Abstract
The transcriptional regulator BRD4 has been shown to be important for the expression of several oncogenes including MYC. Inhibiting of BRD4 has broad antiproliferative activity in different cancer cell types. The small molecule JQ1 blocks the interaction of BRD4 with acetylated histones leading to transcriptional modulation. Depleting BRD4 via engineered bifunctional small molecules named PROTACs (proteolysis targeting chimeras) represents the next-generation approach to JQ1-mediated BRD4 inhibition. PROTACs trigger BRD4 for proteasomale degradation by recruiting E3 ligases. The aim of this study was therefore to validate the importance of BRD4 as a relevant target in colorectal cancer (CRC) cells and to compare the efficacy of BRD4 inhibition with BRD4 degradation on downregulating MYC expression. JQ1 induced a downregulation of both MYC mRNA and MYC protein associated with an antiproliferative phenotype in CRC cells. dBET1 and MZ1 induced degradation of BRD4 followed by a reduction in MYC expression and CRC cell proliferation. In SW480 cells, where dBET1 failed, we found significantly lower levels of the E3 ligase cereblon, which is essential for dBET1-induced BRD4 degradation. To gain mechanistic insight into the unresponsiveness to dBET1, we generated dBET1-resistant LS174t cells and found a strong downregulation of cereblon protein. These findings suggest that inhibition of BRD4 by JQ1 and degradation of BRD4 by dBET1 and MZ1 are powerful tools for reducing MYC expression and CRC cell proliferation. In addition, downregulation of cereblon may be an important mechanism for developing dBET1 resistance, which can be evaded by incubating dBET1-resistant cells with JQ1 or MZ1.
Introduction
Colorectal cancer (CRC) is the third most common cancer type worldwide and is responsible for one million new cases and 500,000 deaths per year [1]. Most CRC develops in a multi-step manner from premalignant precursor lesions after the accumulation of different mutations via the adenoma-carcinoma sequence [2].
Genetic alterations frequently observed in CRC affect the Wnt signaling pathway as one of the key signal transduction pathways regulating growth and development [3]. The proto-oncogene and transcription factor MYC, an important Wnt-target gene, is essential in normal non-transformed cells, where it regulates cell proliferation, metabolism and survival [4]. The oncogene function of MYC causes hyperproliferation, cell-cycle progression and metastasis [5]. Virtually all CRC shows elevated MYC levels and deregulation of MYC target genes. In addition, genesis and progression of CRC is dependent on a constitutively active Wnt pathway and aberrant MYC expression [6], [7]. The addiction of CRC to high-level expression of MYC provides a rationale for its therapeutic targeting [8], [9], [10]. The MYC protein has a leucine zipper and helix-loop-helix motif essential for dimerization and DNA binding. This prevents an effective approach to direct inhibition of MYC [11]. An alternative approaches is indirect inhibition of MYC on transcriptional regulation or modulation of its stability and activity [12], [13], [14].
Bromodomain-containing protein 4 (BRD4) is a member of the BET (bromodomain and extra terminal domain) family of transcriptional regulatory proteins. BRD4 recognizes acetylated lysine residues on histones with its bromodomains and recruits transcriptional regulatory complexes to acetylated chromatin [15]. Because the transcription of the MYC oncogene is dependent on BRD4 [11], [16], different small molecule inhibitors have recently been developed such as JQ1. This cell-permeable small molecule inhibitor occupies the bromodomain pockets of BRD4 thereby preventing binding to acetylated histones and selectively repressing the transcription of MYC oncogene and MYC-dependent genes [17], [18]. Therefore, targeting BRD4 represents a promising therapeutic strategy against CRC.
Beyond inhibition of BRD4 with small-molecule drugs, the next-generation approach is to target its degradation. The advantage of BRD4 degradation instead of inhibition is that it may lead to more potent suppression of MYC oncogene and MYC-dependent genes. Hetero-bifunctional small molecules named proteolysis targeting chimeras or PROTACs with different BET-binding motives have been generated to target specific proteins for selective degradation [19], [20]. PROTACs for mediating BRD4 degradation consist of the bromodomain binder JQ1 with an appended ligand for recruiting an E3 ubiquitin ligase targeting BRD4 for proteasomale degradation [21]. The PROTAC dBET1 contains thalidomide as a binder for the E3 ubiquitin ligase cereblon [21] and the second PROTAC MZ1 contains a binder for the E3 ubiquitin ligase von Hippel-Lindau tumor suppressor (VHL) [22]. Different studies in vivo have shown JQ1 to be an attractive candidate for the selective repression of the MYC oncogene with growth suppression of different solid und hematologic tumors [23]. Data on the anti-tumor activity of dBET1 and MZ1 are presently available for a few hematologic cancer cell lines but not for CRC [23].
Here, we showed that JQ1-mediated inhibition of BRD4 represses expression of MYC mRNA and MYC protein in CRC cells associated with an antiproliferative phenotype. The PROTACs dBET1 and MZ1 demonstrated comparable repressive effects on MYC expression and cell proliferation. Our results underline BRD4 as an important druggable target and the BRD4-targeting small molecules JQ1, dBET1, and MZ1 as promising tools against CRC cells. In addition, we evaluated a potential mechanism of acquired resistance to dBET1 by loss of the E3 ligase cereblon. Our results underline for CRC cells that resistance to dBET1 does not affect the cell response to JQ1 or MZ1, respectively.
Material and methods
Cell culture
Caco2, COLO320, DLD1, and LS174t cells were cultured in RPMI 1640; HCT116 p53+/+, HCT116 p53−/−, and HT29 cells were cultured in McCoy’s; SW480 cells were cultured in DMEM/HamsF12. These cell lines were obtained from American Type Culture Collection, USA (www.atcc.org), Cell Lines Services GmbH, Germany (https://clsgmbh.de), German Collection of Microorganisms and Cell Cultures, Leibniz Institute, Germany (www.dsmz.de). Human dermal fibroblasts (#C-12302) were obtained from Promo Cell, Germany. All media was supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS), 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM glutamine, 50 mM mercaptoethanol, and 1% non-essential amino acids (Invitrogen Life Technologies GmbH, Germany). The cultures were maintained in a humidified atmosphere with 5% CO2 at 37 °C (standard incubator conditions). Cells were validated by short-tandem repeat genotyping (https://clsgmbh.de) prior to starting the experiments. A test for mycoplasma contamination was performed (Minerva Biolabs GmbH). Cells were used for less than 15 passages after revitalization.
Patient-derived intestinal organoid culture
Isolation and culture of patient-derived organoids (T5) was approved by the ethic committee of the University of Würzburg (#142/16-ge). Organoid isolation and culture were performed as described elsewhere [24].
Reagents
JQ1 (Sigma Aldrich), dBET1 (Cayman Chemicals), and MZ1 (Tocris Bioscience) were prepared as stock 2 mM solutions in 100% dimethyl sulfoxide (DMSO; Sigma Aldrich, USA) and aliquots were stored at −20 °C. Additional dilution steps were made with phosphate buffered saline (PBS) or culture medium. Thalidomide and MG132 (both Sigma Aldrich) were prepared as 100 mM (thalidomide) and 10 mM (MG132) stock solution with DMSO, respectively.
Cell proliferation and colony formation assays
To assess cell proliferation, exponentially growing cells (5x103 cells/well in 200 µl culture medium) were cultured in 96-well flat-bottom tissue plates (Greiner bio-one, Germany) and treated the next day with reagents at the indicated concentrations for 72 h under standard incubator conditions. The amount of viable cells was determined by crystal violet (CV) staining (0.3% (w/v) CV in 25% (v/v) methanol) after fixation with methanol. The solubilized crystal violet (with 10% (v/v) acetic acid) was measured by absorbance at 570 nm. To determine colony formation ability, cells (1-2x103) were plated on six well plates. After 48 h, cells were treated with the indicated concentrations of JQ1, dBET1 or MZ1 for 3–5 days. Cells were fixed with methanol and stained with crystal violet.
Quantitative PCR
Total RNA was extracted from 106 cells with Trizol (Invitrogen Life Technologies, Germany), quantified (Nanodrop ND-1000, Thermo Fisher Scientific), and assessed for RNA integrity (Experion, Bio-Rad Laboratories). RNA (0.7 µg) was reverse transcribed using the iScriptcDNA synthesis kit from BioRad (25 °C for 5 min, 42 °C for 30 min, 85 °C for 5 min). qPCR was performed with MESA Green qPCRMasterMix Kit containing Meteor Taq hotstart polymerase (Eurogentic GmbH) on a CFX96 real-time PCR system (Bio-Rad) operated by CFX Manager Software (version 3.0). The cycler protocol was 5 min at 95 °C, 40 cycles of 15 sec at 95 °C, 60 sec at 60 °C, and 5 min at 72 °C. Post-amplification melting curves were controlled to exclude primer-dimer artifacts and contaminations. The reference genes PPIA (peptidylprolyl isomerase A) and ACTB (β-actin) were used for normalization and mRNA expression levels of MYC and BRD4 were calculated with the ΔΔ Cq method. PPIA (PubMed ID: NM_021130.3), forward: GCTGGACCCAACACAAATGG, reverse: CAAACACCACATGCTTGCCA (82 bp); ACTB (NM_001101), forward: CCTTGCCATCCTAAAAGCC, reverse: CACGAAAGCAATGCTATCAC (96 bp); MYC (NM_002467.4), forward: CACCAGCAGCGACTCTGA, reverse: GATCCAGACTCTGACCTTTTG (102 bp); BRD4 (NM_058243.2), forward: CTGACAGCGAAGACTCCGAAA, reverse: GGTGATGATGGTGCTTCTTCT (90 bp).
RNA sequencing
For global gene expression analysis, total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen) with on-column digestion of DNA. Poly-A+ RNA was purified from total RNA with the NEBNextPoly(A) mRNA Magnetic Isolation Module (NEB). RNA sequencing libraries were prepared using the NEB Next Ultra RNA Library Prep Kit for Illumina (NEB) according to manufacturer’s instructions. Library quality and concentration were determined using the Experion chip electrophoresis system (Bio-Rad). Libraries were sequenced on an Illumina Genome Analyzer IIx following to the manufacturer’s instructions.
Immunoblot
One million cells each were lysed in pre-cooled RIPA buffer (Pierce, Thermo Fisher Scientific) containing phosphatase and proteinase inhibitors and 2.5 mmol/l dithiothreitol (Sigma-Aldrich). Equal amounts of proteins (15 µg) were loaded on a 10% polyacrylamide gel (SDS-PAGE), electrophoresed and then blotted by semi-dry transfer onto a nitrocellulose membrane (Schleicher & Schuell). After a blocking step with 5% non-fat milk (Merck KGaA) for 1 h, the membranes were incubated with the indicated antibodies overnight at 4 °C. After washing with PBS, membranes were incubated with 1:10,000 diluted horseradish peroxidaseconjugated goat anti-rabbit or rabbit anti-mouse (both antibodies from Dako, Denmark) for 60 min at room temperature. The peroxidase-conjugated monoclonal mouse antibodies anti-GAPDH (clone GAPDH-71.1) and anti-β-actin (clone AC-15) were used as loading control (at 1:10,000 dilution, Sigma-Aldrich). lmmunoblots were visualized by highly sensitive chemiluminescent detection reagent (Amersham, GE Healthcare, UK). All primary antibodies were used at a 1:1,000 dilution in TBST (5 mM TRIS, 15 mM NaCl, 0.1% Tween 20, pH 7.5) with 5% non-fat milk (Merck KGaA, Germany) for western blots. Anti-BRD4 (E2A7X), anti-cereblon (D8H3S), and anti-VHL (#68547) were purchased from Cell Signaling Technology and anti-MCY (Y69) were purchased from Abcam.
Immunofluorescence
LS174t cells (2x104 cells per chamber) were seeded onto 12-chamber culture slides (ibidi GmbH, Germany) and treated the next day with 1 µM dBET1 or MZ1 for 2 h. Cells were fixed with 100% methanol (5 min), permeabilized with 0.1% Triton X-100 for 5 min and blocked with 5% BSA/0.1% Tween-20 in PBS for 1 h. For visualization, cells were stained with anti-Brd4 antibody (EPR5150) conjugated with Alexa Fluor594 (Abcam) for 1 h at a 1:100 dilution. Nuclear DNA was labelled with DAPI (shown in blue). Images were taken with a confocal laser scanning microscope (Zeiss LSM 780).
siRNA transfection
VHL siRNA (ID: 138745), MYC siRNA (ID: 106821) and silencer negative control siRNA (AM4611) were purchased from Ambion, ThermoFisher. Lipofectamine reagent (life technology) was used for transfections according to the manufactureŕs instructions. Cells were harvested 72 h after transfection.
Statistical analysis
Experiments were performed at least three times with replicate samples. Data are plotted as means ± SD (standard deviation). The means were compared using analysis of variance (ANOVA) plus Bonferroni’s t-test. A P-value of < 0.05 indicated a statistically significant result (*p < 0.05, **p < 0.01, ***p < 0.001).
Results
JQ1-mediated inhibition of BRD4 function blocks transcription of MYC and MYC-related gene sets in CRC cells
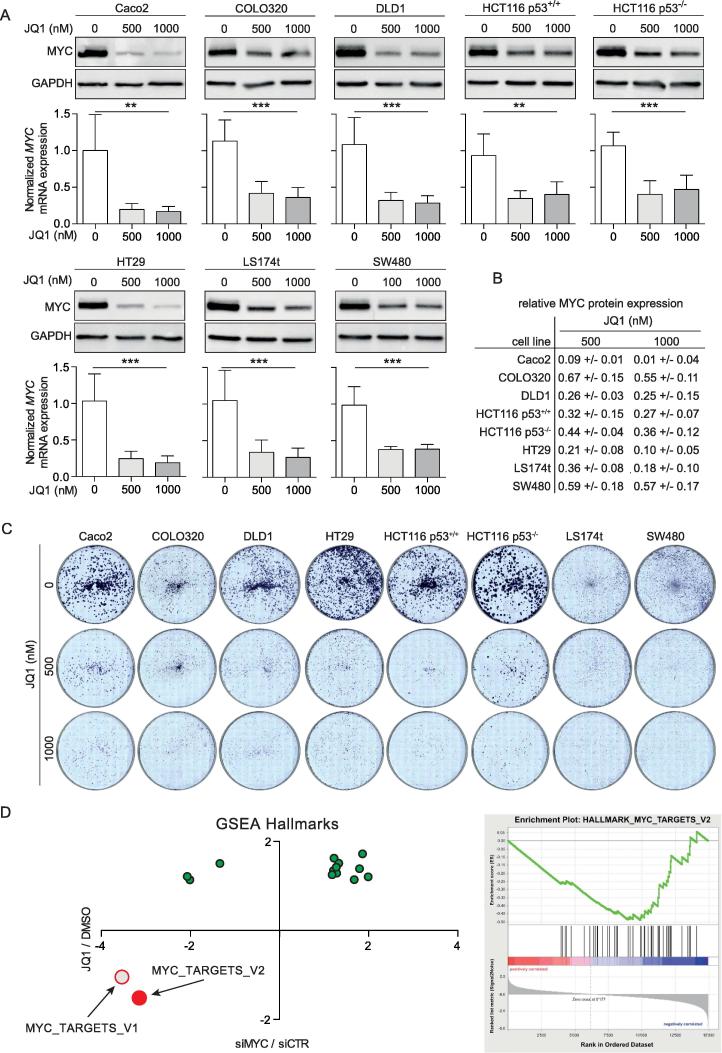
To determine the effect of BRD4 inhibition by JQ1 on MYC expression in CRC cells, we treated eight human CRC cell lines harboring common mutations [25] with JQ1. After 24 h of JQ1 incubation, a clear reduction in cellular MYC mRNA and MYC protein expression was obvious compared to untreated control cells. The level of MYC mRNA reduction ranged between 50 and 75% and the level of MYC protein declined by more than 50% in six of the eight CRC cell lines, and by 55% and 57% in COLO320 and SW480 cells, respectively (Figure 1A, B). In addition, treatment with JQ1 dramatically reduced colony formation in all eight CRC cell lines (Figure 1C).
Figure 1.
JQ1 suppresses expression of MYC and MYC related gene sets with restrained cell proliferation. (A) Western blot of MYC protein (top) and RT-qPCR (bottom) of MYC mRNA expression in eight CRC cell lines treated with 500 and 1000 nM JQ1 for 24 h. Values of relative MYC mRNA expression represent mean ± SD normalized to DMSO-treated control cells. If not otherwise stated, data are representative of 3 independent experiments. (B) Summary of quantified MYC protein levels after JQ1-treatment normalized to corresponding GAPDH control. Data is mean ± SD of 3 independent experiments. (C) Blocking CRC cell growth by 500 nM and 1000 nM JQ1 in crystal violet colony formation assays. Shown are representative images. (D) RNA sequencing and subsequent gene set enrichment analysis (GSEA) of LS174t cells treated with siRNA targeting MYC or 500 nM JQ1. Left panel, GSEA of MYC target genes after 48 h of JQ1 treatment. Right panel, Normalized Enrichment Score (NES; a measure for misregulation) for “hallmark” gene sets that are significantly affected by both MYC depletion and JQ1 treatment (MYC_TARGETS_v2; FDR < 0.25). The set MYC_TARGETS_v1 is significantly repressed by MYC depletion. siMYC: MYC specific siRNA, siCTR: negative control siRNA.
To gain insight into the transcriptional effect of JQ1, LS174t cells were incubated with 500 nM JQ1 for 48 h and total RNA were compared with LS174t cells treated with MYC targeting siRNA. RNA sequencing with subsequent gene set enrichment analysis (GSEA) showed that JQ1 treatment and MYC depletion had qualitatively similar effects on the majority of “hallmark” gene sets, notably on the set MYC-TARGETS_V2 (Figure 1D shows all significantly dysregulated gene sets, software.broadinstitute.org/gsea/index.jsp). The effect of JQ1 on another collection of MYC targets (“V1”) was qualitatively similar but did not reach statistical significance (Figure 1D). These results suggest that targeting BRD4 is a valuable approach to suppressing oncogenic MYC expression and function in CRC cells associated with growth arrest.
The BRD4 PROTACs dBET1 and MZ1 efficiently target BRD4 for degradation by recruiting E3 ligases
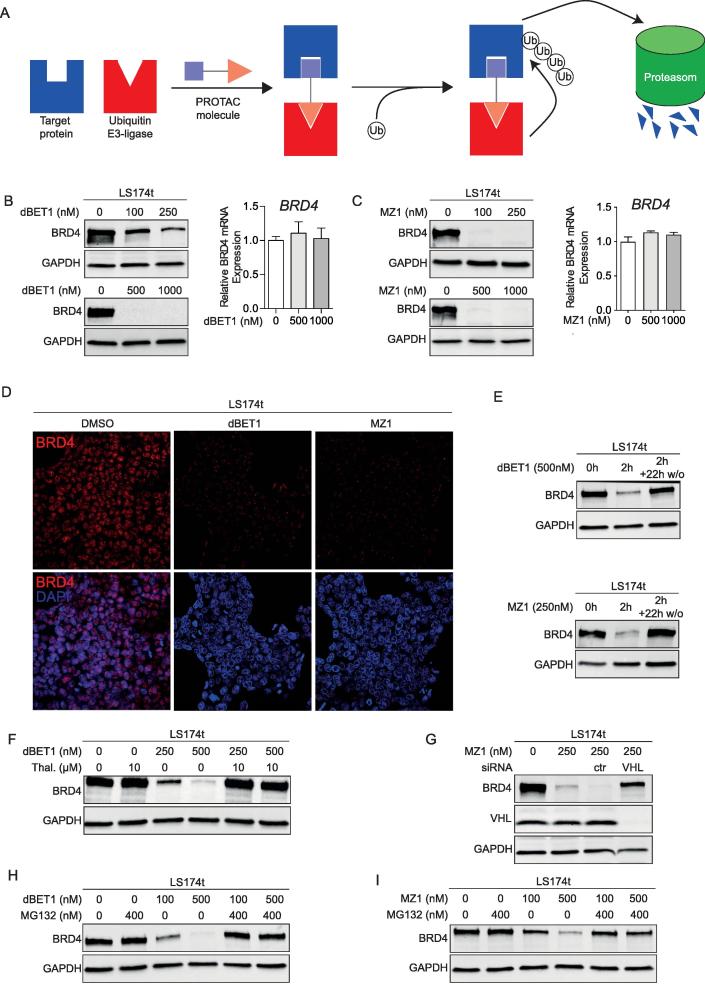
Besides the direct inhibition of BRD4 function with JQ1, PROTAC-mediated degradation of BRD4 by dBET1 or MZ1 provides a next-generation approach for targeting BRD4. Both PROTACs bring BRD4 into close proximity with a specific E3 ligase (cereblon recruiting by dBET1 and VHL recruiting by MZ1) that mediates the ubiquitination of BRD4 with subsequent degradation by the ubiquitin proteasome system (Figure 2A). To investigate the potency of PROTAC-induced BRD4 degradation, LS174t cells were treated with either dBET1 or MZ1 for 24 h. Both PROTACs lead to complete degradation of BRD4 protein, whereas the expression of BRD4 mRNA was not reduced (Figure 2B, C). Notably, both PROTACs demonstrated effective degradation of BRD4 in a concentration-dependent manner, whereas the relevant concentration differs between both. dBET1 and MZ1 directed BRD4 degradation was already apparent within 2 h of incubation (Figure 2D, E). Immunofluorescence staining of BRD4 in LS174t cells treated with dBET1 or MZ1 for 2 h showed a strong reduction in BRD4 signal intensity in the nucleus as well as a clustering in the cytoplasm (Figure 2D). In addition, efficient degradation of BRD4 required the permanent presence of dBET1 or MZ1. The rapid reduction of BRD4 protein induced by dBET1 or MZ1 within 2 h of incubation (Figure 2E) was reversible after removal of PROTACs from cell cultures by changing the culture medium. Within 22 h of removing PROTACs, the physiological level of BRD4 protein recovered (Figure 2E). Shortening the incubation time to 2 h revealed the speed of BRD4 degradation directed by dBET1 and MZ1, although the process of degradation was incomplete within 2 h (Figure 2E) in contrast to 24 h (Figure 2B).
Figure 2.
PROTACs dBET1 and MZ1 promote BRD4 protein degradation in an E3 ligase and proteasome-dependent manner. (A) The function of proteolysis targeting chimera (PROTAC). The bispecific PROTAC molecule bridges E3 ligase and protein of interest to target it for degradation by the ubiquitin proteasome system. (B-C) Western blot of BRD4 protein and RT-qPCR of BRD4 mRNA from LS174t cells treated with dBET1 (B) and MZ1 (C) for 24 h with the indicated concentrations. (D) LS174t cells were treated with 200 nM dBET1 or MZ1 for 2 h and stained with anti-BRD4 antibody. Representative images of immunofluorescence of two independent experiments. (E) Recovery of BRD4 after washout of dBET1 and MZ1. Western blot of BRD4 protein of LS174t cells treated with dBET1 or MZ1 for 2 h and 22 h after washout of dBET1 and MZ1 from cell culture (2 h + 22 h w/o). (F-G) PROTAC-mediated BRD4 degradation requires recruiting of E3 ligase. (F) Cells preincubated with 10 µM thalidomide for 4 h prevent cereblon-mediated BRD4 degradation. (G) Depletion of E3 ligase VHL with specific siRNA rescues cells from MZ1-mediated BRD4 degradation. (H-I) Pretreatment of LS174t cells with proteasome inhibitor MG132 for 4 h rescued BRD4 stability during a 2-hour treatment with dBET1 (H) and MZ1 (I). Shown are representative images of 2–3 independent experiments (E-I).
To validate the requirement for functional E3 ligases recruited by dBET1 and MZ1 for consecutive proteasomal degradation of BRD4, a chemical and genetic editing approach, respectively, was used to hamper recruitment of the E3 ligases cereblon and VHL. Binding of dBET1 to cereblon was blocked by pretreating cells with thalidomide [21], VHL expression was knocked-down using VHL-specific siRNA. BRD4 protein level was not influenced by pretreatment with thalidomide (Figure 2F) and VHL-specific siRNA (not shown). However, pretreatment with thalidomide or VHL-specific siRNA virtually abolished dBET1 or MZ1-induced BRD4 degradation (Figure 2F, G). PROTAC-mediated ubiquitination of BRD4 by PROTAC-recruited E3 ligases requires functional proteasomes. In cells pre-incubated for 4 h with the proteasome inhibitor MG132, BRD4 degradation mediated by dBET1 and MZ1 was rescued (Figure 2H, I). We conclude from these results, that PROTAC-mediated degradation of BRD4 is functional in CRC.
The BRD4 PROTACs dBET1 and MZ1 reduce MYC mRNA and MYC protein levels in CRC cells
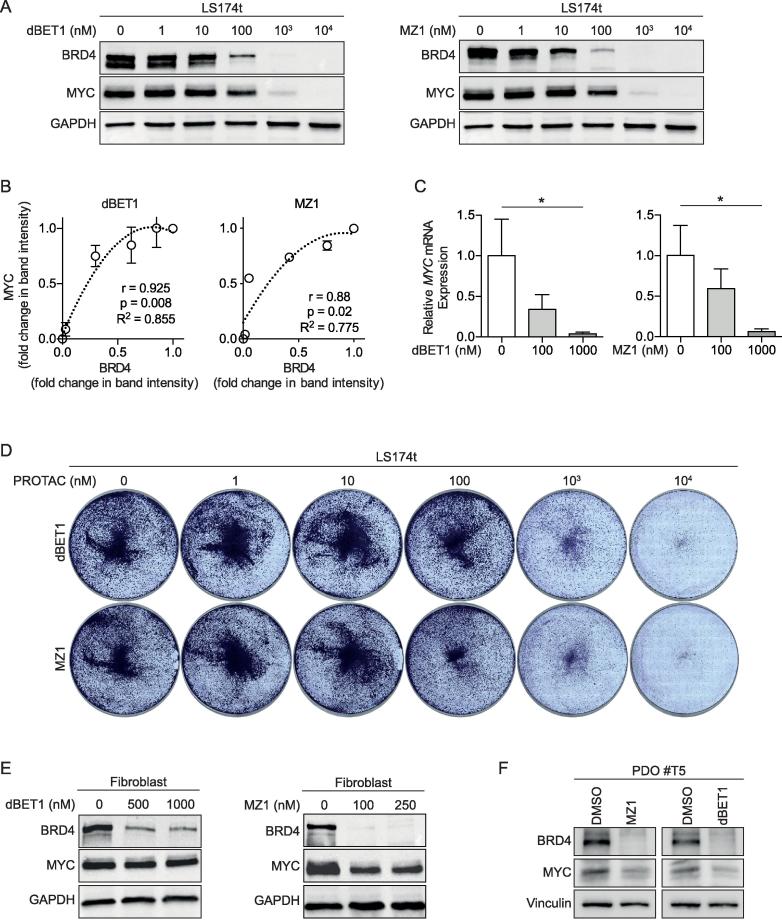
JQ1-mediated inhibition of BRD4 function decreased levels of MYC mRNA and MYC protein (Figure 1A). Next, we analyzed the effectiveness of dBET1 and MZ1 on downregulating MYC expression. For this, LS174t cells were incubated with increasing concentrations (1 nM to 10 µM) of dBET1 or MZ1 for 24 h. BRD4 and MYC protein abundances were evaluated by western blot. At a concentration of 1 µM dBET1 or MZ1, a complete degradation of BRD4 protein and loss of MYC protein was apperent (Figure 3A). The magnitude of MYC protein reduction by the two PROTACs was positively correlated with the rate of BRD4 degradation (Figure 3B). MYC mRNA levels were also reduced, indicating that BRD4 is actively involved in transcriptional regulation of MYC expression (Figure 3C). In correlation with the reduced MYC levels, cell viability was decreased by dBET1 and MZ1, respectively in a dose-dependent manner (Figure 3D). To evaluate the effectiveness of dBET1 and MZ1 for degradation of BRD4 in non-oncogenic cells, we treated normal human dermal fibroblasts with relevant concentration of dBET1 (500/1000 nM) or MZ1 (100/250 nM). Both PROTACs substantially degraded BRD4 in fibroblasts, but strikingly had no or only a weak effect on MYC protein levels (Figure 3E). To test the effect of both PROTACs in a physiological setting we took advantage of a recently established, APC-mutated “Patient Derived Organoid” (POD #T5) [26]. Treatment with relevant concentrations of dBET1 or MZ1 completely abolishes BRD4 expression and significantly reduce MYC levels (Figure 3F). In total this data argues that BRD4 is essential for the translation of oncogenic MYC level but not for physiological levels.
Figure 3.
PROTACs dBET1 and MZ1 target BRD4 for degradation with repression of MYC expression and cell proliferation. (A) Western blot of BRD4 and MYC protein after incubation of LS174t cells with increased concentrations of dBET1 or MZ1 for 24 h. Shown are representative images of two independent experiments. (B) PROTAC-mediated BRD4 degradation correlates with reduction of MYC protein (r: Spearmańs rho coefficient). (C) PROTAC-mediated BRD4 degradation correlates with reduction of MYC mRNA. Shown are results as mean ± SD of three independent experiments. (D) Dose-response effects of dBET1 and MZ1 on LS174t cell growth assessed with crystal violet colony formation assay for a 3-day incubation. Shown are representative images of two independent experiments. (E) Western blot of BRD4 and MYC protein after incubation of fibroblasts with relevant concentrations of dBET1 or MZ1 for 24 h. Representative images of two independent experiments. (F) Western blot of BRD4 and MYC protein after incubation of “Patient Derived Organoid #T5” with relevant concentrations of dBET1 or MZ1 for 24 h.
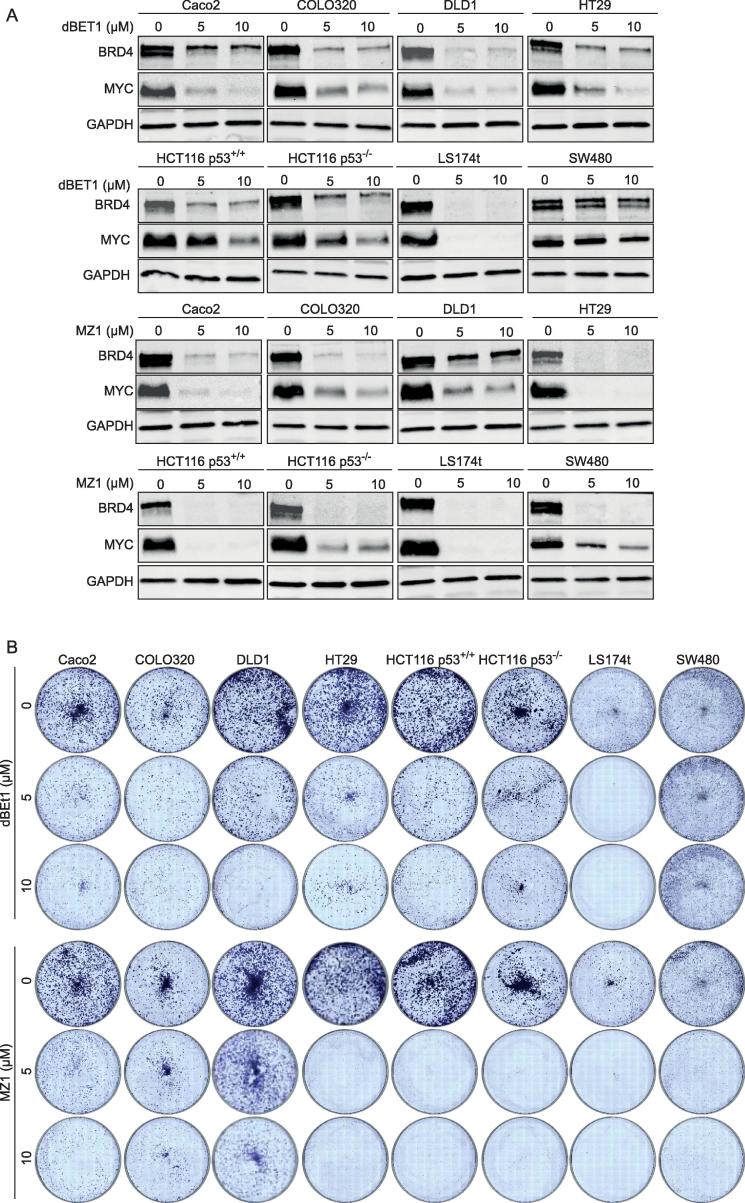
To evaluate the biological activity of dBET1 and MZ1 in a broader range, we treated the seven other CRC cell lines with dBET1 (0.5 and 1.0 µM) or MZ1 (0.1, 0.25, 0,5 and 1.0 µM) and found differences in their effectiveness in depleting BRD4. dBET1 induced a complete degradation of BRD4 in LS174t cells at 500 nM and a weak or no degradation in all other CRC cell lines. In contrast, already 100 nM MZ1 effectively depleted BRD4 in five of eight CRC cell lines but was unable to degrade BRD4 in Caco2, COLO320 and DLD1 cells (Supplementary Figure 1A, B). Interestingly, an increase in concentration of MZ1 to 0.5 and 1.0 µM did not increase the responsiveness of MZ1 to Caco2, COLO320 and DLD1 cells (Supplementary Figure 1C). To evaluate the effect of higher doses of individual PROTAC to degrade BRD4 in more CRC cell lines, we increased the concentration of dBET1 and MZ1 to 5 and 10 µM, respectively. For dBET1, we found a degradation of BRD4 protein in seven of eight cell lines and for MZ1 a profound degradation of BRD4 and downregulation of MYC in seven cell lines (Figure 4A). In dBET1-treated SW480 cells, no potent induced degradation of BRD4 was detectable and in MZ1-treated DLD1 cells, only marginal BRD4 degradation was found (Figure 4A). The different responses of the eight CRC cells lines to dBET1 or MZ1 were also apparent in colony formation assays (Figure 4B). We conclude from these results that CRC cell lines are responsive to at least one of the two tested PROTACs and degradation of BRD4 correlates with reduced MYC levels in CRC cell lines.
Figure 4.
CRC cell lines are responsive to at least one of the two PROTACs dBET1 and MZ1. (A) Western blot of BRD4 protein and MYC protein in eight CRC cell lines treated with 5 and 10 µM dBET1 and MZ1, respectively. (B) Antiproliferative effect of dBET1 and MZ1 in crystal violet colony-forming assay. Shown are representative images of three independent experiments (A, B).
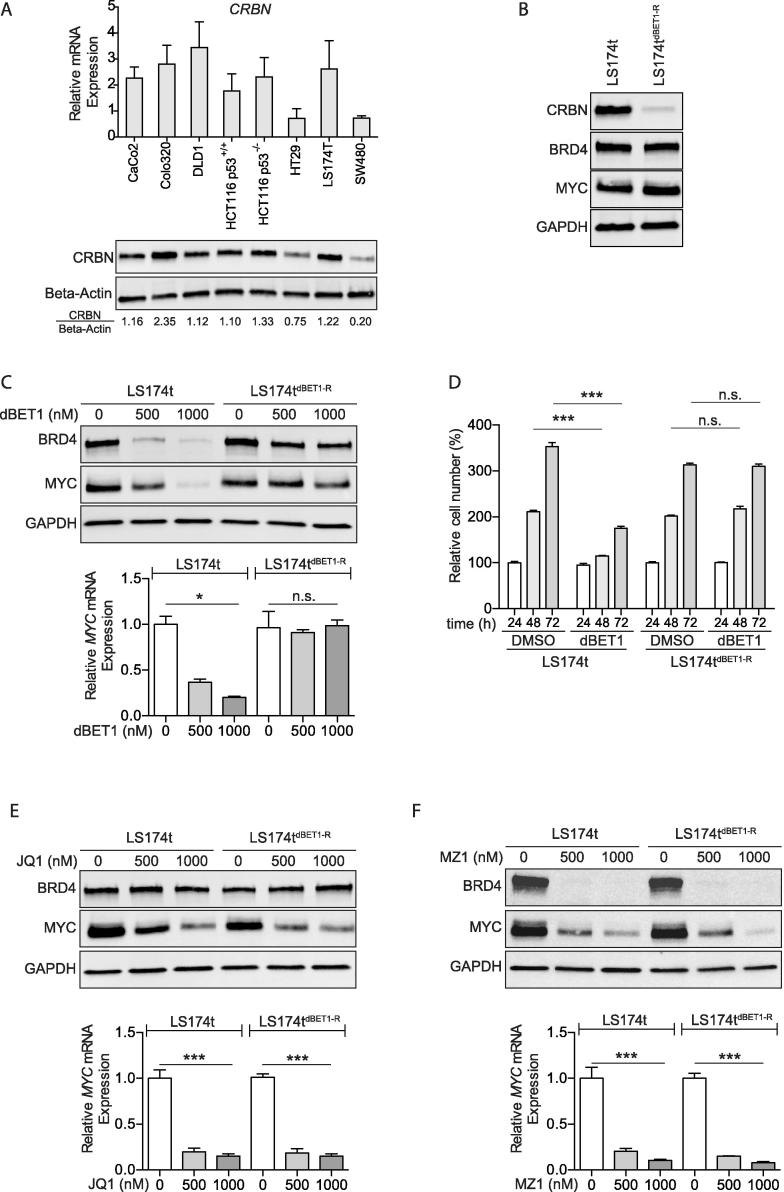
Acquired resistance against PROTAC dBET1 correlates with downregulated expression of E3 ligase cereblon
To analyze the molecular mechanism of dBET1 resistance in SW480 cells (Figure 4A), we first determined the expression of cereblon as the relevant E3 ligase recruited by dBET1 in the eight CRC cell lines. We found lower levels of cereblon (CRBN) mRNA und protein in SW480 compared to all other cell lines (Figure 5A). We speculated that the presence of cereblon is required for dBET1 activity and depletion of cereblon is associated with dBET1 resistance. To test this hypothesis, LS174t cells were exposed to increasing concentrations (25 nM–000 nM) of dBET1 for four months and outgrowing cells were further expanded. The dBET1-resistant LS174t cells, named LS174tdBET1-R, showed a pronounced downregulation of the E3 ligase cereblon (Figure 5B) but unaffected levels of BRD4 protein, MYC mRNA (not shown) and MYC protein in comparison to LS174t parental cells (Figure 5B). The anti-BRD4 effect of dBET1 failed in LS174tdBET1-R cells and levels of BRD4 protein, MYC mRNA and MYC protein were uninfluenced in contrast to LS174t parental cells (Figure 5C). In line with the failed dBET1-mediated degradation of BRD4, the proliferation of LS174tdBET1-R cells was not affected by dBET1 in contrast to parental LS174t cells (Figure 5D). Interestingly, LS174tdBET1-R cells still responded to JQ1 and showed a strong downregulation of MYC mRNA and MYC protein (Figure 5E). In addition, incubation with MZ1 lead to a strong loss of BRD4 and subsequent downregulation of MYC (Figure 5F). Together these data implicate a link between dBET1 sensitivity and level of cereblon expression. An acquired resistance to dBET1 did not result in cross-resistance to JQ1 and MZ1, respectively.
Figure 5.
Acquired resistance to PROTAC dBET1 is characterized by downregulation of E3 ligase cereblon and retained cell response to JQ1 or MZ1. (A) RT-qPCR (top) and Western blot (bottom) of cereblon (CRBN) mRNA and CRBN protein expression in eight CRC cell lines. (B) Western blot of CRBN, BRD4 and MYC protein in LS174tdBET1-R cells and parenteral LS174t cells. (C) Western blot (top) of BRD4 and MYC protein and RT-qPCR (bottom) of MYC mRNA. dBET1-resistant LS174tdBET1-R cells but not LS174t parenteral cells are unresponsive to dBET1. (D) dBET1-resistant LS174tdBET1-R cell proliferation is not affected by dBET1 in contrast to parenteral LS174t cells. Cell proliferation assay performed in biological triplicate (mean ± SD). (E–F) dBET1-resistant LS174tdBET1-R cells are responsive to JQ1 (E) and MZ1 (F) comparable to LS174t parenteral cells. Western blot: Shown are representative images of at least two independent experiments. Other results are shown as mean ± SD of three independent experiments.
Discussion
In this study, we compared the BRD4-targeting drugs JQ1, dBET1 and MZ1 for efficient MYC protein downregulation in CRC cell lines. JQ1 blocks the interaction of BRD4 with acetylated chromatin, whereas dBET1 and MZ1 trigger degradation of BRD4 by recruiting different E3 ligases. We showed that both approaches, BRD4 inhibition and BRD4 degradation, robustly reduced the expression of MYC resulting in a strong inhibitory effect on cell proliferation.
Overexpression of MYC due to mutations in the Wnt signaling pathway is a hallmark of virtually all CRC [3]. We and others have shown that targeting this overexpression can open therapeutic windows for treating specific cancer cells [12], [13], [27], [28]. So far, no direct MYC inhibitors/degraders exist, making this approach difficult. BRD4 represents an important epigenetic modulator that interacts with acetylated histones to change chromatin structure (chromatin remodeling) before transcription can proceed. BRD4 recruits the positive transcription elongation factor b (P-TEFb), which stimulates the phosphorylation of RNA polymerase II (RNA Pol II). Abrogation of the BRD4-mediated recruitment of P-TEFb by BRD4 knockdown represses RNA Pol II-dependent transcription and therefore, the expression of a number of genes involved in cell proliferation. The inhibition of BRD4 switches cancer cells from a pro-proliferative phenotype to an anti-proliferative and pro-apoptotic state [15]. Inhibiting BRD4 function using JQ1 significantly reduces MYC expression and consecutive cell viability. The RNA sequencing analysis we performed uncovered several gene sets that are regulated in the same way by JQ1 as the siRNA-mediated knockdown of MYC. MYC target genes are significantly downregulated by both siRNA and JQ1. We argue that JQ1 inhibits MYC-mediated transcription and the transcription of MYC target genes itself.
Recent study of Sun et al. describes PROTACs as a safe approach without side effects in both small animals (mice and rats) and large animals, e.g. rhesus monkey [29]. The key finding of our study is that loss of BRD4 protein mediated by dBET1 and MZ1, respectively correlated with reduction in MYC mRNA and MYC protein. We tested different concentrations of dBET1 or MZ1 and found that MZ1 induce depletion of BRD4 in lower concentration than dBET1 in CRC cells. To further increase the number of CRC cell lines responsive to dBET1 or MZ1, we tested both PROTACs at higher concentrations of 5 and 10 µM, respectively, and found a degradation effect of BRD4 in seven of eight CRC cell lines. Our study shows that increased concentrations of dBET1 and MZ1 led to an increased number of responsive CRC cell lines. Interestingly, we did not find a reduced capacity of BRD4 protein degradation at high concentrations of dBET1 or MZ1, which is described as the Hook effect [reviewed in [30]). The Hook effect occurs when an excess of PROTAC molecules reduces the formation of PROTAC-mediated degradation complexes by formation of binary complexes of PROTAC and E3 ligase component or PROTAC and target protein.
Further, we clearly showed that long-term treatment with increasing concentrations of dBET1 desensitized LS174t cells to treatment with dBET1. We found that resistance to dBET1 was associated with downregulation of the E3 ligase cereblon in LS174t and SW480 cells. Interestingly, dBET-induced resistance in LS174t cells demonstrated an unaffected response to BRD4 inhibitor JQ1 as well as BRD4-degrading MZ1, indicating that acquired dBET1 resistance is associated with repressed cereblon protein expression whereas BRD4 protein and its transcriptional regulatory function was not affected by dBET1. Recently Zhang and coworkers showed a comparable phenotype for an ovarian cancer cell line with acquired resistance to a cereblon-degrading PROTAC after long-term treatment. The PROTAC-resistant cells demonstrated genomic alterations influencing cereblon expression [31].
For CRC cells, we found a positive correlation between the rate of BRD4 degradation induced by dBET1 or MZ1 and the magnitude of MYC protein reduction. However, the generality of this finding seems unclear. For example, Fong et al. described JQ1-resistant leukemia stem cells in which reduced BRD4 protein levels did not alter MYC protein levels, indicating an alternative BRD4-independent mechanism that regulates the MYC transcription in these cells [32]. It seems that downregulation of cereblon has no adverse effects on cell function of dBET1-resistant cells. Our observations that dBET1-resistant LS174t cells are responsive to JQ1 or MZ1 may have possible therapeutic implications. Zhu et al. showed that patients with multiple myeloma had a downregulation of E3 ligase cereblon in the case of acquired resistance to the thalidomide-derivate lenalidomide [33]. Therefore, downregulation of cereblon appears to be an effective strategy of different types of tumor cells to evade a cereblon-involved drug response. We described here for dBET1-resistant LS174t cells the same molecular mechanism of downregulation of cereblon to evade the effect of dBET1. To optimize development of BRD4 PROTACs, one essential aspect is choosing the optimal E3 ligase for PROTAC engagement. The best E3 ligase to maintain PROTAC function is one that plays a crucial role in cancer cell biology. Its downregulation should be lethal for the cells that might inhibit the generation of resistance against PROTAC. An alternative approach to avoiding resistance to a PROTAC might be the use of several PROTACS recruiting different E3 ligases. The use of a PROTAC with multiple binders for recruiting different E3 ligases might also be effective in preventing the development of resistance [30].
Conclusions
In summary, BRD4 appears to be an important druggable protein target in CRC cells. Both inhibition of BRD4 by JQ1 and degradation of BRD4 by the PROTACs dBET1 and MZ1 were identified as successful tools for reducing MYC expression and restraining CRC cell proliferation. In contrast to JQ1, PROTAC-induced degradation of BRD4 is dependent on both the function of E3 ligase and proteasome activity. We found that long-term incubation of cells with high concentrations of the PROTAC dBET1 developed resistance to dBET1 by downregulation of expression of the E3 ligase cereblon.
Author contribution
C. O. and A. W. designed the study, analyzed and interpreted data and wrote the first draft of the manuscript. S. S., C. K., N. M., J. K. performed the experiments and analyzed and interpreted data, N.M. performed immunofluorescence and confocal microscopy, W.E. and P. W. performed RNA sequencing and data analysis. C.T.G. contributed to the study design, provided important advice and critically revised the manuscript. All authors critically reviewed the manuscript and approved the final version.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors are grateful to Manuela Hofmann, Monika Koospal, and Bettina Pörsch for their skillful technical assistance with the experiments and Mohammed Hankir for proofreading the manuscript.
Funding
The study was supported by funds from the Else-Kröner-Fresenius Foundation (EKFS 2015_A57 to AW) and the lnterdisciplinary Centre for Clinical Research (IZKF) of the University of Würzburg (B-186 and B-335 to AW, B-344 to CO). This publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding program “Open Access Publishing”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.10.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Effectiveness of dBET1 and MZ1 to deplete BRD4 in CRC cell lines. Western blot of BRD4 protein and MYC protein in eight CRC cell lines treated with A) 500 and 1000 nM dBET1; B) 100 and 250 nM MZ1; C) 500 and 1000 nM MZ1. Shown are representative images of two independent experiments.
References
- 1.Siegel R.L. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang C.V. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eilers M., Eisenman R.N. Myc's broad reach. Genes Dev. 2008;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansom O.J. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446(7136):676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 7.Dow L.E. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161(7):1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucek L. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27(5):504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soucek L., Nasi S., Evan G.I. Omomyc expression in skin prevents Myc-induced papillomatosis. Cell Death Differ. 2004;11(9):1038–1045. doi: 10.1038/sj.cdd.4401443. [DOI] [PubMed] [Google Scholar]
- 10.Soucek L. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuber J. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiegering A. CIP2A influences survival in colon cancer and is critical for maintaining Myc expression. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0075292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiegering A. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 2015;5(7):768–781. doi: 10.1158/2159-8290.CD-14-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikary S., Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6(8):635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 15.Donati B., Lorenzini E., Ciarrocchi A. BRD4 and cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17(1):164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranello L. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165(2):357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmore J.E. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carmony K.C., Kim K.B. PROTAC-induced proteolytic targeting. Methods Mol Biol. 2012;832:627–638. doi: 10.1007/978-1-61779-474-2_44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan Y. Targeting Brd4 for cancer therapy: inhibitors and degraders. Medchemcomm. 2018;9(11):1779–1802. doi: 10.1039/c8md00198g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter G.E. Drug development phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zengerle M., Chan K.H., Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol. 2015;10(8):1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pervaiz M., Mishra P., Gunther S. Bromodomain drug discovery – the past, the present, and the future. Chem Rec. 2018;18(12):1808–1817. doi: 10.1002/tcr.201800074. [DOI] [PubMed] [Google Scholar]
- 24.Vlachogiannis G. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg K.C.G. Multi-omics of 34 colorectal cancer cell lines – a resource for biomedical studies. Mol Cancer. 2017;16(1):116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van de Wetering M. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peter S. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6(12):1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegering A. The impact of pyrvinium pamoate on colon cancer cell viability. Int J Colorectal Dis. 2014;29(10):1189–1198. doi: 10.1007/s00384-014-1975-y. [DOI] [PubMed] [Google Scholar]
- 29.Sun X. A chemical approach for global protein knockdown from mice to non-human primates. Cell Discov. 2019;5:10. doi: 10.1038/s41421-018-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An S., Fu L. Small-molecule PROTACs: an emerging and promising approach for the development of targeted therapy drugs. EBioMedicine. 2018;36:553–562. doi: 10.1016/j.ebiom.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L. Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther. 2019;18(7):1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- 32.Fong C.Y. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525(7570):538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Y.X. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118(18):4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effectiveness of dBET1 and MZ1 to deplete BRD4 in CRC cell lines. Western blot of BRD4 protein and MYC protein in eight CRC cell lines treated with A) 500 and 1000 nM dBET1; B) 100 and 250 nM MZ1; C) 500 and 1000 nM MZ1. Shown are representative images of two independent experiments.