JGP study describes the spatiotemporal dynamics of proteins and lipids involved in the exocytosis of the matrix metalloproteinase MMP-9 from breast cancer cells.
Abstract
JGP study describes the spatiotemporal dynamics of proteins and lipids involved in the exocytosis of the matrix metalloproteinase MMP-9 from breast cancer cells.
The metastasis, or spread, of cancer cells to other parts of the body begins with the breakdown and remodeling of the extracellular matrix surrounding the primary tumor. This is achieved, in part, by matrix metalloproteinases (MMPs), a family of matrix-degrading enzymes that are secreted from cancer cells via a highly regulated process involving dozens of protein and lipid factors. In this issue of JGP, Stephens et al. reveal the complex dynamics of many of these factors at individual sites of MMP exocytosis in breast cancer cells (1).
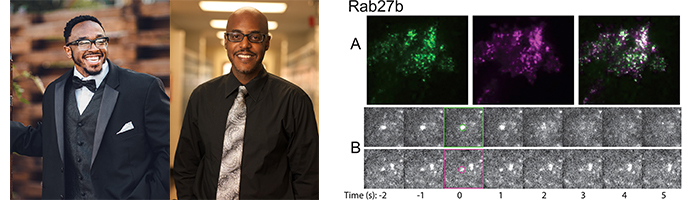
Dominique Stephens (left), Dinari Harris (center), and colleagues used TIRF microscopy to identify factors associated with secretory vesicles containing the enzyme MMP-9 in breast cancer cells and monitored their dynamics over the course of exocytosis. In the example shown, Rab27b (magenta) colocalizes with MMP-9 (green) at steady state (top images). Both proteins rapidly disperse from exocytic sites following membrane fusion (bottom images).
Increased expression of two closely related MMPs, MMP-9 and MMP-2, is found in a wide variety of cancers and is associated with poor prognosis. Secretory vesicles containing these enzymes are trafficked to the plasma membrane of cancer cells, where they can fuse and release their cargo into the extracellular space. This process, and the numerous factors regulating it, have been extensively studied using traditional biochemical methods. “But what’s been lacking in the field is an understanding of the real-time dynamics of these factors at the sites of exocytosis,” says Dinari Harris, an assistant professor at Howard University.
This type of information is more readily available for the factors that regulate secretion from neuronal cells (2,3). These factors, or “organizational elements,” can be broadly classified into three distinct groups: scaffolding proteins, such as Rab GTPases and SNARE proteins, that link secretory vesicles to the plasma membrane; lipid molecules within the membranes themselves; and components of the actin cytoskeleton.
Harris and colleagues, including first author Dominique Stephens, used two-color TIRF microscopy to assess the colocalization of ∼150 different proteins with MMP-9-containing secretory vesicles and saw that a similar set of organizational elements are involved in the secretion of MMP-9 from breast cancer cells.
Multiple Rab GTPases, for example, were found to be associated with MMP-9-positive vesicles, including two closely related isoforms, Rab27a and Rab27b, that have previously been linked to both exocytosis and tumor cell metastasis (4,5). Despite their similarity to each other, Stephens et al. found evidence to suggest that Rab27a and Rab27b may have distinct roles in MMP-9 secretion. By analyzing the kinetics of individual exocytic events, the researchers discovered that some MMP-9-containing vesicles fuse with the plasma membrane as soon as they reach the cell cortex, while others dock at the membrane for a short period before undergoing fusion. Overexpressing Rab27a appears to promote the former type of fusion, whereas overexpressing Rab27b promotes the latter type. Harris plans to investigate this potential specificity using CRISPR/Cas9 to knockout each of the two isoforms.
Stephens et al. found that Rab GTPases, along with many other scaffolding proteins, assemble on MMP-9-containing vesicles docked at the plasma membrane and then quickly dissipate as soon as the vesicle undergoes fusion. In contrast, amphiphysin1 and syndapin1—two BAR domain–containing proteins commonly associated with endocytosis—were transiently recruited to exocytic sites at the moment of fusion. The researchers also saw a similarly transient accumulation of the regulatory phospholipid PIP2. “That suggests that there is some reason for PIP2 recruitment at exocytic sites,” Harris explains.
Harris and colleagues are now investigating the localization and dynamics of other lipids, as well as the behavior of the actin cytoskeleton, the third class of organizational element known to regulate exocytosis. “The bigger question then is how these different elements interact and communicate to cause vesicle fusion and the release of MMP-9 from breast cancer cells,” Harris says.
References
- 1.Stephens D.C., et al. J. Gen. Physiol. 2019 doi: 10.1085/jgp.201812299. [DOI] [Google Scholar]
- 2.Larson B.T., et al. Mol. Biol. Cell. 2014 doi: 10.1091/mbc.e14-02-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Somasundaram A., and Taraska J.W. Mol. Biol. Cell. 2018 doi: 10.1091/mbc.E17-12-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong W., et al. . 2015. Discov. Med. 20:357–367. [PubMed] [Google Scholar]
- 5.Hendrix A., et al. J. Natl. Cancer Inst. 2010 doi: 10.1093/jnci/djq153. [DOI] [Google Scholar]


