Abstract
Abnormal expression of Holliday junction recognition protein (HJURP) in several types of tumor cells plays a vital role in the formation and progression of tumors. Few studies have investigated the role of HJURP in prostate cancer (PCa). The aim of this study was to analyze the expression levels of HJURP in PCa and to establish the association with clinicopathological data. Reverse transcription quantitative polymerase chain reaction and immunohistochemical analysis were used to detect the expression levels of HJURP in benign and PCa prostate tissues. The Taylor dataset was statistically analyzed to determine if HJURP expression levels were associated with PCa clinicopathological data. HJURP was overexpressed in PCa tissues compared with benign prostate tissues. Statistical analysis of the Taylor dataset indicated that upregulation of HJURP was significantly associated with positive prostate-specific antigen (PSA) levels (P=0.004), high Gleason score (P=0.005), advanced pathological stage (P=0.007), metastasis (P<0.001) and PSA failure (P<0.001). Higher HJURP mRNA expression levels were significantly associated with shorter biochemical recurrence (BCR)-free survival (P<0.001). To the best of our knowledge, this study is the first report of HJURP upregulation in PCa tissues. Upregulation of HJURP may predict BCR-free survival and HJURP may be an oncogene that impacts the prognosis of patients with PCa.
Keywords: prostate cancer, Holliday junction recognition protein, biochemical recurrence-free survival, immunohistochemistry, genome stability
Introduction
Almost one-fifth of new cancer cases among American males are prostate cancer (PCa), and ~10% of these led to death in 2018 (1). Despite many efficacious treatments for localized PCa, such as radical prostatectomy (RP) or external-beam radiotherapy, 27–53% of patients exhibit biochemical recurrence (BCR), which may cause an increased risk of distant metastasis and mortality (2). Furthermore, the current predictive models of PCa following surgery have demonstrated limitations and ineffectiveness in daily clinical practice. PCa is a heterogeneous and multifocal cancer, which suggests that patients with PCa with the same prostate-specific antigen (PSA) levels and tumor stage may have a different prognosis (3). Notably, there are no critical predictive methods available to accurately evaluate PCa recurrence or predict the prognosis of individuals. New methods are therefore required to predict BCR in patients with PCa and to provide an effective method for personalized treatment for patients with PCa.
Numerous studies have shown that during mitosis, disfunction or destabilization of centromeres results in chromosome deletion, anomaly separation and aneuploidy, which may also lead to chromosome instability and cancer development (4–7). Centromere protein A (CENP-A) is a crucial centromere protein that is required to maintain the functional centromere and ensure chromosomes separate correctly during mitosis (8) However, many studies have shown that the abnormal expression of CENP-A may lead to chromosome instability and cancer progression (5,6). Further research on centromere disfunction may aid the identification of a new mechanism involved in cancer formation and progression (7).
As the CENP-A chaperone, Holliday junction recognition protein (HJURP) mediates CENP-A deposition at centromeres (9). An increasing body of evidence suggests that HJURP is expressed in normal cells, and also abnormally expressed in cancer cell lines (10,11). Notably, HJURP is overexpressed in lung, ovarian, breast cancer and glioblastoma cells and is associated with poor prognosis (11). Therefore, HJURP may be used as a tumor detection tool for a variety of types of cancer (12). Research indicates that using small interfering RNA against HJURP in cancer cells results in chromosome instability and contributes to cell cycle arrest (13). Findings have demonstrated that the regulation of HJURP is vital for promoting genome stability (6). HJURP has also been considered as a new therapeutic target of some anticancer drugs (13). Thus, these findings suggest that HJURP might have a crucial role in the generation and progression of cancer.
To the best of our knowledge, no study to date has assessed the expression pattern of HJURP in PCa. Therefore, the aim of the present study was to analyze the expression of HJURP in PCa and to assess its association with clinicopathological data.
Materials and methods
Clinical samples
This study was approved by the Research Ethics Committee of Guangzhou First People's Hospital (Guangzhou, China). All patients provided written informed consent. The specimens were handled and made anonymous according to ethical and legal standards.
All patient information and clinical features in this study are presented in Table I. For reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis, a total of 22 PCa and 10 benign prostate tissues were used, which were collected and frozen during RP and transurethral resection of the prostate between January 2012 and December 2016 at Guangzhou First People's Hospital. No patients recruited in this study had received chemotherapy or radiotherapy prior to surgery. Furthermore, immunohistochemical analysis was performed on tissue microarrays (TMAs), which included PCa tissues (n=99) and benign prostate tissues (n=81). The TMAs were obtained from Shanghai Outdo Biotech Co., Ltd., (cat. no. HPro-Ade180PG-01). Owing to some sample tissues falling off during immunohistochemical analysis and others being defective or missing related clinical information, 40 samples were excluded and 140 (65 PCa and 75 benign prostate tissues) samples were subsequently used. Furthermore, an online PCa dataset (Taylor dataset) from http://cbio.mskcc.org/cancergenomics/prostate/ was downloaded for statistical analysis (14). The Taylor dataset contains 150 PCa and 21 benign prostate tissues with detailed clinical information.
Table I.
Detailed clinical information of the patients included in the study.
| Clinical features | RT-qPCR | Immunohistochemistry | Taylor Dataset |
|---|---|---|---|
| Benign tissue, n | 10 | 81 | 29 |
| Prostate cancer, n | 22 | 99 | 150 |
| Age in years, mean ± SD | 72.27±6.94 | 70.71±8.00 | 58.34±7.07 |
| <66 | 4 | 26 | 25 |
| ≥66 | 18 | 73 | 125 |
| Serum prostate-specific antigen, n | |||
| <4 ng/ml | 9 | – | 24 |
| 4–10 ng/ml | 3 | – | 81 |
| ≥10 ng/ml | 10 | – | 42 |
| Gleason score, n | |||
| ≤6 | 8 | 26 | 41 |
| 7 | 3 | 44 | 76 |
| ≥8 | 11 | 28 | 22 |
| Pathological stage, n | |||
| T2 | 22 | 70 | 86 |
| ≥T3A | 0 | 29 | 55 |
‘−’ denotes a lack of relative information. RT-qPCR, reverse transcription-quantitative PCR.
RNA extraction and reverse RT-qPCR analysis
Total RNA was extracted from cultured prostate cells (~5×106 cells) with the RNeasy mini kit (Qiagen, Germany). The Invitrogen SuperScript III First-Strand System (Invitrogen; Thermo Fisher Scientific, Inc.) was used for RT with random primers (Hexadeoxyribonucleotide mixture; pd (N)6; cat. no. 3801; Takara Biotechnology Co., Ltd., Dalian, China). Reverse transcription was performed as follows: 37°C for 15 min and 85°C for 5 sec. mRNA expression was detected using SYBR Green PCR mix (Toyobo Co., Ltd.) and normalized to β-actin as the internal control. The following primers were used: HJURP forward, 5′-CACAAAGCCATCAAGCATCATC-3′ and reverse, 5′-TCAGAGCAGGGTATGAAGTTCT-3′; and β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse, 3′-GGGCACGAAGGCTCATCATT-5′. QPCR was performed on a MyiQ.2 Two-Color Real-Time qPCR Detection System (Bio-Rad Laboratories, Inc.), and the thermocycling conditions were as follows: 48°C for 30 min, 95°C for 1 min, followed by 40 cycles at 95°C for 15 sec, 52°C for 30 sec and 72°C for 30 sec. All assays were carried out in triplicate. CT values were determined using the IQ5 software (Bio-Rad Laboratories, Inc.). Relative quantification of target mRNA expression was evaluated using the 2−ΔΔCq method (15). The mean ± standard deviation (SD) was calculated from three independent experiments.
Immunohistochemistry
Immunohistochemistry and the immunoreactivity scores (IRS) were used to evaluate the expression levels and subcellular localization of HJURP protein in PCa tissues. The specimens were fixed in 10% neutral buffered formalin and were subsequently processed by gradient dehydration in ethanol, embedded in paraffin and sectioned into 4-µm sections for hematoxylin and eosin or immunohistochemistry staining with the DAKO EnVision System (Dako Diagnostics). Following antigen retrieval with citric acid buffer at 95°C for 8 min, the sections were blocked with peroxidase at room temperature for 15 min and 10% goat serum (cat. no. BA1056; Wuhan Boster Biological Technology, Ltd.) at room temperature for 30 min. Subsequently, the sections were incubated overnight with a rabbit anti-HJRUP primary antibody (cat. no. PAB20427; dilution, 1:50; Abnova Corporation) at 4°C. Following three washes (5 min each) in phosphate buffer saline, the sections were incubated with an avidin-conjugated goat anti-rabbit secondary antibody (undiluted; cat. no. BA1056; Wuhan Boster Biological Technology Ltd.) at room temperature for 30 min. Streptavidin-peroxidase-labeled polymer (50 µl for 15 min at room temperature) and substrate-chromogen (100 µl for 2 min at room temperature) were used to observe the staining of the target protein. Negative controls were obtained by omitting the primary antibody. Light microscopy at ×400 magnification was used to examine the sections.
Immunostaining was scored by two experienced independent pathologists who were blinded to the clinicopathological data and clinical outcomes of the patients. The scores of the two pathologists were compared, and any discrepancy between the scores were dealt with by re-examination of the staining by both pathologists to achieve a consensus score. The immunolabeling of cancer cells and stromal cells was evaluated separately. The number of positive-staining cells in ten representative microscopic fields was counted, and the percentage of positive cells was calculated. Given the homogeneity of the staining of the target proteins, tumor specimens were scored in a semi-quantitative manner. The percentage scoring of immunoreactive tumor cells was as follows: 0, 0–5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; and 4, >75%. The staining intensity was visually scored and stratified as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. An IRS value for HJURP staining was obtained for each case by adding the percentage and the intensity score.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM Corp.). Unpaired two-tailed Student's t test was used to analyze the results and the data were expressed as the mean ± SD. The survival analysis was performed by Kaplan-Meier curves and the groups were compared by log-rank (Mantel-Cox) test. P<0.05 was considered to indicate a statistically significant difference.
Results
HJURP is overexpressed in PCa tissues
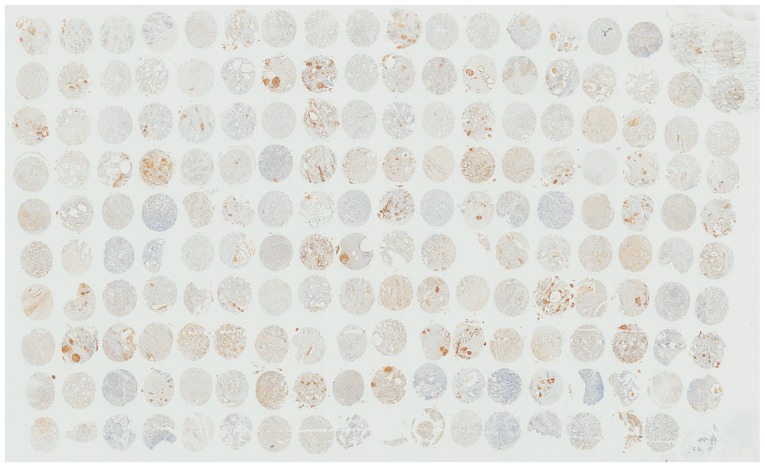
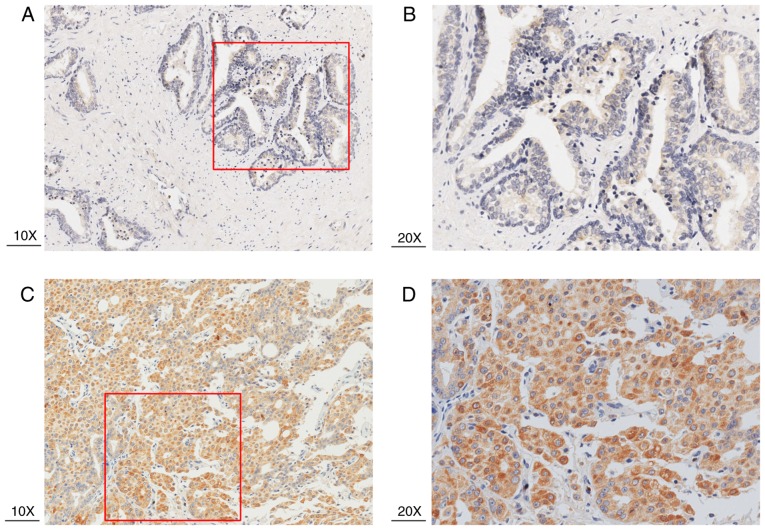
RT-qPCR and immunohistochemistry results revealed that HJURP was upregulated in PCa tissues. As indicated by the RT-qPCR results in Fig. 1, the mRNA expression levels of HJURP were higher in PCa compared with benign prostate tissues (P<0.001). Furthermore, the protein expression levels of HJURP in PCa were investigated using immunohistochemical analysis. The panoramic view examined with immunohistochemical analysis of TMA is presented in Fig. 2. As demonstrated in Fig. 3, IRS values revealed that the protein expression levels of HJURP were higher in PCa compared with benign prostate tissues (P<0.001), which was in accordance with the results of RT-qPCR and with statistical analysis of the Taylor dataset (P<0.001; Table II). As shown in Fig. 4A-D, immunohistochemical staining of HJURP protein revealed that it was predominantly expressed in the cytoplasm. Notably, weak or negative staining was found in the benign prostate tissues (Fig. 4A and B).
Figure 1.
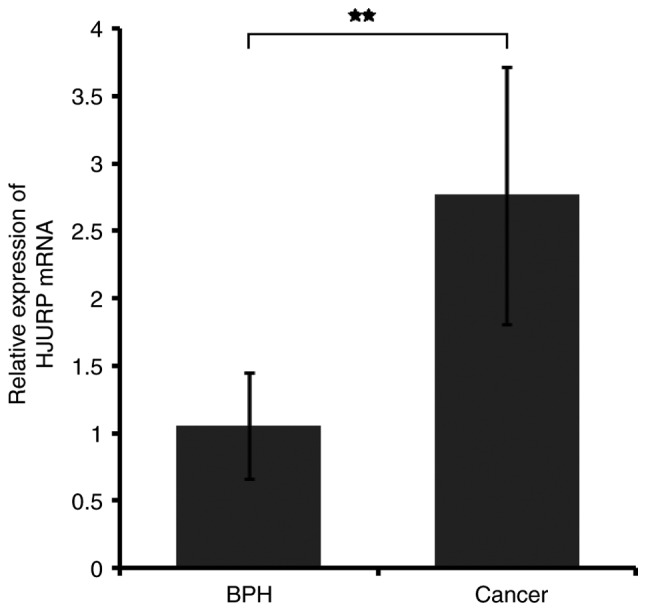
HJURP mRNA is upregulated in prostate cancer. **P<0.001. HJURP, Holliday junction recognition protein; BPT, benign prostate tissue.
Figure 2.
Panoramic view of tissue microarray samples, stained for HJURP. HJURP, Holliday junction recognition protein.
Figure 3.
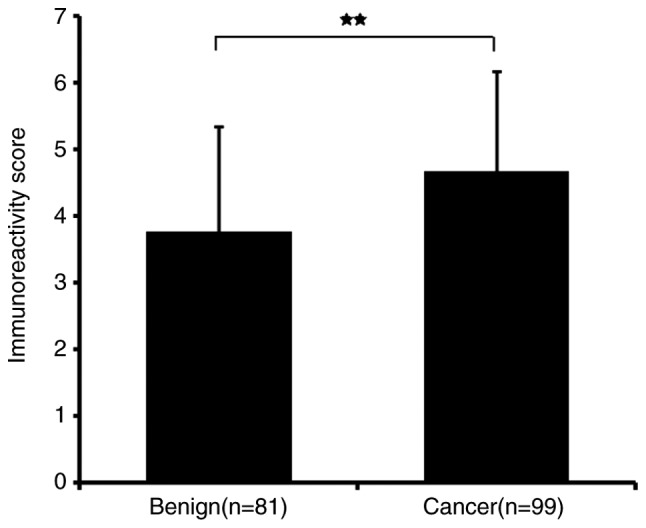
HJURP protein is upregulated in prostate cancer tissues. **P<0.001. HJURP, Holliday junction recognition protein; BPT, benign prostate tissue.
Table II.
Association of Holliday junction recognition protein mRNA expression with clinicopathological characteristics in the Taylor dataset.
| HJURP | |||
|---|---|---|---|
| Clinical features | Cases, n | Mean ± SD | P-value |
| Tissues | <0.001 | ||
| Benign | 29 | 7.10±0.130 | |
| Cancer | 150 | 7.31±0.245 | |
| Age, years | 0.805 | ||
| <60 | 93 | 7.31±0.261 | |
| ≥60 | 57 | 7.30±0.219 | |
| Serum prostate-specific antigen, ng/ml | 0.004 | ||
| <10 | 105 | 7.25±0.201 | |
| ≥10 | 42 | 7.40±0.279 | |
| Gleason score | 0.005 | ||
| <8 | 117 | 7.25±0.184 | |
| ≥8 | 22 | 7.44±0.288 | |
| Clinical stage | 0.731 | ||
| <T2a | 80 | 7.29±0.199 | |
| ≥T2a | 65 | 7.30±0.287 | |
| Pathological stage | 0.007 | ||
| T2a-T2c | 86 | 7.24±0.162 | |
| T3a-T4 | 55 | 7.35±0.266 | |
| Metastasis | <0.001 | ||
| No | 122 | 7.24±0.165 | |
| Yes | 28 | 7.60±0.311 | |
| Prostate-specific antigen failure | <0.001 | ||
| Negative | 104 | 7.23±0.168 | |
| Positive | 36 | 7.42±0.260 | |
HJURP, Holliday junction recognition protein.
Figure 4.
Immunohistochemical analysis using tissue microarray samples of HJURP in PCa and benign prostrate tissues. (A and B) HJURP protein expression was weak or negative in benign prostate tissues (C and D) HJURP was highly expressed in PCa tissues. HJURP, Holliday junction recognition protein; PCa, prostate cancer.
Association of HJURP mRNA expression with the clinicopathological characteristics of PCa
The association of HJURP mRNA expression levels with clinicopathological features was determined (Table II). It was also demonstrated that PSA levels ≥10, a Gleason score ≥8, pathological stage T3a-T4 (16–18), metastasis and PSA failure, including biochemical relapse (BCR) of prostate cancer, which is an important endpoint that can indicate recurrent prostate cancer (19), were associated with higher mRNA expression levels of HJURP compared with PSA <10 (P=0.004), a Gleason score <8 (P=0.005), pathological stage T2a-T2c (P=0.007), no metastasis (P<0.001) and no PSA failure (P<0.001; Table II).
High HJURP expression is associated with reduced BCR-free survival time
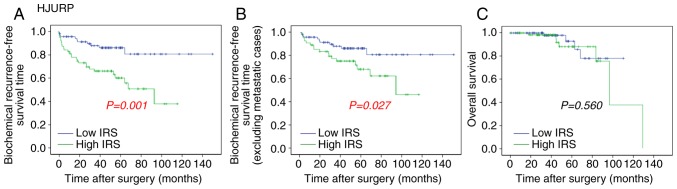
The association of HJURP expression with overall survival and BCR-free survival of patients with PCa was analyzed by the Kaplan-Meier method using the Taylor dataset (Fig. 5). The median value (HJURP median expression =7.22) of HJURP expression level in PCa was used as the cut-off point to separate the patients into high (n=75) and low (n=65) HJURP expression level groups. Comparison between PCa patients with high HJURP expression and low HJURP expression indicated that there was a significant statistical difference in the BCR-free survival time between these groups (P=0.001; Fig. 5A). A statistically significant difference was also observed in the BCR-free survival when patients with metastasis were excluded (P=0.027; Fig. 5B). However, there was no significant difference found between HJURP expression and overall survival (P=0.560; Fig. 5C).
Figure 5.
Association of HJURP expression with overall survival and BCR-free survival in patients with prostate cancer. Data was analyzed using the Kaplan-Meier method. (A) Patients in the high HJURP expression group had a shorter BCR-free survival time compared with the low HJURP expression group. (B) Excluding patients with metastasis, the high HJURP expression group had shorter BCR-free survival time compared with the low HJURP expression group. (C) No association was found between HJURP expression and overall survival. HJURP, Holliday junction recognition protein; BCR, biochemical recurrence; IRS, immunoreactivity score.
Discussion
Although many patients with PCa achieve long-term survival after RP, the prognosis of late stage PCa is currently unsatisfactory. In general, BCR and metastasis lead to cancer-associated mortality. Despite close follow-up, the low sensitivity of tumor indices and biomarkers impedes the evaluation of the progression of PCa (2,3). Therefore, there is a need for the identification of novel and effective prognostic biomarkers to improve the clinical management of PCa which may help to lower mortality and establish personalized treatment for patients with PCa (3).
Accumulating evidence shows that HJURP may be an oncogene that has a crucial role in PCa (10–12). To the best of our knowledge, the current study is the first to report the role of HJURP expression in PCa. The findings demonstrated that HJURP was upregulated in PCa tissues compared with benign prostate tissues, which was in accordance with the Taylor dataset. Upregulation of HJURP has also been identified in other types of cancer, including brain, breast, ovarian and lung cancer, suggesting that HJURP may be involved in tumor progression (10–13). According to the analysis of the Taylor dataset in the present study, upregulation of HJURP was associated with aggressive tumor progression in patients with PCa. In addition, Kaplan-Meier results indicated that high HJURP mRNA expression was associated with shorter BCR-free survival time, and hence, HJURP expression may be useful to determine BCR-free survival in patients with PCa. It has been previously reported that upregulation of HJURP mRNA is associated with the progesterone and estrogen negative status, which has relatively poor prognosis and aggressive behavior in breast cancer (11). Similar results have also been identified in lung cancer and glioma reports (12,13).
Studies suggest that members of the CENP family can become functionally disordered during cell division giving rise to chromosome instability, segregation defects and cancer development (20–22). It has previously been reported that abnormal expression of CENP can lead to cancer progression and prognosis (20–22). The CENP-A chaperone, HJURP plays a significant role in localizing CENP-A on the centromere facilitating accurate chromosome segregation (9). Previous studies have demonstrated that knockdown or inhibition of HJURP in cancer cells strongly affects CENP-A deposition, centrosomes and chromosome stability and contributes to cell cycle arrest and cell death (11–13). These findings suggest that HJURP may serve a key role in cell proliferation, and the abnormal HJURP expression may have an influence on chromosome stability and cell oncogenesis.
During cell division, DNA damage or double-strand breaks (DSB) can initiate the DNA damage response (DDR). Notably, the DDR causes cell cycle arrest or induces cell death, which is beneficial for DNA repair and for preventing genomic instability (23,24). Recent studies have revealed cancer cells might have some mechanisms to resist the DDR process protecting cancer cells from chromosomal instability and contributing to tumorigenesis (25,26). Many DSB repair proteins play a role in inhibiting the DDR in cancer cells and are significantly associated with poor prognosis (27). During the DSB process, HJURP is upregulated after DNA damage induction, interacts with proteins mutS homolog 5 and/or NBS1, and mediates DNA repair via homologous recombination (13). Therefore, HJURP may play a critical role in DNA repair mechanisms and abnormal HJURP expression may suppress the DDR process, which maintains chromosome stability in cancer cells (13). Thus, the strict regulation of HJURP during the cell cycle is a key factor associated with genomic stability.
Further research into this subject is required, including western blot analysis to confirm the findings that HJURP expression is increased in PCa tissues. Cell line experiments to investigate proliferation, migration, invasion and clonal formation are required to unravel the biological mechanism of HJURP in patients with PCa. Future work will further investigate the interaction between HJURP expression and PCa development.
To conclude, the results of the present study indicate that HJURP was upregulated in PCa tissues and may play a crucial role in the prognosis of PCa. High expression of HJURP was also associated, with poor clinicopathological features of PCa, and may predict BCR-free survival time in patients. Although the results presented in this study require further validation and the biological mechanism of HJURP in PCa needs elucidation, the present study extends our knowledge about the prognostic value of HJURP in PCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81571427 and 81600620), the Science and Technology Project of Guangdong Province (grant no. 201607010398 and 2016A020215018), the Science and Technology Project of Huadu District (grant no. HD15CXY005) and Projects of Guangdong Key Laboratory of Clinical Molecular Medicine and Diagnostics.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Author's contributions
WZ, ZH, YW and YL participated in study design, coordination, analysis and interpretation of data, obtained funding and supervised the study. YC, YL and BW performed the majority of the experiments and statistical analysis and drafted the manuscript. JY, DY, JL and SZ performed certain experiments and sample collection. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Guangzhou First People's Hospital (Guangzhou, China). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Artibani W, Porcaro AB, De Marco V, Cerruto MA, Siracusano S. Management of biochemical recurrence after primary curative treatment for prostate cancer: A review. Urol Int. 2018;100:251–262. doi: 10.1159/000481438. [DOI] [PubMed] [Google Scholar]
- 3.Spahn M, Boxler S, Joniau S, Moschini M, Tombal B, Karnes RJ. What is the need for prostatic biomarkers in prostate cancer management? Curr Urol Rep. 2015;16:70. doi: 10.1007/s11934-015-0545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaither TL, Merrett SL, Pun MJ, Scott KC. Centromeric barrier disruption leads to mitotic defects in schizosaccharomyces pombe. G3 (Bethesda) 2014;4:633–642. doi: 10.1534/g3.114.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, Karpen GH. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun. 2016;7:12619. doi: 10.1038/ncomms12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdaasdonk JS, Bloom K. Centromeres: Unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foltz DR, Jansen LE, Bailey AO, Yates JR, III, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valente V, Serafim RB, de Oliveira LC, Adorni FS, Torrieri R, Tirapelli DP, Espreafico EM, Oba-Shinjo SM, Marie SK, Paçó-Larson ML, Carlotti CG., Jr Modulation of HJURP (Holliday Junction-Recognizing Protein) levels is correlated with glioblastoma cells survival. PLoS One. 2013;8:e62200. doi: 10.1371/journal.pone.0062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, Bayani N, Blakely EA, Gray JW, Mao JH. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouam S, Moreau T, Broët P. Identifying common prognostic factors in genomic cancer studies: A novel index for censored outcomes. BMC Bioinformatics. 2010;11:150. doi: 10.1186/1471-2105-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 14.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Brimo F, Montironi R, Egevad L, Erbersdobler A, Lin DW, Nelson JB, Rubin MA, van der Kwast T, Amin M, Epstein JI. Contemporary grading for prostate cancer: Implications for patient care. Eur Urol. 2013;63:892–901. doi: 10.1016/j.eururo.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Kweldam CF, van Leenders GJ, van der Kwast T. Grading of prostate cancer: A work in progress. Histopathology. 2019;74:146–160. doi: 10.1111/his.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soares R, Eden CG. Surgical treatment of high-risk prostate cancer. Minerva Urol Nefrol. 2015;67:33–46. [PubMed] [Google Scholar]
- 19.Giacalone NJ, Wu J, Chen MH, Renshaw A, Loffredo M, Kantoff PW, D'Amico AV. Prostate-specific antigen failure and risk of death within comorbidity subgroups among men with unfavorable-risk prostate cancer treated in a randomized trial. J Clin Oncol. 2016;34:3781–3786. doi: 10.1200/JCO.2016.68.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao WT, Feng Y, Li ML, Liu GL, Li MZ, Zeng MS, Song LB. Overexpression of centromere protein H is significantly associated with breast cancer progression and overall patient survival. Chin J Cancer. 2011;30:627–637. doi: 10.5732/cjc.010.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai Y, Liu L, Zeng T, Zhu YH, Li J, Chen L, Li Y, Yuan YF, Ma S, Guan XY. Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma. Biochem Biophys Res Commun. 2013;436:711–718. doi: 10.1016/j.bbrc.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Georgoulis A, Vorgias CE, Chrousos GP, Rogakou EP. Genome instability and γH2AX. Int J Mol Sci. 2017;18:E1979. doi: 10.3390/ijms18091979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaria P, Robert C, Rassool FV. DNA double-strand break response in stem cells: Mechanisms to maintain genomic integrity. Biochim Biophys Acta. 2013;1830:2345–2353. doi: 10.1016/j.bbagen.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Majidinia M, Yousefi B. DNA damage response regulation by microRNAs as a therapeutic target in cancer. DNA Repair (Amst) 2016;47:1–11. doi: 10.1016/j.dnarep.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Raschellà G, Melino G, Malewicz M. New factors in mammalian DNA repair-the chromatin connection. Oncogene. 2017;36:4673–4681. doi: 10.1038/onc.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhen Y, Xiao R, Chen X, Yuan C, Sun Y, Li J. A non-synonymous polymorphism is associated with progression from chronic hepatitis B virus infection to hepatocellular carcinoma in a Chinese population. Onco Targets Ther. 2018;11:563–569. doi: 10.2147/OTT.S153538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.