Twomey et al. examine recent structural and functional data that have provided insight into AMPA receptor modulation by TARPs.
Abstract
Fast excitatory neurotransmission is mediated by the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) subtype of ionotropic glutamate receptor (AMPAR). AMPARs initiate depolarization of the postsynaptic neuron by allowing cations to enter through their ion channel pores in response to binding of the neurotransmitter glutamate. AMPAR function is dramatically affected by auxiliary subunits, which are regulatory proteins that form various complexes with AMPARs throughout the brain. The most well-studied auxiliary subunits are the transmembrane AMPAR regulatory proteins (TARPs), which alter the assembly, trafficking, localization, kinetics, and pharmacology of AMPARs. Recent structural and functional studies of TARPs and the TARP-fold germ cell-specific gene 1-like (GSG1L) subunit have provided important glimpses into how auxiliary subunits regulate the function of synaptic complexes. In this review, we put these recent structures in the context of new functional findings in order to gain insight into the determinants of AMPAR regulation by TARPs. We thus reveal why TARPs display a broad range of effects despite their conserved modular architecture.
Introduction
Glutamate (Glu) is the principle excitatory neurotransmitter in the central nervous system (CNS) and is thus critical for sensory and cognitive functions (Traynelis et al., 2010; Kumar and Mayer, 2013). It dictates communication between neurons via synapses, electrochemical junctions between neuronal cells, where the action potentials approaching presynaptic terminals trigger the vesicular release of Glu into the synaptic cleft. Glu then diffuses across the synaptic cleft to the postsynaptic terminal, where it binds to specialized membrane proteins called ionotropic Glu receptors (iGluRs). Upon binding of their agonist, Glu, these tetrameric ligand-gated ion channels allow cations to flow into the postsynaptic cell through their pores (Twomey and Sobolevsky, 2018; Wollmuth, 2018). The corresponding postsynaptic currents depolarize the postsynaptic membrane, thus creating an electrical signal that can be further transmitted along the neuronal network. Generally speaking, as iGluRs dictate excitatory neurotransmission throughout the CNS, dysregulation of these proteins results in a broad range of neuropathological conditions, including abnormal mental development, psychiatric disorders, memory loss (Alzheimer’s disease), movement disorders (Parkinson’s), motor neuron disease, as well as seizures and neuronal damage (Kwak and Weiss, 2006; Bowie, 2008; Volk et al., 2015).
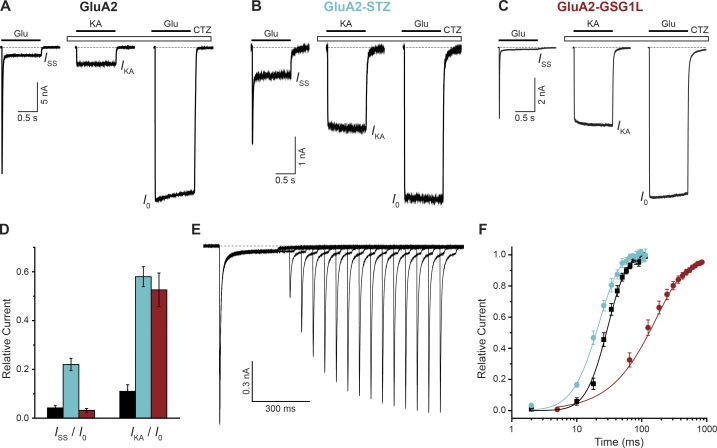
A subtype of iGluRs, AMPARs, is responsible for the initial, fast component of postsynaptic density depolarizing currents (Chater and Goda, 2014; Henley and Wilkinson, 2016). The AMPAR gating process is very rapid and includes three major components (Fig. 1 A; Twomey and Sobolevsky, 2018). Upon Glu binding, AMPARs undergo submillisecond-timescale activation that results in a sharply increasing inward current and thus membrane depolarization. During prolonged exposure to Glu, the receptors enter a desensitized state where they are still Glu bound, but the ion channel is in a nonconducting state to protect the neuronal cell from excessive depolarization. This desensitization gating process underlines the reduction of the AMPAR-mediated current to a steady-state ISS value that is only 1–5% of the maximum current value I0 (Fig. 1 A). After removal of Glu, AMPARs deactivate, or return to the closed state, resulting in the current returning to zero (dashed line in Fig. 1 A).
Figure 1.
Functional effects of auxiliary subunits. (A–C) Representative whole-cell currents recorded at −60 mV membrane potential from HEK cells expressing GluA2 (A), GluA2-STZ (B), or GluA2-GSG1L (C) in response to 1-s applications of the full agonist Glu alone (3 mM) or application of the partial agonist kainate (KA; 0.5 mM) or Glu in the continuous presence of the positive allosteric modulator cyclothiazide (CTZ; 30 µM). (D) Fraction of nondesensitized receptors (ISS/I0) and KA efficacy (IKA/I0) for GluA2 (black), GluA2-STZ (blue), and GluA2-GSG1L (red). (E) Two-pulse protocol monitoring recovery from desensitization for GluA2-GSG1L activated by Glu. (F) Mean recovery from desensitization measured using the protocol illustrated in E for GluA2 (black), GluA2-STZ (blue), and GluA2-GSG1L (red). The curves through the points are fits with the Hodgkin-Huxley equation with the time constant of recovery from desensitization 15.3 ± 1.1 ms for GluA2, 13.9 ± 0.9 ms for GluA2-STZ, and 164 ± 11 ms for GluA2-GSG1L. Error bars represent SEM. Modified from Twomey et al. (2016, 2017b).
Typically, transmembrane AMPAR regulatory proteins (TARPs) regulate AMPAR function in vivo (Chen et al., 2000; Yamazaki et al., 2004; Nakagawa et al., 2005; Priel et al., 2005; Tomita et al., 2005; Turetsky et al., 2005; Bedoukian et al., 2006; Zhang et al., 2006; Howe, 2015). An exemplar TARP is the subtype γ2 subunit (TARPγ2) or stargazin (STZ), which positively modulates AMPAR gating such that it favors the open state of the receptor. AMPAR–STZ complexes thus have increased steady-state currents (Fig. 1 B) compared with AMPARs alone (Fig. 1 A). Pharmacologically, the open state of AMPARs can also be stabilized by the binding of positive allosteric modulators, such as cyclothiazide (Patneau et al., 1993). An additional effect of TARPs on AMPARs is an increased efficacy of partial agonists, such as kainate (Fig. 1, A and B; Howe, 2015). Binding of a TARP relative, GSG1L subunit (Schwenk et al., 2012; Shanks et al., 2012), to AMPARs also results in an increased efficacy of partial agonists, but this has an apparently opposite effect on the open state, as it reduces the steady-state current (Fig. 1, C and D; Twomey et al., 2017b). The negative effect of GSG1L on AMPAR activation is further signified by a dramatically reduced rate of recovery from desensitization, a measure of how long it takes for the receptors to return from the desensitized state to a state in which their ability to activate is restored (Fig. 1 E). Compared with AMPARs alone, GluA2–GSG1L complexes show a greatly increased mean recovery time (Fig. 1 F). Conversely, STZ causes a slight increase in the apparent rate of recovery from desensitization (Priel et al., 2005; Turetsky et al., 2005; Twomey et al., 2016). This effect, however, is likely due to STZ promoting the open state, while the microscopic rate of recovery from desensitization remains unaltered (Carbone and Plested, 2016).
In this review, we discuss recent advances in structural and functional work on AMPAR–TARP complexes. We describe the architecture of AMPARs and how TARPs assemble around them. We compare sequences of claudins and claudin-fold TARPs and discuss how, based on recent structural, biochemical, and biophysical studies, the auxiliary subunits with common topology can exert a multitude of regulatory effects depending on specific features of protein binding interfaces and loop regions. Finally, we speculate on future directions of the field and how exploration of the diverse array of AMPAR regulatory subunits will resolve the fine tuning of AMPAR function throughout the CNS.
Architecture of AMPARs and AMPAR–TARP complexes
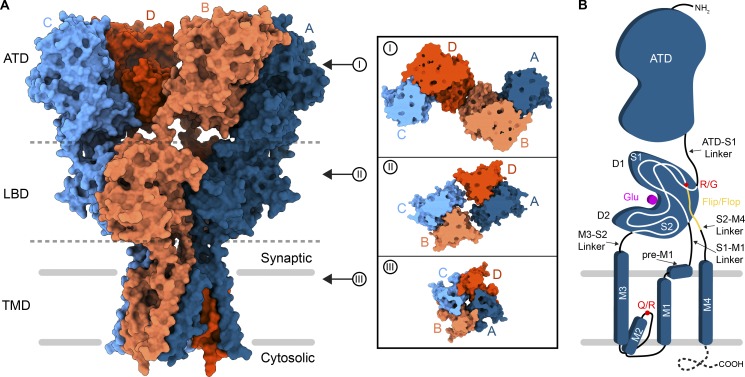
Structurally, AMPARs are made up of variable combinations of four multidomain subunits, GluA1–GluA4, which are arranged in an overall “Y” shape that is composed of three layers (Fig. 2; Mayer, 2016; Twomey and Sobolevsky, 2018). At the top of the Y is the layer of amino-terminal domains (ATDs) that are required for receptor assembly, trafficking, and regulation. Below the ATD layer is the layer of ligand-binding domains (LBDs), where each clamshell-shaped LBD contains a binding site for Glu. Together, the ATDs and LBDs make up the extracellular domain (ECD), which extends into the synaptic cleft. Focusing on a single subunit (Fig. 2 B), the LBD is composed of two polypeptide stretches, S1 and S2, separated by elements of the transmembrane domain (TMD). The TMD has three membrane-spanning helices, M1, M3, and M4, and a reentrant M2 loop between M1 and M3. Binding of the agonist Glu to the LBD is communicated to the TMD by nature of the LBD–TMD linkers S1–M1, M3–S2, and S2–M4. The C-terminal region of S2 leading to the S2–M4 linker appears in two different isoforms, flip or flop, depending on alternative splicing (Sommer et al., 1990). Both flip and flop LBD isoforms have unique gating kinetics and show different responses to allosteric modulators (Partin et al., 1996; Seeburg, 1996). Upstream of the flip/flop site is the R/G mRNA editing site, which can also influence AMPAR gating kinetics and regulation by allosteric modulators (Lomeli et al., 1994). M2 and M3 from the four subunits line a channel for cations to enter the cell from extracellular space. At the tip of the M2 loop is the Q/R site, where the GluA2 AMPAR subunit is mRNA edited from Gln to Arg (Hume et al., 1991). In contrast to assemblies of unedited subunits, AMPARs that contain R-edited GluA2 subunits are impermeable to divalent cations and do not undergo polyamine block (Huettner, 2015; Wollmuth, 2018). The C-termini of AMPAR subunits are typically either truncated in the constructs used for structure determination or unresolved, presumably due to structural heterogeneity, form a putative C-terminal domain (CTD) that is involved in synaptic localization, trafficking, mobility, and receptor regulation.
Figure 2.
Topology of AMPARs. (A) Structure of a homotetrameric AMPAR composed of GluA2 subunits (PDB accession no. 5WEO) in surface representation viewed parallel to the membrane. Synaptic and cytosolic spaces around the membrane (gray bars) are marked. Each GluA2 subunit is colored individually (A, dark blue; B, coral; C, light blue; D, dark orange). Each domain layer (ATD, LBD, and TMD) is labeled and separated by a dashed gray line. Insets mark top-down views of slices into the surface of each domain layer. (B) Topology of a single AMPAR subunit. The dashed line at the C-terminus indicates that the structure of the CTD is not yet determined, either because it is excluded from constructs or because of conformational heterogeneity.
Overall, AMPARs exhibit a unique arrangement of four subunits along their three-layer architecture, which has an overall twofold rotational symmetry despite representing a tetrameric assembly (Sobolevsky et al., 2009). In the ATD layer, there are two local dimers formed by the A/B and C/D subunit pairs (Fig. 2 A, inset I). The ATD layer cross-dimer interface is formed between the B and D subunits. Below, in the LBD layer, the dimer pairs swap, and the cross-dimer interface is formed between the A and C subunits, linking the local A/D and B/C dimers (Fig. 2 A, inset II). The TMD is pseudo-fourfold symmetric (Fig. 2 A, inset III). This unique arrangement of AMPAR subunits keeps four of them together but gives the assembly an exceptional conformational flexibility (Sobolevsky, 2015).
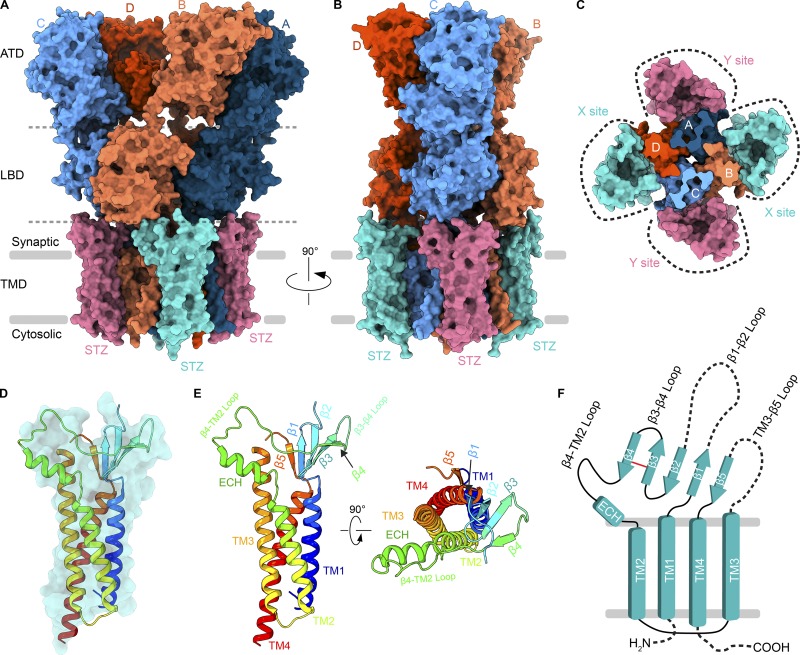
In neurons, however, this core AMPAR structure is typically decorated by regulatory proteins that alter AMPAR gating kinetics and pharmacology, in addition to receptor subunit composition, trafficking, and synaptic localization. Recent proteomics studies have identified >30 AMPAR regulatory proteins, which are structurally and functionally diverse (Schwenk et al., 2012; Shanks et al., 2012). In this review, we focus on the structure and function of claudin-fold auxiliary subunits, including TARPs, and advances in understanding their modulation of AMPAR function associated with recent developments in cryo-EM (Twomey et al., 2016, 2017a,b, 2018; Zhao et al., 2016, 2019; Chen et al., 2017; Herguedas et al., 2019). TARPs assemble around AMPARs at variable stoichiometry (Shi et al., 2009; Kim et al., 2010; Hastie et al., 2013; Twomey et al., 2016) but appear to share conserved assembly interfaces along the AMPAR TMD. Based on available structures, an AMPAR can at maximum interact with four TARPs at a time (Fig. 3, A–C; Zhao et al., 2016, 2019; Twomey et al., 2017a). The primary point of AMPAR–TARP assembly is the TMD, where auxiliary subunits act as a scaffold around the receptor. Four interfaces are available for the assembly, and they can be broken down into two groups. X sites of TARP binding use common interfaces with AMPAR subunit pairs D/C or B/A and Y sites with subunit pairs A/D and C/B (Fig. 3 C). TARPs have a claudin-like fold (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019); their TMDs are a helical bundle composed of four helices (Fig. 3, D and E), and their ECD comprises a β-sheet, where variable size loops between β-strands extend toward the AMPAR LBDs. The majority of the β-sheet is made up of four β-strands between transmembrane helix 1 (TM1) and TM2. A short cytosolic loop links TM2 and TM3, and before TM4 is β5. A conserved disulfide bridge (in STZ, between C66 and C76) strengthens the interaction between β3 and β4 strands. In STZ, a second, additional disulfide bridge between Cys39 near the start of the β1-β2 loop and Cys67 in β3 further strengthens stability of the auxiliary subunit head domain. The TM3-β5 and β1-β2 loops have yet to be well resolved structurally. Nevertheless, the β1-β2, β4-TM2, and TM3-β5 loops of TARP are positioned optimally to modulate AMPAR activity through their interaction with LBDs and LBD–TMD linkers. Depending on the position (X or Y site), different LBD interfaces are available for interaction with the TARP. For example, the extracellular loops of a TARP occupying the X site can contact LBDs from subunits A or B (or C or D) that belong to different local LBD dimers (Fig. 3 A). In contrast, TARPs occupying the Y site can contact LBDs from subunits B or C (or D or A) that belong to the same local LBD dimer (Fig. 3 B).
Figure 3.
Architecture of an AMPAR–TARP complex. (A and B) Surface representation of an AMPAR (GluA2 homotetramer) bound to four STZ molecules viewed parallel to the membrane (PDB accession no. 5WEO). Each GluA2 subunit is colored individually (A, dark blue; B, coral; C, light blue; D, dark orange). Each domain layer (ATD, LBD, and TMD) is labeled and separated by a dashed gray line. STZ subunits are colored teal or pink. (C) Top-down view of the TMD region, with two different sets of STZ or TARP assembly points (X or Y sites) around the AMPAR TMD marked by dashed lines. (D) Semitransparent surface and structure of a STZ subunit shown in ribbon, rainbow colored from N-terminus (blue) to C-terminus (red). (E) STZ structure shown as ribbon as in D, but with structural features labeled and also rotated 90° for a top-down view. (F) Topology of the STZ subunit. Dashed lines represent areas not clearly resolved in structural studies.
Structural conservation and variability among regulatory subunits
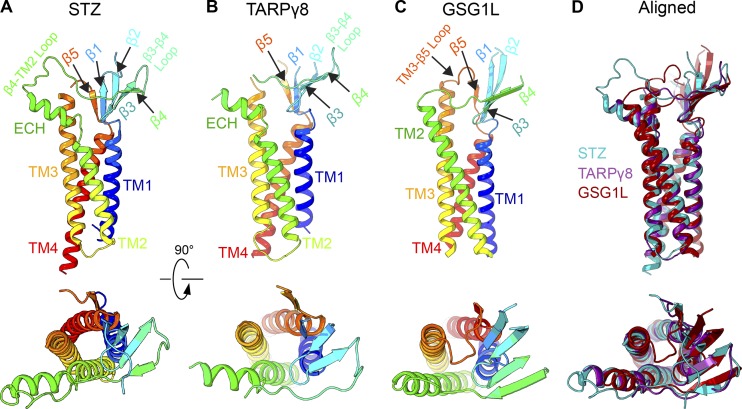
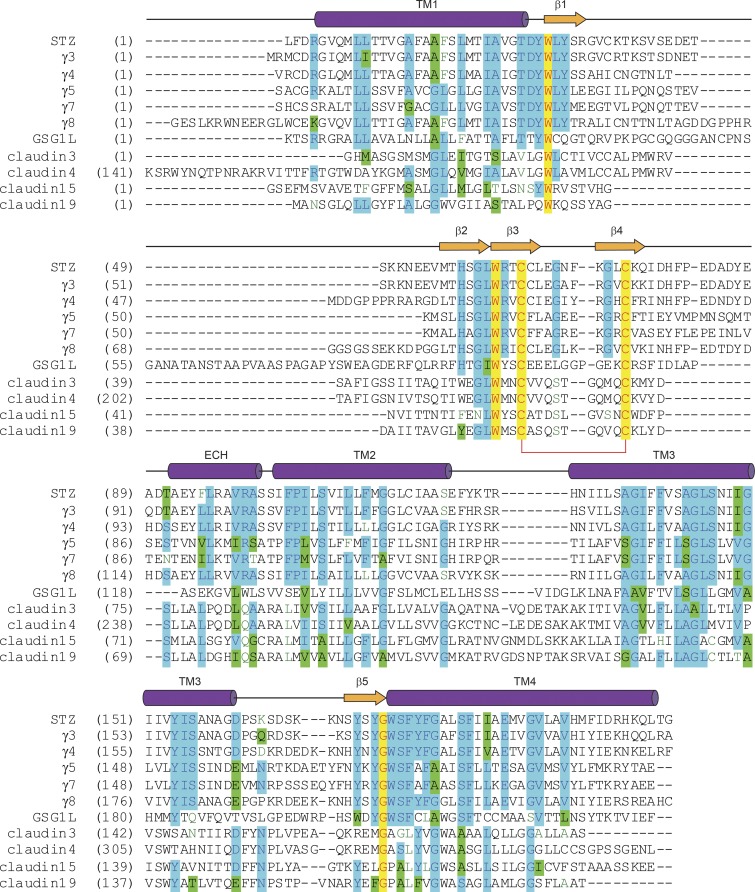
In addition to STZ, the structures of TARPγ8 (Herguedas et al., 2019) and GSG1L (Twomey et al., 2017b) in complex with AMPARs have been elucidated using cryo-EM. The high-resolution crystal structures of the homologous claudins (Suzuki et al., 2014; Saitoh et al., 2015) guided model building of these AMPAR regulatory subunits. Bulky side chains of residues in the TMD, typically better resolved in cryo-EM maps than other domains of auxiliary subunits, served as references and further helped accurate model building. The four-TM-helical bundle and β-sheet are conserved across STZ, TARPγ8, and GSG1L (Fig. 4). Both STZ and TARPγ8 have an extracellular helix (ECH) preceding TM2 (Fig. 4, A and B). Based on sequence conservation, the ECH is likely present in all TARPs (Fig. 5). Notably, GSG1L lacks the ECH but has a longer TM2 (Fig. 4, C and D). Interestingly, claudins, which share the overall fold with TARPs and GSG1L, also display variability in this structural element, with claudins 3, 4, and 19 lacking the ECH and claudin 15 having it present (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019). It is possible that the lack of ECH in claudins 3, 4, and 19 could be due to crystal packing. An important distinction between claudins and claudin-fold AMPAR auxiliary subunits is various degrees of TM3 helix bending. While TARPs and GSG1L have a straight α-helical TM3, it bends in claudins, and the extent of bending defines the type of claudin–claudin interactions and affects the morphology and adhesiveness of the tight junctions (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019).
Figure 4.
Structures of TARPs and GSG1L. (A-C) Structures of STZ or TARPγ2 (PDB accession no. 5WEO), TARPγ8 (PDB accession no. 6QKC), and GSG1L (PDB accession no. 5VHY) shown in ribbon representation and rainbow colored from N-terminus (blue) to C-terminus (red) viewed parallel to the membrane (top row) or perpendicular to the membrane (bottom row). Structural elements are labeled. (D) Superposition of STZ (teal), TARPγ8 (purple), and GSG1L (red) viewed parallel to the membrane (top) or perpendicular to the membrane (bottom).
Figure 5.
Sequence alignment for claudins and claudin-fold AMPAR auxiliary subunits. The secondary structure of STZ is shown above the sequence alignment as cylinders (α-helices), arrows (β-strands), or lines (loops). Completely conserved residues are highlighted in yellow. Mostly conserved residues are highlighted in blue (or green for homologous residues). Conserved cysteines forming a disulfide bridge between β3 and β4 are connected by a red bracket. The C-terminal residues are excluded.
Within the conserved β-sheet of STZ, TARPγ8, and GSG1L, all three subunits have a disulfide bridge between β3 and β4. Sequence conservation suggests that this general topology is conserved across all TARP subunits and claudins (Fig. 5). The second disulfide bridge in STZ makes it distinct from TARPγ8 and GSG1L, as well as claudins. Notably, the loop between β4 and the ECH in TARPs is longer than in claudins, which may account for why the ECH in TARPs is oriented away from the helical bundle (Fig. 4, A and B), while the ECH in claudin 15 leads directly toward β4 (Suzuki et al., 2014); there is a need for this region to have a more direct link here by nature of fewer amino acids, similar to the more claudin-like subunit GSG1L (Fig. 4 C). The extracellular region of TARPs controls AMPAR channel properties, while the CTD regulates receptor trafficking (Tomita et al., 2005). A more recent study, however, suggests that the C-termini of TARPs can also affect complex assembly and AMPAR gating (Ben-Yaacov et al., 2017). The C-termini of these proteins have eluded structural studies because they are truncated from constructs or because of their conformational heterogeneity.
Since the overall topology of the TARP and GSG1L regulatory subunits is grossly conserved, it seems that the variability in the loops between β strands and TM helices (Fig. 5) gives the auxiliary subunits their specific effects on AMPAR gating (Fig. 1). To understand how these loops alter AMPAR function, chimeras between auxiliary subunits were constructed (Riva et al., 2017; Twomey et al., 2017b). For example, replacing the β1-β2 loop of GSG1L with the shorter loop from STZ largely eliminated GSG1L’s effect on AMPAR recovery from desensitization (Twomey et al., 2017b). However, doing the opposite to STZ did not confer the dramatic slowing of recovery from desensitization to the corresponding complex. This indicates that while the β1-β2 loop is important for slowing of recovery from desensitization by GSG1L, the β-sheet scaffold could also be important; β1 and β2 in GSG1L are extended compared with that in STZ (Fig. 4, C and D) and may contribute to the loop’s function. Similarly, the β4-TM2 loop appears to alter the STZ function of increasing the AMPAR ion channel open probability. Overall, these chimeric studies showed that the combined actions of moieties in the GSG1L extracellular region between TM1 and TM2 are responsible for GSG1L’s effects on AMPAR desensitization. An important note is that these STZ–GSG1L chimeras maintained the increased AMPAR partial agonist efficacy, which is a hallmark of AMPAR–TARP complexes. This suggests that partial agonist efficacy can be used to gauge proper complex assembly, possibly applicable to a broader range of AMPAR auxiliary subunits.
Chimeras between STZ and TARPγ8 further show the importance of the fine differences between TARP subunits in the ECD (Riva et al., 2017). TARPγ8 affects AMPARs similarly to STZ but has a more pronounced effect on slowing entry into desensitization. Switching the β1-β2 loop between STZ and TARPγ8 asymmetrically alters AMPAR desensitization. The β1-β2 loop is minimally conserved between different TARPs and GSG1L and varies in length significantly (Fig. 5). Based on the chimeras between STZ and GSG1L, as well as STZ and TARPγ8, the β1-β2 loop appears to have unique effects on desensitization kinetics. However, given the variability between subunits in this region, studies need to be performed across TARP subunits in order to clarify the functions of this loop (Fig. 5). Nevertheless, it is likely that the β1-β2 loop acts in concert with the other highly variable β4-TM2 and TM3-β5 loops in the TARP ECD (Fig. 5), because altering these individual loops between chimeric constructs does not completely change the TARP effects on AMPAR gating. Indeed, placing the TARPγ8 β1-β2 loop into STZ with a GS linker instead of the wild-type TM3-β5 loop had extreme effects on slowing down the entry into desensitization and increasing the steady-state currents (Riva et al., 2017).
Interfaces between AMPARs and TARPs
The diverse effects that TARPs have on AMPAR gating kinetics and pharmacology arise from fine differences in the structures of these regulatory subunits. Ultimately, alterations in the TARP ECD loops allow different sites in the AMPAR ECD to be modulated. The TARP ECD is juxtaposed to the AMPAR LBD (Fig. 6 A) and does not have access to the ATD. Structurally, the primary interface between TARPs and AMPARs is in the TMD (Fig. 4, A and B; Twomey et al., 2016; Zhao et al., 2016), though assembly of the complexes is dependent on the TARP CTD (Ben-Yaacov et al., 2017). The TMD interface between TARPs and AMPARs is mediated by TM3 and TM4 from the TARP and M1, M2, and M4 of the AMPAR. AMPARs have many cross-subunit interfaces (Fig. 2), and TARPs extend complexity of the synaptic complex topology by acting as a scaffold in the TMD and interacting with M1 and M2 from one AMPAR subunit and M4 from a neighboring subunit (Fig. 6 B); X site TARPs interface between subunits A and B or C and D in the TMD, while Y site TARPs interface between subunits B and C or A and D in the TMD. Since most native AMPARs are heteromers made up of combinations of subunits GluA1–GluA4 (Traynelis et al., 2010; Wollmuth, 2018; Zhao et al., 2019), the X and Y assembly sites are likely unique based on the heteromeric combination.
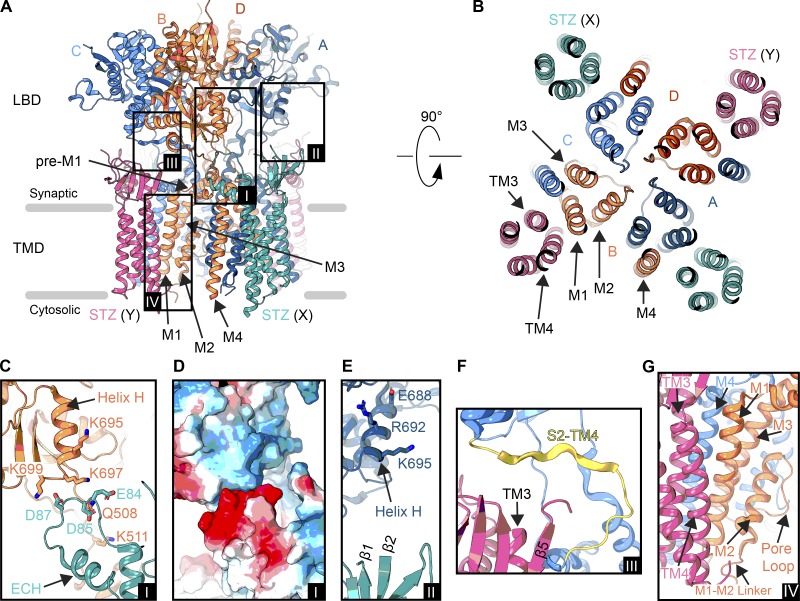
Figure 6.
Interfaces in an AMPAR–STZ complex. (A) Structure of an AMPAR (GluA2 homotetramer) bound to four STZ molecules in ribbon representation viewed parallel to the membrane (PDB accession no. 5WEO). The ATD has been excluded. Each GluA2 subunit is colored individually (A, dark blue; B, coral; C, light blue; D, dark orange), and STZ subunits are colored teal (X site) or pink (Y site). Boxed are regions illustrated in C–G. (B) Extracellular view on the TMD from right above the Q/R site. Membrane segments of one AMPAR subunit (B subunit M1-M4) and one STZ subunit (Y site, TM1-4) are labeled. (C) Interface between the LBD of the B subunit from the AMPAR and the β4-TM2 loop preceding the ECH of a X-site STZ molecule (A, inset I). (D) Electrostatic surface for the region shown in C, blue being positively charged, red negatively charged, and white neutral (A, inset I). (E) Potential interaction between an X-site TARP β1-β2 loop and A subunit LBD helix H (A, inset II). (F) Interface between a Y-site TARP TM3-β5 loop and S2-TM4 linker in the C subunit (A, inset III). (G) AMPAR–TARP interface in the TMD (A, inset IV).
Depending on what position a TARP subunit occupies, its ECD has access to different sites on the core AMPAR. Using the GluA2–STZ complex as an example, an X site STZ likely participates in an electrostatic interaction (Twomey et al., 2016; Zhao et al., 2016) that modulates AMPAR gating kinetics (Fig. 6, C and D). The STZ β4-TM2 loop contains acidic residues E84, D85, and D87 that precede the ECH; this acidic patch is conserved in all TARPs except TARPγ5 and TARPγ7, in addition to GSG1L, which also has a longer TM2 and no ECH when compared with the current TARP structures (Fig. 4 D). In the open-state structure (Twomey et al., 2017a), which represents a higher quality cryo-EM reconstruction of the GluA2–STZ complex than the original closed-state structures (Twomey et al., 2016; Zhao et al., 2016), these three acidic residues sit opposite a basic stretch, comprised of K695, K697, and K699, around helix H in the AMPAR B or D subunit LBDs (Fig. 6, C and D). Charge reversal substitution of K697 and K699, part of the “KGK” motif, nullifies the effects of STZ on AMPAR gating kinetics (Dawe et al., 2016). The exact role of the acidic residues in the STZ β4-TM2 loop, however, remains unclear, as neutralizing some of them resulted in stronger instead of weaker modulation of AMPAR gating by STZ (Riva et al., 2017). An X-site STZ molecule also has access to the LBD in the opposite LBD local dimer by nature of the β1-β2 loop (Fig. 6 E). While this STZ can contact the LBD helix H in the B or D subunits by nature of the β4-TM2 loop, helix H from subunits A or C (in opposite local dimer pairs, see Fig. 2 A inset II) can also be contacted by the β1-β2 loop based on the positioning of the β1 and β2 strands. While this has not been directly seen in TARP complexes, it has been visualized in cryo-EM density for the GluA2–GSG1L complex (Twomey et al., 2017b). Therefore, a regulatory subunit occupying an X site acts as a structural scaffold by further linking the AMPAR subunits in the TMD and LBD and can potentially interact with the two LBD local dimers at once. Thus, the modifications of the environment around the LBD helix H (Dawe et al., 2016) could be altering TARP effects via the β1-β2 loop and not the β4-TM2 loop.
In terms of Y-site occupancy, TARP subunit β1-β2 loops pose an interesting challenge. For example, GSG1L does not seem to assemble in the Y site. We posited that this is due to its extended β1 and β2 strands (Fig. 4), as well as a long β1-β2 loop (roughly 30 amino acids longer than that of STZ), which do not allow assembly at these sites, because the GSG1L ECD would clash with the B/C or A/D LBD dimers (Twomey et al., 2017b). A similar mode of assembly was observed for TARPγ8, although only two of four AMPAR subunits were concatenated with TARPγ8 in the heteromeric GluA1/2, whereas all four AMPAR subunits concatenated with GSG1L in homomeric GluA2. TARPs assembled at the Y site, however, can interact with the AMPAR S2-TM4 linker of subunits A or C through the TM3-β5 loop (Fig. 6 F; Zhao et al., 2016, 2019; Chen et al., 2017; Twomey et al., 2017a; Herguedas et al., 2019). This includes the location of the flip/flop splice site (Fig. 2 B) and may explain why TARPs differentially modulate receptors containing flip/flop splice variants (Tomita et al., 2006; Kott et al., 2007; Milstein and Nicoll, 2008). In addition, altering the stoichiometry of TARPs around differentially spliced flip/flop AMPARs alters the TARP effects on receptor dynamics (Dawe et al., 2019), suggesting that occupancy of X or Y sites may provide different access to the S2–M4 linker. While a key role in modulating AMPAR gating by auxiliary subunits was postulated for both S2–M4 and S1–M1 LBD–TMD linkers based on mutagenesis studies (Riva et al., 2017), the corresponding molecular interactions are yet to be resolved structurally.
All of the discussed TARP ECD interactions are connected to the TMD, which interacts through TM3 and TM4 with M1, M2, and M4 of the core AMPAR (Fig. 6 G). While TM3 of the TARP interacts with M4 of the AMPAR, TM4 juxtaposed to M1 and can modulate M2 movement through interaction with the M1–M2 linker. The AMPAR ion channel is lined by both M3 and M2, with the selectivity filter formed by the extended, nonhelical region of M2. Since the selectivity filter coordinates polyamines and polyamine toxin tails (Twomey et al., 2018), the TM4–M2 interactions could be the reason why TARPs increase polyamine permeation through AMPARs and attenuate polyamine block (Soto et al., 2007; Jackson et al., 2011; Brown et al., 2018). The AMPAR–TARP TMD interface, while overall similar, is unique per TARP subunit. This is exemplified by the design of a noncompetitive inhibitor, LY3130481, that selectivity inhibits AMPAR-TARPγ8 complexes, although specificity can be switched to other TARP-containing complexes by point mutations in the TMD (Kato et al., 2016). To clarify this mechanism of action, and perhaps pave a route toward synthesis of new synaptic complex-specific small-molecule modulators, high-resolution structural studies are needed.
Outlook
While the recent wealth of structural information on TARP/TARP-like complexes (Twomey et al., 2016, 2017a,b, 2018; Zhao et al., 2016, 2019; Chen et al., 2017; Herguedas et al., 2019) and claudins (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019) has provided new insights into potential modulatory interfaces, the exact interactions are in fact ambiguous. The loops are often only visible at low thresholds in cryo-EM density maps, indicating that the loops are conformationally heterogeneous in the states that have been captured in structural studies and are not tightly bound. In addition, direct structural information is limited to TARPγ2, TARPγ8, and GSG1L. It could be that TARP loops alter how AMPARs transition between various gating states. Indeed, the dynamics of AMPARs are altered in the presence of TARPs; FRET and cross-linking studies have shown that AMPARs are overall more compact in the presence of TARPs (Shaikh et al., 2016; Baranovic and Plested, 2018). Furthermore, FRET has shown that TARPs stabilize the closure of AMPAR LBDs (MacLean et al., 2014). This suggests that the effects TARPs have on AMPAR gating kinetics could be due to lowering the energetic barrier for channel opening and increasing the efficacy of agonists and partial agonists by promoting the active state of the LBDs. Perhaps this class of auxiliary subunits alters AMPAR behavior through a concerted effort to alter LBD opening/closing by further coupling the LBD to AMPAR TMD elements. What is clear, however, is that, while individual moieties in TARP structures may have dominant effects on AMPAR function, it is through combined elements in both the ECD and TMD that TARPs exert their regulatory effects on AMPARs.
While higher resolution cryo-EM structures would be excellent for revealing the details of these interfaces and also how conserved the regulatory interfaces are across the entire TARP family, biophysical approaches such as atomic force microscopy and FRET, in combination with computational simulations and electrophysiology, may be more informative in piecing together how TARPs modulate AMPAR activity. Certainly, many key questions still remain. How do TARPs and TARP stoichiometry alter gating transitions? How do TARPs alter agonist accessibility and LBD closure? What roles do cytosolic elements from AMPARs and TARPs play in gating, trafficking, and assembly? Why are there so many TARP subunits, and how unique is each subunit? Can AMPARs interact with a heteromeric mixture of TARPs? Which position, X or Y, is the dominant regulatory point?
The family of AMPAR regulatory subunits also exists beyond TARPs, and many of them regulate AMPAR assembly, homeostasis, trafficking, and localization (Schwenk et al., 2009, 2012; Kalashnikova et al., 2010; von Engelhardt et al., 2010; Gill et al., 2011; Shanks et al., 2012; Erlenhardt et al., 2016; Brechet et al., 2017; Brown et al., 2018). Thus far, these complexes remain structurally unexplored and enigmatic. We anticipate that the coming years will be an exciting time for defining how these complexes fine-tune AMPAR function throughout the CNS.
Acknowledgments
Lesley C. Anson served as editor.
A.I. Sobolevsky is supported by the National Institutes of Health (grants R01 CA206573, R01 NS083660, and R01 NS107253), the National Science Foundation (grant 1818213), and the Irma T. Hirschl Career Scientist Award.
The authors declare no competing financial interests.
References
- Baranovic J., and Plested A.J.. 2018. Auxiliary subunits keep AMPA receptors compact during activation and desensitization. eLife. 7: e40548 10.7554/eLife.40548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoukian M.A., Weeks A.M., and Partin K.M.. 2006. Different domains of the AMPA receptor direct stargazin-mediated trafficking and stargazin-mediated modulation of kinetics. J. Biol. Chem. 281:23908–23921. 10.1074/jbc.M600679200 [DOI] [PubMed] [Google Scholar]
- Ben-Yaacov A., Gillor M., Haham T., Parsai A., Qneibi M., and Stern-Bach Y.. 2017. Molecular Mechanism of AMPA Receptor Modulation by TARP/Stargazin. Neuron. 93:1126–1137.e1124. [DOI] [PubMed] [Google Scholar]
- Bowie D. 2008. Ionotropic glutamate receptors & CNS disorders. CNS Neurol. Disord. Drug Targets. 7:129–143. 10.2174/187152708784083821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechet A., Buchert R., Schwenk J., Boudkkazi S., Zolles G., Siquier-Pernet K., Schaber I., Bildl W., Saadi A., Bole-Feysot C., et al. . 2017. AMPA-receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nat. Commun. 8:15910 10.1038/ncomms15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.M.G.E., McGuire H., and Bowie D.. 2018. Stargazin and cornichon-3 relieve polyamine block of AMPA receptors by enhancing blocker permeation. J. Gen. Physiol. 150:67–82. 10.1085/jgp.201711895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A.L., and Plested A.J.. 2016. Superactivation of AMPA receptors by auxiliary proteins. Nat. Commun. 7:10178 10.1038/ncomms10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater T.E., and Goda Y.. 2014. The role of AMPA receptors in postsynaptic mechanisms of synaptic plasticity. Front. Cell. Neurosci. 8:401 10.3389/fncel.2014.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chetkovich D.M., Petralia R.S., Sweeney N.T., Kawasaki Y., Wenthold R.J., Bredt D.S., and Nicoll R.A.. 2000. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 408:936–943. 10.1038/35050030 [DOI] [PubMed] [Google Scholar]
- Chen S., Zhao Y., Wang Y., Shekhar M., Tajkhorshid E., and Gouaux E.. 2017. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 170:1234–1246.e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe G.B., Musgaard M., Aurousseau M.R.P., Nayeem N., Green T., Biggin P.C., and Bowie D.. 2016. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 89:1264–1276. 10.1016/j.neuron.2016.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe G.B., Kadir M.F., Venskutonyte R., Perozzo A.M., Yan Y., Alexander R.P.D., Navarrete C., Santander E.A., Arsenault M., Fuentes C., et al. . 2019. Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors. Neuron. 102:976–992.e975. [DOI] [PubMed] [Google Scholar]
- Erlenhardt N., Yu H., Abiraman K., Yamasaki T., Wadiche J.I., Tomita S., and Bredt D.S.. 2016. Porcupine Controls Hippocampal AMPAR Levels, Composition, and Synaptic Transmission. Cell Reports. 14:782–794. 10.1016/j.celrep.2015.12.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M.B., Kato A.S., Roberts M.F., Yu H., Wang H., Tomita S., and Bredt D.S.. 2011. Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J. Neurosci. 31:6928–6938. 10.1523/JNEUROSCI.6271-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie P., Ulbrich M.H., Wang H.L., Arant R.J., Lau A.G., Zhang Z., Isacoff E.Y., and Chen L.. 2013. AMPA receptor/TARP stoichiometry visualized by single-molecule subunit counting. Proc. Natl. Acad. Sci. USA. 110:5163–5168. 10.1073/pnas.1218765110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley J.M., and Wilkinson K.A.. 2016. Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 17:337–350. 10.1038/nrn.2016.37 [DOI] [PubMed] [Google Scholar]
- Herguedas B., Watson J.F., Ho H., Cais O., García-Nafría J., and Greger I.H.. 2019. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Science. 364:eaav9011. 10.1126/science.aav9011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J.R. 2015. Modulation of non-NMDA receptor gating by auxiliary subunits. J. Physiol. 593:61–72. 10.1113/jphysiol.2014.273904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettner J.E. 2015. Glutamate receptor pores. J. Physiol. 593:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume R.I., Dingledine R., and Heinemann S.F.. 1991. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 253:1028–1031. 10.1126/science.1653450 [DOI] [PubMed] [Google Scholar]
- Jackson A.C., Milstein A.D., Soto D., Farrant M., Cull-Candy S.G., and Nicoll R.A.. 2011. Probing TARP modulation of AMPA receptor conductance with polyamine toxins. J. Neurosci. 31:7511–7520. 10.1523/JNEUROSCI.6688-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E., Lorca R.A., Kaur I., Barisone G.A., Li B., Ishimaru T., Trimmer J.S., Mohapatra D.P., and Díaz E.. 2010. SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron. 65:80–93. 10.1016/j.neuron.2009.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A.S., Burris K.D., Gardinier K.M., Gernert D.L., Porter W.J., Reel J., Ding C., Tu Y., Schober D.A., Lee M.R., et al. . 2016. Forebrain-selective AMPA-receptor antagonism guided by TARP γ-8 as an antiepileptic mechanism. Nat. Med. 22:1496–1501. 10.1038/nm.4221 [DOI] [PubMed] [Google Scholar]
- Kim K.S., Yan D., and Tomita S.. 2010. Assembly and stoichiometry of the AMPA receptor and transmembrane AMPA receptor regulatory protein complex. J. Neurosci. 30:1064–1072. 10.1523/JNEUROSCI.3909-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kott S., Werner M., Körber C., and Hollmann M.. 2007. Electrophysiological properties of AMPA receptors are differentially modulated depending on the associated member of the TARP family. J. Neurosci. 27:3780–3789. 10.1523/JNEUROSCI.4185-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., and Mayer M.L.. 2013. Functional insights from glutamate receptor ion channel structures. Annu. Rev. Physiol. 75:313–337. 10.1146/annurev-physiol-030212-183711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S., and Weiss J.H.. 2006. Calcium-permeable AMPA channels in neurodegenerative disease and ischemia. Curr. Opin. Neurobiol. 16:281–287. 10.1016/j.conb.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J.R., Kuner T., Monyer H., Higuchi M., Bach A., and Seeburg P.H.. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 266:1709–1713. 10.1126/science.7992055 [DOI] [PubMed] [Google Scholar]
- MacLean D.M., Ramaswamy S.S., Du M., Howe J.R., and Jayaraman V.. 2014. Stargazin promotes closure of the AMPA receptor ligand-binding domain. J. Gen. Physiol. 144:503–512. 10.1085/jgp.201411287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L. 2016. Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 41:119–127. 10.1016/j.sbi.2016.07.002 [DOI] [PubMed] [Google Scholar]
- Milstein A.D., and Nicoll R.A.. 2008. Regulation of AMPA receptor gating and pharmacology by TARP auxiliary subunits. Trends Pharmacol. Sci. 29:333–339. 10.1016/j.tips.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Cheng Y., Ramm E., Sheng M., and Walz T.. 2005. Structure and different conformational states of native AMPA receptor complexes. Nature. 433:545–549. 10.1038/nature03328 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Irie K., Tanaka H., Nishikawa K., Suzuki H., Saitoh Y., Tamura A., Tsukita S., and Fujiyoshi Y.. 2019. Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat. Commun. 10:816 10.1038/s41467-019-08760-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partin K.M., Fleck M.W., and Mayer M.L.. 1996. AMPA receptor flip/flop mutants affecting deactivation, desensitization, and modulation by cyclothiazide, aniracetam, and thiocyanate. J. Neurosci. 16:6634–6647. 10.1523/JNEUROSCI.16-21-06634.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patneau D.K., Vyklicky L. Jr., and Mayer M.L.. 1993. Hippocampal neurons exhibit cyclothiazide-sensitive rapidly desensitizing responses to kainate. J. Neurosci. 13:3496–3509. 10.1523/JNEUROSCI.13-08-03496.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A., Kolleker A., Ayalon G., Gillor M., Osten P., and Stern-Bach Y.. 2005. Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J. Neurosci. 25:2682–2686. 10.1523/JNEUROSCI.4834-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva I., Eibl C., Volkmer R., Carbone A.L., and Plested A.J.. 2017. Control of AMPA receptor activity by the extracellular loops of auxiliary proteins. eLife. 6:e28680 10.7554/eLife.28680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y., Suzuki H., Tani K., Nishikawa K., Irie K., Ogura Y., Tamura A., Tsukita S., and Fujiyoshi Y.. 2015. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 347:775–778. 10.1126/science.1261833 [DOI] [PubMed] [Google Scholar]
- Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A., Heimrich B., Chisaka O., Jonas P., Schulte U., Fakler B., and Klöcker N.. 2009. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 323:1313–1319. 10.1126/science.1167852 [DOI] [PubMed] [Google Scholar]
- Schwenk J., Harmel N., Brechet A., Zolles G., Berkefeld H., Müller C.S., Bildl W., Baehrens D., Hüber B., Kulik A., et al. . 2012. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 74:621–633. 10.1016/j.neuron.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Seeburg P.H. 1996. The role of RNA editing in controlling glutamate receptor channel properties. J. Neurochem. 66:1–5. 10.1046/j.1471-4159.1996.66010001.x [DOI] [PubMed] [Google Scholar]
- Shaikh S.A., Dolino D.M., Lee G., Chatterjee S., MacLean D.M., Flatebo C., Landes C.F., and Jayaraman V.. 2016. Stargazin Modulation of AMPA Receptors. Cell Reports. 17:328–335. 10.1016/j.celrep.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks N.F., Savas J.N., Maruo T., Cais O., Hirao A., Oe S., Ghosh A., Noda Y., Greger I.H., Yates J.R. III, and Nakagawa T.. 2012. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Reports. 1:590–598. 10.1016/j.celrep.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Lu W., Milstein A.D., and Nicoll R.A.. 2009. The stoichiometry of AMPA receptors and TARPs varies by neuronal cell type. Neuron. 62:633–640. 10.1016/j.neuron.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I. 2015. Structure and gating of tetrameric glutamate receptors. J. Physiol. 593:29–38. 10.1113/jphysiol.2013.264911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky A.I., Rosconi M.P., and Gouaux E.. 2009. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 462:745–756. 10.1038/nature08624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Keinänen K., Verdoorn T.A., Wisden W., Burnashev N., Herb A., Köhler M., Takagi T., Sakmann B., and Seeburg P.H.. 1990. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 249:1580–1585. 10.1126/science.1699275 [DOI] [PubMed] [Google Scholar]
- Soto D., Coombs I.D., Kelly L., Farrant M., and Cull-Candy S.G.. 2007. Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat. Neurosci. 10:1260–1267. 10.1038/nn1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Nishizawa T., Tani K., Yamazaki Y., Tamura A., Ishitani R., Dohmae N., Tsukita S., Nureki O., and Fujiyoshi Y.. 2014. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science. 344:304–307. 10.1126/science.1248571 [DOI] [PubMed] [Google Scholar]
- Tomita S., Adesnik H., Sekiguchi M., Zhang W., Wada K., Howe J.R., Nicoll R.A., and Bredt D.S.. 2005. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 435:1052–1058. 10.1038/nature03624 [DOI] [PubMed] [Google Scholar]
- Tomita S., Sekiguchi M., Wada K., Nicoll R.A., and Bredt D.S.. 2006. Stargazin controls the pharmacology of AMPA receptor potentiators. Proc. Natl. Acad. Sci. USA. 103:10064–10067. 10.1073/pnas.0603128103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., and Dingledine R.. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 62:405–496. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetsky D., Garringer E., and Patneau D.K.. 2005. Stargazin modulates native AMPA receptor functional properties by two distinct mechanisms. J. Neurosci. 25:7438–7448. 10.1523/JNEUROSCI.1108-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., and Sobolevsky A.I.. 2018. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry. 57:267–276. 10.1021/acs.biochem.7b00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Grassucci R.A., Frank J., and Sobolevsky A.I.. 2016. Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 353:83–86. 10.1126/science.aaf8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Grassucci R.A., Frank J., and Sobolevsky A.I.. 2017a Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 549:60–65. 10.1038/nature23479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Grassucci R.A., Frank J., and Sobolevsky A.I.. 2017b Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 94:569–580.e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey E.C., Yelshanskaya M.V., Vassilevski A.A., and Sobolevsky A.I.. 2018. Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Neuron. 99:956–968.e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk L., Chiu S.L., Sharma K., and Huganir R.L.. 2015. Glutamate synapses in human cognitive disorders. Annu. Rev. Neurosci. 38:127–149. 10.1146/annurev-neuro-071714-033821 [DOI] [PubMed] [Google Scholar]
- von Engelhardt J., Mack V., Sprengel R., Kavenstock N., Li K.W., Stern-Bach Y., Smit A.B., Seeburg P.H., and Monyer H.. 2010. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 327:1518–1522. 10.1126/science.1184178 [DOI] [PubMed] [Google Scholar]
- Wollmuth L.P. 2018. Ion permeation in ionotropic glutamate receptors: Still dynamic after all these years. Curr. Opin. Physiol. 2:36–41. 10.1016/j.cophys.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M., Ohno-Shosaku T., Fukaya M., Kano M., Watanabe M., and Sakimura K.. 2004. A novel action of stargazin as an enhancer of AMPA receptor activity. Neurosci. Res. 50:369–374. 10.1016/j.neures.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Zhang W., Robert A., Vogensen S.B., and Howe J.R.. 2006. The relationship between agonist potency and AMPA receptor kinetics. Biophys. J. 91:1336–1346. 10.1529/biophysj.106.084426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen S., Yoshioka C., Baconguis I., and Gouaux E.. 2016. Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 536:108–111. 10.1038/nature18961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen S., Swensen A.C., Qian W.J., and Gouaux E.. 2019. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science. 364:355–362. 10.1126/science.aaw8250 [DOI] [PMC free article] [PubMed] [Google Scholar]