Abstract
mir-100-let-7a-2-mir-125b-1 cluster host gene (MIR100HG), which is located on chromosome 11q24.1, is a polycistronic microRNA host gene. MIR100HG overexpression in colorectal cancer (CRC) has been demonstrated to be associated with cetuximab resistance; however, the role of MIR100HG in CRC metastasis remains unclear. The present study aimed to investigate the impact of aberrant MIR100HG expression on metastasis and prognosis in patients with CRC. The results from reverse transcription-quantitative PCR demonstrated that MIR100HG expression was higher in CRC tissues compared with in corresponding normal mucosa tissues. In particular, MIR100HG expression was higher in advanced CRC compared with in early stage CRC. Furthermore, the results from Kaplan-Meier analysis followed by a log-rank test revealed that patients with CRC and high MIR100HG expression exhibited poorer disease-free survival and overall survival compared with patients with CRC and lower MIR100HG expression. Furthermore, results from in vitro Transwell assays and in vivo animal assays demonstrated that upregulated MIR100HG expression promoted CRC cell migration and invasion and the formation of liver metastatic colonies in mice. In conclusion, the present study demonstrated that MIR100HG overexpression may contribute to the progression of CRC and may predict a poorer prognosis in patients with CRC. MIR100HG may therefore be considered as a novel therapeutic target and a prognostic biomarker in patients with CRC.
Keywords: colorectal cancer, long non-coding RNA, MIR100HG, metastasis
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed type of cancer worldwide, and represents a leading cause of mortality in men and women (1,2). Tumor invasion and distant metastasis are the main causes of cancer-associated mortality in patients with CRC (3). The liver is the most frequent site of metastatic spread in CRC, and 15–25% of patients with CRC present with liver metastases at the time of diagnosis (4). Although improvements in screening tests, chemotherapy, surgical techniques and multidisciplinary approaches have substantially decreased the morbidity and mortality rates of patients with CRC in the last decades (5,6), the prognosis of patients with advanced-stage CRC remains unsatisfactory due to frequent metastasis and CRC recurrence (7). Understanding the underlying mechanisms of CRC, particularly during colorectal liver metastasis, is therefore crucial to improve the clinical outcomes and overall survival of patients.
Long non-coding RNAs (lncRNAs) are defined as transcripts of >200 nucleotides in length that are not translated into proteins, and account for much of the transcribed genome (8). lncRNAs can promote or inhibit the expression of various protein-coding genes through interactions with other cellular macromolecules, including DNA, RNA and proteins (8). Numerous studies have reported that lncRNAs are crucial for the regulation of major physiological and pathological processes, including cell growth, apoptosis, stem-cell pluripotency, cancer invasion, metastasis, development, differentiation and the immune response, and serve a role in early diagnosis, targeted therapy and drug resistance of cancers (9–15). A previous study has demonstrated that the lncRNA mir-100-let-7a-2-mir-125b-1 cluster host gene (MIR100HG) is significantly associated with cetuximab resistance (15); however, other functional roles of MIR100HG in the development of cancer, particularly CRC, remain unknown and require further investigation.
In the present study, MIR100HG expression in CRC tissues and paired normal mucosae was assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). In addition, the association between MIR100HG expression and the clinicopathological characteristics of patients with CRC was assessed. Furthermore, Kaplan-Meier analysis with the log-rank test was used to evaluate the prognostic value of MIR100HG in patients with CRC. In addition, in vitro and in vivo cell function assays were performed to investigate the impact of MIR100HG on CRC cell migration and invasion and to determine the potential underlying mechanisms of CRC progression. The findings of the present study suggested that MIR100HG may be involved in CRC invasion and liver metastasis.
Materials and methods
Patients and specimens
A total of 116 paired CRC tissues and matched normal mucosae were collected from patients with CRC who underwent tissue biopsy at the Department of Gastroenterology, Shanghai General Hospital of Nanjing Medical University between September 2010 and March 2016. Patients comprised 60 men and 56 women (mean age, 64 years; age range, 32–89 years). All specimen were histopathologically confirmed as CRC. No patients had received local or other related anticancer therapies prior to tumor biopsy. The tumor grade and clinical stages were classified according to the guidelines of the American Joint Committee on Cancer (AJCC) (16). The present study was approved by the Ethics Committee of Shanghai General Hospital of Nanjing Medical University, and written informed consent was obtained from all patients prior to enrollment in the present study.
RNA extraction and RT-qPCR
Total RNA was isolated from fresh CRC and adjacent normal mucosae tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). First strand cDNA was synthesized from 2 µg RNA using a Prime-Script PCR reagent kit (Takara Bio, Inc.) according to the manufacturer's protocol. By using the LightCycler®480 system (Roche Applied Science), the SYBR Premix Ex-Taq II kit (Takara Bio, Inc.) was applied to perform qPCR. The cycling conditions were as follows: Initial denaturation (2 min at 95°C) followed by 40 cycles of denaturation (10 sec at 95°C), annealing (30 sec at 59°C), elongation (30 sec at 72°C) and a final extension (30 sec at 72°C). The amplified samples were then maintained at 4°C. The primer sequences used for MIR100HG detection were as follows: MIR100HG forward, 5′-CCCAGTGCAAGGACAAAGA-3′ and reverse, 5′-GCAGAGGAGGTGTCTTCAGG-3′; and GAPDH forward, 5′-GGGAAATTCAACGGCACAGT-3′ and reverse, 5′-AGATGGTGATGGGCTTCCC-3′. The relative expression levels of MIR100HG were normalized to the endogenous control GAPDH and expressed as 2−ΔΔCq (17).
Cell culture and transfection
The human CRC LoVo, DLD-1, RKO, SW48, HCT8 and HCT116 cell lines, and the normal intestinal mucous epithelium FHC cell line were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences. All cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin and placed at 37°C in a humidified incubator containing 5% CO2.
The CRC cell line LOVo overexpressing MIR100HG was established by transfection with lentiviral vectors encoding human MIR100HG (PCDH-MIR100HG). In addition, MIR100HG-knockdown CRC cell line HCT116 were constructed by transfection with MIR100HG short hairpin RNA (shRNA). The sequences used for overexpression and knockdown were previously described (18).
In vitro Transwell assay
By using the two transfected CRC cell lines LoVo and HCT116 and their control groups, transwell assays were used to assess tumor cell migration and invasion. A Transwell 24-well Boyden chamber with a polycarbonate membrane (pore size, 8.0 µm; Corning Inc.) was used for cell migration (without Matrigel® coating; BD Biosciences) and invasion (with Matrigel® coating, prepared on ice) assays, according to the manufacturer's protocol. Briefly, 5×105 cells/ml were seeded in 200 µl serum-free DMEM medium in the upper chamber, whereas DMEM medium supplemented with 500 µl 10% FBS was added to the lower chamber. Following incubation for 24 h, cells were fixed with 4% polyoxymethylene and stained with 0.1% crystal violet at room temperature. Cells that had moved to the lower side of the membrane were counted in 10 visual fields and the average value was calculated. Cells images were captured using a light microscope (Olympus Corporation; magnification, ×200). Each experiment was performed independently three times.
In vivo metastatic assay
Nude mice were purchased from Shanghai Research Center for Model Organisms, Inc. Mice had an average weight of 20 g. Mice were placed in plastic cages with airtight air filter at the temperature of 18–22°C, humidity of 40–60%, under a light/dark cycle of 10/14 h each day, and had free access to food and water. For the in vivo cell metastatic assay, following anesthesia with ether inhalation, MIR100HG-overexpressing CRC cells LoVo, sh-MIR100HG knockdown CRC cells HCT116 and their control groups were injected (200 µl at the density of 1×106/ml in PBS) into the tail vein of nude mice (4 weeks old male BALB/C nude mice; n=3 for each group). After 4 weeks, mice were euthanized by cervical dislocation, and livers were collected and immediately fixed with 4% formaldehyde at room temperature for 24–48 h. After paraffin embedding, liver tissue was cut into 4–7 µm-thick sections. After hematoxylin-eosin staining at room temperature for 3–5 min, the tumor colonies formed in the livers were detected by light microscopy and quantified using GraphPad Prim 7 (GraphPad Software, Inc.). All experimental procedures and animal studies involving mice were in accordance with the Shanghai General Hospital of Nanjing Medical University Animal Care and Use guidelines. Ethical approval for the animal study was obtained from the Ethics Committee of Shanghai General Hospital of Nanjing Medical University.
Statistical analysis
Each experiment was performed independently three times. Data were expressed as the means ± standard deviation and were analyzed using SPSS v22.0 software (IBM Corp.). Student's t-test or one-way analysis of variance followed by Student-Newman-Keuls post hoc test were used to analyze the differences in MIR100HG expression among the continuous variables. A χ2 test or Fisher's exact test was appropriately used to determine the statistical significance between MIR100HG expression and the patients' clinicopathological characteristics. Kaplan-Meier analysis and the log-rank test were used for the comparison of patients' survival curves. The hazard ratio with 95% confidence interval in the Cox proportional hazards model was calculated to measure the hazard risk of individual factors for disease-free survival (DFS) and overall survival (OS). All graphs were plotted using GraphPad Prism v5.0 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
High MIR100HG expression is associated with an aggressive phenotype in patients with CRC
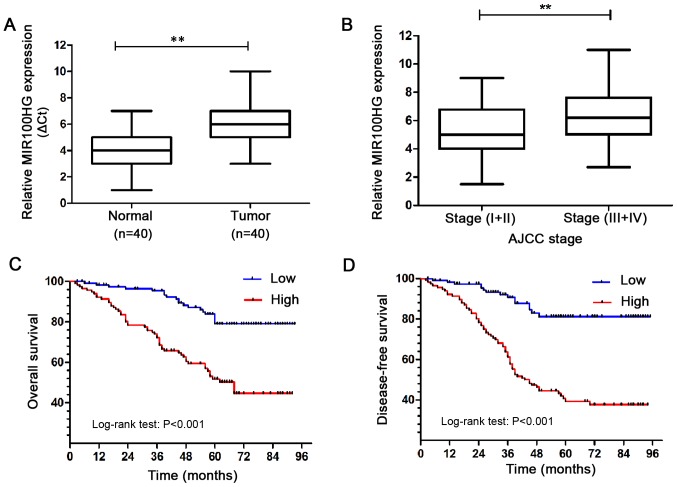
A total of 40 paired CRC and normal specimen were randomly selected from the 116 patients to detect MIR100HG expression by RT-qPCR. The results demonstrated that MIR100HG expression was significantly increased in CRC tissues compared with adjacent normal colon mucosae (Fig. 1A). Furthermore, MIR100HG expression was examined in CRC tissues at different stages. The results revealed that MIR100HG expression was significantly higher in advanced-stage (III+IV) CRC tissues compared with low stage (I+II) CRC tissues (Fig. 1B). These results demonstrated that MIR100HG was significantly elevated in CRC and may be associated with an aggressive phenotype in patients with CRC.
Figure 1.
MIR100HG expression was significantly upregulated in CRC tissues and could predict a poor prognosis. (A) Expression levels of MIR100H in 40 paired CRC and adjacent noncancerous tissues. (B) MIR100HG expression level in low clinical stage (I–II) CRC tissues and advanced stage (III–IV) CRC tissues was analyzed. Kaplan-Meier analysis and log-rank test of the (C) overall survival and (D) disease-free survival in patients with CRC according to MIR100HG expression level. AJCC, American Joint Committee on Cancer; CRC, colorectal cancer; MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene. **P<0.01 vs. normal tissue.
Subsequently, the association between MIR100HG expression and TNM stage in the 116 patients with CRC was investigated (19). MIR100HG expression was first evaluated in the 116 paired samples by RT-qPCR. A mean value of 5.55 was calculated. CRC samples with an expression level of MIR100HG ≥5.55 and <5.55 were classified in the high and low expression groups, respectively. Briefly, 75.0% (87/116) of specimens exhibited low MIR100HG expression in normal colorectal mucosa tissues. Furthermore, 25.0% (29/116) of normal colorectal mucosa tissues presented with high MIR100HG expression, whereas 68.1% (79/116) of CRC samples presented with high MIR100HG expression (Table I). In addition, high MIR100HG expression was positively associated with T stage, lymph node metastasis, distant metastasis, AJCC stage and histological differentiation in CRC samples (Table II). These results further indicated that MIR100HG overexpression may serve a role in CRC metastasis and in the prognosis of patients with CRC.
Table I.
MIR100HG expression in paired normal colorectal mucosa and tumor tissues.
| Relative MIR100HG expression | ||||
|---|---|---|---|---|
| Tissue samples | Number | Low, n (%) | High, n (%) | P-value |
| Normal tissues | 116 | 87 (75.0) | 29 (25.0) | <0.001 |
| Tumor tissue | 116 | 37 (31.9) | 79 (68.1) | |
MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene.
Table II.
Association between MIR100HG expression and clinicopathological characteristics of patients with colorectal cancer.
| Relative MIR100HG expression | ||||
|---|---|---|---|---|
| Variables | Number (%) | Low, n (n=37, %) | High, n (n=79, %) | P-value |
| Age, years | 0.596 | |||
| <65 | 54 (46.2) | 20 (54.1) | 34 (43.0) | |
| ≥65 | 62 (53.8) | 17 (45.9) | 45 (57.0) | |
| Sex | 0.629 | |||
| Male | 60 (51.7) | 16 (43.2) | 44 (55.7) | |
| Female | 56 (48.3) | 21 (56.8) | 35 (44.3) | |
| Tumor size, cm | 0.067 | |||
| <3 | 62 (53.4) | 25 (67.6) | 37 (46.8) | |
| ≥3 | 54 (46.6) | 12 (32.4) | 42 (53.2) | |
| Location | 0.416 | |||
| Right | 29 (25.0) | 6 (16.2) | 23 (29.1) | |
| Transverse | 27 (23.3) | 17 (45.9) | 10 (12.7) | |
| Left | 60 (51.7) | 14 (37.9) | 46 (58.2) | |
| AJCC stage | 0.024a | |||
| I–II | 45 (38.8) | 24 (64.9) | 21 (26.6) | |
| III–IV | 71 (61.2) | 13 (35.1) | 58 (73.4) | |
| pT stage | 0.048a | |||
| T1 | 17 (14.6) | 10 (27.0) | 7 (8.9) | |
| T2 | 46 (39.7) | 12 (32.4) | 34 (43.0) | |
| T3 | 37 (31.9) | 11 (29.8) | 26 (32.9) | |
| T4 | 16 (13.8) | 4 (10.8) | 12 (15.2) | |
| pN stage | 0.020a | |||
| N0 | 45 (38.8) | 24 (64.9) | 21 (26.6) | |
| N1-2 | 71 (61.2) | 13 (35.1) | 58 (73.4) | |
| pM stage | 0.023a | |||
| M0 | 101 (87.1) | 37 (100) | 64 (81.0) | |
| M1 | 15 (12.9) | 0 (0.0) | 15 (19.0) | |
| Differentiation | 0.035a | |||
| Well | 31 (26.7) | 19 (51.4) | 12 (15.2) | |
| Moderate | 40 (34.5) | 10 (27.0) | 30 (38.0) | |
| Poor | 45 (38.8) | 8 (21.6) | 37 (46.8) | |
| Vascular invasion | 0.862 | |||
| Yes | 3 (2.6) | 0 (0.0) | 3 (3.8) | |
| No | 113 (97.4) | 37 (100) | 76 (96.2) | |
P<0.05. AJCC, American Joint Committee on Cancer; MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene.
MIR100HG upregulation predicts an unfavorable prognosis and poor survival for patients with CRC
The prognostic value of MIR100HG expression in CRC was evaluated by assessing the DFS and OS of the 116 patients with CRC using Kaplan-Meier analysis and log-rank test. The results demonstrated that the DFS and OS of patients with CRC and high MIR100HG expression were shorter compared with those of patients with CRC and low MIR100HG expression (Fig. 1C and D). These results suggested that high MIR100HG expression may predict a poor prognosis and survival. Furthermore, the results from univariate and multivariate Cox regression analyses demonstrated that MIR100HG expression, AJCC stage, T classification, N classification, M classification and tumor differentiation may be considered as independent prognostic factors for DFS and OS (Tables III and IV). These findings suggested that high MIR100HG expression in CRC tissues may be considered as a prognostic factor for patients with CRC.
Table III.
Univariate and multivariate analyses of disease-free survival in patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| MIR100HG | 3.658 | 1.956–6.893 | 0.001a | 4.105 | 1.809–6.842 | 0.001a |
| Age | 1.165 | 0.758–2.345 | 0.456 | – | – | – |
| Sex | 1.208 | 0.765–1.998 | 0.547 | – | – | – |
| Tumor size | 1.077 | 0.657–1.954 | 0.658 | – | – | – |
| Location | 0.895 | 0.384–1.848 | 0.506 | – | – | – |
| AJCC Stage | 4.924 | 1.264–5.831 | 0.002a | 5.264 | 1.249–6.048 | 0.001a |
| T classification | 1.216 | 1.851–2.536 | 0.035a | 1.768 | 1.959–2.833 | 0.027a |
| N classification | 1.659 | 1.181–2.611 | 0.032a | 1.937 | 1.349–3.117 | 0.022a |
| M classification | 3.786 | 1.854–4.594 | 0.003a | 3.853 | 2.432–4.897 | 0.001a |
| Differentiation | 1.821 | 1.125–2.543 | 0.021a | 2.018 | 1.275–3.958 | 0.023a |
| Vascular invasion | 1.354 | 0.872–1.685 | 0.183 | – | – | – |
P<0.05 indicates that the 95% CI of the HR did not include 1. AJCC, American Joint Committee on Cancer; HR, hazard ratio; MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene.
Table IV.
Univariate and multivariate analyses of overall survival in patients with colorectal cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| MIR100HG | 3.241 | 2.843–7.231 | <0.001 | 3.533 | 1.877–5.958 | <0.001 |
| Age | 1.072 | 0.694–2.017 | 0.442 | – | – | – |
| Sex | 1.128 | 0.911–1.333 | 0.526 | – | – | – |
| Tumor size | 0.987 | 0.733–2.142 | 0.083 | – | – | – |
| Location | 1.035 | 0.767–1.842 | 0.723 | – | – | – |
| AJCC Stage | 3.951 | 2.523–5.015 | <0.001 | 4.017 | 2.349–3.483 | <0.001 |
| T classification | 1.212 | 1.017–2.335 | 0.047a | 2.012 | 1.954–2.874 | 0.031a |
| N classification | 1.537 | 1.061–4.938 | 0.043a | 1.977 | 1.575–3.213 | 0.044a |
| M classification | 3.596 | 2.427–4.938 | <0.001 | 3.672 | 2.731–5.376 | <0.001 |
| Differentiation | 2.017 | 1.029–3.240 | 0.033a | 1.996 | 1.233–2.841 | 0.042a |
| Vascular invasion | 1.282 | 1.134–2.047 | 0.097 | – | – | – |
P<0.05 indicates that the 95% CI of the HR did not include 1. AJCC, American Joint Committee on Cancer; HR, hazard ratio; MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene.
Association between MIR100HG expression and clinicopathological characteristics
To further confirm the clinical significance of MIR100HG expression in CRC, the association between MIR100HG expression and the clinicopathological characteristics of patients with CRC was assessed (Table II). The results demonstrated that high MIR100HG expression was significantly associated with AJCC stage (P=0.024), pathological T stage (P=0.048), N stage (P=0.020), M stage (P=0.023) and tumor differentiation (P=0.035). However, no association was identified between high MIR100HG expression and other clinical parameters, including age, sex, tumor size, tumor location and vascular invasion status (P>0.05; Table II). These findings suggested that high MIR100HG expression may be associated with CRC migration and invasion.
Effect of MIR100HG on CRC cell migration and invasion in vitro and in vivo
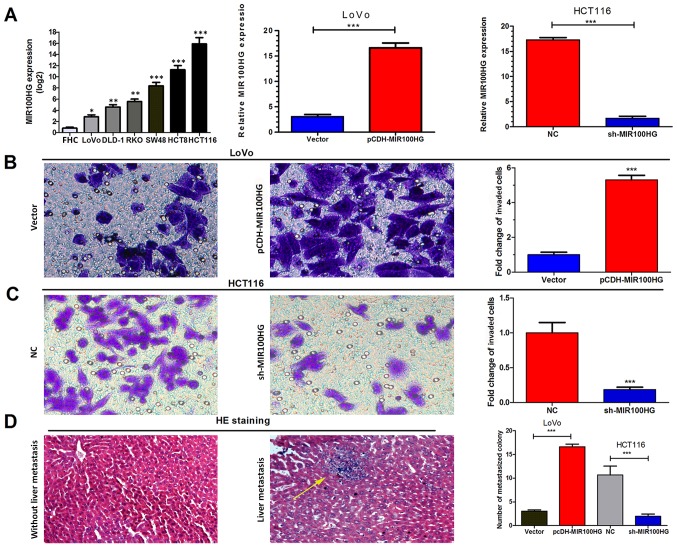
To further investigate the role of MIR100HG in CRC cells, MIR100HG expression in various CRC cell lines (LoVo, DLD-1, RKO, SW48, HCT8 and HCT116) and the normal intestinal mucous epithelium FHC cell line was assessed by RT-qPCR. The results demonstrated that CRC cell lines exhibited higher MIR100HG expression compared with the FHC cell line (Fig. 2A). Subsequently, the HCT116 cell line, which exhibited the highest expression levels of MIR100HG, was selected to establish a cell line with knockdown of MIR100HG. Conversely, the LoVo cell line, which displayed the lowest MIR100HG expression level, was selected to establish an overexpression model for MIR100HG. The results of RT-qPCR analysis demonstrated that the transfections were successful (Fig. 2A).
Figure 2.
MIR100HG facilitated CRC cell migration and invasion in vitro and liver metastasis in vivo. (A) Relative expression levels of MIR100HG in six colorectal cell lines and the normal intestinal mucous epithelium cell line FHC. Transfection efficiency in LoVo and HCT116 cell lines. *P<0.05, **P<0.01 and ***P<0.001 vs. FHC. (B and C) Impact of MIR100HG overexpression or knockdown on CRC cell migration and invasion. Magnification, ×200. (D) Impact of MIR100HG overexpression and knockdown on liver metastasis in nude mice in vivo. Left panel: Without liver metastasis. Right panel: With liver metastasis. HE staining of metastatic colonies formed in the liver. Magnification, ×100. The yellow arrow indicates metastatic colonies. NC, negative control; sh, short hairpin RNA; MIR100HG, mir-100-let-7a-2-mir-125b-1 cluster host gene; HE, hematoxylin and eosin. ***P<0.001 vs. control.
In vitro Transwell assays were used to assess the function of MIR100HG in CRC progression. MIR100HG overexpression significantly enhanced the migration and invasion of LoVo cells (P<0.001; Fig. 2B), whereas MIR100HG-knockdown significantly inhibited the migration and invasion of HCT116 cells (P<0.001; Fig. 2C). These results suggested that MIR100HG overexpression may promote the aggressive phenotype of CRC cells in vitro.
Subsequently, by injecting CRC cells LoVo and HCT116 into tail veins of nude mice, the impact of MIR100HG on the migratory and invasive capacities of CRC cells was investigated. After 4 weeks, the results demonstrated that higher and lower numbers of liver metastatic colonies were formed in the MIR100HG-overexpression and knockdown groups, respectively, compared with their corresponding controls (P<0.001; Fig. 2D). MIR100HG overexpression may therefore enhance the migratory and invasive capacities of CRC cells in vitro and in vivo.
Discussion
Recent studies have reported that distant metastasis remains the main cause of mortality in patients with CRC, and that the liver is the most common metastatic site (4,6). In total, 15–25% of patients with CRC have liver metastases at the time of diagnosis (20). In addition, 30–50% of patients with CRC who undergo radical resection will have local and systemic recurrences, and their risk of recurrence following metastatic resection is ~75% (4–7). It is therefore crucial to understand the mechanism of CRC and metastasis development. The present study demonstrated that MIR100HG expression was higher in CRC tissues compared with adjacent normal mucosa, and that MIR100HG overexpression was associated with poor prognosis and survival in patients with CRC. In addition, MIR100HG overexpression enhanced the migratory and invasive capacities of CRC cells in vitro and liver metastasis in vivo, which indicated that MIR100HG expression may be crucial in CRC progression, particularly in colorectal liver metastasis.
MIR100HG, which is located on chromosome 11q24.1, is a polycistronic micro RNA host gene that is associated with the progression of several types of tumor (21–23). It has been reported that MIR100HG overexpression can predict a poor prognosis and is associated with metastasis in cervical cancer (21). In addition, MIR100HG overexpression is associated with OS in patients with oral cavity cancer (22). MIR100HG is also overexpressed in acute megakaryoblastic leukemia (23), and aberrant MIR100HG expression has been reported in patients with CRC and resistant to cetuximab (24). However, the clinical significance and prognostic value of MIR100HG expression in CRC remains unclear. The present study elucidated MIR100HG expression in CRC samples, examined its association with clinicopathological characteristics and distant metastasis in patients with CRC, and determined whether it could be considered as a prognosis factor.
The results of RT-qPCR demonstrated that MIR100HG expression was increased in CRC tissues compared with normal tissues. Furthermore, MIR100HG expression in advanced stage (III–IV) CRC tissues was significantly increased compared with low stage (I–II) CRC tissues, which was consistent with a previous study from The Cancer Genome Atlas data repository (24). Subsequently, the association between high MIR100HG expression and the clinicopathological characteristics of patients with CRC was investigated. Previous studies on acute megakaryoblastic leukemia, early-stage cervical cancer and head and neck squamous cell carcinoma have reported that MIR100HG overexpression accelerates tumor malignancy and is associated with poor prognosis and survival (21–23). Similarly, the present study reported that MIR100HG overexpression was associated with poor prognosis and survival in patients with CRC. Furthermore, univariate and multivariate Cox regression analyses demonstrated that MIR100HG expression, AJCC stage, N classification, M classification and tumor differentiation were significantly associated with DFS and OS. Subsequently, in vitro cell function assays investigated the impact of MIR100HG on the migratory and invasive capacities of CRC cells. The results demonstrated that MIR100HG upregulation enhanced CRC cell migration and invasion in vitro, and liver metastasis in vivo. These findings suggested that MIR100HG may have crucial roles in CRC invasion and liver metastasis.
In conclusion, the present study reported the crucial roles of MIR100HG in CRC progression, and indicated that MIR100HG may serve as a novel prognostic biomarker for CRC. Furthermore, future manipulation of MIR100HG levels may represent a potential treatment strategy for patients with CRC, particularly those with colorectal liver metastasis. It has been reported that MIR100HG is the host gene of the microRNA (miR)-100/let-7a-2/miR-125b-1 cluster on chromosome 11, and that MIR100HG overexpression can upregulate miR-100 and miR-125b (24). The present study did not examine the impact of MIR100HG on miR-100 and miR-125b-1 expression levels. Future studies will therefore examine the effect of modulating MIR100HG expression on miR-100 and miR-125b expression levels, and will investigate how MIR100HG could promote CRC development and distant metastasis.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Zhejiang Medical Technology Plan Project (grant no. 2017ZD003), the Natural Science Foundation of Jiangsu Province, China (grant no. BK20161168) and the Social Development Project of Xuzhou, China (grant no. KC17109).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LL and XT conceived the study and wrote the manuscript. WL and FY performed the experiments. XZ and WC analyzed and interpreted the data.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Shanghai General Hospital of Nanjing Medical University. All patients provided written informed consent prior to the present study. Animal studies involving mice were in accordance with the Shanghai General Hospital of Nanjing Medical University Animal Care and Use guidelines. Ethical approval was obtained from the Ethics Committee of Shanghai General Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Bhullar DS, Barriuso J, Mullamitha S, Saunders MP, O'Dwyer ST, Aziz O. Biomarker concordance between primary colorectal cancer and its metastases. EBioMedicine. 2019;40:363–374. doi: 10.1016/j.ebiom.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–65. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et al. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer. 2018;124:2785–800. doi: 10.1002/cncr.31551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 7.van der Valk MJM, Hilling DE, Bastiaannet E, Meershoek-Klein Kranenbarg E, Beets GL, Figueiredo NL, Habr-Gama A, Perez RO, Renehan AG, van de Velde CJH, IWWD Consortium. Long-term outcomes of clinical complete responders after neoadjuvant treatment for rectal cancer in the International Watch & Wait Database (IWWD): An international multicentre registry study. Lancet. 2018;391:2537–2545. doi: 10.1016/S0140-6736(18)31078-X. [DOI] [PubMed] [Google Scholar]
- 8.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leisegang MS, Fork C, Josipovic I, Richter FM, Preussner J, Hu J, Miller MJ, Epah J, Hofmann P, Gunther S, et al. Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation. 2017;136:65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, Plath K. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin-Bejar O, Mas AM, Gonzalez J, Martinez D, Athie A, Morales X, Galduroz M, Raimondi I, Grossi E, Guo S, et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017;18:202. doi: 10.1186/s13059-017-1331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nature Cell Biol. 2017;19:1105–15. doi: 10.1038/ncb3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, Wang W, Liu S, Liu W, Ma Y, et al. Self-Recognition of an inducible host LncRNA by RIG-I feedback restricts innate immune response. Cell. 2018;173:906–19.e13. doi: 10.1016/j.cell.2018.03.064. [DOI] [PubMed] [Google Scholar]
- 14.Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–98. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Lv G, Wang B, Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503:2555–2562. doi: 10.1016/j.bbrc.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 16.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L, Yang Q, Zhu H, Nie J, Wu C, Jiao B. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Dis. 2018;9:805. doi: 10.1038/s41419-018-0869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pages F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 20.van der Geest LG, Lam-Boer J, Koopman M, Verhoef C, Elferink MA, de Wilt JH. Nationwide trends in incidence, treatment and survival of colorectal cancer patients with synchronous metastases. Clin Exp Metastasis. 2015;32:457–465. doi: 10.1007/s10585-015-9719-0. [DOI] [PubMed] [Google Scholar]
- 21.Shang C, Zhu W, Liu T, Wang W, Huang G, Huang J, Zhao P, Zhao Y, Yao S. Characterization of long non-coding RNA expression profiles in lymph node metastasis of early-stage cervical cancer. Oncol Rep. 2016;35:3185–3197. doi: 10.3892/or.2016.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins OM, Titus AJ, Salas LA, Gui J, Eliot M, Butler RA, Sturgis EM, Li G, Kelsey KT, Christensen BC. MicroRNA-related genetic variants associated with survival of head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2019;28:127–136. doi: 10.1158/1055-9965.EPI-18-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13:171. doi: 10.1186/1476-4598-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.