Dear Editor,
In humans, the β-like globin genes are encoded from a single locus comprising five globin genes (ε-, Gγ-, Aγ-, δ-, and β-globin in sequence) and their expression is under critical developmental control (Fig. 1a).1 The γ-globin genes (Gγ- and Aγ-) are expressed during fetal life, and then replaced by adult β-globin after birth.1 Mutations in the β-globin gene cause β-hemoglobinopathies such as sickle cell disease (SCD) and β-thalassemia.1–3 The clinical severity of SCD and β-thalassemia can be mitigated by elevated fetal hemoglobin (HbF) levels, which have been found in individuals with the benign hereditary persistence of fetal hemoglobin (HPFH) syndrome.1,3 Thus, reactivating the expression of γ-globin genes is an attractive treatment strategy for β-hemoglobinopathies, and understanding the mechanisms of the γ- to β-globin switch is crucial for designing target interventions to reactivate γ-globin expression.
Fig. 1.
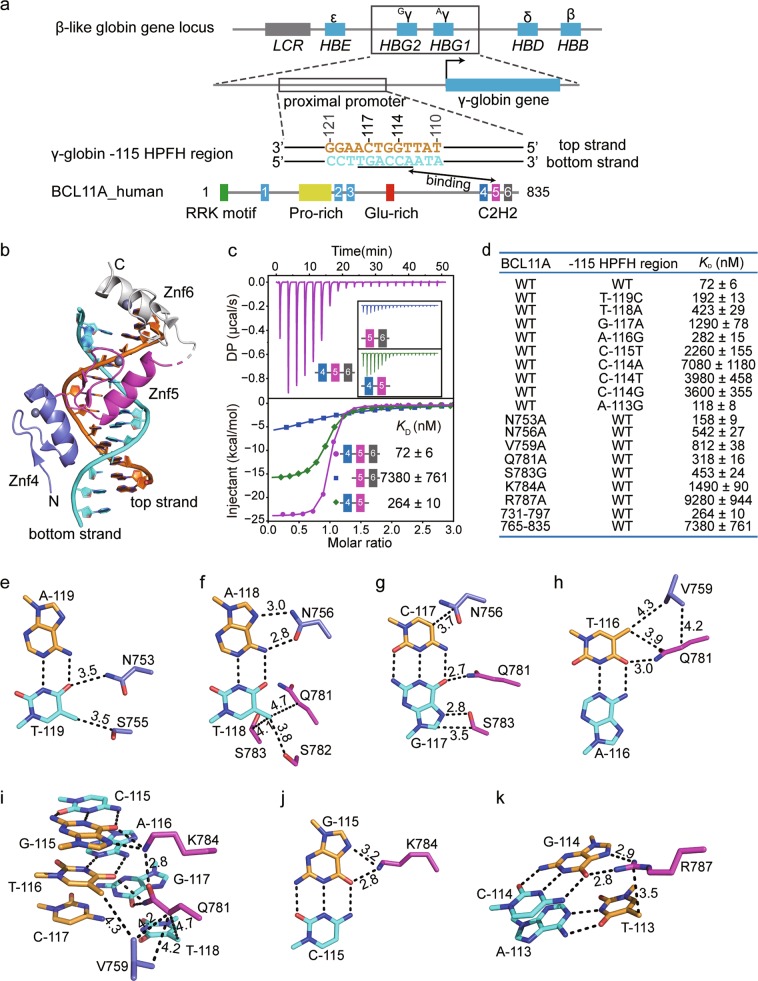
Interactions between Znf4-5 of BCL11A and the γ-globin −115 HPFH region sequence. a Top panel: schematic representation of the β-like globin gene locus, in which the γ-globin genes and their proximal promoters are magnified. Middle panel: schematic representation of the γ-globin −115 HPFH region sequence derived from the proximal promoter located ~115 bp upstream of the transcription start sites of γ-globin genes. The strand that contains the TGACCA (underlined) sequence is referred to as the bottom strand, and the complementary strand is referred to as the top strand. The top and bottom strands are colored orange and cyan, respectively. The two HPFH sites G:C−117 and C:G−114 are labeled by number above the base pairs. Bottom panel: schematic representation of the domain architecture of human BCL11A. The interaction between Znf4-6 of BCL11A and the γ-globin −115 HPFH region sequence is highlighted. b The overall structure of Znf4-6 of BCL11A in complex with the γ-globin −115 HPFH region sequence is shown in cartoon representation. Znf4, Znf5 and Znf6 are colored slate, magenta and gray, respectively. The color codes of the top and bottom strands are defined as described in a. c The raw ITC titration data of Znf4-5, Znf5-6, and Znf4-6 of BCL11A with the γ-globin −115 HPFH region sequence and their fitting curves are shown. KD, dissociation constant; DP, differential power. d Dissociation constants (KD) for the indicated interactions obtained by ITC assays. The indicated mutations in the γ-globin −115 HPFH region sequence (abbreviated as −115 HPFH region) are base-pair mutations. e-k Detailed base-specific interactions between Znf4-5 of BCL11A and the γ-globin −115 HPFH region sequence. The hydrogen-bonding and van der Waals interactions are depicted as black dashed lines with distances measured in Å. The intramolecular interactions among Val759, Gln781, and Lys784 are also shown in i
Rare genetic variations that lead to substantial elevations of HbF in HPFH are located ~115 bp (referred to as the γ-globin −115 HPFH region) (Fig. 1a) and ~200 bp upstream of the transcription start sites of the duplicated γ-globin genes.1,3 The variations located in the γ-globin −115 HPFH region include single base substitutions at −117 (G−117A) and −114 (C−114A/T/G) or a 13-bp deletion (Fig. 1a).4–8 Furthermore, genome-wide association studies (GWAS) of HPFH identified the transcription factor B-cell lymphoma/leukemia 11A (BCL11A) as the major HbF silencer (Fig. 1a);9,10 however, the mechanism by which BCL11A represses γ-globin expression has not been fully elucidated. Recently, two groups independently reported that BCL11A directly binds to the γ-globin −115 HPFH region through its C-terminal three tandem C2H2 zinc fingers (Znf4-6),11,12 establishing a direct link between BCL11A and the γ-globin gene promoters.
In this study, we investigated the molecular mechanism by which Znf4-6 of BCL11A recognizes the γ-globin −115 HPFH region. We crystallized Znf4-6 (residues 731–835) bound to a 12-bp double-stranded oligo (CCTTGACCAATA, from −121 to −110) (Fig. 1a; Supplementary information, Table S1),11,12 designated as the γ-globin −115 HPFH region sequence. To facilitate crystal packing, the oligo was synthesized with 5′-overhang. The protein-DNA complex structure was refined to 2.50 Å in P3221 group (Supplementary information, Table S2).
There are two BCL11A proteins (designated chains A and B), each binding to a DNA duplex (Supplementary information, Fig. S1), in an asymmetric unit. We did not observe the electron densities for Znf6 in chain B; by contrast, although the electron densities of Znf6 could be clearly traced in chain A, there are rare contacts between Znf6 and the DNA duplex (Fig. 1b). These observations suggest that BCL11A specifically recognizes the γ-globin −115 HPFH region sequence with its Znf4-5 but not Znf6. Next, we measured the binding affinities of Znf4-5 (residues 731–797), Znf5-6 (residues 765–835), and Znf4-6 to the 12-bp double-stranded oligo (Supplementary information, Table S1) using isothermal titration calorimetry (ITC). The ITC results illustrate that Znf4-6 binds to the γ-globin −115 HPFH region sequence with a KD value of ~72 nM. Deleting Znf6 (Znf4-5 construct) decreased the binding affinity by ~3.7-fold, whereas deleting Znf4 (Znf5-6 construct) decreased the affinity by > 100-fold (Fig. 1c, d; Supplementary information, Table S3). This further validates that Znf4-5 of BCL11A makes major contribution to the specific recognition of the γ-globin −115 HPFH region sequence. These in vitro observations are also consistent with the data obtained via an in vivo assay, in which deletion of Znf6 had little influence on repression of γ-globin expression compared to deletion of Znf4-6 and Znf5-6.11
Because the electron densities of chain A are clearer than those of chain B (Supplementary information, Fig. S1), we mainly performed structural analysis based on chain A and its corresponding DNA duplex. We refer to the strand that contains the TGACCA sequence as the bottom strand and the complementary strand as the top strand (Fig. 1a). Both Znf4 and Znf5 adopt a canonical C2H2 zinc finger fold, which consists of two β-strands and one C-terminal α-helix, coordinating one zinc ion (Fig. 1b; Supplementary information, Fig. S2).13 They wrap around the DNA duplex with their α-helices inserted into the major groove, contacting 7 bp (−119)(TTGACCA)(−113) from the γ-globin −115 HPFH region sequence in total. In particular, Znf4 makes contact with the 4-bp TTGA sequence, and Znf5 makes direct contact with all 6 bp of the TGACCA motif. The conventional C2H2 zinc finger DNA recognition mode has been established in which three recognition amino acids (at positions −7, −4, and −1 when the first zinc-coordinating histidine was numbered 0) in each finger mediate DNA base contacts primarily with three bases on one DNA strand, whereas the residue at position −5 may also have adaptive interaction with the bases in both DNA strands.14 In our structure, Znf4-5 makes plentiful hydrogen-bonding and van der Waals contacts with both DNA strands by employing the residues at these positions, contributing to the binding affinity and specificity.
Both in vivo and in vitro binding motifs of BCL11A were previously reported,11,12 illustrating that the TGACC motif is the rigid core sequence, whereas T−119 and A−113 are variable. We found that the O4 atom of T−119 has a weak hydrogen-bonding interaction with Asn753 (Fig. 1e). The N753A mutation of Znf4-6 mildly increased the KD to 158 nM (a 2.2-fold weaker binding) (Fig. 1d; Supplementary information, Fig. S3 and Table S3); the same effect was observed for the T−119C mutation (a 2.7-fold weaker binding) (Fig. 1d; Supplementary information, Fig. S4 and Table S3). These observations indicated that T:A−119 is not an essential base pair for BCL11A recognition.
T:A−118 is base-specifically recognized by Asn756, the amide group of Asn756 donates one hydrogen bond to the adenine N7 atom and accepts one hydrogen bond from the N6 atom of A−118 (Fig. 1f). This Asn-Ade recognition mode is often found in C2H2 zinc fingers.13 Accordingly, the N756A mutation reduced binding to the γ-globin −115 HPFH sequence by ~7.5-fold (Fig. 1d; Supplementary information, Fig. S3 and Table S3). Similarly, substitution of the T:A−118 base pair by A:T decreased the binding affinity by ~5.9-fold (Fig. 1d; Supplementary information, Fig. S4 and Table S3), further confirming the importance of these hydrogen-bonding interactions. In addition to the recognition of A−118, we also observed that the C5 methyl group of T−118 has van der Waals contacts with the Cβ group of Ser783, Cγ group of Gln781, and hydroxyl group of Ser782 (Fig. 1f). Thus, the T:A−118 position is specifically recognized.
The pathogenic G−117A mutant identified in HPFH syndrome4 suggests that recognition of the G:C−117 base pair should be essential for BCL11A binding to the γ-globin −115 HPFH sequence. The G:C−117 base pair is recognized by Gln781 and Ser783 (Fig. 1g). Specifically, the amide group of Gln781 and the hydroxyl oxygen of Ser783 form direct hydrogen bonds with the O6 and N7 atoms of G−117 (Fig. 1g), respectively. These hydrogen bonds make the G−117 recognition highly specific, and the G−117A mutation possibly abolishes the hydrogen-bonding interaction between G−117 and Gln781 (Supplementary information, Fig. S5a). Consistently, the binding affinity of the G−117A mutant of the γ-globin −115 HPFH region sequence for Znf4-6 was ~17.9-fold weaker than that of the wild type (WT) (Fig. 1d; Supplementary information, Fig. S4 and Table S3).
In addition to the interaction with G−117, Gln781 also recognizes T−116, with its amide group forming an additional hydrogen bond with the O4 atom of T−116 and a van der Waals contact with the C5 methyl group of T−116 (Fig. 1h). Thus, the side chain of Gln781 adopts a conformation to simultaneously contact three bases, T−118, G−117, and T−116 (Fig. 1f–i). Gln781 is sandwiched between Val759 and Lys784, which makes the conformation further stabilized via a network of intramolecular interactions (Fig. 1i). Val759 makes van der Waals contacts with the C5 methyl group of T−116 and the aliphatic side chain of Gln781 (Fig. 1h, i). Furthermore, the side chain of Gln781 forms another hydrogen bond with Lys784 (Fig. 1i), which further stabilizes the conformations of Gln781. Accordingly, when we replaced the A:T−116 base pair with G:C, both its van der Waals and hydrogen-bonding interactions were ablated, resulting in ~3.9-fold lower binding affinity for Znf4-6 (Fig. 1d; Supplementary information, Fig. S4 and Table S3). Moreover, Znf4-6 harboring the mutation of V759A, Q781A, S783G or Q781A together with S783G had reduced binding affinity for the γ-globin −115 HPFH region sequence by ~11.3-, ~4.4-, ~6.3- or ~15.4-fold, respectively (Fig. 1d; Supplementary information, Fig. S3 and Table S3), further validating the importance of these interactions.
The C:G−115 base pair is also highly specific and recognized by Lys784, which forms two hydrogen bonds with the O6 and N7 atoms of G−115 (Fig. 1i, j). Replacing this C:G−115 base pair with T:A weakened the interaction with Znf4-6 by ~31.4-fold (Fig. 1d; Supplementary information, Fig. S4 and Table S3). Accordingly, the K784A mutation of Znf4-6 reduced binding affinity by ~20.7-fold compared with that of WT Znf4-6 (Fig. 1d; Supplementary information, Fig. S3 and Table S3). These observations explain the specificity of position −115.
As mentioned earlier, single base substitutions found in HPFH individuals occurred in the G:C−117 and C:G−114 base pairs.4,6–8 Arg787 forms base-specific contacts with the C:G−114 base pair, with its terminal Nη1 and Nη2 groups donating two hydrogen bonds to the O6 and N7 atoms of G−114 (Fig. 1k). Substitution of G−114 with any other nucleotide abolishes the hydrogen bonds or introduces steric clashes with Arg787 (Supplementary information, Fig. S5b). This is also consistent with the fact that HPFH patients and patients carrying the C−114T, C−114A, or C−114G mutation have elevated HbF levels.6–8 Indeed, replacement of the C:G−114 base pair with T:A, A:T, or G:C substantially weakened the interaction with Znf4-6 (Fig. 1d; Supplementary information, Fig. S4 and Table S3). Znf4-6 carrying the R787A mutation exhibited severely reduced affinity for the γ-globin −115 HPFH region sequence (Fig. 1d; Supplementary information, Fig. S3 and Table S3), further confirming the importance of Arg787 for recognizing G−114. Thus, the Arg-Gua recognition pattern confers high specificity to the C:G−114.
BCL11A displays a slight preference for the T:A base pair at the −113 position as demonstrated by previous in vivo an in vitro results.11,12 The preference comes from a van der Waals contact between C5 methyl group of T−113 and the guanidino group of Arg787 (Fig. 1k). Consistently, the A−113G mutation resulted in a 1.6-fold reduction of the binding affinity (Fig. 1d; Supplementary information, Fig. S4 and Table S3). Notably, such methyl-Arg-Gua triad recognition was recently identified as a common mechanism of interaction with 5mCpG and TpG dinucleotides.15
In addition to the aforementioned base-specific interactions, Znf4-5 of BCL11A makes extensive hydrogen-bonding contacts with the phosphate backbone, contributing to the binding affinity of BCL11A for the γ-globin −115 HPFH region sequence. Specifically, Lys749, His760, Tyr777, and His788 interact with the four phosphates 5′ to C−117, T−116, G−114, and T−113 in the top strand (Supplementary information, Fig. S6a, b), respectively. By contrast, Ser755 interacts with the phosphate 5′ to C−120 in the bottom strand (Supplementary information, Fig. S6c).
In summary, our crystal structure provides the molecular basis for recognition of the γ-globin −115 HPFH region sequence by BCL11A. We demonstrate that Znf4-5 is responsible for this specific recognition, interacting with its target sequence in both DNA strands via a network of hydrogen bonding and van der Waals contacts (Fig. 1e–k). HPFH mutations, including G−117A and C−114G/T/A,4,6–8 can induce high production of HbF, which is highly consistent with our observations of solid contacts of Znf4-5 with G−117 and C−114 (Fig. 1g, k). Furthermore, our crystal structure can provide valuable insights for improving gene therapy strategies by either deleting Znf4-5 of BCL11A or the TTGACCA motif and for the development of drugs to disrupt the interaction between BCL11A and the γ-globin gene promoter.
The atomic coordinates and structure factors were deposited in the Protein Data Bank (PDB) under accession number 6KI6.
Supplementary information
Acknowledgements
We thank the staff of the BL18U1 beamline at the National Center for Protein Science Shanghai and Shanghai Synchrotron Radiation Facility, Shanghai, China for assistance during data collection. This work was supported by the Ministry of Science and Technology of China (2016YFA0500700), the National Natural Science Foundation of China (31870760), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB1001).
Author contributions
Y.Y., Y.S. and F.L. conceived and designed the experiments. Y.S. and F.L. supervised the project. Y.Y. performed the crystal screen and ITC experiments. Y.Y. and Z.X. performed protein expression and purification. F.L. determined the crystal structure. B.Z. and C.H. provided valuable advice for manuscript revision. All authors contributed to the data analysis. Y.Y., Y.S. and F.L. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Contributor Information
Yunyu Shi, Email: yyshi@ustc.edu.cn.
Fudong Li, Email: lifudong@ustc.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0221-0.
References
- 1.Wienert B, Martyn GE, Funnell APW, Quinlan KGR, Crossley M. Trends Genet. 2018;34:927–940. doi: 10.1016/j.tig.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Williams TN, Thein SL. Annu. Rev. Genom. Hum. Genet. 2018;19:113–147. doi: 10.1146/annurev-genom-083117-021320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orkin SH, Bauer DE. Annu. Rev. Med. 2019;70:257–271. doi: 10.1146/annurev-med-041817-125507. [DOI] [PubMed] [Google Scholar]
- 4.Collins FS, et al. Nature. 1985;313:325–326. doi: 10.1038/313325a0. [DOI] [PubMed] [Google Scholar]
- 5.Gilman JG, et al. Nucleic Acids Res. 1988;16:10635–10642. doi: 10.1093/nar/16.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zertal-Zidani S, et al. Hemoglobin. 1999;23:159–169. doi: 10.3109/03630269908996160. [DOI] [PubMed] [Google Scholar]
- 7.Motum PI, Deng ZM, Huong L, Trent RJ. Br. J. Haematol. 1994;86:219–221. doi: 10.1111/j.1365-2141.1994.tb03284.x. [DOI] [PubMed] [Google Scholar]
- 8.Fucharoen S, Shimizu K, Fukumaki Y. Nucleic Acids Res. 1990;18:5245–5253. doi: 10.1093/nar/18.17.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menzel S, et al. Nat. Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 10.Uda M, et al. Proc. Natl Acad. Sci. USA. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N, et al. Cell. 2018;173:430–442. doi: 10.1016/j.cell.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martyn GE, et al. Nat. Genet. 2018;50:498–503. doi: 10.1038/s41588-018-0085-0. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe SA, Nekludova L, Pabo CO. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Nucleic Acids Res. 2014;42:4859–4867. doi: 10.1093/nar/gku134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. Genes Dev. 2012;26:2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.