Ma et al. studied the 3-D arrangement of thick filaments in skeletal muscle by x-ray diffraction and electron microscopy and found a correlation between thick filament lattice type (simple or superlattice) and fiber type (fast/slow). This suggests that lattice organization contributes to muscle functional properties.
Abstract
The thick (myosin-containing) filaments of vertebrate skeletal muscle are arranged in a hexagonal lattice, interleaved with an array of thin (actin-containing) filaments with which they interact to produce contraction. X-ray diffraction and EM have shown that there are two types of thick filament lattice. In the simple lattice, all filaments have the same orientation about their long axis, while in the superlattice, nearest neighbors have rotations differing by 0° or 60°. Tetrapods (amphibians, reptiles, birds, and mammals) typically have only a superlattice, while the simple lattice is confined to fish. We have performed x-ray diffraction and electron microscopy of the soleus (SOL) and extensor digitorum longus (EDL) muscles of the rat and found that while the EDL has a superlattice as expected, the SOL has a simple lattice. The EDL and SOL of the rat are unusual in being essentially pure fast and slow muscles, respectively. The mixed fiber content of most tetrapod muscles and/or lattice disorder may explain why the simple lattice has not been apparent in these vertebrates before. This is supported by only weak simple lattice diffraction in the x-ray pattern of mouse SOL, which has a greater mix of fiber types than rat SOL. We conclude that the simple lattice might be common in tetrapods. The correlation between fiber type and filament lattice arrangement suggests that the lattice arrangement may contribute to the functional properties of a muscle.
Introduction
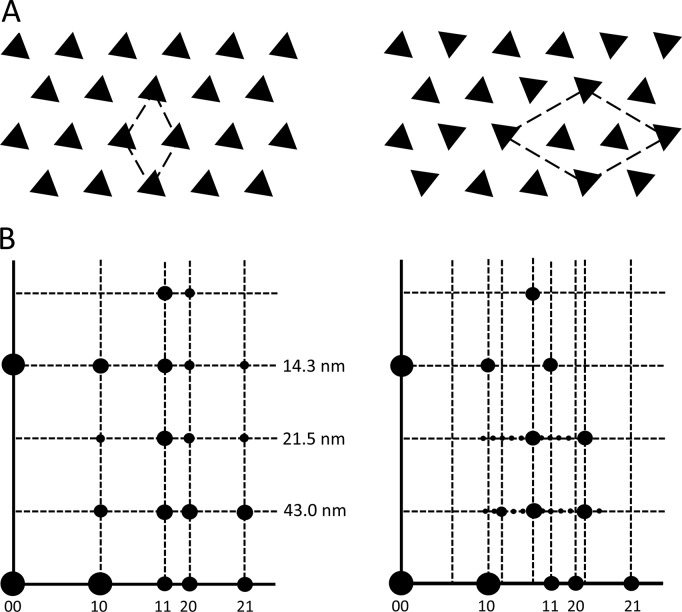
The thick and thin filaments of vertebrate striated muscle are arranged in a double hexagonal lattice, in which each thin filament lies at the trigonal point between three thick filaments (Huxley, 1968). Interaction between myosin heads on the thick filaments and actin subunits of the thin filaments is responsible for the relative filament sliding that generates contraction (Steven et al., 2016). EM combined with x-ray diffraction has shown that the thick filaments are organized in one of two ways (Huxley and Brown, 1967; Luther and Squire, 1980, 2014; Luther et al., 1996). In one, all filaments have the same rotational orientation (a simple lattice), while in the other, nearest neighbors have orientations differing by 0° or 60°, and only next-nearest neighbors have equivalent orientations (a superlattice). These different lattices are recognized in the electron microscope by the orientation of thick filament triangular profiles seen in transverse sections of the bare region of the thick filaments (Fig. 1 A; Luther and Squire, 1980, 2014; Luther et al., 1996). This is the part of the bare zone (Huxley, 1963), just to each side of the M-line (Fig. S1), which lacks both myosin heads and the M-line bridges that link thick filaments to each other (Squire, 1981). The lattices can also be distinguished in x-ray diffraction patterns, where myosin layer lines, arising from pseudohelical organization of the myosin heads (Huxley and Brown, 1967), are sampled either at the same radial positions as the equatorial reflections (simple lattice) or in a more complex pattern (superlattice; Fig. 1 B; Huxley and Brown, 1967; Luther and Squire, 2014). EM analysis has revealed a simple rule for filament orientations in the superlattice: for any group of three nearest neighbor filaments, in a line or in a triangle, if two have the same orientation, then the third is generally rotated by 60° (the no-three-alike rule) and only next-nearest neighbors tend to have equivalent orientations (Luther and Squire, 1980, 2014; Luther et al., 1996).
Figure 1.
Simple and superlattice models. (A) Simple (left) and superlattice (right) models of transverse sections of thick filament bare regions in electron micrographs. (B) Sampling of intensity on myosin layer lines of x-ray diffraction pattern. 10, 11, etc. show positions of reflections on equator. 43.0, 21.5, and 14.3 nm show positions of first, second, and third myosin layer lines. In the simple lattice, note alignment of layer line sampled spots with corresponding equatorial reflections; in the case of the superlattice, the sampling is more complex. Based on Luther et al. (1996) and Harford and Squire (1986), with permission.
The superlattice arrangement was first recognized in x-ray diffraction patterns of frog skeletal (sartorius) muscle (Huxley and Brown, 1967) and was confirmed in electron micrographs of the same muscle (Luther and Squire, 1980), although the specific filament rotations were shown to be different from those suggested by Huxley and Brown (1967). Other tetrapods (amphibians, reptiles, birds, and mammals) examined since then also typically exhibit only a superlattice (Luther et al., 1996). The superlattice is generally not very well ordered and extends over only a small number of unit cells, leading to its description as a “statistical” superlattice (Luther and Squire, 1980). The simple lattice is observed specifically in fish, particularly teleosts (the predominant group of bony fish; Luther et al., 1981, 1996; Luther and Squire, 2014). However, some primitive fish (e.g., hagfish, lampreys, sharks, and rays) have been shown to have a superlattice, suggesting that this form evolved earlier (Luther et al., 1996; Luther and Squire, 2014). Interestingly, sharks and rays exhibit simple and superlattices in the same animal, where they correlate with muscle type, the simple lattice being present in red (slow) fibers and the superlattice in white (fast) fibers (Luther et al., 1996; Luther and Squire, 2014).
Here we study the thick filament lattice arrangement in a mammal (rat) and determine whether it differs between the fast and slow muscles. Most mammals have a mixture of fast and slow fiber types in individual muscles, complicating such analysis. The rat is unusual in that its soleus (SOL) and extensor digitorum longus (EDL) muscles have a relatively homogeneous population of fiber types. The SOL contains 85–95% slow type 1 fibers, with 5–15% fast IIA (Wigston and English, 1992; Li et al., 2019), while the EDL contains ∼100% fast fibers (types IIA, IIB, and IID/X; Eng et al., 2008; Li et al., 2019). Thus it is possible to get relatively pure (i.e., fast or slow muscle) x-ray diffraction patterns and EM images from entire anatomic muscles. The mouse, in contrast, is ∼100% fast in EDL (types IIB/DB/AD,X; Augusto et al., 2004; Luedeke et al., 2004), but in the SOL it is ∼35–40% slow (type I) and 55–65% fast (IIA/D/B/X; Totsuka et al., 2003; Augusto et al., 2004; Luedeke et al., 2004). Our results from rat muscle show that slow fibers have a simple lattice, while the fast fibers have a (relatively poorly ordered) superlattice. This correlation suggests that the specific rotational arrangement of myosin filaments may be one of the determinants of muscle fiber functional properties (e.g., level of tension) in mammals (Luther et al., 1996; Luther and Squire, 2014).
Materials and methods
Muscle preparation
Rats (Sprague-Dawley, 276–300 g, male) and mice (C57BL/6J, 25–30 g, 10–15 wk, male) were euthanized by CO2 asphyxiation followed by cervical dislocation according to Institutional Animal Care and Use Committee–approved protocols of the University of Massachusetts Medical School and Illinois Institute of Technology. The skin was removed, and the hind limbs were separated. The hind limbs were placed in a dish with Ringer’s solution (145 mM NaCl, 2.5 mM KCl, 1.0 mM MgSO4, 1.0 mM CaCl2, 10.0 mM HEPES, and 11 mM glucose, pH 7.4) and perfused with 100% oxygen. For intact muscle experiments, EDL and SOL muscles were rapidly dissected and tied with one suture on each end of the muscle. For skinned muscle experiments, EDL and SOL muscles were pinned to a Sylgard substrate at approximately physiological length during skinning overnight at 4°C with gentle agitation in skinning solution (40 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, 10 mM EGTA, 6.56 mM MgCl2, 5.88 mM Na-ATP, 46.35 mM K-propionate, 15 mM creatine phosphate, and 1% Triton X-100). The muscle was rinsed in relaxing solution (skinning solution lacking detergent) before being placed on the x-ray diffraction apparatus.
X-ray diffraction
Intact or skinned muscles were placed in a specimen chamber containing Ringer’s or relaxing solution and exposed to a collimated x-ray beam of wavelength 0.1033 nm at the BioCAT beam line at the Advanced Photon Source, Argonne National Laboratory (Lemont, IL; Fischetti et al., 2004). All experiments were performed at room temperature (23°C). One suture was attached to a hook inside the chamber, and the other to a dual-model motor/force transducer lever (Aurora Scientific; 300_LR). The muscles were adjusted to optimal length (the length for generating maximal force) by an X-Y-Z positioner attached to the motor/force transducer. For intact muscle experiments, the experimental chamber was oxygenated throughout the experiment. The rat muscle diffraction patterns were recorded on a Mar 165 charge-coupled device detector (Rayonix), and the mouse diffraction patterns on a Pilatus 3 1M detector (Dectris). The diffraction patterns were quadrant folded and background subtracted using the MuscleX software package developed at BioCAT (Jiratrakanvong et al., 2018). The contrast and brightness of the patterns were adjusted in ImageJ (National Institutes of Health) to optimally reveal features of interest. The strong equatorial reflections are shown on a reduced scale for clarity.
EM
Live muscles tied to sticks at rest length were fixed for 1 h at 4°C in 3% paraformaldehyde/0.1% glutaraldehyde followed by 2.5% glutaraldehyde (both in 0.1 M phosphate buffer, pH 7.4), postfixed in 1% OsO4 in 0.1 M sodium cacodylate, dehydrated in an ethanol series, and embedded in Epon. Transverse sections 65 nm thick were cut on a diamond knife using a Leica UC7 ultramicrotome. Sections were stained with uranyl acetate and lead citrate and examined at 120 kV in a Tecnai Spirit transmission electron microscope. Images from myofibrils sectioned in the bare region of the thick filament (Fig. S1) were recorded with a pixel size of 0.41 nm on a Gatan Erlangshen charge-coupled device camera.
Image analysis
To determine thick-filament rotational orientation objectively, an approach was developed based on single-particle analysis methods currently widely used in cryo-EM studies (Frank, 2006). Each triangular filament profile in the transverse image of a bare region was treated as a single particle. Particles were manually selected, boxed, and subjected to 2D classification, and then a class average was computed for each class, all using RELION software (Scheres, 2012). SPIDER software was used for subsequent image processing (Frank et al., 1996). The best 2D class average for each micrograph was used as a reference for projection matching to all the filaments in an image. The rotational angle of a filament that best matched the reference was determined by cross-correlation. This angle defined the rotational orientation of each filament. A triangle was then superposed on each filament in the original image with the determined orientation (Fig. 3, C and E). Finally, the superposed triangles representing each filament were transferred to an empty background to clearly reveal each filament orientation (Fig. 3, D and F).
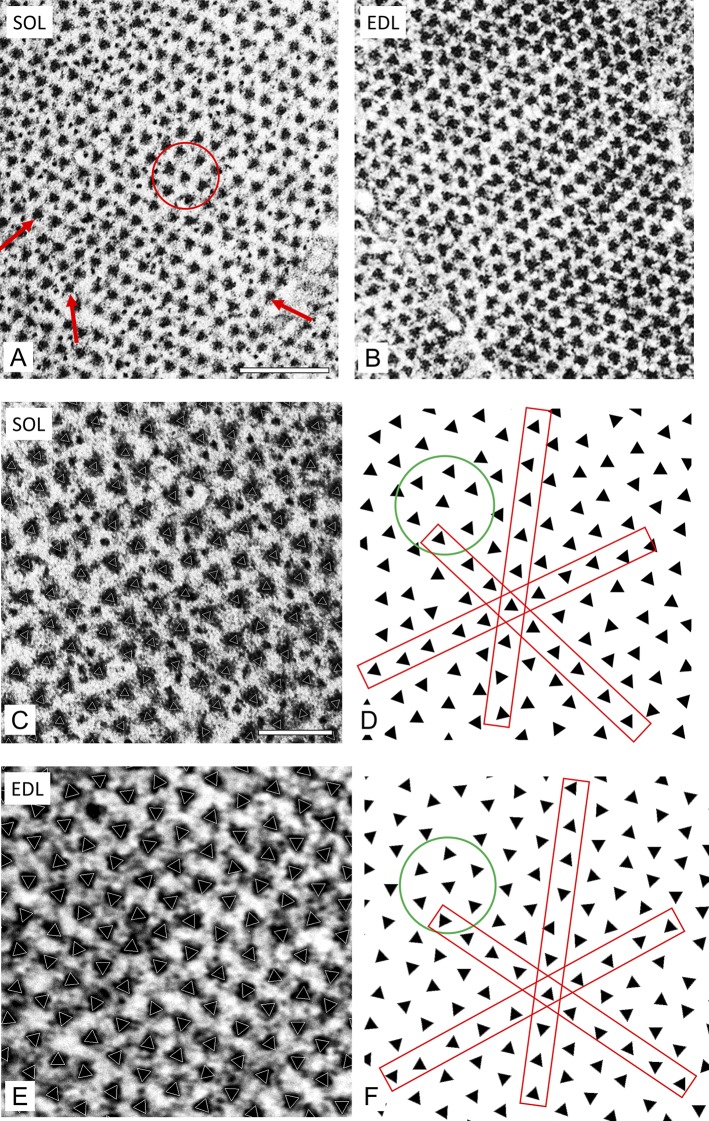
Figure 3.
Transverse sections of bare region of rat SOL and EDL revealing lattice type. (A) SOL shows triangular filament profiles pointing in approximately the same direction along all three lattice planes (arrows) and within a hexagon (circle). This suggests a simple lattice. (B) EDL shows triangular profiles with variable orientations, which are analyzed below. (C–F) SOL and EDL sections at higher magnification analyzed by projection matching. (C and E) Raw images with superposed white triangles representing orientation determined by projection matching (see text). (D and F) Triangles extracted from C and E reveal orientations most clearly. (D) Triangles with similar orientations in all three planes (red rectangles) and in hexagon (circle). (F) Triangles with varying orientations, mostly following the no-three-alike rule. Scale bars, 200 nm (A and B); 100 nm (C–F).
To determine the lattice type, a statistical analysis of the orientations of the triangles in each image was performed. The relative orientations of the triangles in any hexagon of the lattice were determined by selecting one triangle as the central filament and calculating the distance between this triangle and the others in the same image. The nearest-neighbor surrounding filaments were selected as those within a threshold distance of 40 nm (slightly greater than the center-to-center distance of two adjacent triangles), ensuring that only nearest-neighbor filaments of a hexagon were selected. The average deviation angle between a central filament and its six surrounding neighbors (green circles, Fig. 3, D and F) was calculated using the azimuthal angle from the orientation determination above. The sign of the deviation angle was not taken into account. This step was repeated for each triangle in the image, and the distribution of deviation angles was plotted as a histogram. A second distribution was also calculated: the pairwise deviation between one filament and the adjacent filament. To automatically select filaments for this measurement, the distance between the one filament and its neighbor had to be less than the threshold of 40 nm. The deviation between filaments of a pair was then calculated using the orientation angle for the two filaments.
Online supplemental material
Supplemental results and discussion describes x-ray and EM analysis and interpretation of thick filament lattices in mouse SOL and EDL. Fig. S1 illustrates how sections were cut to analyze the bare region of the thick filament by EM. Fig. S2 shows x-ray diffraction of skinned rat and EDL muscles. Fig. S3 shows intensity plots along ML1 of rat SOL and EDL. Fig. S4 is a comparison of superlattice reflections in x-ray patterns of frog sartorius and rat EDL muscles. Fig. S5 is an analysis of thick-filament orientations in rat SOL and EDL (all data). Fig. S6 is a comparison of x-ray diffraction patterns of mouse and rat SOL and EDL. Fig. S7 is a comparison of x-ray diffraction patterns of mouse SOL: wild type, blebbistatin treated, and nebulin deficient. Fig. S8 is a comparison of intensity plots of ML1 in rat and mouse SOL and EDL. Fig. S9 is an analysis of thick-filament orientations in mouse SOL fast and slow fibers. Fig. S10 shows thick-filament orientations in type I/IIA fibers of mouse SOL.
Results
X-ray diffraction
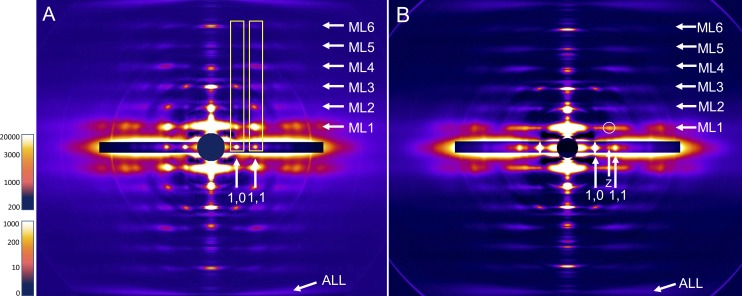
Axial x-ray diffraction patterns were recorded from both intact and freshly skinned rat SOL and EDL muscles under relaxing conditions. Results were similar for both (Figs. 2 and S2), and we describe only intact muscle below. The equator of the patterns showed the typical strong 1,0 and weaker 1,1 reflections of relaxed muscle, consistent with the close association of myosin heads with the thick filament backbone and minimal interaction with actin (Huxley, 1968; Fig. 2). At higher exposure, weaker reflections (e.g., 20, 21; Fig. 1 B) were observed further out on the equator (not visible when scale adjusted for clear 1,0 and 1,1 reflections). The positions of these equatorial reflections reflect the hexagonal arrangement and spacing of myosin filaments, with interleaving actin filaments, but do not provide information on the relative orientations of the myosin filaments. In addition to the equator, the patterns also showed a series of layer lines (ML1–6) indexing on a repeat of 430 Å, reflecting the pseudohelical organization of the myosin heads on the thick filaments (Fig. 2, A and B; cf. Fig. 1 B; Huxley and Brown, 1967). The distribution of intensity along the layer lines was quite different between SOL and EDL. In SOL, lattice sampling produced intensity that was strongest at radial positions corresponding to those of the equatorial reflections (Fig. 2 A). This was most obvious on the first layer line but also evident on higher-order layer lines (Fig. 2 A, rectangular boxes; Fig. S3). This lattice sampling appeared similar to that in the simple lattice x-ray patterns from fish muscle (Squire et al., 2004; Luther and Squire, 2014). In the EDL, the distribution of intensity on the first layer line was relatively continuous, with little sign of lattice sampling, suggesting a rotationally less coherent lattice of thick filaments (Fig. 2 B). Careful inspection, however, revealed weak lattice spots on ML1 that did not align with the equatorial reflections and looked similar to the pattern first described for frog skeletal muscle, which has a superlattice (Fig. S4; Huxley and Brown, 1967; Squire, 1981; Luther and Squire, 2014). Intensity distribution was different on the higher layer lines. We conclude that the SOL and EDL have simple and superlattice thick-filament arrangements, respectively, and that these may be due to the different fiber types (slow and fast) that predominate in these muscles in the rat. The weakness of the superlattice reflections on ML1 in the EDL pattern suggests that the superlattice is significantly disordered (Luther and Squire, 1980, 2014).
Figure 2.
X-ray diffraction patterns of rat SOL (A) and EDL (B) intact muscles. ML1-6 indicate positions of myosin layer lines (repeat 43 nm), and ALL indicates the position of the 5.9-nm actin layer line. Horizontal inserts on equator show shorter exposures to reveal the high-intensity equatorial 1,0 and 1,1 and Z-line reflections. Prominent lattice sampling is seen in SOL at the same positions as equatorial reflections (vertical rectangles aligning with 1,0 and 1,1 reflections). This indicates a simple lattice of myosin filaments. Lattice sampling is largely absent in EDL, except for weak superlattice spot circled. The pattern suggests a local superlattice with disorder. Scale at left is intensity in arbitrary units (upper, equator; lower, rest of pattern).
EM
Rat SOL and EDL muscles were fixed, embedded, and transversely sectioned for examination by transmission EM. Sections were studied in the bare region of the thick filaments, just to each side of the M-line (Fig. S1), where both myosin heads and M-line bridges are absent (Squire, 1981). This region therefore provides the clearest visualization of filament rotational orientation, recognized by the triangular profiles of the thick-filament backbones that reflect their threefold symmetry (Figs. 1 A and 3, A and B; Luther and Squire, 1980). The relative orientations of these profiles provides a direct visualization of the type of filament lattice (Fig. 1 A). However, even with the thin (65-nm) sections that we used, some portion of the M-line bridges and/or myosin heads will be included, as the bare region on each side of the M-line is only ∼30 nm long. The M-line and/or myosin heads presumably account for the additional, low-density material surrounding the thick filaments, which can diminish the clarity of the triangular profiles (Fig. 3, A and B).
In rat SOL, neighboring thick filaments in well-contrasted bare regions typically had approximately the same rotational orientation (Fig. 3 A). This was apparent to the eye by comparing the group of six peripheral and one central filament in a hexagon (Fig. 3 A, circle), or by comparing filament orientation along the rows of filaments in any one of the three lattice directions (Fig. 3 A, arrows). The filament lattice in EDL had a distinctly different appearance, showing nearest neighbors with varying orientations (Fig. 3 B). To provide an objective assessment of filament orientation and to quantify the relative orientations, the rotational angle of each filament was determined computationally using a projection-matching approach (see Materials and methods). A triangle with the orientation thus determined was then superposed on each filament in the image (Fig. 3, C and E). To most clearly visualize filament orientations, the triangles were then displayed without the original micrograph (Fig. 3, D and F). The results confirmed the visual appearance in Fig. 3 (A and B). SOL showed triangles with similar orientations in all three planes (red rectangles, Fig. 3 D) and within individual hexagons (circle; Fig. 3 D), while EDL revealed triangles with varying orientations, which tended to follow the no-three-alike rule (Fig. 3 F; Luther and Squire, 1980, 2014).
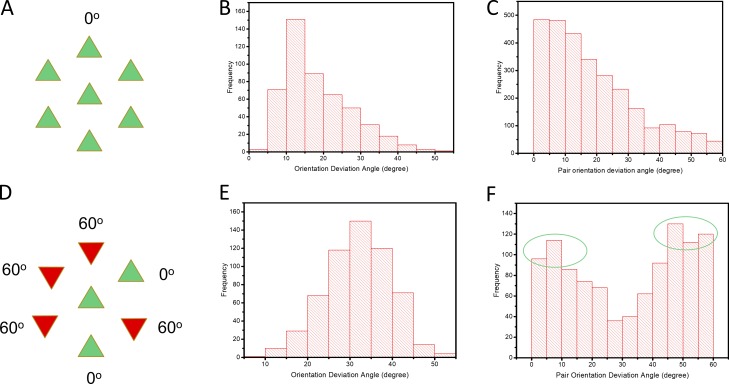
The relative orientations of neighboring filaments were then analyzed statistically by calculating the difference in angle between each filament and its six nearest neighbors and plotting the results as a histogram (Fig. 4). In a simple lattice, all six filaments have the same orientation as the central filament (mean deviation angle 0°; Fig. 4 A). The histogram for the SOL micrograph used in Fig. 3 C showed a peak at ∼12° for the average deviation of the six filaments in a hexagon (Fig. 4 B) and a peak at 0° when considering filaments pairwise (Fig. 4 C). This supports a simple lattice. In a no-three-alike superlattice (Fig. 4 D), two of every three adjacent filaments have the same angle, and the third is rotated by 60°. The predicted mean deviation angle is therefore 30°. The histogram for the EDL micrograph used in Fig. 3 E showed a peak close to 30° for the average deviation of the six filaments in a hexagon (Fig. 4 E), consistent with a no-three-alike superlattice. When considered pairwise, the distribution showed two peaks of similar height, centered near 0° and 60° (Fig. 4 F), exactly as predicted for the no-three-alike superlattice, where a filament has either the same orientation as its neighbor or differs from it by 60°.
Figure 4.
Analysis of filament orientations in one micrograph each of SOL and EDL. (A and D) Filament orientations in simple lattice (A; all filaments have same orientation; mean deviation angle = 0°) and no-three-alike superlattice (D; in any group of three filaments, in a line or forming a triangle, one filament is rotated by 60° with respect to the other two, which are identical; mean deviation angle for any three adjacent filaments = 30°). (B) Histogram for SOL micrograph showing mean deviation angle for the six filaments surrounding the central filament in a hexagon. The histogram peaks near 10°, close to the expected 0° for a simple lattice. (C) Deviation angle for adjacent filaments in SOL considered pairwise. The histogram peaks near to 0°, as expected for a simple lattice. (E) Mean deviation angle for the six filaments surrounding the central filament in a hexagon for EDL micrograph. The peak is close to 30°, as expected for a no-three-alike superlattice. (F) Deviation angle for adjacent filaments in EDL considered pairwise. There are two peaks, close to 0 and 60°, as expected for a no-three-alike superlattice (see D). Note: The relatively broad spread of the angle data is due partly to imperfections in the filament lattice, as seen in bending of the lines of filaments (Fig. 3), and partly to imperfections in particle picking due to thin filament proximity to thick filaments. Both factors affected the accuracy of angle determination. See also Fig. S5.
A similar comparison of filament orientations was performed in eight micrographs for SOL (3,959 filaments) and eight micrographs for EDL (2,850 filaments). Micrographs were selected for the best visibility of bare-region triangular profiles. The aggregate results are shown in Fig. S5 and support the conclusions from the individual micrographs shown in Fig. 4. Thus, EM supports the simple lattices and superlattices suggested by the x-ray diffraction patterns of the rat SOL and EDL muscles, respectively.
Discussion
Previous EM and x-ray studies showed that vertebrate skeletal muscles have one of two types of thick filament lattice: simple or super. Comparison of different species suggested that tetrapods had only superlattices and that simple lattices were confined to fish (Luther et al., 1996; Luther and Squire, 2014). Our observations show that the soleus muscle of the rat (a tetrapod) also has a simple lattice. They also suggest that this is related to the slow fiber type that predominates in this muscle, as the EDL (a fast muscle) has a superlattice. It will be interesting to see whether other muscles of these rodents also show a correlation between fiber type and lattice type, and whether this association applies to other mammals and to the other tetrapod groups (amphibians, reptiles, and birds).
Why has a correlation between fiber type and lattice type not previously been recognized in tetrapods? The main reason is probably the limited number of tetrapod skeletal muscles that have been examined by x-ray diffraction, primarily frog sartorius (Huxley and Brown, 1967; Reconditi, 2006; Linari et al., 2015) and rabbit psoas (Xu et al., 2003), both fast muscles (Gutmann, 1966; Salviati et al., 1982; Hämäläinen and Pette, 1993; Lutz et al., 1998). To our knowledge, only three “pure” tetrapod slow muscles have previously been examined by x-ray diffraction (most muscles are not pure, but have a mixture of fast and slow fibers; Schiaffino and Reggiani, 2011). A study of rat SOL and EDL, under conditions designed to phosphorylate or dephosphorylate the regulatory light chains, showed little to no lattice sampling on ML1 of SOL, and made no conclusion about the type of myosin filament lattice present (Yamaguchi et al., 2016); the conditions used for these experiments may have caused significant disordering of the myosin heads, thus weakening the myosin layer lines and their sampling. A comparison of rat SOL and psoas muscles focused on cross-bridge behavior and made no mention of lattice structure (Horiuti et al., 1997). Likewise, a comparison of chicken slow (anterior latissimus dorsi) and fast (posterior latissimus dorsi) muscles centered on myosin head proximity to thin filaments but did not study myosin layer lines or comment on the type of filament lattice (Matsubara et al., 1991). Given this paucity of x-ray data on fast and slow muscles, and the focus on other aspects of structure when they have been studied, it is possible that a correlation between fiber and lattice type is common but has simply been missed. The relatively pure composition of the rat SOL (∼90% slow) and EDL (∼100% fast), however, made the lattice/fiber type correlation immediately obvious in our x-ray patterns (Fig. 2).
The importance of a pure fiber composition for detecting lattice type is supported by a previous x-ray study, which was designed to compare the structures of the skeletal muscles of amphibia and mammals (Iwamoto et al., 2003). The muscles chosen were frog sartorius and mouse diaphragm. Mouse diaphragm is thought to consist almost entirely (98%) of fast-twitch fatigue-resistant (FFR) type IIA fibers (Coirault et al., 1995). While frog sartorius showed a superlattice, as previously found (Huxley and Brown, 1967), the mouse diaphragm showed evidence for a simple lattice (the only other tetrapod muscle reported to have shown this), with lattice sampling similar to that in rat SOL (Fig. 2 A) and fish (Luther and Squire, 2014). This finding suggests that the simple lattice may correlate not specifically with slow muscle, as we initially hypothesized based on our rat studies, but with fatigue-resistant fibers, which includes both FFR and slow muscles (Schiaffino and Reggiani, 2011). Because other mouse muscles were not included in their comparative study (Iwamoto et al., 2003), it was not recognized that the simple/superlattice dichotomy might relate to fiber type, as our results suggest, rather than animal group (mammal vs. amphibian).
While rat SOL and EDL are typically regarded as slow and fast muscles, respectively, there is still some fiber type mixing. Does the interpretation of our x-ray results hold, given this added layer of complexity? Rat SOL and EDL have been analyzed for both myosin heavy chain and fiber type. Labeling with anti-fast and anti-slow myosin antibodies yielded a ratio of 90% slow and 10% fast in the SOL of young rats (∼12 wk) and 95% slow and 5% fast fibers at 1 yr old (Wigston and English, 1992). This is roughly consistent with myosin heavy chain type determined by SDS-PAGE, which yielded 80% type I (slow) and 20% type IIA (fast) fibers at ∼12 wk (Eng et al., 2008). Similar results are obtained based on myosin ATPase activity and immunocytochemistry (Soukup et al., 2002). By mass spectrometry, rat SOL (age not reported) had 88% type I fibers and 12% type IIA fibers (Li et al., 2019). Overall, these data imply that rat SOL has 80–95% slow, 5–20% fast IIA fibers, and no fast IIB/X/D fibers. Assuming that type I and IIA fibers both have a simple lattice, as discussed above, the lattice sampling from this mixed muscle is consistent with our x-ray data. The rat EDL also has a mixed fiber content: 64% IIB, 25% IIX, and 11% IIA (from myosin type by SDS-PAGE; Eng et al., 2008); 76% IIB, 19% IIA, and 5% type I (Soukup et al., 2002); and 70% IIB, 24% IID/X and 5% IIA by mass spectrometry (Li et al., 2019). IIB/X/D are all considered to be fast fibers (Schiaffino and Reggiani, 2011). With only ∼10% or less IIA fibers, the expected small amount of simple lattice sampling would be difficult to detect in EDL, consistent with our x-ray data. We conclude that our rat diffraction data are consistent with the idea that slow type I and fast type IIA fibers have a simple lattice while fast fibers (IIB/X/D) have a superlattice.
The dearth of x-ray data on tetrapod lattice types is mirrored by a lack of EM data. To our knowledge, no previous EM study has addressed the thick filament lattices present in the different fiber types of tetrapods, and a simple lattice has not previously been observed by EM in any tetrapod (the study of mouse diaphragm (Iwamoto et al., 2003) did not use EM, and it will be important to confirm the x-ray interpretation of the lattice in this muscle by direct EM observation). It is therefore possible that a simple lattice structure in slow and FFR fibers has simply been overlooked. Early EM studies of fish muscle were similarly limited (to teleosts, the predominant group of bony fish) and revealed only simple lattices (Luther et al., 1981). When comparison was widened to include other, more primitive groups of fish (e.g., hagfish, lampreys, sharks, and rays) superlattices were found to be common (Luther et al., 1996; Luther and Squire, 2014). A comparable, more detailed survey might show that simple lattices are common in tetrapods.
Our combined EM and x-ray data suggest that the type of thick-filament lattice is related to muscle fiber type in the rat. Further work will be necessary to determine whether this correlation extends to other rodents and other tetrapods (including other mammals). We have made a first step to answer this question by studying the SOL and EDL muscles of the mouse. Overall, there is reasonable agreement with the conclusions from the rat (see Supplemental results and discussion and Figs. S6, S7, S8, S9, and S10). However, the results are less definitive due to the mixed fiber content of mouse SOL, the difficulty of identifying different fiber types in the SOL by EM, and the apparently greater disorder of the mouse lattices compared with rat. Nevertheless, the mouse data are broadly consistent with the conclusion that low-fatigue fibers (type I and IIA) have a simple lattice while fatigable fibers (type IIB, D, X) have a superlattice. In no case is the lattice perfect, but in both SOL and EDL it is substantially disordered (i.e., of limited extent).
Strikingly, sharks and rays in which white and red fibers were compared by EM showed a lattice/fiber type correlation (Luther et al., 1996; Luther and Squire, 2014) similar to that in the rat. Red fibers (slow) had a simple lattice while white fibers (fast) had a superlattice. This supports the notion that lattice type is connected to fiber type, and suggests that it may occur over a range of vertebrates, including tetrapods as well as fish. However, the correlation is not perfect, as no such connection is found in teleosts, where red and white fibers both have simple lattices (Luther et al., 1995, 1996).
Differences in the physiology of slow and fast fibers have a profound impact on the functioning of skeletal muscle. Slow (type I) fibers play an essential role in postural muscles and muscles that function continuously, without break, for the life of the organism (e.g., respiratory muscles of larger species; Polla et al., 2004; Schiaffino and Reggiani, 2011). They contract more slowly and generate less tension than fast fibers but are highly resistant to fatigue (Schiaffino and Reggiani, 2011). Fast, fatigable fibers (IIB/D/X) are complementary, adding to muscle response when brief but powerful contractions are required (Rome et al., 1988; Schiaffino and Reggiani, 2011). Fast, fatigue-resistant fibers (type IIA) have intermediate properties (Schiaffino and Reggiani, 2011); although classically described as fast, they are in many ways more akin to slow fibers and quite distinct from IIB/D/X (Schiaffino and Reggiani, 2011). Differences in mitochondrial content (and the oxidative metabolism that this supports), and in myosin heavy and light chain isoforms (with associated differences in actin-activated myosin ATPase activity) underlie much of this diversity in function. Our results raise the possibility that the structural organization of the myosin filaments might also contribute to the different contractile properties. The proper functioning of relatively slowly contracting/fatigue-resistant muscles (types I, IIA), designed for prolonged activity, may depend in part on high mechanical efficiency of contraction (Schiaffino and Reggiani, 2011). The arrangement of thick filaments in simple or superlattices will affect the ease with which myosin heads attach to actin filament target zones, which could impact the contractile properties (e.g., speed, tension) of a fiber (Luther et al., 1996; Luther and Squire, 2014). Thus the 3D geometry of actin-myosin interactions defined by the simple lattice might favor efficiency, while the superlattice may enable a greater number of actin-myosin interactions, contributing to the greater force production of fast, fatigable fibers (Luther et al., 1995, 1996; Squire et al., 2006; Luther and Squire, 2014).
What is responsible for generating the two lattice types found in the different fiber types of rats and mice (and possibly other tetrapods)? Thick-filament cross-linking proteins, such as those of the M-line, are likely to be involved, as these provide a means of setting thick filaments with specific orientations relative to each other. M-line proteins specific to particular fiber types may interact differently with thick filaments to generate one or another type of lattice (Schoenauer et al., 2008; Luther and Squire, 2014; Lange et al., 2019). For example, M-protein, primarily responsible for the central stripe of the M-line (M1) and present only in fast (type IIB) fibers (Obermann et al., 1996), could be the linker that defines relative thick-filament orientation in a superlattice. When absent, as in slow and type IIA fibers, the thick filaments may default to a simple lattice arrangement. Alternatively, the lattice type could be dependent on the type of myosin present in the thick filament, which is known to vary with fiber type, as discussed earlier. In this case, the thick-filament bridging proteins of the M-line may be the same in different fibers (e.g., myomesin is present in the M-lines of all fiber types; Schoenauer et al., 2008; Lange et al., 2019), but interact differently with the thick filaments (Pask et al., 1994), depending on their myosin heavy chain type. Another thick filament structural protein with isoforms specific to different fiber types is myosin binding protein C (MyBP-C). Slow MyBP-C occurs in both fast (IIA and IIB) and slow (type I) fibers, while fast MyBP-C is present only in fast type IIB fibers (and absent from fast type IIA fibers; Li et al., 2019). MyBP-C has been observed to bridge thick filaments to neighboring thin filaments (Luther et al., 2011), and thus can indirectly link thick filaments to their neighbors. The different forms of MyBP-C in different fiber types might link filaments differently, contributing to formation of the two types of lattice. If so, this would represent a novel function of MyBP-C, a protein whose role in skeletal muscle is not yet well understood (Wang et al., 2018; Li et al., 2019; Robinett et al., 2019). MyBP-C is in fact long enough to directly link thick filaments, which could more directly influence lattice formation, although there is so far no evidence that such connections exist in the myofibril (Luther et al., 2011). If this speculation is correct, disease mutations in skeletal MyBP-C and M-line proteins could affect phenotype at least in part through their putative role in defining filament lattice type.
Supplementary Material
Acknowledgments
Henk L. Granzier served as editor.
We thank Drs. John Woodhead, Peter Reiser, and Elizabeth Ehler for discussions and Michael Previs for sharing unpublished mass spectrometry data with us.
This research was supported in part by National Institutes of Health grants AR072036 (to R. Craig), AR067279 (to R. Craig and D. Warshaw), and HL139883 (to R. Moss and R. Craig) and used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. Use of the BioCAT Beamline 18ID was supported by National Institute of General Medical Sciences grant P41 GM103622. Use of the Pilatus 3 1M detector was provided by National Institute of General Medical Sciences grant 1S10OD018090-01. We are grateful to the Core Electron Microscope Facility at UMass Medical School and its staff for use of their resources and acknowledge funding from National Institutes of Health grant 1S10OD021580-01 for the ultramicrotome. The content of this work is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: W. Ma carried out the x-ray diffraction experiments and analysis; K.H. Lee carried out the electron microscopy; S. Yang carried out the image analysis; T. Irving contributed to analysis and interpretation of the data and to funding acquisition; R. Craig wrote the paper, with input from all co-authors, and contributed to analysis and interpretation of the data and to funding acquisition.
References
- Augusto V., Padovani C.R., and Campos G.E.R.. 2004. Skeletal muscle fiber types in C57BL6J mice. Braz. J. Morphol. Sci. 21:89–94. [Google Scholar]
- Coirault C., Chemla D., Pery-Man N., Suard I., and Lecarpentier Y.. 1995. Effects of fatigue on force-velocity relation of diaphragm. Energetic implications. Am. J. Respir. Crit. Care Med. 151:123–128. 10.1164/ajrccm.151.1.7812541 [DOI] [PubMed] [Google Scholar]
- Eng C.M., Smallwood L.H., Rainiero M.P., Lahey M., Ward S.R., and Lieber R.L.. 2008. Scaling of muscle architecture and fiber types in the rat hindlimb. J. Exp. Biol. 211:2336–2345. 10.1242/jeb.017640 [DOI] [PubMed] [Google Scholar]
- Fischetti R., Stepanov S., Rosenbaum G., Barrea R., Black E., Gore D., Heurich R., Kondrashkina E., Kropf A.J., Wang S., et al. 2004. The BioCAT undulator beamline 18ID: a facility for biological non-crystalline diffraction and X-ray absorption spectroscopy at the Advanced Photon Source. J. Synchrotron Radiat. 11:399–405. 10.1107/S0909049504016760 [DOI] [PubMed] [Google Scholar]
- Frank J. 2006. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state. 2nd edition. Oxford University Press, New York. [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A.. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199. 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Gutmann E. 1966. “Slow” and “Fast” Muscle Fibers. MCV Q. 2:78–81. [Google Scholar]
- Hämäläinen N., and Pette D.. 1993. The histochemical profiles of fast fiber types IIB, IID, and IIA in skeletal muscles of mouse, rat, and rabbit. J. Histochem. Cytochem. 41:733–743. 10.1177/41.5.8468455 [DOI] [PubMed] [Google Scholar]
- Harford J., and Squire J.. 1986. “Crystalline” myosin cross-bridge array in relaxed bony fish muscle. Low-angle x-ray diffraction from plaice fin muscle and its interpretation. Biophys. J. 50:145–155. 10.1016/S0006-3495(86)83447-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuti K., Yagi N., and Takemori S.. 1997. Mechanical study of rat soleus muscle using caged ATP and X-ray diffraction: high ADP affinity of slow cross-bridges. J. Physiol. 502:433–447. 10.1111/j.1469-7793.1997.433bk.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley H.E. 1963. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J. Mol. Biol. 7:281–308. 10.1016/S0022-2836(63)80008-X [DOI] [PubMed] [Google Scholar]
- Huxley H.E. 1968. Structural difference between resting and rigor muscle; evidence from intensity changes in the low-angle equatorial x-ray diagram. J. Mol. Biol. 37:507–520. 10.1016/0022-2836(68)90118-6 [DOI] [PubMed] [Google Scholar]
- Huxley H.E., and Brown W.. 1967. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 30:383–434. 10.1016/S0022-2836(67)80046-9 [DOI] [PubMed] [Google Scholar]
- Iwamoto H., Wakayama J., Fujisawa T., and Yagi N.. 2003. Static and dynamic x-ray diffraction recordings from living mammalian and amphibian skeletal muscles. Biophys. J. 85:2492–2506. 10.1016/S0006-3495(03)74672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiratrakanvong J., Shao J., Menendez M., Li X., Li J., Ma W., Agam G., and Irving T.. 2018. MuscleX: software suite for diffraction X-ray imaging V1.13.1. Available at: https://zenodo.org/record/3360909#.XbdFaWZOkuU.
- Lange S., Pinotsis N., Agarkova I., and Ehler E.. 2019. The M-band: The underestimated part of the sarcomere. Biochim. Biophys. Acta Mol. Cell Res. 10.1016/j.bbamcr.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Nelson S.R., Rahmanseresht S., Braet F., Cornachione A.S., Previs S.B., O’Leary T.S., McNamara J.W., Rassier D.E., Sadayappan S., et al. 2019. Skeletal MyBP-C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc. Natl. Acad. Sci. U.S.A. 116(43):21882–21892. 10.1073/pnas.1910549116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linari M., Brunello E., Reconditi M., Fusi L., Caremani M., Narayanan T., Piazzesi G., Lombardi V., and Irving M.. 2015. Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature. 528:276–279. 10.1038/nature15727 [DOI] [PubMed] [Google Scholar]
- Luedeke J.D., McCall R.D., Dillaman R.M., and Kinsey S.T.. 2004. Properties of slow- and fast-twitch skeletal muscle from mice with an inherited capacity for hypoxic exercise. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 138:373–382. 10.1016/j.cbpb.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Luther P.K., and Squire J.M.. 1980. Three-dimensional structure of the vertebrate muscle A-band. II. The myosin filament superlattice. J. Mol. Biol. 141:409–439. 10.1016/0022-2836(80)90254-5 [DOI] [PubMed] [Google Scholar]
- Luther P.K., and Squire J.M.. 2014. The intriguing dual lattices of the myosin filaments in vertebrate striated muscles: evolution and advantage. Biology (Basel). 3:846–865. 10.3390/biology3040846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther P.K., Munro P.M., and Squire J.M.. 1981. Three-dimensional structure of the vertebrate muscle A-band. III. M-region structure and myosin filament symmetry. J. Mol. Biol. 151:703–730. 10.1016/0022-2836(81)90430-7 [DOI] [PubMed] [Google Scholar]
- Luther P.K., Munro P.M.G., and Squire J.M.. 1995. Muscle ultrastructure in the teleost fish. Micron. 26:431–459. 10.1016/0968-4328(95)00015-1 [DOI] [Google Scholar]
- Luther P.K., Squire J.M., and Forey P.L.. 1996. Evolution of myosin filament arrangements in vertebrate skeletal muscle. J. Morphol. 229:325–335. [DOI] [PubMed] [Google Scholar]
- Luther P.K., Winkler H., Taylor K., Zoghbi M.E., Craig R., Padrón R., Squire J.M., and Liu J.. 2011. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. USA. 108:11423–11428. 10.1073/pnas.1103216108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz G.J., Bremner S., Lajevardi N., Lieber R.L., and Rome L.C.. 1998. Quantitative analysis of muscle fibre type and myosin heavy chain distribution in the frog hindlimb: implications for locomotory design. J. Muscle Res. Cell Motil. 19:717–731. 10.1023/A:1005466432372 [DOI] [PubMed] [Google Scholar]
- Matsubara I., Yagi N., Saeki Y., and Kurihara S.. 1991. Cross-bridge movement in fast and slow skeletal muscles of the chick. J. Physiol. 441:113–120. 10.1113/jphysiol.1991.sp018741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann W.M., Gautel M., Steiner F., van der Ven P.F., Weber K., and Fürst D.O.. 1996. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J. Cell Biol. 134:1441–1453. 10.1083/jcb.134.6.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask H.T., Jones K.L., Luther P.K., and Squire J.M.. 1994. M-band structure, M-bridge interactions and contraction speed in vertebrate cardiac muscles. J. Muscle Res. Cell Motil. 15:633–645. 10.1007/BF00121071 [DOI] [PubMed] [Google Scholar]
- Polla B., D’Antona G., Bottinelli R., and Reggiani C.. 2004. Respiratory muscle fibres: specialisation and plasticity. Thorax. 59:808–817. 10.1136/thx.2003.009894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reconditi M. 2006. Recent improvements in small angle x-ray diffraction for the study of muscle physiology. Rep. Prog. Phys. 69:2709–2759. 10.1088/0034-4885/69/10/R01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett J.C., Hanft L.M., Geist J., Kontrogianni-Konstantopoulos A., and McDonald K.S.. 2019. Regulation of myofilament force and loaded shortening by skeletal myosin binding protein C. J. Gen. Physiol. 151:645–659. 10.1085/jgp.201812200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome L.C., Funke R.P., Alexander R.M., Lutz G., Aldridge H., Scott F., and Freadman M.. 1988. Why animals have different muscle fibre types. Nature. 335:824–827. 10.1038/335824a0 [DOI] [PubMed] [Google Scholar]
- Salviati G., Betto R., and Danieli Betto D.. 1982. Polymorphism of myofibrillar proteins of rabbit skeletal-muscle fibres. An electrophoretic study of single fibres. Biochem. J. 207:261–272. 10.1042/bj2070261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180:519–530. 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., and Reggiani C.. 2011. Fiber types in mammalian skeletal muscles. Physiol. Rev. 91:1447–1531. 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- Schoenauer R., Lange S., Hirschy A., Ehler E., Perriard J.C., and Agarkova I.. 2008. Myomesin 3, a novel structural component of the M-band in striated muscle. J. Mol. Biol. 376:338–351. 10.1016/j.jmb.2007.11.048 [DOI] [PubMed] [Google Scholar]
- Soukup T., Zacharová G., and Smerdu V.. 2002. Fibre type composition of soleus and extensor digitorum longus muscles in normal female inbred Lewis rats. Acta Histochem. 104:399–405. 10.1078/0065-1281-00660 [DOI] [PubMed] [Google Scholar]
- Squire J. 1981. The Structural Basis of Muscular Contraction. Plenum Press, New York: 10.1007/978-1-4613-3183-4 [DOI] [Google Scholar]
- Squire J.M., Bekyarova T., Farman G., Gore D., Rajkumar G., Knupp C., Lucaveche C., Reedy M.C., Reedy M.K., and Irving T.C.. 2006. The myosin filament superlattice in the flight muscles of flies: A-band lattice optimisation for stretch-activation? J. Mol. Biol. 361:823–838. 10.1016/j.jmb.2006.06.072 [DOI] [PubMed] [Google Scholar]
- Squire J.M., Roessle M., and Knupp C.. 2004. New X-ray diffraction observations on vertebrate muscle: organisation of C-protein (MyBP-C) and troponin and evidence for unknown structures in the vertebrate A-band. J. Mol. Biol. 343:1345–1363. 10.1016/j.jmb.2004.08.084 [DOI] [PubMed] [Google Scholar]
- Steven A.C., Baumeister W., Johnson L.N., and Perham R.N.. 2016. Molecular Biology of Assemblies and Machines. Garland Science, New York, London. [Google Scholar]
- Totsuka Y., Nagao Y., Horii T., Yonekawa H., Imai H., Hatta H., Izaike Y., Tokunaga T., and Atomi Y.. 2003. Physical performance and soleus muscle fiber composition in wild-derived and laboratory inbred mouse strains. J. Appl. Physiol. 95:720–727. 10.1152/japplphysiol.00946.2002 [DOI] [PubMed] [Google Scholar]
- Wang L., Geist J., Grogan A., Hu L.R., and Kontrogianni-Konstantopoulos A.. 2018. Thick Filament Protein Network, Functions, and Disease Association. Compr. Physiol. 8:631–709. 10.1002/cphy.c170023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigston D.J., and English A.W.. 1992. Fiber-type proportions in mammalian soleus muscle during postnatal development. J. Neurobiol. 23:61–70. 10.1002/neu.480230107 [DOI] [PubMed] [Google Scholar]
- Xu S., Offer G., Gu J., White H.D., and Yu L.C.. 2003. Temperature and ligand dependence of conformation and helical order in myosin filaments. Biochemistry. 42:390–401. 10.1021/bi026085t [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Takemori S., Kimura M., Nakahara N., Ohno T., Yamazawa T., Yokomizo S., Akiyama N., and Yagi N.. 2016. Approaches to physical fitness and sports medicine through X-ray diffraction analysis of striated muscle. J. Phys. Fit. Sports Med. 5:47–55. 10.7600/jpfsm.5.47 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.