Abstract
Drinking water outbreaks occur worldwide and may be caused by several factors, including raw water contamination, treatment deficiencies, and distribution network failure. This study describes two drinking water outbreaks in Finland in 2016 (outbreak I) and 2018 (outbreak II). Both outbreaks caused approximately 450 illness cases and were due to drinking water pipe breakage and subsequent wastewater intrusion into the distribution system. In both outbreaks, the sapovirus was found in patient samples as the main causative agent. In addition, adenoviruses and Dientamoeba fragilis (outbreak I), and noroviruses, astroviruses, enterotoxigenic and enterohemorragic Escherichia coli (ETEC and EHEC, respectively) and Plesiomonas shigelloides (outbreak II) were detected in patient samples. Water samples were analyzed for the selected pathogens largely based on the results of patient samples. In addition, traditional fecal indicator bacteria and host-specific microbial source tracking (MST) markers (GenBac3 and HF183) were analyzed from water. In drinking water, sapovirus and enteropathogenic E. coli (EPEC) were found in outbreak II. The MST markers proved useful in the detection of contamination and to ensure the success of contaminant removal from the water distribution system. As mitigation actions, boil water advisory, alternative drinking water sources and chlorination were organized to restrict the outbreaks and to clean the contaminated distribution network. This study highlights the emerging role of sapoviruses as a waterborne pathogen and warrants the need for testing of multiple viruses during outbreak investigation.
Keywords: waterborne outbreak, enteric viruses, contamination, drinking water, wastewater, sapovirus, microbial source tracking, fecal indicators, Dientamoeba fragilis
1. Introduction
The drinking water contaminated with pathogenic microbes may cause large community outbreaks with up to thousands of illness cases in both developing and developed countries. Several factors may cause a drinking water outbreak. Raw water contamination, treatment deficiencies, and distribution network failure are among the most common causes [1]. In addition, waterborne outbreaks have been associated with climatic conditions, especially with increased precipitation and heavy rainfall events [1,2,3,4]. The source of the contamination is most commonly wastewater which may harbor a large number of diverse pathogenic microbes.
In Finland, a food and waterborne outbreak surveillance system has revealed several waterborne outbreaks every year since 1997. In these outbreaks, norovirus has been the most common causative agent followed by Campylobacter [5,6]. In addition to noroviruses, the potential waterborne spread of other enteric viruses, such as adenoviruses [7,8], sapoviruses [9,10], enteroviruses [8], astroviruses [11] and rotaviruses [8] have been reported in Finland.
Sapoviruses are close relatives to noroviruses and the clinical symptoms of sapovirus gastroenteritis are indistinguishable from those caused by noroviruses. Though, in general, the clinical severity of sapovirus-associated disease is milder than that for norovirus and rotavirus [12]. Sapoviruses are common in wastewater [13,14], and due to the availability of improved methodologies, these viruses are also now being analyzed and detected more often. An increasing number of reports related to outbreaks and sporadic cases caused by sapovirus have been described, highlighting the emerging role of sapoviruses as a public health concern [15,16,17,18,19,20,21].
Traditionally, the microbiological quality of drinking water has been estimated by using fecal indicator bacteria (FIB), such as Escherichia coli, intestinal enterococci and Clostridium perfringens. These FIB are part of the normal flora in the intestinal tract of humans and other warm-blooded animals, and thus they are consistently present in wastewater. However, the capability of these indicators to measure water quality and predict waterborne outbreaks has been questioned [22,23,24]. Therefore, more specific and sensitive fecal indicators of water quality have been explored. Potential candidates are the genetic markers from the group of Bacteroidales, such as general Bacteroidales genetic marker (GenBac3) [25] and the host-specific HF183 marker [26], used as targets in quantitative PCR (qPCR) assays for the detection of fecal contamination and human wastewater pollution, respectively. Although the qPCR assays are often designed to target the ribosomal RNA gene (rDNA), it has been proven that the detection frequency of fecal bacteria in water can be enhanced by targeting the assays to rRNA transcripts instead of rDNA [27,28]. While Bacteroidales assays are widely applied in studies of microbial source tracking (MST) in surface waters [29], their use as part of community-wide waterborne outbreak investigations is rare [10]. Thus, more data to assess the suitability of these new indicators as a tool to describe drinking water contamination episodes, to detect drinking water quality deficiencies and their application in processes securing good drinking water quality, is needed.
This study describes two waterborne outbreaks both caused by the intrusion of wastewater into a drinking water distribution system due to pipe breakage. Causative agents of outbreaks were determined through investigations of patient and water samples and the suitability of both traditional FIB and new candidates (GenBac3 and HF183) to provide water quality information was evaluated.
2. Materials and Methods
2.1. Outbreak Descriptions and Samples
This study describes two drinking water outbreaks in Finland in October 2016 (outbreak I) and January 2018 (outbreak II). Both outbreaks were initially caused by the drinking water pipe breakage and subsequent wastewater intrusion into the distribution system. Information regarding the outbreaks was collected from the local investigation reports, including retrospective questionnaires, and personal communications. The outbreaks were defined as waterborne outbreaks with a strong strength of association based on classification criteria presented previously [30,31].
2.1.1. Outbreak I
In outbreak I, the cause of the contamination was a maintenance well containing the air release valves of both drinking water and wastewater pipes (Figure 1). The air release valve of the wastewater pipe allowed wastewater to leak and accumulate into the maintenance well. Due to pipe breakage on the road construction site on 12th October 2016, the under pressure in the drinking water network caused the wastewater inflow from the maintenance well through the air release valve into the drinking water distribution system. The pipe breakage was detected and repaired immediately but the cross-contamination in maintenance well was detected only after six days on 18th October 2016.
Figure 1.
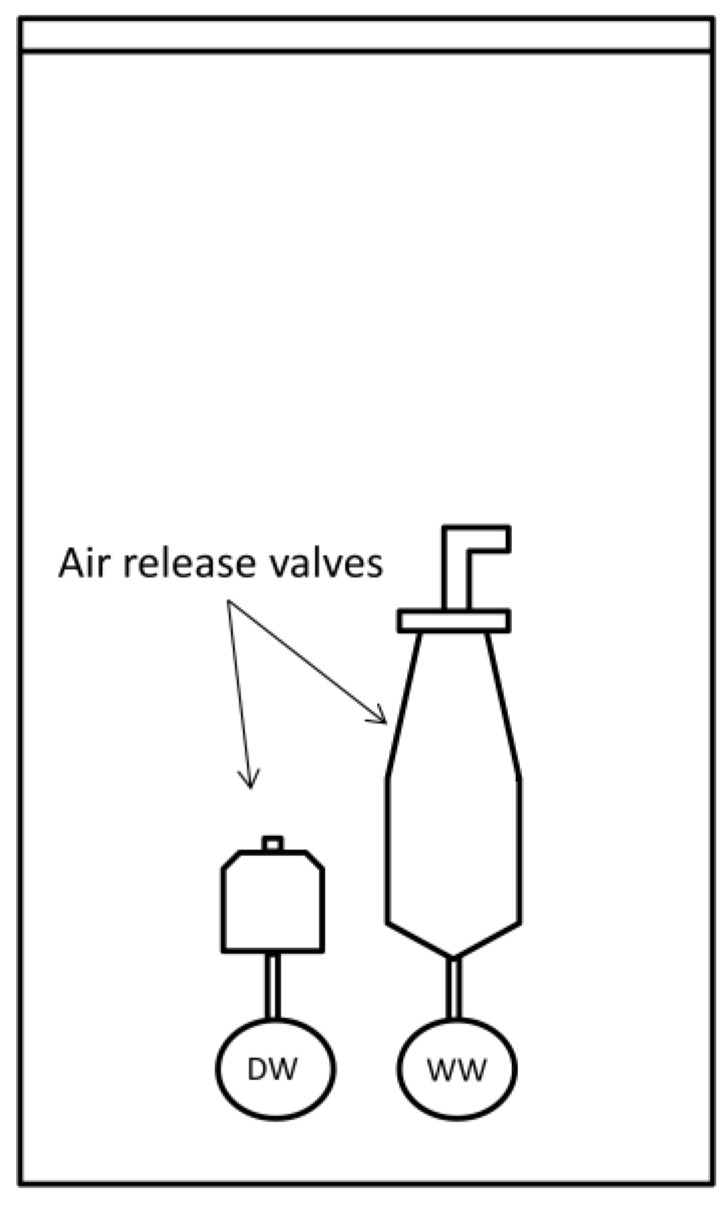
Schematic presentation of the maintenance well containing air release valves of drinking water (DW) pipe and wastewater (WW) pipe.
Drinking water originating from the groundwater source was flocculated with KMnO4, pH was adjusted with NaOH followed by clarification and sand filtration through three sand basins and finally UV-treated prior to distribution. Drinking water was not chlorinated. Mitigation actions during the outbreak included the boil water advisory for two months (from 16th October to 16th December 2016) and chlorination for 50 days (from 16th October to 5th December 2016). The target chlorine levels were as follows: first 2 mg/L for 3 days, then 4.5 mg/L for 3 days and finally 1 mg/L for 44 days. In addition, alternative water sources were arranged for the water users during the outbreak.
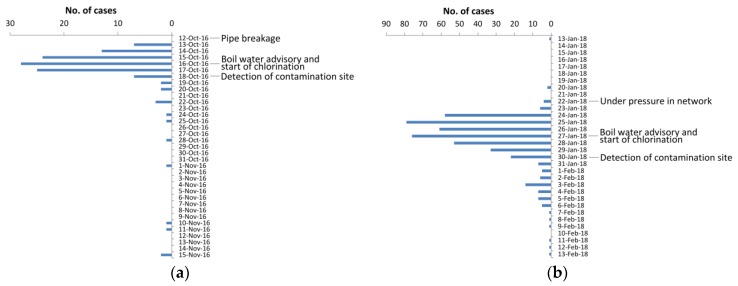
The drinking water contamination affected approximately 790 people. In order to estimate the magnitude of illness, questionnaires were sent to the households of the contaminated area. The response rate was 62% (294/471 households). In the analysis, only one response per household was included. Thus, in total, 115 symptomatic cases of 283 respondents were observed (Figure 2a). When respondents’ family members with gastrointestinal illness were taken into account, the estimated number of patients was 458. According to a questionnaire study, the first patients appeared one day after the pipe breakage. The median duration of the symptoms was one to two days and the most frequently reported symptoms included abdominal pain (94%, 101/107), nausea (91%, 100/110), diarrhea (89%, 100/112), abdominal swelling (83%, 86/104), muscular pain (66%, 64/97), vomiting (53%, 52/98) and fever (46%, 42/91). The symptoms suggested a viral point source outbreak with a rapid increase of cases followed by a fast decrease after the mitigation actions (Figure 2a). In the acute phase of the outbreak, stool samples were collected from patients between 19th October and 3rd November 2016, and were analyzed in clinical laboratories with routine tests for enteric viruses, pathogenic bacteria, and protozoans (Table 1).
Figure 2.
Epidemic curves in (a) outbreak I and (b) outbreak II, including key actions.
Table 1.
The number of positive patient samples/no. of studied samples.
| Microbe | Outbreak I | Outbreak II |
|---|---|---|
| Sapovirus 1 | 16/31 | 11/33 |
| Norovirus 1 | 0/31 | 16/33 |
| Adenovirus 1 | 3/31 | 0/33 |
| Astrovirus 1 | 0/31 | 7/33 |
| Rotavirus 1 | 0/31 | 0/33 |
| ETEC 1 | Nd 3 | 2/34 |
| EHEC 1 | Nd | 1/34 |
| Plesiomonas shigelloides 2 | 0/37 | 1/34 |
| Yersinia spp., Salmonella spp., Campylobacter spp. 2 | 0/37 | 0/34 |
| Cryptosporidium spp., Giardia lamblia, Entamoeba histolytica 1 | 0/5 | 0/38 |
| Dientamoeba fragilis 1 | 2/5 | Nd |
1 Analyzed using PCR/RT-PCR; 2 Analyzed using cultivation; 3 Nd, not done.
FIB (E. coli, coliform bacteria, intestinal enterococci, and C. perfringens) were analyzed in local environmental laboratories from the water samples collected between 15th October 2016 and 27th January 2017. Drinking water samples were taken for pathogen analyses only after start of the chlorination on 24th October 2016 (n = 1) and 26th October 2016 (n = 3). Pathogen analyses for the water samples were selected and prioritized based on results from patients taking into account the available water volume. The early water samples were analyzed only for sapoviruses and protozoans (Cryptosporidium spp., Giardia lamblia, Entamoeba histolytica, and Dientamoeba fragilis). On 28th November 2016, a raw water sample (dead-end ultrafiltration, DEUF), drinking water samples (n = 3, DEUF), biofilm samples from water meters (n = 9) and a sample from the contamination site (maintenance well) were collected and analyzed for sapo- and adenoviruses, MST markers (GenBac3 and HF183), E. coli, coliform bacteria, C. perfringens and protozoans. Moreover, a sewage sample from the municipal wastewater treatment plant was collected on 26th October 2016 and analyzed for sapoviruses and protozoans.
2.1.2. Outbreak II
In outbreak II, both a drinking water pipe and a wastewater pipe were broken at the same site. It was suggested that the drinking water pipe had leaked for several months near the wastewater pipe, and eventually, this caused a collapse of the waterlogged soil and the breakage of the sewer. The under pressure event in the drinking water distribution system during the search of the leakage on 22nd January 2018 most probably caused the inflow of wastewater from the contamination site into the drinking water network. The contamination site with broken pipes was detected eight days after the assumed contamination event on 30th January 2018.
Drinking water originating from the groundwater source was alkalized and UV-treated prior to distribution. Drinking water was not chlorinated. Mitigation actions during the outbreak included the boil water advisory for four weeks (from 27th January to 23rd February 2018) and chlorination for six weeks (from 27th January to 10th March 2018) with chlorine levels ranging from <1 mg/L–2 mg/L and including 3–5 days intensive chlorination with chlorine levels 5 mg/L–10 mg/L (started on 6th February 2018). In addition, alternative water sources were arranged for six weeks (from 29th January to 11th March 2018).
The drinking water contamination affected approximately 4000 people. During January–February, 463 persons with gastrointestinal illness contacted local primary health care. Some cases occurred already before the assumed under pressure event, but most of the patient cases appeared from 24th January–30th January 2018 (Figure 2b). Symptoms lasted on average for two days and included diarrhea (76%, 352/463), vomiting (65%, 299/463) and fever (32%, 150/463). Stool samples were collected during the acute phase of the outbreak and were analyzed in clinical laboratories with routine tests for enteric viruses, pathogenic bacteria, and protozoans (Table 1). Since the clinical laboratory method did not distinguish between norovirus genogroups, seven samples were further analyzed by the genogroup-specific real-time RT-PCR [32].
FIB (E. coli, coliform bacteria, intestinal enterococci, and C. perfringens) were analyzed from drinking water samples collected between 27th January 2018 and 5th March 2018 in a local environmental laboratory. Drinking water samples for pathogen (sapo-, noro- and adenovirus, pathogenic E. coli strains, Campylobacter spp., Giardia spp. and Cryptosporidium spp.) and MST marker (GenBac3 and HF183) analyses were taken before chlorination on 27th January 2018 (n = 1), after the initial low level chlorination (<1 mg/L) on 29th January 2018 and 6th February 2018 (n = 2) and after the intensive chlorination on 14th February 2018 (n = 3, DEUF). A surface water sample from the contamination site and a biofilm sample from water meter were collected on 31st January and were analyzed for the selected microbes (Table 2).
Table 2.
Occurrence (the number of positives/no. samples) and counts (genome copies (GC), most probable number (MPN) or colony forming units (CFU)/100 mL) of studied microbes in water, biofilm, sewage and contamination site samples in outbreak I.
| Microbe | Raw Water | Water Before Chlorination | Water After Chlorination | Biofilms | Sewage | Contamination Site |
|---|---|---|---|---|---|---|
| Sapovirus | 0/1 | Nd 1 | 0/7 | 0/9 | 1/1 | 1/1 (1.0 × 107) |
| Adenovirus | 0/1 | Nd | 0/3 | 0/9 | Nd | 1/1 (5.0 × 106) |
| Giardia lamblia, Entamoeba histolytica, Cryptosporidium spp. and Dientamoeba fragilis | 0/1 | Nd | 0/7 | 0/9 | Dientamoeba fragilis | Dientamoeba fragilis |
| GenBac3 (rDNA) | 0/1 | Nd | 0/3 | 0/9 | Nd | 1/1 (4.5 × 107) |
| GenBac3 (rRNA) | 1/1 (8.0 × 100) | Nd | 1/3 (7.0×100) | 0/9 | Nd | 1/1 (7.5 × 108) |
| HF183 (rDNA) | 0/1 | Nd | 0/3 | 0/9 | Nd | 1/1 (6.3 × 106) |
| HF183 (rRNA) | 0/1 | Nd | 0/3 | 0/9 | Nd | 1/1 (2.7 × 107) |
| E. coli | 0/1 | 1/2 | 0/91 | 0/9 | Nd | 1/1 (2.8 × 105) |
| Coliform bacteria | 1/1 (3.0 × 10−2) | 1/2 | 3/91 (1.0 – 4.0×100) | 2/9 (1.0 – 2.0 × 101) | Nd | 1/1 (1.2 × 106) |
| Intestinal enterococci | 0/1 | Nd | 0/29 | 0/9 | Nd | Nd |
| C. perfringens | 0/1 | Nd | 0/12 | 0/9 | Nd | 1/1 (2.0 × 104) |
1 Nd, not done.
2.2. Environmental Investigation
2.2.1. Sample Collection and Concentration
Water was collected into sampling bottles or large volume (100–200 L) samples were taken using dead-end ultrafiltration (DEUF) method [33]. After the water sampling, sodium thiosulphate was used to inactivate the chlorine from the samples during the transport prior to microbiological analyses. In the DEUF method, water samples were collected using ASAHI Rexeed-25A (Asahi Kasei Medical Co., Ltd., Tokyo, Japan) ultrafilters with an average flow rate of 3 L/min. Backflush of the ultrafilters was performed with 500 mL of backflush solution (0.5% Tween 80, 0.01% sodium polyphosphate and 0.001% Y-30 antifoam emulsion). The secondary concentration of DEUF eluates was performed by filtration through Millipore Express PLUS membrane filters (outbreak I, pore size 0.22 μm, Merck KGaA, Darmstadt, Germany) or Nuclepore polycarbonate (PC) filters (outbreak II, pore size 0.4 μm, Whatman, Kent, UK) and/or polyethylene glycol (PEG) precipitation (Table S1). In PEG precipitation, the sample (pH 7–7.5) was mixed with 1% BSA (only for drinking water samples), 0.9 M NaCl and 12% PEG8000 and kept for at least 2 h at 4 °C. After incubation, the sample was centrifuged 10 000× g for 30 min at 4 °C and the pellet was suspended in PBS. Biofilm from water meters was detached and collected as previously described [34]. Before further analyses, all biofilm samples were sonicated for 1 min in 40 kHz (Branson Ultrasonics, Danbury, USA). Biofilm samples were concentrated by filtration through PC filters and PEG precipitation of the filtrate.
2.2.2. Detection of Enteric Virus Genomes
Enteric viruses were analyzed in raw water and drinking water samples either with low volume (1–2 L) adsorption-elution methods or a large volume DEUF method. Low volume samples were concentrated using disc filters (Sartolon polyamide, Sartorius, Göttingen, Germany; Zetapor, Amf-Cuno, Meriden, USA or Nanoceram, Argonide, Sanford, USA) as previously described [35] or modified from Maunula et al. [36], Schultz et al. [37] and Kim and Ko [38]. Samples from contamination sites were analyzed from a volume of 400 mL by PEG precipitation (outbreak I) or extracted directly from a volume of 2.5 mL (outbreak II). Viral RNA and DNA were extracted from the low volume concentrates using the High Pure Viral RNA Kit and High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Mannheim, Germany), respectively, or the Nuclisens Minimag system (bioMerieux, Marcy-l’Etoile, France). In addition, High Pure Viral Nucleic Acid Large Volume Kit (Roche Diagnostics GmbH) was used with PEG precipitates and directly extracted samples. A sewage sample from municipal wastewater treatment plant was directly treated with Nuclisens kit and the nucleic acid was further purified using OneStep™ PCR Inhibitor Removal (Zymo Research, Irvine, USA). Extractions were made according to the manufacturers’ instructions. Extracted nucleic acids were stored at −75 °C.
For noroviruses, the real-time RT-qPCR assays were carried out in one step, separately for genogroups I and II, using the TaqMan® Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, Austin, TX, USA) as well as primers and probes as previously described [35,39]. For sapoviruses, the real-time RT-qPCR assays were carried using the same protocol with noroviruses [39] or using the QuantiTect probe RT-PCR kit (Qiagen, Hilden, Germany) with a slightly modified norovirus protocol [40]. Sapovirus primers and probes were according to the study by Oka et al. [41] or van Maarseveen et al. [42]. Adenoviruses were detected using primers and a probe described by Jothikumar et al. [43] with the real-time qPCR assay as described previously [44]. The adenovirus real-time qPCR program was 95 °C for 10 min, followed by 45 cycles at 95 °C for 15 s and 60 °C for 1 min. The virus assays were carried out using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Foster City, USA) or the RotorGene PCR cycler (Qiagen). Quantification of genome copies (GC) of each virus was done using standard curves generated with serially diluted gBlocks® Gene Fragments (Integrated DNA Technologies, Leuven, Belgium). The standard curves were included in each run.
The quality of virus extraction was controlled by positive and negative process controls through all stages of the analytical steps. Spiked mengovirus strain VMC0 and human adenovirus 40 (ATCC VR-931) were used as a positive process controls and sterile deionized water as a negative process control. External amplification controls (EACs) were used to control norovirus GI and GII RT-PCR inhibition in samples as previously described [35]. No inhibition was detected in samples tested negative in norovirus analysis.
Genotyping of sapovirus and norovirus was performed with conventional RT-PCR using One-Step RT-PCR kit (Qiagen). Sapovirus genome was amplified using primers p289 and p290 [45]. Norovirus RNA was amplified in polymerase region A according to Vinjé et al. [46]. The amplicons visualized in gel electrophoresis were sent to Sanger sequencing in the Institute of Biotechnology. Sequences were assigned using the Norovirus Genotyping Tool [47] or with NCBI database using BLAST (basic local alignment search tool).
2.2.3. Enumeration of Indicator Bacteria
Standard methods were used to enumerate E. coli, coliform bacteria, intestinal enterococci, and C. perfringens count from water and biofilm samples. In brief, E. coli and coliform bacteria were analyzed using membrane filtration with LES Endo medium [48] and Chromocult Coliform Agar medium [49] or by using the most probable number (MPN) method based on Colilert-18 QuantiTray [50]. The counts of intestinal enterococci were analyzed using the membrane filtration on Slanetz and Bartley medium [51] or Enterolert (IDEXX Laboratories Inc, Westbrook, USA). Vegetative cells and spores of C. perfringens were enumerated on tryptose sulfite cycloserine agar following the international standard [52].
2.2.4. Detection of Microbial Source Tracking (MST) Markers
MST markers were analyzed from nucleic acids extracted from samples of raw water, drinking water and biofilms of water meters either using DEUF method or PC filters. Samples from contamination sites were extracted directly. The nucleic acids were extracted using Chemagic DNA Plant kit (Perkin Elmer, Waltham, USA). Complementary DNA was synthesized as previously described (outbreak I) [34] or by using Superscript IV VILO (outbreak II, Thermo Fisher Scientific, Waltham, USA). MST markers (GenBac3 and HF183) were quantified using DNA-based qPCR assays and RNA-based RT-qPCR assays as described earlier by Pitkänen et al. [27]. The assays were carried out with the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) using standard curves generated with serially diluted gBlocks® Gene Fragments (Integrated DNA Technologies).
2.2.5. Detection of Bacterial Pathogens
The presence/absence of thermotolerant Campylobacter spp. was determined using culture-based selective enrichment methods following the principles of the international standard [53]. Pathogenic E. coli strains (ETEC, EPEC, EHEC, and EAEC) were analyzed from nucleic acid aliquots with in-house PCR method in a clinical laboratory [54].
2.2.6. Detection of Protozoans
In outbreak I, the aliquots of nucleic acids extracted with the Nuclisens Minimag system or Chemagic DNA Plant kit were sent to protozoan (G. lamblia, E. histolytica, Cryptosporidium spp. and D. fragilis) analysis in the United Medix Laboratories Ltd. In outbreak II, Giardia spp. and Cryptosporidium spp. were analyzed from drinking water with qPCR using primers and probes described in Hill et al. [55] and Jothikumar et al. [56], respectively, from nucleic acid subsamples. Samples from the contamination site and water meter biofilms were analyzed using the immunomagnetic separation method (IMS) based on standard ISO 15553 [57]. In brief, the sample was centrifuged (15 min, 1100 g) and IMS (Dynabeads G/C Combo, IDEXX laboratories Inc) was done for pellet in the volume of 10 ml. Samples were stained with FITC and DAPI (EasyStain, bioMerieux) and analyzed with epifluorescence microscopy.
3. Results
3.1. Clinical Findings
Sapoviruses were found from patients’ stool samples in both outbreaks (Table 1). In outbreak II, sapovirus GIV was detected in one patient sample subjected for sequencing. Moreover, adenoviruses were detected in outbreak I and noroviruses and astroviruses in outbreak II. Noroviruses were not detected in outbreak I. In outbreak II, noroviruses were detected more frequently than sapoviruses. Twelve of the sixteen norovirus positive samples were sequenced successfully and identified as genotypes GI.P7 (n = 11) and GI.P6 (n = 1). In addition, seven out of 16 norovirus positive samples were further analyzed by the genogroup-specific real-time RT-PCR. Of these samples, norovirus GI was detected in all seven samples and norovirus GII in one of seven samples. Sporadic bacterial infections (outbreak II) and D. fragilis (outbreak I) were also found in patient samples.
3.2. Environmental Investigations
In outbreak I, only E. coli and coliform bacteria were analyzed before the start of the chlorination and were detected in one of the two water samples (Table 2). In addition, low counts of coliform bacteria were detected in three out of 91 water samples taken after chlorination on 17th October 2016 and 19th October 2016, and two out of nine biofilm samples on 28th November 2016. Water samples were collected for pathogen and MST-marker analyses only after chlorination. Traces of GenBac3 rRNA were found from one of the three samples on 28th November 2016. In the sample taken from the contamination site, high numbers of both pathogens and indicators were detected. Typing of sapovirus was unsuccessful for contamination site sample. The raw water sample was positive only for GenBac3 rRNA and coliform bacteria. Sewage sample taken from the municipal wastewater treatment plant on 26th October 2016 was positive for sapovirus (genotype GI.2, accession number MK689409) and D. fragilis.
In outbreak II, samples were taken before and after the start of the chlorination. Low E. coli and intestinal enterococci counts as well as both MST markers (GenBac3 and HF183) were detected from the water before chlorination (Table 3). In a sample taken after the start of the chlorination, sapovirus and genes of enteropathogenic E. coli (EPEC) were detected from the drinking water. Sapovirus genotyping was attempted but failed most probably due to the small number of viruses in the sample. Findings of fecal microbes in drinking water, however, led to the decision to perform intensive chlorination. After intensive chlorination, intestinal enterococci were detected in two out of 48 water samples taken from the same site on 15th February and 22nd February 2018. Also, small numbers of GenBac3 rDNA and rRNA copies were detected in three water samples on 15th February 2018. The sample taken from the contamination site on 31st January 2018 contained the same pathogens than detected from the patients and high levels of fecal indicators. Typing of sapovirus was unsuccessful for the contamination site sample. A biofilm sample from water meter on 31st January 2018 was positive only for GenBac3 rDNA and rRNA.
Table 3.
Occurrence (the number of positives/no. samples) and counts (GC, MPN or CFU/100 mL) of studied microbes in water, biofilm and contamination site samples in outbreak II.
| Microbe | Water before Chlorination | Water after Chlorination | Water after Intensive Chlorination | Biofilms | Contamination Site |
|---|---|---|---|---|---|
| Sapovirus | 0/1 | 1/2 (<LOQ 2) | 0/3 | 0/1 | 1/1 (1.9 × 105) |
| Norovirus GI | 0/1 | 0/2 | Nd | 0/1 | 1/1 (2.3 × 105) |
| Norovirus GII | 0/1 | 0/2 | Nd | 0/1 | 1/1 (2.8 × 103) |
| Adenovirus | 0/1 | 0/1 | Nd | Nd | 0/1 |
| ETEC, EPEC, EHEC and EAEC 3 | 0/1 | 1/2 (EPEC) | 0/3 | 0/1 | 1/1 (EHEC, ETEC, EAEC) |
| Campylobacter spp. | 0/1 | 0/2 | Nd | Nd | 0/1 |
| Giardia spp. and Cryptosporidium spp. | 0/1 | 0/2 | 0/3 | 0/1 | 0/1 |
| GenBac3 (rDNA) | 1/1 (5.9 × 103) | 1/2 (6.6 × 103) | 3/3 (<LOQ) | 1/1 (3.1 × 102) | 1/1 (6.3 × 1010) |
| GenBac3 (rRNA) | 1/1 (4.4 × 105) | 1/2 (1.9 × 105) | 3/3 (<LOQ) | 1/1 (1.7 × 104) | 1/1 (1.1 × 1013) |
| HF183 (rDNA) | 1/1 (7.7 × 102) | 1/2 (9.0 × 102) | 0/3 | 0/1 | 1/1 (6.1 × 109) |
| HF183 (rRNA) | 1/1 (5.1 × 103) | 1/2 (3.8 × 102) | 0/3 | 0/1 | 1/1 (8.0 × 1012) |
| E. coli | 1/5 (2.0 × 100) | 0/19 | 0/48 | 0/1 | 1/1 (5.5 × 105) |
| Coliform bacteria | 1/5 (1.2 × 101) | 1/19 (3.0 × 100) | 0/48 | 0/1 | 1/1 (1.7 × 106) |
| Intestinal enterococci | 2/5 (1.0 – 2.0 × 100) | 2/19 (1.0–2.0 × 100) | 2/48 (4.0 × 10−1 – 1.0 × 100) | 0/1 | 1/1 (9.0 × 104) |
| C. perfringens | Nd 1 | 1/9 (1.0 × 10−1) | 0/22 | 0/1 | 1/1 (5.9 × 103) |
1 Nd, not done; 2 LOQ, limit of quantitation; 3 Qualitative analysis.
4. Discussion
This study presents two waterborne outbreaks caused by drinking water pipe breakage and subsequent contamination of the distribution network. The sudden onset of symptoms and clinical picture of the illness fitted symptoms of viral infection [12]. Stool samples collected from patients confirmed that most of the clinical cases were due to enteric virus infections and sapoviruses were found from patients’ samples in both outbreaks. Sapovirus genotype GI.2 was detected from a sewage sample in outbreak I and sapovirus GIV in a one patient sample in outbreak II. Genotype GI.2 is one of the predominant genotypes worldwide and sapovirus GIV predominated in several countries in 2007 [12]. Unfortunately, patient samples were not sequenced more comprehensively to determine sapovirus genotypes. In many countries, including Finland, norovirus has been the most common causative agent in waterborne outbreaks [5,6], while the linkage of sapovirus infections to possible waterborne spread and outbreaks is rare [9,10]. To our knowledge, this is the first outbreak study worldwide describing the detection of sapovirus in drinking water. In the future, the significance of this emerging virus may increase and thus testing for sapovirus is important to include in waterborne outbreak investigations.
In both outbreaks, untreated municipal wastewater entered into the drinking water distribution network. Raw wastewater reflects the infection burden among the population and can contain a wide variety of pathogens. Water samples taken from the contamination sites contained the same pathogens that were detected from patient samples. However, of these pathogens only sapovirus and EPEC were detected in drinking water in outbreak II. In outbreak I, no water samples were obtained for pathogen analyses before start of the chlorination, which is presumably the main reason behind the non-detection of pathogens from drinking water. However, the first samples taken before chlorination in outbreak I were positive for coliform bacteria indicating the deficiency in the water quality. In outbreak investigations, it is important to collect enough water before mitigation actions for possible future use, in this case e.g., for sapovirus analysis. However, the pathogen sampling should not delay the actions necessary to prevent further spread of infections.
Overall, pathogens are not analyzed as comprehensively as fecal indicator bacteria (E. coli and intestinal enterococci) in environmental investigations of outbreaks. This is partly due to their higher cost compared to indicator analyses and the need for expert laboratories to conduct the tests. Even though FIB has often been insufficient to prove the safety of water [58,59,60,61], in this study, these indicators were able to detect the water contamination in both outbreaks. In outbreak I, coliform bacteria and in outbreak II, coliform bacteria, E. coli, intestinal enterococci, and C. perfringens were detected in drinking water. Noteworthy, sporadic findings of intestinal enterococci were detected in water even after intensive chlorination in outbreak II. These findings support the use of traditional FIB in water quality assessments during outbreak investigation. However, the value of indicators in the prediction of water contamination seems to be case-specific and may require massive contamination as was the situation in the outbreaks described herein and in previous outbreaks described by Kauppinen et al. [35].
In this study, the suitability of molecular qPCR assays for fecal source tracking markers (HF183 and GenBac3), along with the traditional FIB was evaluated during waterborne outbreak investigations. The use of genetic source identifiers may provide more sensitive detection of the contamination especially when the assays are targeted to rRNA transcripts in addition to the rDNA [27]. Further, by using a host-specific marker, such as HF183 it is possible to identify the source of the contamination. In contamination sites, HF183 and GenBac3 numbers were comparable or higher than the numbers of pathogenic viruses. Moreover, the markers targeting to host-specific sequences from Bacteroidales clearly outnumbered traditional FIB in contamination site samples and thus could be considered for use as specific and sensitive fecal indicators of drinking water quality. Particularly, the human-specific marker HF183 showed promising results and the findings in water were in concordance with pathogen findings. On the other hand, GenBac3 prove to be a very sensitive marker and small GenBac3 copy numbers were found in drinking water after chlorination in both outbreaks and even after intensive chlorination in outbreak II. Interestingly, Diston et al. [62] found in a Swiss groundwater study that genetic markers of Bacteroidales are sensitive indicators, but due to the higher presence of these markers compared to enteric viruses may overestimate the risk from enteric viral pathogens. Thus, more data is needed for the correct interpretation of the significance of GenBac3 marker detection after intensive chlorination in terms of health risk assessment.
Mitigation actions, including boil water advisory, providing an alternative drinking water source and chlorination of the drinking water network, were conducted in both outbreaks and proved efficient in controlling the outbreaks. Previous studies have shown the long persistence of enteric viruses and protozoans in drinking water distribution systems in cases without proper treatment or removal of the contamination source [35,63,64,65]. Even though chlorine has been shown to be an efficient decontaminant in the drinking water distribution system [65], the possible stagnant locations (i.e., dead-ends) in the network and deposits accumulated on the inner surfaces of the old pipes may hamper the success of the chlorination. These factors may explain the sporadic microbial findings in water samples followed chlorination. Therefore, it is important to allow sufficient time for chlorination and to ascertain the purity of the water with microbiological analyses as was carried out in these outbreaks. The aging water infrastructure [66] and improper drinking water pipeline construction practices pose a major challenge for water supply and may compromise drinking water safety even more often in the future.
D. fragilis detection from patient samples induced media headlines and health concerns among the water consumers. The questionable pathogenesis of this parasite [67,68] initiated a more throughout epidemiological investigation (unpublished results). Lack of knowledge related to the drinking waterborne transmission of D. fragilis increased the uncertainty of crowds and up kept the media attention on the topic over a prolonged time.
5. Conclusions
To our knowledge, this is the first outbreak study describing the detection of sapovirus in drinking water. Further, herein we proved the suitability of source tracking identifiers to be applied in waterborne outbreak investigation along with pathogens and water quality indicator analyses. Main conclusions are as follows:
This study highlights the importance of sapovirus as a waterborne pathogen, and warrants the need for testing of multiple pathogens during outbreak investigation
The MST markers proved useful in the detection of contamination and especially HF183 findings were in concordance with the pathogen results, supporting its use in drinking water outbreak investigations
Boil water advisory, alternative drinking water source and chlorination were effective mitigation actions during the outbreaks
The role of D. fragilis as human pathogen and its drinking waterborne transmission potential requires further studies
Acknowledgments
The authors acknowledge the work of personnel at both waterworks to mitigate the extent of outbreaks and for identification of the contamination sources during their technical investigations. We acknowledge the help from health officers Miia Valkonen, Kaisa Nivola and Riitta Laaksonen for their valuable work during the outbreak investigations, understanding the importance of pathogen detection and throughout environmental investigations as well as providing information regarding the outbreaks. We also acknowledge the help of the personnel from the Expert Microbiology unit and Infectious Disease Control and Vaccinations unit of the Finnish Institute for Health and Welfare. The collaborating environmental and clinical laboratories are acknowledged for providing the microbiological analysis results.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-4601/16/22/4376/s1, Table S1: Secondary concentration procedures for target microbes analyzed with the DEUF method.
Author Contributions
Conceptualization, A.K.; methodology, A.K., T.P., H.A.-H., L.M., A.-M.H. and R.R.-F.; investigation, A.K., T.P., H.A.-H., L.M., A.-M.H., R.R.-F. and I.T.M.; data curation, A.K.; writing—original draft preparation, A.K.; writing—review and editing, A.K., T.P., H.A.-H., L.M., A.-M.H., R.R.-F. and I.T.M.; visualization, A.K.; project administration, T.P. and I.T.M.; funding acquisition, T.P. and I.T.M.
Funding
This work was supported by the Finnish Ministry of Social Affairs and Health (STM/1329/2018).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Moreira N.A., Bondelind M. Safe drinking water and waterborne outbreaks. J. Water Health. 2017;15:83–96. doi: 10.2166/wh.2016.103. [DOI] [PubMed] [Google Scholar]
- 2.Curriero F.C., Patz J.A., Rose J.B., Lele S. The association between extreme precipitation and waterborne disease outbreaks in the United States, 1948–1994. Am. J. Public Health. 2001;91:1194–1199. doi: 10.2105/AJPH.91.8.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vantarakis A., Mellou K., Spala G., Kokkinos P., Alamanos Y. A gastroenteritis outbreak caused by noroviruses in Greece. Int. J. Environ. Res. Public Health. 2011;8:3468–3478. doi: 10.3390/ijerph8083468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallender E.K., Ailes E.C., Yoder J.S., Roberts V.A., Brunkard J.M. Contributing factors to disease outbreaks associated with untreated groundwater. Groundwater. 2014;52:886–897. doi: 10.1111/gwat.12121. [DOI] [PubMed] [Google Scholar]
- 5.Zacheus O., Miettinen I.T. Increased information on waterborne outbreaks through efficient notification system enforces actions towards safe drinking water. J. Water Health. 2011;9:763–772. doi: 10.2166/wh.2011.021. [DOI] [PubMed] [Google Scholar]
- 6.Guzman-Herrador B., Carlander A., Ethelberg S., de Blasio B.F., Kuusi M., Lund V., Löfdahl M., MacDonald E., Nichols G., Schönning C., et al. Waterborne outbreaks in the Nordic countries, 1998 to 2012. Eurosurveillance. 2015;20:21160. doi: 10.2807/1560-7917.ES2015.20.24.21160. [DOI] [PubMed] [Google Scholar]
- 7.Kukkula M., Arstila P., Klossner M.L., Maunula L., Bonsdorff C.H., Jaatinen P. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 1997;29:415–418. doi: 10.3109/00365549709011840. [DOI] [PubMed] [Google Scholar]
- 8.Maunula L., Klemola P., Kauppinen A., Söderberg K., Nguyen T., Pitkänen T., Kaijalainen S., Simonen M.L., Miettinen I.T., Lappalainen M., et al. Enteric viruses in a large waterborne outbreak of acute gastroenteritis in Finland. Food Environ. Virol. 2009;1:31–36. doi: 10.1007/s12560-008-9004-3. [DOI] [Google Scholar]
- 9.Räsänen S., Lappalainen S., Kaikkonen S., Hämäläinen M., Salminen M., Vesikari T. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol. Infect. 2010;138:1227–1234. doi: 10.1017/S0950268809991671. [DOI] [PubMed] [Google Scholar]
- 10.Jalava K., Rintala H., Ollgren J., Maunula L., Gomez-Alvarez V., Revez J., Palander M., Antikainen J., Kauppinen A., Räsänen P., et al. Novel microbiological and spatial statistical methods to improve strength of epidemiological evidence in a community-wide waterborne outbreak. PLoS ONE. 2014;9:e104713. doi: 10.1371/journal.pone.0104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maunula L., Kalso S., von Bonsdorff C.H., Pönkä A. Wading pool water contaminated with both noroviruses and astroviruses as the source of a gastroenteritis outbreak. Epidemiol. Infect. 2004;132:737–743. doi: 10.1017/S0950268804002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oka T., Wang Q., Katayama K., Saif L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015;28:32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hata A., Kitajima M., Katayama H. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J. Appl. Microbiol. 2013;114:545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- 14.Fioretti J.M., Rocha M.S., Fumian T.M., Ginuino A., da Silva T.P., de Assis M.R., Rodrigues J.D.S., Carvalho-Costa F.A., Miagostovich M.P. Occurrence of human sapoviruses in wastewater and stool samples in Rio De Janeiro, Brazil. J. Appl. Microbiol. 2016;121:855–862. doi: 10.1111/jam.13205. [DOI] [PubMed] [Google Scholar]
- 15.Pang X.L., Lee B.E., Tyrrell G.J., Preiksaitis J.K. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004–2007. J. Infect. Dis. 2009;199:547–551. doi: 10.1086/596210. [DOI] [PubMed] [Google Scholar]
- 16.Svraka S., Vennema H., van der Veer B., Hedlund K., Thorhagen M., Siebenga J., Duizer E., Koopmans M. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J. Clin. Microbiol. 2010;48:2191–2198. doi: 10.1128/JCM.02427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey S.K., Phathammavong O., Nguyen T.D., Thongprachum A., Chan-It W., Okitsu S., Mizuguchi M., Ushijima H. Seasonal pattern and genotype distribution of sapovirus infection in Japan, 2003–2009. Epidemiol. Infect. 2012;140:74–77. doi: 10.1017/S0950268811000240. [DOI] [PubMed] [Google Scholar]
- 18.Lee L.E., Cebelinski E.A., Fuller C., Keene W.E., Smith K., Vinje J., Besser J.M. Sapovirus outbreaks in long-term care facilities, Oregon and Minnesota, USA, 2002–2009. Emerg. Infect. Dis. 2012;18:873–876. doi: 10.3201/eid1805.111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hergens M.P., Öhd J.N., Alm E., Askling H.H., Helgesson S., Insulander M., Lagerqvist N., Svenungsson B., Tihane M., Tolfvenstam T., et al. Investigation of a food-borne outbreak of gastroenteritis in a school canteen revealed a variant of sapovirus genogroup V not detected by standard PCR, Sollentuna, Sweden, 2016. Eurosurveillance. 2017;22:30543. doi: 10.2807/1560-7917.ES.2017.22.22.30543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Li Y., Kong X., Li H., Zhang Q., Jin M., Wang Y., Duan Z. Two gastroenteritis outbreaks caused by sapovirus in Shenzhen, China. J. Med. Virol. 2018;90:1695–1702. doi: 10.1002/jmv.25236. [DOI] [PubMed] [Google Scholar]
- 21.Varela M.F., Rivadulla E., Lema A., Romalde J.L. Human sapovirus among outpatients with acute gastroenteritis in Spain: A one-year study. Viruses. 2019;11:144. doi: 10.3390/v11020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerba C.P., Goyal S.M., LaBelle R.L., Cech I., Bodgan G.F. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Public Health. 1979;69:1116–1119. doi: 10.2105/AJPH.69.11.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payment P., Trudel M., Plante R. Elimination of viruses and indicator bacteria at each step of treatment during preparation of drinking water at seven water treatment plants. Appl. Environ. Microbiol. 1985;49:1418–1428. doi: 10.1128/aem.49.6.1418-1428.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood V.J., Levine A.D., Scott T.M., Chivukula V., Lukasik J., Farrah S.R., Rose J.B. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 2005;71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siefring S., Varma M., Atikovic E., Wymer L., Haugland R.A. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health. 2008;6:225–237. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- 26.Kildare B.J., Leutenegger C.M., McSwain B.S., Bambic D.G., Rajal V.B., Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 27.Pitkänen T., Ryu H., Elk M., Hokajärvi A.-M., Siponen S., Vepsäläinen A., Räsänen P., Santo Domingo J.W. Detection of fecal bacteria and source tracking identifiers in environmental waters using rRNA-based RT-qPCR and rDNA-based qPCR assays. Environ. Sci. Technol. 2013;47:13611–13620. doi: 10.1021/es403489b. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor V., Pitkänen T., Ryu H., Elk M., Wendell D., Santo Domingo J.W. Distribution of human-specific bacteroidales and fecal indicator bacteria in an urban watershed impacted by sewage pollution, determined using RNA-and DNA-based quantitative PCR assays. Appl. Environ. Microbiol. 2015;81:91–99. doi: 10.1128/AEM.02446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed W., Hughes B., Harwood V. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water. 2016;8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- 30.Communicable Disease Surveillance Centre (CDSC) Strength of association between human illness and water: Revised definitions for use in outbreak investigations. Commun. Dis. Rep. CDR Wkly. 1996;6:65–68. [PubMed] [Google Scholar]
- 31.Tillett H.E., de Louvois J., Wall P.G. Surveillance of outbreaks of waterborne infectious disease: Categorizing levels of evidence. Epidemiol. Infect. 1998;120:37–42. doi: 10.1017/S0950268897008431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loisy F., Atmar R.L., Guillon P., Le Cann P., Pommepuy M., Le Guyader F.S. Real-time RT-PCR for norovirus screening in shellfish. J. Virol. Methods. 2005;123:1–7. doi: 10.1016/j.jviromet.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Mull B., Hill V.R. Recovery of diverse microbes in high turbidity surface water samples using dead-end ultrafiltration. J. Microbiol. Methods. 2012;91:429–433. doi: 10.1016/j.mimet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inkinen J., Jayaprakash B., Siponen S., Hokajärvi A.M., Pursiainen A., Ikonen J., Ryzhikov I., Täubel M., Kauppinen A., Paananen J., et al. Active eukaryotes in drinking water distribution systems of ground and surface waterworks. Microbiome. 2019;7:99. doi: 10.1186/s40168-019-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kauppinen A., Pitkänen T., Miettinen I.T. Persistent norovirus contamination of groundwater supplies in two waterborne outbreaks. Food Environ. Virol. 2018;10:39–50. doi: 10.1007/s12560-017-9320-6. [DOI] [PubMed] [Google Scholar]
- 36.Maunula L., Söderberg K., Vahtera H., Vuorilehto V., von Bonsdorff C., Valtari M., Laakso T., Lahti K. Presence of human noro-and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J. Water Health. 2012;10:87–99. doi: 10.2166/wh.2011.095. [DOI] [PubMed] [Google Scholar]
- 37.Schultz A.C., Perelle S., di Pasquale S., Kovac K., de Medici D., Fach P., Sommer H.M., Hoorfar J. Collaborative validation of a rapid method for efficient virus concentration in bottled water. Int. J. Food Microbiol. 2011;145:5158–5166. doi: 10.1016/j.ijfoodmicro.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 38.Kim M.J., Ko G.P. Quantitative characterization of the inhibitory effects of salt, humic acid, and heavy metals on the recovery of waterborne norovirus by electropositive filters. J. Water Health. 2013;11:613–622. doi: 10.2166/wh.2013.187. [DOI] [PubMed] [Google Scholar]
- 39.Kauppinen A., Miettinen I.T. Persistence of norovirus GII genome in drinking water and wastewater at different temperatures. Pathogens. 2017;6:48. doi: 10.3390/pathogens6040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rönnqvist M., Rättö M., Tuominen P., Salo S., Maunula L. Swabs as a tool for monitoring the presence of norovirus on environmental surfaces in the food industry. J. Food Protect. 2013;76:1421–1428. doi: 10.4315/0362-028X.JFP-12-371. [DOI] [PubMed] [Google Scholar]
- 41.Oka T., Katayama K., Hansman G.S., Kageyama T., Ogawa S., Wu F.T., White P.A., Takeda N. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 2006;78:1347–1353. doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- 42.van Maarseveen N.M., Wessels E., de Brouwer C.S., Vossen A.C., Claas E.C. Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus, Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 2010;49:205–210. doi: 10.1016/j.jcv.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Jothikumar N., Cromeans T.L., Hill V.R., Lu X., Sobsey M.D., Erdman D.D. Quantitative real-time PCR assays for detection of human adenoviruses and identification of serotypes 40 and 41. Appl. Environ. Microbiol. 2005;71:3131–3136. doi: 10.1128/AEM.71.6.3131-3136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kauppinen A., Al-Hello H., Zacheus O., Kilponen J., Maunula L., Huusko S., Lappalainen M., Miettinen I., Blomqvist S., Rimhanen-Finne R. Increase in outbreaks of gastroenteritis linked to bathing water in Finland in summer 2014. Eurosurveillance. 2017;22:30470. doi: 10.2807/1560-7917.ES.2017.22.8.30470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk-and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods. 1999;83:145–154. doi: 10.1016/S0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- 46.Vinjé J., Hamidjaja R.A., Sobsey M.D. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods. 2004;116:109–117. doi: 10.1016/j.jviromet.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Kroneman A., Vennema H., Deforche K., Avoort H.V.D., Peñaranda S., Oberste M.S., Vinjé J., Koopmans M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 48.Finnish Standard Association . Water Quality—Membrane Filtration Technique for the Enumeration of Total Coliform Bacteria. Finnish Standard Association; Helsinki, Finland: 2011. SFS 3016. [Google Scholar]
- 49.International Organization for Standardization . Water Quality—Enumeration of Escherichia Coli and Coliform Bacteria—Part 1: Membrane Filtration Method for Waters with Low Bacterial Background Flora. International Organization for Standardization; Geneva, Switzerland: 2014. ISO 9308-1. [Google Scholar]
- 50.International Organization for Standardization . Water Quality—Enumeration of Escherichia coli and Coliform Bacteria—Part 2: Most Probable Number Method. International Organization for Standardization; Geneva, Switzerland: 2012. ISO 9308-2. [Google Scholar]
- 51.International Organization for Standardization . Water Quality—Detection and Enumeration of Intestinal Enterococci—Part 2: Membrane Filtration Method. International Organization for Standardization; Geneva, Switzerland: 2000. ISO 7899-2. [Google Scholar]
- 52.International Organization for Standardization . Water Quality—Enumeration of Clostridium Perfringens—Method Using Membrane Filtration. International Organization for Standardization; Geneva, Switzerland: 2013. ISO 14189. [Google Scholar]
- 53.International Organization for Standardization . Water Quality—Detection and Enumeration of Thermotolerant Campylobacter Species. International Organization for Standardization; Geneva, Switzerland: 2005. ISO 17995. [Google Scholar]
- 54.Antikainen J., Kantele A., Pakkanen S.H., Lääveri T., Riutta J., Vaara M., Kirveskari J. A quantitative polymerase chain reaction assay for rapid detection of 9 pathogens directly from stools of travelers with diarrhea. Clin. Gastroenterol. Hepatol. 2013;11:1300–1307. doi: 10.1016/j.cgh.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 55.Hill V., Narayanan J., Gallen R., Ferdinand K., Cromeans T., Vinjé J. Development of a nucleic acid extraction procedure for simultaneous recovery of DNA and RNA from diverse microbes in water. Pathogens. 2015;4:335–354. doi: 10.3390/pathogens4020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jothikumar N., da Silva A.J., Moura I., Qvarnstrom Y., Hill V.R. Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J. Med. Microbiol. 2008;57:1099–1105. doi: 10.1099/jmm.0.2008/001461-0. [DOI] [PubMed] [Google Scholar]
- 57.International Organization for Standardization . Water Quality—Isolation and Identification of Cryptosporidium oocysts and Giardia cysts from Water. International Organization for Standardization; Geneva, Switzerland: 2006. ISO 15553. [Google Scholar]
- 58.Bosch A., Lucena F., Diez J.M., Gajardo R., Blasi M., Jofre J. Waterborne viruses associated with hepatitis outbreak. J. Am. Water Work. Assoc. 1991;83:80–83. doi: 10.1002/j.1551-8833.1991.tb07119.x. [DOI] [Google Scholar]
- 59.Maunula L., Miettinen I.T., von Bonsdorff C. Norovirus outbreaks from drinking water. Emerg. Infect. Dis. 2005;11:1716–1721. doi: 10.3201/eid1111.050487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinelli D., Prato R., Chironna M., Sallustio A., Caputi G., Conversano M., Atti M.C.D., D’Ancona F.P., Germinario C.A., Quarto M. Large outbreak of viral gastroenteritis caused by contaminated drinking water in Apulia, Italy, May–October 2006. Eurosurveillance. 2007;12:3176. doi: 10.2807/esw.12.16.03176-en. [DOI] [PubMed] [Google Scholar]
- 61.Blanco A., Guix S., Fuster N., Fuentes C., Bartolomé R., Cornejo T., Pintó R.M., Bosch A. Norovirus in bottled water associated with gastroenteritis outbreak, Spain, 2016. Emerg. Infect. Dis. 2017;23:1531–1534. doi: 10.3201/eid2309.161489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diston D., Sinreich M., Zimmermann S., Baumgartner A., Felleisen R. Evaluation of molecular-and culture-dependent MST markers to detect fecal contamination and indicate viral presence in good quality groundwater. Environ. Sci. Technol. 2015;49:7142–7151. doi: 10.1021/acs.est.5b00515. [DOI] [PubMed] [Google Scholar]
- 63.Lehtola M.J., Torvinen E., Kusnetsov J., Pitkänen T., Maunula L., von Bonsdorff C., Martikainen P.J., Wilks S.A., Keevil C.W., Miettinen I.T. Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Appl. Environ. Microbiol. 2007;73:2854–2859. doi: 10.1128/AEM.02916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helmi K., Skraber S., Gantzer C., Willame R., Hoffmann L., Cauchie H.M. Interactions of Cryptosporidium parvum, Giardia lamblia, vaccinal poliovirus type 1, and bacteriophages phiX174 and MS2 with a drinking water biofilm and a wastewater biofilm. Appl. Environ. Microbiol. 2008;74:2079–2088. doi: 10.1128/AEM.02495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kauppinen A., Ikonen J., Pursiainen A., Pitkänen T., Miettinen I.T. Decontamination of a drinking water pipeline system contaminated with adenovirus and Escherichia coli utilizing peracetic acid and chlorine. J. Water Health. 2012;10:406–418. doi: 10.2166/wh.2012.003. [DOI] [PubMed] [Google Scholar]
- 66.Allen M., Clark R., Cotruvo J.A., Grigg N. Drinking water and public health in an era of aging distribution infrastructure. Public Work. Manag. Policy. 2018;23:301–309. doi: 10.1177/1087724X18788368. [DOI] [Google Scholar]
- 67.Barratt J.L., Harkness J., Marriott D., Ellis J.T., Stark D. A review of Dientamoeba fragilis carriage in humans: Several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes. 2011;2:3–12. doi: 10.4161/gmic.2.1.14755. [DOI] [PubMed] [Google Scholar]
- 68.Wong Z.W., Faulder K., Robinson J.L. Does Dientamoeba fragilis cause diarrhea? A systematic review. Parasitol. Res. 2018;117:971–980. doi: 10.1007/s00436-018-5771-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.