Abstract
Regulation of the Hippo signaling pathway is essential for normal organ growth and tissue homeostasis. The proteins that act to regulate this pathway are important for ensuring proper function and cellular location. Deubiquitinases (DUBs) are a family of proteases that act upon many proteins. While ubiquitinases add ubiquitin and target proteins for degradation, DUBs act by removing ubiquitin (Ub) moieties. Changes in ubiquitin chain topology results in the stabilization of proteins, membrane trafficking, and the alteration of cellular localization. While the roles of these proteins have been well established in a cancer setting, their convergence in cancer is still under investigation. In this review, we discuss the roles that DUBs play in the regulation of the Hippo signaling pathway for homeostasis and disease.
Keywords: Cancer, Deubiquitinase, Hippo signaling pathway, Ubiquitinase, YAP/TAZ
Introduction
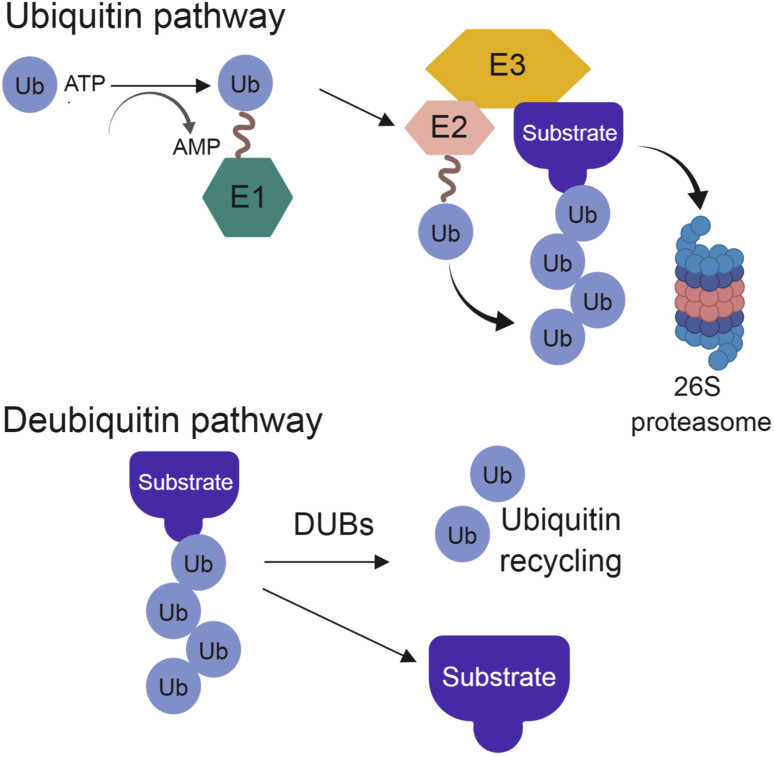
The addition of ubiquitin, the most common post-translational modification, marks proteins for degradation, alters their function, or changes their signaling processes.1 The linkages and types of ubiquitin chains determine the fates of target proteins. The conjugation of ubiquitin to substrates is a 3-step process. The initial step requires an E1 activating enzyme, which is catalyzed in an ATP-dependent manner. The second step covalently links the ubiquitin to an E2 intermediate enzyme. The final step, which confers the specificity of this system, transfers the ubiquitin moiety to an E3 ligase.2 The E3 ligase then links ubiquitin to its substrate, driving the alterations of its target protein (Fig. 1).
Figure 1.
Ubiquitination and Deubiquitinating pathway overview.
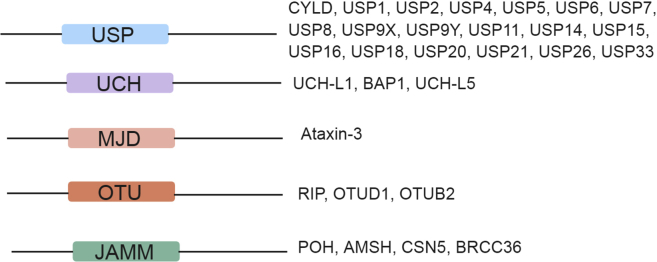
As with most cellular processes, the addition of ubiquitin is a reversible process. The deubiquitinating of enzymes allows for the removal of ubiquitin from proteins, changes their cellular localization, rescues them from degradation and alters their function (Fig. 1). The human genome encodes for ∼100 deubiquitinases (DUBs) in five major families (Fig. 2).3 The largest family of deubiquitinating enzymes is the ubiquitin specific proteases (USPs). USPs have a highly conserved USP domain, which allows direct binding to ubiquitin.4, 5, 6 The second family of DUBs, ubiquitin c-terminal hydrolases (UCHs), is named for their C-terminal domain. These DUBs preferentially bind to small leaving groups and are thought to be involved in processing trapped ubiquitin.7 The ovarian-tumor related proteases (OTUs), contain a conserved cysteine domain that allows for their DUB activity.8 The Josephin family of DUBs is closely related to the USP and UCH families, but has a conserved Josephin domain. The final family of DUBs is the JAMM/MPN + metalloproteases. This family of proteases often requires a protein partner to be enzymatically active.9 A comprehensive list of the known DUB family members was reviewed by Nijman et al and is summarized in Fig. 2.10 DUBs function by processing ubiquitin gene products, remodeling poly-ubiquitin chains, and reversing the process of ubiquitin addition to target proteins. The activity of DUBs, like other enzymatic proteins, is tightly controlled by cleavage specificity, ubiquitin recognition, linkage preference, positioning specificity, and substrate and/or protein recognition.11 There are a large number of reviews covering the activities and structures of deubiquitinating enzymes in the existing literature.6, 8, 12, 13, 14 Here, we review the known deubiquitinating enzymes that function to activate or suppress the Hippo signaling pathway and how these interactions may be altered in cancer.
Figure 2.
DUB family members.
The role of deubiquitinating enzymes in cancer
Deubuiquitinating enzymes play an important role in cancer from supporting primary lesions to driving tumor cell dissemination and metastasis. Through genomic analysis a much-needed understanding of how these enzymes can be used as markers for cancer aggressiveness has been gained. Ge et al recently reviewed the importance of understanding the DUB landscape by using integrative genomics analysis.15 To target DUBs therapeutically, a large amount of data has been compiled which demonstrates the importance of these enzymes in cancer.
BRCA-1 associated protein 1 (BAP1), is a member of the UCH family of DUBs and is a potent tumor suppressor. BAP1 is associated with hematopoietic cell development. Loss of BAP1 can cause myeloid neoplasia's and myelodysplastic syndrome.16 CYLD, a gene that was initially discovered in 2000 and shown to have tumor suppressive functions, controls the activity of nuclear factor kappa B subunit 1 (NF-kB).17 Upon receptor stimulation by nuclear factor-kappa B essential modulator (NEMO), TNF receptor (TNFR) associated factors (TRAF-2) and (TRAF-6) are modified by ubiquitination to activate IƙB kinase and target these proteins for degradation. The removal of ubiquitin moieties by CYLD from NEMO, TRAF-2, and TRAF-6 reduces NF-kB signaling, resulting in a tumor suppressive function of CYLD.18, 19, 20 In Ewing Sarcoma, which commonly occurs due to fusion proteins that lead to transformation, USP6 has been shown to be fused to Osteoblast Cadherin 11.21 The constitutive activation of USP6 can lead to actin remodeling, endocytosis, and membrane trafficking, allowing it to perform oncogenic functions.22, 23
The OTU family of DUBs can act on various proteins affecting their functions. OTU-A20 has been shown to remove Ub from several NF-kB signaling enzymes, reducing the lifespan of these proteins. The reduction of these enzymes can lead to B-cell and T-cell lymphomas, as well as Burkett's lymphoma.24 This enzyme has also been shown to have tumor-suppressive functions related to the control of inflammation.25 USP9 can stabilize Wnt signaling effector β-catenin, driving the formation of many cancers. USP9 also has the ability to remove Ub from SMAD4, restoring its functionality and tumor promoting activities.26 Loss of p53 is known to promote tumorigenesis in various cancer types. The loss of USP22 can reduce the transcription of Myc and p53 by removing Ub from H2B.27 USP3 and USP21 also have functions in chromatin modification, leading to aberrant gene transcription.28 Loss of USP3 results in defects of the cell cycle, while USP21 can promote regenerative functions in hepatocytes. Whether these gene transcription alterations occur in known tumor suppressor or oncogenic promoter regions requires further study. USP1 has been shown to be involved in the Fanconi Anemia pathway, leading to a reduction in DNA repair.3 Increased DNA damage can lead to genomic instability, a hallmark of cancer initiation and progression. USP2A has both tumor suppressive and oncogenic functions. USP2A deubiquitinates MDM2/MDMX leading to increased p53 protein levels.29 It also binds to Aurora-A, a protein essential for centrosome duplication, ensuring proper mitosis.30 Overexpression of USP2A has been seen in prostate cancer due to its androgen regulation.31 A summary of the tumor suppressive or oncogenic function of these DUBs can be found in Table 1.
Table 1.
Functional relevance of deubiquitinating enzymes in tumor-suppression or oncogenesis.
| Tumor suppressive | Main function |
|---|---|
| CYLD | Regulation of NF-kB; germline mutations cause malignant transformation and skin tumors17 |
| BAP1 | Part of polycomb-group proteins that regulate cell fate determination, stem cell pluripotency, and developmental processes32, 33 |
| USP2A | Interferon-mediated signaling34 |
| Oncogenic | |
| USP6 | Chromosomal translocations present in Ewing sarcoma, leading to aneurysmal bone cysts35 |
| OTU-A20 | Control of NF-kB protein levels; TNF-induced apoptosis |
| USP9X | Control of Hippo signaling pathway components; stabilization of beta-catenin in colon cancer36 |
| USP22 | Regulation of histone acetylation complexes37 |
| USP3 | Chromatin modification |
| USP21 | Chromatin modification |
| USP1 | DNA damage repair pathways |
| USP2A | Interacts with MDM2 complexes |
Hippo signaling pathway
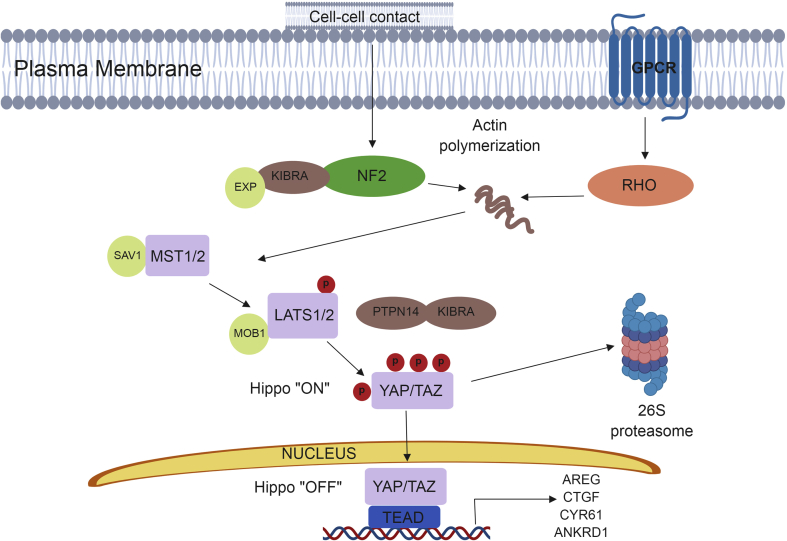
The Hippo signaling pathway, an evolutionarily conserved pathway, was first identified in Drosophila melanogaster and found to control cell proliferation, apoptosis, and organ size. This pathway is necessary for normal tissue growth and repair, and disruptions to or aberrations of Hippo signaling are involved in tumorigenesis.38, 39, 40, 41 The progression of breast cancer can be driven by the activation of its effector molecules, YAP (yes-associated protein) and TAZ (transcriptional co-activator with PDZ binding motif). Following nuclear translocation, YAP and TAZ activity promotes various phenotypes associated with breast cancer progression including; epithelial to mesenchymal transition, epithelial stem cell regeneration, and therapeutic resistance.42 Hippo signaling is involved in regulating important cellular processes including cell cycle progression and apoptosis, therefore a tight regulation of this pathway is necessary to ensure cellular homeostasis. Through activation by various external stimuli, Hippo signaling results in a serine–threonine kinase cascade.43 In mammals, mammalian Ste20-like serine/threonine kinase 1/2 (MST1/2) is phosphorylated. Following protein–protein interactions with Salvador (SAV), large tumor-suppressor kinase 1/2 (LATS1/2) are then phosphorylated.44, 45, 46, 47 In concert with protein tyrosine phosphatase and kidney and brain expressed protein (KIBRA) LATS1/2 phosphorylates the key effectors of Hippo signaling, YAP and TAZ.46, 48 YAP and TAZ activity is regulated via interactions with 14-3-3 proteins and sequestering of YAP/TAZ in the cytoplasm. When phosphorylated βTrCP is recruited to YAP/TAZ, this leads to ubiquitination and subsequent proteasomal degradation (Fig. 3).48
Figure 3.
Schematic of Hippo signaling pathway.
The Hippo signaling pathway and cancer
The Hippo signaling pathway plays a central role in the occurrence and control of cancer phenotypes. Hyper-activation of YAP/TAZ leads to uncontrolled cell proliferation, a major aspect of neoplasia. Insensitive to apoptosis is also central to carcinogenesis. Overexpression of YAP has been shown to block TNF-alpha and FAS-induced apoptosis in the liver, driving cell survival.49 Competition between healthy cells and neoplastic cells can lead to drug resistance, cancer progression, and metastasis. Overexpression of Yki, a YAP homolog present in Drosophila, has the ability to give a survival advantage to cells, leading to drug resistance.50 The maintenance of stem cells is also a major hallmark of cancer. YAP and TEA domain family member 1 (TEAD1) have been shown to be enriched in stem cells.51 YAP/TAZ has also been shown to promote the pluripotency of embryonic stem cells, as well as the control of mesenchymal stem cell differentiation.52, 53, 54 The major regulators of the Hippo signaling pathway have been shown to be dysregulated in a multitude of cancers and many reviews offer in-depth explanations of the various alterations seen in the Hippo signaling pathway and cancer.55, 56, 57, 58 While it is widely understood how the ubiquitin-proteasome system regulates the Hippo signaling pathway,59 much less is known about the DUBs that act on this pathway.
Inhibitory DUBs of the Hippo signaling pathway
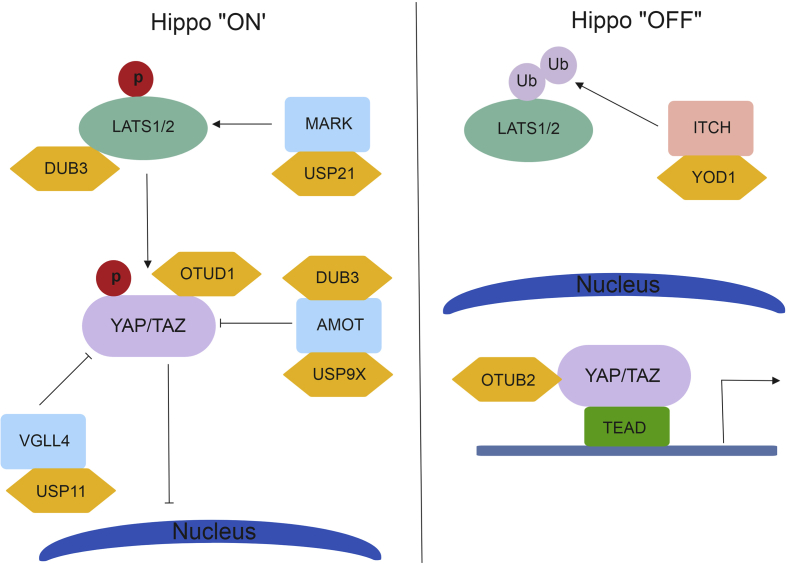
YOD1 is a member of the ovarian tumor family of DUBs. YOD1 cleaves both poly-ubiquitin and di-ubiquitin chains and is associated with endoplasmic reticulum associated degradation for misfolded proteins.60 In 2017 Kim and Jho et al showed that YOD1 can remove ubiquitin from the E3 ligase ITCH, resulting in ITCH stabilization (Fig. 4). The WW domain of ITCH interacts with the PPxY motif of LATS1/2 and promotes the degradation of LATS1/2 proteins.61 This reduction in LATS protein levels results in the translocation of YAP/TAZ to the nucleus and drive the transcription of genes involved in cell cycle progression. The co-expression of YOD1 and YAP in liver cancer patients proves that targeting this DUB may be used as a therapeutic intervention.
Figure 4.
Schematic of DUBs and their role in Hippo signaling.
OTUB2, a member of the ovarian tumor family, has been shown to stimulate cancer stem cell traits and sustain circulating tumor cells and metastasis. Recently, OTUB2 has been shown to bind directly to and activate YAP/TAZ.62 For this interaction to occur, OTUB2 must be sumoylated. The sumoylation of OTUB2 allows it to interact with the sumoylation interaction motif found in YAP/TAZ. This allows for YAP/TAZ to interact with TEAD transcription factors and drive tumorigenic phenotypes both in vitro and in vivo. Targeting OTUB2 sumoylation could reduce its ability to interact with YAP/TAZ, resulting in YAP/TAZ degradation.
OTUD1 was recently shown to regulate YAP in a Hippo-independent manner.63 K63-linked polyubiquitination of YAP allows nuclear translocation and interaction with TEAD family transcription factors. OTUD1 can remove these ubiquitin moieties, resulting in YAP cytoplasmic retention and reduction in growth promoting phenotypes.
Activating DUBs of the Hippo signaling pathway
USP9X, a member of the ubiquitin-specific protease family of DUBs controls angiomotin (AMOT) levels in vitro. AMOT is a junction-associated scaffold protein known to be involved in the Hippo signaling pathway.64 The PPxY motif in AMOT binds to and represses the function of YAP/TAZ through interaction with the YAP/TAZ WW domain, allowing for the degradation of these proteins.65 In the absence of AMOT, YAP/TAZ are able to translocate to the nucleus and drive target gene expression. USP9X binds to and removes ubiquitin from AMOT, leading to a reduction in YAP/TAZ activity. This reduction in YAP/TAZ leads to a decrease in tumorigenic potential.66
DUB3 is a member of the USP family and belongs to a subfamily of cytokine-inducible DUBS.67 DUB3 can directly interact with AMOT and LATS to regulate YAP. Nguyen et al showed that AMOT and LATS1/2 are required for the DUB3 repression of YAP. While DUB3 can also act on and stabilize ITCH protein levels, its interaction with AMOT and LATS1/2 confers control of YAP turnover.68 By removing ubiquitin from both of these proteins, there is dual repression of YAP nuclear translocation.
USP11 has been recently shown that control VGLL4 protein stability.69 VGLL4 is known to block the interaction between YAP and TEAD to reduce cell proliferation and migration.70 By removing ubiquitin moieties from VGLL4, USP11 allows for increased expression of VGLL4, modulating the interaction between YAP-TEAD complexes.
USP21 has been shown to control YAP levels by increasing microtubule affinity-regulating kinase (MARK).71 MARK proteins serve as regulators of LATS1/2 kinase.72 The loss of USP21 increases MARK turnover, resulting in increased YAP/TAZ nuclear localization. The binding of YAP/TAZ to TEAD can then drive oncogenic transformation through transcription of genes associated with cell-cycle progression and anti-apoptosis.
Discussion
Many studies reveal that Hippo pathway components contribute to cancer; however, it is not known why mutations are so rare. Due to the low amount of germline mutations in this pathway (with the exception of neurofibromatosis type II (NF2) and TAZ) external signaling nodes are crucial to control this pathway. Other signaling pathways that converge on the Hippo pathway may harbor genetic mutations, which in turn can change the fate of Hippo signaling genes through bypassing the normal kinase cascade. Understanding of these molecular triggers could help define the genetic landscape of Hippo dysregulation in cancer.
The extracellular matrix has the ability to regulate YAP/TAZ activity.73 At high or low density there are actin and cytoskeletal reorganization, mechanical cues that YAP/TAZ can sense. Merlin, Expanded, KIBRA, and CRUMBS interact with the cytoskeleton and drive Hippo signaling. As previously mentioned, AMOT has the ability to inhibit YAP/TAZ activity by binding to tight junctions, independent of their phosphorylation status.65, 74 The most prominent pathways that regulate Hippo signaling cascades are GPCRs, epidermal growth factor, bone-morphogenic protein, TGF-β, WNT, and Notch. The crosstalk with these signaling pathways can increase or decrease YAP/TAZ activity.
Based on what is known about external crosstalk, finding novel genes related to Hippo signaling is needed. Understanding of how the ubiquitin proteasome system regulates this pathway may lead to new avenues for therapeutic intervention in cancer. Understanding of how ubiquitin, more directly E3 ligases, interact with Hippo signaling has been well studied. The lack of knowledge about DUBs and their effect on this pathway offers an important starting point for new drugs. In this review we summarized what is known about the DUB enzymes that activate or repress this pathway. Further studies on the post-translational modifications (more specifically the DUBs that act on this pathway) could lead to potential therapeutic leverage points in cancer progression and metastasis.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) Grant #P30 CA016056, Roswell Park Alliance Foundation, the National Cancer Institute (NCI) R01 CA207504 and the American Cancer Society Research Scholar Grant RSG-14-214-01-TBE (to J.Z.).
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Chen Z.J., Sun L.J. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Nijman S.M., Huang T.T., Dirac A.M. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Hu M., Li P., Li M. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 5.Hu M., Li P., Song L. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 2005;24(21):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komander D., Lord C.J., Scheel H. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29(4):451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Johnston S.C., Riddle S.M., Cohen R.E., Hill C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999;18(14):3877–3887. doi: 10.1093/emboj/18.14.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1–3):189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Cao S., Engilberge S., Girard E., Gabel F., Franzetti B., Maupin-Furlow J.A. Structural insight into ubiquitin-like protein recognition and oligomeric states of JAMM/MPN+ proteases. Structure. 2017;25(6):823–833 e826. doi: 10.1016/j.str.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nijman S.M., Luna-Vargas M.P., Velds A. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 11.He M., Zhou Z., Shah A.A. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 2016;6:62. doi: 10.1186/s13578-016-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman K.E., Huang T.T. In a class of its own: a new family of deubiquitinases promotes genome stability. Mol Cell. 2018;70(1):1–3. doi: 10.1016/j.molcel.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Pfoh R., Lacdao I.K., Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer. 2015;22(1):T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson K.D. DUBs at a glance. J Cell Sci. 2009;122(Pt 14):2325–2329. doi: 10.1242/jcs.041046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge Z., Leighton J.S., Wang Y. Integrated genomic analysis of the ubiquitin pathway across cancer types. Cell Rep. 2018;23(1):213–226 e213. doi: 10.1016/j.celrep.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey A., Seshasayee D., Noubade R. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337(6101):1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bignell G.R., Warren W., Seal S. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25(2):160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 18.Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 19.Kovalenko A., Chable-Bessia C., Cantarella G., Israel A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424(6950):801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp T.R., Nijman S.M., Dirac A.M., Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira A.M., Hsi B.L., Weremowicz S. USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res. 2004;64(6):1920–1923. doi: 10.1158/0008-5472.can-03-2827. [DOI] [PubMed] [Google Scholar]
- 22.Martinu L., Masuda-Robens J.M., Robertson S.E., Santy L.C., Casanova J.E., Chou M.M. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol Cell Biol. 2004;24(22):9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda-Robens J.M., Kutney S.N., Qi H., Chou M.M. The TRE17 oncogene encodes a component of a novel effector pathway for Rho GTPases Cdc42 and Rac1 and stimulates actin remodeling. Mol Cell Biol. 2003;23(6):2151–2161. doi: 10.1128/MCB.23.6.2151-2161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durkop H., Hirsch B., Hahn C., Foss H.D., Stein H. Differential expression and function of A20 and TRAF1 in Hodgkin lymphoma and anaplastic large cell lymphoma and their induction by CD30 stimulation. J Pathol. 2003;200(2):229–239. doi: 10.1002/path.1351. [DOI] [PubMed] [Google Scholar]
- 25.Shembade N., Harhaj E. A20 inhibition of NFkappaB and inflammation: targeting E2:E3 ubiquitin enzyme complexes. Cell Cycle. 2010;9(13):2481–2482. doi: 10.4161/cc.9.13.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupont S., Mamidi A., Cordenonsi M. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X.Y., Varthi M., Sykes S.M. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29(1):102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto-Fernandez A., Kessler B.M. DUBbing cancer: deubiquitylating enzymes involved in epigenetics, DNA damage and the cell cycle as therapeutic targets. Front Genet. 2016;7:133. doi: 10.3389/fgene.2016.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson L.F., Sparks A., Allende-Vega N., Xirodimas D.P., Lane D.P., Saville M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26(4):976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Y., Solomon L.R., Pereda-Lopez A. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J Biol Chem. 2011;286(45):38960–38968. doi: 10.1074/jbc.M111.231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graner E., Tang D., Rossi S. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5(3):253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 32.Scheuermann J.C., de Ayala Alonso A.G., Oktaba K. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaytan de Ayala Alonso A., Gutierrez L., Fritsch C., Papp B., Beuchle D., Muller J. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics. 2007;176(4):2099–2108. doi: 10.1534/genetics.107.075739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y., Zhao P., Liu J. Deubiquitinase USP2a sustains interferons antiviral activity by restricting ubiquitination of activated STAT1 in the nucleus. PLoS Pathog. 2016;12(7) doi: 10.1371/journal.ppat.1005764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussain S., Zhang Y., Galardy P.J. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8(11):1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 36.Murray R.Z., Jolly L.A., Wood S.A. The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and β-catenin. Mol Biol Cell. 2004;15(4):1591–1599. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melo-Cardenas J., Zhang Y., Zhang D.D., Fang D. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget. 2016;7(28):44848–44856. doi: 10.18632/oncotarget.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azzolin L., Panciera T., Soligo S. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158(1):157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Bai H., Zhang N., Xu Y. Yes-associated protein regulates the hepatic response after bile duct ligation. Hepatology. 2012;56(3):1097–1107. doi: 10.1002/hep.25769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai J., Zhang N., Zheng Y., de Wilde R.F., Maitra A., Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24(21):2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q., Zhang N., Gray R.S. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev. 2014;28(5):432–437. doi: 10.1101/gad.233676.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humtsoe J.O., Kramer R.H. Differential epidermal growth factor receptor signaling regulates anchorage-independent growth by modulation of the PI3K/AKT pathway. Oncogene. 2010;29(8):1214–1226. doi: 10.1038/onc.2009.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C.L., Gajewski K.M., Hamaratoglu F. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107(36):15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hergovich A. Mammalian Hippo signalling: a kinase network regulated by protein-protein interactions. Biochem Soc Trans. 2012;40(1):124–128. doi: 10.1042/BST20110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson G.R., Kannan B., Shoyab M., Stromberg K. Amphiregulin induces tyrosine phosphorylation of the epidermal growth factor receptor and p185erbB2. Evidence that amphiregulin acts exclusively through the epidermal growth factor receptor at the surface of human epithelial cells. J Biol Chem. 1993;268(4):2924–2931. [PubMed] [Google Scholar]
- 46.Lahaye D.H., Camps M.G., Van Zoelen E.J. Central role of epidermal growth factor (EGF) receptor density in anchorage-independent growth of normal rat kidney cells. FEBS Lett. 1999;446(2–3):256–260. doi: 10.1016/s0014-5793(99)00216-1. [DOI] [PubMed] [Google Scholar]
- 47.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24(50):7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 48.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camargo F.D., Gokhale S., Johnnidis J.B. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17(23):2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 50.de Beco S., Ziosi M., Johnston L.A. New frontiers in cell competition. Dev Dynam. 2012;241(5):831–841. doi: 10.1002/dvdy.23783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordenonsi M., Zanconato F., Azzolin L. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 52.Mo J.S., Park H.W., Guan K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15(6):642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullen A.C. Hippo tips the TGF-β scale in favor of pluripotency. Cell Stem Cell. 2014;14(1):6–8. doi: 10.1016/j.stem.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinhardt A.A., Gayyed M.F., Klein A.P. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39(11):1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harvey K.F., Zhang X., Thomas D.M. The Hippo pathway and human cancer. Nat Rev Canc. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 56.Park J.H., Shin J.E., Park H.W. The role of Hippo pathway in cancer stem cell biology. Mol Cells. 2018;41(2):83–92. doi: 10.14348/molcells.2018.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zygulska A.L., Krzemieniecki K., Pierzchalski P. Hippo pathway - brief overview of its relevance in cancer. J Physiol Pharmacol. 2017;68(3):311–335. [PubMed] [Google Scholar]
- 58.Janse van Rensburg H.J., Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signal. 2016;28(11):1761–1772. doi: 10.1016/j.cellsig.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Kim Y., Jho E.H. Regulation of the Hippo signaling pathway by ubiquitin modification. BMB Rep. 2018;51(3):143–150. doi: 10.5483/BMBRep.2018.51.3.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papadopoulos C., Kirchner P., Bug M. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36(2):135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho K.C., Zhou Z., She Y.M., Chun A., Cyr T.D., Yang X. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability. Proc Natl Acad Sci U S A. 2011;108(12):4870–4875. doi: 10.1073/pnas.1101273108. [corrected] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z., Du J., Wang S. OTUB2 promotes cancer metastasis via Hippo-independent activation of YAP and TAZ. Mol Cell. 2019;73(1):7–21 e27. doi: 10.1016/j.molcel.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 63.Yao F., Zhou Z., Kim J. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun. 2018;9(1):2269. doi: 10.1038/s41467-018-04620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao B., Li L., Lu Q. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25(1):51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan S.W., Lim C.J., Chong Y.F., Pobbati A.V., Huang C., Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286(9):7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toloczko A., Guo F., Yuen H.F. Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the Hippo pathway. Cancer Res. 2017;77(18):4921–4933. doi: 10.1158/0008-5472.CAN-16-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen H.T., Kugler J.M., Cohen S.M. DUB3 deubiquitylating enzymes regulate Hippo pathway activity by regulating the stability of ITCH, LATS and AMOT proteins. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang E., Shen B., Mu X. Ubiquitin-specific protease 11 (USP11) functions as a tumor suppressor through deubiquitinating and stabilizing VGLL4 protein. Am J Cancer Res. 2016;6(12):2901–2909. [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W., Gao Y., Li P. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24(3):331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen H.T., Kugler J.M., Loya A.C., Cohen S.M. USP21 regulates Hippo pathway activity by mediating MARK protein turnover. Oncotarget. 2017;8(38):64095–64105. doi: 10.18632/oncotarget.19322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohseni M., Sun J., Lau A. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16(1):108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bae J.S., Kim S.M., Lee H. The Hippo signaling pathway provides novel anti-cancer drug targets. Oncotarget. 2017;8(9):16084–16098. doi: 10.18632/oncotarget.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W., Huang J., Chen J. Angiomotin-like proteins associate with and negatively regulate YAP1. J Biol Chem. 2011;286(6):4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]