Abstract
Rationale: Disruption of alveologenesis is associated with severe pediatric lung disorders, including bronchopulmonary dysplasia (BPD). Although c-KIT+ endothelial cell (EC) progenitors are abundant in embryonic and neonatal lungs, their role in alveolar septation and the therapeutic potential of these cells remain unknown.
Objectives: To determine whether c-KIT+ EC progenitors stimulate alveologenesis in the neonatal lung.
Methods: We used single-cell RNA sequencing of neonatal human and mouse lung tissues, immunostaining, and FACS analysis to identify transcriptional and signaling networks shared by human and mouse pulmonary c-KIT+ EC progenitors. A mouse model of perinatal hyperoxia-induced lung injury was used to identify molecular mechanisms that are critical for the survival, proliferation, and engraftment of c-KIT+ EC progenitors in the neonatal lung.
Measurements and Main Results: Pulmonary c-KIT+ EC progenitors expressing PECAM-1, CD34, VE-Cadherin, FLK1, and TIE2 lacked mature arterial, venal, and lymphatic cell-surface markers. The transcriptomic signature of c-KIT+ ECs was conserved in mouse and human lungs and enriched in FOXF1-regulated transcriptional targets. Expression of FOXF1 and c-KIT was decreased in the lungs of infants with BPD. In the mouse, neonatal hyperoxia decreased the number of c-KIT+ EC progenitors. Haploinsufficiency or endothelial-specific deletion of Foxf1 in mice increased apoptosis and decreased proliferation of c-KIT+ ECs. Inactivation of either Foxf1 or c-Kit caused alveolar simplification. Adoptive transfer of c-KIT+ ECs into the neonatal circulation increased lung angiogenesis and prevented alveolar simplification in neonatal mice exposed to hyperoxia.
Conclusions: Cell therapy involving c-KIT+ EC progenitors can be beneficial for the treatment of BPD.
Keywords: bronchopulmonary dysplasia, FOXF1, C-KIT, endothelial progenitor cells
At a Glance Commentary
Scientific Knowledge on the Subject
Inhibition of neonatal lung angiogenesis is an important pathophysiological mechanism through which hyperoxia causes alveolar simplification in bronchopulmonary dysplasia (BPD). c-KIT is expressed in endothelial progenitor cells, which are abundant in embryonic and neonatal lungs. Administration of stem cell factor, a ligand of c-KIT, improves angiogenesis and alveolar septation after hyperoxia in newborn rats. Although stem cell factor/c-KIT signaling stimulates alveologenesis, the transcriptional networks that are critical for the survival and proliferation of pulmonary endothelial progenitor cells remain unknown.
What This Study Adds to the Field
Using single-cell RNA sequencing of human and mouse neonatal lung tissue, we found that FOXF1 and its transcriptional target genes, including c-KIT, were increased in pulmonary endothelial progenitor cells. FOXF1 and c-KIT were decreased in lungs of infants with BPD. In mice, pulmonary endothelial progenitor cells were highly sensitive to injury by high oxygen concentrations and dependent on FOXF1 and c-KIT to maintain cell survival and proliferation. Genetic deletion of either Foxf1 or c-Kit inhibited neonatal angiogenesis and disrupted alveolar septation. Transplantation of pulmonary endothelial progenitor cells into the neonatal circulation improved lung angiogenesis and protected newborn mice from hyperoxia-induced alveolar simplification, supporting efforts to prevent BPD by restoring endothelial progenitor cells.
Lung morphogenesis is dependent on the formation of an extensive capillary network mediated by reciprocal signaling among endothelial, epithelial, and mesenchymal cells that regulates alveolar formation (1–3). Before and during the saccular stage of lung development, alveolar capillaries are embedded in a thick mesenchyme, limiting interactions with alveolar epithelial cells. During sacculation and septation, the mesenchyme thins, the alveolar surface area increases, and capillary networks are formed in close apposition to the alveolar walls. During angiogenesis, basement membranes form between endothelial and type I alveolar epithelial cells to support efficient gas exchange (4, 5). Prematurity, oxygen exposure, and mechanical ventilation contribute to the pathogenesis of bronchopulmonary dysplasia (BPD), disrupting neonatal lung angiogenesis and alveologenesis, and causing alveolar simplification. Multiple cellular and molecular mechanissms, including apoptosis of resident lung cells, diminished cellular proliferation, infection, and inflammation, contribute to reduced angiogenesis, abnormal epithelial differentiation, and altered surfactant homeostasis in patients with BPD (6–11).
Inhibition of neonatal lung angiogenesis is an important pathophysiological mechanism through which hyperoxia causes alveolar simplification (12–14). Endothelial cells (ECs) produce a diversity of signaling molecules, including PDGFs (platelet-derived growth factors), fibroblast growth factors, hepatocyte growth factor, WNTs (proteins from the int/Wingless family, members of Wnt [Wingless-related integration site]-signaling pathways), and matrix metalloproteinases, that regulate tissue remodeling and deposition of elastin and collagens into the alveolar wall by myofibroblasts (3–5). VEGF-A (vascular endothelial growth factor A) which is secreted primarily by epithelial cells, signals through VEGFR2 (VEGF receptor 2) tyrosine kinase (KDR or FLK1) to stimulate endothelial proliferation, survival, and migration, accelerating neonatal lung angiogenesis (9, 11). Recombinant VEGF or intratracheal adenovirus-mediated VEGF gene transfer was shown to restore angiogenesis, increase alveolar septation, and improve survival in a rat BPD model (12, 13). VEGF signaling during angiogenesis is dependent on multiple angiogenic transcription factors from the FOX (forkhead box transcription factor), HOX (homeobox transcription factor), KLF (Krüppel-like transcription factor), SOX (Sry-related HMG box transcription factor), GATA (GATA-binding transcription factor), and ETS (E26 transformation-specific transcription factor) families (2, 3, 15), and FOXF1 (Forkhead Box F1) is a key regulator of pulmonary angiogenesis (1, 3, 16). Heterozygous mutations in the FOXF1 gene are associated with alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV) (17, 18), a severe congenital disorder associated with the loss of alveolar capillaries, malposition of pulmonary veins, and lung hypoplasia, causing respiratory insufficiency after birth (19). Patients with ACDMPV and delayed clinical presentations have alveolar simplification and pulmonary vasculature abnormalities (19). Foxf1−/− mice die in utero due to vascular defects in the yolk sac and allantois (20). Foxf1 heterozygous mice (Foxf1+/−) have diminished numbers of alveolar capillaries and are susceptible to lung injury by toxic and inflammatory insults (21–23). FOXF1 stimulates endothelial proliferation during embryogenesis and repair after partial pneumonectomy, regulating the expression of genes that are critical for the VEGF, PDGF, NOTCH, and Angiopoietin/TIE2 signaling pathways (24–26).
Stem cell factor (SCF) is produced by multiple cell types and signals through c-KIT tyrosine kinase to increase endothelial proliferation and cell survival (27, 28). Administration of SCF increases neonatal angiogenesis and improves alveolar septation after hyperoxia in newborn rats (29). c-KIT is expressed in EC progenitors (c-KIT+/PECAM-1+/CD45− or c-KIT+ ECs), which are capable of self-renewal and differentiation into mature ECs in many organs (27, 28). Although c-KIT+ ECs are present in embryonic and neonatal lungs (30), and SCF/c-KIT signaling stimulates alveologenesis in rodent BPD models (29), the molecular mechanisms that regulate SCF/c-KIT signaling in EC progenitors remain unknown. The potential of c-KIT+ ECs for treating neonatal pulmonary disorders is unclear.
In the present study, we used single-cell RNA sequencing (RNA-seq) of human and mouse neonatal lung tissue to identify an evolutionarily conserved gene expression signature in pulmonary c-KIT+ EC progenitors, and demonstrate that c-KIT+ ECs require FOXF1 and c-KIT to stimulate postnatal angiogenesis and alveologenesis.
Methods
Transgenic Mice and Neonatal Hyperoxia
The Foxf1+/− mouse line was described previously (31). Kitw-sh mice were purchased from The Jackson Laboratory (C57Bl/6 background). Foxf1fl/fl mice (24) were bred with Pdgfb-CreER mice (32) to generate Pdgfb-CreER Foxf1fl/fl offspring. In Pdgfb-CreER Foxf1fl/fl mice, tamoxifen causes Foxf1 deletion in ECs but not in other cell types (24, 26). Tamoxifen (3 mg; Sigma) was given intraperitoneally at Postnatal Day 0.5 (P0.5) and P2.5. Neonatal hyperoxia protocols and measurements of lung functions were performed as described previously (33, 34) and in the online supplement. Animal studies were approved by the Animal Care and Use Committee of the Cincinnati Children’s Research Foundation.
Flow Cytometry and Adoptive Transfer of EC Progenitors
FACS was performed after enzyme digestion of lung tissue as described previously (35, 36). Apoptotic ECs were detected with the use of an APC Annexin V apoptosis detection kit (eBioscience). 7-AAD (eBioscience) was used for labeling of necrotic cells. Bromodeoxyuridine incorporation was measured as described previously (24). The antibodies used for FACS are listed in the online supplement. The protocol used for intracellular labeling with cell fixation and permeabilization was described previously (34). Stained cells were separated using a five-laser FACSAria II (BD Biosciences). For adoptive transfer of EC progenitors, c-KIT+ ECs (c-KIT+ CD31+ CD45−) and c-KIT− ECs (c-KIT– CD31+ CD45−) were FACS-sorted from lung tissue of donor P3 mice expressing the tdTomato transgene (The Jackson Laboratory) and injected into the facial vein of P3 recipient mice.
Single-Cell RNA-seq Analysis
RNA-seq analyses (using the Drop-seq platform) of human P1 and mouse P7 lungs were performed at Cincinnati Children’s Hospital Medical Center. The raw data are available at LungMAP (https://www.lungmap.net/breath-entity-page/?entityType=noneandentityId=andentityLabel=andexperimentTypes[]=LMXT0000000016) and interpreted data can be searched at LungGENS (https://research.cchmc.org/pbge/lunggens/celltype_dp.html?tps=pnd1andspe=MO). Analyses, data filtering, and normalization criteria are described in the online supplement. Seurat (37) was used to detect highly variable genes and perform principle component analysis–based dimension reduction. Reduced dimensions were used for cell cluster identification using the Jaccard-Louvain clustering algorithm (38). A binomial test–based method (38) was used to identify differentially expressed genes. Human donor lung tissue and slides from BPD lungs were provided by the LungMAP Human Tissue Core at the University of Rochester. Tissue for LungMAP was obtained via the nonprofit United Network for Organ Sharing, International Institute for Advancement of Medicine and National Disease Research Interchange. Consent for use of tissue was overseen by the University of Rochester Research Subjects Review Board (RSR B00056775) and the Cincinnati Children’s Hospital Medical Center Institutional Review Board (#20180852).
Real-Time qRT-PCR
qRT-PCR was performed using a StepOnePlus Real-Time PCR system (Applied Biosystems) with TaqMan primers (see Table E1 in the online supplement) as described previously (39–41). Expression levels were normalized to β-actin mRNA.
Immunostaining
Lung sections were stained with hematoxylin and eosin (H&E) or used for immunostaining as described previously (42–44). The primary antibodies and detection methods used are listed in the online supplement. Morphometric measurements were obtained as described previously (33). Colocalization experiments were performed as described previously (45–47).
Chromatin Immunoprecipitation Sequencing
Cross-linked chromatin was sonicated to 200–300 bp fragments and used for chromatin immunoprecipitation sequencing (ChIP-seq) (48, 49). Data analysis was performed using the BioWardrobe platform as described previously (25).
Statistical Analysis
One-way ANOVA and Student’s t test were used to determine statistical significance. Right-skewed measurements were log transformed to meet normality assumptions before analysis. P < 0.05 was considered significant. Values for all measurements were expressed as mean ± SD.
Results
Distinct and Evolutionarily Conserved Gene Expression Signature of Pulmonary c-KIT+ EC Progenitors
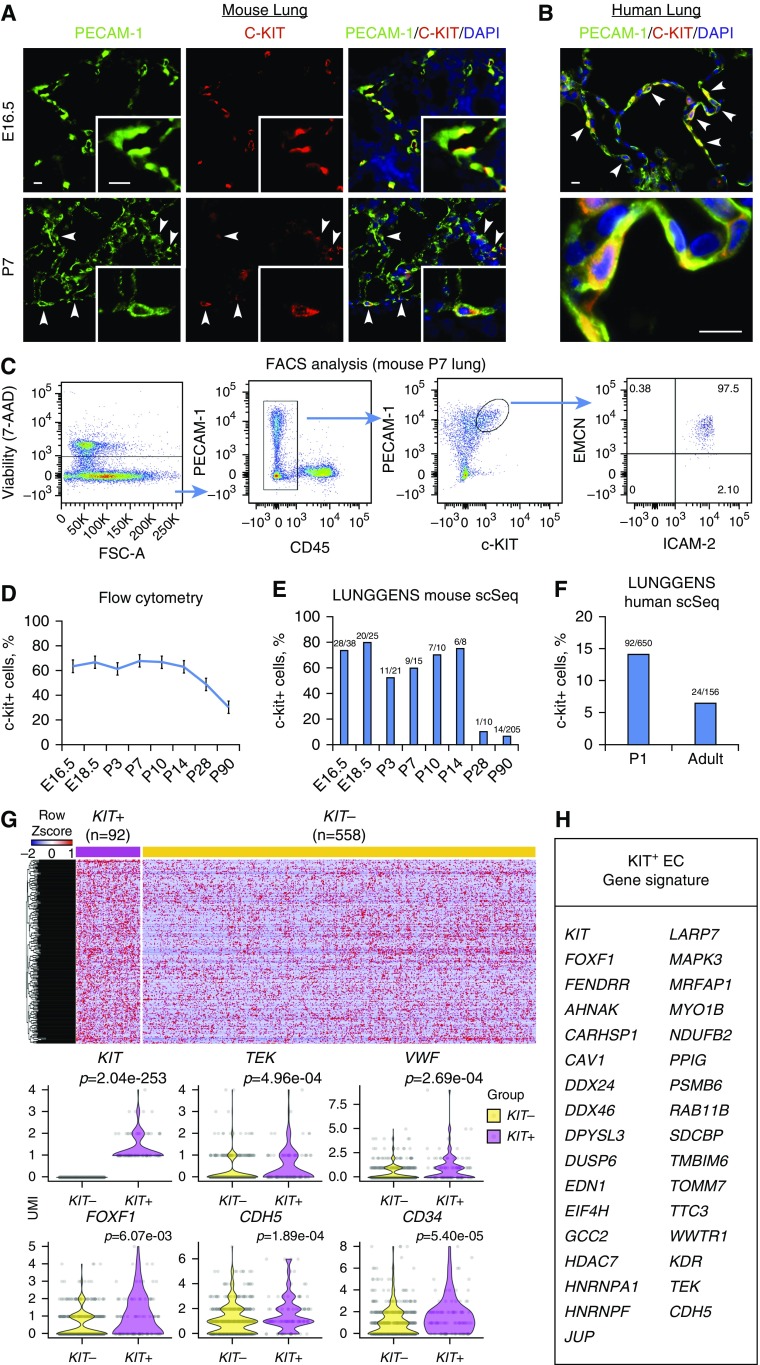
c-KIT+ ECs (c-KIT+ PECAM-1(CD31)+ CD45−) are abundant in fetal lung tissue (30), where they serve as progenitors for EC lineages (27, 28). We examined cell-surface markers and gene expression signatures of c-KIT+ ECs in developing mouse and human lungs. Consistent with published studies (28, 30), c-KIT+ ECs were frequently detected in the endothelium of pulmonary blood vessels and lung parenchyma of embryos and neonates, as shown by colocalization of c-KIT with PECAM-1 (Figures 1A and E1A). c-KIT+ ECs were also found in human neonatal lung (Figures 1B and E1B). Based on FACS analysis of neonatal mouse lung tissue, c-KIT+ ECs stained for endothelial markers PECAM-1, ICAM-2, and Endomucin (Figure 1C), but lacked cell-surface molecules expressed in pericytes (CD140b), fibroblasts (CD140a), hematopoietic cells (CD45), and epithelial cells (CD326) (Figure E2). c-KIT+ ECs did not express arterial NRP1, venous EphB4, or the lymphatic marker LYVE-1, but stained for CD34 (Figure E2), a cell-surface molecule that is expressed in pulmonary capillary ECs (1, 3). c-KIT+ ECs were abundant during embryonic and postnatal periods of lung development, but their numbers decreased in the adult lung as shown by FACS (Figure 1D), consistent with the observed numbers of c-KIT+ ECs identified by single-cell RNA-seq analyses (Figure 1E). The fraction of c-KIT+ ECs was smaller in human lung than in mouse lung and decreased with age (Figure 1F).
Figure 1.
The unique gene expression signature of c-KIT+ endothelial cell (EC) progenitors. (A and B) Immunostaining shows that c-KIT+ ECs are present in lung tissue from wild-type E16.5 and Postnatal Day 7 (P7) mice (A), and in normal human neonatal lung (B). Arrowheads indicate c-KIT+ ECs. Scale bars, 10 μm. (C) FACS analysis of collagenase-digested P7 lungs shows that c-KIT+ ECs express PECAM-1, ICAM-2, and endomucin (EMCN), and do not express CD45. (D) The percentage of pulmonary c-KIT+ ECs (c-KIT+ PECAM-1+ CD45−) was determined by FACS at different time points during murine lung development (n = 3–5 mice in each group). (E and F) c-KIT+ ECs were decreased in mouse and human adult lungs. The percentages of ECs expressing KIT mRNA were calculated using single-cell RNA-sequencing datasets obtained from the Lung Gene Expression Analysis Web Portal (https://research.cchmc.org/pbge/lunggens/mainportal.html). Ratios between c-KIT+ ECs and total ECs are shown for each time point. (G and H) Bioinformatic analysis identified 92 c-KIT+ and 558 c-KIT− ECs obtained from human P1 lung, showing significant differences in expression of 335 genes (P < 0.05). Heatmap and violin plots were used to identify gene expression differences between human lung c-KIT+ and c-KIT− ECs. FOXF1 target genes from gene expression overlap between human and mouse c-KIT+ ECs are shown in H. 7-AAD = 7-amino-actinomycin D; FSC = forward scatter; UMI = unique molecular identifier.
To identify gene signatures in c-KIT+ ECs during alveologenesis, we undertook a bioinformatic analysis of the single-cell RNA-seq (Drop-seq platform) data from human newborn lungs. A comparison of 92 c-KIT+ and 558 c-KIT− human lung ECs revealed significant differences in the expression of 335 genes (P < 0.05) (Figure 1G). Differentially expressed genes were functionally classified according to gene ontology. Functional categories of “angiogenesis,” “VEGF signaling,” “stem cell development,” and “cell cycle” were significantly enriched in the subset of upregulated c-KIT+ EC genes (Figure E3A). Single-cell RNA-seq of mouse neonatal lung tissue (383 c-KIT+ ECs and 667 c-KIT− ECs) identified gene expression changes characteristic of mouse c-KIT+ EC progenitors (Figures E4A and E4B). Functional categories identified by RNA expression were similar in human and mouse c-KIT+ ECs (Figures E3A and E3B). After a cross-comparison between human and mouse datasets, we identified 152 genes whose expression differed significantly between c-KIT+ and c-KIT− ECs (Figure 1H and Table E2). Multiple transcription factors, including ERG, SOX18, KLF6, SOX7, and FOXF1 (the latter being required for development of the alveolar microvasculature [16, 17, 21]), were differentially expressed in c-KIT+ versus c-KIT− ECs (Table E2). Expression of known FOXF1 transcriptional target genes, such as TEK, CDH5, and KDR (24, 26), was increased in both human and mouse c-KIT+ ECs (Figure 1H and Table E2). Additional FOXF1 targets in the c-KIT+ EC gene signature were identified in a ChIP-seq analysis of fetal lung endothelial MFLM-91 U cells (50). FOXF1 DNA-binding regions were found within 2 kb of the transcription initiation site of 29 genes identified in the c-KIT+ EC signature gene list (Figure 1H and Table E3), including those that are critical for VEGF signaling (Mapk3 [Erk1], Dusp6, and Psmb6) and EC function (Ahnak, Cav1, Jup, and Sdcbp) (Figure E5). FOXF1 binding was frequently associated with H3K4me3 methylation marks (Figure E5), predicting transcriptionally active promoters and enhancers (25). FOXF1 binding was detected in the bidirectional promoter of Foxf1 and the Foxf1-adjacent noncoding RNA Fendrr (Figure E6 and Table E3), consistent with autoregulation of the Foxf1 gene.
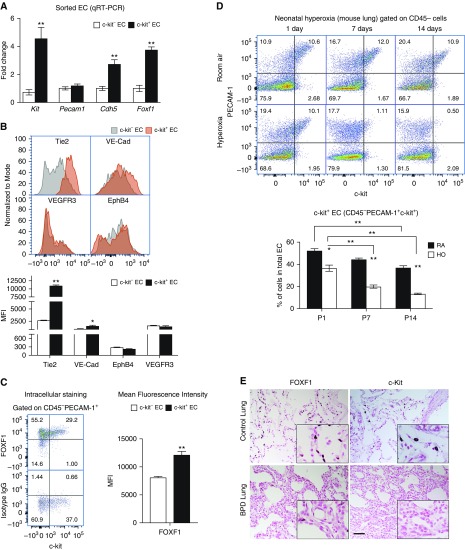
We isolated murine c-KIT+ and c-KIT− ECs from P7 lungs using FACS and performed qRT-PCR. Consistent with the single-cell RNA-seq analysis, Foxf1, Kit, and Cdh5 mRNAs were increased in c-KIT+ ECs, whereas Pecam1 mRNA was unchanged (Figure 2A). Cell-surface expression of VE-Cadherin (CDH5) and TIE-2 (TEK) was increased in c-KIT+ ECs, whereas EPHB4 and VEGFR3 were similar in c-KIT+ and c-KIT− ECs (Figure 2B). Intracellular staining for FOXF1 protein was examined by FACS, which showed increased FOXF1 in c-KIT+ ECs (Figures 2C and E7). Thus, c-KIT+ EC progenitors have a unique gene expression signature, which is enriched in FOXF1 and its transcriptional target genes. The c-KIT+ EC progenitor signature is conserved in murine and human lungs.
Figure 2.
Neonatal hyperoxia decreases c-KIT+ endothelial cells (ECs) in lung tissue. (A) RNA was prepared from FACS-sorted c-KIT+ and c-KIT− ECs from Postnatal Day 7 (P7) mouse lungs and analyzed by qRT-PCR. Increased mRNAs of Foxf1, Kit, and Cdh5 were found in c-KIT+ ECs (n = 3 mice in each group). Pecam1 mRNA was not changed. Expression levels were normalized to β-actin mRNA. (B) FACS analysis of P7 mouse lungs shows that c-KIT+ ECs have increased cell-surface expression of TIE2 and VE-Cadherin. Cell-surface staining of EphB4 and VEGFR3 was unaltered in c-KIT+ ECs. The mean fluorescence intensity (MFI) was calculated using n = 3 mice in each group. (C) FACS analysis shows increased amounts of intracellular FOXF1 protein in c-KIT+ ECs. Isotype control IgG was used as a control for intracellular FOXF1 staining. (D) Neonatal hyperoxia (HO) decreases the percentage of pulmonary c-KIT+ ECs in a time-dependent manner. Percentages were calculated from total number of pulmonary ECs (PECAM-1+ CD45−) using FACS analysis of wild-type lungs. Lungs of mice exposed to room air (RA) were used as controls (n = 5). *P < 0.05 and **P < 0.01. (E) Immunohistostaining shows reduced FOXF1 and c-KIT in lungs of patients with BPD (n = 7) compared with donor neonatal lungs (n = 6). Scale bar, 50 μm. BPD = bronchopulmonary dysplasia.
Neonatal Hyperoxia Decreases Pulmonary c-KIT+ EC Progenitors
To examine whether hyperoxic lung injury affects c-KIT+ EC progenitors, newborn mice were exposed to 85% O2 for 3 weeks followed by recovery in room air (Figure E8A). Consistent with published studies (33), neonatal hyperoxia caused alveolar simplification and altered lung function, increasing respiratory system resistance and elastance, and decreasing lung compliance (Figures E8B and E8C). Alveolar simplification was associated with reduced numbers of endothelial and epithelial cells as shown by FACS (Figures E9A and E9B). The percentage of c-KIT+ ECs was selectively decreased in a time-dependent manner after hyperoxia (Figure 2D). Immunostaining for c-KIT and FOXF1 was decreased in the lungs of patients with BPD compared with normal lungs (Figure 2E). Thus, neonatal hyperoxia decreases pulmonary c-KIT+ ECs.
FOXF1 Is Required for Maintenance of c-KIT+ ECs
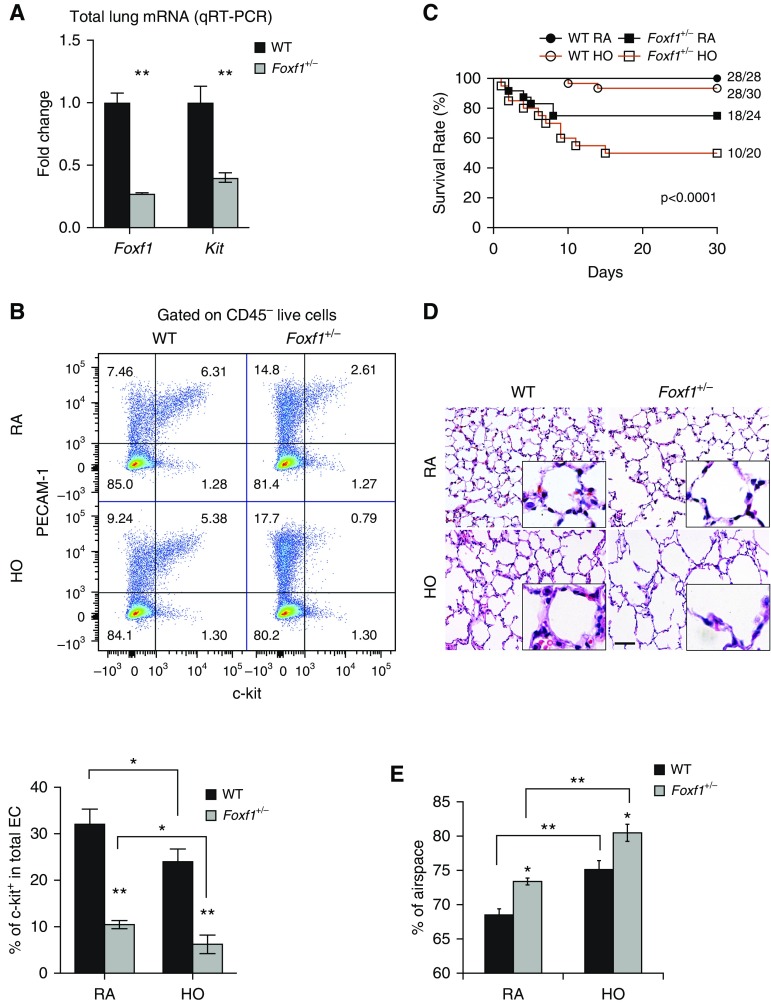
Because expression of FOXF1 was increased in mouse and human c-KIT+ ECs, we examined the requirements for FOXF1 in c-KIT+ ECs using mice haploinsufficient for the Foxf1 gene (Foxf1+/−). Foxf1 mRNA was decreased in Foxf1+/− lungs and associated with decreased c-Kit mRNA (Figure 3A). The percentages of c-KIT+ ECs were decreased in Foxf1+/− lung tissue (Figure 3B). Foxf1+/− mice were susceptible to hyperoxia, as shown by increased mortality (Figure 3C) and increased alveolar simplification (Figures 3D and 3E).
Figure 3.
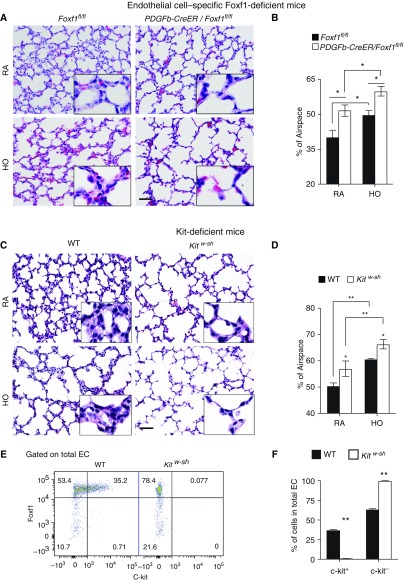
Haploinsufficiency of Foxf1 decreases c-KIT+ endothelial cell (EC) progenitors and causes alveolar simplification. (A) RNA was prepared from lungs of Foxf1+/− and wild-type (WT) Postnatal Day 7 (P7) mice and analyzed by qRT-PCR. Foxf1 and Kit mRNAs were decreased in Foxf1+/− lungs (n = 5 mice). Expression levels were normalized to β-actin mRNA. (B) FACS analysis shows decreased percentages of c-KIT+ ECs in Foxf1+/− lungs compared with WT controls (n = 5). Hyperoxia (HO) exacerbates the loss of c-KIT+ ECs compared with room air (RA)-exposed lungs. (C) Survival of Foxf1+/− and WT mice after hyperoxic lung injury is shown by Kaplan-Meier curves. (D and E) Hematoxylin and eosin staining of mouse P30 lungs shows alveolar simplification in Foxf1+/− mice. Mice were exposed to HO or RA from P1 to P7, followed by RA until harvest at P30. The percentage of airspace was calculated using 10 random lung fields from n = 5 mice per group. Scale bar, 50 μm. *P < 0.05 and **P < 0.01.
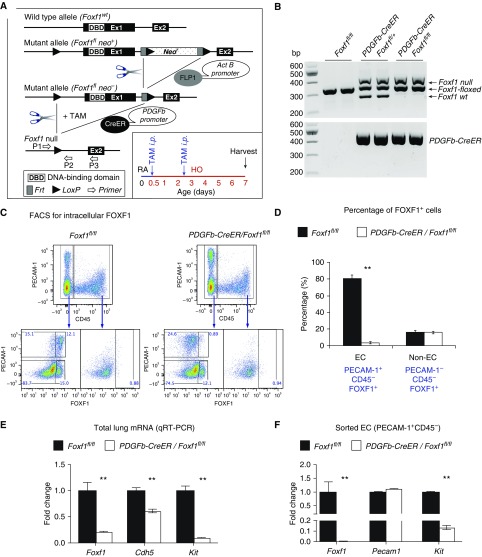
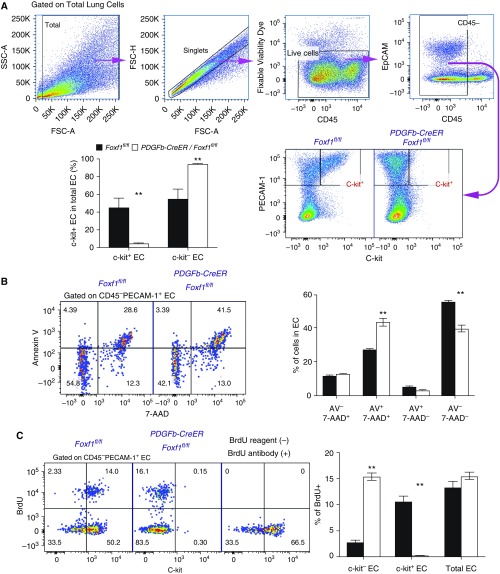
To examine whether FOXF1 acts in a cell-autonomous manner to regulate the maintenance of c-KIT+ ECs during alveologenesis, we deleted Foxf1 using the PDGFb-CreER transgene (Figures 4A and 4B), which specifically targets the EC lineages after tamoxifen administration (24, 26). FOXF1 was decreased in ECs, but not in non-ECs, after neonatal administration of tamoxifen (Figures 4C and 4D). Foxf1 mRNA was associated with decreased c-Kit in whole lung tissue (Figure 4E) and FACS-sorted ECs from PDGFb-CreER Foxf1−/− lungs (Figure 4F). Cell-surface and intracellular c-KIT staining was decreased in ECs after deletion of Foxf1 (Figures E10A and E10B). In contrast, cell-surface expression of SCF, a ligand of c-KIT, was increased in FOXF1-deficient ECs (Figures E11A–E11C). FOXF1-binding sites were found in the mouse c-Kit gene by ChIP-seq (Figure E10C), implicating FOXF1 in the regulation of c-Kit transcription. Thus, deletion of Foxf1 in neonatal pulmonary ECs decreased c-KIT and increased the c-KIT ligand SCF.
Figure 4.
Deletion of Foxf1 from endothelial cells (ECs) inhibits c-KIT. (A) Schematic representation of the generation of PDGFb-Cre/Foxf1−/− mice and tamoxifen (TAM) treatment of newborn pups. Foxf1-floxed mice were bred with PDGFb-CreER mice to generate PDGFb-Cre/Foxf1fl/fl offspring. LoxP sites were introduced to the Foxf1 gene flanking Exon 1, containing the DNA-binding domain. TAM was given intraperitoneally at Postnatal Day 0.5 (P0.5) and P2.5. Mice were exposed to hyperoxia (HO) or room air (RA) for 7 days. (B) PCR of genomic DNA was used to identify the Cre transgene and Foxf1-null, floxed, and wild-type alleles. (C and D) FACS shows that intracellular FOXF1 staining was decreased in ECs (PECAM-1+ CD45−) but not in non-ECs (PECAM-1− CD45−), in PDGFb-Cre/Foxf1−/− P7 lungs. The percentages of FOXF1+ cells were calculated using FACS (n = 3 mice). (E and F) qRT-PCR shows that Foxf1 and Kit mRNAs were decreased in lung tissue (E) and FACS-sorted ECs (F) obtained from TAM-treated PDGFb-Cre/Foxf1−/− lungs compared with Foxf1fl/fl controls (n = 3–5 mice in each group). Expression levels were normalized to β-actin mRNA. **P < 0.01.
Deletion of Foxf1 or c-Kit Decreases EC Progenitors and Causes Alveolar Simplification
The percentages of c-KIT+ EC progenitors were decreased after Foxf1 was deleted in neonatal pulmonary ECs (Figure 5A). The decrease in c-KIT+ ECs correlated with the degree of Foxf1 gene deletion and was more severe after deletion of both Foxf1 alleles (Figures E10D and E10E). SCF/c-KIT signaling in ECs is critical for cell survival and proliferation (28, 29). Consistent with published studies (21), FACS analysis for annexin V and 7-AAD showed increased EC death in Pdgfb-CreER Foxf1−/− (Figure 5B) and Foxf1+/− (Figure E12) lungs. Increased EC death in FOXF1-deficient mice was associated with decreased PECAM-1 immunostaining in alveolar septa (Figure E13). Deletion of Foxf1 selectively diminished bromodeoxyuridine incorporation in c-KIT+ ECs (Figure 5C). Interestingly, Pdgfb-CreER Foxf1−/− mice developed alveolar simplification even in the absence of hyperoxia (Figures 6A and 6B). Hyperoxia exacerbated the alveolar simplification in FOXF1-deficient mice (Figures 6A and 6B). c-KIT–deficient Kitw-sh mice also exhibited alveolar simplification in the presence or absence of hyperoxia (Figures 6C and 6D). PECAM-1 immunostaining and the percentages of ECs and epithelial cells identified by FACS were reduced in lung tissue from Kitw-sh mice (Figures E14A and E14B). ECs in Kitw-sh lungs lacked c-KIT (Figures 6E and 6F and E14A). Thus, deletion of either Foxf1 or c-Kit reduced the number of EC progenitors and caused alveolar simplification in the neonatal lung.
Figure 5.
Deletion of Foxf1 from endothelial cells (ECs) causes apoptosis, reduces cell proliferation, and decreases c-KIT+ EC progenitors. (A) Dot plots show the gating strategy to identify live c-KIT+ ECs by FACS analysis. The percentage of c-KIT+ ECs was decreased in tamoxifen-treated PDGFb-Cre/Foxf1−/− lungs compared with Foxf1fl/fl controls (n = 5). Tamoxifen was given at Postnatal Day 0.5 (P0.5) and P2.5, and lungs were harvested at P5. (B) FACS analysis shows a significant increase in cell death (Annexin V+ 7AAD+ cells) among the EC population of tamoxifen-treated PDGFb-Cre/Foxf1−/− lungs (n = 5 mice in each group). (C) Bromodeoxyuridine (BrdU) incorporation is decreased in c-KIT+ ECs from tamoxifen-treated PDGFb-Cre/Foxf1−/− lungs. Proliferation is increased in c-KIT− ECs. BrdU was given 2 hours before mouse harvest (n = 5). Mice without BrdU were used as controls for specificity of the BrdU antibody. **P < 0.01. 7-AAD = 7-amino-actinomycin D; FSC = forward scatter; SSC = side scatter.
Figure 6.
Deletion of Foxf1 from endothelial cells (ECs) causes alveolar simplification. (A–D) Hematoxylin and eosin staining of lung paraffin sections shows alveolar simplification in tamoxifen-treated PDGFb-Cre/Foxf1−/− mice (A and B) and Kitw-sh mice (C and D). Mice were exposed to hyperoxia (HO) or room air (RA) from Postnatal Day 1 (P1) and harvested at P7. Percentages of airspace were calculated using 10 random lung fields (n = 5 mice per group). Scale bars, 50 μm. (E and F) FACS analysis shows the lack of c-KIT staining in ECs of Kitw-sh lungs (n = 3). *P < 0.05 and **P < 0.01. WT = wild type.
Adoptive Transfer of c-KIT+ EC Progenitors Protects against Alveolar Simplification after Neonatal Hyperoxia
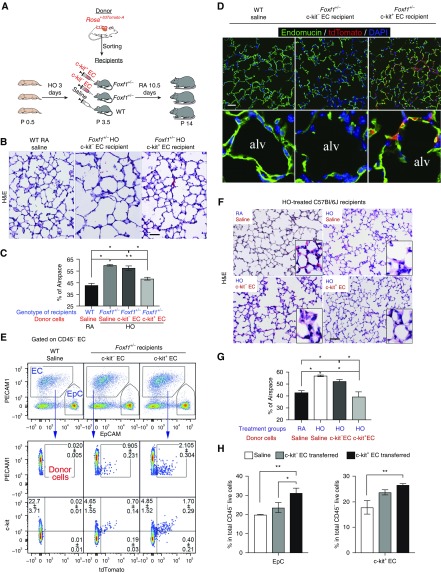
To establish a causal relationship between the loss of c-KIT+ EC progenitors and alveolar simplification, we used adoptive transfer of c-KIT+ ECs to prevent alveolar simplification after hyperoxia. c-KIT+ ECs were FACS-sorted from donor mice expressing tdTomato and transferred to hyperoxia-treated Foxf1+/− recipient mice (Figure 7A). Adoptive transfer of donor c-KIT+ ECs decreased alveolar simplification (Figures 7B and 7C) and increased capillary density in Foxf1+/− recipient mice, whereas c-KIT− ECs had no effect (Figures E15A and E15B). The protective effects of c-KIT+ ECs were associated with engraftment of these cells into the alveolar microvasculature of Foxf1+/− mice as demonstrated by colocalization of tdTomato and endomucin (Figure 7D) and FACS analysis (Figure 7E). The majority of donor c-KIT+ ECs maintained c-KIT on the cell surface (Figure 7E). In contrast, c-KIT− ECs were inefficient in engraftment into the peripheral microvasculature (Figures 7D and 7E). Neither c-KIT+ nor c-KIT− ECs integrated into the endothelium of large pulmonary arteries, veins, or lymphatic vessels (Figure E16B). There were no tdTomato+ cells among the epithelial and stromal cells of Foxf1+/− recipient lungs (Figure E16A).
Figure 7.
Adoptive transfer of c-KIT+ endothelial cell (EC) progenitors protects hyperoxia-treated lungs from alveolar simplification. (A) Schematic representation of hyperoxia (HO) treatment of Foxf1+/− newborn mice and adoptive transfer of c-KIT+ ECs. Cells were FACS-sorted from Postnatal Day 3 (P3) lungs of Rosa/tdTomato donor mice and injected intravenously into Foxf1+/− P3 recipient mice. (B and C) Hematoxylin and eosin (H&E) staining shows alveolar simplification in lungs of HO-treated Foxf1+/− mice. Mice were exposed to HO for 3 days (P1–P3) and lungs were harvested at P14. The percentage of airspace was calculated using 10 random lung fields and n = 3–5 mice per group. Adoptive transfer of c-KIT+ ECs decreases alveolar simplification compared with c-KIT− ECs or saline. (D) Immunostaining shows that exogenous c-KIT+ ECs labeled by tdTomato (red) integrated into alveolar microvasculature stained by endomucin (green). Endomucin staining is decreased in alveoli (alv) of Foxf1+/− mice. Adoptive transfer of c-KIT+ ECs increases endomucin staining in Foxf1+/− lungs. Representative images from n = 6–8 mice in each group are shown. (E) FACS analysis shows the presence of tdTomato+ ECs in Foxf1+/− lungs 7 days after adoptive transfer. Foxf1+/− lungs had more exogenous ECs after adoptive transfer of c-KIT+ ECs compared with c-KIT− ECs (n = 5 mice, P < 0.05). EpC, epithelial cells. (F and G) Adoptive transfer of c-KIT+ ECs prevented alveolar simplification in wild-type (WT) C57Bl/6 mice. Mice were exposed to HO between P1 and P3. Donor cells were injected intravenously at P3. (F and G) H&E staining (F) and measurement of airspace (G) were performed using P14 lungs. The percentage of airspace was calculated using 10 random lung fields (n = 5 mice per group). (H) FACS analysis shows that adoptive transfer of c-KIT+ ECs to HO-treated WT mice increases c-KIT+ ECs and epithelial cells (EpCam+ CD45− PECAM1−) in the lung tissue. *P < 0.05 and **P < 0.01. Scale bars, 50 μm.
We also performed adoptive transfer of c-KIT+ ECs using a BPD-like mouse model in which wild-type newborn mice were exposed to hyperoxia for 3 days (from P1 to P3). The 3-day hyperoxia exposure was sufficient to cause alveolar simplification, but did not cause arterial wall remodeling or right ventricular hypertrophy (Figures 7F, 7G, and E17A–E17C). Adoptive transfer of donor c-KIT+ ECs to hyperoxia-treated wild-type recipient mice decreased alveolar simplification (Figures 7F and 7G) and increased the total numbers of lung epithelial and c-KIT+ ECs identified by FACS analysis (Figure 7H). Capillary density in hyperoxia-treated lungs was increased after adoptive transfer of c-KIT+ ECs as demonstrated by immunostaining for endomucin (Figures E18A, E18B, and E19) and lung angiography using intravenous injection of isolectin B4, which labels the luminal surface of perfused blood vessels (Figures E18C and E19). Donor ECs expressing tdTomato were detected in the alveolar microvasculature by scanning confocal microscopy (Figure E20) and FACS analysis (Figures E21A and E21B). The percentages of tdTomato-labeled ECs gradually increased until P18, followed by a decline from P30 to P60 (Figures E21A and E21B). Consistent with increased numbers of donor cells at P18, tdTomato+ ECs showed greater proliferation than tdTomato− recipient ECs, as shown by FACS analysis for DNA content (Figures E21C and E21D). Taken together, these results show that c-KIT+ ECs efficiently integrated into the alveolar microvasculature, increased neonatal lung angiogenesis, and protected recipient mice from alveolar simplification induced by hyperoxia or caused by the loss of Foxf1.
Discussion
Disruption of pulmonary angiogenesis is a key mechanism that contributes to the alveolar simplification associated with BPD (12–14). In the present study, we demonstrated that c-KIT+ EC progenitors engrafted into the neonatal alveolar microvasculature and inhibited alveolar simplification caused by hyperoxia or deletion of Foxf1. Pulmonary EC progenitors express VEGFR2 (FLK1) and TIE2. Considering that administration of either VEGF-A or Angiopoietin-1, a ligand of TIE2, restored capillary density, improved alveologenesis, and increased survival after hyperoxia (12), activation of the VEGF/FLK1 and Angiopoietin-1/TIE2 signaling pathways may contribute to the increased angiogenesis and alveologenesis seen after engraftment of c-KIT+ ECs. Consistent with these observations, the gene expression signature in c-KIT+ EC progenitors wass enriched in genes associated with cellular proliferation, angiogenesis, and VEGF signaling. A c-KIT+ EC gene expression signature was conserved in mouse and human lungs, suggesting that cell-surface markers identified in our studies, such as CD34, CD36, CD93, CD105, and CD119, can be used to purify c-KIT+ EC progenitors from human lung tissue. Because c-KIT+ EC progenitors were decreased in the lungs of patients with BPD, increasing c-KIT+ ECs by transplantation of donor or embryonic stem cell/induced pluripotent stem cell–derived c-KIT+ ECs may be considered in the future development of therapies for BPD. Based on our findings, c-KIT+ ECs may represent specialized microvascular EC progenitors, as these cells engraft into alveolar capillaries but not into large pulmonary blood or lymphatic vessels. After adoptive transfer of c-KIT+ ECs, the number of donor-derived cells increased within the first weeks of life but did not persist into adulthood, suggesting that the progenitor potential of c-KIT+ ECs may be limited to postnatal angiogenesis and alveolarization.
Herein, we demonstrate that c-KIT+ ECs are dependent on FOXF1, a transcription factor that regulates a network of genes that are critical for neonatal pulmonary angiogenesis and alveolarization (16, 17, 21). Genetic deletion of one Foxf1 allele in mice reduced pulmonary angiogenesis and increased the mortality of Foxf1+/− pups in the early postnatal period (21). Inactivating mutations in FOXF1 cause ACDMPV, a fatal congenital disorder in newborns and infants that is characterized by respiratory insufficiency due to loss of capillaries in the alveolar septa (17, 18). We found that c-KIT+ EC progenitors were decreased in the lungs of Foxf1+/− mice, and that hyperoxia caused a nearly complete loss of c-KIT+ ECs but did not alter c-KIT− ECs in Foxf1+/− lungs, indicating that c-KIT+ ECs may be more sensitive to hyperoxia. Loss of c-KIT+ ECs during hyperoxia is likely to depend on FOXF1 and occur in a cell-autonomous manner, as endothelial-specific inactivation of Foxf1 was sufficient to decrease the number of c-KIT+ ECs, causing increased apoptosis and impaired proliferation. Although oxygen therapy is required to support infants with ACDMPV, findings in Foxf1+/− mice indicate that oxygen therapy may selectively reduce c-KIT+ EC progenitors in patients with ACDMPV, further inhibiting pulmonary angiogenesis and exacerbating respiratory insufficiency after birth. Because c-KIT+ ECs efficiently engraft into the neonatal alveolar microvasculature and stimulate angiogenesis, it is possible that cell transplantation of c-KIT+ ECs could be beneficial for patients with ACDMPV. Because c-KIT+ EC progenitors are hypersensitive to high oxygen concentrations, prenatal genetic screening for FOXF1 mutations and the early use of extracorporeal membrane oxygenation in patients with ACDMPV might be considered to avoid the loss of c-KIT+ endothelial progenitors and excessive lung remodeling before c-KIT+ EC transplantation for future therapies.
In summary, pulmonary EC progenitors are dependent on FOXF1 and c-KIT to promote alveolar septation. c-KIT+ ECs are highly sensitive to injury by high oxygen concentrations and undergo apoptosis during neonatal hyperoxia. c-KIT+ ECs engraft into the alveolar microvasculature, stimulate neonatal lung angiogenesis, and protect the lung from alveolar simplification caused by hyperoxia. These findings support further study of the use of donor c-KIT+ ECs and embryonic stem cell/induced pluripotent stem cell–derived EC progenitors for therapy of pediatric disorders associated with decreased pulmonary angiogenesis and alveolarization.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Y. Cheng for assistance with data mining, Dr. L. Fei for statistical analyses, and Dr. G. Pryhuber for providing human lung tissue. Donor tissues were obtained through United Network Sharing. The authors are grateful to the families who have generously given their precious tissues to support research.
Footnotes
Supported by LungMAP grants HL84151, HL141174, and HL123490 (to V.V.K.); HL132849 (to T.V.K.); HL122642 and HL124745 (to Y.X. and J.A.W.); and HL122638 and HL122700 (to J.A.W.).
Author Contributions: X.R. and V.V.K. designed the study. X.R., V.U., G.W., C.B., and Y.Z. conducted experiments. M.G. and Y.X. conducted bioinformatic analyses. J.A.W., T.V.K., and V.V.K. wrote the manuscript with input from all authors.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201812-2312OC on June 24, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev. 2019;99:513–554. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV. Transcription factors regulating embryonic development of pulmonary vasculature. Adv Anat Embryol Cell Biol. 2018;228:1–20. doi: 10.1007/978-3-319-68483-3_1. [DOI] [PubMed] [Google Scholar]
- 4.Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 5.Maeda Y, Davé V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 7.Vohr BR, Wright LL, Dusick AM, Mele L, Verter J, Steichen JJ, et al. Neurodevelopmental and functional outcomes of extremely low birth weight infants in the National Institute of Child Health and Human Development Neonatal Research Network, 1993-1994. Pediatrics. 2000;105:1216–1226. doi: 10.1542/peds.105.6.1216. [DOI] [PubMed] [Google Scholar]
- 8.Jobe AH. Antenatal factors and the development of bronchopulmonary dysplasia. Semin Neonatol. 2003;8:9–17. doi: 10.1016/s1084-2756(02)00188-4. [DOI] [PubMed] [Google Scholar]
- 9.Stenmark KR, Abman SH. Lung vascular development: implications for the pathogenesis of bronchopulmonary dysplasia. Annu Rev Physiol. 2005;67:623–661. doi: 10.1146/annurev.physiol.67.040403.102229. [DOI] [PubMed] [Google Scholar]
- 10.Bokodi G, Treszl A, Kovács L, Tulassay T, Vásárhelyi B. Dysplasia: a review. Pediatr Pulmonol. 2007;42:952–961. doi: 10.1002/ppul.20689. [DOI] [PubMed] [Google Scholar]
- 11.Thébaud B, Abman SH. Bronchopulmonary dysplasia: where have all the vessels gone? Roles of angiogenic growth factors in chronic lung disease. Am J Respir Crit Care Med. 2007;175:978–985. doi: 10.1164/rccm.200611-1660PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thébaud B, Ladha F, Michelakis ED, Sawicka M, Thurston G, Eaton F, et al. Vascular endothelial growth factor gene therapy increases survival, promotes lung angiogenesis, and prevents alveolar damage in hyperoxia-induced lung injury: evidence that angiogenesis participates in alveolarization. Circulation. 2005;112:2477–2486. doi: 10.1161/CIRCULATIONAHA.105.541524. [DOI] [PubMed] [Google Scholar]
- 13.Kunig AM, Balasubramaniam V, Markham NE, Morgan D, Montgomery G, Grover TR, et al. Recombinant human VEGF treatment enhances alveolarization after hyperoxic lung injury in neonatal rats. Am J Physiol Lung Cell Mol Physiol. 2005;289:L529–L535. doi: 10.1152/ajplung.00336.2004. [DOI] [PubMed] [Google Scholar]
- 14.Baker CD, Abman SH. Impaired pulmonary vascular development in bronchopulmonary dysplasia. Neonatology. 2015;107:344–351. doi: 10.1159/000381129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16:180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dharmadhikari AV, Szafranski P, Kalinichenko VV, Stankiewicz P. Genomic and epigenetic complexity of the FOXF1 locus in 16q24.1: implications for development and disease. Curr Genomics. 2015;16:107–116. doi: 10.2174/1389202916666150122223252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [Published erratum appears in Am J Hum Genet 85:537.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen P, Yang Y, Navarro C, Silva I, Szafranski P, Kolodziejska KE, et al. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Hum Mutat. 2013;34:801–811. doi: 10.1002/humu.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. Am J Respir Crit Care Med. 2011;184:172–179. doi: 10.1164/rccm.201010-1697CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahlapuu M, Ormestad M, Enerbäck S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 21.Kalinichenko VV, Lim L, Stolz DB, Shin B, Rausa FM, Clark J, et al. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 22.Kalinichenko VV, Zhou Y, Shin B, Stolz DB, Watkins SC, Whitsett JA, et al. Wild-type levels of the mouse Forkhead Box f1 gene are essential for lung repair. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1253–L1265. doi: 10.1152/ajplung.00463.2001. [DOI] [PubMed] [Google Scholar]
- 23.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol. 2008;39:390–399. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res. 2014;115:709–720. doi: 10.1161/CIRCRESAHA.115.304382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep. 2017;7:10690. doi: 10.1038/s41598-017-11175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal. 2016;9:ra40. doi: 10.1126/scisignal.aad1899. [DOI] [PubMed] [Google Scholar]
- 27.Liu Q, Huang X, Zhang H, Tian X, He L, Yang R, et al. c-kit(+) cells adopt vascular endothelial but not epithelial cell fates during lung maintenance and repair. Nat Med. 2015;21:866–868. doi: 10.1038/nm.3888. [DOI] [PubMed] [Google Scholar]
- 28.Fang S, Wei J, Pentinmikko N, Leinonen H, Salven P. Generation of functional blood vessels from a single c-kit+ adult vascular endothelial stem cell. PLoS Biol. 2012;10:e1001407. doi: 10.1371/journal.pbio.1001407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miranda LF, Rodrigues CO, Ramachandran S, Torres E, Huang J, Klim J, et al. Stem cell factor improves lung recovery in rats following neonatal hyperoxia-induced lung injury. Pediatr Res. 2013;74:682–688. doi: 10.1038/pr.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, Suzuki S, Fujino N, Ota C, Yamada M, Suzuki T, et al. c-Kit immunoexpression delineates a putative endothelial progenitor cell population in developing human lungs. Am J Physiol Lung Cell Mol Physiol. 2014;306:L855–L865. doi: 10.1152/ajplung.00211.2013. [DOI] [PubMed] [Google Scholar]
- 31.Sen P, Dharmadhikari AV, Majewski T, Mohammad MA, Kalin TV, Zabielska J, et al. Comparative analyses of lung transcriptomes in patients with alveolar capillary dysplasia with misalignment of pulmonary veins and in foxf1 heterozygous knockout mice. PLoS One. 2014;9:e94390. doi: 10.1371/journal.pone.0094390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claxton S, Kostourou V, Jadeja S, Chambon P, Hodivala-Dilke K, Fruttiger M. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 2008;46:74–80. doi: 10.1002/dvg.20367. [DOI] [PubMed] [Google Scholar]
- 33.Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, et al. Foxm1 regulates resolution of hyperoxic lung injury in newborns. Am J Respir Cell Mol Biol. 2015;52:611–621. doi: 10.1165/rcmb.2014-0091OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren X, Shah TA, Ustiyan V, Zhang Y, Shinn J, Chen G, et al. FOXM1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Mol Cell Biol. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balli D, Ren X, Chou FS, Cross E, Zhang Y, Kalinichenko VV, et al. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene. 2012;31:3875–3888. doi: 10.1038/onc.2011.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323, e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4:e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang IC, Snyder J, Zhang Y, Lander J, Nakafuku Y, Lin J, et al. Foxm1 mediates cross talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol Cell Biol. 2012;32:3838–3850. doi: 10.1128/MCB.00355-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras(G12D.) Oncogene. 2014;33:5391–5396. doi: 10.1038/onc.2013.475. [DOI] [PubMed] [Google Scholar]
- 42.Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, et al. Functional characterization of evolutionarily conserved DNA regions in forkhead box f1 gene locus. J Biol Chem. 2005;280:37908–37916. doi: 10.1074/jbc.M506531200. [DOI] [PubMed] [Google Scholar]
- 43.Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expr Patterns. 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, et al. Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 2003;11:149–162. doi: 10.3727/000000003108749044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, et al. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Dev Biol. 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ustiyan V, Wert SE, Ikegami M, Wang IC, Kalin TV, Whitsett JA, et al. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Dev Biol. 2012;370:198–212. doi: 10.1016/j.ydbio.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ustiyan V, Zhang Y, Perl AK, Whitsett JA, Kalin TV, Kalinichenko VV. β-catenin and Kras/Foxm1 signaling pathway are critical to restrict Sox9 in basal cells during pulmonary branching morphogenesis. Dev Dyn. 2016;245:590–604. doi: 10.1002/dvdy.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, et al. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J Biol Chem. 2013;288:28477–28487. doi: 10.1074/jbc.M113.478974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem. 2015;290:7563–7575. doi: 10.1074/jbc.M114.609487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akeson AL, Wetzel B, Thompson FY, Brooks SK, Paradis H, Gendron RL, et al. Embryonic vasculogenesis by endothelial precursor cells derived from lung mesenchyme. Dev Dyn. 2000;217:11–23. doi: 10.1002/(SICI)1097-0177(200001)217:1<11::AID-DVDY2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.