | 0.631 ± 1.1 | 4.231 ± 4.3 | −3.1 | 0.044 |
| Rectum | Verrucomicrobia | 2.623 ± 6.2 | 1.099 ± 3.4 | 5.5 | 0.001 |
| Rectum | Tenericutes | 2.622 ± 6.6 | 0.197 ± 0.4 | 3.5 | 0.020 |
| Rectum | Fusobacteria | 0.241 ± 0.6 | 3.739 ± 9.8 | −3.4 | 0.001 |
Values are mean relative abundance ± SEMs, n = 8–11 per diet group. Adjusted P values from Wald test indicate differences between log2 FC values, calculated by DeSeq2 where reference group is HM. Phyla that were significant with an adjusted P value < 0.05 are shown. FC, fold change; HM, human milk; MF, milk formula.
TABLE 2.
Relative abundance of genera affected by human milk or milk formula in piglets at postnatal day 211
| Regions | Genus | HM | MF | Log2 FC | P adj |
|---|---|---|---|---|---|
| Duodenum | Campylobacter | 2.11 ± 3.5 | 0.128 ± 0.1 | 3.2 | 0.037 |
| Duodenum | f__Flavobacteriaceae | 0.12 ± 0.2 | 0.762 ± 0.9 | −3.6 | 0.037 |
| Jejunum | Campylobacter | 1.72 ± 3.8 | 0.067 ± 0.1 | 4.7 | 0.023 |
| Jejunum | f__Peptostreptococcaceae | 0.36 ± 1.0 | 0.817 ± 1.7 | −1.9 | 0.048 |
| Ileum | Campylobacter | 0.202 ± 0.3 | 0.018 ± 0.04 | 4.0 | 0.015 |
| Ileum | Veillonella | 1.161 ± 2.2 | 1.624 ± 2.4 | −2.4 | 0.015 |
| Ileum | Clostridium | 0.73 ± 1.9 | 2.43 ± 5.2 | −3.9 | 0.003 |
| Ileum | f__Clostridiaceae | 0.52 ± 0.6 | 4.727 ± 8.4 | −3.3 | 0.005 |
| Ileum | Sarcina | 0.005 ± 0.02 | 0.091 ± 0.2 | −4.5 | 0.015 |
| Ileum | Turicibacter | 0.079 ± 0.2 | 2.412 ± 4.5 | −5.4 | 0.002 |
| Cecum | c__Bacteroidia | 5.194 ± 9.1 | 2.328 ± 7.7 | 7.8 | 0.001 |
| Cecum | c__Mollicutes | 3.137 ± 6.1 | 0.153 ± 0.4 | 6.4 | 0.001 |
| Cecum | Dorea | 2.637 ± 2.1 | 0.25 ± 0.3 | 3.5 | 0.001 |
| Cecum | Actinobacillus | 1.99 ± 1.7 | 0.495 ± 0.7 | 2.2 | 0.005 |
| Cecum | Prevotella | 1.553 ± 2.8 | 0.015 ± 0.1 | 5.9 | 0.009 |
| Cecum | f__S24-7 | 1.505 ± 3.9 | 0.523 ± 1.7 | 5.7 | 0.005 |
| Cecum | Campylobacter | 0.575 ± 0.9 | 0.039 ± 0.1 | 5.5 | 0.005 |
| Cecum | Haemophilus | 0.428 ± 0.7 | 0.058 ± 0.1 | 2.7 | 0.013 |
| Cecum | Anaerovibrio | 0.176 ± 0.4 | 0.001 ± 0.002 | 7.6 | 0.015 |
| Cecum | f__Prevotellaceae | 0.069 ± 0.1 | 0.001 ± 0.002 | 6.0 | 0.045 |
| Cecum | Fusobacterium | 0.546 ± 1.4 | 3.935 ± 7.3 | −3.4 | 0.016 |
| Cecum | Turicibacter | 0.022 ± 0.1 | 0.558 ± 0.8 | −5.2 | 0.001 |
| Cecum | rc4-4 | 0.016 ± 0.02 | 0.159 ± 0.3 | −3.7 | 0.006 |
| Proximal colon | c__Bacteroidia | 3.634 ± 7.5 | 1.455 ± 4.6 | 5.9 | 0.008 |
| Proximal colon | Dorea | 3.305 ± 3.1 | 0.34 ± 0.3 | 3.8 | 0.001 |
| Proximal colon | Actinobacillus | 1.249 ± 1.9 | 0.178 ± 0.3 | 3.1 | 0.001 |
| Proximal colon | f__S24-7 | 0.635 ± 1.3 | 0.608 ± 1.9 | 5.0 | 0.044 |
| Proximal colon | Campylobacter | 0.394 ± 0.8 | 0.01 ± 0.03 | 6.1 | 0.012 |
| Proximal colon | f__Prevotellaceae | 0.124 ± 0.3 | 0.00 ± 0.00 | 22 | 0.001 |
| Proximal colon | Anaerovibrio | 0.062 ± 0.1 | 0.00 ± 0.00 | 22 | 0.001 |
| Proximal colon | Turicibacter | 0.013 ± 0.03 | 0.67 ± 1.3 | −5.4 | 0.001 |
| Proximal colon | rc4-4 | 0.01 ± 0.02 | 0.148 ± 0.2 | −4.0 | 0.019 |
| Distal colon | Dorea | 1.907 ± 1.3 | 0.577 ± 0.7 | 1.8 | 0.011 |
| Distal colon | Actinobacillus | 0.89 ± 1.6 | 0.02 ± 0.02 | 5.2 | 0.001 |
| Distal colon | Campylobacter | 0.743 ± 1.5 | 0.023 ± 0.1 | 5.1 | 0.004 |
| Distal colon | Prevotella | 0.341 ± 0.6 | 0.083 ± 0.3 | 8.7 | 0.001 |
| Distal colon | Haemophilus | 0.141 ± 0.3 | 0.004 ± 0.009 | 5.2 | 0.003 |
| Distal colon | Oscillospira | 1.151 ± 0.4 | 4.317 ± 5.8 | −1.3 | 0.018 |
| Distal colon | Ruminococcus | 0.322 ± 0.7 | 0.86 ± 1.3 | −2.6 | 0.002 |
| Distal colon | Turicibacter | 0.003 ± 0.005 | 0.381 ± 1.0 | −4.6 | 0.003 |
| Rectum | Dorea | 3.103 ± 3.4 | 0.64 ± 0.6 | 2.5 | 0.005 |
| Rectum | Akkermansia | 2.623 ± 6.2 | 1.099 ± 3.4 | 5.7 | 0.001 |
| Rectum | c__Bacteroidia | 1.884 ± 3.8 | 0.102 ± 0.3 | 4.6 | 0.019 |
| Rectum | f__S24-7 | 1.538 ± 2.5 | 0.067 ± 0.1 | 5.2 | 0.003 |
| Rectum | f__Erysipelotrichaceae | 0.762 ± 1.3 | 0.347 ± 0.8 | 2.7 | 0.017 |
| Rectum | Streptococcus | 0.622 ± 1.1 | 1.018 ± 1.1 | −1.9 | 0.019 |
| Rectum | Fusobacterium | 0.241 ± 0.6 | 3.739 ± 9.8 | −3.1 | 0.007 |
| Rectum | Eubacterium | 0.118 ± 0.1 | 1.222 ± 1.8 | −2.7 | 0.028 |
| Rectum | Turicibacter | 0.006 ± 0.01 | 0.172 ± 0.5 | −2.6 | 0.046 |
| Rectum | f__Mogibacteriaceae | 0.001 ± 0.003 | 0.056 ± 0.1 | −5.3 | 0.001 |
Values are mean relative abundance ± SEMs, n = 8–11 per diet group. Adjusted P values from Wald test indicate differences between log2 FC values, calculated by DeSeq2 where reference group is HM. Genera that were significant with an adjusted P value < 0.05 are shown. c__, Class; f__, Family; FC, fold change; HM, human milk; MF, milk formula.
At PND 51, there were a smaller number of taxa significantly affected by diet; however, in the large intestinal lumen phyla taxonomic level changes were still observed. In HM-fed piglets, Proteabacteria phyla were lower in the cecum and distal colon lumen, while Tenericutes was more abundant in PC lumen, relative to the MF-fed piglets (Table 3). No dietary differences were observed in the duodenal lumen; however, several genera were significantly altered by diet throughout the remaining regions of the GI tract (Table 4). Turicibacter, which was significant across many luminal regions at PND 21, was not altered by diet in any region at PND 51. Within this time point, multiple unspecified genera were identified, which are indicated with family or class designations in Table 4. YRC22 and an unspecified genus within the 4C0d-2 class are decreased in the HM-fed group from the cecal to the rectal lumen. Furthermore, an unidentified genus from class Bacteroidia was more abundant in HM-fed piglets than in MF-fed piglets in the distal colon lumen (Table 4).
TABLE 3.
Relative abundance of phyla affected by human milk or milk formula in piglets at postnatal day 511
| Regions | Phylum | HM | MF | log2 FC | P adj |
|---|---|---|---|---|---|
| Cecum | Proteobacteria | 1.749 ± 1.7 | 4.879 ± 3.9 | −1.4 | 0.011 |
| Proximal colon | Tenericutes | 0.478 ± 0.7 | 0.199 ± 0.2 | 1.5 | 0.024 |
| Proximal colon | Actinobacteria | 0.01 ± 0.01 | 0.05 ± 0.1 | −1.8 | 0.024 |
| Distal colon | Proteobacteria | 1.569 ± 1.2 | 5.032 ± 4.7 | −1.4 | 0.043 |
| Rectum | Firmicutes | 31.691 ± 11.0 | 27.24 ± 8.7 | 0.9 | 0.033 |
| Rectum | Lentisphaerae | 0.859 ± 1.2 | 0.103 ± 0.2 | 2.8 | 0.033 |
Values are mean relative abundance ± SEMs, n = 8–11 per diet group. Adjusted P values from Wald test indicate differences between log2 FC values, calculated by DeSeq2 where reference group is HM. Phyla that were significant with an adjusted P value < 0.05 are shown. Animal numbers are n = 8–15 per diet group. FC, fold change; HM, human milk; MF, milk formula.
TABLE 4.
Relative abundance of genera affected by human milk or milk formula in piglets at postnatal day 511
| Regions | Genus | HM | MF | log2 FC | P adj |
|---|---|---|---|---|---|
| Jejunum | Streptococcus | 0.158 ± 0.4 | 0.58 ± 0.8 | −3.0 | 0.034 |
| Jejunum | Moraxella | 0.044 ± 0.1 | 1.226 ± 3.2 | −4.6 | 0.034 |
| Ileum | f__Enterobacteriaceae | 0.559 ± 0.9 | 1.25 ± 4.8 | 5.5 | 0.001 |
| Ileum | Streptococcus | 0.166 ± 0.4 | 1.234 ± 2.6 | −4.0 | 0.001 |
| Cecum | c__Chloroplast | 0.337 ± 0.8 | 0.03 ± 0.05 | 3.1 | 0.010 |
| Cecum | Sutterella | 1.169 ± 1.3 | 3.37 ± 3.5 | −1.7 | 0.035 |
| Cecum | Faecalibacterium | 0.152 ± 0.1 | 0.643 ± 0.9 | −2.1 | 0.009 |
| Cecum | CF231 | 0.003 ± 0.009 | 0.58 ± 1.2 | −7.7 | 0.001 |
| Cecum | YRC22 | 0.002 ± 0.007 | 2.915 ± 3.9 | −10.1 | 0.001 |
| Cecum | c__4C0d-2 | ND | 1.895 ± 3.6 | −11.6 | 0.001 |
| Proximal colon | Faecalibacterium | 0.091 ± 0.1 | 0.307 ± 0.4 | −1.7 | 0.048 |
| Proximal colon | Collinsella | 0.01 ± 0.01 | 0.05 ± 0.1 | −2.2 | 0.048 |
| Proximal colon | CF231 | 0.003 ± 0.008 | 1.289 ± 2.9 | −7.9 | 0.007 |
| Proximal colon | YRC22 | 0.001 ± 0.003 | 3.662 ± 5.3 | −11.4 | 0.001 |
| Proximal colon | c__4C0d-2 | ND | 0.386 ± 0.8 | −7.6 | 0.002 |
| Distal colon | c__Bacteroidia | 13.468 ± 6.7 | 6.189 ± 5.1 | 1.5 | 0.012 |
| Distal colon | c__RF3 | 0.029 ± 0.05 | 1.067 ± 3.4 | −2.1 | 0.049 |
| Distal colon | YRC22 | ND | 2.419 ± 3.5 | −12.3 | 0.001 |
| Distal colon | c__4C0d-2 | ND | 0.445 ± 0.9 | −23.6 | 0.001 |
| Distal colon | c__Alphaproteobacteria | ND | 1.003 ± 3.2 | −7.2 | 0.001 |
| Rectum | YRC22 | 0.037 ± 0.1 | 2.893 ± 4.0 | −6.7 | 0.011 |
| Rectum | c__4C0d-2 | 0.003 ± 0.006 | 0.738 ± 1.0 | −7.6 | 0.001 |
Values are mean relative abundance ± SEMs, n = 8–15 per diet group. Adjusted P values from Wald test indicate differences between log2 FC values, calculated by DeSeq2 where reference group is HM. Genera that were significant with an adjusted P value < 0.05 are shown. c__, Class; f__, Family; FC, fold change; HM, human milk; MF, milk formula; ND, not detected.
Discussion
Several studies have shown differences in microbiota composition between HM-fed and formula-fed infants using fecal samples (11, 12, 28–31). The current study examined bioregional differences in the microbiota of piglets fed either a HM or MF diet. The piglet model has been described to be a good model for infant gut and immune system development (32) and, indeed, has been utilized to study the effects of different diets on the microbiota of various regions of the GI tract (11, 33–35). These studies have been limited, however, to 1 or 2 sections within the porcine GI tract. Moreover, when utilizing piglets for neonatal diet studies, sow-reared (SR) piglets are usually utilized as a control. Specifically relating to microbiota research, this poses a serious limitation, as SR piglets generally never leave the farm, whereas the treated piglets were transferred to a vivarium setting. The study we conducted is unique in that we utilized samples from piglets that received donor HM and at day 21 compared microbiota differences directly to piglets fed MF. Piglets used for this study were housed in the same location; thus, changes in microbiota composition are attributed to diet and not environmental influences. Interestingly, the microbiota composition differed 4 weeks after weaning from the neonatal diet (PND 51), suggesting persistent effects of a neonatal diet on the microbiota composition. Moreover, our results suggest that piglets fed HM are closer to infants in their microbiota composition.
The piglet microbiota across the 7 GI sections has been well-characterized by Crespo-Piazuelo et al. (32); thus, only dietary differences are discussed. Additionally, the bacterial and host proteins across the porcine GI tract has also been recently described (33). However, to our knowledge, the current study is the first exhaustive investigation of all 7 regions of the piglet GI tract following a neonatal diet intervention and during the neonatal versus postweaned conditions. We observed that MF-fed piglets had increased microbial diversity and richness across the luminal regions compared to HM-fed piglets, which is in agreements with infants who receive formula (11). For example, a study of Chinese infants showed that formula-fed infants have higher microbiota richness, as measured by the Shannon and Chao1 indices (28). Another study from a Canadian birth cohort also described an inverse relationship between breastfeeding and microbial diversity at 3 months of age (36). Finally, infants fed formula with added prebiotics and probiotics showed higher alpha-diversity (as measured by the Shannon index) at both 6 and 12 weeks of age compared to breast-fed babies (37). We have previously published on the small intestinal microbiota of piglets receiving either a dairy or soy formula compared to sow-fed (SF) piglets (38). When compared to the SF piglets, the formula-fed piglets displayed lowered richness and evenness within the duodenum. Additionally, within the jejunum, microbial diversity was also higher in the SF piglets than in their formula-fed counterparts (38). This was in agreement with a study by Wang et al. (39), which compared formula-fed piglets with (FP) or without prebiotics (FF) to SF piglets, and found that the SF group had the highest alpha-diversity at PND 7, the prebiotic-fed had the lowest, and the FF were in the middle. Thus, SF piglets have a higher alpha-diversity compared to FF piglets, whereas in our study HM-fed piglets had a lower alpha-diversity, which is similar to findings from breast-fed infants. This further highlights how our controlled environmental conditions of the HM-fed piglet model are closer to those of infants fed with human milk. The diversity observed in the FP group suggests that the FP diet interacts with the microbiota similar to HM-fed group. Most recently, higher microbial diversity at 4 months of life has been associated with an increased risk of developing celiac disease later in life (40). However, a different report indicated that higher microbial diversity at 6–9 months of age is associated with alleviating symptoms of atopic eczema, further suggesting that microbiota development during early life possibly plays a role in immune programming (41).
The Bray–Curtis measurements performed for beta-diversity utilized quantitative sequence abundances to map distances between samples by diet. We observed the clustering of bioregional microbiota between the small and large intestinal lumen. We also found the greatest dietary differences within the large intestinal lumen. These findings are in agreement with a study of a milk fat globule membrane and lactoferrin test diet fed to piglets, which found significant diet effects in the ascending colon and rectum (42). Indeed, the beta-diversity segregation of samples by early diet has been seen in human infants as well. Exclusively breastfed infants have been shown to segregate from those nonexclusively breastfed both prior to and after the introduction of solid foods (43, 44).
Specific bacteria were found to be significantly affected by diet across multiple luminal regions. Previous studies have demonstrated that microbes found within the small intestine are more likely to induce an IgA response than those found in the large intestine (colon) (45–47). The microbiota of the colon is likely to be involved in immune tolerance mechanisms (48, 49). Furthermore, adaptive immune responses were shaped by lymph node drainage to different compartments of the GI tract (45–47). Interestingly, in our previous publication, we have shown a CTB–IgA response in small intestinal contents (19) but no CTB–IgA response was observed in the colon (unpublished Yeruva laboratory data). Additionally, in the HM-fed piglets, Turicibacter was decreased, which has previously been found to be positively correlated with nutrient digestibility (50) and is significantly higher in SR piglets (51). In a rat hypertension model, Turicibacter-induced T-cell activation and polarization in mesenteric lymph nodes, resulting in an increased Th17/IL-17 and reduced Treg cell response (52). Interestingly, in HM-fed piglets, Turicibacter was decreased, in comparison to MF-fed piglets. Furthermore, a gut commensal, Prevotella histicola, has been shown to decrease Th1/Th17 cells and increase the CD4 + FoxP3 + regulatory T-cells (53). Our study shows that Prevotella is more abundant in HM-fed piglets than in MF-fed piglets. Thus, it is possible that some of the commensal bacteria observed in the HM-fed group exhibit a protective immune response compared to the MF-fed group. We have previously shown that FF piglets have altered GI morphology compared to SR piglets, while we found no differences between the HM- and MF-fed groups (19, 54). Some of these bacteria, however, may be influencing the GI tract gene expression and immune cell composition, which is the scope of future studies. In this paper, we are reporting on the luminal microbiota, which may not interface with the intestinal epithelial cells. Bacteria within the mucosa, which can interface with the epithelial layer, may play a larger role in the development of the GI immune system. The aim here is to describe the bioregional microbiota differences while examining the bacteria found within the mucosal layer would enhance our understanding of the diet effects on mucosal gut microbiota. In addition, the DNA isolation kit used in the study does not include the step for mechanical disruption of the bacteria. Thus, it is possible that some of the microbiota, such as Lactobacillus species, present in the small and large intestines of piglets were not detected. Future studies comparing the DNA isolation procedures are needed to determine the differences in microbiota composition.
The major differences found in this study were from the lumen of the large intestine, which appears to be similar to the infant fecal microbiota composition. Thus, examining the fecal material from infants can be a good proxy to the infant distal end of the GI tract. At 3 months of age, breastfed babies have a higher percentage of Bacteroidetes and a lower percentage of Firmicutes and Verrucomicrobia compared with FF infants (55). It is interesting to note that the opposite findings were noted in our study, as Verrucomicrobia was higher at the PND 21 time point in the HM-fed piglets. In a study by Poroyko et al. (51) in which MF-fed and SR piglets were studied, Bacteroides and Parabacteroides were significantly higher, whereas Prevotella,Oscillibacter, and Turicibacter were significantly lower, in the MF-fed group. Turicibacter was significantly higher in the MF-fed group at the weaning time point across multiple regions: the ileum, cecum, PC, DC, and rectum. Prevotella, in contrast, was significantly lower in the MF-fed group at this time point in the cecum and DC. In addition, we found unspecified genera in the Bacteroidia class were more abundant in the HM-fed group within the cecum, PC, and rectal lumen, relative to the MF-fed group. Studies conducted worldwide, such as in Norway (56), Sweden (57), Canada (58), Italy (59), Switzerland (60), Bangladesh (61), the United States (62), Malawi, and Finland (63), show that some of the healthy breast-fed infants have lower amounts of Bifidobacterium species than others. The most recent report suggests that at least 3 compositionally distinct neonatal gut microbiota might be present, with a high relative abundance of Bacteroides or Bifidobacteria, and higher abundances of Bifidobacterium and Bacteroides (64). Interestingly, when MF-fed piglets were compared to SR piglets, more abundant Bacteroides were observed in MF-fed piglets, which is the opposite of what is seen in infants. Moreover, we observed a higher abundance of genera from class Bacteroidia in the large intestine, and also Bacteroides genera in feces, in HM-fed piglets (19) relative to MF-fed piglets.
In summary, our data suggest that piglets fed with human milk are closer to infants fed breastmilk. These data further highlight Dutch microbiologist Lourens Baas Becking's hypothesis, “everything is everywhere, but the environment selects” (65). Finally, the complex mixture of breastmilk could be key to shaping the gut microbiota. Future studies are needed to understand how microbial colonization impacts immune programming and health outcomes later in life.
Supplementary Material
Acknowledgments
The authors thank Matt Ferguson, Bobby Fay, and Trae Pittman for vivarium help with the piglets.
The authors’ responsibilities were as follows—LY: designed the research; LRB, KM, LY: wrote the manuscript; AKB: conducted technical aspects of the research; SVC, BDP, KS: helped with microbiota data acquisition and analysis; LY: had primary responsibility for the final content and for editing the manuscript; and all authors: read and approved the final manuscript.
Notes
This study was funded by the USDA Agricultural Research Service (project 6026-51000-010-05S).
Authors' disclosures: LY is funded by NIH P20GM121293. LRB, KM, BDP, AKB, SVC, KS, no conflicts of interest.
Supplemental Tables 1–3 and Supplemental Figure 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: CTB, cholera toxin subunit B; DC, distal colon; FF, formula-fed piglets without prebiotics; FP, formula-fed piglets with prebiotics; GI, gastrointestinal; HM, human breast milk; HMO, human milk oligosaccharide; MF, milk formula; PC, proximal colon; PERMANOVA, permutational multivariate ANOVA; PND, postnatal day; OTU, operational taxonomic units; rRNA, ribosomal RNA; SF, sow-fed; SR, sow-reared.
References
- 1. Beaudry M, Dufour R, Marcoux S. Relation between infant feeding and infections during the first six months of life. J Pediatr. 1995;126(2):191–7. [DOI] [PubMed] [Google Scholar]
- 2. Beaudry M, Dufour R, Marcoux S. Breast feeding and protection against infection in industrialized countries. Arch Pediatr. 1996;3(Suppl 1):126s–7s. [DOI] [PubMed] [Google Scholar]
- 3. Hanson LA, Korotkova M. The role of breastfeeding in prevention of neonatal infection. Semin Neonatol. 2002;7(4):275–81. [DOI] [PubMed] [Google Scholar]
- 4. Hanson LA, Korotkova M, Telemo E. Breast-feeding, infant formulas, and the immune system. Ann Allergy Asthma Immunol. 2003;90(6) Suppl 3:59–63. [DOI] [PubMed] [Google Scholar]
- 5. Hanson LA, Hofvander Y, Lindquist B, Zetterstrom R. Breast-feeding as a protection against gastroenteritis and other infections. Acta Paediatr Scand. 1985;74(5):641–2. [DOI] [PubMed] [Google Scholar]
- 6. Meier PP. Human milk and clinical outcomes in preterm infants. Nestle Nutr Inst Workshop Ser. 2019;90:163–74. [DOI] [PubMed] [Google Scholar]
- 7. Ayechu-Muruzabal V, van Stigt AH, Mank M, Willemsen LEM, Stahl B, Garssen J, Van't Land B. Diversity of human milk oligosaccharides and effects on early life immune development. Front Pediatr. 2018;6:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comstock SS, Li M, Wang M, Monaco MH, Kuhlenschmidt TB, Kuhlenschmidt MS, Donovan SM. Dietary human milk oligosaccharides but not prebiotic oligosaccharides increase circulating natural killer cell and mesenteric lymph node memory T cell populations in noninfected and rotavirus-infected neonatal piglets. J Nutr. 2017;147(6):1041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doherty AM, Lodge CJ, Dharmage SC, Dai X, Bode L, Lowe AJ. Human milk oligosaccharides and associations with immune-mediated disease and infection in childhood: a systematic review. Front Pediatr. 2018;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8(2):143–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe. 2011;17(6):478–82. [DOI] [PubMed] [Google Scholar]
- 13. Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, Xu G, Davis JCC, Lebrilla CB, Henrick BM et al.. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. 2017;2(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ et al.. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360(6390):795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heinritz SN, Mosenthin R, Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr Res Rev. 2013;26(2):191–209. [DOI] [PubMed] [Google Scholar]
- 16. Moughan PJ, Birtles MJ, Cranwell PD, Smith WC, Pedraza M. The piglet as a model animal for studying aspects of digestion and absorption in milk-fed human infants. World Rev Nutr Diet. 1992;67:40–113. [DOI] [PubMed] [Google Scholar]
- 17. Darragh AJ, Moughan PJ. The three-week-old piglet as a model animal for studying protein digestion in human infants. J Pediatr Gastroenterol Nutr. 1995;21(4):387–93. [DOI] [PubMed] [Google Scholar]
- 18. Helm RM, Golden C, McMahon M, Thampi P, Badger TM, Nagarajan S. Diet regulates the development of gut-associated lymphoid tissue in neonatal piglets. Neonatology. 2007;91(4):248–55. [DOI] [PubMed] [Google Scholar]
- 19. Miklavcic JJ, Badger TM, Bowlin AK, Matazel KS, Cleves MA, LeRoith T, Saraf MK, Chintapalli SV, Piccolo BD, Shankar K et al.. Human breast-milk feeding enhances the humoral and cell-mediated immune response in neonatal piglets. J Nutr. 2018; 148 (11): 1860-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guevarra RB, Hong SH, Cho JH, Kim BR, Shin J, Lee JH, Kang BN, Kim YH, Wattanaphansak S, Isaacson RE et al.. The dynamics of the piglet gut microbiome during the weaning transition in association with health and nutrition. J Anim Sci Biotechnol. 2018;9:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Council NR. Nutrient Requirements of Swine: Eleventh Revised Edition. Washington, DC: The National Academies Press; 2012. [Google Scholar]
- 22. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Piccolo BD, Wankhade UD, Chintapalli SV, Bhattacharyya S, Chunqiao L, Shankar K. Dynamic assessment of microbial ecology (DAME): a web app for interactive analysis and visualization of microbial sequencing data. Bioinformatics. 2018;34(6):1050–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fan W, Huo G, Li X, Yang L, Duan C. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. J Microbiol Biotechnol. 2014;24(2):133–43. [DOI] [PubMed] [Google Scholar]
- 29. Fanaro S, Vigi V. Infant formulas supplemented with prebiotics: Intestinal microbiota and immune responses. Minerva Pediatr. 2008;60(3):327–35. [PubMed] [Google Scholar]
- 30. Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13(4):r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I, Donovan SM. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60(6):825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Crespo-Piazuelo D, Estelle J, Revilla M, Criado-Mesas L, Ramayo-Caldas Y, Ovilo C, Fernandez AI, Ballester M, Folch JM. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci Rep. 2018;8(1):12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tröscher-Mußotter J, Tilocca B, Stefanski V, Seifert J. Analysis of the bacterial and host proteins along and across the porcine gastrointestinal tract. Proteomes. 2019;7(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M, Bauer LL, Chen X, Wang M, Kuhlenschmidt TB, Kuhlenschmidt MS, Fahey GC Jr., Donovan SM. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. J Nutr. 2012;142(4):681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alizadeh A, Akbari P, Difilippo E, Schols HA, Ulfman LH, Schoterman MH, Garssen J, Fink-Gremmels J, Braber S. The piglet as a model for studying dietary components in infant diets: effects of galacto-oligosaccharides on intestinal functions. Br J Nutr. 2016;115(4):605–18. [DOI] [PubMed] [Google Scholar]
- 36. Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, Sears MR, Mandhane PJ, Turvey SE, Subbarao P et al.. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016;123(6):983–93. [DOI] [PubMed] [Google Scholar]
- 37. Simeoni U, Berger B, Junick J, Blaut M, Pecquet S, Rezzonico E, Grathwohl D, Sprenger N, Brussow H, Szajewska H et al.. Gut microbiota analysis reveals a marked shift to bifidobacteria by a starter infant formula containing a synbiotic of bovine milk-derived oligosaccharides and Bifidobacterium animalis subsp. lactis CNCM I-3446. Environ Microbiol. 2016;18(7):2185–95. [DOI] [PubMed] [Google Scholar]
- 38. Piccolo BD, Mercer KE, Bhattacharyya S, Bowlin AK, Saraf MK, Pack L, Chintapalli SV, Shankar K, Adams SH, Badger TM et al.. Early postnatal diets affect the bioregional small intestine microbiome and ileal metabolome in neonatal pigs. J Nutr. 2017;147(8):1499–509. [DOI] [PubMed] [Google Scholar]
- 39. Wang M, Radlowski EC, Monaco MH, Fahey GC Jr., Gaskins HR, Donovan SM. Mode of delivery and early nutrition modulate microbial colonization and fermentation products in neonatal piglets. J Nutr. 2013;143(6):795–803. [DOI] [PubMed] [Google Scholar]
- 40. Olivares M, Walker AW, Capilla A, Benitez-Paez A, Palau F, Parkhill J, Castillejo G, Sanz Y. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome. 2018;6(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015;70(2):241–4. [DOI] [PubMed] [Google Scholar]
- 42. Berding K, Wang M, Monaco MH, Alexander LS, Mudd AT, Chichlowski M, Waworuntu RV, Berg BM, Miller MJ, Dilger RN et al.. Prebiotics and bioactive milk fractions affect gut development, microbiota and neurotransmitter expression in piglets. J Pediatr Gastroenterol Nutr. 2016;63(6):688–97. [DOI] [PubMed] [Google Scholar]
- 43. Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D et al.. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157(Pt 5):1385–92. [DOI] [PubMed] [Google Scholar]
- 44. Thompson AL, Monteagudo-Mera A, Cadenas MB, Lampl ML, Azcarate-Peril MA. Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front Cell Infect Microbiol. 2015;5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Esterhazy D, Canesso MCC, Mesin L, Muller PA, de Castro TBR, Lockhart A, ElJalby M, Faria AMC, Mucida D. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature. 2019;569:126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Houston SA, Cerovic V, Thomson C, Brewer J, Mowat AM, Milling S. The lymph nodes draining the small intestine and colon are anatomically separate and immunologically distinct. Mucosal Immunol. 2016;9(2):468–78. [DOI] [PubMed] [Google Scholar]
- 47. Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B et al.. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–85. [DOI] [PubMed] [Google Scholar]
- 49. Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187(2):733–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niu Q, Li P, Hao S, Kim SW, Du T, Hua J, Huang R. Characteristics of gut microbiota in sows and their relationship with apparent nutrient digestibility. Int J Mol Sci. 2019;20(4):E870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poroyko V, White JR, Wang M, Donovan S, Alverdy J, Liu DC, Morowitz MJ. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One. 2010;5(8):e12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toral M, Robles-Vera I, de la Visitacion N, Romero M, Sanchez M, Gomez-Guzman M, Rodriguez-Nogales A, Yang T, Jimenez R, Algieri F et al.. Role of the immune system in vascular function and blood pressure control induced by fecal microbiota transplantation in rats. Acta Physiol (Oxf). 2019:e13285. [DOI] [PubMed] [Google Scholar]
- 53. Mangalam A, Shahi SK, Luckey D, Karau M, Marietta E, Luo N, Choung RS, Ju J, Sompallae R, Gibson-Corley K et al.. Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell Rep. 2017;20(6):1269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yeruva L, Spencer NE, Saraf MK, Hennings L, Bowlin AK, Cleves MA, Mercer K, Chintapalli SV, Shankar K, Rank RG et al.. Formula diet alters small intestine morphology, microbial abundance and reduces VE-cadherin and IL-10 expression in neonatal porcine model. BMC Gastroenterol. 2016;16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Donovan SM, Wang M, Li M, Friedberg I, Schwartz SL, Chapkin RS. Host-microbe interactions in the neonatal intestine: role of human milk oligosaccharides. Adv Nutr. 2012;3(3):450S–5S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol. 2014;87(1):280–90. [DOI] [PubMed] [Google Scholar]
- 57. Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44(6):842–50. [DOI] [PubMed] [Google Scholar]
- 58. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL et al.. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185(5):385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roos S, Dicksved J, Tarasco V, Locatelli E, Ricceri F, Grandin U, Savino F. 454 pyrosequencing analysis on faecal samples from a randomized DBPC trial of colicky infants treated with Lactobacillus reuteri DSM 17938. PLoS One. 2013;8(2):e56710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7(8):e44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134(2):e362–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB et al.. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grzeskowiak L, Collado MC, Mangani C, Maleta K, Laitinen K, Ashorn P, Isolauri E, Salminen S. Distinct gut microbiota in southeastern African and northern European infants. J Pediatr Gastroenterol Nutr. 2012;54(6):812–6. [DOI] [PubMed] [Google Scholar]
- 64. Borewicz K, Suarez-Diez M, Hechler C, Beijers R, de Weerth C, Arts I, Penders J, Thijs C, Nauta A, Lindner C et al.. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci Rep. 2019;9(1):2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. de Wit R, Bouvier T. “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say?. Environ Microbiol. 2006;8(4):755–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.