Abstract
The human HEAT repeat-containing protein 1 (HEATR1), consisting of 2144 amino acids, is a member of the UTP10 family and contains one HEAT repeat at its C-terminal. HEATR1 has been reported to regulate cytotoxic T lymphocytes and rRNA synthesis, while its functions in tumors are poorly understood. Here, we found that HEATR1 competed with Keap1 for binding to p62/sequestosome 1 (SQSTM1), resulted in up-regulation of Keap1, which then inhibited Nrf2 signaling in pancreatic cancer cells. HEATR1 knockdown enhanced proliferation and gemcitabine resistance of pancreatic cancer cells. Moreover, HEATR1 deficiency significantly improved xenografts growth and led to gemcitabine resistance in pancreatic cancer cell-derived xenografts through up-regulating Nrf2 signaling. By analyzing tumor tissue samples from pancreatic cancer patients, we found that low expression of HEATR1 was closely correlated with poor prognosis and clinicopathological features. Collectively, we suggest that HEATR1 deficiency promotes proliferation and gemcitabine resistance of pancreatic cancer through up-regulating Nrf2 signaling, indicating that HEATR1 may be a promising therapeutic target for pancreatic cancer.
Keywords: HEATR1, Nrf2, Pancreatic cancer, Gemcitabine resistance
Abbreviations: HEATR1, human HEAT repeat-containing protein 1; SQSTM1, sequestosome 1; PDAC, pancreatic ductal adenocarcinoma; Nrf2, nuclear factor erythroid 2-related factor 2; Keap1, Kelch-like ECH-associated protein 1; NQO1, NAD(P)H quinone oxidoreductase 1; HO-1, heme oxygenase-1; mRNA, messenger RNA; shRNA, short hairpin RNA; qRT-PCR, quantitative reverse transcription-PCR
Highlights
-
•
HEATR1 inhibited Nrf2 signaling in pancreatic cancer cells.
-
•
HEATR1 inhibited Nrf2 signaling through competing with Keap1 for p62 binding in pancreatic cancer cells.
-
•
HEATR1 deficiency promoted pancreatic cancer proliferation and gemcitabine resistance by up-regulating Nrf2 signaling.
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death worldwide. The prognosis of PDAC patients is poor with a 5-year survival rate of less than 5% [1,2]. While most chemotherapy regimens utilize gemcitabine as the first-line treatment for pancreatic cancer, frequently occurrence of drug resistance limit its use [3]. Thus, exploring the molecular mechanisms contributing to gemcitabine resistance may develop more effective therapeutic strategies.
The human HEAT repeat-containing protein 1 (HEATR1) protein, consisting of 2144 amino acids, has one HEAT repeat which is also found in other proteins such as PP2A, elongation factor-3 and huntingtin at its C-terminal end [4]. Previous studies suggest that HEATR1 regulate cytotoxic T lymphocytes and rRNA synthesis [5,6]. Currently, the functions of HEATR1 protein in tumors are rarely reported. One study showed that pancreatic cancer tissue exhibited lower HEATR1 protein level when compared with normal pancreatic tissues and HEATR1 deficiency led to chemoresistance to anticancer drugs through enhancing the activity of AKT in pancreatic cancer cells [7], indicating that HEATR1 had tumor suppressive effects. However, another study showed that HEATR1 depletion led to cell-cycle arrest of U2OS cells by disruption of nucleolar structure and activation of p53-dependent cell cycle checkpoint pathway [8], indicating that HEATR1 had tumor promoting effects. Therefore, the roles of HEATR1 in tumors are controversial and further studies are needed.
Nuclear factor erythroid-2-related factor 2 (Nrf2), a transcription factor, participates in protecting cells from electrophilic or oxidative stresses through regulating expression of cytoprotective and antioxidant genes [9]. Under normal conditions, Nrf2 is bound to Kelch-like ECH-associated protein 1 (Keap1) in the cytoplasm, and degraded by E3 ubiquitin-proteasome. Therefore, intracellular Nrf2 protein levels are maintained at a low level. Under stressed conditions, Keap1 goes through a conformational change and Nrf2 dissociates from it. As a consequence, the degradation of Nrf2 is reduced, leading to accumulation of Nrf2 in the nucleus, promoting the transcription of its downstream antioxidant and cytoprotective genes [10]. Recently, emerging evidences show that overexpression of Nrf2 is involved in cell proliferation and chemoresistance in various cancers [[11], [12], [13], [14]]. Here, we reported a novel mechanism of HEATR1 on the regulation of Nrf2 signaling pathway. We found that HEATR1 inhibited the activity of Nrf2 via competing with Keap1 for p62 binding. Moreover, HEATR1 deficiency could promote pancreatic cancer proliferation and gemcitabine resistance. In addition, we also demonstrated that low expression of HEATR1 was closely related to poor prognosis and clinicopathology features of pancreatic cancer patients. These finding indicate that HEATR1 may be a promising therapeutic target for pancreatic cancer.
2. Materials and methods
2.1. Materials
MTT (purity>98%), cycloheximide (purity>93%), gemcitabine (purity>98%) and primary antibody for lamin A were purchased from Sigma-Aldrich (St. Louis, USA). MG132 (purity>97%) was purchased from Selleck Chemicals (Houston, USA). KI696 was obtained from MedChemExpress (NJ, USA). Primary antibodies for HEATR1, NQO1, HO-1 and Kras were obtained from Santa Cruz Biotechnology (Texas, USA). Primary antibody for p62 (SQSTM1) was obtained from ABclonal (Wuhan, China). Primary antibodies for Keap1, Nrf2 and cleaved caspase-3 were purchased from Cell Signaling Technology (Danvers, USA). Primary antibodies for ubiquitin, β-actin, and Ki67 were purchased from Bioworld (Minnesota, USA). Primary antibodies for HA and FLAG were purchased from proteintech group (IL, USA).
2.2. Cell culture and RNA interference
Panc-1, MiaPaCa-2, A549, H460 and HEK293T cells were obtained from Cell Bank of the Chinese Academic of Sciences (Shanghai, China). Panc-1, MiaPaCa-2, A549 and HEK293T cells were cultured in completed DMEM medium (Gibco). H460 cells were cultured in completed RPMI-1640 medium (Gibco). All cells were cultured under a humidified 5% CO2 environment at 37 °C. The lentiviral particles encoding non-targeting, HEATR1-targeting, Nrf2-targeting, Keap1-targeting and p62-targeting shRNAs (Sigma-Aldrich, St. Louis, USA) were incubated with cells for 72 h and cells infected with lentiviral particles were selected by using puromycin. The HEATR1, Nrf2, Keap1 and p62 protein levels were detected by using western blot.
2.3. Construction of recombinant DNA molecules
HEATR1, p62, KrasG12C and KrasG12D plasmids were obtained from Sangon Biotech (Shanghai, China). Wild-type and truncated mutants of HEATR1 were subcloned into pCMV-FLAG plasmids (Beyotime, Shanghai, China). Wild-type and truncated mutants of p62 were subcloned into pCMV-HA plasmids (Beyotime, Shanghai, China). All plasmids transfection experiments were performed by using ExFect transfection reagent (Vazyme, Nanjing, China) according to manufacturer's protocol.
2.4. Real-time quantitative PCR
RNA samples were reverse-transcribed to cDNA and then Real-time PCR was performed with the Light-Cycler1 96 Real-Time PCR System (Roche) using AceQ qPCR SYBR Green Master Mix (Vazyme). The primer sequences used in this study were shown in Supplementary Table S1.
2.5. Western blot and immunoprecipitation
Whole-cell and nuclear protein samples were extracted. Western blot was performed according to a standard protocol. For immunoprecipitation, the protein samples were incubated with indicated antibodies overnight at 4 °C, and then incubated with protein A + G agarose beads (Beyotime, Shanghai, China) for another 4 h at 4 °C. Immunoprecipitation mixtures were detected by using western blot with indicated primary antibodies.
2.6. CHX-chase analysis
Firstly, 25 μM of cycloheximide were incubated with cells to inhibit protein synthesis. Then, cell protein samples at indicated time points were extracted and detected by using western blot with specific primary antibodies for Nrf2 and β-actin.
2.7. MTT assay
Cells stably expressing the indicated shRNA were seeded into 96-well plates (5000 cells/well), then treated with various concentrations of gemcitabine for 24 h. MTT assay was performed following the manufacturer's protocol. The absorbance was measured by using a microplate reader at 570 nm.
2.8. Cell growth assay
Cells stably expressing the indicated shRNA were seeded into 6-well plates (10000 cells/well). The number of viable cells per well were measured daily.
2.9. Colony formation assay
About 500 cells stably expressing the indicated shRNA were seeded into 35 mm culture dishes and incubated for 14 days. 4% formaldehyde was used to fixate Cells for 20 min. Then, 0.5% crystal violet was used to stain cells.
2.10. Immunofluorescence staining
Immunofluorescence staining was performed as described previously [15]. Images were acquired by inverted fluorescence microscope (Nikon, Japan).
2.11. Measurement of intracellular ROS level
Intracellular ROS level was detected by using ROS assay kit (Beyotime, Shanghai, China) according to the manufacturer's instructions. The fluorescence intensity was measured by using microplate reader at Ex./Em. = 488/525 nm.
2.12. Tumor xenograft experiment
Female BALB/c nude mice (6 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). All protocols for mice were approved by the Animal Ethics Committee of China Pharmaceutical University. Mice were injected with the non-targeting shRNA (shControl)-, Nrf2-targeting shRNA (shNrf2)-, HEATR1-targeting shRNA (shHEATR1)-, or HEATR1/Nrf2-targeting shRNA (shHEATR1/Nrf2)-transfected Panc-1 cells (200 μl, 2 × 106 cells) in the subdermal space. Body weight and tumor volume were measured every week (n = 5/each group). Tumor volume = (m × m × n)/2 (m, the smallest diameter; n, the largest diameter). For the gemcitabine treatment, Mice were injected with the shControl-, shNrf2-, shHEATR1-, or shHEATR1/Nrf2-transfected Panc-1 cells (200 μl, 2 × 106 cells) in the subdermal space. Once the tumors reached 80–100 mm3, the mice were treated with PBS or gemcitabine (50 mg/kg, Once every four days, intraperitoneally) for 24 days (n = 5/each group). Body weight and tumor volume were measured every four days.
2.13. Immunohistochemistry staining
Immunohistochemical staining assay was performed by using immunohistochemistry kit (Maixin Biotech, Fuzhou, China) according to the manufacturer's protocol. All sections were photographed by using inverted fluorescence microscope (Nikon, Japan).
2.14. Human pancreatic cancer tissue microarray
A human pancreatic adenocarcinoma tissues microarray was purchased from Shanghai Outdo Biotech (Shanghai, China), which contained pancreatic adenocarcinoma and paired adjacent pancreatic tissues. All patients had been pathologically diagnosed with pancreatic cancer. Immunohistochemistry staining was used to analyze HEATR1 protein levels in human pancreatic adenocarcinoma tissues and paired adjacent pancreatic tissues. The staining of tumor tissues was observed under microscope, and the staining intensity was evaluated (score 0 = none staining; score 1 = Weak/light yellow staining; score 2 = Moderate/light brown staining; score 3 = Strong/dark brown staining). The intensity of HEATR1 staining was scored from 0 to 3 and grouped into low expression (score = 0, 1) and high expression (score = 2, 3). Scoring was evaluated by investigators who were blinded to the clinical information.
2.15. Statistical analysis
For patient results, the χ2 test was used to analyze the correlation between HEATR1 expression and clinicopathologic variables. Kaplan-Meier analysis was performed for overall survival analyses. Overall survival curve was calculated according to Kaplan-Meier analysis. Multivariable Cox proportional hazards regression model was performed to analyze the relative risk for patient poor outcome. The other results were expressed as mean ± SD and were representative of three independent experiments. Statistical analysis was performed with the t-test for two groups or one-way ANOVA for multiple groups by using SPSS statistical software. P < 0.05 was considered significant.
3. Results
3.1. HEATR1 inhibited Nrf2 protein levels in pancreatic cancer cells
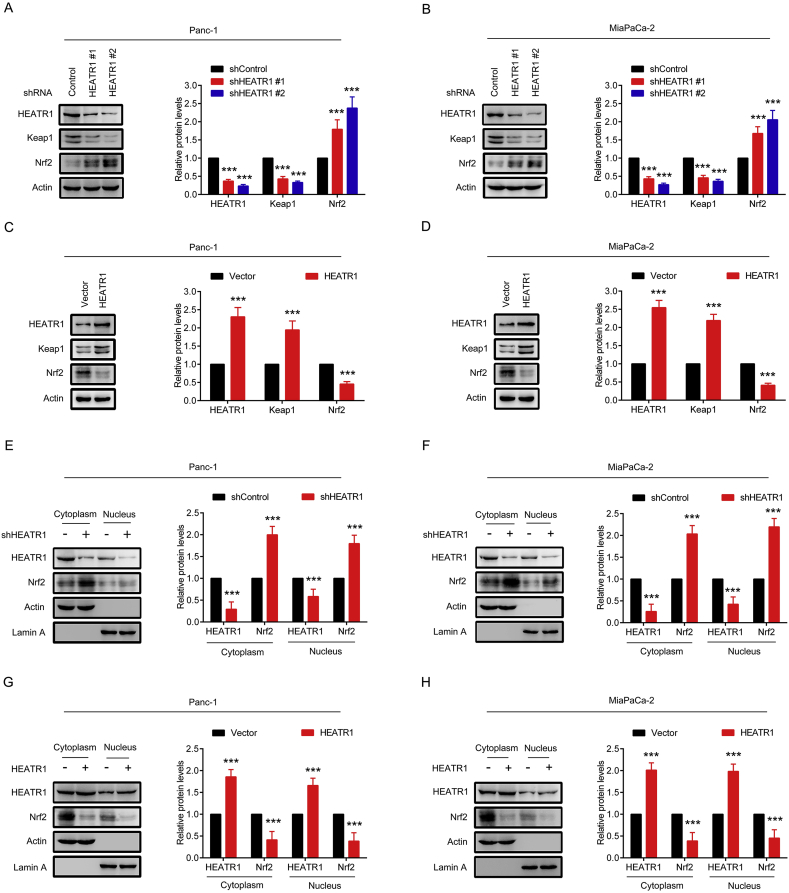
To identify whether HEATR1 was involved in regulating Nrf2 signaling, the Nrf2 and Keap1 protein levels in pancreatic cancer cells were detected after HEATR1 knockdown or overexpression. As shown in Fig. 1A–B and Supplementary Figs. S1A–B, HEATR1 protein levels in Panc-1 and MiaPaCa-2 cells with shHEATR1 transfection were significantly decreased. Moreover, the silence efficiency of shHEATR1 #2 was stronger than that of shHEATR1 #1 (Fig. 1A–B). Therefore, shHEATR1 #2 was used to silence HEATR1 in the following experiments. In addition, HEATR1 knockdown markedly reduced Keap1 protein levels and increased Nrf2 protein levels in Panc-1 and MiaPaCa-2 cells, while Keap1 protein levels were significantly up-regulated and Nrf2 protein levels were down-regulated in cells with overexpression of HEATR1 (Fig. 1A–D). Notably, cytoplasmic and nuclear Nrf2 protein levels were markedly up-regulated in cells with HEATR1 knockdown and down-regulated in cells with overexpression of HEATR1 (Fig. 1E–H). These data demonstrated that HEATR1 inhibited Nrf2 protein levels in pancreatic cancer cells.
Fig. 1.
HEATR1 inhibited Nrf2 protein levels in pancreatic cancer cells. (A–B) Western blot analysis of HEATR1, Keap1 and Nrf2 protein levels in Panc-1 and MiaPaCa-2 cells stably expressing control shRNA or two specific HEATR1 shRNA. ***P < 0.001 compared with shControl group. (C–D) Western blot analysis of HEATR1, Keap1 and Nrf2 protein levels in Panc-1 and MiaPaCa-2 cells stably expressing vector or HEATR1. ***P < 0.001 compared with vector group. (E–H) The distribution of HEATR1 and Nrf2 in Panc-1 and MiaPaCa-2 cells with HEATR1 knockdown or overexpression. ***P < 0.001 compared with shControl group or vector group. Data were expressed as mean ± SD, and the results were representative of three independent experiments.
3.2. HEATR1 inhibited the expression of Nrf2 target genes in pancreatic cancer cells
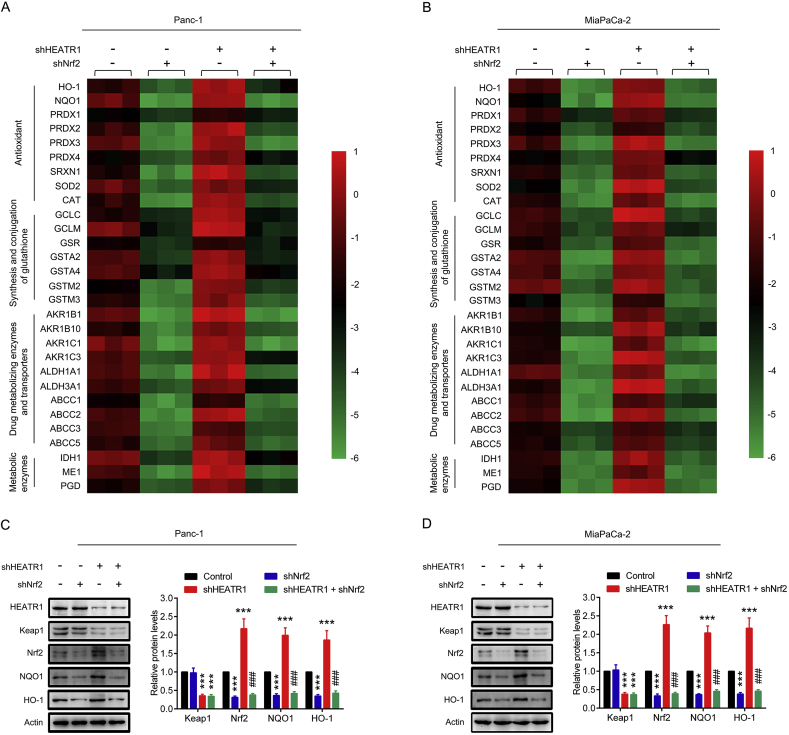
We next investigated whether HEATR1 could inhibit Nrf2 target genes levels in pancreatic cancer cells. Indeed, qRT-PCR results demonstrated that HEATR1 knockdown significantly promoted the mRNA levels of Nrf2 target genes in Panc-1 and MiaPaCa-2 cells, while Nrf2 knockdown markedly inhibited HEATR1 depletion-induced up-regulation of these genes (Fig. 2A–B). In addition, HEATR1 knockdown markedly increased NQO1 and HO-1 protein levels and Nrf2 knockdown markedly inhibited HEATR1 depletion-induced up-regulation of NQO1 and HO-1 protein levels in cells (Fig. 2C–D). These data suggested that HEATR1 inhibited the expression of Nrf2 target genes in pancreatic cancer cells.
Fig. 2.
HEATR1 inhibited the expression of Nrf2 target genes in pancreatic cancer cells. (A–B) Effects of HEATR1 on the expression of Nrf2 target genes in Panc-1 and MiaPaCa-2 cells. The colors of the heatmap reflect log2 (expression levels of Nrf2 target genes) in Panc-1 and MiaPaCa-2 cells. (C–D) Western blot analysis of cell extracts from Panc-1 and MiaPaCa-2 cells stably expressing indicated shRNAs. ***P < 0.001 compared with control group. ###P < 0.001 compared with shHEATR1 group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. HEATR1 inhibited Nrf2 protein level through enhancing ubiquitin-proteasome degradation of Nrf2
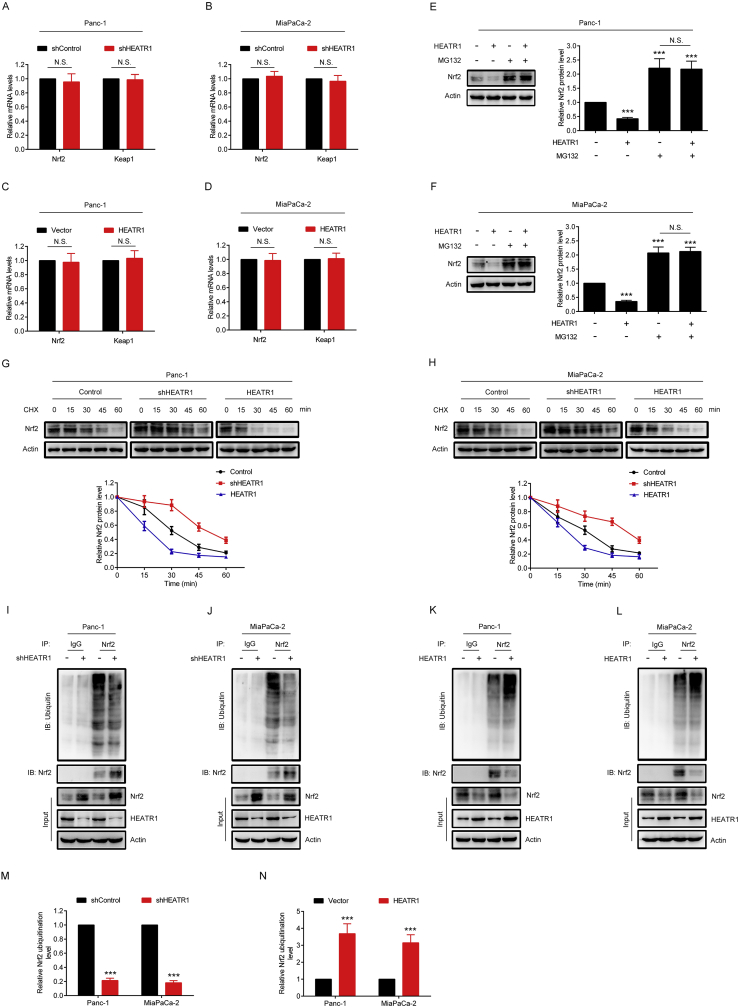
As shown in Fig. 3A–D, knockdown or overexpression of HEATR1 did not significantly alter the mRNA levels of Nrf2 and Keap1, suggesting that HEATR1 inhibited Nrf2 protein level not through a transcriptional mechanism. Therefore, we postulated that HEATR1 might inhibit Nrf2 through enhancing ubiquitin-proteasome degradation of Nrf2. We noted that MG132 (a proteasomal inhibitor) could completely restore HEATR1 overexpression-induced Nrf2 reduction (Fig. 3E–F). Moreover, HEATR1 deficiency and overexpression could significantly prolong and shorten degradation half-life of Nrf2, respectively (Fig. 3G–H), indicating that HEATR1 inhibited Nrf2 through promoting its proteasome-mediated degradation. In the meanwhile, the level of ubiquitin-Nrf2 was decreased in cells with HEATR1 knockdown (Fig. 3I, J, M), indicating that HEATR1 knockdown inhibited ubiquitination of Nrf2. Conversely, the level of ubiquitin-Nrf2 was increased in cells overexpressing HEATR1 (Fig. 3K, L, N), suggesting that HEATR1 overexpression facilitated the ubiquitination of Nrf2. These data demonstrated that HEATR1 inhibited Nrf2 protein level through enhancing ubiquitin-proteasome degradation of Nrf2.
Fig. 3.
HEATR1 inhibited Nrf2 protein level through enhancing ubiquitin-proteasome degradation of Nrf2. (A–D) qRT-PCR. (E–F) Panc-1 and MiaPaCa-2 cells stably expressing vector or HEATR1 were treated with or without MG132 (20 μM) for 4 h, western blot was used to detect Nrf2 protein level. ***P < 0.001 compared with control group. (G–H) CHX-chase analysis. (I–N) Ubiqutin-Nrf2 was determined by immunoprecipitation of Nrf2 with a subsequent western blot analysis with anti-ubiquitin antibody in Panc-1 and MiaPaCa-2 cells with HEATR1 knockdown or overexpression. ***P < 0.001 compared with shControl group or vector group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. N.S., no significant.
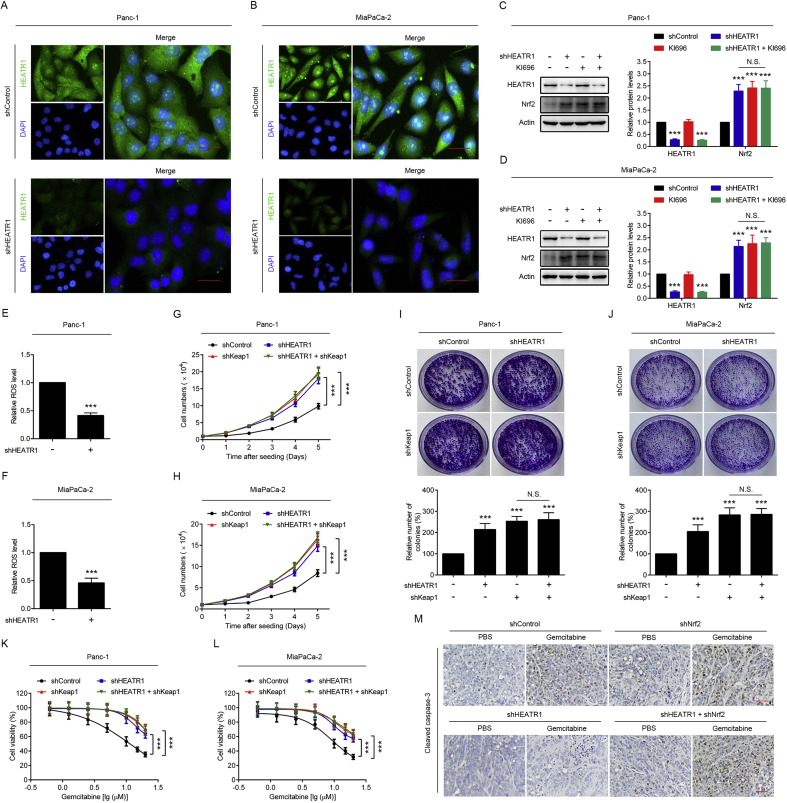
3.4. HEATR1 inhibited Nrf2 signaling in a p62/Keap1-dependent manner
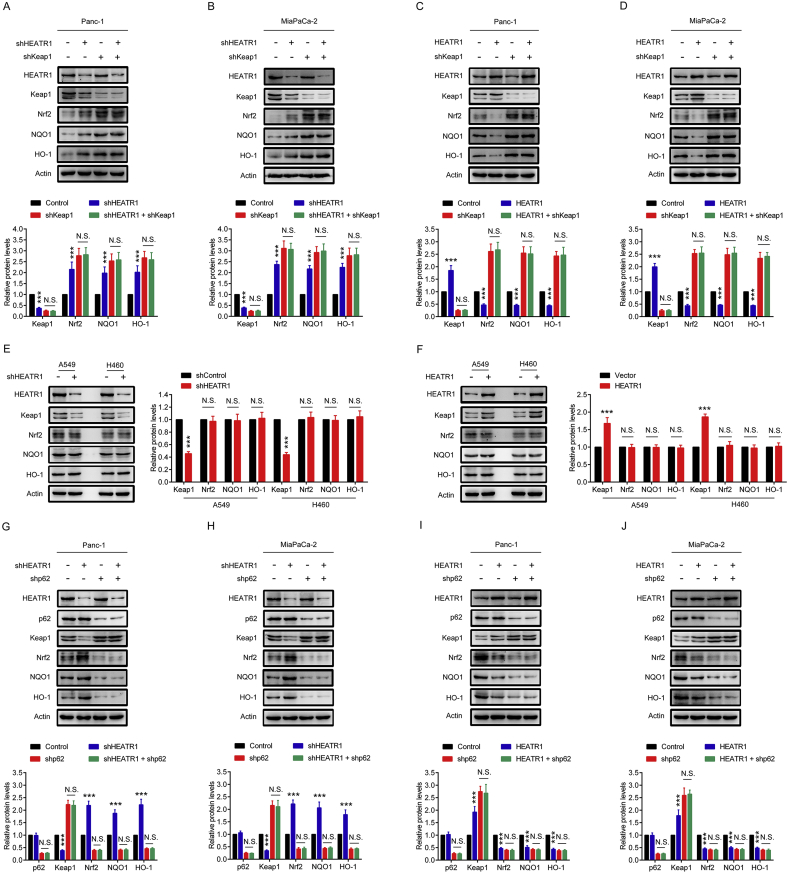
To investigate whether HEATR1-mediated inhibition of Nrf2 was Keap1-dependent, we performed knockdown and overexpression of HEATR1 in Keap1 knockdown cells. As shown in Fig. 4A–D, neither HEATR1 deficiency nor HEATR1 overexpression could significantly alter the Keap1 knockdown-induced up-regulation of Nrf2. Moreover, we found that HEATR1 deficiency did not further promote KI696 (an inhibitor of the Keap1/Nrf2 interaction)-induced up-regulation of Nrf2 in cells (Supplementary Figs. S1C–D). We then tested the effect of HEATR1 on the activation of Nrf2 in two Keap1 mutant A549 and H460 lung cancer cell lines. It was noted that HEATR1 deficiency and overexpression significantly inhibited and promoted mutant Keap1 protein levels in A549 and H460 cells, respectively, while Nrf2 protein levels were not affected (Fig. 4E–F), further demonstrating that HEATR1 negatively regulated Nrf2 in a Keap1-dependent manner. In order to elucidate if p62 played an essential role in HEATR1-mediated regulation of Nrf2 signaling, we performed knockdown and overexpression of HEATR1 in p62 knockdown cells. It was found that Keap1 and Nrf2 protein levels in cells with both HEATR1 and p62 knockdown did not significantly change when compared with cells with only p62 knockdown (Fig. 4G–H). Meanwhile, the overexpression of HEATR1 had no effect on p62 knockdown-induced up-regulation of Keap1 and down-regulation of Nrf2 in these cells (Fig. 4I–J). In addition, we detected the ROS levels in pancreatic cancer cells with or without HEATR1 knockdown, and it was found that HEATR1 knockdown significantly inhibited ROS levels in cells, indicating that HEATR1 deficiency did not up-regulate Nrf2 signaling through increasing ROS levels in cells (Supplementary Figs. S1E–F). Collectively, HEATR1 regulated Nrf2 signaling in a p62/Keap1-dependent manner.
Fig. 4.
HEATR1 inhibited Nrf2 signaling in a p62/Keap1-dependent manner. (A–B) Cell lysates of Panc-1 and MiaPaCa-2 cells stably expressing indicated shRNAs were detected by western blot analysis. ***P < 0.001 compared with control group. (C–D) Panc-1 and MiaPaCa-2 cells with Keap1 knockdown were transfected with empty plasmid vector and HEATR1 overexpression plasmid vector, cell lysates were detected by western blot analysis. ***P < 0.001 compared with control group. (E) A549 and H460 cells were transfected with control shRNA or HEATR1 shRNA, cell lysates were detected by western blot analysis. ***P < 0.001 compared with shControl group. (F) A549 and H460 cells were transfected with empty plasmid vector and HEATR1 overexpression plasmid vector, cell lysates were detected by western blot analysis. ***P < 0.001 compared with vector group. (G–H) Panc-1 and MiaPaCa-2 cells stably expressing the indicated shRNAs, cell lysates were detected by western blot analysis. ***P < 0.001 compared with control group. (I–J) Panc-1 and MiaPaCa-2 cells with p62 knockdown were transfected with empty plasmid vector and HEATR1 overexpression plasmid vector, cell lysates were detected by western blot analysis. ***P < 0.001 compared with control group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. N.S., no significant.
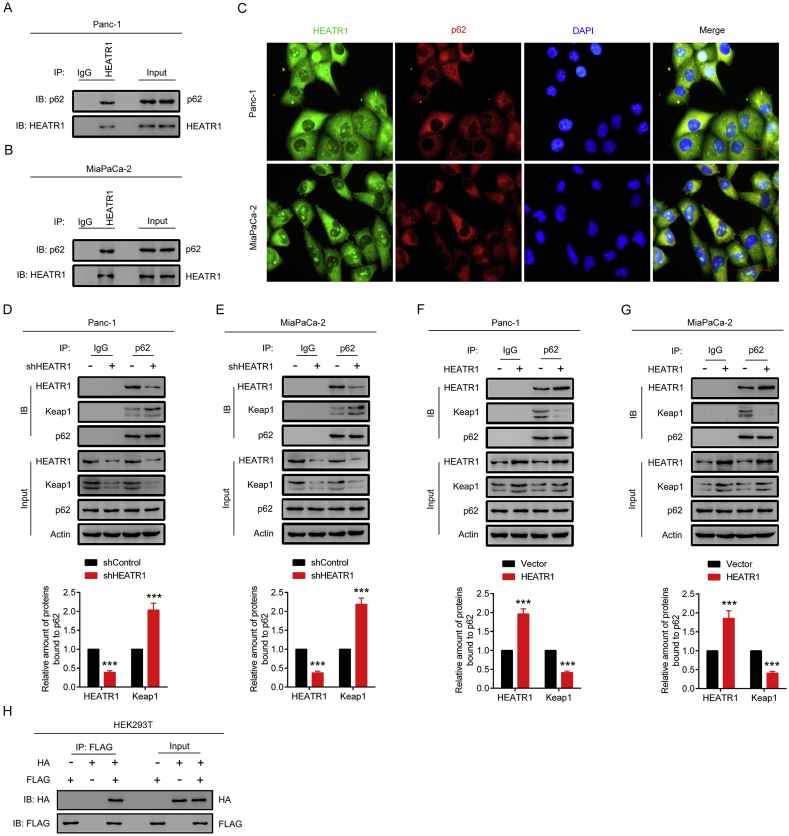
3.5. HEATR1 inhibited Nrf2 signaling through competing with Keap1 for p62 binding
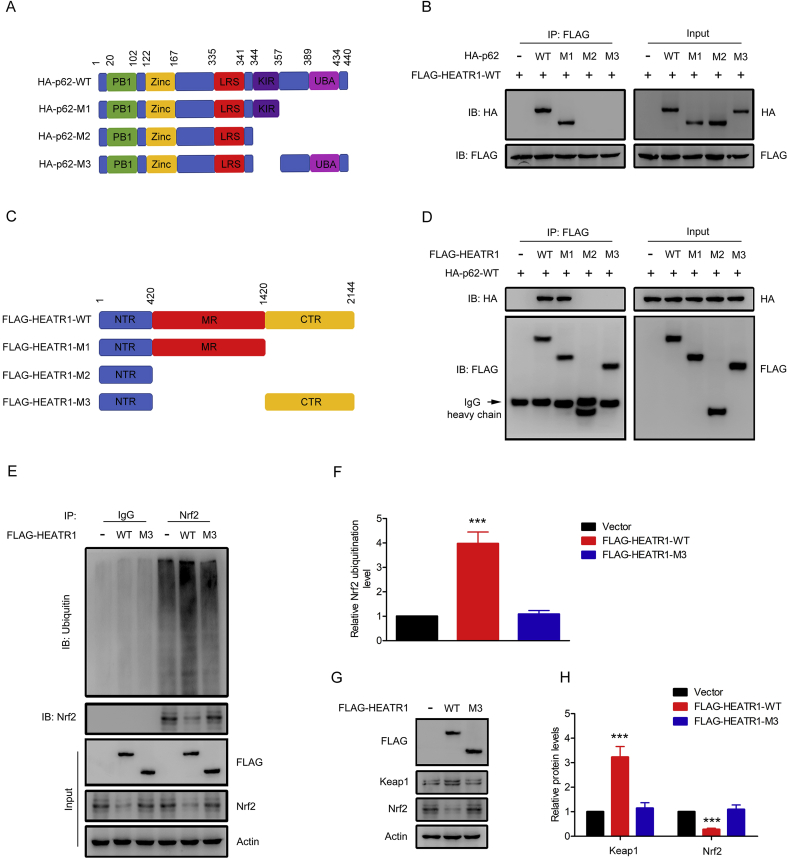
It has been shown that p62 regulates Nrf2 pathway through binding with Keap1. In our study, we found that HEATR1 could not significantly change the protein levels of p62 in Panc-1 and MiaPaCa-2 cells (Fig. 4G–J). However, immunoprecipitation assay revealed that endogenous HEATR1 was able to interact with p62 in Panc-1 and MiaPaCa-2 cells (Fig. 5A–B). Moreover, immunofluorescence assay suggested that endogenous HEATR1 and p62 colocalized in the cytoplasm (Fig. 5C). Interestingly, HEATR1 knockdown increased whereas HEATR1 overexpression decreased the interaction between p62 and Keap1, indicating that HEATR1 competed with Keap1 for binding with p62 (Fig. 5D–G). Moreover, when FLAG-tagged HEATR1 and HA-tagged p62 plasmids were transfected into HEK293T cells, it was found that FLAG-tagged HEATR1 was able to interact with HA-tagged p62 (Fig. 5H). In order to further investigate how HEATR1 disrupted the p62-Keap1 interaction, we constructed truncated mutants of FLAG-tagged HEATR1 and HA-tagged p62 and transfected them into HEK293T cells. As shown in Fig. 6A–B, FLAG-tagged HEATR1 only interacted with the wild type (WT) and truncated mutants containing KIR domain of HA-tagged p62 in HEK293T cells, but not other truncated mutants, suggesting that KIR domain in p62, which is the binding site for Keap1, bound with HEATR1 as well, confirming the competitive binding of Keap1 and HEATR1 with p62. Moreover, HA-tagged p62 only interacted with the WT and truncated mutants containing middle region (aa 420–1420) of FLAG-tagged HEATR1 in HEK293T cells (Fig. 6C–D), suggesting that middle region (aa 420–1420) of HEATR1 played an important role in interacting with p62. In addition, WT FLAG-tagged HEATR1 could significantly increase ubiquitination of Nrf2 and decrease Nrf2 protein level in Panc-1 cells, while ubiquitination of Nrf2 and Nrf2 protein level did not significantly change in Panc-1 cells with overexpression of M3, the FLAG-tagged HEATR1 truncated mutants not containing middle region (aa 420–1420) (Fig. 6E–H). Collectively, our results demonstrated that HEATR1 inhibited Nrf2 signaling through competing with Keap1 for p62 binding.
Fig. 5.
HEATR1 inhibited Nrf2 signaling through competing with Keap1 for p62 binding. (A–B) Endogenous interaction between HEATR1 and p62. Immunoprecipitations were performed using IgG, anti-HEATR1 antibodies, and analyzed by western blotting using the indicated antibodies. (C) Immunofluorescence assay. Scale bars, 20 μm. (D–E) Cell lysates of Panc-1 and MiaPaCa-2 cells stably expressing control shRNA or HEATR1 shRNA were co-immunoprecipitated with IgG or anti-p62 antibody and then analyzed by western blotting. ***P < 0.001 compared with shControl group. (F–G) Cell lysates of Panc-1 and MiaPaCa-2 cells stably expressing vector or HEATR1 were immunoprecipitated with IgG or anti-p62 antibody and then analyzed by western blotting. ***P < 0.001 compared with vector group. (H) FLAG-tagged HEATR1 and HA-tagged p62 plasmids were transfected into HEK293T cells. Immunoprecipitations were performed using anti-FLAG antibody, and analyzed by western blot using the indicated antibodies. Data were expressed as mean ± SD, and the results were representative of three independent experiments.
Fig. 6.
HEATR1 interacted with the KIR domain of p62 via its middle region. (A) Schematic description of WT (wild-type) and truncated mutants p62. PB1, Phox and Bem1p; Zinc, zinc finger; LRS, LC3-recognition sequence; KIR, Keap1-interacting region; UBA, ubiquitin-associated domain. (B) The interaction between HEATR1 and the WT and truncated mutants of p62 were determined by co-immunoprecipitation assay in HEK293T cells. (C) Schematic description of WT (wild-type) and truncated mutants HEATR1. NTR, N-terminal region; MR, Middle region; CTR, C-terminal region. (D) The interaction between p62 and the WT and truncated mutants of HEATR1 were determined by co-immunoprecipitation assay in HEK293T cells. (E–F) Panc-1 cells were transfected with WT and truncated mutants of HEATR1, ubiqutin-Nrf2 was determined by immunoprecipitation of Nrf2 with a subsequent western blot analysis. ***P < 0.001 compared with vector group. (G–H) Panc-1 cells were transfected with WT and truncated mutants of HEATR1, cell lysates were detected by western blot analysis. ***P < 0.001 compared with vector group. Data were expressed as mean ± SD, and the results were representative of three independent experiments.
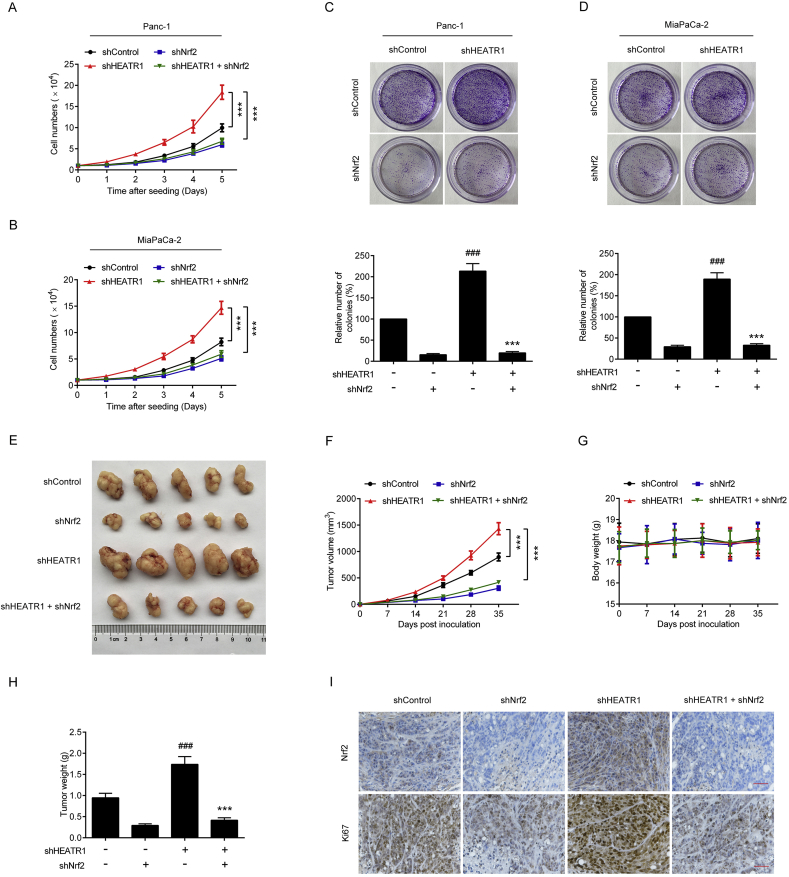
3.6. HEATR1 deficiency promoted proliferation of pancreatic cancer through up-regulating Nrf2 signaling
We next investigated if HEATR1 deficiency promoted proliferation of pancreatic cancer through up-regulating Nrf2 signaling. As shown in Fig. 7A–D, HEATR1 depletion dramatically improved cell growth and colony formation of pancreatic cancer cells, which could be blocked by knockdown of Nrf2. Moreover, HEATR1 depletion did not further promote Keap1 knockdown-induced cell growth and colony formation of pancreatic cancer cells (Supplementary Figs. S1G–J). In in vivo study, Panc-1-shHEATR1 cells-derived xenografts grew more rapidly than Panc-1-shControl cells-derived xenografts (Fig. 7E, F, H). However, knockdown of Nrf2 significantly reduced HEATR1 depletion-induced increase of tumor volume and tumor weight (Fig. 7E, F, H), with no markedly change of body weight observed (Fig. 7G). In addition, Panc-1-shHEATR1 cells-derived xenografts exhibited higher Nrf2 and Ki67 protein levels, and Nrf2 depletion significantly reduced Nrf2 and Ki67 protein levels in Panc-1-shHEATR1 cells-derived xenografts (Fig. 7I). These data suggested that HEATR1 deficiency promoted proliferation of pancreatic cancer through up-regulating Nrf2 signaling.
Fig. 7.
HEATR1 deficiency promoted proliferation of pancreatic cancer through up-regulating Nrf2 signaling. (A–B) Cell growth curves. ***P < 0.001 compared with shHEATR1 group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. (C–D) Colony formation assay. ***P < 0.001 compared with shHEATR1 group. ###P < 0.001 compared with shControl group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. (E–H) Panc-1 cells stably expressing indicated shRNA were injected into the subdermal space of nude mice. Tumor volume (n = 5) and body weight (n = 5) were recorded every week. The tumors were photographed and weighed after removing from nude mice (n = 5). ***P < 0.001 compared with shHEATR1 group. ###P < 0.001 compared with shControl group. (I) Immunohistochemistry staining. Scale bar, 50 μm.
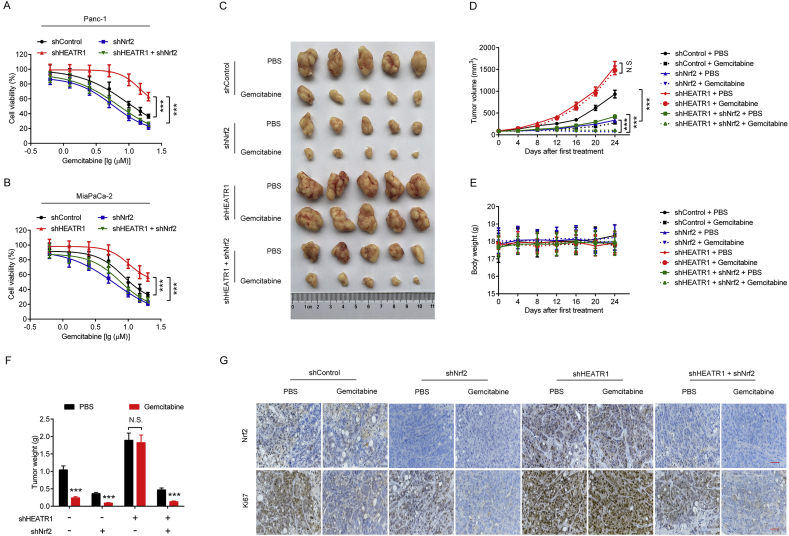
3.7. HEATR1 deficiency promoted gemcitabine resistance of pancreatic cancer through up-regulating Nrf2 signaling
We then investigated if HEATR1 deficiency enhanced gemcitabine resistance in pancreatic cancer. As shown in Fig. 8A–B, Panc-1 and MiaPaCa-2 cells with HEATR1 knockdown exhibited resistance to gemcitabine. To see if HEATR1 deficiency-induced gemcitabine resistance was depended on Nrf2, we depleted Nrf2 with shRNA and found that Nrf2 depletion completely reversed HEATR1 deficiency-induced gemcitabine resistance (Fig. 8A–B). Moreover, HEATR1 depletion did not further promote Keap1 knockdown-induced gemcitabine resistance in cells (Supplementary Fig. S1K-L). In in vivo study, gemcitabine treatment significantly inhibited Panc-1-shControl or -shNrf2 cells-derived xenografts growth (Fig. 8C, D, F). However, Panc-1-shHEATR1 cells-derived xenografts exhibited marginal response to gemcitabine (Fig. 8C, D, F), and Nrf2 knockdown significantly enhanced the sensitivity of Panc-1-shHEATR1 cells-derived xenografts to gemcitabine (Fig. 8C, D, F). Meanwhile, body weight of mice did not markedly change during these experiments (Fig. 8E). Moreover, we found that gemcitabine treatment could not markedly inhibit Ki67 protein level in Panc-1-shHEATR1 cells-derived xenografts (Fig. 8G), which was significantly decreased in Panc-1-shHEATR1/Nrf2 cells-derived xenografts treated with gemcitabine when compared with PBS control (Fig. 8G). In addition, we found that gemcitabine treatment did not significantly increase cleaved caspase-3 protein level in Panc-1-shHEATR1 cells-derived xenografts (Supplementary Fig. S1M). Collectively, HEATR1 deficiency promoted gemcitabine resistance of pancreatic cancer through up-regulating Nrf2 signaling.
Fig. 8.
HEATR1 deficiency promoted gemcitabine resistance of pancreatic cancer through up-regulating Nrf2 signaling. (A–B) MTT assay. ***P < 0.001 compared with shHEATR1 group. Data were expressed as mean ± SD, and the results were representative of three independent experiments. (C–F) Panc-1 cells-derived xenografts expressing indicated shRNA were treated with PBS or gemcitabine. Tumor volume (n = 5) and body weigh (n = 5) were recorded every four days. The tumors were photographed and weighed after removing from nude mice (n = 5). ***P < 0.001 compared with PBS group. (G) Immunohistochemistry staining. Scale bar, 50 μm. N.S., no significant.
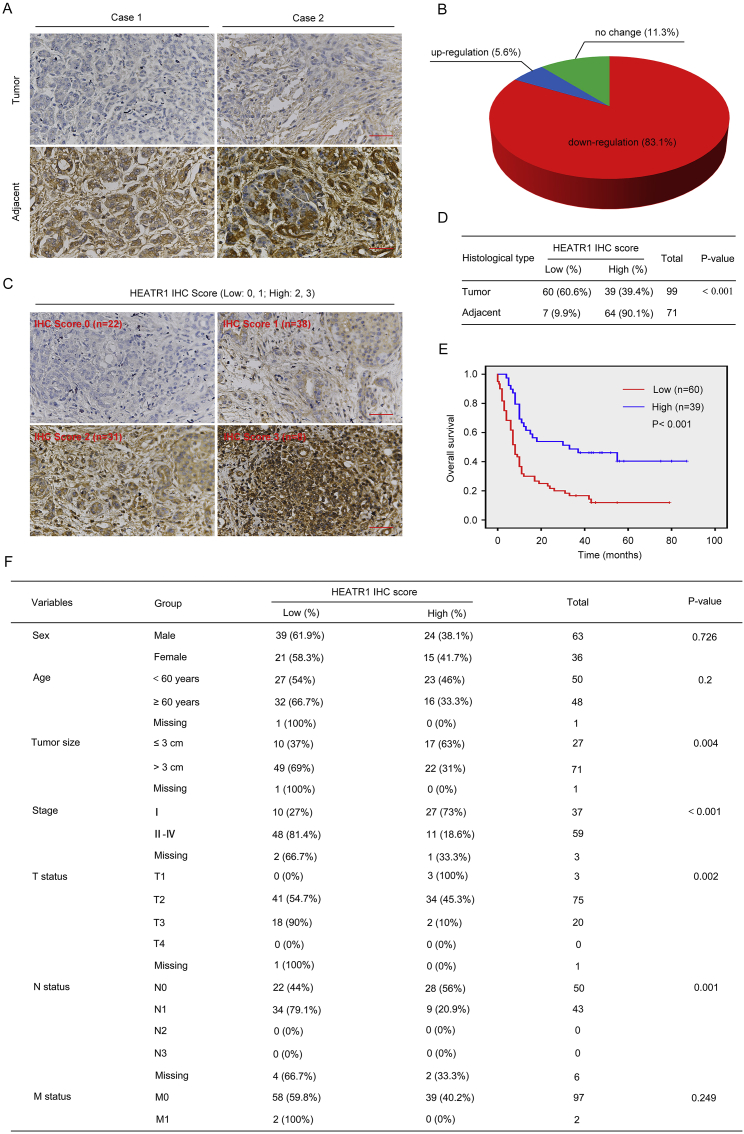
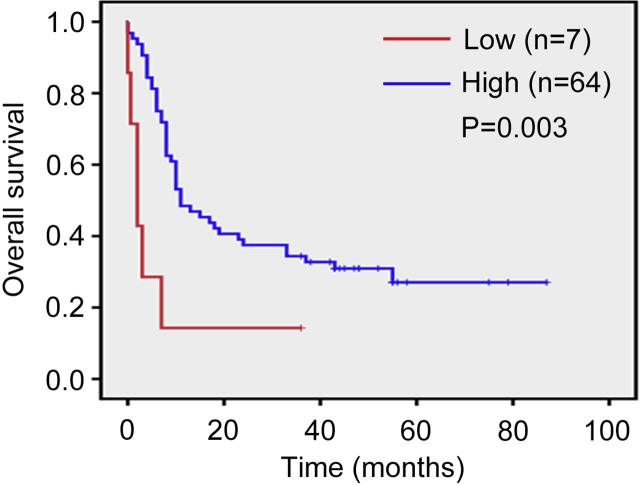
3.8. 8. HEATR1 was down-regulated in primary pancreatic cancer samples and associated with poor prognosis
To investigate if HEATR1 was down-regulated in human pancreatic cancer tissues, we determined the protein level of HEATR1 in tissue microarrays which contained pancreatic cancer tissues and paired adjacent pancreatic tissues by using immunohistochemical analysis. We found that HEATR1 protein level was markedly higher in adjacent pancreatic tissues than pancreatic cancer tissues (Fig. 9A). Moreover, about 83.1% of pancreatic cancer patients had lower HEATR1 protein level in tumor tissues than in adjacent tissues (Fig. 9B). In all pancreatic cancer tissues, 60.6% had low HEATR1 expression (IHC score 0 or 1, Fig. 9D), while in all adjacent tissues, 9.9% had low HEATR1 expression (IHC score 0 or 1, Fig. 9D). By analyzing Kaplan–Meier survival curve, it was found that there was a significant correlation between HEATR1 protein expression (tumor tissues or adjacent tissues) and pancreatic cancer patient survival, with low expression of HEATR1 (IHC score 0 or 1) associated with poor prognosis (Fig. 9E and Supplementary Fig. S2). Importantly, Multivariate analysis indicated that low HEATR1 protein expression (IHC score 0 or 1) was an independent poor prognostic factor (Supplementary Table S2). As shown in Fig. 9F, HEATR1 protein expression had significant correlation with clinicopathological features including tumor size (P = 0.004), clinical stage (P < 0.001), T status (P = 0.002) and N status (P = 0.001).
Fig. 9.
HEATR1 was frequently down-regulated in pancreatic cancer. (A) The protein expression of HEATR1 in tumor and adjacent nontumor tissues. Scale bars, 50 μm. (B) The proportion of patients with down-regulation and up-regulation and no change of HEATR1. (C) Representative IHC images showing assigned intensity scores (0 = none staining; 1 = Weak/light yellow staining; 2 = Moderate/light brown staining; 3 = Strong/dark brown staining) of HEATR1 staining. Scale bars, 50 μm. (D) Correlation between histological type and HEATR1 protein expression. (E) Kaplan–Meier survival analysis of 99 patients based on low (score = 0, 1) or high (score = 2, 3) HEATR1 expression in tumor tissues. (F) Correlation between clinicopathologic variables of pancreatic cancer patients and HEATR1 protein expression. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Chemoresistance remains a major obstacle in cancer chemotherapy. Activation of Nrf2 signaling is associated with chemoresistance in different types of cancers. It has been reported that Nrf2 protects cancer cells from electrophiles and oxidants through promoting the expression of its target genes [16], and activation of Nrf2 promotes proliferation and chemoresistance of malignant pancreatic cancer cells [17]. However, the upstream regulatory mechanism of Nrf2 in pancreatic cancer is unclear. In this study, we found that HEATR1 negatively regulated Nrf2 in pancreatic cancer cells.
It has been shown that p62 is involved in regulating innate immune signaling complexes [18], and its physiological function is clarified through an important finding that Paget disease is caused by mutations within the ubiquitin-binding region of p62 gene [19]. The previous studies have shown that the stability of Nrf2 is regulated by p62 which binds to and recruits Keap1 into autophagosomes, leading to down-regulation of Keap1 protein level and activation of Nrf2 signaling [[20], [21], [22], [23]]. Moreover, these studies suggest that p62 binds to Keap1 through its KIR domain [[21], [22], [23]]. Interestingly, in this study, HEATR1 protein was also found to combine with the KIR domain of p62 protein. Therefore, we asked if HEATR1 competed with Keap1 for p62 binding. Indeed, we found that HEATR1 depletion or overexpression could increase or decrease the interaction between p62 and Keap1, respectively. Therefore, we suggest that HEATR1 negatively regulate the Nrf2 signaling through competing with Keap1 for binding to p62 in pancreatic cancer cells.
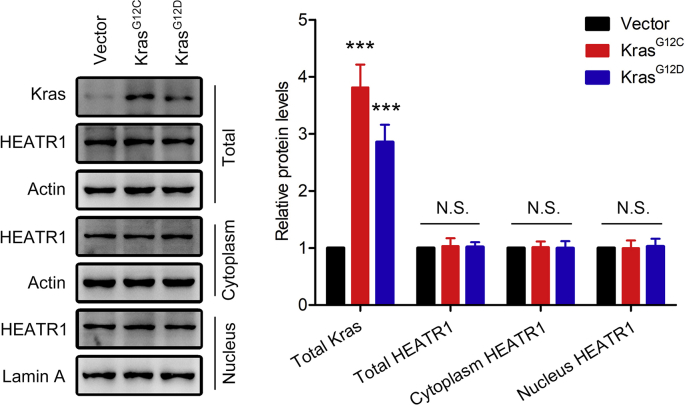
Recently, more and more attention is focused on finding diagnostic and prognostic biomarkers for pancreatic cancer patients [24]. However, existing biomarkers, including CA19-9, are not adequate as early diagnostic markers of pancreatic cancer patients because of low specificity and sensitivity [25]. Liu et al. demonstrated that poor prognosis of pancreatic cancer patients was correlated with down-regulation of HEATR1 protein level and pancreatic cancer cells with depletion of HEATR1 exhibited drug resistance, indicating that HEATR1 might be a potential diagnostic and prognostic biomarker for pancreatic cancer patients [7]. In consistent with previous study, we also demonstrated that HEATR1 was down-regulated in primary pancreatic cancer tissues and predicted poor prognosis, while HEATR1 depletion promoted proliferation and gemcitabine resistance of pancreatic cancer cells. Liu et al. also found that HEATR1 facilitated the interaction between AKT and its inactivating protein PP2A and inhibited the activity of AKT [7]. Except for regulating AKT pathway, we found that HEATR1 could also regulate Nrf2 signaling in pancreatic cancer cells. Moreover, we found that HEATR1 lost its ability to regulate Nrf2 signaling in pancreatic cancer cells with Keap1 or p62 knockdown and HEATR1 was unable to negatively regulate Nrf2 signaling pathway in Keap1 mutant human NSCLC cells (A549 and H460), indicating that HEATR1 regulated Nrf2 signaling pathway through p62-Keap1 axis. Previous studies have reported that β-TrCP and Hrd1 are involved in regulating the activity of Nrf2 in a Keap1-independent manner under certain circumstances [[26], [27], [28]]. It remains to be elucidated whether β-TrCP and Hrd1 were involved in the HEATR1-mediated regulation of Nrf2 signaling pathway. In present, the mechanism of HEATR1 down-regulation in pancreatic cancer is unclear. It has been reported that oncogene Kras is able to regulate the expression of Nrf2 in pancreatic cancer [29]. We asked whether oncogene Kras could regulate HEATR1 protein expression or intracellular localization, and it was found that overexpression of KrasG12C and KrasG12D did not alter the protein expression or intracellular localization of HEATR1 (Supplementary Fig. S3). Therefore, more research is needed to investigate the mechanism of HEATR1 down-regulation in pancreatic cancer.
Interestingly, one recent study found that HEATR1 knockdown inhibited proliferation of U2OS cells and induced p53-dependent cell cycle arrest [8], indicating that HEATR1 had tumor promoting effect. Unlike our finding presented here, they demonstrated that HEATR1 depletion disrupted ribosome biogenesis and activated the p53-p21 checkpoint response through the RPL5/RPL11-MDM2 axis. Here, we found that HEATR1 showed tumor suppressive effect in pancreatic cancer through down-regulating Nrf2 signaling. We speculated that the roles of HEATR1 might be related to tumor types. However, here we did not investigate the effect of HEATR1 on ribosome biogenesis and the p53-p21 checkpoint in pancreatic cancer cells. Thus, it is unclear whether HEATR1 could promote ribosome biogenesis and inhibit p53-p21 checkpoint in pancreatic cancer cells.
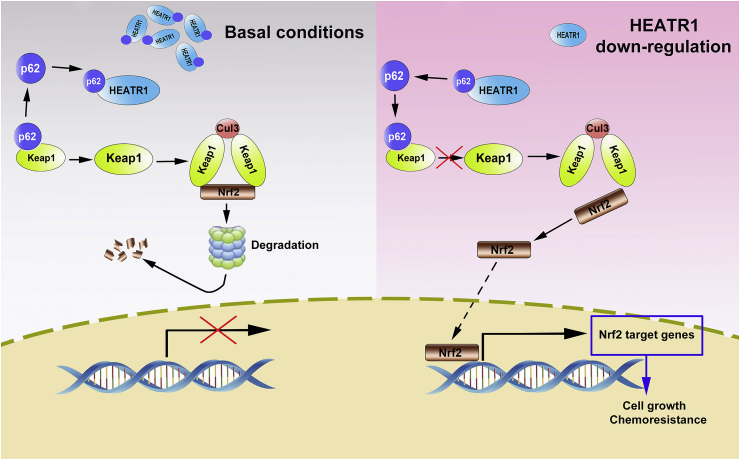
In conclusion, HEATR1 inhibits Nrf2 signaling via competing with Keap1 for p62 binding in pancreatic cancer and HEATR1 deficiency promotes proliferation and gemcitabine resistance of pancreatic cancer through regulating Nrf2 signaling (Fig. 10). As an alternative strategy, Nrf2 inhibitors (such as brusatol [30] and digoxin [15]) may be used in combination with gemcitabine for the treatment of pancreatic cancer patients with low HEATR1 protein levels.
Fig. 10.
Proposed model of regulation of Nrf2 signaling by HEATR1 in pancreatic cancer cells.
Declaration of competing interest
There is no conflict of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81372268, No. 81672816 and No. 81872337), the Program for Jiangsu Province Innovative Research (KYLX16_1120), the Natural Science Foundation for Distinguished Young Scholars of Jiangsu Province (No. BK20130026), the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (No. ZJ11173).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101390.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. S1.
(A-B) Immunofluorescence assay. Scale bar, 20 μm. (C-D) Panc-1 and MiaPaCa-2 cells were treated with or without KI696 (5 μM) for 24 h, and then HEATR1 shRNA were used to knockdown HEATR1 expression in cells. The protein levels of HEATR1 in cells were detected by using western blot. ***P < 0.001 compared with shControl group. (E-F) The levels of ROS in Panc-1 and MiaPaCa-2 cells stably expressing control shRNA or HEATR1 shRNA. ***P < 0.001 compared with shControl group. (G-H) Cell growth curves. ***P < 0.001 compared with shControl group. (I-J) Colony formation assay. ***P < 0.001 compared with shControl group. (K-L) MTT assay. ***P < 0.001 compared with shControl group. (M) Immunohistochemistry staining. Scale bar, 50 μm. Data were expressed as mean ± SD, and the results were representative of three independent experiments. N.S., no significant.
Supplementary Fig. S2.
Kaplan–Meier survival analysis of 71 patients based on low (score = 0, 1) or high (score = 2, 3) HEATR1 expression in adjacent tissues
Supplementary Fig. S3.
Effects of KrasG12C and KrasG12D overexpression plasmids on total HEATR1, cytoplasm HEATR1 and nucleus HEATR1 protein levels in HEK293T cells. Data were expressed as mean ± SD, and the results were representative of three independent experiments. ***P < 0.001 compared with vector group. N.S., no significant.
References
- 1.Stathis A., Moore M.J. Advanced pancreatic carcinoma: current treatment and future challenges. Nat. Rev. Clin. Oncol. 2010;7:163–172. doi: 10.1038/nrclinonc.2009.236. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA A Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.de Sousa C.L., Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura S.H., Hirano T. HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 2016;129:3963–3970. doi: 10.1242/jcs.185710. [DOI] [PubMed] [Google Scholar]
- 5.Wu Z.B., Qiu C., Zhang A.L., Cai L., Lin S.J., Yao Y., Tang Q.S., Xu M., Hua W., Chu Y.W., Mao Y., Zhu J.H., Xu J., Zhou L.F. Glioma-associated antigen HEATR1 induces functional cytotoxic T lymphocytes in patients with glioma. J Immunol Res. 2014;2014:131494. doi: 10.1155/2014/131494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azuma M., Toyama R., Laver E., Dawid I.B. Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J. Biol. Chem. 2006;281:13309–13316. doi: 10.1074/jbc.M601892200. [DOI] [PubMed] [Google Scholar]
- 7.Liu T., Fang Y., Zhang H., Deng M., Gao B., Niu N., Yu J., Lee S., Kim J., Qin B., Xie F., Evans D., Wang L., Lou W., Lou Z. HEATR1 negatively regulates Akt to help sensitize pancreatic cancer cells to chemotherapy. Cancer Res. 2016;76:572–581. doi: 10.1158/0008-5472.CAN-15-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turi Z., Senkyrikova M., Mistrik M., Bartek J., Moudry P. Perturbation of RNA Polymerase I transcription machinery by ablation of HEATR1 triggers the RPL5/RPL11-MDM2-p53 ribosome biogenesis stress checkpoint pathway in human cells. Cell Cycle. 2018;17:92–101. doi: 10.1080/15384101.2017.1403685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuishi Y., Motohashi H., Yamamoto M. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol. 2012;2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh K., Mimura J., Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxidants Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 11.Hong Y.B., Kang H.J., Kwon S.Y., Kim H.J., Kwon K.Y., Cho C.H., Lee J.M., Kallakury B.V., Bae I. Nuclear factor (erythroid-derived 2)-like 2 regulates drug resistance in pancreatic cancer cells. Pancreas. 2010;39:463–472. doi: 10.1097/MPA.0b013e3181c31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma S., Ishii Y., Morishima Y., Yamadori T., Matsuno Y., Haraguchi N., Kikuchi N., Satoh H., Sakamoto T., Hizawa N., Itoh K., Yamamoto M. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 13.Hartikainen J.M., Tengstrom M., Kosma V.M., Kinnula V.L., Mannermaa A., Soini Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012;72:5537–5546. doi: 10.1158/0008-5472.CAN-12-1474. [DOI] [PubMed] [Google Scholar]
- 14.Hu X.F., Yao J., Gao S.G., Wang X.S., Peng X.Q., Yang Y.T., Feng X.S. Nrf2 overexpression predicts prognosis and 5-FU resistance in gastric cancer. Asian Pac. J. Cancer Prev. APJCP. 2013;14:5231–5235. doi: 10.7314/apjcp.2013.14.9.5231. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y., Zhou Y., Yang M., Wang K., Liu Y., Zhang M., Yang Y., Jin C., Wang R., Hu R. Digoxin sensitizes gemcitabine-resistant pancreatic cancer cells to gemcitabine via inhibiting Nrf2 signaling pathway. Redox Biol. 2019;22:101131. doi: 10.1016/j.redox.2019.101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister A., Nedjadi T., Kitteringham N.R., Campbell F., Costello E., Lloyd B., Copple I.M., Williams S., Owen A., Neoptolemos J.P., Goldring C.E., Park B.K. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol. Cancer. 2011;10:37. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanz L., Diaz-Meco M.T., Nakano H., Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hocking L.J., Lucas G.J., Daroszewska A., Mangion J., Olavesen M., Cundy T., Nicholson G.C., Ward L., Bennett S.T., Wuyts W., Van Hul W., Ralston S.H. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget's disease. Hum. Mol. Genet. 2002;11:2735–2739. doi: 10.1093/hmg/11.22.2735. [DOI] [PubMed] [Google Scholar]
- 20.Copple I.M., Lister A., Obeng A.D., Kitteringham N.R., Jenkins R.E., Layfield R., Foster B.J., Goldring C.E., Park B.K. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J. Biol. Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 23.Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costello E., Greenhalf W., Neoptolemos J.P. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat. Rev. Gastroenterol. Hepatol. 2012;9:435–444. doi: 10.1038/nrgastro.2012.119. [DOI] [PubMed] [Google Scholar]
- 25.Misek D.E., Patwa T.H., Lubman D.M., Simeone D.M. Early detection and biomarkers in pancreatic cancer. J. Natl. Compr. Cancer Netw. 2007;5:1034–1041. doi: 10.6004/jnccn.2007.0086. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P.K., Chapman E., Fang D., Zhang D.D. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 2014;28:708–722. doi: 10.1101/gad.238246.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.