Abstract
HSF1 (heat shock factor 1) is an evolutionarily conserved master transcriptional regulator of the heat shock response (HSR) in eukaryotic cells. In response to high temperatures, HSF1 upregulates genes encoding molecular chaperones, also called heat shock proteins, which assist the refolding or degradation of damaged intracellular proteins. Accumulating evidence reveals however that HSF1 participates in several other physiological and pathological processes such as differentiation, immune response, and multidrug resistance, as well as in ageing, neurodegenerative demise, and cancer. To address how HSF1 controls these processes one should systematically analyze its target genes. Here we present a novel database called HSF1Base (hsf1base.org) that contains a nearly comprehensive list of HSF1 target genes identified so far. The list was obtained by manually curating publications on individual HSF1 targets and analyzing relevant high throughput transcriptomic and chromatin immunoprecipitation data derived from the literature and the Yeastract database. To support the biological relevance of HSF1 targets identified by high throughput methods, we performed an enrichment analysis of (potential) HSF1 targets across different tissues/cell types and organisms. We found that general HSF1 functions (targets are expressed in all tissues/cell types) are mostly related to cellular proteostasis. Furthermore, HSF1 targets that are conserved across various animal taxa operate mostly in cellular stress pathways (e.g., autophagy), chromatin remodeling, ribosome biogenesis, and ageing. Together, these data highlight diverse roles for HSF1, expanding far beyond the HSR.
Keywords: ageing, autophagy, cell adhesion, cell cycle, circadian rhythm, chromatin remodeling, heat shock factor 1, heat shock proteins, heat shock response, ribosome biogenesis
1. Introduction
Upon proteotoxic stress, such as high temperatures, elevated oxygen levels, heavy metals, toxins, and bacterial infections, a highly conserved cell protective mechanism called the heat shock response (HSR) is induced to preserve cellular proteostasis [1,2,3]. The HSR leads to a robust activation of genes encoding heat shock proteins (HSPs). HSPs function as molecular chaperones to help refold or degrade damaged proteins, thereby contributing to the protection of cells from protein-damaging factors. The master regulator of the HSR is an evolutionarily conserved heat shock transcription factor, HSF1, which becomes activated via trimerization and phosphorylation, and then translocated into the nucleus to promote the transcription of HSP genes [4,5]. In the proximal enhancer region of its target genes, HSF1 acts through a 5’ regulatory cis element called the heat shock responsive element (HSE). The HSE consists of at least three subsequent inverted repeats, TTCnnGAAnnTTC (where n denotes any arbitrary nucleotide) [6,7,8].
The idea that HSF1 upregulates HSP genes has been initiated from the 2000s. However, several genome-scale high throughput gene expression data have uncovered that numerous non-HSP genes are also under the control of HSF1 [9,10,11,12,13,14,15,16]. Accordingly, HSF1 has been implicated in several fundamental biological processes being independent of the HSR, including metabolism, gametogenesis, development, and ageing, as well as in various pathologies, especially neurodegenerative disorders and cancer [2,17,18,19,20,21,22,23]. These studies have also shown that HSF1-driven transcription strongly depends on the actual type, developmental state, metabolic condition, and phase of the affected cell. Whether HSF1 targets involved in these processes are specific to a given species or conserved throughout evolution has yet determined.
Here, we report the development of a novel database called HSF1base (hsf1base.org) containing a nearly comprehensive list of HSF1 target genes identified to date. In the HSF1base, direct HSF1 targets were identified by manually curating HSF1-related publications on single gene analyses. In addition, relevant high throughput transcriptomic and chromatin immunoprecipitation data were analyzed to support the biological relevance of predicted HSF1 targets. The format of the HSF1base is PSI-MITAB 2.8, an extended version of the PSI-MI tab-delimited format [24]. This uses standardized expression data denoted with a well-defined ID, thereby facilitating the accurate use of the database for systems biology. To illustrate the applicability of the HSF1base, we performed an enrichment meta-analysis for HSF1 target genes in the yeast Saccharomyces cerevisiae, nematode Caenorhabditis elegans, fruit fly Drosophila melanogaster, mouse Mus musculus, rat Rattus norvegicus, and human Homo sapiens. Meta-analysis of HSF1 targets showed that HSF1 regulates both general and tissue- and cell type-specific targets in mice and humans, suggesting cell-specific functions for HSF1 in biological processes such as cell adhesion, mitosis, cell cycle regulation, and ageing. We also identified a relatively large set of evolutionarily conserved HSF1 target genes. Taken together, this study reveals the specificity and conservation of HSF1 regulatory networks across tissues and species, and highlights that the role of HSF1 expands far beyond regulating the HSR.
2. Results
2.1. The HSF1base Database
The HSF1base database is based on 117 manually selected and curated relevant publications (see Table S1). It contains altogether 24,635 HSF1 target gene interactions (i.e., genes that are up- or down-regulated by HSF1 or the regulatory region of which is able to bind HSF1) derived from several model systems such S. cerevisiae, C. elegans, D. melanogaster, M. musculus, R. norvegicus, and human cell lines (Table 1). Note that a given interaction could be obtained with multiple times, i.e., from more than one species. Out of these interactions, 18,356 were considered as direct ones, based on evidence for physical interaction between HSF1 and the corresponding gene. In most cases, data were resulted from genome-scale ChIP-on-chip or ChIP-Seq analyses, where up- or down-regulation of HSF1 targets was not studied just the fact of HSF1-DNA binding.
Table 1.
HSF-1 targets found in the database HSF1base (hsf1base.org).
| Activated | Inhibited | N/D | All | |
|---|---|---|---|---|
| ALL INTERACTIONS | ||||
| All records | 5258 | 1942 | 17,435 | 24,635 |
| HSF1 targets with evidence for direct interaction | 577 | 344 | 17,435 | 18,356 |
| HSF1 targets with evidence for HSF1 dependent regulation | 4681 | 1598 | 0 | 6279 |
| HSF1 targets with evidence for direct interaction and HSF1 dependent regulation | 1869 | 1023 | 0 | 2892 |
| UNIQUE INTERACTIONS | ||||
| All records | 3869 | 1272 | 10,500 | 15,641 |
| HSF1 targets with evidence for direct interaction | 404 | 207 | 10,499 | 11,110 |
| HSF1 targets with evidence for HSF1 dependent regulation | 3877 | 1325 | 0 | 5202 |
| HSF1 targets with evidence for direct interaction and HSF1 dependent regulation | 774 | 547 | 0 | 1321 |
| ALL INTERACTIONS | ||||
| Homo sapiens | 1685 | 958 | 7955 | 10,598 |
| Mus musculus | 135 | 279 | 9315 | 9729 |
| Rattus norvegicus | 465 | 166 | 0 | 631 |
| Caenorhabditis elegans | 2662 | 492 | 0 | 3154 |
| Drosophila melanogaster | 99 | 38 | 54 | 191 |
| Saccharomyces cerevisiae | 211 | 5 | 111 | 327 |
| other | 0 | 3 | 0 | 3 |
| UNIQUE INTERACTIONS | ||||
| Homo sapiens | 876 | 562 | 4182 | 5620 |
| Mus musculus | 83 | 143 | 6153 | 6379 |
| Rattus norvegicus | 454 | 146 | 0 | 600 |
| Caenorhabditis elegans | 2218 | 375 | 0 | 2593 |
| Drosophila melanogaster | 98 | 38 | 54 | 190 |
| Saccharomyces cerevisiae | 140 | 5 | 111 | 256 |
| other | 0 | 3 | 0 | 3 |
A table summarizing the HSF-1 targets found in the database HSF1base (hsf1base.org). The ‘activated’ gene expression is upregulated by HSF-1. The ‘inhibited’ gene expression is downregulated by HSF-1. ‘N/D’ means there is no regulatory information available. ‘All interactions’ contains all entries in the database; duplicates are included. ‘Unique interactions’ does not contain duplicates; every gene is included only once. ‘HSF1 targets with evidence for direct interaction’ are considered as such based on evidence for physical interaction between HSF1 and the corresponding gene. In case of ‘HSF1 targets with evidence for HSF1 dependent regulation’, regulatory information is available, but there is no evidence for physical interaction between HSF1 and the corresponding gene. For ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’, both regulatory information and evidence for physical interaction are available.
Only a relatively small number (921) of direct interactions was further analyzed to provide information about the nature of HSF1 target gene regulatory interactions (positive or negative). HSF1 target genes were directly activated in 577 records while repressed in 344 cases by the protein. In 6279 records, there were no evidence for a direct interaction between HSF1 and its target gene. In these cases, HSF1-dependent regulation of target genes was demonstrated by RNA-seq or microarray studies. Altogether, the HSF1base contains 15,641 unique HSF1 target gene interactions, 11,110 of which were considered as direct targets based on evidence for physical interaction between HSF1 and the gene such as ChIP-on-chip or ChIP-Seq analyses (Table 1). HSF1-dependent gene expression was detected in 1321 direct targets (774 are activated and 547 are inhibited).
2.2. Strategy for Testing the Applicability of the HSF1Base
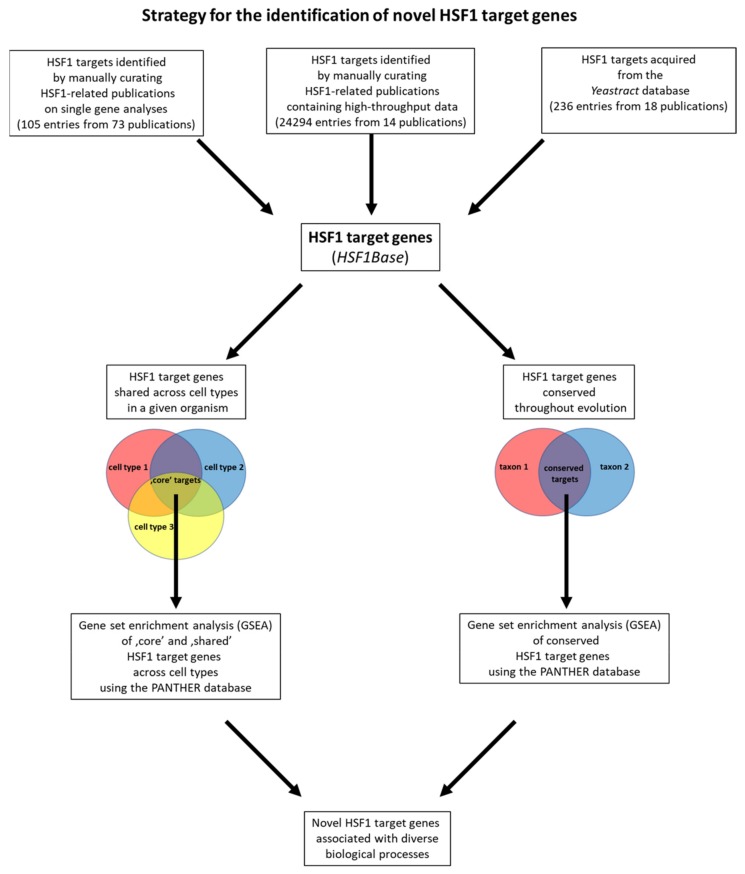
To select potential targets of HSF1 from the primary list containing every hits of high throughput analyses (Table S2), we applied the approach of Webb and colleagues (2016) who analyzed tissue specificity and conservation of FoxO (Forkhead box-O transcription factor, the effector of the insulin/IGF1 (insulin-like growth factor-1) signalling system) target genes identified by ChIP-seq binding data from various mouse cell types and in several model systems [25] (Figure 1). We determined HSF1 targets that had been described parallel in more than one cell types within a single species, and found to be evolutionarily conserved (Tables S3–S5). The resulting ‘shared’ gene sets were further applied to statistical overrepresentation tests, using the Protein Analysis Through Evolutionary Relationships (PANTHER) database [26]. In this way, gene groups were identified which are enriched within a given gene set, and can be linked to a given biological process, molecular function, protein class, PANTHER pathway or Reactome pathway. Within the gene groups we determined, individual genes were further labelled that had not previously been characterized as a HSF1 target in a single-gene-analysis study (Table S6). In addition, individual genes that had been identified to be directly regulated by HSF1 were also selected (Table S6). This approach allowed us to examine general and cell/tissue type-specific functions of HSF1, as well as its conserved and species-specific activities (Figure 1).
Figure 1.
A flowchart depicting our strategy for the identification of novel potential HSF1 target genes. The database (HSF1Base) was built by manually curating HSF1-related publications on single gene analyses (105 entries from 73 publications), HSF1-related publications containing high-throughput data (24,294 entries from 14 publications) and acquiring HSF1 targets from Yeastract database (236 entries from 18 publications). Out of all target genes contained in our database we selected those shared across cell types in each organism. Then, we performed gene set enrichment analysis (GSEA) using PANTHER database on the ‘core’ (shared between all three cell types) and ‘shared’ (shared between at least two cell types) gene sets. We also selected evolutionally conserved target genes and performed GSEA using the PANTHER database on these target genes as well. We then identified novel HSF1 target genes associated with diverse biological processes of interest.
2.3. General and Tissue-Specific HSF1 Target Genes in Human
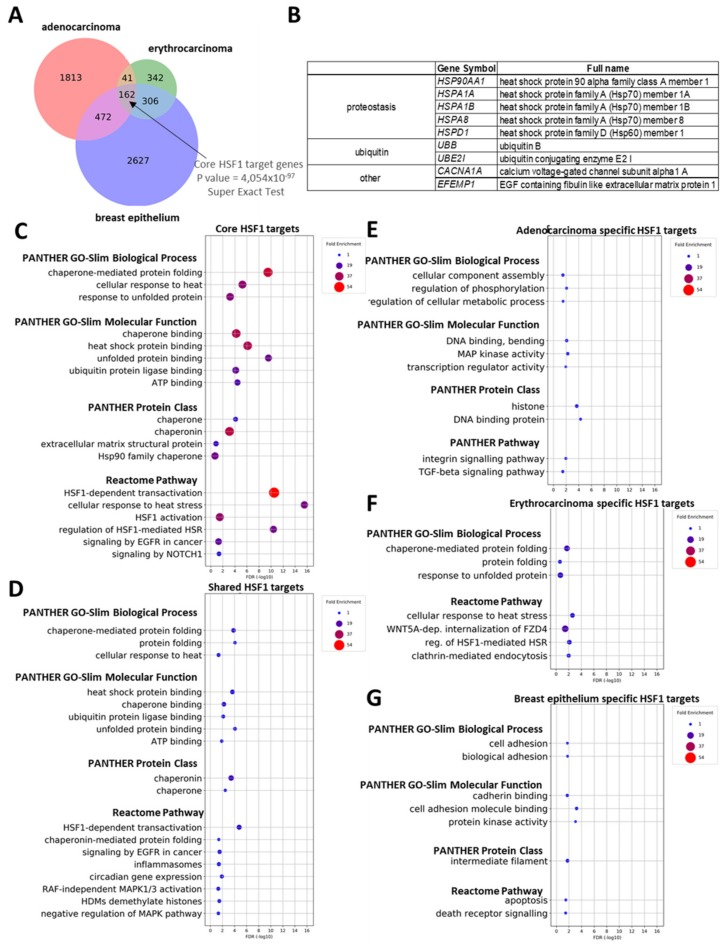
To investigate which HSF1 targets are regulated in a cell type/tissue-specific manner, we compared HSF1 targets obtained from several murine and human cell lines. We observed that while some HSF1 targets are ubiquitously expressed in all human tissues examined, most of them are active in (a) specific cell types(s) only (Figure 2). Comparing the expression of HSF1 targets in each human cell type confirmed that many of these genes are active in specific cell types; e.g., 2488 genes in cervical adenocarcinoma cell lines (HeLa and HF73 cells), 851 genes in erythroleukemia-derived cell lines (K562 cells), and 3567 genes in breast epithelium (HME1, BPE, and MCF7 cell lines) (Figure 2A and Table S3). On the other hand, the expression of numerous other HSF1 target genes, specifically 17.03% of all targets (981 genes), was found in at least two cell types. These genes represent the so-called ‘shared’ HSF1 targets (Figure 2A and Table S3). Interestingly, a significant number of HSF1 target genes (162, 2.81%) are active in all cell types examined so far, representing the ‘core’ HSF1 direct targets (Figure 2A and Table S3). The list of ‘core’ HSF1 targets includes several previously identified target genes of HSF1 (e.g., HYPK [27], STIP1 [28], BAG3 [28], JUN [29], and UBB [30]), as well as targets that have not yet been characterized in detail (e.g., CELSR1, JMJD6, and TBL1X).
Figure 2.
HSF1 transcription factor binds tissue-specific and shared targets in human. (A) Venn diagram depicting overlaps between HSF1 target genes in all cell types analysed (p = 4.05 × 10−97, super exact test; 19,608; all human genes were used as a background); (B) HSF1 target genes involved in ageing that are shared between all three cell types (GenAge database); (C) PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class and Reactome Pathway analysis of the 162 HSF1 target genes shared among all three cell types; (D) PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class and Reactome Pathway analysis of the 981 HSF1 target genes shared between at least two cell types; (E) PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class and PANTHER Pathway analysis of the 1813 adenocarcinoma specific HSF1 target genes; (F) PANTHER GO-Slim Biological Process and Reactome Pathway analysis of the 342, erythrocarcinoma-specific HSF1 target genes; (G) PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class and Reactome Pathway analysis of the 2627, breast epithelium specific HSF1 target genes. Filled circles depict statistically significant enrichments; X axis shows -LOG10 (p-value) p < 0.05; Fisher’s exact test with Benjamini–Hochberg false discovery rate correction (FDR).
We then tested whether cell type/tissue-specific and ‘shared’ HSF1 targets have different functions, using the Protein Analysis Through Evolutionary Relationships database (PANTHER) [26]. Gene set enrichment analysis (GSEA) of ‘core’ HSF1 targets showed that most of the significantly enriched gene ontology (GO) terms are related to biological processes previously associated with a HSF1 function such as chaperone-mediated protein folding (e.g., HSPA1A, HSPA8, HSPA6, DNAJB1, ST13, and FKBP4) and ubiquitin protein ligase binding (e.g., UBB, and UBC) (Figure 2C, Tables S6 and S7). In good accordance with this observation, the most enriched molecular function terms are chaperone binding and unfolded protein binding (Figure 2C). The significantly enriched protein class terms in the ‘core’ HSF1 target gene set indeed were chaperones and chaperonins (Figure 2C). Panther pathway analysis of the ‘core’ gene set showed that the apoptotic pathway, a system known to be regulated by HSF1 [17,31,32,33], was also enriched for these genes (Figure 2C, Table 2, Tables S6 and S7). Similarly, ‘shared’ target genes were enriched for chaperone-related GO terms (Figure 2D, Tables S6 and S7). Taken together, the HSF1 ‘core’ and ‘shared’ target gene sets in three different human cell types contain genes associated with the role HSF1 in maintaining proteostasis. We also found over-represented annotations that cannot be linked to classical functions of HSF1 (Figure 2C, Tables S6 and S7). A representative example is the extracellular matrix structural protein term containing LTBP4, CRELD1, and EFEMP1 genes. These are potential direct targets of HSF1, which have not been characterized in a single-gene-analysis study, and according to the HSF1base, these genes are directly controlled by HSF1. Similarly, novel HSF1 direct targets were found in the term Signalling by NOTCH1 (NOTCH1, TBL1X, and DTX2). Analyzing ‘shared’ HSF1 targets revealed over-represented Reactome pathway terms such as inflammasomes (BCL2L1, SUGT1, APP, MEFV, HSP90AB1, and NFKB2), circadian rhythm (RXRA, NOCT, NCOA2, BHLHE40, ARNTL, SERPINE1, DBP, and TBL1X), RAF-independent MAPK1/3 activation (IL6R, DUSP2, DUSP10, DUSP5, MAP2K2, and DUSP1), HDMs demethylate histones (PHF8, HIST1H4A, ARID5B, KDM6A, KDM4B, JMJD6, and KDM1A) and negative regulation of MAPK signalling (UBB, DUSP2, UBC, DUSP10, KSR1, DUSP5, MAP2K2, and DUSP1) (Figure 2D, Table 2, Table S6 and Table S7). Although most of these genes have not been described as direct HSF1 targets by single-gene-analysis studies, the HSF1base identifies them as genetic factors that are directly regulated by HSF1 (for details, see Table S6). This implies that in these three cell types HSF1 exerts specific functions being independent of the HSR.
Table 2.
Selected list of HSF1 target genes identified in human cell types, using the HSF1Base.
| Human ’Core’ HSF1 Target Genes | ||
|---|---|---|
| Extracellular matrix structural protein | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| creld1 | Yes | activated |
| EFEMP1 | Yes | activated |
| LTBP4 | Yes | activated |
| Signaling by NOTCH1 | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| UBB | No | activated |
| UBC | Yes | activated |
| notch1 | Yes | activated |
| TBL1X | Yes | inhibited |
| DTX2 | Yes | activated |
| Apoptotic pathway | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| HSPA1L | Yes | activated |
| HSPA1A | No | activated |
| hspa1b | No | activated |
| JUN | No | activated |
| HSPA6 | Yes | activated |
| BCL2L1 | Yes | activated |
| FOS | Yes | activated |
| ATF3 | No | activated |
| BAG3 | No | activated |
| HSPA8 | Yes | activated |
| MAP4K1 | Yes | activated |
| NFKB2 | Yes | activated |
| CFLAR | Yes | activated |
| Human ’shared’ HSF1 target genes | ||
| Inflammasomes | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| BCL2L1 | Yes | activated |
| SUGT1 | Yes | activated |
| APP | Yes | activated |
| MEFV | Yes | activated |
| HSP90AB1 | Yes | activated |
| NFKB2 | Yes | activated |
| BMAL1:CLOCK,NPAS2 activates circadian gene expression | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| RXRA | Yes | activated |
| NOCT | Yes | activated |
| NCOA2 | Yes | inhibited |
| BHLHE40 | Yes | activated |
| ARNTL | Yes | inhibited |
| SERPINE1 | No | activated |
| DBP | Yes | activated |
| TBL1X | Yes | inhibited |
| RAF-independent MAPK1/3 activation | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| DUSP2 | Yes | activated |
| DUSP10 | Yes | activated |
| DUSP5 | Yes | activated |
| MAP2K2 | Yes | activated |
| DUSP1 | Yes | activated |
| HDMs demethylate histones | ||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information |
| PHF8 | Yes | activated |
| ARID5B | Yes | activated |
| KDM4B | Yes | activated |
| JMJD6 | Yes | activated |
A table summarizing the HSF1 target genes identified using the database HSF1base. Selected HSF1 target genes are ordered by functional categories, based on PANTHER analyses. A target gene is considered novel if it had not previously been characterized as a HSF1 target in a single-gene-analysis study. Regulatory information is ‘activated’ or ‘inhibited’ if there is evidence for HSF1 dependent up- or down-regulation of the expression of the given gene. ‘N/D’ means there is no regulatory information available.
Results obtained by the PANTHER analysis of cell type-specific HSF1 target genes were different from the ‘shared’ and ‘core’ target genes. For example, target genes specific to cervical adenocarcinoma cells were most enriched for PANTHER molecular function term DNA binding, bending (e.g., HIST2H2BE, NCAPD3, and NUSAP1). Consistently, this gene set was enriched for genes associated with the PANTHER protein class term histone (HIST2H2AA3, H1FX, and HIST2H2BE) and genes associated with Reactome pathways terms such as the TGF-beta signalling pathway (e.g., SMAD3, BMP4, TGFBR1, and SMAD7) and integrin signalling pathway (COL4A6, ITGAM, ITGB4, FN1 and MAPK8). In these cases, the majority of term-linked genes have not been identified as a HSF1 target in single-gene-analysis studies (Figure 2E, Tables S6 and S7).
Contrary to written above, a PANTHER analysis on HSF1 target genes in erythrocarcinoma cells did not reveal cell-specific enrichment of PANTHER terms associated with a function of HSF1 beyond maintaining proteostasis (Figure 2F, Tables S6 and S7). However, overrepresentation of Reactome-pathway-associated genes was observed, including WNT5A-dependent internalization of FZD4 (e.g., AP2A1, FZD4, PRKCB, and CLTC), clathrin-mediated endocytosis (e.g., BIN1 and EPS15) (Figure 2F, Tables S6 and S7). Breast epithelium-specific target genes statistically over-represented for the term of cell adhesion biological process (e.g., cadherin coding genes (CDH13, CDH12, and CDH3), Tensin-1 (TNS1), Nectin-4 (NECTIN4), and Teneurin-2 (TENM2)), and cadherin binding molecular function (e.g., CTNND1, CDH4, PLEC, and CTNND2). In this gene set, the most enriched protein class term was intermediate filament (e.g., catenin delta-encoding genes (CTNND1 and CTNND2) and plakophilin-encoding genes (PKP1 and PKP2)) (Figure 2G, Tables S6 and S7). In sum, these data suggest a novel function for HSF1 in cell adhesion specific to breast epithelium.
2.4. Cell Type-Specific and General Ageing-Related HSF1 Target Genes in Human
Since HSF1 is known to regulate the ageing process [17,18], we tested whether its target genes in any of the three cell types (adenocarcinoma, erythrocarcinoma, and breast epithelium) are associated with longevity. By overlapping the human HSF1 targets with ageing-related genes extracted from the GenAge database [34], we revealed that 117 HSF1 targets (2.08% of all genes) were previously implicated in ageing (Figure 2B and Table S8). These targets include genetic factors for insulin/IGF1 signalling (e.g., IGF1R, IGFBP2, IRS2, INSR, FOXO1, and GSK3B), stress response pathways (e.g., SOD1, PRDX1, HSPA1A, HSPA8, HSPA1B, and HSPA9), and DNA damage repair pathways (e.g., PRKDC, MLH1, POLA1, RAD52, ERCC1, and RECQL4). Out of the 10 ageing-related ‘core’ HSF1 target genes, five codes for heat shock proteins (HSP90AA1, HSPA8, HSPD1, HSPA1B, and HSPA1A) determine components of the ubiquitin-proteasome system (UBE2I and UBB), while the others encode the EGF- (epidermal growth factor) containing fibulin-like extracellular matrix protein 1 (EFEMP1), calcium voltage-gated channel subunit alpha1 A (CACNA1A), and a Jun proto-oncogene, the AP-1 transcription factor subunit (JUN). Taken together, these results convincingly show that many human HSF1 target genes are related to the ageing process.
2.5. ‘Shared’ and Cell Type-Specific HSF1 Target Genes in Mice
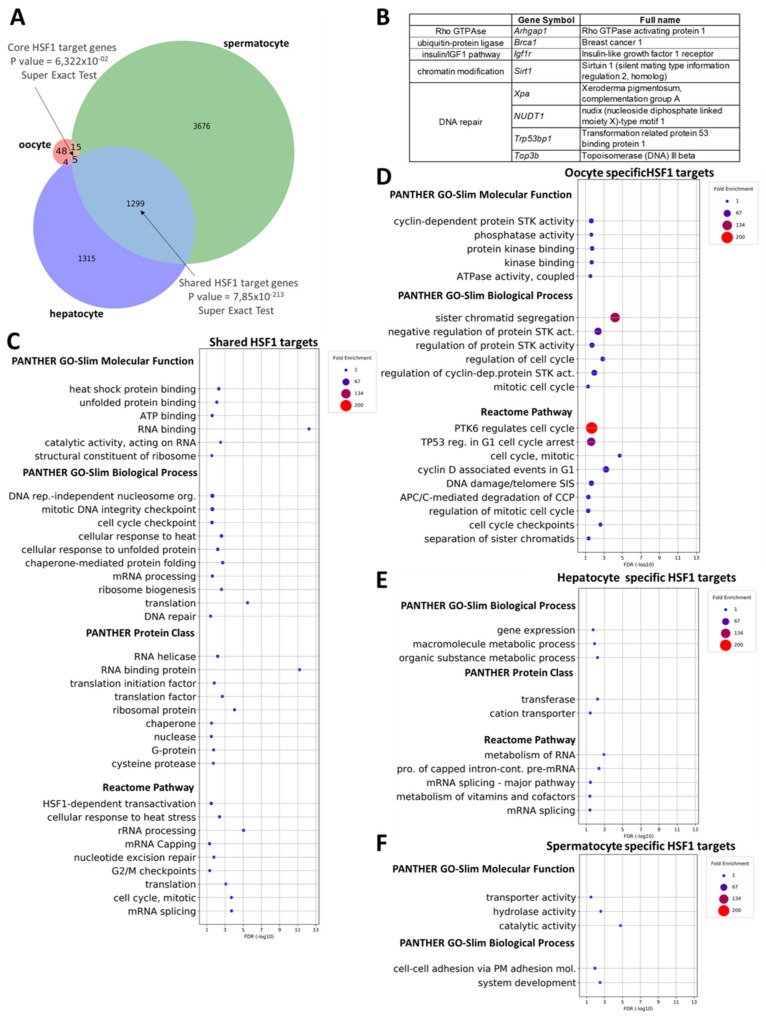
To support the existence of tissue-specific functions for HSF1, we compared HSF1 targets from three different murine (mouse) cell types, spermatocytes, oocytes, and hepatocytes. Similar to human tissues, we observed both ‘shared’ and cell type-specific HSF1 target genes in these cell types (Figure 3A, Tables S4 and S9). We identified numerous cell type-specific potential HSF1 targets: 3676 genes in spermatocytes, 48 genes in oocytes, and 1315 genes in hepatocytes. Interestingly, only five HSF1 target genes were ‘shared’ by the three cell types (Ubc, Tex11, Msh3, Ubb, and Cit). The reason for this is probably the high number of oocyte-specific HSF1 target genes, implying an oocyte-specific function for HSF1. Only 24 HSF1 targets identified in oocytes are ‘shared’ by at least two different cell types, suggesting that HSF1 functions in oocytes significantly differ from the general function of HSF1. Similar to human cell types, we observed a great number (1318; 20.72% of all targets) of ‘shared’ HSF1 targets in at least two different murine cell types (Figure 3A, Tables S4 and S9 ).
Figure 3.
HSF1 binds tissue-specific and shared targets in mouse. (A) Venn-diagram representing overlaps between HSF1 target genes in all cell types analyzed. Overlaps between two datasets (blue and grey) are statistically significant (p = 6.322 × 10−2 and p = 7.85 × 10−213, super exact test; 20,993 unique mouse genes were used as a background); (B) HSF1 targets involved in ageing are shared between the three cell types; (C) PANTHER GO-Slim Molecular Function, PANTHER GO-Slim Biological Process, PANTHER Protein Class and Reactome Pathway analysis of the 1318 HSF1 target genes shared between at least two cell types; (D) PANTHER GO-Slim Molecular Function, PANTHER GO-Slim Biological Process and Reactome Pathway analysis of the 48, oocyte-specific HSF1 target genes; (E) PANTHER GO-Slim Biological Process, PANTHER Protein Class and Reactome Pathway analysis of 1315 hepatocyte-specific HSF1 target genes; (F) PANTHER GO-Slim Molecular Function and PANTHER GO-Slim Biological Process analysis of 3676 spermatocyte-specific HSF1 target genes. Filled circles depict statistically significant enrichment; X axis shows -LOG10 (p-value) p < 0.05; Fisher’s exact test with Benjamini–Hochberg false discovery rate correction (FDR).
PANTHER gene set enrichment analysis (GSEA) of ‘shared’ mouse HSF1 targets revealed that most of the significantly enriched GO terms are related to the classical function of HSF1 in heat shock response (including heat shock protein binding (e.g., Hspa1a, Ptges3, Hspa5, and Stip1) and cellular response to unfolded protein (e.g., Hspa8, Hspa9, and Hspa5)] (Figure 3C, Tables S6 and S9). ‘Shared’ target genes were also enriched for biological process terms associated with RNA metabolism such as RNA binding (e.g., Rpl4, Zfand2a, Setx, Srek1, Rbm27, Polr2d, Nop9, Celf1, and Slbp), ribosome biogenesis (e.g., Rpl7l1, Rpl7a, Rrp1b, and Xpo1) and mRNA processing (e.g., Sltm, Srrm4, Dusp11, Cpsf4, and Pum1) (Figure 3C, Table 3, Tables S6 and S9). Many of these HSF targets have not been identified in a single-gene-analysis experiment, but high throughput analysis has predicted them, except for Zfand2a, as genes repressed directly by HSF1 (Table S6). It is quite intriguing that ‘shared’ HSF1 targets are enriched in several cell cycle checkpoint-associated terms such as mitotic DNA integrity checkpoint (e.g., Topbp1, Orc1, Rad17, and Cdc6) and G2/M checkpoints (Rad1, Cdc6, and Ccnb2), pointing to a role of HSF1 in the regulation of cell cycle (Figure 3C, Table 3, Tables S6 and S9).
Table 3.
Selected list of HSF1 target genes identified in murine cell types, using the HSF1Base.
| Murine ’Shared’ HSF1 Target Genes | |||
|---|---|---|---|
| Ribosome biogenesis | |||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information | |
| RPL7L1 | Yes | inhibited | |
| XPO1 | Yes | inhibited | |
| G2/M Checkpoints | |||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information | |
| TOPBP1 | Yes | inhibited | |
| RAD1 | Yes | inhibited | |
| UBC | No | activated | |
| Oocyte | |||
| Sister chromatid segregation | |||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulatory information | |
| STAG3 | Yes | inhibited | |
| STAG2 | Yes | activated | |
| Spermatocyte | |||
| Cell–cell adhesion via plasma-membrane adhesion molecules | |||
| Official Gene Symbol | Novelty (target not characterized in detail) | Regulated directly by HSF1 | Regulatory information |
| CRB1 | Yes | N/D | N/D |
| TENM4 | Yes | N/D | N/D |
| CDH19 | Yes | N/D | N/D |
| CCDC141 | Yes | N/D | N/D |
| CDH12 | Yes | N/D | N/D |
| CDH18 | Yes | N/D | N/D |
| SDK1 | Yes | N/D | N/D |
| CNTN5 | Yes | N/D | N/D |
| CDH6 | Yes | N/D | N/D |
| CDH26 | Yes | N/D | N/D |
| NECTIN1 | Yes | N/D | N/D |
| CDH15 | Yes | N/D | N/D |
| CDH13 | Yes | N/D | N/D |
| CNTN6 | Yes | N/D | N/D |
| CDH2 | Yes | N/D | N/D |
| NECTIN3 | Yes | N/D | N/D |
| CDH10 | Yes | N/D | N/D |
| MYPN | Yes | N/D | N/D |
| CDH4 | Yes | N/D | N/D |
A table summarizing the HSF1 target genes identified, using the database HSF1base. Selected HSF1 target genes are ordered by functional categories, based on PANTHER analyses. A target gene is considered novel if it had not previously been characterized as a HSF1 target in a single-gene-analysis study. Regulatory information is ‘activated’ or ‘inhibited’ if there is evidence for HSF1 dependent up- or down-regulation of the expression of the given gene. ‘N/D’ means there is no regulatory information available.
Oocyte-specific HSF1 target genes were enriched in biological processes including sister chromatid segregation (e.g., Stag2, Bub1b, Stag3, and Ddx11), negative regulation of protein serine/threonine kinase activity (e.g., Cdkn1b, Dusp1, and Cdkn1c) and several Reactome pathways were associated with mitotic cell cycle regulation (Figure 3D, Tables S6 and S9). These results highlight a role for HSF1 in mitotic cell division and oogenesis.
In the spermatocyte-specific HSF1 target gene set, the most enriched term is cell adhesion via plasma-membrane adhesion molecules. HSF1 target genes associated with this GO term include those coding for cadherins (Cdh4, Cdh19, Cdh15, Cdh26, Cdh2, and Cdh10), nectins (Nectin1 and Nectin3), and teneurin (Tenm4). This unexpected finding can be explained by the fact that samples used for Chip-seq in the source study were contaminated by round spermatids [35]. Round spermatids in turn are associated with Sertoli cells, suggesting that HSF1 may play a role in controlling Sertoli-germ cell adhesion or in sperm-oocyte interaction during fertilization (Figure 3E, Table 3, Tables S6 and S9). In the hepatocyte-specific HSF1 target gene set, a significant enrichment was detected in terms associated with RNA metabolism and hepatic functions such as Reactome pathway terms mRNA splicing (e.g., Polr2g, Clp1, Hnrnp,l and Ddx23) and metabolism of vitamins and cofactors (e.g., Ttpa, Nt5e, Nnmt, and Slc5a6) (Figure 3F, Tables S6 and S9).
2.6. Identifying HSF1 Target Genes Related to Ageing in Mouse
To identify putative HSF1 target genes involved in the ageing process of mice, we overlapped the mouse HSF1 target list with murine ageing-related genes provided by the GenAge database. We observed that there are 44 HSF1 targets (0.7% of all genes) associated with ageing in mouse (Figure 3B and Table S10). These genes involve components of the insulin/IGF1 signalling pathway (e.g., Igf1r and Insr), mTORC1-mediated signalling (e.g., Atm and Mtor), cellular stress response pathways (e.g., Atm, Mtor, Tp53, Prdx1, and Sirt1), and DNA damage repair (Msh2, Neil1, Atm, Tp53bp1, Brca1, Xpa and Ercc4) (Figure 3B and Table S10). The eight ageing-related ‘shared’ HSF1 targets include Igf1r, Brca1, Sirt1, Top3b, Arhgap1, Xpa, NUDT1, and Trp53bp1. Altogether, these results further support the role of HSF1 in the ageing process of mice.
2.7. Novel Conserved HSF1 Target Genes
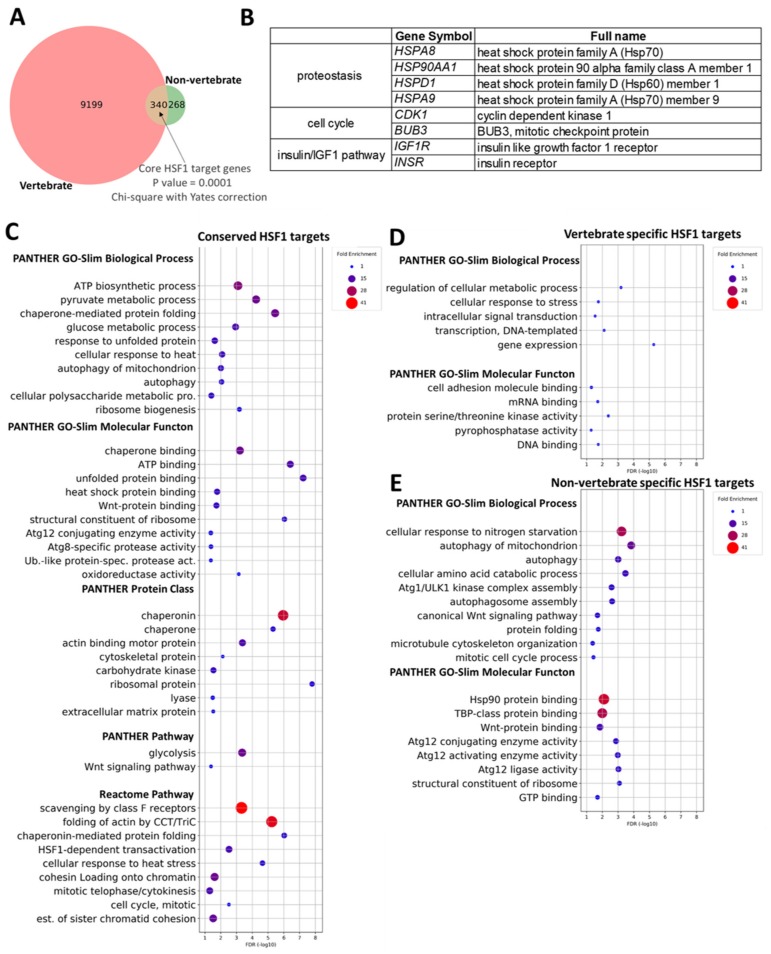
To identify novel potential HSF1 target genes based on conservation throughout evolution, we first performed a cross-species analysis by determining the human orthologues of HSF1 targets in S. cerevisiae, D. melanogaster, C. elegans, R. norvegicus, and M. musculus (see Materials and Methods). In total, 9807 orthologous gene pairs were identified (Figure 4A and Table S5). Next, we divided these genes into two taxonomic subgroups: vertebrates and non-vertebrates. The vertebrate group contains human HSF1 targets and human orthologues of murine HSF1 target genes. The non-vertebrate group contains human orthologues of HSF1 target genes identified in worms, flies, and yeast. We found that some HSF1 target genes (340, 4% of vertebrate and 56% of non-vertebrate target genes) were ‘shared’ by the subgroups, while others were unique to a single subgroup; 9199 out of 9539 (96%) unique to vertebrate, and 268 out of 608 (44%) unique to non-vertebrate (Figure 4A and Table S5).
Figure 4.
HSF1 targets are conserved between vertebrate and non-vertebrate species. (A) Venn-diagram depicting overlaps between HSF1 target genes in vertebrate (H. sapiens, R. norvegicus, and M. musculus) and non-vertebrate (C. elegans, D. melanogaster, and S. cerevisiae) species, (p = 0.0001, Chi-square with Yates correction; all unique vertebrate and non-vertebrate orthologues were used as background; 96,072 genes); (B) examples of HSF1 targets involved in ageing that are shared among groups vertebrate and non-vertebrate; (C) PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class, PANTHER Pathway, and Reactome Pathway analysis of 340 HSF1 target genes conserved between vertebrate and non-vertebrate species; (D) PANTHER GO-Slim Biological Process and PANTHER GO-Slim Molecular Function analysis of 9199 vertebrate-specific HSF1 target genes; (E) PANTHER GO-Slim Biological Process and PANTHER GO-Slim Molecular Function analysis of 268 non-vertebrate-specific HSF1 target genes. Filled circles depict statistically significant enrichment; X axis shows -LOG10 (p-value) p < 0.05; Fisher’s exact test with Benjamini–Hochberg False Discovery Rate correction (FDR).
The ‘shared’ HSF1 targets present in both subgroups were enriched for GO biological process terms associated with the classical role of HSF1, including proteostasis [like chaperone-mediated protein folding (e.g., DNAJB5/dnj-13/SIS1, HSPA5/hsp-3/KAR2, HSPA8/hsp-1, and SGTB/SGT2)], as well as terms such as chaperone binding, chaperonin HSF1-dependent transactivation, and cellular response to heat stress (Figure 4C, Tables S6 and S11). Interestingly, other significantly enriched HSF1 target genes are represented among biological process terms such as mitophagy (degradation of mitochondria by the lysosomal system) (ATG2B/atg-2, FIS1/fis-1, ATG9A/atg-9, RB1CC1/epg-7, and ATG2A/atg-2), Atg8-specific protease activity (atg9a/atg-9, ATG101/epg-9, ATG2A/atg-2, atg2b/atg-2, and RB1CC1/epg-7), ATP biosynthetic process (e.g., PFKL/pkf-1.1, GAPDH/gpd-2, PFKM/pkf-1.1, and PFKP/pkf-1.1), ribosome biogenesis (e.g., RPL35A/rpl-33, RPS27/rps-27, SART1/F19F10.9, RPS7/rps-7, RPS21/rps-21, BTRC/lin-23, and RPS14/rps-14), and molecular function terms such as Wnt-protein binding (e.g., FZD1/mom-5, ROR2/cam-1 and MIR4683/cfz-2) (Figure 4C, Table 4, Tables S6 and S11). In many cases, the mouse orthologue of human-C. elegans orthologue pairs mentioned above was also found in the ‘shared’ target gene set (see for example, Atg8-specific protease activity (ATG9A/Atg9a/atg-9, ATG101/Atg101/epg-9, ATG2A/Atg2a/atg-2, and ATG2B/Atg2b/atg-2)) (Table S6). These results were consistent with the finding that ‘shared’ HSF1 target genes are enriched for PANTHER protein classes such as ribosomal protein and extracellular matrix protein (e.g., LRIG2/sma-10, SART1/F19F10.9, SDC3/Y49E10.10, and ADAMTS1/T19D2.1) and pathways such as Wnt signalling (e.g., FZD1/mom-5, PRKCE/pkc-1, BTRC/lin-23, and PPP3CC/tax-6) (Figure 4C, Table 4, Tables S6 and S11). Hence, HSF1 may play diverse conserved roles beyond its canonical function as a guardian of proteostasis.
Table 4.
Selected list of HSF1 target genes conserved between vertebrate and non-vertebrate species, identified by using HSF1Base.
| HSF1 Target Genes Conserved between Vertebrate and Non-Vertebrate Species | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | Worm | Fly | Yeast | ||||||
| autophagy | |||||||||||
| Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. |
| FIS1 | N/D | N/D | N/D | N/D | N/D | fis-1 | N/D | N/D | N/D | N/D | N/D |
| atg9a | N/D | atg9a | N/D | atg9a | N/D | atg-9 | N/D | N/D | N/D | N/D | N/D |
| ATG2A | N/D | ATG2A | N/D | N/D | N/D | atg-2 | N/D | N/D | N/D | N/D | N/D |
| HSPA8 | act. | HSPA8 | act. | N/D | N/D | HSP-1 | act. | N/D | N/D | N/D | N/D |
| atg2b | N/D | atg2b | N/D | N/D | N/D | atg-2 | N/D | N/D | N/D | N/D | N/D |
| RB1CC1 | N/D | N/D | N/D | N/D | N/D | epg-7 | N/D | N/D | N/D | N/D | N/D |
| ribosome biogenesis | |||||||||||
| Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. |
| RPL35A | N/D | RPL35A | N/D | N/D | N/D | rpl-33 | N/D | N/D | N/D | N/D | N/D |
| RPS27 | N/D | N/D | N/D | N/D | N/D | rps-27 | N/D | N/D | N/D | N/D | N/D |
| SART1 | N/D | SART1 | N/D | N/D | N/D | F19F10.9 | N/D | N/D | N/D | N/D | N/D |
| RPS7 | N/D | N/D | N/D | N/D | N/D | rps-7 | N/D | N/D | N/D | N/D | N/D |
| RPS21 | N/D | RPS21 | N/D | N/D | N/D | rps-21 | N/D | N/D | N/D | N/D | N/D |
| BTRC | N/D | N/D | N/D | N/D | N/D | lin-23 | N/D | N/D | N/D | N/D | N/D |
| RPS14 | act. | N/D | N/D | N/D | N/D | rps-14 | N/D | N/D | N/D | N/D | N/D |
| RPL26 | N/D | RPL26 | N/D | N/D | N/D | rpl-26 | N/D | N/D | N/D | N/D | N/D |
| HEATR3 | N/D | HEATR3 | N/D | N/D | N/D | N/D | N/D | CG10286 | act. | N/D | N/D |
| MRTO4 | N/D | N/D | N/D | MRTO4 | N/D | N/D | N/D | N/D | N/D | MRT4 | act. |
| RPL26L1 | N/D | RPL26 | N/D | N/D | N/D | rpl-26 | N/D | N/D | N/D | N/D | N/D |
| Glycolysis | |||||||||||
| Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. |
| PGAM1 | N/D | PGAM1 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | GPM1 | act. |
| GPI | N/D | Gpi1 | N/D | N/D | N/D | N/D | N/D | Pgi | act. | N/D | N/D |
| PGK1 | N/D | PGK1 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | PGK1 | act. |
| Wnt signalling pathway | |||||||||||
| Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. |
| SMAD1 | N/D | SMAD1 | inh. | N/D | N/D | sma-2 | N/D | N/D | N/D | N/D | N/D |
| PPP3CB | N/D | PPP3CB | inh. | N/D | N/D | tax-6 | N/D | N/D | N/D | N/D | N/D |
| FBXW11 | inh. | FBXW11 | N/D | N/D | N/D | lin-23 | N/D | N/D | N/D | N/D | N/D |
| Establishment of Sister Chromatid Cohesion | |||||||||||
| Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. | Off. gene symb. | Reg. info. |
| SMC3 | N/D | SMC3 | N/D | N/D | N/D | N/D | N/D | N/D | N/D | SMC3 | act. |
A table summarizing the HSF1 target genes identified by using the database HSF1base. Selected HSF1 target genes are ordered by functional categories based on PANTHER analyses. A target gene is considered novel if it had not previously been characterized as a HSF1 target in a single-gene-analysis study. ‘Off. gene symb.’ stands for official gene symbol. ‘Reg. info.’ stands for regulatory information. ‘act.’ stands for activated, ‘inh.’ stands for inhibited. Regulatory information is activated or inhibited if there is evidence for HSF1 dependent up- or down-regulation of the expression of the given gene. ‘N/D’ means there is no regulatory information available.
Next, we analyzed HSF1 target genes being unique to each subgroup to find out whether HSF1 acquired or lost specific functions during evolution. PANTHER analysis of the Vertebrate HSF1 gene set showed only a moderate enrichment for specific terms (e.g., 1.89-fold enrichment in cell adhesion molecule binding consisting of 44 putative HSF1 target genes that encode proteins such as cadherins, nectins, teneurins, and plakophilins). We identified nearly half of these genes (26 out of 44) as HSF targets in both mice and human (Figure 4D, Tables S6 and S11). Nevertheless, the non-vertebrate gene set was significantly enriched in PANTHER terms such as autophagy (e.g., lgg-1, lgg-2, and atg-18) and mitotic cell cycle process (mec-12, mec-7, T08D2.7, and R02F2.1) in C. elegans. A closer look at the aforementioned autophagy-related genes revealed that they were directly inhibited by HSF-1 in C. elegans [9,12].
2.8. Ageing-Related HSF1 Target Genes Conserved Throughout Evolution
We further asked whether ‘shared’ orthologous targets in the groups vertebrate and non-vertebrate are associated with ageing. We found that out of the 340 ‘shared’ target genes, nine (2.6% of the ‘shared’ orthologue pairs) were related to ageing according to the GenAge database. These genes code for heat shock proteins (HSPA8, HSP90AA1, HSPD1, and HSPA9), regulators of cell cycle (CDK1 and BUB3) components of insulin/IGF1 signalling (INSR and IGF1R) and the zinc metallopeptidase ZMPSTE24, highlighting the fact that HSF1 may play a role in ageing via maintaining cellular proteostasis and affecting the insulin/IGF1 pathway (Figure 4B and Table S12).
3. Discussion
In this work, we established a database called HSF1base that contains a comprehensive list of HSF1 target genes identified so far by single gene analyses and high throughput experiments. The PSI-MITAB 2.8 format makes the HSF1base to be an easy-to-use systems-level resource. The database provides 15,641 unique HSF1 gene interactions and contains 1321 HSF1-bound target genes, the expression of which is regulated in a HSF1-dependent manner (774 activated and 547 inhibited by the transcription factor). The target genes derive from several species such as S. cerevisiae, C. elegans, D. melanogaster, M. musculus, R. norvegicus, and H. sapiens. The HSF1base website (hsf1base.org) serves as a graphical interface and provides a user-friendly environment for the scientific community to interactively search, browse or download the database. The database was made in August 2019, and it will be updated regularly (in every half year).
Like most bioinformatics resources relying on high throughput data, the HSF1Base also has several limitations. Some of the HSF1 target gene interactions may not be functional or may function only under specific circumstances. This is caused by the fact that transcriptional activity of HSF1 highly depends on the nature and magnitude of cellular stress, as well as the type and actual state (cell cycle phase, metabolic, and differentiation status) of the affected cell [2,18,36]. It has been shown for example that HSF1 regulates different sets of target genes during cellular stress response, development, and tumorigenesis [12,15,16,37,38]. Thus, users should keep in mind that further experimental validation is required to confirm these interactions. Despite these limitations, useful information can be obtained for researchers working on the terrain of HSF1 biology, heat-shock proteins, and ageing.
According to the HSF1Base, a great number of HSF1 target genes (547 out of 1321) are down-regulated. This number is surprisingly high since a great majority of studies identified HSF1 as a transcriptional activator. In several publications, however, HSF1 has been described as a transcriptional repressor [39,40,41,42,43]. Moreover, in murine fibroblast cells HSF1 has been also shown to play a role in maintaining an open chromatin state in the proximity of IL-6 gene, thereby endorsing the accessibility of other transcription factors to this regulatory region [44]. It is possible that HSF1 represses its target genes through modifying their chromatin structure. HSF1 was also reported to co-regulate a developmental program with E2F/DP transcription factors in C. elegans [12]. Based on this observation it is possible that HSF1 may repress genes in collaborating with other transcription factors.
To illustrate the applicability of the HSF1Base, we analyzed ‘shared’ and cell type-specific targets in human and murine cell types, using PANTHER. Our analysis showed that in both human and mouse cell types the main role of HSF1 is the regulation of proteostasis. This result is consistent with our general knowledge on HSF1 function, and supports relevant information on HSF1 functions that can be obtained using the HSF1Base. However, we also identified putative target genes shared by different cell types in human and mouse which play a role in diverse biological processes such as regulation of the circadian rhythm, chromatin modification, mitotic cell cycle, and RNA metabolism (Figure 1D and Figure 2C, Table 2 and Table S6).
HSF1 is induced upon cell stressors such as UV light, oxidative stress and heat stress, and triggers the synchronization of the circadian clock via directly regulating the core clock gene Per2 [45,46,47]. According to the HSF1Base, some of the putative HSF1 targets associated with the circadian rhythm are upregulated (RXRA, NOCT, BHLHE40, SERPINE1, and DBP) while others are inhibited (NCOA2, ARNTL, and TBL1X) by the transcription factor (Figure 2D, Table 2 and Table S6). Among the eight targets listed above, only SERPINE1 was previously described as a HSF1-regulated direct target gene [48].
It has been shown that after heat stress, HSF1 interacts with HDAC1 and HDCA2 histone deacetylases [49]. Thus, it may also function as a master regulator of stress-induced chromatin deacetylation. In good accordance with this assumption, here we identified several HSF1 targets which code for lysine (KDM6A, KDM4B, and KDM1A) or arginine (JMJD6) demethylases. HSF1 may thus directly activate KDM4B and JMJD6 genes. Based on these data one may predict that HSF1 influences stress-induced chromatin reorganization via transcriptionally controlling the two genes. Indeed, HSF1 binds several (23) histone-related genes, two of which (Hist2h2aa3 and HIST2H2BE) are directly upregulated by HSF1 (Figure 2D, Table 2 and Table S6).
Among the ‘shared’ human HSF1 targets, we identified several genes coding for extracellular and matrix structural proteins (CRELD1, EFEMP1, LTBP4, and LTBP1) (Figure 2D, Table 2 and Table S6). We suggest that HSF1 influences the process of extracellular matrix remodeling. HSF1 has been indeed identified as a key regulator of idiopathic pulmonary fibrosis [50]. Silencing of HSF1 impaired the expression of fibrillar collagen (COL1A1 and COL1A2), and several genes required for the biogenesis of collagen fibers (P4HA2, PLOD1, and FKBP4) in pulmonary fibroblasts. HSF1 was also implicated in pulmonary fibroblasts to regulate extracellular matrix-specific gene expression [50].
We also identified cell type-specific roles for HSF1. In both human breast epithelium and murine spermatocyte (or round spermatids; see before), we found that a significant number of HSF1 target genes are related to cell adhesion (Figure 1G and Figure 2F, Table 3 and Table S6). Products of these target genes include cadherins, teneurins, and nectins, present in both breast epithelium and spermatocyte-specific HSF1 target gene sets. A role of HSF1 in cell adhesion and migration has been observed in several studies. In highly malignant human cancer cell lines, HSF1 activates genes involved in cell adhesion and migration [16]. Moreover, FN1, the gene coding for the extracellular matrix protein fibronectin, was recently identified as a stress-responsive gene regulated by HSF1 [51]. In hepatocellular carcinoma cells, HSF1 modulates E-Cadherin expression through directly regulating Snail1 transcription [52]. Finally, in gastric cancer cells HSF1 overexpression stimulates vimentin, N-cadherin, and Snail1 expression, and decreases the level of E-cadherin [53]. Taken together, it is plausible that HSF1 regulates cadherin, teneurin, and nectin genes directly in human breast epithelium.
In mammalian testis, cadherins, α-, β- or γ-catenin and nectins are important components in Sertoli–Sertoli and Sertoli-germ cell adhesion [54]. Teneurins playing a significant role in cell adhesion and migration are expressed predominantly in the central nervous system [55,56]. Nevertheless, a recent study showed that teneurins are also expressed in adult mouse testes and regulate testicular size and testosterone production [57]. Although the role of HSF1 in spermatogenesis has been studied extensively [17,37,58,59,60], involvement of HSF1-regulated genes encoding cell adhesion molecules in this process has not been described yet.
Comparison of HSF1 target genes in three murine cell types showed that most target genes were found exclusively in oocytes (Figure 3D, Table 3 and Table S6). This suggest a highly specific role for HSF1 during oogenesis. Indeed, in yeast HSF1 was identified as an essential checkpoint component required for mitotic progression [61]. Oocyte-specific functions of HSF1 were also described [38]. The result of gene ontology analysis performed in this study also showed that in oocytes HSF1 binds genes encoding key regulators of mitosis, cytokinesis, and core components of various signaling pathways. Taken together, our analysis supports that HSF1 functions can vary in the context of cell type.
We also analyzed the possible evolutionary conservation of putative HSF1 target genes. The assay revealed that beyond the HSR numerous HSF1 functions emerged across various eukaryotic taxa. According to our analysis, HSF1 targets involve for example several autophagy-related genes (Figure 4C, Table 4 and Table S6). Although regulatory interaction between HSF1 and autophagy has been well documented [62,63,64,65,66], Atg genes identified by our analysis as HSF1 targets (ATG2B, FIS1, ATG9A, RB1CC1, and ATG2A; see Figure 4C, Table S6) had not been examined for control by the transcription factor. Interestingly, our present study pointed to the fact that HSF1 indeed represses several Atg genes in C. elegans. These data were provided by two recent studies, in which the authors used high-throughput methods to uncover targets of CeHSF-1 (C. elegans HSF1) during development [12] and at adulthood followed by heat shock [9]. Both analyses explored the repression of several Atg genes (lgg-1, lgg-2, atg-2, atg-9, atg-11, and atg-18) by HSF-1.
In this study we showed that numerous putative HSF1 target genes associated with ribosome biogenesis term (e.g., RPL35A/rpl-33, SART1/F19F10.9, RPS21/rps-21, RPL26/rpl-26, and HEATR3/ rpl-26) are evolutionarily conserved between mice and nematodes (Figure 4C, Table 4 and Table S6). Moreover, we identified several ribosome biogenesis-associated genes as HSF1 targets in both mouse hepatocytes and spermatocytes (Figure 3E and Figure 2F, Table S7). According to the HSF1base, among these genes RPL7L1 and XPO1 are directly repressed by HSF1. Relationship between HSF1 activity and ribosome biogenesis has become clear in the last years only [67,68]. It is possible that in fast-growing cells HSF1 attenuates cellular stress triggered by orphan (not integrated into a ribosome) ribosomal proteins and rRNAs through inhibiting or activating ribosomal genes.
The role of HSF1 in ageing has been well documented in C. elegans [69,70]. In other model organism, like Drosophila, overexpression of certain HSPs such as mitochondrial Hsp22 extends lifespan [71]. Furthermore, mutant mice defective for co-chaperone (CHIP) activity age faster than normal [72], and certain HSP-encoding genes become upregulated in long-lived mutant mouse strains [73]. Thus, it seems plausible that HSF1 influences ageing via controlling HSP gene activity. However, in C. elegans HSF1 affects also the activity of the longevity pathway TGFβ (transforming growth factor-beta) signaling [39], and autophagy [64]. Moreover, overexpressing a hypomorphic mutant allele of hsf-1 that is not capable of inducing hsp genes also increases the survival of C. elegans [74]. In this study we identified several HSF1 targets that influence ageing independently of the HSR. Such genes encode for example a fibulin-like extracellular matrix protein (EFEMP1) [75,76] in different human tissues, a calcium voltage-gated channel subunit alpha1 A protein (CACNA1A) [77,78,79], and AP-1 transcription factor subunit JUN [80,81,82,83] (Figure 2B, Table S8). Among these factors, only JUN has been identified as a direct HSF1 target [29]. By comparing potential HSF1 targets in different mouse cell types we further identified eight genes that may modulate lifespan in a HSF1-dependent manner (Figure 3B, Table S10). The regulatory relationship between HSF1 and these potential targets has not been examined in single-gene-analysis studies. In addition, by monitoring the evolutionary conservation of potential HSF1 target genes we recognized five other genes that may modulate the rate at which cells age (Figure 4B, Table S12).
4. Materials and Methods
4.1. Origin of Datasets Used
Data for HSF1 target genes were obtained from 117 publications (see Table S1). PANTHER analysis of high throughput data was obtained from 14 publications (see Table S1).
4.2. Statistical Overrepresentation Tests
Statistical overrepresentation tests (PANTHER GO-Slim Biological Process, PANTHER GO-Slim Molecular Function, PANTHER Protein Class, PANTHER Pathways and Reactome pathways) of gene sets were performed using the PANTHER database (http://PANTHERdb.org/, Accessed on 12 September 2019) [84]. Gene lists were uploaded in Ensemble Gene ID format, and default whole genome lists from the appropriate species were used as reference. To analyze statistical significance, Fisher’s exact test with Benjamini–Hochberg False Discovery Rate correction (FDR) was applied [85]. Over-represented terms were plotted by Matplotlib using Python.
4.3. Orthologue Analysis
Mus musculus, Ratus norvegicus, Caenorhabditis elegans, Saccharomyces cerevisiae, and Drosophila melanogaster orthologs of human genes were obtained from the OMA orthology database [86], using Python.
4.4. Venn Diagrams and Statistical Analysis of Overlaps
Venn diagrams were created in Python with Matplotlib. Statistical analysis for the significance of overlaps was performed using super exact test or Chi-square test with Yates correction in R [87]. For tests within a species, all unique genes within the given species were used as background. For the cross-species analysis, only the genes with orthologues (human–C. elegans, human–D. melanogaster, human–R. norvegicus, or human–M. musculus orthologues) were used as a background.
5. Conclusions
In the last two decades a great number of novel functions were assigned to HSF1. The protein has been implicated in physiological and pathological processes such as cell cycle, apoptosis, circadian rhythm, immune response, as well as in ageing, and many age-related diseases, including cancer and neurodegenerative pathologies. The growing amount of high throughput data and the increasing interest in the scientific community on HSF1 justifies the development of a comprehensive, well-organized database of HSF1 targets. We believe that HSF1Base can be used as a resource to discover novel functions of HSF1.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/22/5815/s1. Table S1: List of publications used to establish the database. ‘Relevant publications on individual HSF1 target genes’: List of HSF1-related publications on single gene analyses used. ‘Publications containing high throughput data’: List of publications HSF1-related publications containing high-throughput data used. Table S2: List of HSF1 target genes found in the database. ‘Statistics’: Table summarizing the HSF-1 targets found in Table S2. ‘Putative HSF1 targets’: List of all target genes included in ‘Unique interactions’ (see: Table 1). ‘Direct HSF1 targets’: List of ‘HSF1 targets with evidence for direct interaction’ included in ‘Unique interactions’ (see: Table 1). ‘Targets with HSF1 dep. expr’ (Targets with HSF1 dependent expression): List of ‘HSF1 targets with evidence for HSF1 dependent regulation’ included in ‘Unique interactions’ (see: Table 1). ‘Directly reg. HSF1 targets 1’ (directly regulated HSF1 targets 1): List of ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’ in a single publication (see: Table 1). ‘Directly reg. HSF1 targets 2’ (directly regulated HSF1 targets 2): List of ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’ in a single publication. The evidence for physical interaction between HSF1 and the gene and the regulatory information came from two separate publications (see: Table 1). ‘Directly regulated HSF1 targets’: The union of ‘Directly reg. HSF1 targets 1’ and ‘Directly reg. HSF1 targets 2’. The equivalent of the list of ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’ (see: Table 1). Table S3: HSF1 target genes in human by tissue/cell type. ‘Core targets’: List of HSF1 target genes shared between all three examined cell types in human. ‘Shared targets’: List of HSF1 target genes shared between at least two examined cell types in human. ‘Cervix adenocarcinoma’: List of cervix adenocarcinoma specific HSF1 targets in human. ‘Erythroleukemia cells’: List of erythroleukemia cells specific HSF1 targets in human. ‘Breast epithelium’: List of breast epithelium specific HSF1 targets in human. Table S4: HSF1 target genes in mouse by tissue/cell type. ‘Core targets’: List of HSF1 target genes shared between all three examined cell types in mouse. ‘Shared targets’: List of HSF1 target genes shared between at least two examined cell types in mouse. ‘Oocyte’: List of oocyte specific HSF1 targets in mouse. ‘Spermatocyte’: List of spermatocyte specific HSF1 targets in mouse. ‘Hepatocyte’: List of hepatocyte specific HSF1 targets in mouse. Table S5: Conserved and unique HSF1 target genes in groups vertebrate and non-vertebrate. ‘Vertebrate_Nonvertebrete_shared’: List of HSF1 target genes shared between the examined vertebrate and non-vertebrate species. ‘Vertebrate_unique’: List of HSF1 target genes only found in the examined vertebrate species. ‘Nonvertebrate_unique’: List of HSF1 target genes only found in the examined non-vertebrate species. Table S6: Selected HSF1 target genes over-represented in GO categories. ‘Selected HSF1 targets in human’: List of selected putative HSF1 target genes over-represented in GO categories in human. ‘Selected HSF1 targets in mouse’: List of selected putative HSF1 target genes over-represented in GO categories in mouse. ‘Selected conserved HSF1 targets’: List of selected conserved HSF1 target genes over-represented in GO categories shared between the examined vertebrate and non-vertebrate species. ‘Selected HSF1 targets in Verteb’ (Selected HSF1 targets in Vertebrate): List of selected conserved HSF1 target genes over-represented in GO categories in the examined vertebrate species. ‘HSF1 targets in Non-vertebrate’: List of selected conserved HSF1 target genes over-represented in GO categories in the examined non-vertebrate species. Table S7: PANTHER analyses of human HSF1 target genes by tissue/cell type. ‘Core targets’: PANTHER analyses of human HSF1 target genes shared between all three examined cell types. ‘Shared targets’: PANTHER analyses of human HSF1 target genes shared between at least two examined cell types. ‘Adenocarcinoma’: PANTHER analyses of cervix adenocarcinoma specific human HSF1 target genes. ‘Erythroleukemia’: PANTHER analyses of erythroleukemia specific human HSF1 target genes. ‘Breast epithelium’: PANTHER analyses of breast epithelium specific human HSF1 target genes. Table S8: HSF1 target genes related to ageing in human. ‘All human HSF1 targets’: List of all human HSF1 target genes related to ageing according to the GenAge database. ‘Directly regulated HSF1 targets’: List of human ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’ (see: Table 1) according to the GenAge database. Table S9: PANTHER analyses of mouse HSF1 target genes by tissue / cell type. ‘Core targets’: PANTHER analyses of mouse HSF1 target genes shared between all three examined cell types. ‘Shared targets’: PANTHER analyses of mouse HSF1 target genes shared between at least two examined cell types. ‘Oocyte’: PANTHER analyses of oocyte specific mouse HSF1 target genes. ‘Spermatocyte’: PANTHER analyses of spermatocyte specific mouse HSF1 target genes. ‘Hepatocyte’: PANTHER analyses of hepatocyte specific mouse HSF1 target genes. Table S10: HSF-1 target genes related to ageing in mice. ‘All mouse HSF1 targets’: List of all mouse HSF1 target genes related to ageing according to the GenAge database. ‘Directly regulated HSF1 targets’: List of mouse ‘HSF1 targets with evidence for direct interaction and HSF1 dependent regulation’ (see: Table 1) according to the GenAge database. Table S11: PANTHER analyses of conserved and unique HSF1 target genes in groups vertebrate and non-vertebrate. ‘Vertebrate’: PANTHER analyses of conserved and unique HSF1 target genes in the examined vertebrate species. ‘Non-vertebrate’: PANTHER analyses of conserved and unique HSF1 target genes in the examined non-vertebrate species. Table S12: HSF1 target genes related to ageing by organism and tissue/cell type. ‘Human’: List of HSF1 target genes related to ageing in human. ‘Mouse’: List of HSF1 target genes related to ageing in mouse. ‘Worm’: List of HSF1 target genes related to ageing in C. elegans. ‘Fly’: List of HSF1 target genes related to ageing in D. melanogaster. ‘Yeast’: List of HSF1 target genes related to ageing in S. cerevisiae.
Author Contributions
Conceptualization, J.B., and D.F.; methodology, J.B. and D.F.; software, D.F., and B.B.; formal analysis, B.H., and V.D.; investigation, D.K., T.S. and B.H.; resources, T.V.; data curation, T.S.; writing and original draft preparation, J.B. and T.V.; writing and review and editing, D.K., T.S., B.H., V.D., J.B. and T.V.; visualization, B.H.; funding acquisition, T.V.
Funding
This work was supported by the MEDinPROT Protein Science Research Synergy Program (provided by the Hungarian Academy of Sciences; HAS) and VEKOP (VEKOP-2.3.2-16-2017-00014) grant. This work was completed in the ELTE Institutional Excellence Program supported by the National Research, Development and Innovation Office (NKFIH-1157-8/2019-DT). Grant 2018-1.2.1-NKP-2018-00005 has been implemented with the support provided from the National Research, Development and Innovation. Fund of Hungary, financed under the 2018-1.2.1-NKP funding scheme. B.J and T.V. were also supported by the MTA-ELTE Genetics Research Group (01062).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lindquist S., Craig E.A. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 2.Akerfelt M., Morimoto R.I., Sistonen L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010;11:545–555. doi: 10.1038/nrm2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter K., Haslbeck M., Buchner J. The heat shock response: Life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Morimoto R.I. Cells in stress: Transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Pastor R., Burchfiel E.T., Thiele D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:4–20. doi: 10.1038/nrm.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neef D.W., Jaeger A.M., Thiele D.J. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat. Rev. Drug Discov. 2011;10:930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin J., Ananthan J., Voellmy R. Key features of heat shock regulatory elements. Mol. Cell. Biol. 1988;8:3761–3769. doi: 10.1128/MCB.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C. Heat shock transcription factors: Structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 9.Brunquell J., Morris S., Lu Y., Cheng F., Westerheide S.D. The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genom. 2016;17:559. doi: 10.1186/s12864-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn J.-S., Hu Z., Thiele D.J., Iyer V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birch-Machin I., Gao S., Huen D., McGirr R., White R.A.H., Russell S. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol. 2005;6:R63. doi: 10.1186/gb-2005-6-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Chauve L., Phelps G., Brielmann R.M., Morimoto R.I. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 2016;30:2062–2075. doi: 10.1101/gad.283317.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahat D.B., Salamanca H.H., Duarte F.M., Danko C.G., Lis J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinklein N.D., Murray J.I., Hartman S.J., Botstein D., Myers R.M. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol. Biol. Cell. 2004;15:1254–1261. doi: 10.1091/mbc.e03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Masson F., Razak Z., Kaigo M., Audouard C., Charry C., Cooke H., Westwood J.T., Christians E.S. Identification of heat shock factor 1 molecular and cellular targets during embryonic and adult female meiosis. Mol. Cell. Biol. 2011;31:3410–3423. doi: 10.1128/MCB.05237-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendillo M.L., Santagata S., Koeva M., Bell G.W., Hu R., Tamimi R.M., Fraenkel E., Ince T.A., Whitesell L., Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150:549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barna J., Csermely P., Vellai T. Roles of heat shock factor 1 beyond the heat shock response. Cell. Mol. Life Sci. 2018;75:2897–2916. doi: 10.1007/s00018-018-2836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Labbadia J., Morimoto R.I. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 2017;27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santagata S., Hu R., Lin N.U., Mendillo M.L., Collins L.C., Hankinson S.E., Schnitt S.J., Whitesell L., Tamimi R.M., Lindquist S., et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. USA. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai C., Whitesell L., Rogers A.B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee S., Burns T.F. Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 2017;18:1978. doi: 10.3390/ijms18091978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira L.M.R., Cunha-Oliveira T., Sobral M.C., Abreu P.L., Alpoim M.C., Urbano A.M. Impact of carcinogenic chromium on the cellular response to proteotoxic stress. Int. J. Mol. Sci. 2019;20:4901. doi: 10.3390/ijms20194901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnemolla A., Labbadia J.P., Lazell H., Neueder A., Moussaoui S., Bates G.P. Contesting the dogma of an age-related heat shock response impairment; implications for cardiac-specific age-related disorders. Hum. Mol. Genet. 2014;23:3641–3656. doi: 10.1093/hmg/ddu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perfetto L., Acencio M.L., Bradley G., Cesareni G., Del Toro N., Fazekas D., Hermjakob H., Korcsmaros T., Kuiper M., Lægreid A., et al. CausalTAB: The PSI-MITAB 2.8 updated format for signalling data representation and dissemination. Bioinformatics. 2019;35:3779–3785. doi: 10.1093/bioinformatics/btz132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb A.E., Kundaje A., Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016;15:673–685. doi: 10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Bhattacharyya N.P. Transcription regulation of HYPK by Heat Shock Factor. PLoS ONE. 2014;9:e85552. doi: 10.1371/journal.pone.0085552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song S., Kole S., Precht P., Pazin M.J., Bernier M. Activation of heat shock factor 1 plays a role in pyrrolidine dithiocarbamate-mediated expression of the co-chaperone BAG. Int. J. Biochem. Cell Biol. 2010;42:1856–1863. doi: 10.1016/j.biocel.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawai M., Ishikawa Y., Ota A., Sakurai H. The proto-oncogene JUN is a target of the heat shock transcription factor HSF1. FEBS J. 2013;280:6672–6680. doi: 10.1111/febs.12570. [DOI] [PubMed] [Google Scholar]
- 30.Lecomte S., Desmots F., Le Masson F., Le Goff P., Michel D., Christians E.S., Le Dréan Y. Roles of heat shock factor 1 and 2 in response to proteasome inhibition: Consequence on p53 stability. Oncogene. 2010;29:4216–4224. doi: 10.1038/onc.2010.171. [DOI] [PubMed] [Google Scholar]
- 31.Benderska N., Ivanovska J., Rau T.T., Schulze-Luehrmann J., Mohan S., Chakilam S., Gandesiri M., Ziesché E., Fischer T., Söder S., et al. DAPK-HSF1 interaction as a positive-feedback mechanism stimulating TNF-induced apoptosis in colorectal cancer cells. J. Cell Sci. 2014;127:5273–5287. doi: 10.1242/jcs.157024. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs A.T., Marnett L.J. HSF1-mediated BAG3 expression attenuates apoptosis in 4-hydroxynonenal-treated colon cancer cells via stabilization of anti-apoptotic Bcl-2 proteins. J. Biol. Chem. 2009;284:9176–9183. doi: 10.1074/jbc.M808656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Logan I.R., McNeill H.V., Cook S., Lu X., Meek D.W., Fuller-Pace F.V., Lunec J., Robson C.N. Heat shock factor-1 modulates p53 activity in the transcriptional response to DNA damage. Nucleic Acids Res. 2009;37:2962–2973. doi: 10.1093/nar/gkp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tacutu R., Craig T., Budovsky A., Wuttke D., Lehmann G., Taranukha D., Costa J., Fraifeld V.E., de Magalhães J.P. Human Ageing Genomic Resources: Integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korfanty J., Stokowy T., Widlak P., Gogler-Piglowska A., Handschuh L., Podkowiński J., Vydra N., Naumowicz A., Toma-Jonik A., Widlak W. Crosstalk between HSF1 and HSF2 during the heat shock response in mouse testes. Int. J. Biochem. Cell Biol. 2014;57:76–83. doi: 10.1016/j.biocel.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Vihervaara A., Sistonen L. HSF1 at a glance. J. Cell Sci. 2014;127:261–266. doi: 10.1242/jcs.132605. [DOI] [PubMed] [Google Scholar]
- 37.Akerfelt M., Vihervaara A., Laiho A., Conter A., Christians E.S., Sistonen L., Henriksson E. Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J. Biol. Chem. 2010;285:34469–34476. doi: 10.1074/jbc.M110.157552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vihervaara A., Sergelius C., Vasara J., Blom M.A.H., Elsing A.N., Roos-Mattjus P., Sistonen L. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. USA. 2013;110:E3388–E3397. doi: 10.1073/pnas.1305275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barna J., Princz A., Kosztelnik M., Hargitai B., Takács-Vellai K., Vellai T. Heat shock factor-1 intertwines insulin/IGF-1, TGF-β and cGMP signalling to control development and ageing. BMC Dev. Biol. 2012;12:32. doi: 10.1186/1471-213X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inouye S., Izu H., Takaki E., Suzuki H., Shirai M., Yokota Y., Ichikawa H., Fujimoto M., Nakai A. Impaired IgG production in mice deficient for heat shock transcription factor. J. Biol. Chem. 2004;279:38701–38709. doi: 10.1074/jbc.M405986200. [DOI] [PubMed] [Google Scholar]
- 41.Singh I.S., He J.-R., Calderwood S., Hasday J.D. A high affinity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-alpha gene is a transcriptional repressor. J. Biol. Chem. 2002;277:4981–4988. doi: 10.1074/jbc.M108154200. [DOI] [PubMed] [Google Scholar]
- 42.Xie Y., Zhong R., Chen C., Calderwood S.K. Heat shock factor 1 contains two functional domains that mediate transcriptional repression of the c-fos and c-fms genes. J. Biol. Chem. 2003;278:4687–4698. doi: 10.1074/jbc.M210189200. [DOI] [PubMed] [Google Scholar]
- 43.Wang J., He H., Yu L., Xia H.H.-X., Lin M.C.M., Gu Q., Li M., Zou B., An X., Jiang B., et al. HSF1 down-regulates XAF1 through transcriptional regulation. J. Biol. Chem. 2006;281:2451–2459. doi: 10.1074/jbc.M505890200. [DOI] [PubMed] [Google Scholar]
- 44.Inouye S., Fujimoto M., Nakamura T., Takaki E., Hayashida N., Hai T., Nakai A. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J. Biol. Chem. 2007;282:33210–33217. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- 45.Tamaru T., Hattori M., Honda K., Benjamin I., Ozawa T., Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1: CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS ONE. 2011;6:e24521. doi: 10.1371/journal.pone.0024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamaru T., Hattori M., Ninomiya Y., Kawamura G., Varès G., Honda K., Mishra D.P., Wang B., Benjamin I., Sassone-Corsi P., et al. ROS stress resets circadian clocks to coordinate pro-survival signals. PLoS ONE. 2013;8:e82006. doi: 10.1371/journal.pone.0082006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawamura G., Hattori M., Takamatsu K., Tsukada T., Ninomiya Y., Benjamin I., Sassone-Corsi P., Ozawa T., Tamaru T. Cooperative interaction among BMAL1, HSF1, and p53 protects mammalian cells from UV stress. Commun. Biol. 2018;1:204. doi: 10.1038/s42003-018-0209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S.-J., Sohn H.-Y., Park S.I. TRAIL regulates collagen production through HSF1-dependent Hsp47 expression in activated hepatic stellate cells. Cell Signal. 2013;25:1635–1643. doi: 10.1016/j.cellsig.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Fritah S., Col E., Boyault C., Govin J., Sadoul K., Chiocca S., Christians E., Khochbin S., Jolly C., Vourc’h C. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol. Biol. Cell. 2009;20:4976–4984. doi: 10.1091/mbc.e09-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones M., Wales C.T.K., Gristock R.A., Jacobs A.T. Regulation of extracellular matrix gene expression and biosynthetic pathways in pulmonary fibroblasts by HSF1. FASEB J. 2017;31:6311. [Google Scholar]
- 51.Dhanani K.C.H., Samson W.J., Edkins A.L. Fibronectin is a stress responsive gene regulated by HSF1 in response to geldanamycin. Sci. Rep. 2017;7:17617. doi: 10.1038/s41598-017-18061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D., Sun L., Qin X., Liu T., Zhang S., Liu Y., Li S., Guo K. HSF1 promotes the inhibition of EMT-associated migration by low glucose via directly regulating Snail1 expression in HCC cells. Discov. Med. 2016;22:87–96. [PubMed] [Google Scholar]
- 53.Dai W., Ye J., Zhang Z., Yang L., Ren H., Wu H., Chen J., Ma J., Zhai E., Cai S., et al. Increased expression of heat shock factor 1 (HSF1) is associated with poor survival in gastric cancer patients. Diagn. Pathol. 2018;13:80. doi: 10.1186/s13000-018-0755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao X., Mruk D.D., Cheng C.Y. Intercellular adhesion molecules (ICAMs) and spermatogenesis. Hum. Reprod. Update. 2013;19:167–186. doi: 10.1093/humupd/dms049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young T.R., Leamey C.A. Teneurins: Important regulators of neural circuitry. Int. J. Biochem. Cell Biol. 2009;41:990–993. doi: 10.1016/j.biocel.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 56.Kenzelmann D., Chiquet-Ehrismann R., Tucker R.P. Teneurins, a transmembrane protein family involved in cell communication during neuronal development. Cell Mol. Life Sci. 2007;64:1452–1456. doi: 10.1007/s00018-007-7108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chand D., Colacci M., Dixon K., Kollara A., Brown T.J., Lovejoy D.A. C-terminal region of teneurin-1 co-localizes with the dystroglycan complex in adult mouse testes and regulates testicular size and testosterone production. Histochem. Cell Biol. 2014;141:191–211. doi: 10.1007/s00418-013-1154-1. [DOI] [PubMed] [Google Scholar]
- 58.Izu H., Inouye S., Fujimoto M., Shiraishi K., Naito K., Nakai A. Heat shock transcription factor 1 is involved in quality-control mechanisms in male germ cells. Biol. Reprod. 2004;70:18–24. doi: 10.1095/biolreprod.103.020065. [DOI] [PubMed] [Google Scholar]
- 59.Widłak W., Benedyk K., Vydra N., Głowala M., Scieglińska D., Małusecka E., Nakai A., Krawczyk Z. Expression of a constitutively active mutant of heat shock factor 1 under the control of testis-specific hst70 gene promoter in transgenic mice induces degeneration of seminiferous epithelium. Acta Biochim. Pol. 2003;50:535–541. [PubMed] [Google Scholar]
- 60.Yang L., Wang Y., Zhang Q., Lai Y., Li C., Zhang Q., Huang W., Duan Y., Jiang Z., Li X., et al. Identification of Hsf1 as a novel androgen receptor-regulated gene in mouse Sertoli cells. Mol. Reprod. Dev. 2014;81:514–523. doi: 10.1002/mrd.22318. [DOI] [PubMed] [Google Scholar]
- 61.Lee Y.-J., Kim E.-H., Lee J.S., Jeoung D., Bae S., Kwon S.H., Lee Y.-S. HSF1 as a mitotic regulator: Phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 2008;68:7550–7560. doi: 10.1158/0008-5472.CAN-08-0129. [DOI] [PubMed] [Google Scholar]
- 62.Desai S., Liu Z., Yao J., Patel N., Chen J., Wu Y., Ahn E.E.-Y., Fodstad O., Tan M. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7) J. Biol. Chem. 2013;288:9165–9176. doi: 10.1074/jbc.M112.422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dokladny K., Zuhl M.N., Mandell M., Bhattacharya D., Schneider S., Deretic V., Moseley P.L. Regulatory coordination between two major intracellular homeostatic systems: Heat shock response and autophagy. J. Biol. Chem. 2013;288:14959–14972. doi: 10.1074/jbc.M113.462408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumsta C., Chang J.T., Schmalz J., Hansen M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 2017;8:14337. doi: 10.1038/ncomms14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Watanabe Y., Tsujimura A., Taguchi K., Tanaka M. HSF1 stress response pathway regulates autophagy receptor SQSTM1/p62-associated proteostasis. Autophagy. 2017;13:133–148. doi: 10.1080/15548627.2016.1248018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Samarasinghe B., Wales C.T.K., Taylor F.R., Jacobs A.T. Heat shock factor 1 confers resistance to Hsp90 inhibitors through p62/SQSTM1 expression and promotion of autophagic flux. Biochem. Pharmacol. 2014;87:445–455. doi: 10.1016/j.bcp.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tye B.W., Commins N., Springer M., Pincus D., Churchman L.S. A risk-reward tradeoff of high ribosome production in proliferating cells. BioRxiv. 2018 doi: 10.1101/458810. [DOI] [Google Scholar]
- 68.Albert B., Kos-Braun I.C., Henras A.K., Dez C., Rueda M.P., Zhang X., Gadal O., Kos M., Shore D. A ribosome assembly stress response regulates transcription to maintain proteome homeostasis. Elife. 2019;8 doi: 10.7554/eLife.45002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu A.-L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 70.Morley J.F., Morimoto R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.e03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morrow G., Samson M., Michaud S., Tanguay R.M. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 72.Min J.-N., Whaley R.A., Sharpless N.E., Lockyer P., Portbury A.L., Patterson C. CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 2008;28:4018–4025. doi: 10.1128/MCB.00296-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swindell W.R., Masternak M.M., Kopchick J.J., Conover C.A., Bartke A., Miller R.A. Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech. Ageing Dev. 2009;130:393–400. doi: 10.1016/j.mad.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baird N.A., Douglas P.M., Simic M.S., Grant A.R., Moresco J.J., Wolff S.C., Yates J.R., Manning G., Dillin A. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346:360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McLaughlin P.J., Bakall B., Choi J., Liu Z., Sasaki T., Davis E.C., Marmorstein A.D., Marmorstein L.Y. Lack of fibulin-3 causes early ageing and herniation, but not macular degeneration in mice. Hum. Mol. Genet. 2007;16:3059–3070. doi: 10.1093/hmg/ddm264. [DOI] [PubMed] [Google Scholar]
- 76.Stone E.M., Lotery A.J., Munier F.L., Héon E., Piguet B., Guymer R.H., Vandenburgh K., Cousin P., Nishimura D., Swiderski R.E., et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 77.Ackerman M.J., Clapham D.E. Ion channels--basic science and clinical disease. N. Engl. J. Med. 1997;336:1575–1586. doi: 10.1056/NEJM199705293362207. [DOI] [PubMed] [Google Scholar]
- 78.Fletcher C.F., Lutz C.M., O’Sullivan T.N., Shaughnessy J.D., Hawkes R., Frankel W.N., Copeland N.G., Jenkins N.A. Absence epilepsy in tottering mutant mice is associated with calcium channel defects. Cell. 1996;87:607–617. doi: 10.1016/S0092-8674(00)81381-1. [DOI] [PubMed] [Google Scholar]
- 79.Ophoff R.A., Terwindt G.M., Vergouwe M.N., van Eijk R., Oefner P.J., Hoffman S.M., Lamerdin J.E., Mohrenweiser H.W., Bulman D.E., Ferrari M., et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/S0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- 80.Wang M.C., Bohmann D., Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev. Cell. 2003;5:811–816. doi: 10.1016/S1534-5807(03)00323-X. [DOI] [PubMed] [Google Scholar]
- 81.Oh S.W., Mukhopadhyay A., Svrzikapa N., Jiang F., Davis R.J., Tissenbaum H.A. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grassilli E., Bellesia E., Salomoni P., Croce M.A., Sikora E., Radziszewska E., Tesco G., Vergelli M., Latorraca S., Barbieri D., et al. c-fos/c-jun expression and AP-1 activation in skin fibroblasts from centenarians. Biochem. Biophys. Res. Commun. 1996;226:517–523. doi: 10.1006/bbrc.1996.1387. [DOI] [PubMed] [Google Scholar]
- 83.Hilberg F., Aguzzi A., Howells N., Wagner E.F. c-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 84.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 86.Altenhoff A.M., Glover N.M., Train C.-M., Kaleb K., Warwick Vesztrocy A., Dylus D., de Farias T.M., Zile K., Stevenson C., Long J., et al. The OMA orthology database in 2018: Retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018;46:D477–D485. doi: 10.1093/nar/gkx1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang M., Zhao Y., Zhang B. Efficient Test and Visualization of Multi-Set Intersections. Sci. Rep. 2015;5:16923. doi: 10.1038/srep16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.