Abstract
Macrosolen plants are parasitic shrubs, several of which are important medicinal plants, that are used as folk medicine in some provinces of China. However, reports on Macrosolen are limited. In this study, the complete chloroplast genome sequences of Macrosolen cochinchinensis, Macrosolen tricolor and Macrosolen bibracteolatus are reported. The chloroplast genomes were sequenced by Illumina HiSeq X. The length of the chloroplast genomes ranged from 129,570 bp (M. cochinchinensis) to 126,621 bp (M. tricolor), with a total of 113 genes, including 35 tRNA, eight rRNA, 68 protein-coding genes, and two pseudogenes (ycf1 and rpl2). The simple sequence repeats are mainly comprised of A/T mononucleotide repeats. Comparative genome analyses of the three species detected the most divergent regions in the non-coding spacers. Phylogenetic analyses using maximum parsimony and maximum likelihood strongly supported the idea that Loranthaceae and Viscaceae are monophyletic clades. The data obtained in this study are beneficial for further investigations of Macrosolen in respect to evolution and molecular identification.
Keywords: Macrosolen, Macrosolen cochinchinensis, Macrosolen tricolor, Macrosolen bibracteolatus, Santalales, gene loss, chloroplast genome, phylogenetic relationship
1. Introduction
The traits of trophic specialization in all parasitic plants are described as “parasitic reduction syndrome”. At the genetic level, parasitic reduction syndrome includes the functional and physical reduction of heterotrophs’ plastid genomes, where rampant gene loss and an acceleration of molecular evolutionary rates occur [1,2]. Considering the partial or complete absence of their photosynthetic capacity, parasitic plants have to absorb organic nutrients, inorganic nutrients, and water from their hosts [3]. Most parasitic plants are included in the order Santalales and the families Orobanchaceae and Orchidaceae [2]. The first complete chloroplast genome of a parasitic plant was obtained from Epifagus virginiana, and all of its photosynthesis and energy producing genes have been lost [4]. Petersen et al. reported the complete plastome sequences of one species of Osyris and three species of Viscum. These researchers found that these four species have experienced rearrangements, and a number of protein-coding genes and two tRNA genes have been pseudogenised or completely lost [5]. The complete chloroplast genome of Schoepfia jasminodora has been reported; S. jasminodora represents the early stages of chloroplast genome degradation along with its transition to heterotrophy in related taxa [6]. Li et al. determined the complete chloroplast genome sequences of Taxillus chinensis and Taxillus sutchuenensis. The results showed that all ndh genes, three ribosomal protein genes, seven tRNA genes, four ycf genes, and the infA gene of these two species have been lost [7]. Previous studies have reported that Rafflesia lagascae only contains small fragments of plastid sequences at low coverage depth, and they cannot recover any substantial portions of the chloroplast genome [8]. In the parasitic family Orobanchaceae, the complete chloroplast genomes of some species, including Cistanche deserticola [9], Aureolaria virginica, Lindenbergia philippensis [10], and Lathraea squamaria [11], have been reported. These chloroplast genomes have shown physical and functional gene loss or pseudogenization. The Balanophora plastid genomes of Balanophora laxiflora and Balanophora reflexa [12], at 15.5 kb in size with only 19 genes, are the most reduced plastomes reported thus far, except for the 11.3 and 15.2 kb genomes of two holoparasitic species of Pilostyles [13] and the 12.8 kb genome of the myco-heterotroph Sciaphila thaidanica [14]. Rhopalocnemis phalloides [15], which belongs to the family Balanophoraceae, has also shown highly plastid genome reduction with 18.6 kb in length. In addition, gene loss has also been found in myco-heterotrophs [16], where carbon is obtained from fungi, thus forming mycorrhizal symbiosis with their roots. Photosynthesis-related genes are lost first, followed by housekeeping genes, which eventually results in a highly reduced genome [17].
The chloroplast is an important organelle in plant cells, and it primarily carries out photosynthesis and carbon fixation. The chloroplast genome is independent of nuclear genes, and the chloroplast possesses its own independent transcription and transport system [18,19]. A typical chloroplast genome of most angiosperms consists of four parts, namely a pair of inverted repeats (IRa and IRb), a large single-copy (LSC) region and a small single-copy (SSC) region [20]. The chloroplast genome sequences are highly conserved in gene order and content [21], and they are thus ideal research models for the study of molecular markers [22,23], species identification [24,25,26], and species evolution [27].
Macrosolen plants are parasitic shrubs that belong to the family Loranthaceae. There are approximately 40 species of Macrosolen, and most of them are distributed in Southern and Southeastern Asia, whereas five species of Macrosolen are dispersed in China [28]. Macrosolen cochinchinensis, Macrosolen tricolor, and Macrosolen bibracteolatus have been used as folk medicines in China for a long time. M. cochinchinensis is used to clear heat and fire, remove blood stasis, and relieve pain. M. tricolor is used to dissipate heat and relieve coughing. M. bibracteolatus is used to invigorate the liver and kidney, expel wind, remove dampness, and strengthen tendons and bones [29,30,31]. These species exhibit different medicinal effects. However, they have similar morphologies when they are not in fluorescence (Figure 1), resulting in an extreme difficulty in their identification on the basis of morphological features. The limited reports on Macrosolen hinder the related research and development. In this study, we determined the complete chloroplast genome sequences of M. cochinchinensis, M. tricolor and M. bibracteolatus. To reveal the phylogenetic positions of the three species and the evolution of Macrosolen within Santalales, we conducted phylogenetic trees using the maximum parsimony (MP) and maximum likelihood (ML) methods on the basis of common protein-coding genes from 16 species. Our results can provide important genetic resources for the study of Macrosolen.
Figure 1.
Plant materials of three Macrosolen species. ① Macrosolen cochinchinensis; ② Macrosolen tricolor; and ③ Macrosolen bibracteolatus.
2. Results
2.1. Complete Chloroplast Genomes of Three Macrosolen Species
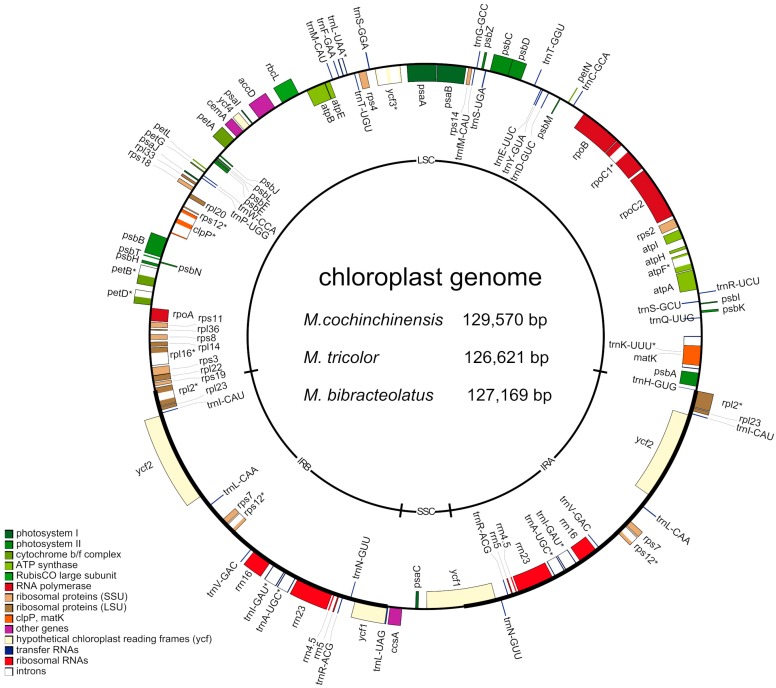
The length of the three studied chloroplast genomes ranged from 129,570 bp (M. cochinchinensis) to 126,621 bp (M. tricolor) with a typical quadripartite structure consisting of a pair of IRs (24,703–25,445 bp) separated by the LSC (70,692–73,052 bp) and the SSC (5320–5724 bp) regions (Figure 2). The three chloroplast genomes were found to highly conserved in GC content, gene content and gene order (Table 1 and Table S1). All three species comprised 113 genes, including 68 protein-coding genes, 35 tRNAs, eight rRNAs and two pseudogenes (rps12 and ycf2). A total of 17 genes were found to be repeated genes, and 79 were found to be unique genes in the chloroplast genomes. Three genes (clpP, ycf3 and rps12) contained two introns, whereas 10 genes (atpF, rpoC1, rpl2, rpl16, petB, petD, trnA-UGC, trnI-GAU, trnK-UUU and trnL-UAA) had only one intron (Table 2 and Table S2).
Figure 2.
Gene map of the complete chloroplast genome of three Macrosolen species. Genes outside the large ring circle are transcribed in a counter-clockwise direction, and genes inside the circle are transcribed clockwise. The same color represents the same category of genes. Deep grey in the inner circle represents GC content, and lighter grey represents A/T content.
Table 1.
Length of chloroplast genome of three Macrosolen species and their base composition.
| Species | M. cochinchinensis | M. tricolor | M. bibracteolatus |
|---|---|---|---|
| Accession No. | MH161424 | MH161425 | MH161423 |
| Genome size (bp) | 129,570 | 126,621 | 127,169 |
| LSC length (bp) | 73,052 | 71,895 | 70,692 |
| SSC length (bp) | 5724 | 5320 | 5587 |
| IRs length (bp) | 25,397 | 24,703 | 25,445 |
| GC content (%) | 37.3 | 37.7 | 37.9 |
| Number of genes | 113 | 113 | 113 |
| Number of protein-coding genes | 68 | 68 | 68 |
| Number of tRNAs | 35 | 35 | 35 |
| Number of rRNAs | 8 | 8 | 8 |
| Number of pseudogenes | 2 | 2 | 2 |
Table 2.
Gene list of chloroplast genome of three Macrosolen species.
| No. | Group of Genes | Gene Names | Number |
|---|---|---|---|
| 1 | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| 2 | Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 3 | Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | 6 |
| 4 | ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | 6 |
| 5 | NADH dehydrogenase | - | 0 |
| 6 | RubisCO large subuni | rbcL | 1 |
| 7 | RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| 8 | Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 ** (×2), rps14, rps18, rps19 | 12(2) |
| 9 | Ribosomal proteins (LSU) | rpl2 * (×2), rpl14, rpl16 *, rpl20, rpl22, rpl23 (×2), rpl33, rpl36 | 10(2) |
| 10 | Proteins of unknown function | ycf1(×2), ycf2(×2), ycf3 **, ycf4 | 6(2) |
| 11 | Transfer RNAs | 35 tRNAs (4 contain an intron, 7 in the IRs) | 35(7) |
| 12 | Ribosomal RNAs | rrn4.5 (×2), rrn5(×2), rrn16 (×2), rrn23 (×2) | 8(4) |
| 13 | Other genes | accD, clpP **, matK, ccsA, cemA | 5 |
* One or two asterisks following genes indicate one or two contained introns, respectively. (×2) indicates that the number of the repeat unit is two. The numbers in parenthesis at the line of ‘Number’ indicate the total number of repeated genes.
2.2. Codon Usage Analyses and RNA Editing Sites
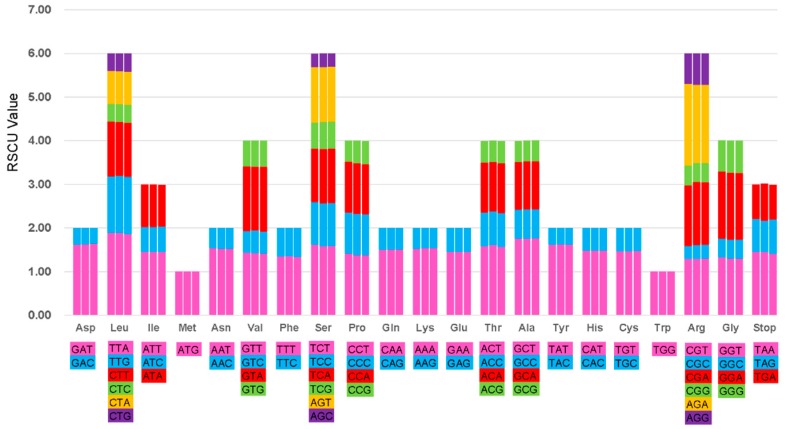
Relative synonymous codon usage (RSCU) is the ratio between the use and expected frequencies for a particular codon and a measure of nonuniform synonymous codon usage in coding sequences [32]. On the basis of the sequences of protein-coding genes, the codon usage frequency was estimated for the chloroplast genome of the three Macrosolen species (Figure 3). All the protein-coding genes were found to consist of 21,581, 21,598 and 21,520 codons in the chloroplast genomes of M. cochinchinensis, M. tricolor and M. bibracteolatus, respectively (Table S3). Figure 3 shows that the RSCU value increased with the increase in the quantity of codons which coded for a specific amino acid. Most of the amino acid codons show preferences except for methionine and tryptophan. Potential RNA editing sites were also predicted for 29 genes in the chloroplast genomes of the three species. A total of 39 RNA editing sites were identified (Table S4). The amino acid conversion from serine (S) to leucine (L) occurred most frequently, whereas that from proline (P) to serine (S) and from threonine (T) to methionine (M) occurred the least.
Figure 3.
Codon content of 20 amino acids and stop codons in all of the protein-coding genes of the chloroplast genomes of three Macrosolen species.
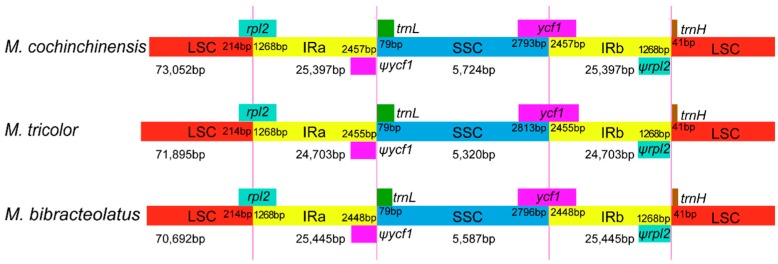
2.3. IR Constriction and Expansion
Figure 4 shows the comparison of the boundaries of the LSC/IR/SSC regions of three Macrosolen species. The LSC/IR/SSC boundaries and gene contents in the chloroplast genomes of the three species were found to be highly conserved, featuring the same sequence structure and differences in length. In the three species, the rpl2 gene, which is a normal functional gene, crossed the LSC/IRa boundary, but the rpl2 pseudogene with a length of 1268 bp formed in the IRb region. The SSC/IRb boundaries of M. cochinchinensis, M. tricolor and M. bibracteolatus were found to be located in the complete ycf1 gene, and their ycf1 pseudogenes with lengths of 2457, 2455 and 2448 bp, respectively, were found to be produced in IRa.
Figure 4.
Comparison of the borders of the large single-copy (LSC), small single-copy (SSC), and inverted repeats (IR) regions among the chloroplast genomes of three Macrosolen species. The number above the gene features means the distance between the ends of genes and the borders sites. These features are not to scale. Ψ: pseudogenes.
2.4. Simple Sequence Repeats (SSRs) and Repeat Structure Analyses
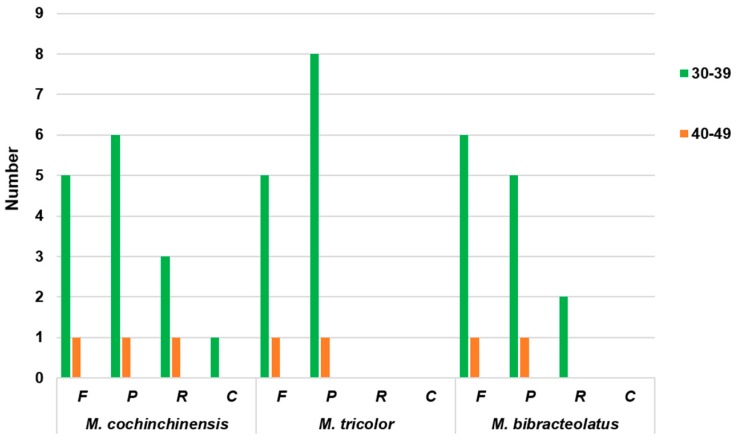
A simple sequence repeat (SSR), which is also known as microsatellite DNA, is a tandem repeat sequence consisting of one to six nucleotide repeat units [22]. SSRs are widely used as molecular markers in species identification, population genetics, and phylogenetic investigations due to their high polymorphism level [33,34]. A total of 238, 226 and 217 SSRs were identified in the chloroplast genomes of M. cochinchinensis, M. tricolor and M. bibracteolatus, respectively (Table 3). Amongst all SSRs, the numbers of mononucleotide repeats were the highest, with values detected at 169, 166 and 162 times in M. cochinchinensis, M. tricolor and M. bibracteolatus, respectively. Amongst these mononucleotide repeats, A/T was found to be the most frequent SSR. In accordance with the number of repeats, mononucleotide and dinucleotide SSRs exhibited a certain base preference that mainly contained A/T units. Long repeat sequences should be >30 bp, and these repeats are mainly distributed in the gene spacer and intron sequences. The result shows that M. cochinchinensis presented the highest number, comprising six forward, seven palindromic, four reverse and one complement repeats (Figure 5). Two types of M. tricolor, comprising six forward and nine palindromic repeats, were present. M. bibracteolatus presented seven forward, six palindromic and two reverse repeats.
Table 3.
Types and amounts of simple sequence repeats (SSRs) in the chloroplast genomes of three Macrosolen species.
| SSR Type | Repeat Unit | Amount | Ratio (%) | ||||
|---|---|---|---|---|---|---|---|
| ① | ② | ③ | ① | ② | ③ | ||
| mono | A/T | 161 | 159 | 153 | 95.3 | 95.8 | 94.4 |
| C/G | 8 | 7 | 9 | 4.7 | 4.2 | 5.6 | |
| di | AC/GT | 5 | 4 | 4 | 9.6 | 8.5 | 9.3 |
| AG/CT | 13 | 14 | 13 | 25 | 29.8 | 30.2 | |
| AT/TA | 34 | 29 | 26 | 64.4 | 61.7 | 60.5 | |
| tri | AAT/ATT | 4 | 4 | 0 | 66.7 | 66.7 | 0 |
| ATC/ATG | 2 | 2 | 2 | 33.3 | 33.3 | 100 | |
| tetra | AAAG/CTTT | 3 | 3 | 3 | 33.3 | 42.9 | 30 |
| AATC/ATTG | 1 | 1 | 0 | 11.1 | 14.3 | 0 | |
| ACAG/CTGT | 1 | 1 | 1 | 11.1 | 14.3 | 10 | |
| AAAT/ATTT | 3 | 1 | 3 | 33.3 | 14.3 | 30 | |
| AATG/ATTC | 1 | 0 | 1 | 11.1 | 0 | 10 | |
| AGAT/ATCT | 0 | 1 | 1 | 0 | 14.3 | 10 | |
| ACAT/ATGT | 0 | 0 | 1 | 0 | 0 | 10 | |
| penta | AATAT/ATATT | 1 | 0 | 0 | 100 | 0 | 0 |
| hexa | ATATCC/ATATGG | 1 | 0 | 0 | 100 | 0 | 0 |
① M. cochinchinensis; ② M. tricolor; and ③ M. bibracteolatus.
Figure 5.
Repeat sequences in the chloroplast genomes of three Macrosolen species. F, P, R, and C indicate the repeat types F (forward), P (palindrome), R (reverse) and C (complement), respectively. Repeats with different lengths are indicated in different colors.
2.5. Comparative Genomic Analyses
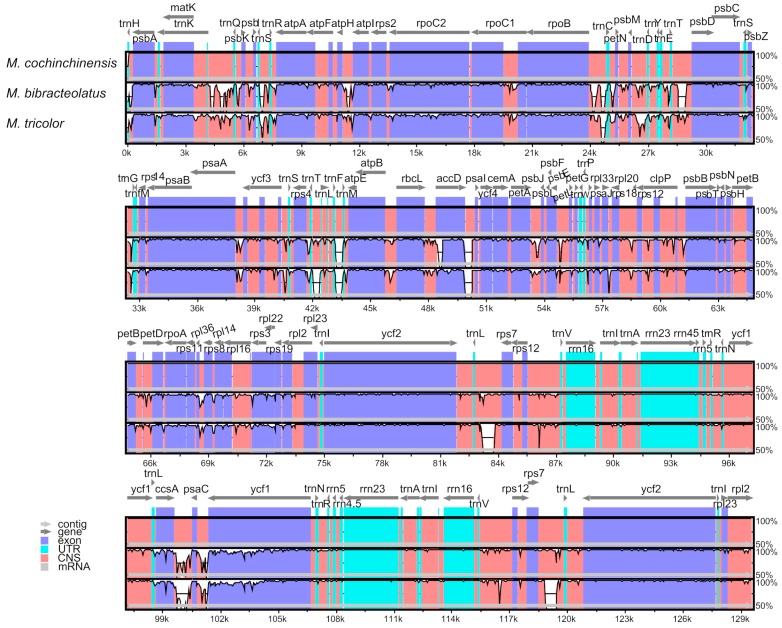
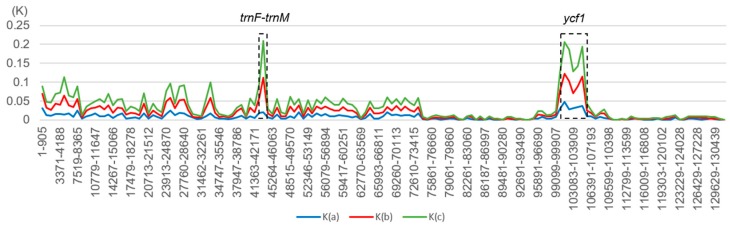
The complete chloroplast of the three chloroplast genomes were compared with that of M. cochinchinensis as a reference using the mVISTA program. As shown in Figure 6, the ycf1 and ccsA genes were found to be the most mutant genes. Except for these genes, the other genes were found to be highly conserved, and most of them showed similarities of >90%. The variations in the coding regions were smaller than those in the noncoding regions. Amongst the three chloroplast genomes, the most divergent regions were found to be localized in the intergenic spacers such as trnF-trnM. The rRNA genes of the three species were highly conservative, and almost no variations were observed. The K values (sequence divergence between species) were calculated, and the sliding windows of the K values were constructed by the DnaSP [35] (Figure 7). Figure 7 shows that the sequence divergence between M. tricolor and M. cochinchinensis was much higher than the other two K values. M. bibracteolatus and M. tricolor showed a small divergence (K < 0.05). The LSC and SSC regions were more divergent than IRs. Two mutational hotspots were found with high K values, and they were located at the LSC and SSC regions. Combined with genes location and the mVISTA result, the two hotspots were found to be trnF-trnM and ycf1.
Figure 6.
Sequence identity plot comparing the three chloroplast genomes with M. cochinchinensis as a reference by using mVISTA. Grey arrows and thick black lines above the alignment indicate genes with their orientation and the position of their IRs, respectively. A cut-off of 70% identity was used for the plots, and the Y-scale represents the percent identity ranging from 50% to 100%.
Figure 7.
Sliding window analyses of the three whole chloroplast genomes. X-axis: position of a window. Y-axis: sequence divergence between species of each window. K(a): K values between M. bibracteolatus and M. tricolor; K(b): K values between M. bibracteolatus and M. cochinchinensis; K(c): K values between M. tricolor and M. cochinchinensis.
2.6. Phylogenetic Analyses
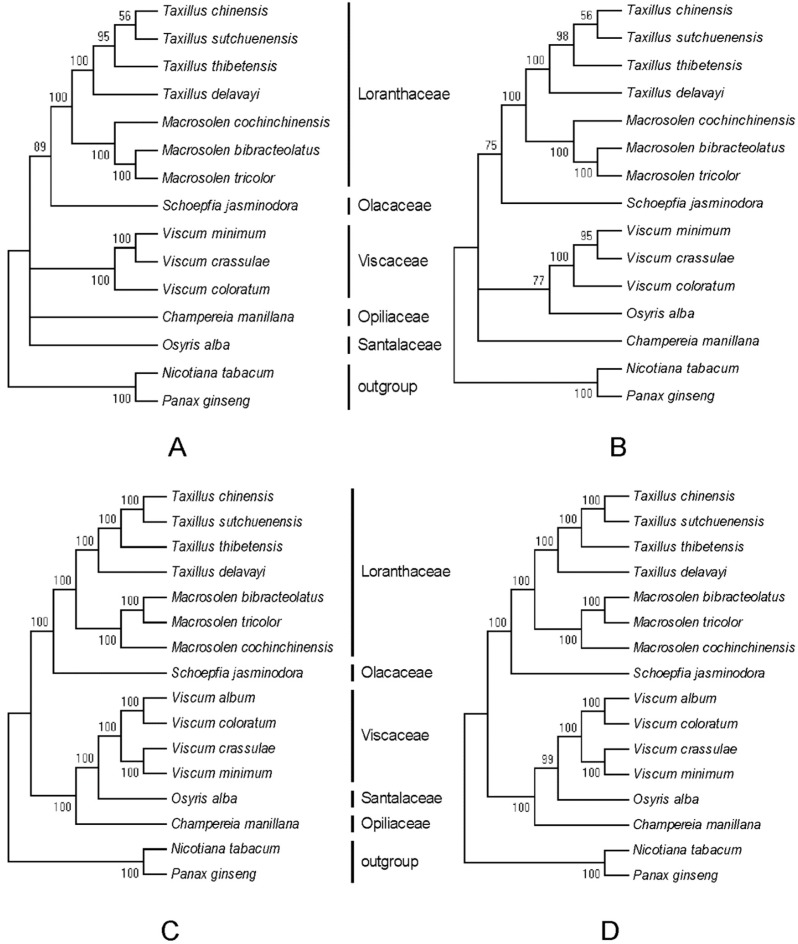
To analyze the phylogenetic relationships of Macrosolen in Santalales, we constructed phylogenetic trees using 58 common protein-coding genes of 16 species and matK genes of 15 species by the MP and ML methods with a bootstrap of 1000 repetitions. The MP and ML trees were the same whether they were constructed by either common protein-coding genes or matK genes (Figure 8). All nodes in all the phylogenetic trees received a >50% bootstrap value. All four phylogenetic trees showed that the three Macrosolen species are sister taxa with respect to S. jasminodora (Olacaceae). M. cochinchinensis, M. tricolor and M. bibracteolatus were gathered into one branch with a well-supported bootstrap value (100%). The three species within the genus Viscum grouped with Osyris alba (Santalaceae) and all Santalales species were clustered within a lineage distinct from the outgroup. As shown in Figure 8, the trees constructed by common protein-coding genes also received a higher bootstrap value than the trees constructed by the matK genes.
Figure 8.
Phylogenetic trees constructed with the matK genes of 15 species by using the maximum parsimony (MP) (A) and maximum likelihood (ML) (B) methods. Phylogenetic trees constructed with 58 common protein-coding genes of 16 species using the MP (C) and ML (D) methods. Numbers at nodes are bootstrap values.
3. Discussion
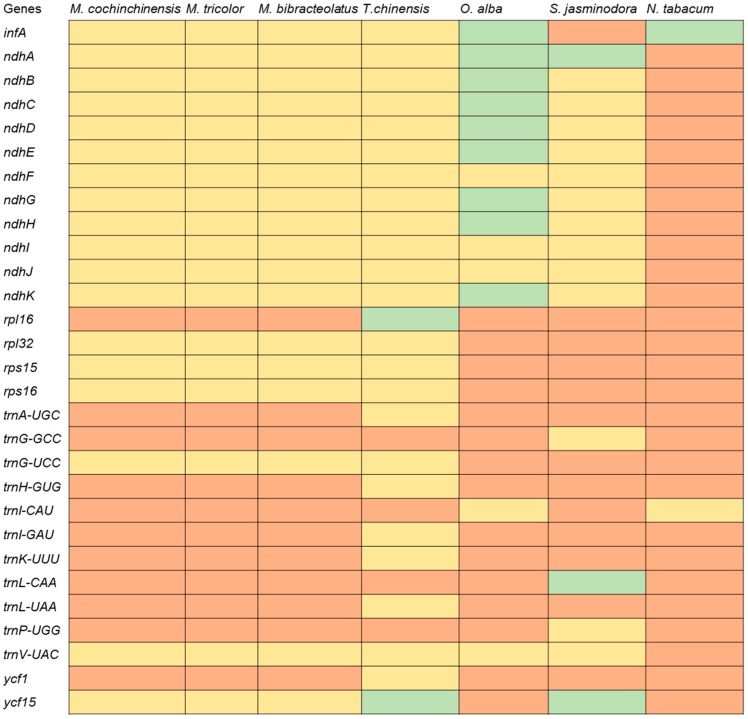
Numerous variations occur in the chloroplast genomes of parasitic plants. However, only a small number of plants within Santalales have been studied. In this study, the complete chloroplast genomes of M. cochinchinensis, M. tricolor and M. bibracteolatus from Santalales were assembled, annotated and analyzed. Compared with the chloroplast genomes of the model plant Nicotiana tabacum, all the ndh genes of the chloroplast genomes were lost amongst the three species, and the infA gene, which codes for a translation initiation factor, was also missing in these species. These cases were similar to those of T. chinensis and T. sutchuenensis [7]. The rpl16 and ycf15 genes were lost in the three species, but they were still present in T. chinensis as pseudogenes (Figure 9). However, compared with the results reported by Shin et al. [36], different gene contents of the chloroplast genome were observed in M. cochinchinensis. These studies have shown that M. cochinchinensis contains the exon 1 fragment of the ndhB gene and a fragment of the infA gene, whereas the rpl36 gene is completely lost. However, the rpl36 gene is still present in the chloroplast genome according to our results. M. cochinchinensis has lost the infA gene and all ndh genes. The number of tRNA genes also differed between the two studies. We annotated 35 tRNA genes, but previous studies only obtained 30 tRNA genes. The evolution of the chloroplast genome in parasitic plants, particularly nonphotosynthetic holoparasites, can lead to significantly reconfigured plastomes [21]. The losses of ndh genes are associated with nutritional status or extensive rearrangements of chloroplast structures [37], and they have occurred in the reported chloroplast genomes of parasitic plants [7]. Our study also showed that ndh genes were lost in the transformation from autotrophy to heterotrophy [38].
Figure 9.
Comparison of the chloroplast genome gene content of six parasitic plants and one model plant (Nicotiana tabacum). The common existing genes in the complete chloroplast genome of the seven species are not listed. Red boxes indicate each gene present, and green boxes indicate that each gene is considered as a pseudogene. The yellow boxes indicate an absent gene.
The Santalales order consists of a small number of autotrophic species and a large number of parasitic species which are root or aerial (stem) parasites [39]. According to the Engler system, Santalales consists of seven families. We downloaded five families belonging to Santalales, which were available in the National Center for Biotechnology Information (NCBI) at that time, and two species as outgroups to analyze the phylogenetic relationships of Macrosolen in Santalales. The present study showed that Loranthaceae is closely related to Olacaceae, whereas Viscaceae is closely related to Santalaceae and Opiliaceae. These results are similar to those of previous studies [13,14]. All the phylogenetic results strongly support that Loranthaceae and Viscaceae diverged independently from each other.
As folk medicine in China, M. cochinchinensis, M. tricolor and M. bibracteolatus have been used to treat diseases for a long time, and their dried stems and branches with leaves are used as medicinal parts. However, Macrosolen species are similar in appearance, especially when they are processed into medicinal slices, thereby causing difficulty in their identification. The identification of parasitic medicinal materials has rarely been reported. Though phytochemical approaches have played an important role in species identification [26], they are inadequate because they are limited to the environment and harvest period. Molecular characterization has shown an improved specificity for plants [23,26]. In our study, mutational hotspots such as the ycf1 gene, the ccsA gene and the trnF-trnM intergenic region are potential sites for identification of Macrosolen species.
4. Materials and Methods
4.1. Plant Materials
All the samples in this study were collected from the Guangxi Province of China. Fresh leaves of M. cochinchinensis and M. tricolor were collected from Qinzhou city, and fresh leaves of M. bibracteolatus were collected from Chongzuo city. The three samples were identified by Yonghua Li, who is from the College of Pharmacy, Guangxi University of Traditional Chinese Medicine. The collected fresh leaves were stored in a −80 °C refrigerator until use.
4.2. DNA Extraction, Sequencing and Assembly
All the methods in this article were based on the methods of Zhou et al. [40]. Total genomic DNA was extracted from samples using the DNeasy Plant Mini Kit with a standard protocol (Qiagen Co., Hilden, Germany). The DNA was sequenced according to the manufacturer’s manual for the Illumina Hiseq X. Approximately 6.2 Gb of raw data from M. cochinchinensis, 6.5 Gb of raw data from M. tricolor, and 6.3 Gb of raw data from M. bibracteolatus were generated with 150 bp paired-end read lengths. The software Trimmomatic (version 0.39, Institute for Biology, Aachen, German) [41] was used to filter the low-quality reads of the raw data, and the Q value was defined as Sanger. Then, all the clean reads were mapped to the database on the basis of their coverage and similarity. Burrows–Wheeler Aligner (BWA-MEM, Wellcome Trust Sanger Institute, Wellcome Genome Campus, Cambridge, UK) was used in chloroplast genome assembly to generate the bam files. The depth was calculated using Samtools (Medical Population Genetics Program, Broad Institute, Cambridge, MA, USA) and plotted using Rscript (with the smoothScatter function). The accuracy of the assembly of the four boundaries (SSC, LSC and IR regions) of the chloroplast sequences was confirmed through PCR and Sanger sequencing using the validated primers listed in Table S5. The assembled complete chloroplast genome sequence of M. cochinchinensis, M. tricolor and M. bibracteolatus were submitted to the NCBI, and the accession numbers were MH161424, MH161425 and MH161423, respectively. The raw data of three species were submitted to the NCBI. The Bioproject ID of this study is PRJNA587349. The SRA accession ID of M. tricolor is SRR10442639, that of M. bibracteolatus is SRR10442640, and that of M. cochinchinensis is SRR10442641.
4.3. Genome Comparison and Phylogenetic Analyses
The whole-genome alignment for the chloroplast genomes of three Macrosolen species were performed and plotted using the mVISTA program (http://genome.lbl.gov/vista/mvista/submit.shtml) [42]. Gene content comparison was analyzed by CPGAVAS2 (Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) [43] and identified by manual correction. To determine the phylogenetic positions of three Macrosolen species within Santalales, we analyzed the chloroplast genomes of 16 species, encompassing 11 other taxa within this lineage, Viscum album (KT003925), V. coloratu (NC_035414), V. crassula (KT070881), V. minimum (KJ512176), Osyris alba (KT070882), Schoepfia jasminodora (KX775962), Champereia manillana (NC_034931), T. chinensis (KY996492), T. sutchuenensis (KY996493), T. delavayi (MH161426), and T. thibetensis (MH161427). The chloroplast genomes of Panax ginseng (AY582139) and N. tabacum (NC_001879) were used as outgroups.
4.4. Other Analyses
On the basis of the study of Zhou et al. [40], we analyzed the complete chloroplast genome of three Macrosolen species, including genome structure analyses (genome length, gene content and GC content), codon usage analyses, RNA editing site prediction, and repeat sequences analyses. The distribution of codon usage was investigated using the CodonW software (University of Texas, Houston, TX, USA) with the RSCU ratio [32]. Potential RNA editing sites were predicted using the Predictive RNA Editor for Plants (PREP-Cp, Center for Plant Science Innovation, University of Nebraska-Lincoln, Lincoln, NE, USA) suite online program [44] with a cutoff value of 0.8. Simple sequence repeats were detected using the MISA software (Pgrc.ipk-gatersleben.de/misa/) [45]. Repeat sequences were identified by REPuter (University of Bielefeld, Bielefeld, Germany) [46].
Abbreviations
| LSC | Large single copy |
| SSC | Small single copy |
| IR | Inverted repeat |
| MP | Maximum parsimony |
| ML | Maximum likelihood |
| RSCU | Relative synonymous codon usage |
| SSR | Simple Sequence Repeats |
| NCBI | National Center for Biotechnology Information |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/22/5812/s1.
Author Contributions
Conceptualization, H.Y. and Y.L.; methodology, L.N., L.W. and J.Z.; formal analyses, L.N., Y.C. and J.Z.; resources, Y.L. and Y.W.; data curation, L.N., Y.C., Z.X. and X.L.; writing—original draft preparation, L.N. and H.Y.; writing—review and editing, L.N. and H.Y.; funding acquisition, H.Y. and Y.L.
Funding
This research was funded by Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2018ZX09711001-008-007), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS) (No. 2016-I2M-3-016) and Guangxi Natural Science Foundation (No. 2013GXNSFAA019120).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wicke S., Naumann J. Molecular evolution of plastid genomes in parasitic flowering plants. Adv. Bot. Res. 2018;85:315–347. [Google Scholar]
- 2.Wang L., Dong W., Zhou S. Structural mutations and reorganizations in chloroplast genomes of flowering plants. Acta Bot. Boreali Occident. Sin. 2012;32:1282–1288. [Google Scholar]
- 3.Huang X., Guan K., Li A. Biological traits and their ecological significances of parasitic plants: A review. Chin. J. Ecol. 2011;30:1838–1844. [Google Scholar]
- 4.Wolfe K.H., Morden C.W., Palmer J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen G., Cuenca A., Seberg O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 2015;7:2520–2532. doi: 10.1093/gbe/evv165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H., Hu J. The complete chloroplast genome of hemiparasitic flowering plant Schoepfia jasminodora. Mitochondrial DNA Part B. 2016;1:767–769. doi: 10.1080/23802359.2016.1238753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Zhou J.G., Chen X.L., Cui Y.X., Xu Z.C., Li Y.H., Song J.Y., Duan B.Z., Yao H. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci. Rep. 2017;7:12834. doi: 10.1038/s41598-017-13401-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molina J., Hazzouri K.M., Nickrent D., Geisler M., Meyer R.S., Pentony M.M., Flowers J.M., Pelser P., Barcelona J., Inovejas S.A., et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol. Biol. Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Zhang T.C., Qiao Q., Ren Z., Zhao J., Yonezawa T., Hasegawa M., Crabbe M.J., Li J., Zhong Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae) PLoS ONE. 2013;8:e58747. doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frailey D.C., Chaluvadi S.R., Vaughn J.N., Coatney C.G., Bennetzen J.L. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol. 2018;18:30. doi: 10.1186/s12870-018-1249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samigullin T.H., Logacheva M.D., Penin A.A., Vallejo-Roman C.M. Complete plastid genome of the recent holoparasite Lathraea squamaria reveals earliest stages of plastome reduction in orobanchaceae. PLoS ONE. 2016;11:e0150718. doi: 10.1371/journal.pone.0150718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su H.J., Barkman T.J., Hao W., Jones S.S., Naumann J., Skippington E., Wafula E.K., Hu J.M., Palmer J.D., de Pamphilis C.W. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc. Natl. Acad. Sci. USA. 2019;116:934–943. doi: 10.1073/pnas.1816822116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellot S., Renner S.S. The plastomes of two species in the endoparasite genus pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol. Evol. 2016;8:189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen G., Zervas A., Pedersen H., Seberg O. Contracted genes and dwarfed plastome in mycoheterotrophic Sciaphila thaidanica (Triuridaceae, Pandanales) Genome Biol. Evol. 2018;10:976–981. doi: 10.1093/gbe/evy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schelkunov M.I., Nuraliev M.S., Logacheva M.D. Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known. Peer J. 2019;7:e7500. doi: 10.7717/peerj.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham S.W., Lam V.K., Merckx V.S. Plastomes on the edge: The evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017;214:48. doi: 10.1111/nph.14398. [DOI] [PubMed] [Google Scholar]
- 17.Lallemand F., Logacheva M., Le Clainche I., Berard A., Zheleznaia E., May M., Jakalski M., Delannoy E., Le Paslier M.C., Selosse M.A. Thirteen new plastid genomes from mixotrophic and autotrophic species provide insights into heterotrophy evolution in Neottieae orchids. Genome Biol. Evol. 2019;11:2457–2467. doi: 10.1093/gbe/evz170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clegg M.T., Gaut B.S., Learn G.H., Jr., Morton B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyall S.D., Brown M.T., Johnson P.J. Ancient invasions: From endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 20.Palmer J.D. Comparative organization of chloroplast genomes. Annu. Rev. Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 21.Wicke S., Schneeweiss G.M., dePamphilis C.W., Müller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell W., Morgante M., Andre C., Hanafey M., Vogel J., Tingey S., Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996;2:225–238. doi: 10.1007/BF00564200. [DOI] [Google Scholar]
- 23.Niu Z., Zhu S., Pan J., Li L., Sun J., Ding X. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017;7:2073. doi: 10.1038/s41598-017-02252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y., Nie J., Xiao L., Hu Z., Wang B. Comparative chloroplast genome analysis of rhubarb botanical origins and the development of specific identification markers. Molecules. 2018;23:2811. doi: 10.3390/molecules23112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Zhou J., Cui Y., Wang Y., Duan B., Yao H. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front. Pharmacol. 2018;9:695. doi: 10.3389/fphar.2018.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu Z., Pan J., Xue Q., Zhu S., Liu W., Ding X. Plastome-wide comparison reveals new SNV resources for the authentication of Dendrobium huoshanense and its corresponding medicinal slice (Huoshan Fengdou) Acta Pharm. Sin. B. 2018;8:466–477. doi: 10.1016/j.apsb.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M., Li Q., Hu Z., Li X., Chen S. The complete Amomum kravanh chloroplast genome sequence and phylogenetic analysis of the commelinids. Molecules. 2017;22:1875. doi: 10.3390/molecules22111875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flora Reipublicae Popularis Sinicae (FRPS) [(accessed on 28 October 2019)]; Available online: http://www.iplant.cn/
- 29.Gong Z., Wang Z., Xu L., Xu G., Wu J. Studies on medicinal plants of Loranthaceae in China. Chin. Wild Plant Resour. 1996;1:11–15. [Google Scholar]
- 30.Zhao Q., Xu Q., Zhang H. Pharmacognostic identification on crude drug of Macrosolen cochinchinensis. Chin. J. Ethnomed. Ethnopharm. 1998;34:1–3. [Google Scholar]
- 31.Li Y., Lu D., Zhao M., Zhu K. Research on the developments and applications for medicinal plants of Loranthaceae in Guangxi. Guangxi Med. J. 2006;28:1695–1698. [Google Scholar]
- 32.Sharp P.M., Li W.H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell W., Morgante M., Mcdevitt R., Vendramin G.G., Rafalski J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akkaya S.M. Length polymorphism of simple sequence repeat DNA in soybean. Genetics. 1992;132:1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 36.Shin H.W., Lee N.S. Understanding plastome evolution in Hemiparasitic Santalales: Complete chloroplast genomes of three species, Dendrotrophe varians, Helixanthera parasitica, and Macrosolen cochinchinensis. PLoS ONE. 2018;13:e0200293. doi: 10.1371/journal.pone.0200293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.T., Kim J.S., Moore M.J., Neubig K.M., Williams N.H., Whitten W.M., Kim J.H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE. 2015;10:e0142215. doi: 10.1371/journal.pone.0142215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mcneal J.R., Kuehl J.V., Boore J.L., Pamphilis C.W.D. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57. doi: 10.1186/1471-2229-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Der J.P., Nickrent D.L. A molecular phylogeny of Santalaceae (Santalales) Syst. Bot. 2008;33:107–116. doi: 10.1600/036364408783887438. [DOI] [Google Scholar]
- 40.Zhou J., Chen X., Cui Y., Sun W., Li Y., Wang Y., Song J., Yao H. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species. Int. J. Mol. Sci. 2017;18:1839. doi: 10.3390/ijms18091839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frazer K.A., Lior P., Alexander P., Rubin E.M., Inna D. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.CPGAVAS2. [(accessed on 30 July 2019)]; Available online: http://47.96.249.172:16018/analyzer/extractSeq.
- 44.Mower J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37:253–259. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misa-Microsatellite Identification Tool. [(accessed on 2 June 2017)]; Available online: Pgrc.ipk-gatersleben.de/misa/
- 46.Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.