Abstract
The assembly of functional neural circuits in vertebrate organisms requires complex mechanisms of self-recognition and self-avoidance. Neurites (axons and dendrites) from the same neuron recognize and avoid self, but engage in synaptic interactions with other neurons. Vertebrate neural self-avoidance requires the expression of distinct repertoires of clustered Protocadherin (Pcdh) cell-surface protein isoforms, which act as cell-surface molecular barcodes that mediate highly specific homophilic self-recognition, followed by repulsion. The generation of sufficiently diverse cell-surface barcodes is achieved by the stochastic and combinatorial activation of a subset of clustered Pcdh promoters in individual neurons. This remarkable mechanism leads to the generation of enormous molecular diversity at the cell surface. Here we review recent studies showing that stochastic expression of individual Pcdhα isoforms is accomplished through an extraordinary mechanism involving the activation of “anti-sense” promoter within Pcdhα “variable” exons, antisense transcription of a long non-coding RNA through the upstream “sense strand” promoter, demethylation of this promoter, binding of the CTCF/cohesin complex and DNA looping to a distant enhancer through a mechanism of chromatin “extrusion”.
Introduction
The human brain is estimated to be comprised of over 80 billion neurons, each of which may have as many as 1000 neurites (dendrites and axons), which assemble into complex neural circuits required for accurate transmission of signals and effective processing of sensory motor, and cognitive information. This process is dependent, in part, upon the ability of neurites to project into distinct regions of the nervous system during development to form specific and highly complex neural networks (Cameron and Rao, 2010; Grueber and Sagasti, 2010). Most importantly, this process requires that as many as 1000 neurites of individual neurons must remain separated from each other (self-avoidance) in order to maximize the formation of functional synaptic connections. Neural self-avoidance in both vertebrates and invertebrates has been shown to require the expression of unique combinations of cell surface homophilic recognition molecules to generate a molecular recognition code, i.e. a single cell surface identity (Zipursky and Grueber, 2013; Zipursky and Sanes, 2010; Mountoufaris et al., 2018).
An important and initially counter-intuitive observation that shed light on the molecular mechanism of neural self-recognition was the observation that the homophilic engagement between certain cell-surface proteins displayed on opposing membranes of neurites (self-avoidance), results in repulsion, rather than adhesion (Zipursky and Grueber, 2013). In an extraordinary example of convergent evolution, the same cell-surface mechanism involving specific homophilic interactions followed by repulsion is used in both Drosophila and vertebrates. However, in Drosophila, the diversity code required for self-avoidance is mediated by an extraordinary example of stochastic alternative splicing of the Down syndrome cell adhesion molecule, Dscam1, pre-mRNA, a process that can generate up to 18,000 distinct extracellular protein isoforms (Zipursky and Grueber, 2013). By contrast, single cell-surface diversity in mammals is generated by stochastic promoter choice in the Protocadherin gene cluster (see Mountoufaris et al., 2018 for recent review). Each of the two alleles of the Pcdh gene cluster generates distinct sets of Pcdh isoforms, which are displayed on the surface of neurons (Mountoufaris et al., 2018). Thus, in both flies and vertebrates, self-avoidance is provided by stochastic expression of multiple protein isoforms. However, this is accomplished by distinct mechanisms: stochastic alternative splicing of Dscam1 pre-mRNA in flies, and stochastic promoter choice of Pcdh genes in vertebrates (Zipursky and Sanes, 2010). Here we focus on recent progress in understanding the detailed molecular mechanisms involved in the generation of Pcdh cell surface diversity in individual mammalian neurons.
The genomic organization of the Pcdh gene cluster
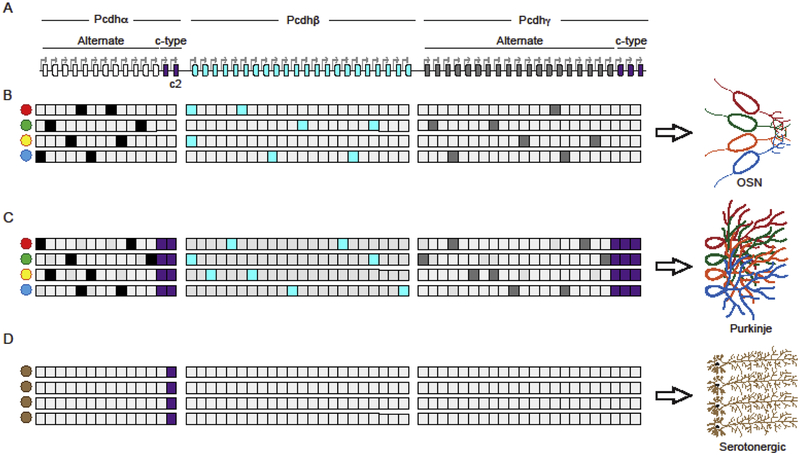
The generation of a Pcdh cell surface recognition code is a consequence of the genomic organization of the Pcdh gene cluster, and a remarkable mechanism of stochastic activation of transcription (promoter choice). Pcdh genes are organized into three closely linked gene clusters (designated as Pcdh α, and β, γ), which together, span nearly 1 million base pairs (bp) of genomic DNA (Wu and Maniatis, 1999) (Figure 1A). These genes (50 in humans and 60 in mouse) are organized into variable and constant exons, and their organization is reminiscent of that of immunoglobin and T-cell receptor gene clusters (Wu and Maniatis, 1999). The variable regions in the Pcdh α and γ clusters are further distinguished as alternate and c-type exons. Each Pcdh alternate exon contains a nearly identical promoter sequence driving its transcription (Tasic et al., 2002; Wang et al., 2002) (Figure 1A). These promoters, together with the promoters from the β cluster, are randomly activated in individual neurons to generate individual cell-specific patterns of Pcdh gene expression (Esumi et al., 2005; Hirano et al., 2012; Kaneko et al., 2006; Mountoufaris et al., 2017, 2018) (Figure 1B and 1C). By contrast, the c-type exon promoters differ in sequence from the alternate promoters and are independently regulated (Figure 1) (Chen et al., 2017; Esumi et al., 2005; Kaneko et al., 2006; Katori et al., 2017, Mountoufaris, et al 2018). In addition to promoter choice, the generation of full-length Pcdh α and γ messenger RNA (mRNA) requires RNA splicing of each variable exon RNA to three constant exons, located as much as 300,000 thousand nucleotides downstream of the startpoint of transcription (Tasic et al., 2002; Wang et al., 2002). In contrast, Pcdhβ mRNA consists of only the variable exon, as the Pcdhβ cluster does not encode a constant region (Wu and Maniatis, 1999). Thus, full length Pcdh α and γ protein contains six extracellular domains, a transmembrane domain-both encoded by the variable exon, and an intracellular domain encoded by the constant exons, while β Pcdhs do not have an intracellular domain. Like the Dscam proteins in flies, the extracellular domain of Pcdh proteins engage in highly specific homophilic interactions, while the intracellular domain is required for repulsion (Matthews et al., 2007; Mountoufaris et al., 2017), the mechanism of which is not understood. Finally, functional diversity of the clustered Pcdhs is generated by a nearly random assembly of Pcdh α, β, and γ cis-dimers in individual neurons, the formation of a cell surface lattice consisting of specific cis/trans homophilic tetramers that join opposing plasma membranes, and a strict specificity requirement whereby a single mismatch can prevent functional lattice formation (Brasch et al., 2019; Rubinstein et al., 2017, Mountoufaris, et al 2018).
Figure 1: Neuron-type specific expression of cluster Pcdh genes (stochastic vs determinative).
(A) Organization of the murine Pcdh gene clusters. White: Pcdhα alternate genes; Aqua: Pcdhβ genes; Grey: Pcdhγ alternate genes; Purple: c-type genes from the Pcdh α and γ clusters. Arrows indicate all the promoters of the Pcdh genes. (B) Example of a stochastic and combinatorial expression of Pcdh genes from all three clusters in olfactory sensory neurons (OSNs). (C) Example of a stochastic and combinatorial expression of Pcdh alternate genes from all three clusters in Purkinje neurons. In these neurons, the c-type are biallelically and constitutively expressed in every cell. (D) Example of a determinative expression of the sole cluster Pcdhαc2 in serotonergic neurons.
The expression of Pcdh genes shown in (B-C) is meant to summarize expression from both chromosomes, as the two chromosomes behave independently with regards to Pcdh promoter choice.
A neuron-type specific Pcdh cell-surface identity code
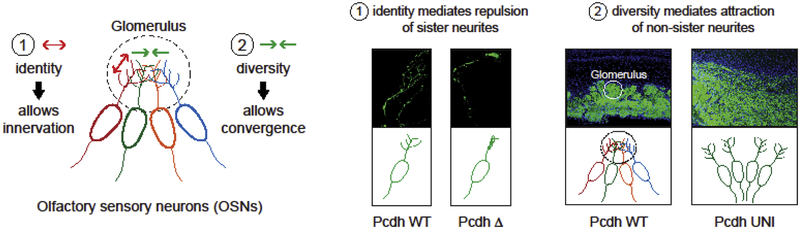
Our understanding of the mechanisms by which a Pcdh cell-surface identity code is generated is based in large measure on recent studies performed in Olfactory Sensory Neurons (OSNs) in mice. Mature OSNs express distinct repertoires of Pcdh isoforms from all three clusters as revealed in single cell RNA sequencing studies (RNAseq) (Mountoufaris et al., 2017; 2018) (Figure 1B). Deletion of all three gene clusters results in a self-avoidance phenotype, whereby terminal axons from a single OSN are unable to recognize self and therefore clump and cross each other (Mountoufaris et al., 2017) (Figure 2, Pcdh WT vs Pcdh Δ). On the other hand, forced overexpression of a specific subset of Pcdh α, β and γ isoforms in every OSN (Pcdh UNI), or in OSN’s expressing the same olfactory receptor, results in repulsion between OSN axons (Figure 2, Pcdh WT vs Pcdh UNI) (Mountoufaris et al., 2017). As a result, OSNs fail to converge to form normal glomeruli in the olfactory bulb, and these mice display defects in odor discrimination (Mountoufaris et al., 2017) (see Moutoufaris et al, 2017 and 2018 for additional details).
Figure 2: Functional role of a Pcdh diversity code in OSNs.
Left: Schematics of how a Pcdh cell-surface diversity code provides: (1) self-recognition of sister neurites (axons and dendrites) to allow them to innervate their territory and (2) attraction of non-sister neurites (axons and dendrites) to allow different neurons to converge to form a glomerulus. Right: Schematics of the phenotypes observed by deletion of the Pcdh gene tri-cluster (Pcdh Δ) and overexpression of a specific subset of Pcdh α, β and γ isoforms in every OSN (Pcdh UNI). The data presented here are from Mountoufaris et al. Science 2017.
Single-cell RT-PCR studies performed in cerebellar Purkinje neurons revealed that the Pcdh α and γ c-type mRNAs are expressed constitutively in every neuron, independently from both allelic chromosomes (Esumi et al., 2005; Kaneko et al., 2006). Importantly, Pcdhγ-deficient mice result in a dendritic self-avoidance phenotype in these cells (Figure 1C) (Lefebvre et al., 2012). Thus, in Purkinje cells, and in OSNs, the Pcdh α and γ alternate promoters, as well as Pcdh β promoters, are randomly chosen (Esumi et al., 2005; Hirano et al., 2012; Kaneko et al., 2006) (Figure 1C). However, in contrast to OSNs, the promoters of the c-type exons are constitutively active in Purkinje cells (Figure 1C) (Esumi et al., 2005; Hirano et al., 2012; Kaneko et al., 2006).
By contrast, recent studies of serotonergic neurons revealed the existence of an additional type of neural specific-transcriptional program generated from the Pcdh gene cluster whereby the sole Pcdhα exon expressed is Pcdhαc2 (Figure 1C). These studies provide evidence that the expression of a single Pcdh isoform in individual neurons of the same type is used by neurons to avoid each other as they project into receptive fields of the brain, a process known as tiling (Chen et al., 2017; Mountoufaris et al., 2018)
These observations, taken together, support the existence of a Pcdh neuron type-specific Pcdh self-recognition code based upon expression of alternate and c-type Pcdh genes in the brain. Understanding how the Pcdh gene cluster is expressed in different neuronal types is therefore critical to understanding how this remarkable gene cluster is required for the assembly of neural circuits. Here we review our current understanding of how Pcdhα alternate and c-type exons are differentially regulated.
Distinct mechanisms of clustered Pcdh gene expression
Enhancer-dependent stochastic expression of Pcdhα isoforms
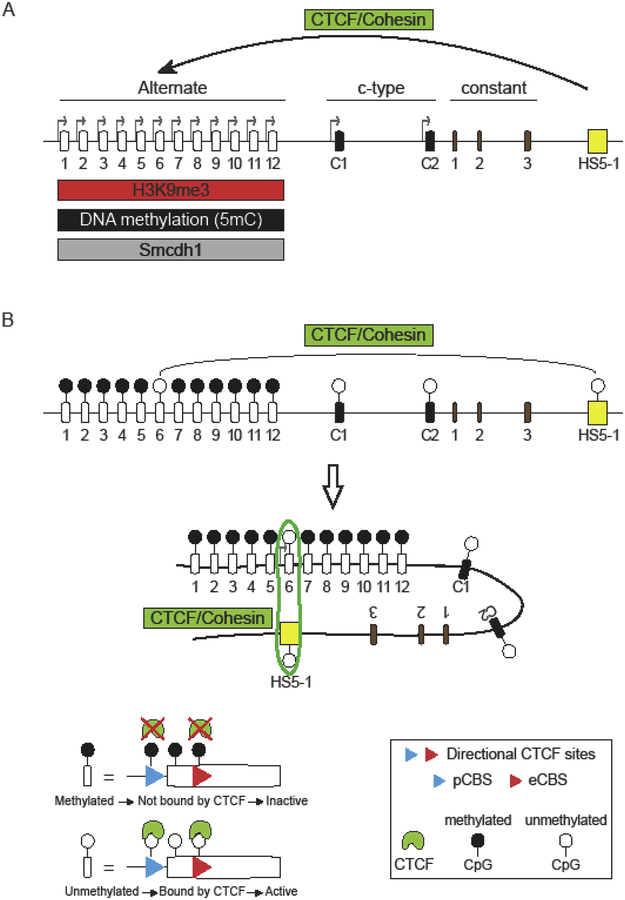
The expression of Pcdhα alternate exons requires long-range DNA looping between individual Pcdhα alternate promoters and a transcriptional enhancer, called HS5–1 (hypersensitivity site 5–1) which is located downstream of the Pcdhα variable and c-type exons (Kehayova et al., 2011; Ribich et al., 2006) (Figure 3A). Importantly, deletion of the HS5–1 enhancer sequence results in a decrease in expression of Pcdhα alternate exons but not Pcdhαc2 (Kehayova et al., 2011; Ribich et al., 2006). Thus, the expression of Pcdhαc2 does not require the HS5–1 enhancer (Figure 3A). Long-range DNA contacts between the HS5–1 enhancer and Pcdhα alternate promoters was shown to be mediated by the CCCTC-binding protein (CTCF) (Guo et al., 2012; Hirayama et al., 2012; Monahan et al., 2012) (Figure 3A). CTCF binds to two sites within and downstream of the alternate exon promoters: one in the promoter (pCBS) and the other in the protein coding sequence of the first exon (eCBS) (Guo et al., 2012; Monahan et al., 2012) (Figure 3B). Similarly spaced CBS sites are also located in the HS5–1 enhancer (L-CBS and R-CBS) and they are also binding sites for CTCF (Guo et al., 2012; Monahan et al., 2012) (Figure 4). A critical requirement for the formation of a functional enhancer/promoter complex is the opposite relative orientations of the CBSs in the HS5–1 enhancer and the Pcdhα promoters (Guo et al., 2015). Remarkably, inversion of the HS5–1 enhancer, which results in enhancer CBSs in the same orientation as those in the Pcdhα promoters, results in a significant decrease in Pcdhα gene cluster expression (Guo et al., 2015). Notably, the Pcdhαc2 exon does not contain a CBS in its promoter nor in its exon. Thus, the absence of a CBSs renders this gene independent of CTCF and the HS5–1 enhancer, as indicated above (Figure 3A).
Figure 3: Differential regulation of cluster Pcdhα alternate and c-type generates distinct transcriptional programs.
(A) Differential regulation of expression of alternate and c-type promoters in the Pcdhα gene cluster. Pcdhα alternate genes are expressed by long-range DNA contacts between their promoters and the HS5–1 enhancer. These contacts are mediated by the CTCF and Cohesin protein complexes. The choice of Pcdhα alternate promoters is also regulated by histone and DNA methylation (H3K9me3 and 5mC, respectively) and the activity of the Smcdh1 protein. (B) Relation between DNA methylation and promoter activation in the Pcdhα cluster: DNA demethylation of CTCF binding sites and, the DNA sequence between them, correlates with CTCF occupancy of its CBS sites, engagement with the HS5–1 enhancer and transcriptional activation of the unmethylated promoter.
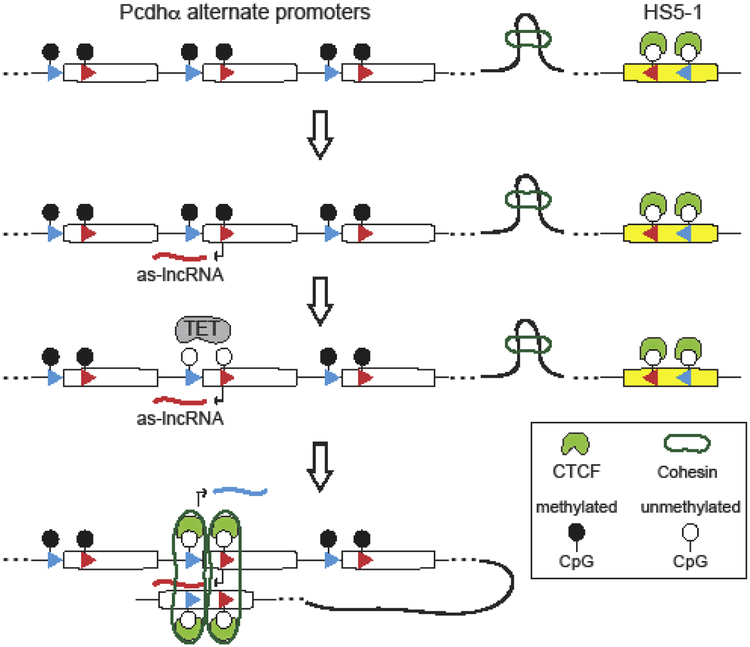
Figure 4: Model of stochastic Pcdhα alternate promoter choice.
Coupling antisense lncRNA transcription to DNA demethylation mediates stochastic promoter choice of clustered Pcdhα alternate promoters by CTCF and Cohesin through DNA loop extrusion. This mechanism ensures that the choice of a Pcdhα alternate promoter occurs independently from the distance between the stochastically chosen promoter and the HS5–1 enhancer.
Antisense lncRNA transcription and DNA demethylation drives stochastic alternate promoter choice
Studies performed in a variety of mouse and human neuroblastoma cell lines that stably express distinct combinations of Pcdhα alternate genes revealed a strong correlation between DNA methylation of the 5 methyl cytosine (5mC) of the CBS sites and, of the DNA sequences located between the two CBS sites, and transcriptional silencing (Guo et al., 2012; Toyoda et al., 2014) (Figure 3A and 3B). DNA methylation of the CBS sites prevents CTCF/Cohesin binding (Guo et al., 2012) and thus HS5–1 enhancer engagement and consequently transcription (Guo et al., 2012) (Figure 3B). By contrast, all of the transcriptionally active alternate exon promoters, are hypomethylated, bound by CTCF and Cohesin proteins, and engaged in DNA looping to the HS5–1 enhancer (Guo et al., 2012) (Figure 2B). Given this correlation, it became apparent that DNA methylation of the CBSs was likely to play a fundamental role in the mechanism of stochastic promoter choice of Pcdhα alternate exons. Moreover, a distinct mechanism is required for the activation of the Pcdhαc2 promoter (Figure 3A). DNA methylation of Pcdh alternate promoters is mediated by the de novo DNA methyltransferase Dnmt3b, which is expressed during early embryogenesis prior to E10 at high levels in neuronal tissues (Toyoda et al., 2014). Deletion of the Dnmt3b in mice results in an increase expression of Pcdh isoforms in individual Purkinje and cortical neurons (Toyoda et al., 2014).
These studies, taken together, clearly suggest a relationship between the timing of DNA methylation and Pcdh promoter activation during neural differentiation. However, several questions remained to be addressed regarding the mechanism of stochastic promoter choice. For example: are all alternate promoters initially unmethylated and bound by CTCF/Cohesin, and does DNA methylation of the inactive promoters occur subsequent to stochastic promoter choice? Alternatively, is the ground state of the DNA of all the alternate promoters methylated, and stochastic promoter choice requires demethylation? Insights into these questions were provided by a “DNA loop extrusion” model for how CTCF and Cohesin scan the genome to mediate enhancer/promoter interactions genome-wide (Fudenberg et al., 2016). According to this model, long-range DNA looping in the mammalian genome occurs as a consequence of stalling Cohesin complexes at DNA sites bound to CTCF, which acts as an extrusion barrier (Fudenberg et al., 2016). That is, Cohesin moves along the chromosome and extrudes DNA through its protein ring structure until it reaches CTCF bound promoters and enhancer sequences, thus resulting in the formation of an enhancer/promoter complex (Fudenberg et al., 2016). In light of this model and given that Pcdhα alternate promoters are organized in tandem, the question was: how can random enhancer/promoter complexes form between the HS5–1 and alternate exon promoters in the Pcdhα cluster in an HS5–1 distance-independent manner? In other words, if all Pcdhα promoters are bound by CTCF, how can they be equally likely to engage the HS5–1 enhancer via the CTSF/Cohesin complex, considering that they are located at a distance varying from about 220,000 to 320,000 nucleotides from the HS5–1 enhancer? In fact, the DNA loop extrusion model would predict that that if the Pcdhα12 promoter is always bound by CTCF, it would be chosen most of the time for enhancer engagement by virtue of its closest proximity to the HS5–1 enhancer.
These issues were addressed, at least in part, using a combination of cell-culture and in vivo studies of OSN differentiation (Canzio et al., 2019). These studies showed that the ground state of a Pcdhα alternate promoter DNA is methylated and transcriptionally repressed in olfactory sensory neuron precursor cells (Figure 4). Thus, DNA methylation of the promoter and the two CBS sites prevents CTCF binding anywhere in the Pcdhα gene cluster. Stochastic promoter activation occurs through a mechanism that involves stochastic transcription of an antisense long noncoding RNA (as-lncRNA) from an antisense promoter located within the protein coding region of each alternate Pcdhα exon located by the exonic CBS site (Canzio et al., 2019) (Figure 4). Stochastic transcriptional activation of this antisense promoter and transcription through the upstream sense strand promoter generates a large multiply-spliced, polyadenylated long non-coding RNA (lncRNA) (Canzio et al., 2019) (Figure 4). This transcriptional read-though leads to the demethylation, de-repression and activation of Pcdhα proximal sense strand promoters and the two CBS sites (Canzio et al., 2019) (Figure 3). DNA demethylation of the CBS sites allows CTCF binding and coordinate CTCF/Cohesin-dependent long-range DNA looping between the demethylated, CTCF bound promoter, and the HS5–1 enhancer (Canzio et al., 2019) (Figure 4). This model is consistent with the DNA loop extrusion model. Thus, coupling antisense transcription to DNA demethylation provides an elegant mechanism to ensure random alternate promoter selection and prevents proximity-bias in Pcdhα alternate promoter choice (Figure 4). While additional experiments are required to dissect the mechanism of coupling antisense lncRNA transcription to Tet-dependent DNA demethylation of Pcdh promoters, we note here a mechanism described in a recent study on transcriptional activation of the tumor suppressor gene, TCF21. As with the Pcdh alternate promoters, TCF21 promoter activation requires transcription of an antisense lncRNA, TARID, whose transcription is initiated at an intronic promoter sequence located within the TCF21 gene (Arab et al., 2014). Transcription of TARID leads to the formation of promoter-associated DNA-RNA hybrid. These “R-loop” structures are recognized by the growth arrest and DNA damage protein 45A, GADD45A, which recruits TET1 to drive TET-mediated DNA demethylation and activation of the TCF21 sense strand promoter (Arab et al., 2019). It is therefore reasonable to propose that a similar mechanism is involved in the antisense lncRNA-dependent transcriptional activation of the Pcdhα alternate exon promoters.
Taken together, these observations suggest that promoter activation mediated by antisense RNA transcription is another major difference between alternate Pcdhα promoters and the promoter of Pcdhαc2, thus providing an example of the molecular mechanisms that underly differential transcriptional activation of alternate and c-type promoters in the Pcdhα gene cluster.
Additional chromatin-based mechanisms of alternate regulation of Pcdh promoters
In addition to DNA methylation, histone H3 Lysine 9 trimethylation (H3K9me3) plays a fundamental role in regulating the expression of cluster Pcdhα genes (Jiang et al., 2017) (Figure 3A). Loss of H3K9me3 by conditional knockout studies of the SET domain bifurcated 1 (Setdb1) results in DNA hypomethylation, increase CTCF binding and increase expression of Pcdh genes (Jiang et al., 2017). Based on these data and the data described above from studies in OSNs, DNA methylation and histone H3 Lysine 9 trimethylation (H3K9me3) cooperate to transcriptionally repress the Pcdhα gene cluster (Figure 3A). Finally, an additional regulator of Pcdhα alternate exons is the structural maintenance of chromosome hinge domain containing 1 protein (Smchd1) (Chen et al., 2015) (Figure 3A). Smchd1 associates with H3K9me3 and antagonizes CTCF binding at alternate Pcdhα genes and the HS5–1 enhancer (Chen et al., 2015). Loss of Smchd1 results in increase expression of all Pcdhα alternate genes but not of Pcdhαc2 (Chen et al., 2015). The mechanisms by which Smchd1 antagonizes CTCF to regulate Pcdh alternate promoter choice remain to be addressed.
Concluding remarks
Studies of the Pcdh gene cluster has provided deep insights into general principles of eukaryotic gene expression mechanisms, from genomic organization, chromatin regulation, enhancer/promoter choice, and transcription. However, many mechanistic questions remain to be answered. For example, how is the expression of Pcdhαc2 selectively activated in serotonergic neurons, while the alternate exons promoters silenced? Upon stochastic activation of an alternate exon promoter by antisense lncRNA transcription, what is the mechanism that prevents other promoters from being turned on? Perhaps most puzzling is that of the activation of the promoters in the Pcdh β and γ gene clusters, as those promoters are also stochastically chosen, but apparently require a distinct mechanism from the one described for the Pcdhα cluster, as these gene clusters do not have exonal antisense promoters, and do not express antisense lncRNA. To address these and other fundamental unanswered questions, we must probe deeply into cell-type specific patterns of Pcdh gene expression throughout the nervous system and couple these studies to state-of-the-art single-cell genomic and microscopy technologies.
Highlights.
In vertebrates, neural self-avoidance requires the clustered Pcdh proteins
Self-avoidance requires stochastic and combinatorial expression of multiple Pcdh isoforms
Stochastic Pcdhα promoter choice is mediated by DNA loop-extrusion by CTCF/Cohesin
Acknowledgments
This work was supported by the NIH grants 4R00GM121815 (D.C.), and 1R01MH108579 and 5R01NS088476 (T.M).
Footnotes
Conflict of interest
The authors declare no competing financial interests.
References
(*) Special interest
(**) Outstanding interest
- Arab K, Karaulanov E, Musheev M, Trnka P, Schaefer A, Grummt I, and Niehrs C (2019). GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M, et al. (2014). Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell 55, 604–614. [DOI] [PubMed] [Google Scholar]
- (*).Brasch J, Goodman KM, Noble AJ, Rapp M, Mannepalli S, Bahna F, Dandey VP, Bepler T, Berger B, Maniatis T, et al. (2019). Visualization of clustered protocadherin neuronal self-recognition complexes. Nature 36, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron S, and Rao Y (2010). Molecular mechanisms of tiling and self-avoidance in neural development. Mol Brain 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**).Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, Duffié R, Monahan K, O’Keeffe S, Simon MD, et al. (2019). Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 177, 639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Hu J, Moore DL, Liu R, Kessans SA, Breslin K, Lucet IS, Keniry A, Leong HS, Parish CL, et al. (2015). Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. U.S.a. 112, 201504232–201504244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**).Chen WV, Nwakeze CL, Denny CA, O’Keeffe S, Rieger MA, Mountoufaris G, Kirner A, Dougherty JD, Hen R, Wu Q, et al. (2017). Pcdhαc2 is required for axonal tiling and assembly of serotonergic circuitries in mice. Science 356, 406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, and Yagi T (2005). Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet 37, 171–176. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, and Mirny LA (2016). Formation of Chromosomal Domains by Loop Extrusion. Cell Reports 15, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, and Sagasti A (2010). Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harbor Perspectives in Biology 2, a001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*).Guo Y, Maniatis T, Monahan K, Myers RM, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, et al. (2012). CTCF/cohesin-mediated DNA looping is required for protocadherin promoter choice. Proceedings of the National Academy of Sciences 109, 21081–21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. (2015). CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. 162, 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj, El N, Dittrich M, and Haaf T (2017). Epigenetic dysregulation of protocadherins in human disease. Semin. Cell Dev. Biol 69, 172–182. [DOI] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, and Zipursky SL (2008). Dscam-mediated cell recognition regulates neural circuit formation. Annu. Rev. Cell Dev. Biol 24, 597–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, and Yagi T (2012). Single-neuron diversity generated by Protocadherin-β cluster in mouse central and peripheral nervous systems. Front. Mol. Neurosci 5, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, and Yagi T (2012). CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Reports 2, 345–357. [DOI] [PubMed] [Google Scholar]
- (*).Jiang Y, Loh Y-HE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, et al. (2017). The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat Genet 159, 1665–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*).Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, and Yagi T (2006). Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. Journal of Biological Chemistry 281, 30551–30560. [DOI] [PubMed] [Google Scholar]
- (**).Katori S, Noguchi-Katori Y, Okayama A, Kawamura Y, Luo W, Sakimura K, Hirabayashi T, Iwasato T, and Yagi T (2017). Protocadherin-αC2 is required for diffuse projections of serotonergic axons. Scientific Reports 7, 15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehayova P, Monahan K, Chen W, and Maniatis T (2011). Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences 108, 17195–17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*).Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, and Sanes JR (2012). Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, and Grueber WB (2007). Dendrite self-avoidance is controlled by Dscam. Cell 129, 593–604. [DOI] [PubMed] [Google Scholar]
- Millard SS, Flanagan JJ, Pappu KS, Wu W, and Zipursky SL (2007). Dscam2 mediates axonal tiling in the Drosophila visual system. Nature 447, 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, and Maniatis T (2012). Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc. Natl. Acad. Sci. U.S.a. 109, 9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**).Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, and Maniatis T (2018). Writing, Reading, and Translating the Clustered Protocadherin Cell Surface Recognition Code for Neural Circuit Assembly. Annu. Rev. Cell Dev. Biol 34, 471–493. [DOI] [PubMed] [Google Scholar]
- (**).Mountoufaris G, Chen WV, Hirabayashi Y, O’Keeffe S, Chevee M, Nwakeze CL, Polleux F, and Maniatis T (2017). Multicluster Pcdh diversity is required for mouse olfactory neural circuit assembly. Science 356, 411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Tasic B, and Maniatis T (2006). Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proceedings of the National Academy of Sciences 103, 19719–19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (*).Rubinstein R, Goodman KM, Maniatis T, Shapiro L, and Honig B (2017). Structural origins of clustered protocadherin-mediated neuronal barcoding. Semin. Cell Dev. Biol 69, 140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**).Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, and Maniatis T (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33. [DOI] [PubMed] [Google Scholar]
- Toyoda S, Okano M, Tarusawa E, Kawaguchi M, Hirabayashi M, Kobayashi T, Toyama T, Oda M, Nakauchi H, Yoshimura Y, et al. (2014). Developmental epigenetic modification regulates stochastic expression of clustered protocadherin genes, generating single neuron diversity. Neuron 82, 94–108. [DOI] [PubMed] [Google Scholar]
- (**).Wang X, Su H, and Bradley A (2002). Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes & Development 16, 1890–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (**).Wu Q, and Maniatis T (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. 97, 779–790. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, and Grueber WB (2013). The molecular basis of self-avoidance. Annu. Rev. Neurosci 36, 547–568. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, and Sanes JR (2010). Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. 143, 343–353. [DOI] [PubMed] [Google Scholar]