Abstract
Background
There is solid evidence that obesity induces the acceleration of liver epigenetic aging. However, unlike easily accessible blood or subcutaneous adipose tissue, little is known about the impact of obesity on epigenetic aging of metabolically active visceral adipose tissue (VAT). Herein, we aimed to test whether obesity accelerates VAT epigenetic aging in subjects with severe obesity.
Results
A significant and positive correlation between chronological age and epigenetic age, estimated with a reduced version of the Horvath’s epigenetic clock, was found in both blood (r = 0.78, p = 9.4 × 10−12) and VAT (r = 0.80, p = 1.1 × 10−12). Epigenetic age acceleration, defined as the residual resulting from regressing epigenetic age on chronological age, was significantly correlated with body mass index (BMI) in VAT (r = 0.29, p = 0.037). Multivariate linear regression analysis showed that, after adjusting for chronological age, sex and metabolic syndrome status, BMI remained significantly associated with epigenetic age acceleration in VAT (beta = 0.15, p = 0.035), equivalent to 2.3 years for each 10 BMI units. Binomial logistic regression showed that BMI-adjusted epigenetic age acceleration in VAT was significantly associated with a higher loss of excess body weight following biliopancreatic diversion with duodenal switch surgery (odds ratio = 1.21; 95% CI = 1.04–1.48; p = 0.03).
Conclusions
Epigenetic age acceleration increases with BMI in VAT, but not in blood, as previously reported in liver. These results suggest that obesity is associated with epigenetic age acceleration of metabolically active tissues. Further studies that deepen the physiological relevance of VAT epigenetic aging will help to better understand the onset of metabolic syndrome and weight loss dynamics following bariatric surgery.
Keywords: Epigenetic aging, Obesity, Visceral adipose tissue, Weight loss, Metabolic syndrome, Bariatric surgery, Epigenetic clock
Background
Obesity is a condition with a complex and heterogenous metabolic phenotype [1, 2]. Apart from its main feature, an excessive body fat accumulation in body fat depots, a plethora of additional harmful metabolic disturbances may appear in patients affected by obesity [3]. Among these, increased fasting plasma triglyceride and glucose levels, reduced HDL-cholesterol and elevated blood pressure, together with increased waist circumference, represent a set criteria often used to identify patients with the metabolic syndrome [4]. The frequency and severity of these comorbidities varies widely among patients with obesity, raising questions about the onset of metabolically unhealthy phenotypes in certain patients [5]. Mounting evidence suggests that, in addition to body mass index (BMI), sex, age or body fat distribution, the prevalence of obesity comorbidities depends on the concurrence of multiple factors, among which genetics and epigenetics would be playing a prominent role [6, 7].
Focusing on epigenetics, we and others have shown that altered DNA methylation in obesity would be associated to increased prevalence of metabolic comorbidities [8–13]. As such, both global methylation differences observed in blood and tissue-specific methylation alterations have been found to be associated with healthy or unhealthy obesity phenotypes [14, 15]. Likewise, given the innate plasticity of DNA methylation at cytosine-phosphate-guanine (CpG) dinucleotides [16], its modulation appears to be mediated through numerous environmental and lifestyle factors, such as diet or metabolic stress, as well as by intrinsic individual features, mainly sex and age [17]. Under this multifactorial scenario, the concept of epigenetic aging emerges as a straightforward approach to illustrate how a complex environment such as obesity may impact the epigenetic signature of human tissues [18].
Methylation levels of a number of CpG sites positively correlate with chronological age [19, 20]. These and other findings led to the construction of various algorithms allowing to estimate epigenetic age, also known as DNA methylation age (DNAm age), a novel parameter able to accurately measure an individual’s age [21–23]. Epigenetic age acceleration, i.e. the deviation of epigenetic age from chronological age, given a specific metabolic condition then reflects the impact of such condition on epigenetic age. That is the case in obesity, where BMI has been found to be associated with epigenetic age acceleration [24]. Interestingly, the positive correlation between BMI and epigenetic age acceleration does occur only in liver, while no association is observed in blood or in other sites, such as muscle or subcutaneous adipose tissue [24].
Given the relevance of liver as a central regulator of metabolism under both physiological and pathological conditions, these results point out to a specific impact of obesity on the epigenetic aging of metabolically active tissues [25]. These findings gain significance when considering that the acceleration of epigenetic age in visceral adipose tissue (VAT), a key tissue in obesity development and progression, has never been analyzed, probably due to its inaccessibility, as compared to more accessible blood or subcutaneous adipose tissue. Under this perspective, epigenetic aging of target tissues may also have an impact on the onset of metabolic syndrome and participate in other major metabolic processes occurring in obesity, such as body weight loss. We then hypothesized that, mirroring liver, BMI is associated with epigenetic age acceleration of VAT, which may explain part of the heterogeneity of obesity phenotypes and/or play a role in the interindividual variability previously observed in weight loss dynamics following bariatric surgery [26].
Results
Phenotype data distribution
After patient exclusions, 24 men and 28 women matched for age, BMI and metabolic syndrome were available for epigenetic aging analysis. Phenotype data passed normality tests. On the one hand, BMI ranged from 40.1 to 81.2 kg/m2 and was significantly higher in men than in women (mean = 54.4, SD = 9.1 vs mean = 48.9, SD = 7.2 kg/m2; p = 0.02) (Table 1). The replication liver dataset showed a wider BMI range (17.4–70.2 kg/m2), with women having a BMI higher than men (mean = 43.7, SD = 12.3 vs mean = 35.9, SD = 14.4 kg/m2; p = 0.04) (Table 1). BMI range in the validation liver subset including only subjects with severe obesity was closer to ours (40.4–70.2 kg/m2) and no difference was observed between men and women (Table 1). On the other hand, chronological age ranged from 18.8 to 54.4 years old in the entire population and no difference was found between men and women (Table 1). By contrast, women in the liver replication dataset, which ranged from 23.0 to 83.0 years old, were significantly younger than men (mean = 45.2, SD = 10.3 vs mean = 55.6, SD = 17.3; p = 0.01). Such a difference faded out in the subset of individuals with obesity (Table 1). As expected, no significant differences in BMI or chronological age were found within each sex group between matched individuals with and without metabolic syndrome (Additional file 1: Table S1).
Table 1.
Data summary of the cohorts used in this study
| Variable | Study participants (n = 52) | Liver (n = 62) | Liver (obesity) (n = 40) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Men | Women | p | Men | Women | p | Men | Women | p | |||||||
| (n = 24) | (n = 28) | (n = 17) | (n = 45) | (n = 6) | (n = 34) | ||||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||||
| Chronological age | 35.0 | 10.6 | 32.9 | 5.7 | 0.36 | 55.6 | 17.3 | 45.2 | 10.3 | 0.01 | 51.2 | 10.0 | 43.6 | 8.9 | 0.07 |
| BMI | 54.4 | 9.1 | 48.9 | 7.2 | 0.02 | 35.9 | 14.4 | 43.7 | 12.3 | 0.04 | 54.1 | 4.7 | 49.5 | 6.8 | 0.12 |
Epigenetic age acceleration of VAT correlates with BMI
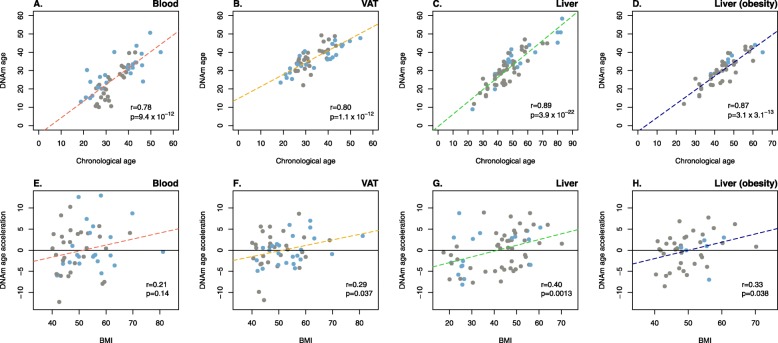
A strongly significant and positive correlation between chronological age and epigenetic age was found in both blood (r = 0.78, p = 9.4 × 10−12) (Fig. 1a) and VAT (r = 0.80, p = 1.1 × 10−12) (Fig. 1b). These results were similar to those previously obtained in liver [24], and successfully replicated herein with our reduced version of the epigenetic clock (r = 0.89, p = 3.9 × 10−22) (Fig. 1c), as well as to those in the liver of subjects with obesity (r = 0.87, p = 3.3 × 10−13) (Fig. 1d). Residuals resulting from regressing epigenetic age on chronological age were then used as a measurement of epigenetic age acceleration, whose association with BMI was tested. Epigenetic age acceleration in blood was not correlated with BMI (r = 0.21, p = 0.14) (Fig. 1e), as previously reported [24]. In contrast, a significant and positive correlation was found between epigenetic age acceleration and BMI in VAT (r = 0.29, p = 0.037) (Fig. 1f). A positive correlation was also observed in liver (r = 0.40, p = 0.0013) (Fig. 1g), where we were able to consistently reproduce previously reported experimental findings [24]. Results in the validation dataset showed that BMI correlated with epigenetic age acceleration in the liver of subjects with severe obesity to a similar extent to what we observed in VAT (r = 0.33, p = 0.038) (Fig. 1h). With the entire population of 52 individuals, a medium Pearson correlation coefficient of 0.38 is needed to attain a statistical power of 0.8. We also analyzed the impact of BMI on epigenetic age acceleration after excluding younger (10th percentile, ~ 24 years old) and older (90th percentile, ~ 44 years old) participants. The new results in the so-called middle-age dataset (n = 42, 14 men and 28 women) still showed a significant and even stronger correlation between BMI and epigenetic age acceleration in VAT (r = 0.34, p = 0.028), whereas results in blood revealed a non-significant and low correlation (r = 0.036, p = 0.82) (Additional file 1: Figure S1).
Fig. 1.
The acceleration of epigenetic aging in VAT correlates with BMI. First row of panels represents the correlation between chronological age and DNA methylation (DNAm) age, estimated with the reduced version of Horvath’s epigenetic clock [21], in blood (a, red dashed line), visceral adipose tissue—VAT—(b, yellow dashed line), liver (c, green dashed line), and in the liver of subjects with severe obesity (d, blue dashed line). Second row of panels from e to h show the correlation between residuals from regressing DNAm age on chronological age, and body mass index (BMI) in blood (e), VAT (f), liver (g), and in the liver of subjects with obesity (h). Residuals above zero (horizontal black line) stand for an acceleration of DNAm age. r and p stand for Pearson correlation coefficients and p values, respectively. Blue and gray dots refer to men and women, respectively
BMI is consistently associated with epigenetic age acceleration of VAT
Multivariate linear regression models, adjusted by chronological age, sex and metabolic syndrome, confirmed that epigenetic age acceleration was not dependent on BMI in blood (beta = 0.16; 95% CI = − 0.04–0.36; p = 0.12) (Table 2). Our results also showed that BMI remained significantly associated with epigenetic age acceleration in VAT (beta = 0.15; 95% CI = 0.04–0.28; p = 0.03) (Table 2), as well as in liver (beta = 0.16; 95% CI = 0.07–0.25; p = 5.6 × 10−4) and in the liver of subjects with obesity (beta = 0.24; 95% CI = 0.04–0.45; p = 0.02). With beta estimates representing the acceleration of epigenetic age in years by unit change of BMI and after correcting for chronological age, the observed epigenetic age acceleration in VAT was equivalent to 2.20 years for each 10 BMI units, similar to the additional 2.28 years observed in liver, and the 3.04 years in the liver of subjects with obesity (Table 2).
Table 2.
Association of BMI with epigenetic age acceleration in blood, VAT and liver
| Variable | Blood | Visceral adipose tissue | Liver | Liver (obesity) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | p | β | 95% CI | p | β | 95% CI | p | β | 95% CI | p | ||||
| Chronological age | 0.87 | 0.67–1.07 | 1.48 × 10−11 | 0.66 | 0.53–0.80 | 2.63 × 10−13 | 0.71 | 0.62–0.81 | < 2 × 10−16 | 0.80 | 0.65–0.95 | 7.56 × 10−13 | |||
| BMI | 0.16 | − 0.04–0.36 | 0.12 | 0.15 | 0.04–0.28 | 0.03 | 0.16 | 0.07–0.25 | 5.6 × 10−4 | 0.24 | 0.04–0.45 | 0.02 | |||
| Sex | − 3.11 | − 6.54–0.33 | 0.08 | 2.89 | 0.59–5.20 | 0.02 | − 1.47 | − 4.07–1.13 | 0.26 | − 0.28 | − 4.12–3.56 | 0.88 | |||
| Metabolic syndrome | 0.47 | − 2.75–3.69 | 0.77 | 1.42 | − 0.74–3.58 | 0.19 | - | - | - | - | - | - | |||
| Adjusted R2 | 0.64 | 0.67 | 0.82 | 0.78 | |||||||||||
| Age acceleration | 1.83 | 2.20 | 2.28 | 3.04 | |||||||||||
Estimates (β), 95% confidence intervals (95%CI), and p values (p) are from multivariate linear regression models of DNA methylation age acceleration adjusted by chronological age, body mass index (BMI), sex and metabolic syndrome. β represents the acceleration of epigenetic age in years by unit change of dependent variable. Age acceleration stands for the increase of epigenetic age in years for each 10-point increase in BMI
Epigenetic age acceleration of VAT correlates with BMI only in men
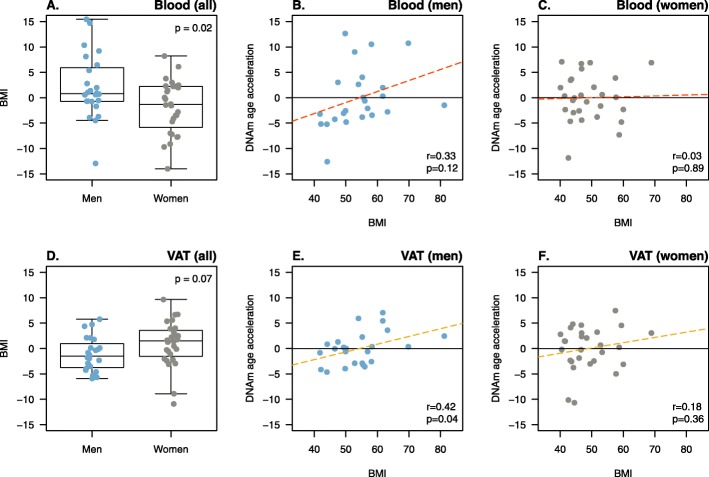
In view that results from multivariate regression models showed a significant association between sex and epigenetic age acceleration (Table 2), the latter was compared between men and women, and its correlation with BMI was analyzed separately. Significant sex differences were observed in blood, with men having a higher epigenetic age acceleration than women (2.1 vs − 1.8, p = 0.02) (Fig. 2a). However, BMI was not correlated with epigenetic age acceleration in blood, neither in men (r = 0.33, p = 0.12) (Fig. 2b) nor in women (r = 0.03, p = 0.89) (Fig. 2c). Although no difference was found in epigenetic age acceleration between men and women in VAT (− 1.11 vs 0.95, p = 0.07) (Fig. 2d), BMI was significantly and positively correlated with epigenetic age acceleration in men (r = 0.42, p = 0.04) (Fig. 2c), but not in women (r = 0.19, p = 0.35) (Fig. 2d). With a population of 24 men and 28 women, Pearson correlation coefficients larger than 0.5 are needed to attain a statistical power of 0.8. By contrast, a significant correlation between BMI and liver epigenetic age acceleration was observed only in women (r = 0.45, p = 0.002), but not in men (r = 0.38, p = 0.13). Similar results were found in the liver obesity group in both women (r = 0.42, p = 0.013) and men (r = − 0.05, p = 0.93) (Additional file 1: Figure S2).
Fig. 2.
The acceleration of epigenetic aging in VAT correlates with BMI in men. Panels a and d compare the distribution of DNA methylation (DNAm) age acceleration, defined as the residual from regressing DNAm age on chronological age, between men and women in blood (a) and visceral adipose tissue—VAT—(d). Boxplots represent the distribution of phenotype data with median (dark horizontal line) and interquartile range (box), and p stands for the p values obtained in Student’s t test for independent samples. The correlation between DNAm age acceleration and body mass index (BMI) in blood (b and c; red dashed line) and VAT (e and f, yellow dashed line) is shown separately in men and women. Residuals over zero (horizontal black line) stand for an acceleration of DNAm age, while r and p refer to Pearson correlation coefficients and p values, respectively. Blue and gray dots refer to men and women, respectively.
Epigenetic age acceleration of VAT is associated with postsurgery weight loss
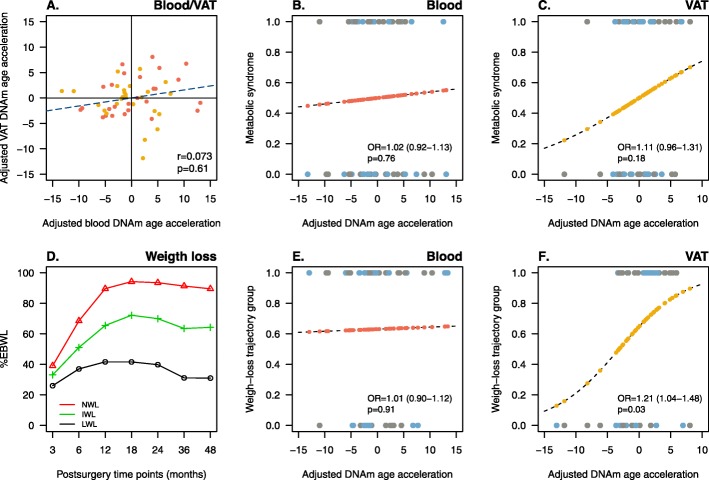
The measurement of epigenetic age acceleration adjusted by chronological age, sex and BMI in blood and VAT was not significantly correlated with each other (Fig. 3a). Binomial logistic regression was further used to test whether adjusted epigenetic age acceleration was associated with metabolic syndrome and/or with weight loss trajectory groups. On the one hand, results from the linear trend test did not show an association between the adjusted epigenetic age acceleration with metabolic syndrome, neither in blood (OR = 1.02; 95% CI = 0.92–1.13; p = 0.76) (Fig. 3b) nor in VAT (OR = 1.11; 95% CI =0.96–1.31; p = 0.18) (Fig. 3c). On the other hand, weight loss clustering procedure resulted in three trajectory groups depending on the percentage of excess body weight loss (%EBWL) as follows: normal weight loss (NWL), intermediate weight loss (IWL), and low weight loss (LWL), representing 65%, 30%, and 5% of patients (Fig. 3d). Patients from IWL and LWL groups were reassigned into a unique group (ILWL). While no association was found in blood (OR = 1.01; 95% CI = 0.90–1.12; p = 0.91) (Fig. 3e), the probability of belonging to the NWL group significantly increased with the adjusted epigenetic age acceleration of VAT (OR = 1.21; 95% CI = 1.04–1.48; p = 0.03) (Fig. 3f). In other words, patients showing higher epigenetic age acceleration in VAT exhibited a more pronounced weight loss response to bariatric surgery. Whether the adjusted epigenetic age acceleration was associated with metabolic syndrome and/or with weight loss trajectory groups was further tested separately in men and women with no significant result (data not shown).
Fig. 3.
The acceleration of epigenetic aging in VAT is associated with a more pronounced weight loss response following bariatric surgery. Panel a shows the correlation between DNA methylation (DNAm) age acceleration adjusted by sex and body mass index (BMI) between blood and visceral adipose tissue—VAT. Red and yellow dots represent participants with and without metabolic syndrome. r and p refer to Pearson correlation coefficients and p values, respectively. Panels b and c show the predicted probability (from 0 to 1), obtained by binomial logistic regression, of each participant to have a healthy (0) or an unhealthy (1) obesity phenotype based on its adjusted DNAm age acceleration in blood (red dots, b) and VAT (yellow dots, c). Panel d shows the three weight-loss clusters resulting from estimating postsurgery excess body weight loss (%EBWL) trajectories for each participant over a follow-up period of 4 years in the traj R package. Red, green, and black lines stand for normal (NWL), intermediate (IWL) and low (LWL) weight-loss trajectory clusters, representing 65%, 30%, and 5% of participants. IWL and LWL clusters were reassigned into a unique group (ILWL). Panels e and f show the predicted probability (from 0 to 1), obtained by binomial logistic regression, of each participant to belongs to the NWL group (1) or to the ILWL group (0), based on its adjusted DNAm age acceleration in blood (red dots, e) and VAT (yellow dots, f). OR is the odds ratio with 95% confidence intervals and p is the p value for the linear trend of association. Blue and gray dots refer to men and women, respectively
Discussion
This is, to our knowledge, the first study to show the association between BMI and epigenetic age acceleration of VAT. Concretely, the main finding of the present work revealed that increasing BMI in obesity is positively correlated with epigenetic age acceleration in VAT. Importantly, this study also replicates the absence of association between BMI and epigenetic age acceleration in blood [24]. The present results also revealed that VAT epigenetic aging is more strongly related to BMI in men, as compared to women. Additional findings suggested that VAT epigenetic age acceleration may not have a major association with features of the metabolic syndrome in obesity, but a potential and significant effect on the evolution of body weight loss following bariatric surgery.
Previously, Horvath et al. [24] carried out an elegant study to test whether obesity may increase the acceleration of tissue aging. The original version of the epigenetic clock [21], herein used in its reduced form, was utilized to test whether BMI correlated with epigenetic age acceleration in various tissues. Results were very enlightening since they suggested a tissue-specific effect of obesity on the acceleration of epigenetic aging. Concretely, BMI was highly correlated with accelerated liver aging, whereas no effect was observed in blood and, more importantly, nor in subcutaneous adipose tissue. Since fat depots are submitted to an important metabolic stress during weight gain, we found particularly interesting and paradoxical the fact that epigenetic aging of subcutaneous adipose tissue was not altered by increasing BMI, and we decided to investigate whether a distinct effect of obesity on VAT was taking place. We hypothesized that BMI relates to accelerated epigenetic aging of metabolically active tissues, such as VAT and liver, and that such accelerated aging may in part be responsible for the shifting from healthy to unhealthy obesity phenotypes. Although the latter part of the hypothesis was not fully supported by results, our findings are still of interest. In particular, our results support a specific impact of obesity on the epigenetic aging of key metabolic tissues. As mentioned above, the significant correlation showed herein between BMI and epigenetic age acceleration in VAT mirrored in some way that previously observed in liver [28]. Both the similar effect size of BMI on epigenetic age acceleration and the estimated epigenetic aging in years found in both VAT and liver further supported the hypothesis of a tissue-specific dysregulation of methylation [15, 29, 30].
Some methodological differences between the previous study in the liver and the present analysis need to be clearly pointed out. First, we had to use a reduced version of the Horvath’s clock due to technical constraints. However, the successful replication of previous results supported the use of our modified version of the epigenetic clock. Concretely, almost identical correlation coefficients between chronological and epigenetic age were observed in both studies, while correlation coefficients with BMI in liver showed the same magnitude and direction than in Horvath’s study [24]. Second, unlike in the liver study, all the patients from the present work had severe obesity, with or without associated metabolic disturbances, which somehow limited the BMI study range. Nevertheless, the correlation between BMI and epigenetic age acceleration in VAT and liver from patients with obesity was again highly similar, thus supporting the validity of this study. Second, the decision of matching pairs for age, BMI and metabolic syndrome within each sex further limited the spectrum of eligible participants, leading to a narrower range of age, as compared to the Horvath’s study. Both BMI and age ranges may influence the final results, as well as sex representation [28, 31]. Our results showed that men had higher epigenetic aging rates than women in blood, which has been previously linked to a lower morbidity but higher mortality in men than in women [28]. On the other hand, while the impact of BMI on liver epigenetic aging has been previously observed in men and women [24], herein association in VAT was seen only in men. Though, the correlation between BMI and epigenetic age acceleration in liver was only significant in women in the discovery dataset in [24]. Herein, we also observed a significant correlation between BMI and liver epigenetic age acceleration only in women, which may suggest a different impact of BMI on liver and VAT epigenetic aging between men and women. Although the present results did not show a significant association between the adjusted measurement of epigenetic age acceleration and the presence or absence of metabolic syndrome, these results still suggest a sex-specific impact of obesity on VAT epigenetic aging, which could explain, in part, the distinct development of obesity comorbidities between men and women, rather than waist circumference or visceral fat accumulation per se [32]. In any case, these results should be taken with caution, because the effect sizes observed, together with the number of samples from each sex, could lead to uncertain results. We acknowledged that this represents a limitation of the present study. Similarly, the level of statistical power achieved in the entire dataset does not allow us to derive statements regarding the association between BMI and epigenetic aging as consistent as desired. This fact, together with the heterogeneous and inconsistent results previously reported in blood [22, 24, 31], prevent us from ascertaining an actual lack of association between BMI and the epigenetic age acceleration in blood. In spite of reducing the total number of subjects, we tried to support our findings by reanalyzing blood and VAT samples in a dataset without extreme age values, as previously shown [31]. Interestingly, a stronger relationship between BMI and epigenetic age acceleration in VAT was observed, as well as the near absence of association in blood, supporting results observed in the entire dataset. Yet, further studies in larger cohorts designed to capture a broad BMI and age spectrum, as well as to attain a reasonable degree of statistical power, are still required to elucidate the actual impact of BMI on the epigenetic age acceleration in blood and VAT. Taken together, although the specific nature of our cohort somehow limits the number of participants, we consider it worth to be studied, since the findings drawn from its analysis add valuable insights to the discussion on the role of BMI on epigenetic aging.
As just mentioned, another relevant finding of the present work that deserves to be highlighted is the absence of association between VAT epigenetic aging and metabolic syndrome. This is important since it does not support the second term of the main hypothesis, that is, the development of obesity comorbidities through an acceleration of VAT epigenetic age. Among others, a potential explanation of this result is the complexity of the chosen composite endpoint, that is, the presence or absence of metabolic syndrome defined as the sum of a number of metabolic disturbances [4]. Another rather unexpected result was the positive association between the acceleration of VAT epigenetic aging and postsurgery weight loss trajectories, especially when BMI is negatively associated with the percentage of excess body weight loss following bariatric surgery [26]. However, these latter results may suggest that bariatric surgery would yield more beneficial outcomes in those patients with aged VAT. Previous studies have already reported a deep epigenetic remodeling after different weight loss interventions in adipose tissue [33, 34]. In view of that, these results could imply that a more intense methylation remodeling may be taking place in an aged VAT following bariatric surgery, leading to a tissue rejuvenation. Since our longitudinal weight-loss study is still not finished, it is not yet possible to have access to postsurgery VAT methylation data that would allow us to establish a causal relationship. It is also worth noting that this is a whole-tissue analysis and that knowing which cell type is responsible for the accelerated aging of VAT might help understand the link with the response to weight loss. In this regard, senescence of adipocytes is a hallmark of adipose tissue aging, and it would be expected that senescent mature adipocytes would mobilize less fat during weight loss [35]. However, the observed accelerated aging might be associated with senescence of another cell type which might impair lipid mobilization. This could be cells from the stromal vascular fraction involved in adipose tissue remodeling [36], e.g. immune cells or endothelial cells of the vascular bed which could reduce blood flow to the tissue [37]. Further studies are in process and will help understand the actual impact of bariatric surgery on VAT epigenetic aging.
Conclusions
In conclusion, our results seem to corroborate that obesity accelerates epigenetic aging of metabolically active tissues, such as VAT and liver. Likewise, these results suggest that epigenetic age acceleration in blood does not correlate with BMI in obesity. Moreover, BMI seems to have a more pronounced effect on epigenetic age acceleration in men than in women. Finally, while not having an effect on metabolic syndrome development, the acceleration of VAT epigenetic aging seems to play an important role in weight loss dynamics following bariatric surgery.
Methods
Study participants
A total of 56 patients, 28 men and 28 women with severe obesity (BMI > 40 kg/m2) and undergoing bariatric surgery (biliopancreatic diversion with duodenal switch) at the Quebec Heart and Lung Institute, were selected to participate in the present study. Patients were matched within each sex for age, BMI and the presence or absence of metabolic syndrome. Omental samples (VAT) were obtained during the course of surgery and blood samples were collected preoperatively. The surgical protocol, blood and VAT sample collection, and the standardized procedures to measure anthropometric and metabolic parameters are described elsewhere [38]. Patients were diagnosed with the metabolic syndrome when three or more criteria of the National Cholesterol Education Program Adult Treatment Panel III guidelines were present [4]. Waist circumference, blood pressure, HDL-cholesterol, plasma triglycerides and fasting glucose levels were measured preoperatively and used to identify those with the metabolic syndrome. Severe obesity was defined as BMI > 40 kg/m2, calculated as weight in kilograms divided by height in meters squared. Due to the lack of accurate phenotype data, one patient and its matching pair were excluded from further analyses.
Genome-wide DNA methylation analysis
Genomic DNA of the 56 study participants was extracted from 200 mg of VAT using the DNeasy Blood & Tissue kit (QIAGEN, Mississauga, Ontario, Canada) and isolated from the blood buffy coat using the GenElute™ Blood Genomic DNA kit (Sigma, St Louis, MO, USA). Following quantification of DNA using both NanoDrop Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and PicoGreen DNA methods, DNA (1 μg) was bisulfite converted and quantitative genome-wide methylation analysis was conducted using Infinium HumanMethylation450 (450k) and EPIC platforms (Illumina, San Diego, CA) interrogating over 485,000 and 850,000 CpG sites at single-nucleotide resolution, respectively. Methylation arrays were processed at the McGill University and Génome Québec Innovation Centre (Montreal, Canada) according to the manufacturer’s instructions (Illumina, San Diego, CA). Methylation data was preprocessed and normalized using the minfi R package [39]. Before background correction and normalization, the 450k and EPIC arrays were combined and integrated into a virtual 450k array, leaving 453,093 CpG sites for further statistical analyses. The single-sample Noob (ssNoob) method was the preferred normalization procedure, as previously recommended when integrating data from multiple Infinium methylation arrays [40]. Methylation levels (beta values; β) were estimated as the ratio of signal intensity of the methylated alleles to the sum of methylated and unmethylated intensity signals of the alleles (β value = C/(T+C)). The β values varied from 0 (no methylation) to 1 (100% methylation). The overall correlation across 453,093 CpG sites between 450 k and EPIC arrays was very high in blood and VAT (r = 0.992 in both tissues). One sample did not fulfill methylation quality control criteria and was excluded, together with its matching pair, from further analyses.
Epigenetic clock
Epigenetic age was estimated for each patient in VAT and blood according to the Horvath’s epigenetic clock [21], currently the gold-standard for determining epigenetic age in humans [41]. Because part of methylation data was obtained from the EPIC array, which does not include the whole dataset of 353 CpG sites used to build the original epigenetic clock, we estimated epigenetic age by using a reduced version of it, as previously done [42]. The final dataset used to estimate epigenetic age consisted on a set of 336 CpG sites, 4.8% less than in the original dataset. The correlation among CpG sites included in the epigenetic clock was similarly strong in blood (r = 0.992) and VAT (r = 0.991) between 450k and EPIC arrays, with a total of 323 CpG sites in blood (96.1%) and 318 CpG sites in VAT (94.6%) showing a mean β value difference lower than 0.1 (Additional file 1: Figure S3). Aiming to analyze whether this modification could cause a lack of accuracy, we replicated previous results obtained with the original epigenetic clock in liver [24]. Publicly available methylation data from 62 liver samples (GSE48325) [27] was used as a replication dataset. A subset of 40 liver samples including only subjects with severe obesity (BMI > 40 kg/m2) was also used for comparison purposes. The reduced version of the epigenetic clock was built in R using publicly available data (https://horvath.genetics.ucla.edu/html/dnamage/) [21].
Weight loss trajectories
Postsurgery weight loss trajectories were estimated for each participant by identifying clusters of individual longitudinal weight loss data implemented in the traj R package [43]. Briefly, the traj procedure uses a factor analysis to select nonredundant measurements, followed by a cluster analysis to identify subsets of patients with similar weight loss trajectories [44]. Weight loss data from 46 patients was available for trajectory group assessment. Body weight was measured during postoperative visits or phone calls thorough a follow-up period of four years, and a total of seven postsurgery time points at 3, 6, 12, 18, 24, 36, and 48 months were used for cluster estimation. Postsurgery weight loss was defined as the percentage of excess body weight loss (%EBWL), calculated as the difference between actual body weight loss (initial BMI minus actual BMI) and ideal body weight loss (initial BMI minus ideal BMI fixed at 25 kg/m2) [45]. Resulting groups allowed the categorization of patients as a function of their %EBWL.
Statistics
Phenotype data was checked for normality with the Kolmogorov-Smirnov test. Two-group comparisons were tested with Student’s t test for paired and independent samples, as appropriate. Epigenetic age acceleration was defined as the residual resulting from regressing epigenetic age on chronological age, and its correlation with BMI was tested in both VAT and blood using Pearson correlation coefficients. A multivariate linear regression model including chronological age, sex and metabolic syndrome was further used to test the association between BMI and epigenetic age acceleration. Binomial logistic regression was used to predict the probability that a patient falls into a healthy or unhealthy phenotype (presence or absence of metabolic syndrome), as well as into a weight loss trajectory group, both set as dichotomous variables. Epigenetic age acceleration adjusted by chronological age, sex and BMI was set as a continuous variable in logistic regression. Regression models and the rest of statistical calculations were performed in R (https://www.R-project.org) [46]. Power calculations were performed in G*Power [47].
Supplementary information
Additional file 1: Table S1. Data summary of participants in the study cohort. Figure S1. The acceleration of epigenetic aging in VAT correlates with BMI in middle-aged subjects. Figure S2. The acceleration of epigenetic age in liver correlates with BMI in women. Figure S3. Correlation among CpG sites included in the epigenetic clock between 450k and EPIC arrays.
Acknowledgements
We thank all participants for their collaboration, and the surgeons of the Bariatric Surgery Center of the Quebec Heart and Lung Institute (Odette Lescelleur, Laurent Biertho, Annie Lafortune) for their invaluable collaboration in clinical care and patient recruitment. We are grateful to Paule Marceau for data management, Catherine Raymond for DNA extraction and technical assistance, and the staff of the Quebec Heart and Lung Institute Biobank for patient consent and sample processing. We also sincerely thank our long-term collaborator Picard Marceau, with whom this project was initiated.
Abbreviations
- %EBWL
Percentage of excess body weight loss
- BMI
Body mass index
- CpG
Cytosine-phosphate-guanine dinucleotides
- DNAm age
DNA methylation age
- IWL
Intermediate weight loss
- LWL
Low weight loss
- NWL
Normal weight loss
- VAT
Visceral adipose tissue
Authors’ contributions
JTM performed statistical analysis, interpreted the data and wrote the manuscript; MCV and FG conceived and designed the research; AT participated in the elaboration of the study design and critically reviewed the manuscript. FSH, SL, FJ and SM participated in clinical care, patient recruitment, and tissue sampling. All authors read and approved the final manuscript.
Funding
This study was supported by a grant-in-aid from the Heart and Stroke Foundation of Canada (G-17-0016627) and by the Canada Research Chair in Genomics Applied to Nutrition and Metabolic Health, held by MCV. JTM received a postdoctoral fellowship from the Fonds de Recherche de Québec-Santé.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by Laval University and Quebec Heart and Lung Institute ethics committees and was performed in accordance with the principles of the Declaration of Helsinki. All participants provided written, informed consent.
Consent for publication
Not applicable.
Competing interests
AT receives research funding from Johnson & Johnson Medical companies, Pfizer and Medtronic for studies on bariatric surgery. The rest of authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juan de Toro-Martín, Email: juan.de-toro-martin.1@ulaval.ca.
Frédéric Guénard, Email: Frederic.Guenard@fsaa.ulaval.ca.
André Tchernof, Email: andre.tchernof@criucpq.ulaval.ca.
Frédéric-Simon Hould, Email: Frederic.Hould@fmed.ulaval.ca.
Stéfane Lebel, Email: stefane.lebel@fmed.ulaval.ca.
François Julien, Email: francois.julien@criucpq.ulaval.ca.
Simon Marceau, Email: simon.marceau@fmed.ulaval.ca.
Marie-Claude Vohl, Email: marie-claude.vohl@fsaa.ulaval.ca.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13148-019-0754-6.
References
- 1.van Vliet-Ostaptchouk JV, Nuotio M-L, Slagter SN, Doiron D, Fischer K, Foco L, et al. The prevalence of metabolic syndrome and metabolically healthy obesity in Europe: a collaborative analysis of ten large cohort studies. BMC Endocr Disord. 2014;14:9. doi: 10.1186/1472-6823-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinnouho G-M, Czernichow S, Dugravot A, Nabi H, Brunner EJ, Kivimaki M, et al. Metabolically healthy obesity and the risk of cardiovascular disease and type 2 diabetes: the Whitehall II cohort study. Eur Heart J. 2015;36(9):551–559. doi: 10.1093/eurheartj/ehu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tchernof A, Després J-P. Pathophysiology of human visceral obesity: an update. Physiol Rev. 2013;93(1):359–404. doi: 10.1152/physrev.00033.2011. [DOI] [PubMed] [Google Scholar]
- 4.Grundy SM, Cleeman JI, Daniels SR, Donato KA, Eckel RH, Franklin BA, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112(17):2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 5.Hamer M, Stamatakis E. Metabolically healthy obesity and risk of all-cause and cardiovascular disease mortality. J Clin Endocrinol Metab. 2012;97(7):2482–2488. doi: 10.1210/jc.2011-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh S, Bouchard C. Convergence between biological, behavioural and genetic determinants of obesity. Nat Rev Genet. 2017;18(12):731–748. doi: 10.1038/nrg.2017.72. [DOI] [PubMed] [Google Scholar]
- 7.Pigeyre M, Yazdi FT, Kaur Y, Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin Sci. 2016;130(12):943. doi: 10.1042/CS20160136. [DOI] [PubMed] [Google Scholar]
- 8.Crujeiras AB, Diaz-Lagares A, Moreno-Navarrete JM, Sandoval J, Hervas D, Gomez A, et al. Genome-wide DNA methylation pattern in visceral adipose tissue differentiates insulin-resistant from insulin-sensitive obese subjects. Transl Res. 2016;178:13–24.e5. doi: 10.1016/j.trsl.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 9.De Toro-Martín J, Guénard F, Tchernof A, Deshaies Y, Pérusse L, Hould FS, et al. Methylation quantitative trait loci within the TOMM20 gene are associated with metabolic syndrome-related lipid alterations in severely obese subjects. Diabetol Metab Syndr. 2016;8(1):55. doi: 10.1186/s13098-016-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Toro-Martín J, Guénard F, Tchernof A, Deshaies Y, Pérusse L, Biron S, et al. A CpG-SNP located within the ARPC3 gene promoter is associated with hypertriglyceridemia in severely obese patients. Ann Nutr Metab. 2016;68(3):203–212. doi: 10.1159/000445358. [DOI] [PubMed] [Google Scholar]
- 11.Guénard F, Tchernof A, Deshaies Y, Pérusse L, Biron S, Lescelleur O, et al. Differential methylation in visceral adipose tissue of obese men discordant for metabolic disturbances. Physiol Genomics. 2014;46(6):216–222. doi: 10.1152/physiolgenomics.00160.2013. [DOI] [PubMed] [Google Scholar]
- 12.Ribel-Madsen R, Fraga MF, Jacobsen S, Bork-Jensen J, Lara E, Calvanese V, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. Dahlman IA, editor. PLoS One. 2012;7(12):e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turcot V, Tchernof A, Deshaies Y, Pérusse L, Bélisle A, Marceau S, et al. LINE-1 methylation in visceral adipose tissue of severely obese individuals is associated with metabolic syndrome status and related phenotypes. Clin Epigenetics. 2012;4(1):10. doi: 10.1186/1868-7083-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crujeiras AB, Diaz-Lagares A, Sandoval J, Milagro FI, Navas-Carretero S, Carreira MC, et al. DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: a genome-wide analysis from non-obese and obese patients. Sci Rep. 2017;7:41903. doi: 10.1038/srep41903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guénard F, Deshaies Y, Hould F-S, Lebel S, Tchernof A, Marceau P, et al. Use of blood as a surrogate model for the assessment of visceral adipose tissue methylation profiles associated with the metabolic syndrome in men. J Mol Genet Med. 2016;10(01):1–8. doi: 10.4172/1747-0862.1000198. [DOI] [Google Scholar]
- 16.Ravlić S, Škrobot Vidaček N, Nanić L, Laganović M, Slade N, Jelaković B, et al. Mechanisms of fetal epigenetics that determine telomere dynamics and health span in adulthood. Mech Ageing Dev. 2018;174:55–62. doi: 10.1016/j.mad.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Z, Zheng L, Almeida FA. Epigenetic reprogramming in metabolic disorders: nutritional factors and beyond. J Nutr Biochem. 2018;54:1–10. doi: 10.1016/j.jnutbio.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvestrini V, Sell C, Lorenzini A. Obesity may accelerate the aging process. Front Endocrinol (Lausanne) 2019;10:266. doi: 10.3389/fendo.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CPG island context. Schübeler D, editor. PLoS Genet. 2009;5(8):e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MPM, van Eijk K, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci. 2014;111(43):15538–43. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019;29(5):1028–1044. doi: 10.1016/j.cmet.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Toro-Martín J, Guénard F, Tchernof A, Pérusse L, Marceau S, Vohl, M-C. Polygenic risk score for predicting weight loss after bariatric surgery. JCI Insight. 2018;3(17):e122011. [DOI] [PMC free article] [PubMed]
- 27.Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, et al. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9(4):506–518. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y-TT, Chu S, Loucks EB, Lin C-LL, Eaton CB, Buka SL, et al. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics. 2016;11(3):227–236. doi: 10.1080/15592294.2016.1146853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nevalainen T, Kananen L, Marttila S, Jylhävä J, Mononen N, Kähönen M, et al. Obesity accelerates epigenetic aging in middle-aged but not in elderly individuals. Clin Epigenetics. 2017;9(1):20. doi: 10.1186/s13148-016-0301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grundy SM, Williams C, Vega GL. Upper body fat predicts metabolic syndrome similarly in men and women. Eur J Clin Investig. 2018;48(7):e12941. doi: 10.1111/eci.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gensous N, Franceschi C, Santoro A, Milazzo M, Garagnani P, Bacalini MG. The impact of caloric restriction on the epigenetic signatures of aging. Int J Mol Sci. 2019;20(8):2022. doi: 10.3390/ijms20082022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rönn T, Volkov P, Davegårdh C, Dayeh T, Hall E, Olsson AH, et al. A six months exercise intervention influences the genome-wide dna methylation pattern in human adipose tissue. Greally JM, editor. PLoS Genet. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tchkonia T, Morbeck DE, Von Zglinicki T, Van Deursen J, Lustgarten J, Scrable H, et al. Fat tissue, aging, and cellular senescence. Aging Cell. 2010;9(5):667–684. doi: 10.1111/j.1474-9726.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakowa N, Trieu N, Flehmig G, Lohmann T, Schön MR, Dietrich A, et al. Telomere length differences between subcutaneous and visceral adipose tissue in humans. Biochem Biophys Res Commun. 2015;457(3):426–432. doi: 10.1016/j.bbrc.2014.12.122. [DOI] [PubMed] [Google Scholar]
- 37.Sun K, Kusminski CM, Scherer PE. Adipose tissue remodeling and obesity. J Clin Invest. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biertho L, Lebel S, Marceau S, Hould F-SS, Julien F, Biron S. Biliopancreatic diversion with duodenal switch: surgical technique and perioperative care. Surg Clin North Am. 2016;96:815–826. doi: 10.1016/j.suc.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 39.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortin J-P, Triche TJ, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2016;33(4):btw691. doi: 10.1093/bioinformatics/btw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 42.McEwen LM, Jones MJ, Lin DTS, Edgar RD, Husquin LT, MacIsaac JL, et al. Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array. Clin Epigenetics. 2018;10(1):123. doi: 10.1186/s13148-018-0556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sylvestre M-P, McCusker J, Cole M, Regeasse A, Belzile E, Abrahamowicz M. Classification of patterns of delirium severity scores over time in an elderly population. Int Psychogeriatr. 2006;18(4):667–680. doi: 10.1017/S1041610206003334. [DOI] [PubMed] [Google Scholar]
- 44.Leffondré K, Abrahamowicz M, Regeasse A, Hawker GA, Badley EM, McCusker J, et al. Statistical measures were proposed for identifying longitudinal patterns of change in quantitative health indicators. J Clin Epidemiol. 2004;57(10):1049–1062. doi: 10.1016/j.jclinepi.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Marceau P, Biron S, Marceau S, Hould F-S, Lebel S, Lescelleur O, et al. Long-term metabolic outcomes 5 to 20 years after biliopancreatic diversion. Obes Surg. 2015;25(9):1584–1593. doi: 10.1007/s11695-015-1599-5. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 47.Faul F, Erdfelder E, Lang A-G, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/BF03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Data summary of participants in the study cohort. Figure S1. The acceleration of epigenetic aging in VAT correlates with BMI in middle-aged subjects. Figure S2. The acceleration of epigenetic age in liver correlates with BMI in women. Figure S3. Correlation among CpG sites included in the epigenetic clock between 450k and EPIC arrays.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.