Abstract
Aberrant gene expression underlies drug addiction. Therefore, studying the regulation of gene expression in drug addiction may provide mechanistic insights into this disease, for which there are still only limited treatments. Recently, the three-dimensional (3D) compaction and organization of linear DNA in the nucleus has been recognized as having a major influence on gene transcription. Here, we review its roles in both basic brain function and neuropsychiatric disorders, while also highlighting its emerging implications in drug addiction. Unraveling the 3D organization of chromosomes in drug addiction is adding to our understanding of this disease and has the potential to trigger novel approaches for better diagnosis and therapy.
Introduction
Substance use disorders, or drug addiction, are characterized by maladaptive neural plasticity in response to drugs of abuse that persists throughout the lifetime of an individual. Susceptibility to addiction is believed to be the convergence of genetic predisposition and exposure to environmental risk factors, which results in dysregulation of specific genes [1,2]. Recent research into the molecular basis of drug addiction has focused on epigenetic mechanisms, such as posttranslational histone modifications and covalent DNA modifications, and demonstrated that drugs of abuse lead to widespread alterations across the epigenome, which are often associated with aberrant gene transcriptions in key brain regions of the reward system [3,4]. However, these have been predominantly studied on the level of one-dimensional linear genome.
In each eukaryotic cell, billions of DNA nucleotides span about two meters when stretched linearly, requiring extensive three-dimensional (3D) folding to fit into a nuclear space of only several micrometers in diameter. Through the mobilization of distal (i.e. up to several million bases away) regulatory elements into spatial proximity of their target genes, 3D genomic organization regulates cell type-specific transcriptional programs throughout development and culminates into discrete organizational profiles in mature cells [5–7]. The discovery of such 3D chromosome interactions points to a functional interplay between genome topology and gene expression, revolutionizing the current view of transcriptional regulation. However, its implications in brain function, particularly in relation to substance use disorders, remain largely unknown [8–12].
Three-dimensional chromosome organization and brain function
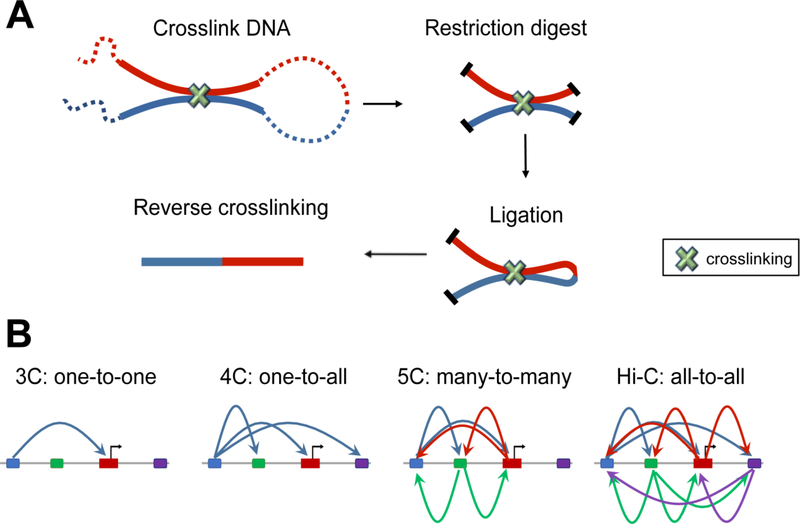
The recent development of chromosome conformation capture (3C)-based techniques [13–15] has complemented traditional microscopy-based studies in characterizing 3D genomic architecture with much higher throughput and genomic resolution than previously possible. The basic strategy of these techniques is nuclear proximity ligation, which allows for the detection of linearly distant genomic segments that reside within spatial proximity of each other in vivo (Figure 1). Over the years, these methods have evolved to focus on interactions at different scales, which include between a single pair of genomic loci (3C), between one locus and all other genomic loci (4C), between all restriction fragments within a given region (5C), or between all possible pairs of fragments across the genome (Hi-C).
Figure 1. Chromosome conformation capture based techniques.
(A) In Chromosome conformation capture (3C), three dimentional proximal DNA fragments are crosslinked and digested while in the nucleus. Open ends of DNA fragments are then repaired and chimeric fragments are ligated, which allows the detection of linearly-distant genomic segments that reside in spatial proximity. (B) The 3C-based techniques have evolved to focus on interactions between a single pair of genomic loci (3C), between one locus and all other genomic loci (4C), between all restriction fragments within a given region (5C), or between all possible pairs of fragments across the genome (Hi-C).
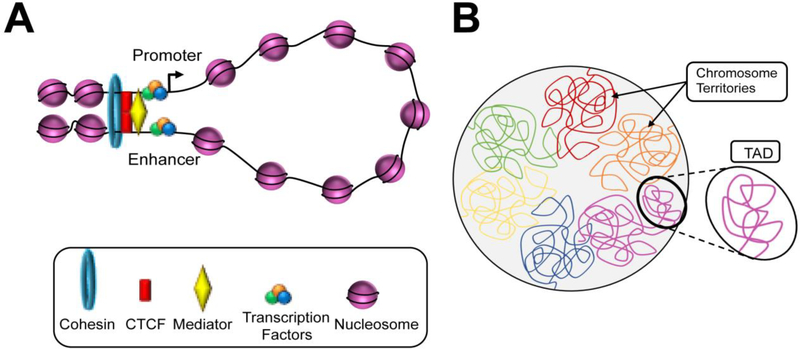
It is now understood that the genome is highly organized at several hierarchical 3D levels superimposed above nucleosomal organization (Figure 2). First, each chromosome occupies a defined nuclear volume known as a “chromosome territory” [16]. Additionally, the genome as a whole can be partitioned into two spatial compartments: an active “A” compartment and an inactive “B” compartment [15]. Located in the core of the nucleus, the “A” compartment is characterized by a high density of transcription factories which promotes the expression of chromosomal regions that are typically open, gene-rich, and high in GC content. In contrast, the “B” compartment is characterized by compact, gene-poor genomic regions that are minimally expressed. The “B” compartment exists in the nuclear periphery and interacts with nuclear lamina, a network of lamina proteins on the inner membrane of the nucleus. Such interactions with the lamina lead to formation of lamina-associated domains (LADs), which typically contain heterochromatic segments and function to anchor inactive chromosomal regions to the periphery of the nucleus [17]. Furthermore, the borders of LADs are defined by enrichment of active promoters, CpG islands, and the insulator protein CCCTC-binding factor (CTCF), which may prevent the mobilization of silenced elements into the nuclear core [17]. Additionally, each chromosome is comprised of a number of topologically associating domains (TADs), which contain chromosomal regions that preferentially interact with each other in cis and can be hundreds of kilobases to several megabases in length. Furthermore, TADs are maintained throughout cell divisions, and consistent between different cell types [18–20]; due to their high degree of conservation across species, these chromatin domains are considered to be the fundamental units of chromosome folding. Finally, the lowest level of 3D genomic organization occurs via DNA looping which, through the activity of architectural proteins (e.g. CTCF), enables distal genomic segments (e.g. enhancers) to be positioned into close spatial proximity of target genes and influence their expression [5–7].
Figure 2. Three-dimensional chromosome organization.
(A) Locally, at the submegabase scale, chromatin loop brings the distant cis-regulartory elements (e.g. enhancers) into close spatial proximity with its target promoter to regulate transcription. Architectural proteins and complexes including CTCF, mediator, and cohesin facilitate the formation of DNA loops. (B) On a larger scale, multiple loci undergoing regulatory interactions are grouped into topologically associated domains (TADs). The sequences within the same TADs have a high degree of interaction, which correlates with coordinated gene expression. Furthermore, all TADs are assembled into two spatial compartments (not shown). Lastly, each chromosome tends to locate in a discrete area within the nucleus, namely chromosome territories
Though only a handful of studies have investigated higher-order genome organization in the brain, they have all highlighted its essential roles in brain function [8–12]. One of the first pieces of evidence was revealed nearly 70 years ago with the demonstration of nuclear reorganization in motor neurons following electrical stimulation of the feline hypoglossal nucleus [21,22]. It was also found that during neural cell differentiation, respective sets of genes associated with cell identity undergo repositioning from peripheral heterochromatin domains to the inner compartment of the nucleus [23]. Using Hi-C to profile the chromosomal connectome, it was recently demonstrated that neuron- and astrocyte-like cells undergo cell type-specific 3D architecture remodeling during differentiation from human induced pluripotent stem cells [24]. Similarly, the role of 3D nuclear organization has been examined in the context of olfactory and retinal sensory neurons [25–27]. Though heterochromatin normally resides at the nuclear periphery, whereas euchromatin situates toward the nuclear interior, it was found their organization in rod photoreceptor neurons of nocturnal retinas is inverted [27,28]. The dense heterochromatin localized in the nuclear center may therefore serve as collecting lenses to facilitate light transduction efficiency, which provides an example on how nuclear architecture implicates in neuron function. In the mouse nose, odorants are detected by more than 1,000 different olfactory receptors (ORs) with each olfactory sensory neuron expressing a single receptor allele. ORs are used in combination to detect odorants, which explains how a myriad of odorants can be discriminated. From a series of studies from Lomvardas’ group, it was found that selective chromatin-mediated silencing and spatial positioning of OR genes ensure aggregation of inactive OR genes into heterochromatic foci which allows such monogenic and monoallelic expression of a single OR gene in olfactory sensory neurons [29–31]. Similarly, the expression of trace amine-associated receptors (TAARs), whose expression resembles that of ORs, is accompanied by an escape of the encoding genes from peripheral heterochromatin to the permissive interior chromatin environment [26]. Furthermore, it was demonstrated that the transcription factor and adapter protein that bind to intergenic olfactory receptor enhancers (or “Greek islands”) also mediate trans-chromosomal interactions so to facilitate multi-chromosomal super-enhancer that is associated with the active single olfactory receptor gene [32]. Recently, a single neuron 3D structure was characterized by using diploid chromatin conformation capture (Dip-C) [28], which further illustrated multiple aggregates of OR genes and enhancers from different chromosomes in the olfactory sensory neuron. Combined together, these studies provide convincing evidence for the link between chromatin organization and neuronal function.
Upon neuronal activation, the expression of many genes, particularly immediate early genes, require changes in chromatin accessibility [33] and formation of DNA loops involving regulatory regions such as enhancers [34]. Furthermore, neuronal activity induces the transcription of regulatory sequences to produce noncoding enhancer RNAs (eRNAs) [35], which may also participate in DNA looping to regulate transcription factor binding at target genes [36]. Such enhancer activities are often co-regulated to drive the coordinated expression of subsets of gene [37] or perform combinatorial functions to ensure proper expression in response to a variety of stimuli [38]. Chromosomal organization, therefore, is a crucial modulator of neuronal gene expression in the brain. Unsurprisingly, the malfunction of key molecules involved in chromosomal looping is associated with several brain disorders. For example, mutations of cohesin-related genes cause the neurodevelopmental disorder Cornelia de Lange syndrome (CdLS) [39,40], while mutations to the gene encoding for CTCF have been implicated in the etiology of microcephaly and intellectual disabilities [41].
Three-dimensional chromosome organization in neuropsychiatric disorders
The implications of changes to 3D genome organization in neuropsychiatric disorders have also recently been examined [8,9]. For example, it was recently discovered that all known disease-associated short tandem repeats, such as those responsible for Fragile X syndrome, are located at the boundaries of 3D chromatin domains and that topological disruption of these boundaries may contribute the etiology of disease [42]. Using a FISH assay, it was also found that accumulation of H2BGFP (green fluorescent protein-tagged histone H2B) in hippocampal CA1 neurons of transgenic mice disrupts chromatin compaction and nuclear lamina interaction [43], which is also associated with detrimental effects on cognitive functions. Though this does not affect neuronal viability, it is accompanied with specific transcriptional changes in serotonin and dopamine signaling pathways, suggesting that some neuropsychiatric disorder-associated genomic loci may be vulnerable to changes in higher-order genome organization.
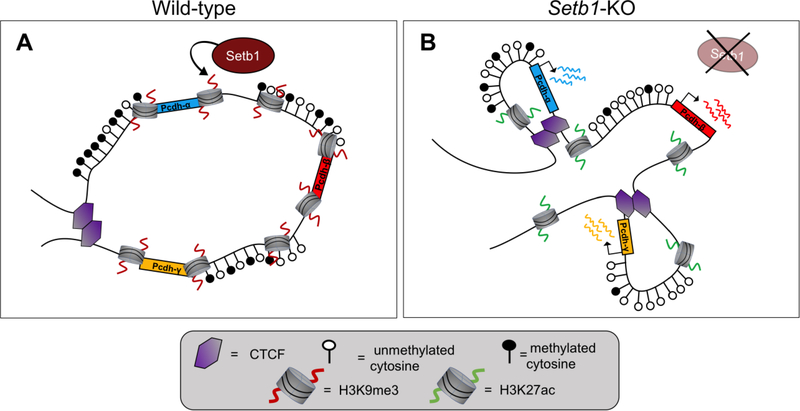
Akbarian and colleagues have done extensive investigations into 3D genomic organization in schizophrenia. From their work, it was found that the expression of NMDA receptor subunit GRIN2B/Grin2b is regulated by H3K9 methyltransferase SETDB1 through a conserved cis-regulatory element which forms activity-dependent DNA loopings encompassing over 500 kb of linear sequences; dysregulation of this locus affects mood-related behaviors and contributes to cognitive defects [44,45]. The group also reported destabilization of a megabase-scale neuron-specific topological chromatin domain following conditional knockout of Setdb1 in mice, which was accompanied by increased CTCF binding, DNA hypomethylation, histone hyperacetylation, and up-regulated gene expression [46] (Figure 3). Similarly, they discovered that conserved chromosomal looping governs the expression of GAD1/Gad1 in the prefrontal cortex (PFC), and that these structures are significantly weakened in humans with schizophrenia [47]. Furthermore, increased DNA looping of a putative MEF2C (encoding for a neurodevelopmental factor associated with schizophrenia) super-enhancer to risk loci over 500 kb away occurs in induced stem cell-derived cultured neurons of subjects with schizophrenia, and binding sites for MEF2C are enriched in cis-regulatory enhancer and promoter regions marked by aberrant H3K4 hypermethylation in postmortem PFC samples [48].
Figure 3. Setb1 repressor complex regulates a large topologically associated domain (megaTAD) formation and shields from excessive CTCF binding in the neuronal genome.
(A) Setb1 maintains H3K9me3, which acts in tandem with high levels of DNA cytosine methylation to create a heterochromatic gene environment in mature neurons. (B) Ablation of Setb1 activity disrupts formation of this megaTAD. Setb1-KO neurons display decreased H3K9me3, increased H3K27ac. This results in increased CTCF binding at cryptic binding sites, decreased cytosine methylation, and upregulated expression of subsets of clustered Pcdh genes (genes Pcdhα, Pcdhβ, and Pcdhγ are shown in blue, red, and amber respectively).
Given the aforementioned dynamic 3D structure changes during neural development [23,24,26] and a wide spectrum of neuropsychiatric disorders (e.g. schizophrenia) with developmental etiologies, it is reasoned that studying chromosome conformations in the developing brain would provide novel genetic insights of these neuropsychiatric disorders. Indeed, through Hi-C mapping generated during human brain corticogenesis, Won et al. not only recognized many novel long range enhancer-promoter interactions, but also displayed several candidate schizophrenia risk genes/pathways by integrating this 3D interactome with non-coding variants identified in schizophrenia genome wide association studies (GWAS) [49]. A recent report also found that risk variants for schizophrenia are overrepresented in genomic regions decorated with nucleosomal marks associated with active transcription in the dorsolateral PFC and anterior cingulate cortex of subjects with schizophrenia [50]. Additionally, these 3D genomic changes associated with schizophrenia risk sequences are also highly enriched in neurons instead of glial cells [24]. Together, these studies highlight the epigenomic mechanisms by which the genome confers liability to the development of schizophrenia and how disruption to genomic architecture may contribute to neuropsychiatric disorders in general.
Three-dimensional chromosome organization in drug addiction
Addiction is regarded as a malfunctioning of neural plasticity. Several genes implicated in this process have been shown to undergo 3D genome organization changes in various contexts. For example, Bdnf, a well-studied gene in addiction, detaches from the peripheral nuclear lamina in rat hippocampal neurons after kainate-induced seizures [51]. The resulting translocation of Bdnf to the open chromatin environment of the nucleus center is believed to allow for its exposure to transcription factors in the inner “A” compartment, which facilitate promoter-enhancer interactions and gene activation [51]. Similarly, in mice, a single dose of cocaine induces translocation of Sig-R1 from the endoplasmic reticulum to the nuclear envelope, resulting in increased Sig-R1 interactions with emerin (a nuclear envelope transmembrane protein) and recruitment of chromatin compacting factors [52]. Interestingly, these alterations were found to specifically decrease expression of monoamine oxidase B, providing a possible mechanism of enhanced dopaminergic responses following drug exposure that involves 3D chromatin reorganization in the nuclear periphery. Furthermore, Arc (an immediate early gene product that is associated with addiction) was found to accumulate to the nucleus of striatal medium spiny neurons following cocaine-induced rapid upregulation. Arc downregulates phospho-Ser10-histone H3 (an important component of nucleosomal response) and is suspected to dull the decompaction of heterochromatic regions [53]. These results are consistent with findings from other studies that have demonstrated the impact of nucleosome remodeling on addiction-related behaviors and learning and memory processes [54,55].
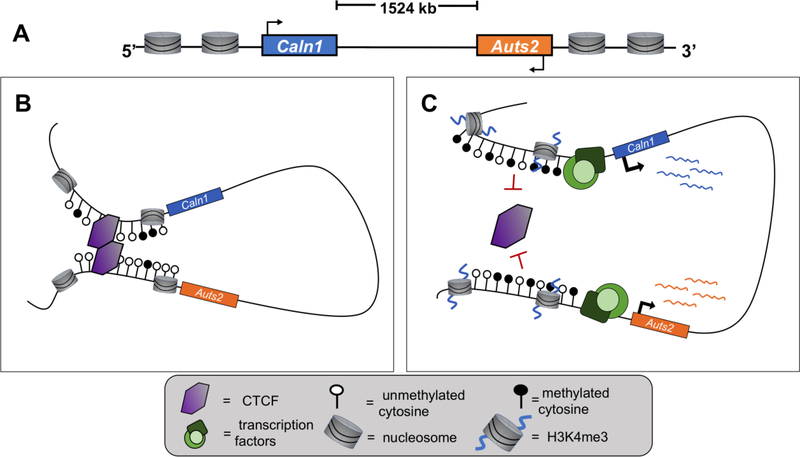
Currently, there is still limited evidence that directly demonstrates the higher-order 3D genomic structures in addiction. In one study, however, Engmann et al. used chromosome conformation capture-based techniques (3C, 4C) to demonstrate cocaine-induced destabilization of a DNA loop that spans 1524 kb in the mouse nucleus accumbens, a central brain region involved in reward behaviors [56] (Figure 4). It was found that chromosomal looping which brings Auts2 and Caln1 within close proximity of each other is disrupted following repeated cocaine exposure. This was accompanied by a range of epigenetic changes, such as altered levels of H3K4me3 and DNA methylation, as well as disruption to CTCF binding at these loci. Importantly, these alterations occur in a neuronal subtype-specific manner, preferentially affecting D2-like medium spiny neurons (MSNs) of the nucleus accumbens, but not D1-like MSNs. Additionally, these effects were only observed in male mice, but not females. These results highlight the need for further investigation into the molecular divergences between the sexes that may contribute to differential susceptibility to drug addiction, as well as the specific roles of neuronal subtypes in brain regions containing heterogeneous cell types.
Figure 4. Cocaine-induced disruption of DNA loop encompassing Auts2 and Caln1.
(A) Linear representation of Auts2 (orange) and Caln1 (blue) genes separated by ~1524 kilobases in rodents and humans. (B) DNA looping encompassing Auts2 and Caln1 under baseline conditions in the brain. Low levels DNA methylation upstream of Auts2 and Caln1 allow for CTCF binding and homodimerization, resulting in DNA loop formation. Auts2 and Caln1 are within close 3D space of each other and both genes remain minimally expressed. (C) Repeated cocaine exposure disrupts Auts2-Caln1 interactions. Increased levels of DNA methylation upstream of Auts2 and Caln1 prevents CTCF binding in regulatory regions and prevents DNA loop formation. The associated increased levels of H3K4me3 facilitate the recruitment of transcription factors and therefore upregulation of both genes’ expression.
Conclusion
The 3D folding of chromosomes has been shown to have a major influence on gene transcription by facilitating interactions between gene promoters and distal non-coding regulatory elements. Emerging evidence indicates its roles in both basic brain function as well as in neuropsychiatric disorders. However, how 3D genomic architecture is organized and functions in drug addiction is still unclear.
Though there are still few studies that directly investigate 3D looping in relation to substances of abuse, previous observations suggest that 3D nuclear reorganization in drug addiction could be widespread. For example, numerous studies have demonstrated cocaine-induced epigenetic changes across intergenic non-coding regions (e.g. [57]). It is predicted that these epigenetic changes are associated with hundreds of thousands of regulatory elements located great distances away from their target genes. This indeed has been demonstrated by a study on the DNA loop connecting Auts2 and Caln1 genes, which bypasses 1524 kb linear DNA sequence; the epigenetic alterations on the non-coding DNA sequences within this loop are associated with cocaine-induced dissociation of both genes and their expression changes [56].
With coding sequences representing about 3% of the genome, the function of the vast majority of genome that is comprised of non-coding sequences remains elusive. We expect that the study of 3D chromatin organization will provide insights into regulation of gene transcription in drug addiction. For example, it is currently still obscure as to what the gene targets are, if any, for the large number of intergenic regions that display drug-induced epigenetic alterations. To address this question, future studies should take advantage of evolving tools for investigating 3D nuclear organization [58], such as recent advances in Hi-C methods that specifically probe 3D interactions at promoters and allow for characterization of the promoter interactome with unprecedented resolution [59]. Furthermore, GWAS have identified numerous SNP variants in addiction-relevant gene loci, many of which occur in noncoding regions (e.g. [60,61]). It has been shown that these SNP sites are highly enriched at intergenic regulatory regions, such as enhancers, which suggests a role for 3D chromatin organization in the pathogenesis (e.g. [46,49,62]). Therefore, elucidating higher-order genome organization in drug addiction should help us to prioritize candidate gene targets and decode disease-relevant pathways.
Although the study of 3D genome in the brain holds great promise, there are several questions that will need to be addressed moving forward. One technical challenge is the heterogeneous cell population of even brain micronuclei. As 3D chromosomal organization is highly cell-type specific, it is essential to probe the higher-order genomic architecture within the contexts of defined cell types. Though neuronal NeuN labeling and neuronal subtype fluorescent protein-expressing transgenic mice have shown success in this approach [46,56], isolation processes usually result in the loss of most genetic material. It is still a challenge to study the 3D genome in small brain structures (e.g. nucleus accumbens) at a global level (e.g. Hi-C), which usually requires far more cells than a single brain can provide. Furthermore, given the enormous number of chromatin interactions across the genome, thorough characterization of 3D genomic organization requires significant sequencing depth of coverage. This also demands advanced bioinformatic analytical tools that may be developed through creative collaborations between biologists and bioinformaticians. Fortunately, technological advances are now allowing us to identify the spatial organization within a single cell [63] and across both space and time (i.e. the 4th dimension) [58]. Through the application of these and other tools, further investigation of 3D chromosome organization in drug addiction should reveal novel molecular insights into this disease that may have the potential to trigger novel approaches for better diagnosis and therapy.
Highlights.
Three-dimensional (3D) chromosome architecture is highly organized.
3D chromosome organization plays important roles in basic brain function
Altered 3D interactions are associated with neuropsychiatric disorders
Cocaine disrupts a particular DNA loop formation in the brain.
Acknowledgements
We thank all our lab members for help and advice. Preparation of this review was supported by grants from the National Institute on Drug Abuse. JMC is supported by a Legacy Fellowship and a Neuroscience Fellowship from Florida State University. The authors apologize to any colleagues whose work was not referenced due to space limations
Footnotes
Declaration of Conflicts
None to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Hyman SE, Malenka RC, Nestler EJ: Neural mechanisms of addiction: The role of reward-related learning and memory. Annu Rev Neurosci (2006) 29(565–598. [DOI] [PubMed] [Google Scholar]
- 2.Koob GF, Volkow ND: Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry (2016) 3(8):760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng J, Nestler EJ: Epigenetic mechanisms of drug addiction. Curr Opin Neurobiol (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robison AJ, Nestler EJ: Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci (2011) 12(11):623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furlan-Magaril M, Varnai C, Nagano T, Fraser P: 3d genome architecture from populations to single cells. Current opinion in genetics & development (2015) 31(36–41. [DOI] [PubMed] [Google Scholar]
- 6.Cattoni DI, Valeri A, Le Gall A, Nollmann M: A matter of scale: How emerging technologies are redefining our view of chromosome architecture. Trends in genetics : TIG (2015) 31(8):454–464. [DOI] [PubMed] [Google Scholar]
- 7.Bonev B, Cavalli G: Organization and function of the 3d genome. Nat Rev Genet (2016) 17(11):661–678. [DOI] [PubMed] [Google Scholar]
- 8.Rajarajan P, Gil SE, Brennand KJ, Akbarian S: Spatial genome organization and cognition. Nat Rev Neurosci (2016) 17(11):681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medrano-Fernandez A, Barco A: Nuclear organization and 3d chromatin architecture in cognition and neuropsychiatric disorders. Mol Brain (2016) 9(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilczynski GM: Significance of higher-order chromatin architecture for neuronal function and dysfunction. Neuropharmacology (2014) 80(28–33. [DOI] [PubMed] [Google Scholar]
- 11.Watson LA, Tsai LH: In the loop: How chromatin topology links genome structure to function in mechanisms underlying learning and memory. Curr Opin Neurobiol (2017) 43(48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y, Yamashita T: Spatial organization of genome architecture in neuronal development and disease. Neurochemistry international (2017). [DOI] [PubMed] [Google Scholar]
- 13.Schmitt AD, Hu M, Ren B: Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol (2016) 17(12):743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denker A, de Laat W: The second decade of 3c technologies: Detailed insights into nuclear organization. Genes Dev (2016) 30(12):1357–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R et al. : Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (2009) 326(5950):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer T, Cremer M: Chromosome territories. Cold Spring Harb Perspect Biol (2010) 2(3):a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B: Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature (2008) 453(7197):948–951. [DOI] [PubMed] [Google Scholar]
- 18.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B: Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012) 485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B: A map of the cis-regulatory sequences in the mouse genome. Nature (2012) 488(7409):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL: A 3d map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell (2014) 159(7):1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr ML, Bertram EG: A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature (1949) 163(4148):676. [DOI] [PubMed] [Google Scholar]
- 22.Barr ML, Bertram EG: The behaviour of nuclear structures during depletion and restoration of nissl material in motor neurons. Journal of anatomy (1951) 85(2):171–181. [PMC free article] [PubMed] [Google Scholar]
- 23.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M et al. : Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell (2010) 38(4):603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casino C, Powell S, Yashaswini C, LaMarca EA, Kassim B, Javidfar B et al. : Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science (2018) 362(6420).•This is a representative report of the PsychENCODE Consortium. The authors found that the chromosomal connectome is subjected to developmental regulated changes in neurons and glial cells. The 3D changes that coincide with schizophrenia relevant risk regions are highly enriched in neurons instead of glial cells.
- 25.Armelin-Correa LM, Gutiyama LM, Brandt DY, Malnic B: Nuclear compartmentalization of odorant receptor genes. Proc Natl Acad Sci U S A (2014) 111(7):2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon KH, Ragoczy T, Lu Z, Kondoh K, Kuang D, Groudine M, Buck LB: Olfactory receptor genes expressed in distinct lineages are sequestered in different nuclear compartments. Proc Natl Acad Sci U S A (2015) 112(18):E2403–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B: Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell (2009) 137(2):356–368. [DOI] [PubMed] [Google Scholar]
- 28.Tan L, Xing D, Daley N, Xie XS: Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat Struct Mol Biol (2019) 26(4):297–307.••Here, the authors described the unusual 3D chromatin organization of single retinal and olfactory sensory neurons. Using diploid chromatin conformation capture, they found that rod photoreceptors display inverted radial distribution of euchromatin and heterochromatin, which is in sharp contrast to the genomic organization of most other cell types. While olfactory sensory neurons do not display such inverted organization, they found that there is a distinct concentration of olfactory receptor genes in the A compartment of the nucleus, which is unlike non-neuronal cell types. This paper importantly highlights the differences in genomic organization among single cells and the functional relevance it may have in neurons.
- 29.Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, Barnea G et al. : An epigenetic signature for monoallelic olfactory receptor expression. Cell (2011) 145(4):555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S: Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell (2012) 151(4):724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, Mosley CP, Ahituv N, Lomvardas S: Enhancer interaction networks as a means for singular olfactory receptor expression. Cell (2014) 159(3):543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monahan K, Horta A, Lomvardas S: Lhx2- and ldb1-mediated trans interactions regulate olfactory receptor choice. Nature (2019) 565(7740):448–453.•• Using in situ Hi-C on fluorescent-activated cell sorted olfactory sensory neurons, the researchers discovered that LXH2 and LDB1 regulate trans interactions involving a series of intergenic enhancers, known as ‘Greek islands’, which ultimately participate in a multi-chromosomal super-enhancer to regulate the expression of a single olfactory receptor gene. While most 3D genomic interactions between regulatory elements and target genes occur in cis, this study demonstrates the important role that trans-chromosomal interactions plays in the regulation of gene expression.
- 33.Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, Kim D, Ming GL, Song H: Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci (2017) 20(3):476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME: Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci (2014) 17(10):1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E et al. : Widespread transcription at neuronal activity-regulated enhancers. Nature (2010) 465(7295):182–U165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK: Enhancer rna facilitates nelf release from immediate early genes. Mol Cell (2014) 56(1):29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermunt MW, Reinink P, Korving J, de Bruijn E, Creyghton PM, Basak O, Geeven G, Toonen PW, Lansu N, Meunier C, van Heesch S et al. : Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep (2014) 9(2):767–779. [DOI] [PubMed] [Google Scholar]
- 38.Joo JY, Schaukowitch K, Farbiak L, Kilaru G, Kim TK: Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat Neurosci (2016) 19(1):75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ et al. : Cornelia de lange syndrome is caused by mutations in nipbl, the human homolog of drosophila melanogaster nipped-b. Nat Genet (2004) 36(6):631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T: Nipbl, encoding a homolog of fungal scc2-type sister chromatid cohesion proteins and fly nipped-b, is mutated in cornelia de lange syndrome. Nat Genet (2004) 36(6):636–641. [DOI] [PubMed] [Google Scholar]
- 41.Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, Vasileiou G et al. : De novo mutations in the genome organizer ctcf cause intellectual disability. American journal of human genetics (2013) 93(1):124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun JH, Zhou L, Emerson DJ, Phyo SA, Titus KR, Gong W, Gilgenast TG, Beagan JA, Davidson BL, Tassone F, Phillips-Cremins JE: Disease-associated short tandem repeats co-localize with chromatin domain boundaries. Cell (2018) 175(1):224–238 e215.•In this study, it was revealed that disease-associated short tandem repeats (daSTRs), such as those involved in Fragile X syndrome, are specifically localized to topological boundaries of chromatin domains. For example, the 3D chromatin structure at the FMR1 locus is shown to be disrupted due to localization and expansion of such repeats in this region. This is correlated with FRM1 silencing and decreased CTCF occupancy, providing novel mechanistic insight into the etiology of Fragile X syndrome and possibly other daSTRs.
- 43.Ito S, Magalska A, Alcaraz-Iborra M, Lopez-Atalaya JP, Rovira V, Contreras-Moreira B, Lipinski M, Olivares R, Martinez-Hernandez J, Ruszczycki B, Lujan R et al. : Loss of neuronal 3d chromatin organization causes transcriptional and behavioural deficits related to serotonergic dysfunction. Nat Commun (2014) 5(4450. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, Straubhaar J, Martin G, Akbarian S: Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the nmda receptor subunit nr2b. J Neurosci (2010) 30(21):7152–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY, Mitchell AC, Mao W, Whittle C, Dincer A, Jakovcevski M et al. : Conserved higher-order chromatin regulates nmda receptor gene expression and cognition. Neuron (2014) 84(5):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, Labonte B et al. : The methyltransferase setdb1 regulates a large neuron-specific topological chromatin domain. Nat Genet (2017) 49(8):1239–1250.••By using conditional knockout in defined neurons in the mouse brain, the authors demonstrated that the SETDB1 repressor complex regulates a megabase scale topological associated domain. This was accompanied by alterations to CTCF binding, DNA methylation, histone modifications, and gene expression, highlighting the dynamic relationship between various epigenetic processes and how they converge to regulate gene expression.
- 47.Bharadwaj R, Jiang Y, Mao W, Jakovcevski M, Dincer A, Krueger W, Garbett K, Whittle C, Tushir JS, Liu J, Sequeira A et al. : Conserved chromosome 2q31 conformations are associated with transcriptional regulation of gad1 gaba synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci (2013) 33(29):11839–11851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell AC, Javidfar B, Pothula V, Ibi D, Shen EY, Peter CJ, Bicks LK, Fehr T, Jiang Y, Brennand KJ, Neve RL et al. : Mef2c transcription factor is associated with the genetic and epigenetic risk architecture of schizophrenia and improves cognition in mice. Mol Psychiatry (2018) 23(1):123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C et al. : Chromosome conformation elucidates regulatory relationships in developing human brain. Nature (2016) 538(7626):523–527.•The authors constructed Hi-C libraries from mid-gestation developing human cerebral cortex. They identified numerous novel enhancer-promoter interactions during corticogenesis. By integrating this chromatin contacts with non-coding risk variants identified in schizophrenia GWAS studies, they also recognized multiple disease risk genes and pathways. This demonstrates how analysis of 3D genome architecture provides insights into gene regulation and disease pathogenesis.
- 50.Girdhar K, Hoffman GE, Jiang Y, Brown L, Kundakovic M, Hauberg ME, Francoeur NJ, Wang YC, Shah H, Kavanagh DH, Zharovsky E et al. : Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat Neurosci (2018) 21(8):1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walczak A, Szczepankiewicz AA, Ruszczycki B, Magalska A, Zamlynska K, Dzwonek J, Wilczek E, Zybura-Broda K, Rylski M, Malinowska M, Dabrowski M et al. : Novel higher-order epigenetic regulation of the bdnf gene upon seizures. J Neurosci (2013) 33(6):2507–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai SY, Chuang JY, Tsai MS, Wang XF, Xi ZX, Hung JJ, Chang WC, Bonci A, Su TP: Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope. Proc Natl Acad Sci U S A (2015) 112(47):E6562–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salery M, Dos Santos M, Saint-Jour E, Moumne L, Pages C, Kappes V, Parnaudeau S, Caboche J, Vanhoutte P: Activity-regulated cytoskeleton-associated protein accumulates in the nucleus in response to cocaine and acts as a brake on chromatin remodeling and long-term behavioral alterations. Biol Psychiatry (2017) 81(7):573–584.•A novel role for Arc, which is generally considered a cytoskeletal effector protein, was revealed following exposure to cocaine. Here, the authors demonstrated that Arc also localizes to the nucleus, where it acts to prevent decompaction of the heterochromatin, which is typically observed following acute and chronic cocaine exposure. Interestingly, Arc was shown to have a role in the sensitization to cocaine over repeated exposures, indicating a possible role in the susceptibility to drug addiction.
- 54.Vogel-Ciernia A, Matheos DP, Barrett RM, Kramar EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, Jia Y et al. : The neuron-specific chromatin regulatory subunit baf53b is necessary for synaptic plasticity and memory. Nat Neurosci (2013) 16(5):552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun H, Martin JA, Werner CT, Wang ZJ, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, Koo JW, Gancarz AM et al. : Baz1b in nucleus accumbens regulates reward-related behaviors in response to distinct emotional stimuli. J Neurosci (2016) 36(14):3954–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engmann O, Labonte B, Mitchell A, Bashtrykov P, Calipari ES, Rosenbluh C, Loh YE, Walker DM, Burek D, Hamilton PJ, Issler O et al. : Cocaine-induced chromatin modifications associate with increased expression and three-dimensional looping of auts2. Biol Psychiatry (2017) 82(11):794–805.••Using 3C and 4C approaches, this study is the first to demonstrate cocaine-induced disruption of neuronal three-dimensional genome organization at loci involved in responses to drugs of abuse. Moreover, it was revealed that these changes occur in a cell- and sex-type specific manner. This study provides a framework for future investigations into the functional consequences of drugs of abuse on 3D chromatin architecture.
- 57.Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C et al. : Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol (2014) 15(4):R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, Politz JCR et al. : The 4d nucleome project. Nature (2017) 549(7671):219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schoenfelder S, Javierre BM, Furlan-Magaril M, Wingett SW, Fraser P: Promoter capture hi-c: High-resolution, genome-wide profiling of promoter interactions. J Vis Exp (2018) 136). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hancock DB, Markunas CA, Bierut LJ, Johnson EO: Human genetics of addiction: New insights and future directions. Curr Psychiatry Rep (2018) 20(2):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, Abdellaoui A, Nivard MG, Baselmans BML, Ong JS, Ip HF et al. : Gwas of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci (2018) 21(9):1161–1170.• In this largest GWAS for lifetime cannabis use, the authors identified numerous genome-wide significant independent single nucleotide polymorphisms and genes, which provides new insights into the etiology of cannabis use and its relation to mental health.
- 62.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Varnai C, Thiecke MJ, Burden F et al. : Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell (2016) 167(5):1369–1384 e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P: Single-cell hi-c reveals cell-to-cell variability in chromosome structure. Nature (2013) 502(7469):59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]