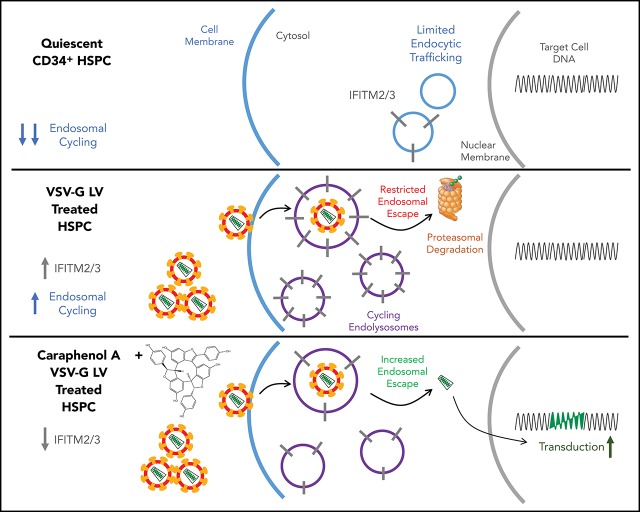
Key Points
The cyclic resveratrol trimer caraphenol A safely enhances lentiviral vector gene delivery to hematopoietic stem and progenitor cells.
Caraphenol A decreases interferon-induced transmembrane protein-mediated restriction in an endosomal trafficking-dependent manner.
Abstract
Therapeutic gene delivery to hematopoietic stem cells (HSCs) holds great potential as a life-saving treatment of monogenic, oncologic, and infectious diseases. However, clinical gene therapy is severely limited by intrinsic HSC resistance to modification with lentiviral vectors (LVs), thus requiring high doses or repeat LV administration to achieve therapeutic gene correction. Here we show that temporary coapplication of the cyclic resveratrol trimer caraphenol A enhances LV gene delivery efficiency to human and nonhuman primate hematopoietic stem and progenitor cells with integrating and nonintegrating LVs. Although significant ex vivo, this effect was most dramatically observed in human lineages derived from HSCs transplanted into immunodeficient mice. We further show that caraphenol A relieves restriction of LV transduction by altering the levels of interferon-induced transmembrane (IFITM) proteins IFITM2 and IFITM3 and their association with late endosomes, thus augmenting LV core endosomal escape. Caraphenol A-mediated IFITM downregulation did not alter the LV integration pattern or bias lineage differentiation. Taken together, these findings compellingly demonstrate that the pharmacologic modification of intrinsic immune restriction factors is a promising and nontoxic approach for improving LV-mediated gene therapy.
Visual Abstract
Introduction
Genetic modification of hematopoietic stem cells (HSCs) by γ-retroviral or lentiviral vectors (LVs) has shown efficacy in treating several hematologic disorders.1-6 A critical factor in determining treatment effectiveness remains the degree of modification of true repopulating HSCs.7,8 Transduction-enhancing techniques, including culture with HSC-enhancing cytokines,9-11 high multiplicity of infection (MOI), repeat LV administration,9,10 alternate LV envelope pseudotyping,12-14 or addition of transduction-enhancing small molecules15-17 have all been shown to improve gene delivery. However, the predominant underlying mechanism of HSC resistance to LV gene delivery remains an open question.9,18-21
Along with LV transduction resistance, hematopoietic stem and progenitor cells (HSPCs) are resistant to infection by many viruses and intracellular bacteria.22-25 Recent findings have highlighted the role of constitutive interferon-stimulated gene expression in pluripotent and multipotent cell types.26 Interferon-regulated innate effectors, especially the interferon-induced transmembrane (IFITM) family of proteins, provide an intrinsic defense against pathogens that rely on cellular endosomes for entry and transport. The IFITM proteins were first identified as antiviral effectors against vesicular stomatitis virus (VSV)27 and can restrict VSV G protein pseudotyped (VSV-G) LV transduction28,29 as well as regulate cellular growth, adhesion, and development.30,31 We recently showed that IFITM3 protein expression limits gene delivery efficiency with VSV-G pseudotyped LVs in HPSCs, and that IFITM restriction is pharmacologically overcome by the mammalian target of rapamycin (mTOR) inhibitor rapamycin.32 However, as an immunosuppressive compound with many effects, rapamycin can induce unwanted outcomes including cell expansion delay.15,33 Staurosporine and the IFITM3-modulating cyclosporines also have LV transduction enhancer activity, but can have undesirable cytotoxic effects.17,34 The differing subcellular trafficking strategy used by VSV-G pseudotyped LVs results in LV encountering distinct restriction factors from HIV-1 trafficking that may affect integration and alter latency.29,35
We report the identification and evaluation of caraphenol A, an HSPC noncytotoxic compound able to transiently reduce IFITM protein expression and association with endosomes in cell lines and human HSPCs. We show that caraphenol A treatment significantly improved HSC gene delivery at both low and high LV doses without altering LV integration patterns. This enhancement translates to lasting improvements in gene marking efficiency in vivo.
Methods
Compounds
Resveratrol, prostaglandin-E2 (PGE-2), and rapamycin were commercially purchased (Calbiochem, Millipore-Sigma, CAT#554325, #538904, #553210). Caraphenol A was synthesized as previously published,36 and naturally derived caraphenol A and α-viniferin were purified as described in the supplemental Methods, available on the Blood Web site.
Lentiviral vector
Third-generation VSV-G pseudotyped pRRL-SIN-MND-EGFP LV, termed LV, was generated as described,37 and stocks were produced and titered as described.15,16,38
CD34+ cell isolation and LV transduction
Umbilical cord blood (UCB) CD34+ cells were isolated as described15 from UCB generously donated from the Cleveland Cord Blood Center (Cleveland, OH), frozen granulocyte colony-stimulating factor mobilized peripheral blood (mPB) CD34+ cells were purchased from the Co-Operative Center for Excellence in Hematology at the Fred Hutchinson Cancer Research Center (Seattle, WA), and nonhuman primate CD34+ cells were isolated by bone marrow aspiration from rhesus macaques at the Wisconsin National Primate Research Center (Madison, WI). All approved human and nonhuman protocols are available on request. Isolation, transduction, and culture protocols are provided in detail in the supplemental Methods.
Mouse transplantation
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were obtained from Jackson Laboratory and maintained at The Scripps Research Institute under approved institutional protocols (available on request). CD34+ cell transplantation, posttransplant procedures, and analysis were previously described,15 and are further detailed in the supplemental Methods.
Results
Caraphenol A enhances gene delivery to cell lines and hematopoietic stem and progenitor cells ex vivo
Naturally occurring resveratrol oligomers are reported to have anti-inflammatory, anti-acetylcholinesterase, and anticancer properties and are the focus of significant isolation and synthesis efforts.39-48 α-viniferin, a plant-derived cyclic resveratrol trimer, is present in elevated quantities during and after fungal infection.46 It has been shown to inhibit signal transducer and activation of transcription-1 signaling, as well as cyclooxygenase-2 and inducible nitric oxide synthase activity in mammalian cells,40,41 indicating an ability to modulate innate immune sensing. In addition, resveratrol has been shown to enhance engraftment and promote expansion of mouse and human UCB-HSPCs.49,50
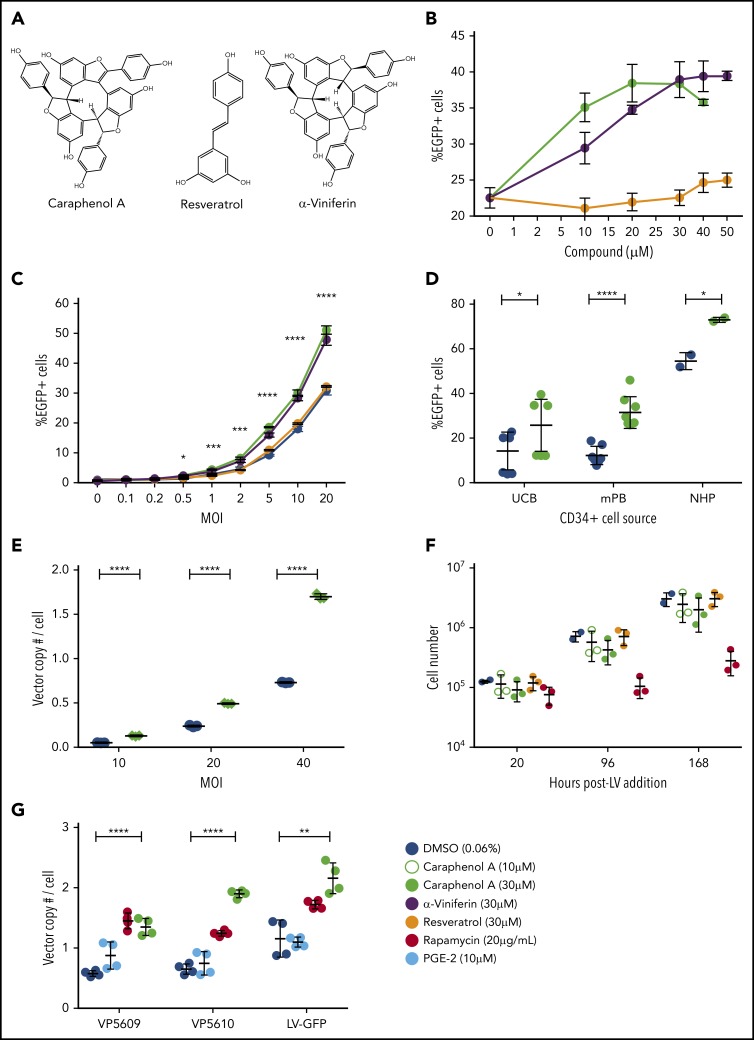
Therefore, we examined resveratrol, α-viniferin, and caraphenol A (Figure 1A), a closely related resveratrol cyclotrimer of higher oxidation state, for their capacity to augment VSV-G pseudotyped pRRL-SIN-MND-EGFP LV transduction. Caraphenol A and α-viniferin, but not resveratrol, enhanced LV gene delivery to HeLa, but not HEK293T, cells. HeLa cells responded in a dose-dependent manner across a range of MOIs, increasing EGFP marking frequency about twofold at doses between 30 and 50 μM (Figure 1B-C; supplemental Figure 1A). Twofold or greater enhancement in gene marking frequency was observed in UCB-, mPB-, and nonhuman primate-CD34+ HSPCs (Figure 1D) after caraphenol A treatment. Transduction improvement was most significant in mPB-CD34+ HSPCs, a clinically important cell type reported to be more resistant to transduction than UCB-CD34+ HSPCs.51 Caraphenol A improved integrated vector copy number (VCN) at several MOIs in UCB-CD34+ HSPCs (Figure 1E), indicating treatment may improve delivery even at high LV doses used in some clinical trial protocols.2,3,52-54 As a result of the observation that α-viniferin has more of an effect on HeLa cell growth (supplemental Figure 1B), caraphenol A was chosen for further studies. Although synthetic caraphenol A was used initially,36 similar LV transduction enhancement was seen with plant-derived, HPLC-purified compound (supplemental Figure 1C-G).
Figure 1.
Caraphenol A enhances LV gene marking in HeLa and primary hematopoietic cells. (A) Chemical structure of caraphenol A, α-viniferin, and resveratrol. (B) HeLa cells were transduced with pRRL-SIN-MND-EGFP (termed LV) at a MOI of 10, in the presence of DMSO (diluent control = no compound) or indicated concentrations of resveratrol (closed orange circles), α-viniferin (closed purple circles), or caraphenol A (closed green circles) over the course of 8 hours, before removal of caraphenol A and LV and then ex vivo culture. Cells were analyzed 5 days later by flow cytometry for EGFP expression (n = 3 independent experiments). (C) HeLa cells were transduced as earlier, with various MOIs of LV in the presence of 30 μM of each compound or DMSO (closed blue circles) for 8 hours and analyzed as earlier (n = 3). Data are shown as linear plots (mean ± standard deviation [SD]). *P < .032, **P < .0021, ****P < 0002, ****P < .0001 by 2-tailed Student t test comparing percentage EGFP expression in caraphenol A- and DMSO-treated cells. (D) LV transduction of CD34+ human UCB (n = 6 donors), human granulocyte colony-stimulating factor mobilized peripheral blood (mPB) (n = 6 donors), and nonhuman primate bone marrow aspirate (n = 2 donors) cells. CD34+ cells were transduced with LV (MOI = 8) in the absence or presence of 30 μM caraphenol A for 20 hours before LV and compound removal and expansion. Cells were analyzed 7 days later by flow cytometry for EGFP expression. Data presented as dot plots (mean ± SD) *P < .0406, ****P < .0001 by 2-tailed Student t test. (E) Average VCN in UCB-CD34+ (n = 3 donors) at increasing MOI of LV treatment with either DMSO vehicle control or caraphenol A at 30 μM. VCN was calculated as a ratio of copy number of integrated LV Gag sequences per RNAse P copies. Data presented as dot plots (mean ± SD). ****P < .0001 by 2-tailed Student t test. (F) Effects of caraphenol A (10 μM, open green circles or 30 μM, closed green circles), resveratrol (30 μM, closed orange circles), and rapamycin (20 μg/mL, closed red circles) on proliferation of mPB-CD34+ cells compared with DMSO (0.06%, closed blue circles; n = 3 donors, n = 2 for DMSO). Proliferation of UCB-CD34+ cells after compound treatment are shown in supplemental Figure 2A. Data presented as dot plots (mean ± SD). (G) VCN in mPB-CD34+ cells (n = 4 donors) of 2 separate batches of clinical-grade CL204i-EF1α-hγc-OPT SIN-lentiviral vectors (VP5609 and VP5610) developed for the treatment of SCID-X1, incorporating an internal EF1α promoter for human interleukin 2Rγc. Cell were transduced with SCID-X1 or LV at an MOI = 15 in the presence of DMSO, 30 μM caraphenol A, 20 μg/mL rapamycin, or 10 μM PGE-2 (closed light blue circles). Data presented as mean ± SD. **P = .0024, ****P < .0001 by Student t test, comparing DMSO with caraphenol A.
Rapamycin treatment has been shown to relieve LV transduction resistance in mouse and human HSPCs.15,55,56 Critically, however, rapamycin also slows proliferation of treated cells ex vivo, which might then slow progenitor expansion required for rapid leukocyte recovery in clinical transplant settings.15,55-57 In contrast, caraphenol A treatment of mPB- or UCB-CD34+ HSPCs did not reduce proliferation at a range of concentrations (Figure 1F; supplemental Figure 2A). Caraphenol A also showed no effect on cell viability (supplemental Figure 2B), plating efficiency, or lineage differentiation by colony-forming unit assay (supplemental Figure 2C-D).
A recent report identified PGE-2 as a LV transduction enhancer that improved gene delivery at a postfusion step.16 We observed a slight increase in UCB-CD34+ cell transduction with PGE-2 alone, and additive transduction enhancement when cells were coincubated with PGE-2 and caraphenol A (supplemental Figure 2E). Cotreatment of UCB-CD34+ HSPCs with rapamycin and caraphenol A or PGE-2 and rapamycin further enhanced transduction, but reduced cell viability (supplemental Figure 2F).
VCN analysis of transduction of 2 separate clinical-grade LVs used for correction of X-linked severe combined immunodeficiency (X-SCID)58 showed that caraphenol A treatment improves therapeutic gene delivery to mPB-CD34+ HSPCs (Figure 1G). These findings confirm that caraphenol A treatment enhances LV transduction in a range of hematopoietic tissue types with a favorable toxicity profile.
Caraphenol A-treated HSCs maintain improved gene marking in vivo without altering lentiviral integration profiles
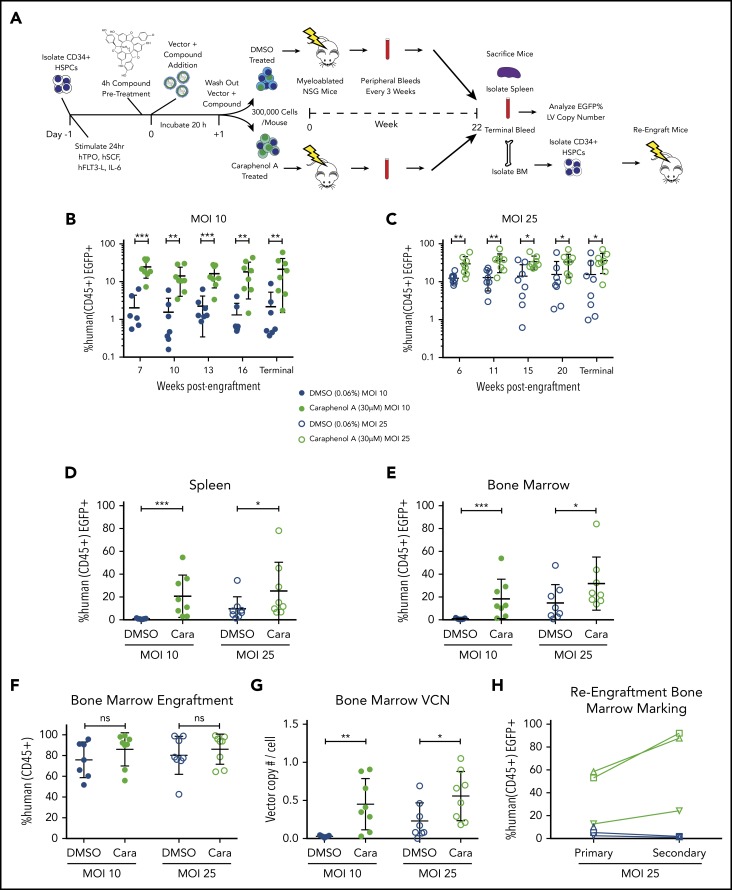
To test whether gene delivery enhancement by caraphenol A is observed in vivo, we transduced pooled human UCB-CD34+ HSPCs pretreated for 4 hours (total transduction period = 24 h) with 30 μM caraphenol A or 0.06% DMSO (dimethyl sulfoxide; vehicle control). We then transplanted 3 × 105 cells each into irradiated NSG mice (Figure 2A). Two separate cohorts (n = 8 mice per treatment) were established with CD34+ HSPCs transduced with either low (MOI = 10) or high (MOI = 25) LV doses, and EGFP marking in peripheral blood (PB) was measured approximately every 3 weeks (supplemental Figure 3A). All mice were healthy throughout the engraftment period, with no adverse events noted. Unlike other transduction enhancers, no effect on short-term human CD45+ (hCD45+) PB levels was observed with caraphenol A treatment (supplemental Figure 3B).15-17 Significant enhancement of EGFP marking levels in human hCD45+ cells in mouse PB after caraphenol A treatment was observed in low (Figure 2B) and high (Figure 2C) MOI cohorts. In both, mice transplanted with DMSO-treated CD34+ cells showed EGFP marking waning over time, whereas mice receiving caraphenol A-treated CD34+ cells retained stable EGFP marking.
Figure 2.
Caraphenol A improves gene delivery to human HSCs in mice. (A) Experimental set-up of mouse transplant experiments. NSG mice were irradiated with 2.40 Gy. UCB-CD34+ cells from a pool of donors were thawed and prestimulated for 24 hours before a 4-hour incubation of caraphenol A or DMSO (n = 8 mice per treatment and MOI, 32 mice total) and LV, MOI = 10 (MOI 10) or MOI = 25 (MOI 25). Incubation with UCB-CD34+ cells lasted 20 hours, after which, 3 × 105 cells per mouse were injected retro-orbitally, and the remaining UCB-CD34+ cells were cultured ex vivo. Transgene expression was measured 7 and 14 days posttransduction. Peripheral blood samples were obtained and evaluated every 3 to 5 weeks after an initial 6- to 7-week engraftment period. Mice were euthanized at 22 weeks (terminal) and peripheral blood, bone marrows, and spleens were obtained. For re-engraftment studies, CD34+ cells were isolated from MOI 25 cohort bone marrows (n = 3 from each treatment group), and 1 × 105 cells were injected into irradiated NSG mice. Re-engraftment and gene marking were determined after 12 weeks. (B) Percentage human CD45+ EGFP+ cells in peripheral blood of UCB-CD34+ cell-engrafted NSG mice transduced with LV at either MOI = 10, (C) or MOI = 25, treatments as per the legend, throughout indicated points during the study period. Human cells were gated from the total leukocyte population and analyzed for EGFP expression. Dot plots presented (mean ± SD) with the y-axis in log10 scale. *P < .028, **P < .0042, ***P < .0006 by 2-tailed Mann-Whitney U test. Percentage human CD45+ EGFP+ cells in spleen (D) and bone marrow (E) of UCB-CD34+ cell engrafted NSG mice at the terminal time, comparing EGFP+ expression in caraphenol A- (green circles) and DMSO-treated (blue circles) mice at 2 MOIs. Dot plots presented (mean ± SD) comparing spleen MOI 10 (closed circles; ***P = .0003), MOI 25 (open circles; *P = .049) and bone marrow MOI 10 (closed circles; ***P = .0006), MOI 25 (open circles; *P = .042) by 2-tailed Mann-Whitney U test. (F) Comparison of donor engraftment in bone marrow at terminal time, as measured by total proportion of leukocytes that were mCD45− hCD45+. Dot plots presented (mean ± SD), n.s., not significant. (G) VCN of human cells from bone marrow of caraphenol A and DMSO-treated cohorts, 22 weeks after ex vivo LV transduction and compound treatment. VCN was recorded as a ratio of integrated Gag sequences per RNase P sequence. Dot plots presented (mean ± SD), comparing caraphenol A- with DMSO-treated mice at MOI 10 (closed circles; **P = .0022), and MOI 25 (open circles; *P = .022) by 2-tailed Mann-Whitney U test. (H) Percentage human CD45+ EGFP+ cells in bone marrow of NSG mice receiving UCB-CD34+ cells at terminal points of primary (left, 22 weeks) and secondary (right, 12 weeks) transplant, comparing EGFP+ expression in caraphenol A and DMSO mice at MOI 25. Data presented as dot plots, each representing individual mice and change from primary to secondary transplant.
Mice were euthanized at 22 weeks, and EGFP marking was assessed in PB, bone marrow, and spleen (Figure 2B-E), and human cell engraftment and VCN were assessed in bone marrow (Figure 2F-G). One mouse in the low MOI DMSO-treated group was excluded from analyses because of human bone marrow engraftment below 2%. Consistent with PB and ex vivo observations, caraphenol A treatment significantly increased EGFP marking of hCD45+ spleen and bone marrow cells (Figure 2D-E). No significant difference in human cell engraftment was observed in caraphenol A- vs DMSO-treated cells in either cohort (Figure 2F). Interestingly, VCN values from the bone marrow of mice transplanted with caraphenol A-treated CD34+ HSPCs showed ∼10-fold higher levels compared with DMSO-treated controls ex vivo (MOI 10 VCN = 0.05 ex vivo vs 0.5 in vivo; Figure 1E vs Figure 2G). Hematopoietic lineage analysis of bone marrow subsets showed increased EGFP marking in myeloid, T-, and B-cell subsets (supplemental Figure 3C) in mice receiving caraphenol A-treated cells. No skewing of myeloid, T, or B subsets was observed, suggesting that enhanced EGFP marking was not a result of dysregulated hematopoiesis (supplemental Figure 3D).
To study the effect of caraphenol A on gene marking in long-term repopulating (LTR) HSCs, we established a third cohort of humanized mice from high-dose (MOI = 25) LV-treated cells. Caraphenol A treatment again increased the EGFP marking considerably more in vivo after 22 weeks than from initial ex vivo observations (supplemental Figure 4A). As before, caraphenol A produced no significant effect on engraftment or lineage frequency (supplemental Figure 4B-C). Subsequently, 1 × 105 CD34+ cells isolated from the bone marrow of the lowest-, middle-, and highest-frequency EGFP marked mice from the caraphenol A- and DMSO-treated cells were engrafted into secondary recipient mice. Because of the lower number of hCD34+ cells used; engraftment was less robust after 12 weeks (supplemental Figure 4E). Percentage EGFP marking dropped in cells originating from DMSO-treated CD34+ primary mice, but was dramatically increased in caraphenol A-treated CD34+ cell secondary mice (Figure 2H). Both caraphenol A- and DMSO-treated mice could regenerate B-, T-, and myeloid cell lineages after secondary transplantation (supplemental Figure 4F-G), indicating LTR-HSC engraftment.
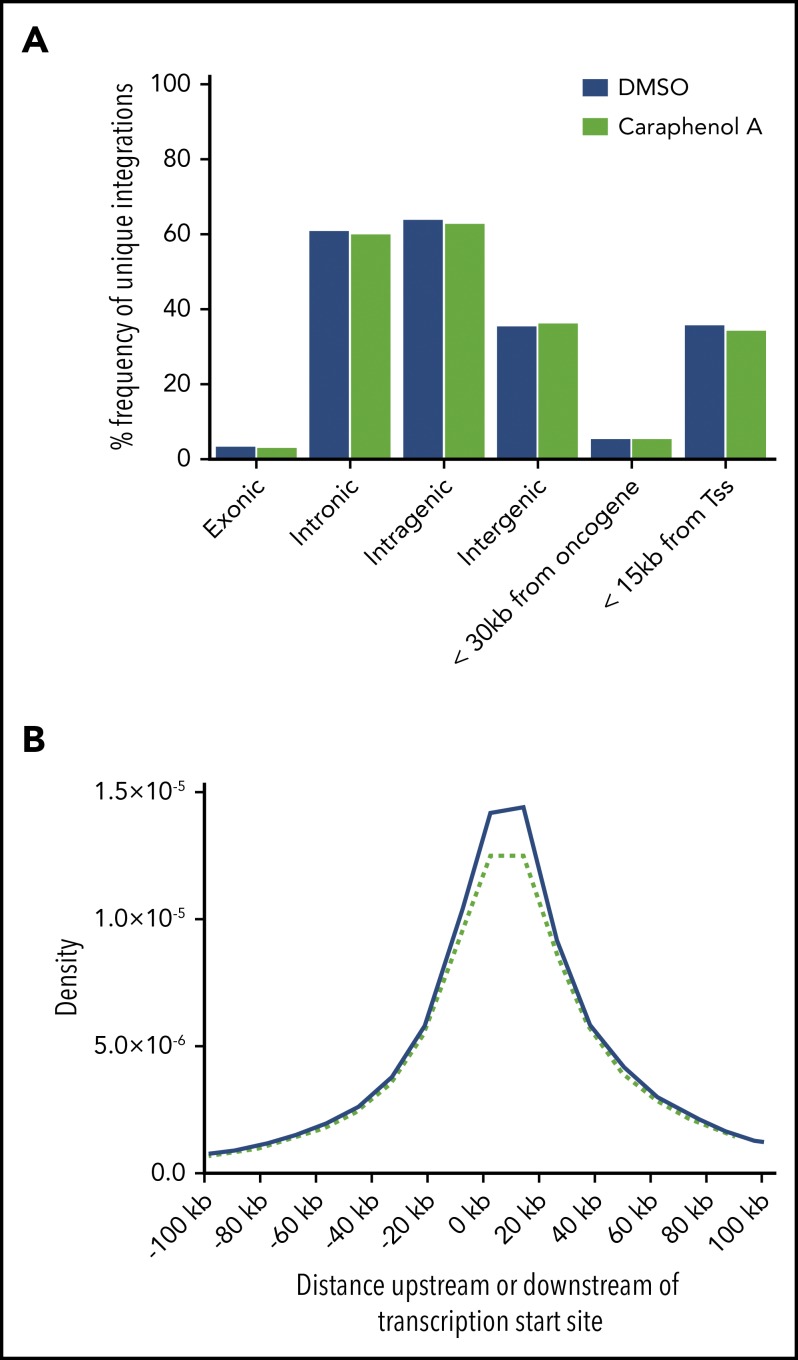
With the observation that VCN increased with caraphenol A treatment, and given the reported effects of resveratrol and other polyphenols on global patterns of gene regulation,59 we investigated whether caraphenol A treatment altered patterns of LV integration. High-throughput retroviral integration site analysis60 was performed on human cells from bone marrow and spleen samples harvested from both the low and high MOI cohorts. Unique integrations were identified across all samples, and no significant differences were observed in the frequency of chromosomal insertion between treatment groups relative to genomic features or known oncogenes. However, caraphenol A treatment appeared to reduce integrations in close proximity to transcription start sites only slightly (Figure 3A-B). Together, these findings suggest that caraphenol A treatment enhances gene delivery frequency ex vivo, in LTR-HSCs, without biasing LV integration.
Figure 3.
Caraphenol A treatment during CD34+ cell LV transduction does not affect patterns of LV integration observed in human cells relative to DMSO-treated controls. A composite of individual integration events obtained from bone marrow and spleen samples (n = 8 mice per treatment and 2 MOIs, 32 mice total; see Figure 2A, at the terminal end point). (A) Frequency of unique integration events occurring relative to indicated genomic features, or within 30 kb of known oncogenes as a function of caraphenol A (green bars) or DMSO treatment (blue bars). (B) Probability density function for integration to occur within 100 kb of transcription start sites (Tss) for caraphenol A-treated (green dashed line) compared with DMSO-treated (blue solid line) bone marrow and spleen cells.
Caraphenol A treatment facilitates lentiviral escape from endosomes
We next investigated the step by which caraphenol A facilitates vector entry. Caraphenol A did not alter the frequency or density of the VSV-G receptor LDL-R in UCB-CD34+ HSPCs (supplemental Figure 5A). Similarly, transduction with LVs pseudotyped with measles virus glycoproteins was not significantly enhanced (supplemental Figure 5B). Measles virus has been proposed to fuse at the cell surface,61,62 distinct from the clathrin-mediated and pH-dependent endocytosis mechanism used by VSV-G.14,63 Caraphenol A treatment also did not increase AAV6-mediated gene delivery in UCB-CD34+ HSPCs (supplemental Figure 5C). These results suggest that caraphenol A specifically enhances enveloped vector entry routes that require pH-dependent endocytosis.
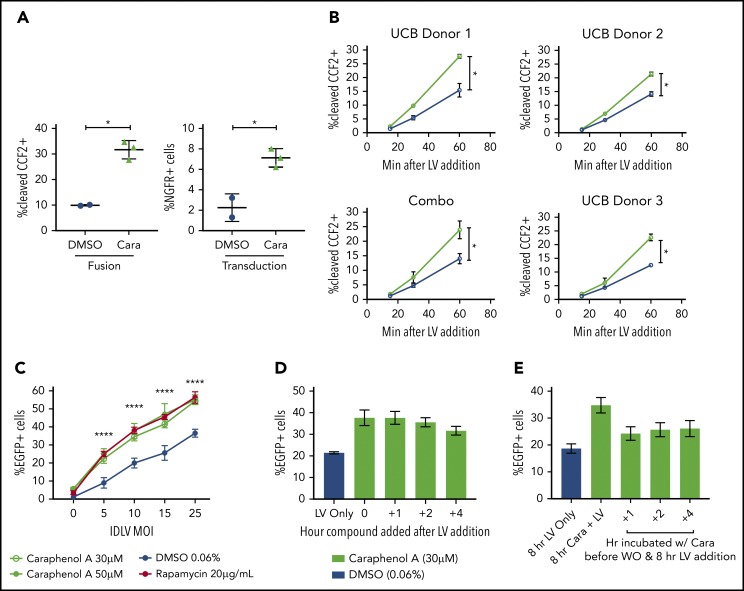
Caraphenol A treatment of UCB-CD34+ HSPCs increased LV-endosomal membrane fusion and escape of LV cores to the cytoplasm, as measured by the BLaM-Vpr assay64 (Figure 4A). A modified kinetic BLaM-Vpr assay65 demonstrated that UCB-CD34+ HSPCs pretreated with caraphenol A showed an immediate twofold increase in fusion that was maintained throughout, indicating that the rate of LV cytoplasmic entry had increased (Figure 4B). mPB-CD34+ HSPCs also showed an enhanced rate of LV fusion with caraphenol A treatment, which later translated to increased transduction (supplemental Figure 5D-E). Measurements of both early- and late-stage reverse transcription products by quantitative polymerase chain reaction showed two- to threefold increases on LV and caraphenol A addition, whereas the ratio of early to late products remained the same per UCB donor (supplemental Figure 5F-H). This indicates that caraphenol A increases the total pool of reverse transcribed viral DNA, but has no effect on reverse transcription efficiency in UCB-CD34+ HSPCs.66 Caraphenol A also increased integration-deficient LV transduction of mPB-CD34+ HSPCs (Figure 4C), consistent with an effect early in LV entry, before genomic integration. Caraphenol A treatment showed maximal transduction enhancement in HeLa cells when LV and caraphenol A were added simultaneously. However, caraphenol A could be added up to 4 hours after LV, or washed out before LV addition, and still resulted in some enhancement (Figure 4D-E). These findings localize the effect of caraphenol A on enhancing LV escape from the endosomes, with maximal effect during simultaneous addition of caraphenol A and vector.
Figure 4.
Concurrent caraphenol A treatment improves LV endosomal escape into the cytoplasm. (A) UCB-CD34+ cells (n = 3 donors) were transduced in the presence of caraphenol A (Cara, 30 μM, green symbols) or DMSO (0.06%, blue symbols) with LV, MOI 15, carrying the enzyme-Vpr protein (LV-Vpr). After a 6-hour transduction, cells were loaded with 7-hydroxycoumarin cephalosporin fluorescein–acetoxymethyl (CCF2-AM) substrate, and LV entry was quantified by flow cytometric detection of cells exhibiting cleaved CCF2. Transduction was measured by nerve growth factor receptor (NGFR) expression 7 days later. Data presented as dot plots (mean ± SD). Fusion, **P = .0039; transduction, *P = .016 by 2-tailed Student t test. (B) UCB-CD34+ cells (n = 3 donors) were LV-Vpr-transduced in the presence of DMSO or caraphenol A, as above, and then transferred to 4°C at indicated points before loading with CCF2-AM substrate at 12°C for overnight, then analyzed as earlier. Data presented as linear plots (mean ± SD). Combo plot indicates mean ± SD of 3 donors combined. Small plots (closed circles) indicate mean ± SD of technical replicates (n = 2 cultures) from each donor. Combination figure (*P = .0406, slopes of linear regression significantly different). (C) mPB-CD34+ cells (n = 2 donors) were pretreated with either caraphenol A (30 µM, green open circles; 50 µM, green closed symbols), rapamycin (20 µg/mL red closed symbols), or 0.06% DMSO (blue closed symbols) for 4 hours and transduced with varying doses of integration-deficient LV. Percentage EGFP expression was measured 3 days later by flow cytometric analysis. Data are shown as linear plots (mean ± SD). ****P < .0001 by 2-tailed Student t test, comparing percentage EGFP expression in 30 µM caraphenol A and 0.6% DMSO-treated cells. (D) HeLa cells (n = 5 cultures) were treated with LV at MOI 10 for 8 hours and caraphenol A (30 μM, green bars) was added at indicated points after LV addition. DMSO (0.06%, blue bar) with LV only was added as a separate control. Compounds and LV were washed out 8 hours after LV addition, and cells were analyzed for EGFP expression by flow cytometry 5 days later. Data presented as bar graphs (mean ± SD). (E) Cells were pretreated with caraphenol A (30 μM, green bars) for indicated lengths of time before washout of compound and exposure to LV-GFP for an additional 8 hours. After transduction, LV and any remaining compound was removed, and cells were analyzed as in panel D. Bar graphs presented (mean ± SD).
LV restriction by IFITM2/3 proteins is relieved by caraphenol A treatment
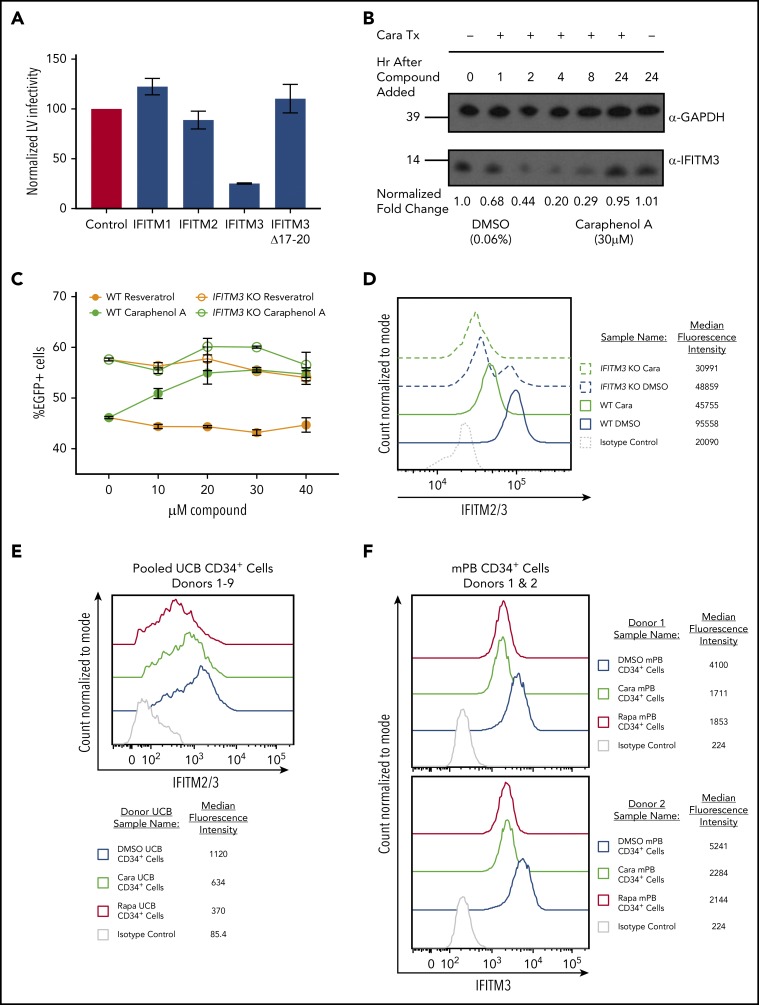
The IFITM family of proteins are potent restrictors of viruses that use pH-dependent fusion for endosomal escape and are intrinsically expressed in pluripotent and multipotent tissue types.26,28-30,67-72 To confirm the effect of IFITM proteins on LV transduction, we ectopically expressed IFITM1, IFITM2, or IFITM3 in HEK293T cells (supplemental Figure 6A-B) and compared transduction efficiency with LV (Figure 5A). Although IFITM1 ectopic expression had little effect, IFITM2- and IFITM3-expressing cells had reduced transduction, with IFITM3 exhibiting the most inhibition. After introducing the Δ17-20 mutation into IFITM3, which redirects IFITM3 localization to the cell periphery,73 we observed a loss of LV restriction, confirming that IFITM3 localization has a significant role in restricting VSV-G-mediated LV transduction.
Figure 5.
IFITM2/3 proteins restrict LV transduction and are downregulated by caraphenol A. (A) HEK 293T cells stably expressing pQCXIP-FLAG-IFITM1, pQCXIP-FLAG-IFITM2, pQCXIP-FLAG-IFITM3, or pQCXIP-FLAG-IFITM3 Δ17-20 were used for evaluating LV IFITM restriction. Each 293T IFITM cell line was transduced with LV at MOI 13, then analyzed for EGFP expression 24 hours later by flow cytometry. EGFP levels were normalized to EGFP of LV transduced pQCXIP-FLAG 293T (no IFITMs) control set to 100 and results presented as bar graphs (mean ± SD; n = 5 independent experiments). (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot analysis of cell lysates generated from HeLa cells treated with caraphenol A (30 μM) or DMSO (0.06%) for the indicated period. Immunoblotting was performed with α-IFITM3-specific antibody and α-GAPDH as a loading control. Numbers indicate location and size (kD) of protein standards in ladder. Image provided is a representative blot of 3 experiments. (C) TZM-bl (wild-type and IFITM3 KO, n = 2 cultures from 2 independent experiments) were seeded and transduced with LV at MOI 10 in the presence of the indicated dose of caraphenol A (green lines) or resveratrol (orange lines) for 8 hours before LV and compound removal. Cells were evaluated for EGFP expression by flow cytometry 5 days later. Wild-type cells (closed circles) demonstrated significant transduction enhancements response to caraphenol A (DMSO vs caraphenol A, P < .0001, 2-tailed Student t test), but not resveratrol (not significant, Student t test). IFITM3 KO cells showed no significant difference compared with DMSO with caraphenol A treatment. (D) TZM-bl wild-type (solid lines) and IFITM3 KO (dotted lines) cells were treated for 4 hours with caraphenol A (30 μM, green) or DMSO (0.06%, blue), then fixed, permeabilized, and analyzed for intracellular IFITM2/3 expression by flow cytometry (n = 3 independent experiments). Expression reported as median fluorescence intensity of IFITM2/3+ cells. (E) UCB-CD34+ cells (n = 9 donors pooled) were prestimulated and then treated with either caraphenol A (30 μM) or DMSO (0.06%) for 4 hours. Cells were then fixed, permeabilized, and immunostained for IFITM2/3 expression before analysis by flow cytometry. Expression reported as percentage of IFITM2/3+ cells compared with isotype control and median fluorescence intensity of IFITM2/3+ cells. (F) mPB-CD34+ cells (n = 2 donors) were prestimulated (see "Methods") and then treated with caraphenol A (30 µM, green line), rapamycin (20 µg/mL red line), or 0.06% DMSO (blue line) for 4 hours followed by a 2-hour addition of 15 MOI integration-deficient LV (Figure 4C). Cells were then fixed, permeabilized, and immunostained for IFITM3 expression before analysis by flow cytometry. Expression reported as median fluorescence intensity of IFITM2/3+ cells.
Next, we evaluated the effect of caraphenol A treatment on IFITM2/3 expression in HeLa cells. We observed a high level of IFITM3 protein expression by western blot that was reduced by up to 80% after 4 hours of caraphenol A treatment (Figure 5B; supplemental Figure 6C). Longer incubation with caraphenol A showed a recovery of IFITM3 protein levels with a return to baseline after 24 hours of continuous incubation. Quantitative polymerase chain reaction evaluating IFITM3 mRNA expression in HeLa cells, under the above treatment conditions, showed no significant changes (supplemental Figure 6D), indicating caraphenol A’s effect is likely posttranslational. Unlike caraphenol A, PGE-2 treatment did not alter IFITM3 protein levels (supplemental Figure 6E). Caraphenol A reduced levels of IFITM2 and IFITM3, but not IFITM1, proteins in HEK 293T cells ectopically expressing each, as determined by flow cytometry (supplemental Figure 6F-G).
We next sought to determine whether IFITM3 expression was required for caraphenol A-mediated LV transduction enhancement. IFITM3 was knocked out in HeLa-derived TZM-bl cells by directed IFITM3 CRISRP/Cas9 disruption and homologous recombination.32 These cells showed a complete loss of IFITM3 protein with no observed effect on IFITM2 levels (supplemental Figure 6H). Caraphenol A treatment of wild-type (WT) TZM-bl cells exhibited a 1.5-fold increase in transduction, whereas resveratrol showed no effect (Figure 5C). In contrast, IFITM3 knockout (KO) cells demonstrated increased baseline LV transduction, and caraphenol A treatment led to only a slight additional transduction enhancement, potentially as a result of downregulation of IFITM2 (Figure 5D). In addition, we observed that vacuolar ATPase inhibitor bafilomycin A1 rescued the caraphenol A-mediated downregulation of IFITM2 and IFITM3 (supplemental Figure 6I), indicating that endolysosomal acidification contributes to IFITM protein degradation.32
IFITM protein expression in mPB-CD34+ HSPCs from several donors was evaluated by western blot for IFITM1, IFITM2, and IFITM3 expression. No appreciable IFITM1 was observed, but IFITM2/3 were seen in all tested donors (supplemental Figure 6J). Flow cytometric analysis of pooled (n = 9) donor UCB-CD34+ HSPCs and mPB-CD34+ HSPCs from 2 different donors treated with caraphenol A showed a consistent 50% reduction in median fluorescence intensity in IFITM2/3+ cells (Figure 5E-F). These findings imply that caraphenol A reduces IFITM2/3 expression in therapeutically relevant CD34+ cells.
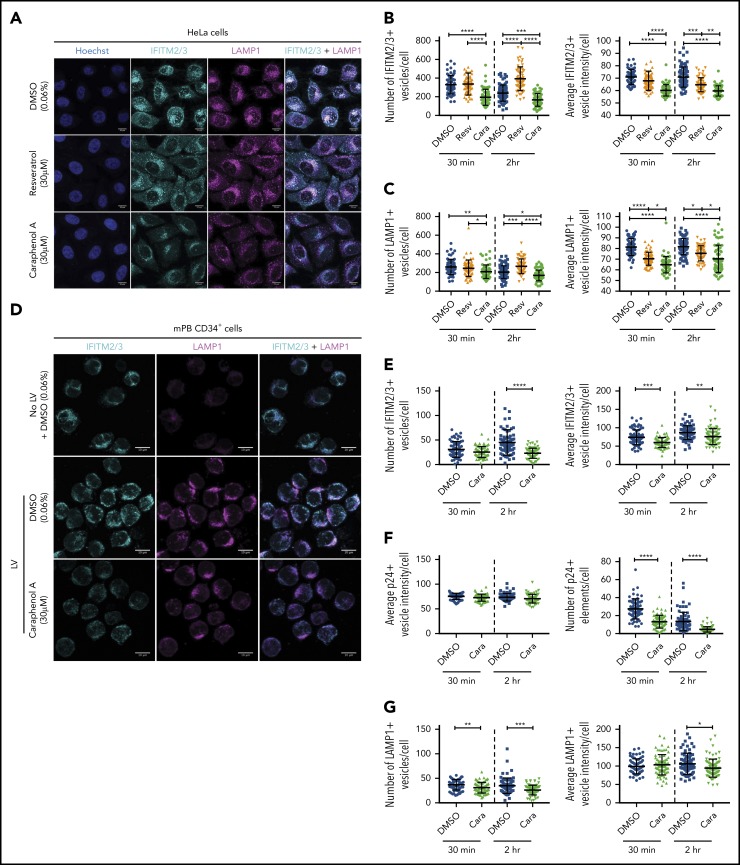
To evaluate the effect of caraphenol A on IFITM2/3 protein localization in the endosome, we evaluated HeLa cells and mPB-CD34+ HSPCs by confocal microscopy. HeLa cells treated with caraphenol A and LV showed a reduction in and relocation of IFITM2/3 signal from the cell periphery to the perinuclear region, a result not observed with resveratrol or DMSO treatment (Figure 6A). Three-dimensional analysis and quantification of IFITM2/3-containing endosomes demonstrated a highly significant reduction in number and staining intensity compared with resveratrol- or DMSO-treated cells at 30 minutes and 2 hours after LV addition (Figure 6B). In contrast to Hela cells, mPB-CD34+ HSPCs exhibited limited endosomal activity without LV addition (Figure 6D). Within 30 minutes of LV addition, lysosomal-associated membrane protein-1 (LAMP1+) endosomes were apparent and the IFITM2/3 signal appreciated in the endosomal compartment (Figure 6D). As seen in HeLa cells, a reduction and relocalization of the IFITM2/3 signal in caraphenol A-treated mPB-CD34+ HSPCs was observed (Figure 6D). The kinetics of the effect were different than HeLa cells, as a significant reduction in IFITM2/3+ vesicle number was not observed until 2 hours after LV addition (Figure 6E), although a significant reduction in IFTIM2/3 staining intensity per endosome was observed at both points. In support of the above, we observed no difference in p24+ (LV) signal intensity per endosome, but a reduction in the number of p24+-containing endosomes with caraphenol A treatment (Figure 6F; supplemental Figure 7A), indicating similar initial vector uptake but increased LV fusion and escape from endosomes. In addition, we noted differing effects on various endosomal compartments, as caraphenol A treatment had no effect on the number of early endosomal antigen-1-containing vesicles (supplemental Figure 7B-C). However, a moderate but significant reduction in the number and staining intensity of late endosomes carrying LAMP1 after caraphenol A treatment was observed in both HeLa (Figure 6C) cells and mPB-CD34+ HSPCs (Figure 6G). We also detected an overall increase in endosomal pH, as determined by LysoSensor pH-sensitive dextran (supplemental Figure 7D).
Figure 6.
Caraphenol A alters expression and subcellular localization of IFITM2/3 protein and late endosomes. (A) Representative images of HeLa cells obtained by confocal microscopy using 63× oil immersion objective lens at room temperature. HeLa cells were plated and treated for 4 hours with caraphenol A (30 μM), resveratrol (30 μM), or DMSO (0.06%) before addition of LV, MOI of 15, for 30 minutes or 2 hours. Cells were then fixed, permeabilized, and immunostained using α-IFITM2/3, α-LAMP1 antibodies, and Hoechst 33342 nuclei stain. Scale bars are 10 microns. Images were collected on Zeiss Zen Software and analyzed using Imaris InCell software, identifying the number and mean intensity of IFITM2/3 (B) and LAMP1 (C) stained vesicles per cell. At least 50 cells were imaged per condition and plotted as dot plots (caraphenol A, green symbols; resveratrol, orange symbols; DMSO, blue symbols; mean ± SD). *P < .032, **P < .0021, ***P < .0002, ****P < .0001 by Kruskal-Wallis test with Dunn’s multiple comparison correction. (D) Representative images of mPB-CD34+ HSPC that were thawed, prestimulated for 48 hours, and then either immediately imaged without LV and DMSO (0.06%) (top), or then treated for 4 hours with either DMSO (middle, 0.06%) or caraphenol A (bottom, 30 μM) before addition of LV for 30 minutes or 2 hours. Cells were fixed, stained, and analyzed as above for HeLa cells for IFITM2/3 (E), p24 protein (F), and LAMP1+ (G) vesicle number and mean vesicle intensity (mean ± SD).
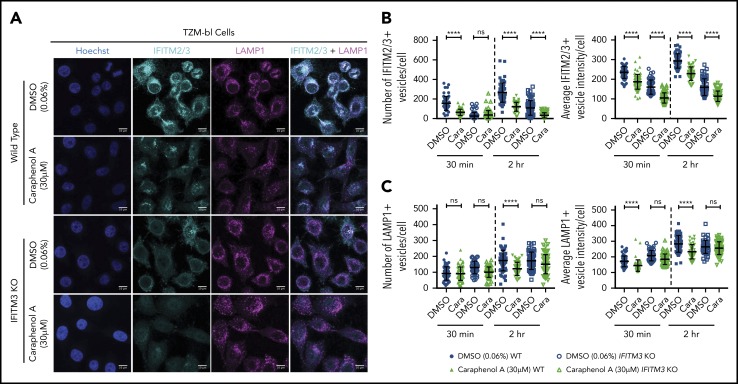
Given that caraphenol A reduced the number of LAMP1+ vesicles and aided LV endosomal escape in both cell types tested, we next investigated whether this effect was dependent on IFITM3. Both IFITM3 KO and WT TZM-bl cells were evaluated as described previously in the presence of LV and caraphenol A or DMSO (Figure 7A). Caraphenol A reduced the number and intensity of IFITM2/3-containing endosomes in WT TZM-bl cells and of IFITM2 in IFITM3 KO cells (Figure 7B). In contrast to WT cells, the reduction in LAMP1+ vesicle number and staining intensity was not observed in the IFITM3 KO cells after caraphenol A treatment (Figure 7C). These results support the finding that effects of caraphenol A on the late endosome compartment are dependent on the presence of IFITM3 proteins.
Figure 7.
Caraphenol A effect on late endosome is dependent on IFITM3 expression. (A) Representative images of WT TZM-bl and IFITM3 KO cells imaged by confocal microscopy, as above. Cells were plated and treated for 4 hours with caraphenol A (30 μM) or DMSO (0.06%) before addition of LV for 30 minutes or 2 hours. Cells were then fixed, permeabilized, and immunostained using α-IFITM2/3, α-LAMP1 antibodies, and Hoechst 33342 nuclei stain. Scale bars are 10 microns. Images were collected and analyzed as previously described, identifying the number and mean intensity of IFITM2/3- (B) and LAMP1-stained (C) vesicles per cell. Data plotted as dot plots comparing WT (closed symbols) to IFITM3 KO (open symbols) treated with caraphenol A (green symbols) or DMSO (blue symbols, mean ± SD). *P < .032, **P < .0021, ***P < .0002, ****P < .0001 by Kruskal-Wallis test with Dunn’s multiple comparison correction.
Discussion
An ongoing barrier to LV-mediated gene therapy for hematologic disorders is the well-documented transduction resistance of HSCs, previously attributed to cell cycle quiescence,74 lack of vector receptor,19 or proteasome-mediated inhibition.9 We report here that short-term treatment of CD34+ HSPCs with the resveratrol cyclotrimer, caraphenol A, increases transduction by clinical, laboratory-grade, and nonintegrating LVs, indicating the potential to improve both gene delivery and gene-editing methodologies. Importantly, we observed stable marking in all lineages derived from human LT-HSCs in serially engrafted NSG mice, and comparable LV integration profiles to DMSO treatment. The HSPC LV restriction mechanism modulated by caraphenol A was found to be the reduction and relocalization of endosomal IFITM2/3 proteins.
We and others have previously demonstrated that short-term rapamycin and cyclosporine treatment enhanced transduction of CD34+ HSPCs, resulting in a lasting improvement in gene marking in vivo.15,17,32,55,56 Similar to caraphenol A, both rapamycin and cyclosporin H downregulated IFITM2/3,17,32 indicating this pathway may be a major barrier to HSC LV-mediated gene therapy. However, these compounds can be cytostatic with LV treatment in vivo and, in the case of rapamycin, a delay in proliferation of hematopoietic progenitors is observed in culture. This may translate to delayed progenitor expansion required for leukocyte recovery in clinical transplant settings.57
Caraphenol A does not delay expansion of CD34+ HSPCs with LV treatment (Figure 1F; supplemental Figure 2A), further implying that LV transduction effect can be uncoupled from pathways that influence engraftment and cell cycle delay. Similar to rapamycin, resveratrol has been demonstrated to induce autophagy through the inhibition of mTOR in an ULK1-, SIRT1-, and AMPK-dependent manner.75-77 However, it remains unclear whether caraphenol A also acts as an mTOR inhibitor. We previously showed that gene delivery enhancement by rapamycin is independent of autophagy induction and is likely achieved through inhibition of mTOR complex (mTORC) 2, rather than mTORC1,32 suggesting that mTORC2 inhibition may represent a common means for improving transduction. In contrast, PGE-2 does not alter IFITM2/3 expression (supplemental Figure 6E) and is additive when used with caraphenol A for LV transduction, which suggests a mechanism of enhancement distinct from caraphenol A, rapamycin, and cyclosporin H.
The surprising finding that initiation of endosomal activity in mPB-CD34+ HSPC and relocation of IFITM2/3 staining from peripheral to perinuclear endosomes depends on LV addition implies that endosomal-IFITM-lysosomal restriction activity remains dormant in quiescent CD34+ HSPCs until vector binding and entry. The observation that the caraphenol A-dependent effect of decreasing late-stage LAMP1+ endosomes was not observed in IFITM3 KO cells suggests a link between the presence of IFITM3 and late endosomal activity. This finding is further supported by a recent independent analysis in IFITM KO cell lines78 and is consistent with the observed higher cellular pH observed after caraphenol A treatment, indicating a reduction in acidic cellular compartments. VSV-G triggers vector/host membrane fusion at a defined pH, a separate event from LV core escape into the cytoplasm.62 IFITM proteins are reported to interact with vacuolar ATPase, altering acidification of the endolysosomal lumen,79 which may trap virions in vesicles intended for degradation.80 Determining whether the transduction enhancement effect of caraphenol A is related to increased vector escape from a normally degradative trafficking pathway or diversion to compartments favorable to vector fusion requires further study. The finding that complete whole-cell IFITM3 downregulation was not needed for transduction enhancement indicates that proper IFITM localization, rather than overall expression, may be a critical barrier to gene delivery.17,32
Because of the role of IFITM proteins in cellular protection from pathogens,81,82 long-term reduction of IFITM2/3 expression in CD34+ HSPCs would be unfavorable. Transient inhibition of IFITM2/3 through short-term ex vivo compound pulsing may be a safer strategy for increasing LV gene delivery. However, IFITM expression abnormalities have been observed in various cancers, and IFITM gene disruption has been associated with tumor malignancy and growth.31,83,84 The differing trafficking strategy used by VSV-G pseudotyped LVs, compared with HIV-1, exposes LV to distinct restriction factors that may affect integration and alter latency.29,35 Thus, an open question is whether transient pharmacologic IFITM modulation is a genetically safe approach. Our findings of IFITM2/3 protein recovery after transient caraphenol A treatment that show no altered LV integration profile or lineage frequency in primary or secondary transplantation recipients are therefore encouraging.
One promising finding was the observed transduction improvement in vivo, beyond that seen ex vivo with the same applied dose of LV (Figure 1E vs Figure 2G). Serial transplantation studies demonstrated a further increase in EGFP marking frequency in transplanted cells originally treated with caraphenol A, unlike the decrease observed in DMSO-treated HSPCs. A recent report highlights that constitutive interferon-stimulated gene expression is a unique phenotype found in pluripotent and multipotent tissue in the absence of IFN signaling.26 An explanation consistent with our findings is a higher overall expression of IFITM proteins in LTR-HSCs compared with HSPCs. As a consequence, LTR-HSCs may have a more pronounced response to caraphenol A IFITM downregulation, although additional experiments evaluating IFITM expression in these cells is required.
We have described novel small molecules that significantly improve LV gene delivery to HSCs via a cellular mechanism involving disruption of IFITM2/3-mediated LV restriction. Importantly, IFITM2/3 modulation by caraphenol A did not result in an unusual integration profile or lineage abnormalities in vivo. Moreover, we provide evidence that additional non-IFITM2/3 LV restriction pathways are operating in HSPCs that, when countered, further increase transduction efficacy when combined with compounds that target IFITM2/3. Gene delivery strategies that minimize treatment time, expense, complexity, and vector concentrations are of critical importance for advancing the fields of gene therapy and cellular engineering to the clinic. We anticipate that the use of caraphenol A may help achieve these goals, whereas our findings may help to uncover additional restriction pathways that facilitate enhanced gene delivery to stem cells.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the following entities and individuals for reagents and advice: Nathan E. Wright for achieving the laboratory synthesis of caraphenol A and Columbia University for providing samples of synthetic caraphenol A; Mary Laughlin and The Cleveland Cord Blood Center, Cleveland, OH, for their generous supply of human cord blood; David Rawlings, Seattle, WA, for helpful advice; Chen Liang, McGill University, for the gift of IFITM plasmids; Gabor Verse, Bluebird Bio, Boston, MA, for helpful advice and reagents; Rocket Pharma, LLC, NY, for advice; San Diego, CA, CFAR for p24 determination; Scott Henderson for technical advice on microscopy data collection and analysis; Geraldine Goebrecht for her helpful editing; and The Cell Processing and Procurement Core, Fred Hutchinson Cancer Research Center, for adult CD34 cells, Seattle, WA.
Research reported in this publication was supported by the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute; NIH National Institute of General Medical Sciences; NIH National Institute of Allergy and Infectious Diseases; and the Office of Research Infrastructure Programs of the National Institutes of Health, National Cancer Institute; National Science Foundation; and the Fred Hutchinson Cancer Research Center (FHCRC), under award numbers CA042056 (D.L.B.), U54GM103368 (B.E.T.), P30 AI036214 CFAR Core, R01HL116221 (B.E.T.), U54DK106829, P51RR000167 (I.S.), NSF/DGE-1346837 fellowship and a Turner B. and Lesly Starr Shelton Endowment Scholarship (C.M.G.), 5T32AI007354 (K.H.), 5T32AI007354 (N.D.T.), F30 HL137563 (S.O.), FHCRC Development (J.E.A.), HL115128 (H.-P.K.), HL098489 (H.-P.K.), AI096111 (H.-P.K.), the José Carreras/E. Donnall Thomas Endowed Chair for Cancer Research, the Fred Hutch Endowed Chair for Cell and Gene Therapy (H.-P.K.), and P30 CA015704. Work in the laboratory of A.A.C. is supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The original synthesis of caraphenol A was principally supported by grants from the NIH National Institute of General Medical Sciences (R01GM84994) and the Research Corporation for Science Advancement (S.A.S.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and National Science Foundation. This is publication 29720 from The Scripps Research Institute.
Footnotes
For original data, please contact Bruce Torbett (betorbet@scripps.edu).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.O., N.D.T., and K.H. performed most of the experiments and analyzed data; S.O. and B.E.T. wrote the manuscript; O.G. and E.S. performed western blotting for IFITM proteins; K.G.H., H.-P.K., and J.E.A. designed and interpreted integration site analysis experiments; K.G.H., L.E.S. G.S.-H., and R.R.C. performed integration site analysis sample processing, next-generation sequencing, and analysis; G.S. and A.A.C. generated IFITM3 KO cell lines, evaluated IFITM expression in primary cells, performed IFITM LV restriction experiments and western blotting for IFITM proteins, and generated graphics; C.M.G. and D.L.B. advised on and performed purification of natural compounds; L.E.S. conducted bioinformatics analysis and generated graphics; S.D. and I.S. performed and advised on experiments involving nonhuman primate CD34+ cells; B.Y.R. and B.P.S. generated and advised on the use of clinical X-SCID vectors; S.A.S. and B.E.T. conceptualized the study; S.A.S. generated initial synthesis of caraphenol A, identified key structural analogs worth further study, and advised on purification of commercial natural compounds; and all authors contributed text, discussed results, and edited the manuscript.
Brian Sorrentino died on 16 November 2018.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bruce E. Torbett, The Scripps Research Institute, Department of Immunology and Microbiology, 10550 N. Torrey Pines Rd, IMM 115, La Jolla, CA 92037; e-mail: betorbet@scripps.edu.
REFERENCES
- 1.Aiuti A, Cattaneo F, Galimberti S, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447-458. [DOI] [PubMed] [Google Scholar]
- 2.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341(6148):1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341(6148):1233158. [DOI] [PubMed] [Google Scholar]
- 4.Adair JE, Becker PS, Chandrasekaran D, et al. Gene therapy for Fanconi anemia in Seattle: Clinical experience and next steps. Blood. 2016;128(22):3510. [Google Scholar]
- 5.Thompson AA, Walters MC, Kwiatkowski J, et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N Engl J Med. 2018;378(16):1479-1493. [DOI] [PubMed] [Google Scholar]
- 6.Ribeil J-A, Hacein-Bey-bina S, Payen E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med. 2017;376(9):848-855. [DOI] [PubMed] [Google Scholar]
- 7.Biffi A. Hematopoietic stem cell gene therapy for storage disease: current and new indications. Mol Ther. 2017;25(5):1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar SRP, Markusic DM, Biswas M, High KA, Herzog RW. Clinical development of gene therapy: results and lessons from recent successes. Mol Ther Methods Clin Dev. 2016;3:16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santoni de Sio FR, Cascio P, Zingale A, Gasparini M, Naldini L. Proteasome activity restricts lentiviral gene transfer into hematopoietic stem cells and is down-regulated by cytokines that enhance transduction. Blood. 2006;107(11):4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ailles L, Schmidt M, Santoni de Sio FR, et al. Molecular evidence of lentiviral vector-mediated gene transfer into human self-renewing, multi-potent, long-term NOD/SCID repopulating hematopoietic cells. Mol Ther. 2002;6(5):615-626. [PubMed] [Google Scholar]
- 11.Mazurier F, Gan OI, McKenzie JL, Doedens M, Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103(2):545-552. [DOI] [PubMed] [Google Scholar]
- 12.Girard-Gagnepain A, Amirache F, Costa C, et al. Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood. 2014;124(8):1221-1231. [DOI] [PubMed] [Google Scholar]
- 13.Lévy C, Amirache F, Girard-Gagnepain A, et al. Measles virus envelope pseudotyped lentiviral vectors transduce quiescent human HSCs at an efficiency without precedent. Blood Adv. 2017;1(23):2088-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozog S, Chen CX, Simpson E, et al. CD46 null packaging cell line improves measles lentiviral vector production and gene delivery to hematopoietic stem and progenitor cells. Mol Ther Methods Clin Dev. 2019;13:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang CX, Sather BD, Wang X, et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124(6):913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heffner GC, Bonner M, Christiansen L, et al. Prostaglandin E2 increases lentiviral vector transduction efficiency of adult human hematopoietic stem and progenitor cells. Mol Ther. 2018;26(1):320-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrillo C, Thorne LG, Unali G, et al. Cyclosporine H Overcomes innate immune restrictions to improve lentiviral transduction and gene editing in human hematopoietic stem cells. Cell Stem Cell. 2018;23(6):820-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajaste-Rudnitski A, Naldini L. Cellular innate immunity and restriction of viral infection: implications for lentiviral gene therapy in human hematopoietic cells. Hum Gene Ther. 2015;26(4):201-209. [DOI] [PubMed] [Google Scholar]
- 19.Amirache F, Lévy C, Costa C, et al. Mystery solved: VSV-G-LVs do not allow efficient gene transfer into unstimulated T cells, B cells, and HSCs because they lack the LDL receptor. Blood. 2014;123(9):1422-1424. [DOI] [PubMed] [Google Scholar]
- 20.Shen H, Cheng T, Preffer FI, et al. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J Virol. 1999;73(1):728-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117(2):473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villa NY, Bais S, Chan WM, et al. Ex vivo virotherapy with myxoma virus does not impair hematopoietic stem and progenitor cells. Cytotherapy. 2016;18(3):465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin DO, Goff SP. HIV-1 is restricted prior to integration of viral DNA in primary cord-derived human CD34+ cells. J Virol. 2015;89(15):8096-8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolb-Mäurer A, Wilhelm M, Weissinger F, Bröcker E-B, Goebel W. Interaction of human hematopoietic stem cells with bacterial pathogens. Blood. 2002;100(10):3703-3709. [DOI] [PubMed] [Google Scholar]
- 25.Pascutti MF, Erkelens MN, Nolte MA. Impact of viral infections on hematopoiesis: from beneficial to detrimental effects on bone marrow output. Front Immunol. 2016;7:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Dao Thi VL, Huang Y, et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172(3):423-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alber D, Staeheli P. Partial inhibition of vesicular stomatitis virus by the interferon-induced human 9-27 protein. J Interferon Cytokine Res. 1996;16(5):375-380. [DOI] [PubMed] [Google Scholar]
- 28.Hornick AL, Li N, Oakland M, McCray PB Jr, Sinn PL. Human, pig, and mouse interferon-induced transmembrane proteins partially restrict pseudotyped lentiviral vectors. Hum Gene Ther. 2016;27(5):354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roesch F, OhAinle M, Emerman M. A CRISPR screen for factors regulating SAMHD1 degradation identifies IFITMs as potent inhibitors of lentiviral particle delivery. Retrovirology. 2018;15(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi G, Schwartz O, Compton AA. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology. 2017;14(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res. 2011;31(1):183-197. [DOI] [PubMed] [Google Scholar]
- 32.Shi G, Ozog S, Torbett BE, Compton AA. mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc Natl Acad Sci USA. 2018;115(43):E10069-E10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis G, Christiansen L, McKenzie J, et al. Staurosporine increases lentiviral vector transduction efficiency of human hematopoietic stem and progenitor cells. Mol Ther Methods Clin Dev. 2018;9:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu D, Wang W, Yoder A, Spear M, Wu Y. The HIV envelope but not VSV glycoprotein is capable of mediating HIV latent infection of resting CD4 T cells. PLoS Pathog. 2009;5(10):e1000633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright NE, Snyder SA. 9-Membered carbocycle formation: development of distinct Friedel-Crafts cyclizations and application to a scalable total synthesis of (±)-caraphenol A. Angew Chem Int Ed Engl. 2014;53(13):3409-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swan CH, Bühler B, Steinberger P, Tschan MP, Barbas CF III, Torbett BE. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery [published correction appears in Gene Ther. 2007;14(7):626]. Gene Ther. 2006;13(20):1480-1492. [DOI] [PubMed] [Google Scholar]
- 38.Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4(4):495-505. [DOI] [PubMed] [Google Scholar]
- 39.Bobrowska-Hägerstrand M, Lillås M, Mrówczyñska L, et al. Resveratrol oligomers are potent MRP1 transport inhibitors. Anticancer Res. 2006;26(3A 3a):2081-2084. [PubMed] [Google Scholar]
- 40.Chung EY, Kim BH, Lee MK, et al. Anti-inflammatory effect of the oligomeric stilbene alpha-Viniferin and its mode of the action through inhibition of cyclooxygenase-2 and inducible nitric oxide synthase. Planta Med. 2003;69(8):710-714. [DOI] [PubMed] [Google Scholar]
- 41.Chung EY, Roh E, Kwak JA, et al. alpha-Viniferin suppresses the signal transducer and activation of transcription-1 (STAT-1)-inducible inflammatory genes in interferon-gamma-stimulated macrophages. J Pharmacol Sci. 2010;112(4):405-414. [DOI] [PubMed] [Google Scholar]
- 42.González-Sarrías A, Gromek S, Niesen D, Seeram NP, Henry GE. Resveratrol oligomers isolated from Carex species inhibit growth of human colon tumorigenic cells mediated by cell cycle arrest. J Agric Food Chem. 2011;59(16):8632-8638. [DOI] [PubMed] [Google Scholar]
- 43.Kim DH, Kim SH, Kim HJ, Jin C, Chung KC, Rhim H. Stilbene derivatives as human 5-HT(6) receptor antagonists from the root of Caragana sinica. Biol Pharm Bull. 2010;33(12):2024-2028. [DOI] [PubMed] [Google Scholar]
- 44.Yan T, Wang T, Wei W, et al. Polyphenolic acetylcholinesterase inhibitors from Hopea chinensis. Planta Med. 2012;78(10):1015-1019. [DOI] [PubMed] [Google Scholar]
- 45.Quideau S, Deffieux D, Douat-Casassus C, Pouységu L. Plant polyphenols: chemical properties, biological activities, and synthesis. Angew Chem Int Ed Engl. 2011;50(3):586-621. [DOI] [PubMed] [Google Scholar]
- 46.Keylor MH, Matsuura BS, Stephenson CRJ. Chemistry and biology of resveratrol-derived natural products. Chem Rev. 2015;115(17):8976-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder SA, Gollner A, Chiriac MI. Regioselective reactions for programmable resveratrol oligomer synthesis. Nature. 2011;474(7352):461-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keylor MH, Matsuura BS, Griesser M, et al. Synthesis of resveratrol tetramers via a stereoconvergent radical equilibrium. Science. 2016;354(6317):1260-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimmelé P, Lofek-Czubek S, Ghaffari S. Resveratrol increases the bone marrow hematopoietic stem and progenitor cell capacity. Am J Hematol. 2014;89(12):E235-E238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz N, Ehrnström B, Schambach A, Schwarzer A, Modlich U, Schiedlmeier B. Comparison of different cytokine conditions reveals resveratrol as a new molecule for ex vivo cultivation of cord blood-derived hematopoietic stem cells. Stem Cells Transl Med. 2015;4(9):1064-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pollok KE, van Der Loo JC, Cooper RJ, et al. Differential transduction efficiency of SCID-repopulating cells derived from umbilical cord blood and granulocyte colony-stimulating factor-mobilized peripheral blood. Hum Gene Ther. 2001;12(17):2095-2108. [DOI] [PubMed] [Google Scholar]
- 52.DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2(36):36ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467(7313):318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818-823. [DOI] [PubMed] [Google Scholar]
- 55.Petrillo C, Cesana D, Piras F, et al. Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol Ther. 2015;23(2):352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Torres-Coronado M, Gu A, et al. Enhanced genetic modification of adult growth factor mobilized peripheral blood hematopoietic stem and progenitor cells with rapamycin. Stem Cells Transl Med. 2014;3(10):1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Blanc K, Barrett AJ, Schaffer M, et al. Lymphocyte recovery is a major determinant of outcome after matched unrelated myeloablative transplantation for myelogenous malignancies. Biol Blood Marrow Transplant. 2009;15(9):1108-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Ravin SS, Wu X, Moir S, et al. Lentiviral hematopoietic stem cell gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med. 2016;8(335):335ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berman AY, Motechin RA, Wiesenfeld MY, Holz MK. The therapeutic potential of resveratrol: a review of clinical trials. NPJ Precis Oncol. 2017;1:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beard BC, Adair JE, Trobridge GD, Kiem HP. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol Biol. 2014;1185:321-344. [DOI] [PubMed] [Google Scholar]
- 61.Takeuchi K, Miyajima N, Nagata N, Takeda M, Tashiro M. Wild-type measles virus induces large syncytium formation in primary human small airway epithelial cells by a SLAM(CD150)-independent mechanism. Virus Res. 2003;94(1):11-16. [DOI] [PubMed] [Google Scholar]
- 62.Gonçalves-Carneiro D, McKeating JA, Bailey D. The Measles virus receptor SLAMF1 can mediate particle endocytosis. J Virol. 2017;91(7):e02255-e02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Blanc I, Luyet P-P, Pons V, et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat Cell Biol. 2005;7(7):653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20(11):1151-1154. [DOI] [PubMed] [Google Scholar]
- 65.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137(3):433-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol Biol. 2009;485:55-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang IC, Bailey CC, Weyer JL, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7(1):e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feeley EM, Sims JS, John SP, et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7(10):e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weidner JM, Jiang D, Pan X-B, Chang J, Block TM, Guo J-T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J Virol. 2010;84(24):12646-12657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerlach T, Hensen L, Matrosovich T, et al. pH optimum of hemagglutinin-mediated membrane fusion determines sensitivity of influenza A viruses to the interferon-induced antiviral state and IFITMs. J Virol. 2017;91(11):e00246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu Y, Zhou Z, Wang H, et al. IFITM1 suppresses expression of human endogenous retroviruses in human embryonic stem cells. FEBS Open Bio. 2017;7(8):1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grow EJ, Flynn RA, Chavez SL, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522(7555):221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jia R, Pan Q, Ding S, et al. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J Virol. 2012;86(24):13697-13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sutton RE, Reitsma MJ, Uchida N, Brown PO. Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vectors is cell cycle dependent. J Virol. 1999;73(5):3649-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park D, Jeong H, Lee MN, et al. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci Rep. 2016;6(1):21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5(2):e9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Puissant A, Robert G, Fenouille N, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70(3):1042-1052. [DOI] [PubMed] [Google Scholar]
- 78.Spence JS, He R, Hoffmann H-H, et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol. 2019;15(3):259-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wee YS, Roundy KM, Weis JJ, Weis JH. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 2012;18(6):834-845. [DOI] [PubMed] [Google Scholar]
- 80.Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425(24):4937-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Everitt AR, Clare S, Pertel T, et al. ; MOSAIC Investigators . IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allen EK, Randolph AG, Bhangale T, et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat Med. 2017;23(8):975-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang D, Wang H, He H, Niu H, Li Y. Interferon induced transmembrane protein 3 regulates the growth and invasion of human lung adenocarcinoma. Thorac Cancer. 2017;8(4):337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang M, Gao H, Chen P, Jia J, Wu S. Knockdown of interferon-induced transmembrane protein 3 expression suppresses breast cancer cell growth and colony formation and affects the cell cycle. Oncol Rep. 2013;30(1):171-178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.