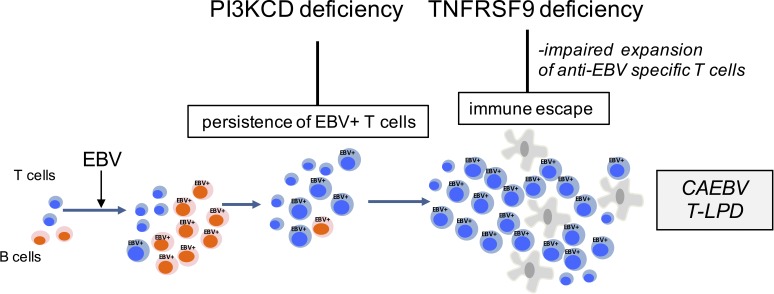
Identification of biallelic loss-of-function mutations in TNFRSF9 and PIK3CD in a kindred with chronic active Epstein-Barr virus infection of T cells (CAEBV) suggests that CAEBV is the consequence of factors providing growth advantage to EBV-infected T cells combined with defective cell immunity toward EBV-infected cells.
Abstract
Infection of T cells by Epstein-Barr virus (EBV) causes chronic active EBV infection (CAEBV) characterized by T cell lymphoproliferative disorders (T-LPD) of unclear etiology. Here, we identified two homozygous biallelic loss-of-function mutations in PIK3CD and TNFRSF9 in a patient who developed a fatal CAEBV. The mutation in TNFRSF9 gene coding CD137/4-1BB, a costimulatory molecule expressed by antigen-specific activated T cells, resulted in a complete loss of CD137 expression and impaired T cell expansion toward CD137 ligand–expressing cells. Isolated as observed in one sibling, CD137 deficiency resulted in persistent EBV-infected T cells but without clinical manifestations. The mutation in PIK3CD gene that encodes the catalytic subunit p110δ of the PI3K significantly reduced its kinase activity. Deficient T cells for PIK3CD exhibited reduced AKT signaling, while calcium flux, RAS-MAPK activation, and proliferation were increased, suggestive of an imbalance between the PLCγ1 and PI3K pathways. These skewed signals in T cells may sustain accumulation of EBV-infected T cells, a process controlled by the CD137–CD137L pathway, highlighting its critical role in immunity to EBV.
Graphical Abstract
Introduction
The Epstein-Barr virus (EBV) is an oncogenic virus that infects most of humans with a marked tropism for epithelial cells and B lymphocytes (Taylor et al., 2015). The primary infection is self-limited, while latent EBV-infected B cells persist lifelong. In immune-compromised hosts, primary infection and persistence of proliferative EBV-infected B cells result in severe and often fatal lymphoproliferative diseases, including hemophagocytic lymphohistiocytosis (HLH) and nonmalignant and malignant B cell proliferations (Taylor et al., 2015; Tangye et al., 2017; Latour and Winter, 2018). Several inherited forms of susceptibility to develop EBV-driven B cell lymphoproliferative disorders have been identified, including gene defects in SH2D1A, CORO1A, CTPS1, MAGT1, ITK, CD27, and CD70 (Cohen, 2015; Tangye et al., 2017; Latour and Winter, 2018). Heterozygous gain-of-function (GOF) mutations in PIK3CD have also been associated with impaired immune control of EBV-infected B cells (Coulter et al., 2017; Edwards et al., 2019). Patients with these primary immunodeficiencies are characterized by their inability to mount efficient virus-specific T cell responses to eliminate EBV-infected B cells. Studies of these primary immunodeficiencies allowed the identification of several key factors required for T cell expansion and cell cytotoxicity and highlighted the role of T–B interactions in the control of EBV-infected B cells.
EBV can also occasionally infect T lymphocytes and/or natural killer (NK) cells, leading to EBV-driven T/NK cell lymphoproliferative diseases, also termed as chronic active EBV infection (CAEBV; Fujiwara et al., 2014; Park and Ko, 2014). T/NK cell lymphoproliferative diseases are rare diseases of childhood and young adults often observed in populations from Asia and Central South America. CAEBV is characterized by persistence of EBV-infected T or/and NK cells that may progress to life-threatening lymphoproliferative disorders including T and NK lymphomas frequently associated with HLH symptoms (Kimura et al., 2001, 2012). Patients typically exhibit a high EBV load that can persist over years. They also often develop characteristic cutaneous manifestations, namely hypersensitivity to mosquito bites and hydroa vacciniforme–like lymphoma (Okano et al., 2005; Quintanilla-Martinez et al., 2013). The pathogenesis of T/NK lymphoproliferative disorders is poorly understood. It is associated in some cases with somatic mutations in DDX3X (Okuno et al., 2019). However, it is strongly suspected that these patients may suffer from an immune defect that results in inefficient CD8+ T cell responses toward EBV-infected T cells (Fujiwara et al., 2014; Taylor et al., 2015). Of note, few cases of B cell type of CAEBV associated with B cell lymphoproliferations, chronic viremia, and HLH have been also reported (Kimura and Cohen, 2017). It is not clear whether these cases differ from “classical” EBV-associated B cell disorders.
Herein, we report the first identification of germline mutations causing CAEBV. We identified two homozygous loss-of-function (LOF) mutations in PIK3CD and TNFRSF9 in a patient presenting lethal CAEBV. Studies of the functional consequences of these mutations indicate that occurrence of CAEBV may be viewed as a consequence of factors providing a growth advantage to EBV-infected T cells combined with defective cell immunity toward EBV-infected cells.
Results
Identification of a family with a susceptibility to infection of T cells by EBV
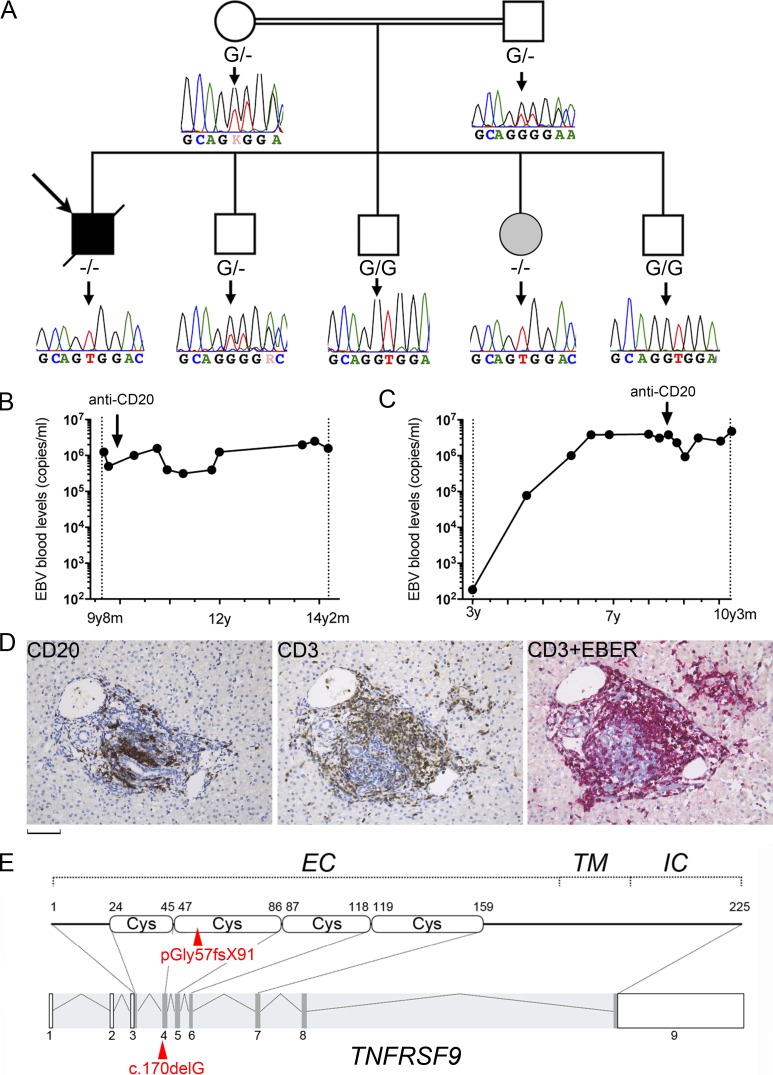
We investigated the case of a male child born to consanguineous parents originating from Pakistan diagnosed with an EBV-associated T cell lymphoproliferative disease (Fig. 1 A). He had recurrent upper respiratory tract and skin infections that started at 3 mo of age necessitating recurrent hospitalizations. Those included episodes of panaritium (at the age of 6, 7, and 9.5 yr) and boils requiring antibiotic intravenous infusion. Candida albicans, Staphylococcus aureus, and HSV were found in skin specimens. At the age of 10 yr, he received a prophylactic antibiotic treatment, and since then, he had not developed infections (with the exception of EBV infection [see below]) requiring hospitalization and antibiotic intravenous infusion. At the age of 9 yr and 10 mo, he came to our attention for persistent fever, weight loss, hepatosplenomegaly, and recurrent lymphadenopathies associated with persistent high blood load of EBV (6.5 × 106 copies/ml; Fig. 1 B). Episodes resolved spontaneously without treatment in a few weeks. High EBV loads were also detected in a liver biopsy (data not shown). Immunohistochemistry of the liver biopsy showed mild portal infiltration with CD3+ CD8+ mononuclear cells (Fig. 1 D). In situ hybridization with Epstein-Barr encoding region (EBER) probe was mainly positive in CD3+ cells, while most of CD20+ B cells were negative indicative of liver infiltration by EBV-infected T cells. Hydroa vacciniforme–like lesions also contained accumulation of EBV-containing CD8+ T cells that hybridized with EBER (Fig. S1). The presence of EBV in T cells was also examined by PCR on purified CD3+ cells from blood showing a high EBV load (5.3 × 106 copies/µg DNA) in CD3+ cells similar to the level detected in whole blood at that time (5.5 × 106 copies/µg DNA), whereas purified B cells were negative. Blood EBV load remained stably high over the years (between 5.5 × 106 and 6.5 × 106 copies/ml) despite treatment with the anti-CD20 monoclonal antibody (Fig. 1 B). Between the ages of 10 and 14 yr, symptoms recurred one or twice a year. At the age of 14 yr, he developed a fatal acute episode of HLH.
Figure 1.
Identification of a homozygous mutation in TNFRSF9 in two siblings with chronic EBV viremia and EBV-infected T cells. (A) Family pedigree with DNA electropherograms and the TNFRSF9 c.170DelG (−) genotype of each individual. The proband is indicated by an arrow. The healthy sister with chronic EBV corresponds to the gray circle. (B and C) EBV blood loads in the patient (B) and his sister (C) with EBV copies at different time points (black circles; y, years; m, months). Arrows indicate anti-CD20 treatments. (D) Patient liver biopsy. Staining with anti-CD3 (CD3) or anti-CD20 (CD20) showing portal and sinusoidal inflammatory infiltrate composed of CD3+ T lymphocytes and CD20+ B cells. Double staining with anti-CD3 antibody and EBER probe (CD3+EBER) detects EBV in the nucleus of most of CD3+ T cells. Magnification, ×200. Scale bar, 60 µm. (E) Schematic representation of TNFRSF9 intron–exon organization (coding regions in dark gray) and protein structure (extracellular [EC], transmembrane [TM], and intracytoplasmic [IC] domains) above. The mutation is indicated in red.
Immunological investigations showed no major abnormalities. The patient had normal T cell counts with fluctuating CD8+ lymphopenia associated with decreased naive CD8+ T cells and an increased proportion of memory CD8+ T cells (Table 1). Prior to anti-CD20 therapy, B cell counts were normal, but there was a nearly complete lack of circulating memory and switched B cells. Ig levels were normal to high for IgG and low to normal for IgM and IgA. Low IgM levels persisted after recovery of normal B cell numbers following anti-CD20 treatment. NK, invariant NKT (iNKT), and mucosal-associated invariant T cell counts were reduced. T cell proliferation in response to CD3, PHA, and antigen stimulations was elevated. Based on the severity of the clinical phenotype, a gene defect leading to T and B cell immunodeficiency was suspected to be the cause of his condition.
Table 1. Immunological features of PBMCs from the patient and his sister.
| Patient | Sister | ||
|---|---|---|---|
| Age-matched normal values | 10/12/14 yr | 6/10 yr | |
| Leukocytes (cells mm−3) | (4,400–15,500) | 6,800/ND/4,900 | 6,600/6,000 |
| Neutrophils (cells mm−3) | (1,800–8,000) | 1,400/ND/2,200 | 3,800/ND |
| Monocytes (cells mm−3) | (200–1,000) | 400/ND/200 | 400/ND |
| Lymphocytes (cells mm−3) | (1,900–3,700) | 1,900/2,000/2,200 | 2,100/1,543 |
| T cells | |||
| CD3+ (cells mm−3) | (1,200–2,600) | 1,501/1,440/1,474 | 1,596/1,096 |
| CD4+ (cells mm−3) | (650–1,500) | 1,102/1,140/1,056 | 987/663 |
| CD8+ (cells mm−3) | (370–1,100) | 342/260/396 | 462/370 |
| CD4/CD8 ratio | (0.9–2.6) | 3.2/4.4/2.7 | 2.1/2.9 |
| TCRγ/δ (%) | (0.2–14) | 4/ND/ND | 1.4/ND |
| CD31+CD45RA+/CD4+ (recent naive thymic emigrant; %) | (43–55) | 23/35/34 | 50 |
| CD45RO+/CD4+ (memory; %) | (13/30) | 69/53/52 | 32/40 |
| CCR7+CD45RA+/CD8+ (naive; %) | (52/68) | 23/29/25 | 62/58 |
| CCR7+CD45RA−/CD8+ (central memory; %) | (2–4) | 6/5/4 | 4/4.5 |
| CCR7−CD27−CD45RA−/CD8+ (effector memory; %) | (11/20) | 67/48/67 | 17/21 |
| CCR7+CD27−CD45RA+/CD8+ (exhausted effector memory; %) | (1–18) | 6/18/8 | 0.7 |
| CD127lowCD25+/CD4+ (regulatory; %) | (4–20) | 5.2/ND/ND | 6.6/ND |
| Va7.2+CD161+/CD3+ (MAIT; %) | (1–8) | 0.2/0.7/0.1 | 2.9/ND |
| Va24+Vb11+CD161+/CD3+ (iNKT; %) | (>0.02) | 0.06/0.09/0.05 | 0.03/ND |
| T cell proliferation (cpm 10−3) | |||
| PHA (6.25 mg ml−1) | (>50) | 150/ND/ND | 75/ND |
| OKT3 (10 ng ml−1) | (>30) | 65.5/ND/ND | ND |
| Candida | (>10) | 23/ND/ND | ND |
| Tetanus toxoid | (>10) | 12/ND/ND | ND |
| NK cells | |||
| CD16+CD56+ (cells mm−3) | (100–480) | 57/40/2 | 147/123 |
| CD16+CD56+ (%) | (4–17) | 3/2/1 | 7/8 |
| B cells | |||
| CD19+ (cells mm−3) | (270–860) | 342/520/704 | 357/332 |
| CD19+ (%) | (13–27) | 18/26/32 | 17/23 |
| CD21+CD27+/CD19+ (memory; %) | (>10) | 2/1/1 | 3/1 |
| IgD−IgM−/CD19+ CD21+CD27+ (switched; %) | (21–49) | ND/0.3/0.2 | ND |
| Ig levels (g liter−1; 16 yr old) | |||
| IgG | (6.6–12.8) | 13.9/14.21/8.92 | ND/12.5 |
| IgM | (0.5–2.1) | 0.99/0.74/0.28 | ND/0.27 |
| IgA | (0.7–3.4) | 0.69/0.56/0.45 | ND/0.81 |
Different immunological parameters of the patient were tested from blood and PBMCs: numbers of blood cell populations, different T cell subsets, B cell subsets, and NK cells from PBMCs (tested by flow cytometry), T cell proliferation in response to different stimuli (evaluated by incorporation of [3H]thymidine), and serum (Ig subclasses). Values in bold correspond to values below the age-matched normal values. MAIT, mucosal-associated invariant T cell.
The other healthy siblings were also evaluated repeatedly for the presence of EBV during the study. All had EBV-positive serology with IgG anti-VCA and anti-EBNA. No detectable EBV load was found in the three brothers. In contrast, the asymptomatic 8-yr-old sister exhibited a persistent and elevated blood EBV load (>106 copies/ml) once she encountered EBV at the age of 6 yr (Fig. 1 C). This high EBV load was only slightly reduced by treatment with anti-CD20, indicating that persistent EBV-infected cells were not B lymphocytes. This was confirmed by PCR from purified CD3+ cells, which detected a high content of EBV (5.9 × 106 copies/µg DNA), as well as by EBER staining of peripheral blood mononuclear cells (PBMCs; not shown). Even though she maintained a persistent high viremia, she remained asymptomatic since. She had normal immunological parameters, except that the proportion of memory B cells that was low before the anti-CD20 treatment remained low 2 yr after treatment (Table 1). These results suggest that the sister is affected by the same immunodeficiency, with the possibility of an up-to-now incomplete clinical penetrance.
Identification of a homozygous LOF mutation in TNFRSF9 in both siblings with EBV-infected T cells
To identify a gene defect responsible for the disease in the patient and his sister, whole-exome sequencing (WES) was performed in all family members. Because of the consanguinity, an autosomal recessive inheritance pattern of the gene defect was applied to filter WES data with a focus on homozygous genetic variations. One significant homozygous variation in TNFRSF9 was identified in exon 4 (NM_001561.5), corresponding to a frameshift point deletion that results in a premature stop codon (chromosome 1, g.7998819delC, c.170delG, p.G57fsX91; Fig. 1 E). It predicted to remove a large part of the extracellular, transmembrane, and intracytoplasmic domains of the protein. Sanger sequencing confirmed the WES results; only the patient and his sister were homozygous carriers of the mutation, while the parents were heterozygous (Fig. 1 A). Other siblings had heterozygous or WT genotypes. This mutation was not present in public WES and whole-genome sequencing databases and our own databases at IMAGINE Institute.
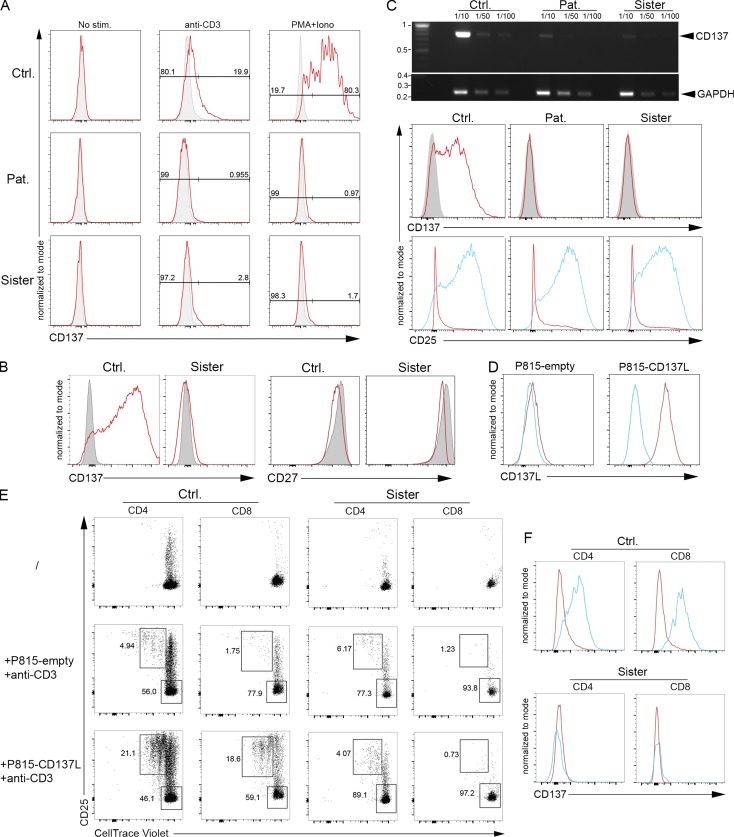
TNFRSF9 encodes CD137 (also known as 4-1BB), which belongs to the TNF receptor (TNFR) family (Wortzman et al., 2013). CD137 is a costimulatory molecule that is only expressed by activated T cells. When engaged by its ligand (CD137L), CD137 enhances antigen-specific T cell responses, including proliferation, IL-2 secretion, survival, and cytolytic activity (DeBenedette et al., 1997, 1999; Shuford et al., 1997; Wen et al., 2002; Bukczynski et al., 2004). As reported, CD137 expression was only detectable on control T cells that were activated with anti-CD3 antibody, anti-CD3/CD28–coated beads, PHA, or PMA + ionomycin, but not on resting T cells (Fig. 2, A–C). In contrast, T cells of the patient and his sister failed to express CD137 in these conditions. The expression of the full transcript of CD137 was also examined in PHA-activated T cells by semiquantitative RT-PCR and found to be markedly decreased in cells of the patient and his sister, while cells were activated similarly as control cells based on CD25 expression up-regulation (Fig. 2 C). This suggests that the CD137 transcript is subject to nonsense mutation–mediated RNA decay. Furthermore, no alternative spliced CD137 transcripts were detected in cells from the patient and his sister. These findings indicate that the premature stop codon p.G57fsX91 led to the absence of CD137 expression.
Figure 2.
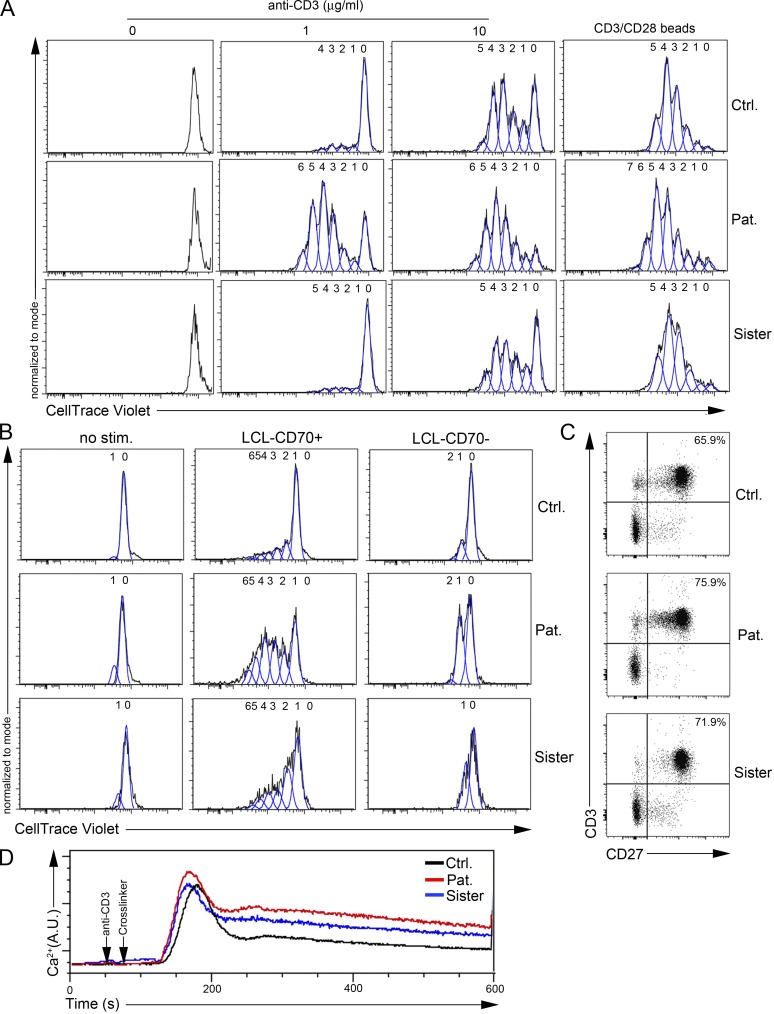
The G57fsX91 mutation prevents CD137 expression in activated T cells. (A) FACS histograms of CD137 of T cell blasts from the patient (Pat.), his sister, and a control (Ctrl.) unstimulated (No stim.) or stimulated with immobilized anti-CD3 antibody or PMA+ionomycin (PMA+Iono) for 7 d. Isotype control in gray. (B) FACS histograms of CD137 and CD27 of PBMCs unstimulated (gray) or stimulated with anti-CD3/CD28–coated beads (red) for 3 d. (C) Expression of CD137 full-length transcript (768 pb) and GAPDH (258 pb) by qRT-PCR in 72 h PHA-stimulated blasts. GAPDH as normalization controls for cDNA samples diluted as indicated, with the DNA ladder (kilobases) on the right. Bottom: FACS histograms of CD137 (isotype control in gray) and CD25 (non- and PHA-stimulated cells in red and blue, respectively). (D) FACS histograms of CD137L expression on P815 expressing CD137L (P815-CD137L) or not (P815-empty). Isotype control is shown in blue. (E) FACS dot-plots of proliferation assays of PBMCs labeled with the CellTrace Violet dye and cocultured or not (/) for 5 d with irradiated P815-CD137L or P815-empty cells preincubated with anti-CD3. Staining with anti-CD25 antibody as activation marker and gating on CD4+ or CD8+ T cells. Proliferating and nonproliferating T cells in the upper left and right lower gates, respectively. (F) FACS histograms of CD137 in proliferating (blue) or nonproliferating (red) CD4+ or CD8+ T cells gated from E. Numbers in A and E correspond the percentage of cells in gates. (D) One representative experiment of three. (E and F) experiments done in duplicate with the same results. One replicate is shown.
The costimulatory function of CD137 was next examined in T cells of the asymptomatic mutated sister only, since no material was available to test her affected brother. T cell proliferation was assessed following coculture with mouse mastocytoma P815 cells stably expressing or not CD137L (P815-CD137L) in the presence of soluble anti-CD3 antibody at a low concentration (Wen et al., 2002; Fig. 2 D). Coculture with P815-CD137L cells, but not with CD137L-negative P815 (P815-empty), triggered proliferation and up-regulation of the activation marker CD25 of both control CD4+ and CD8+ T cells from a healthy donor (Fig. 2 E, left panels). Importantly, dividing control CD25+ T cells expressed CD137, whereas nonproliferating cells were negative for CD137 (Fig. 2 F, upper panels). In contrast, T cells of the sister failed to proliferate when cocultured with CD137L-expressing P815 (Fig. 2 E, right panels), while CD137 was not detected (Fig. 2 F, lower panels). However, CD3/CD28-dependent proliferation of T cells of the sister (as well as the patient) was preserved and thus unaffected by CD137 deficiency (Fig. 6 E and Fig. 7, A and B).
Figure 6.
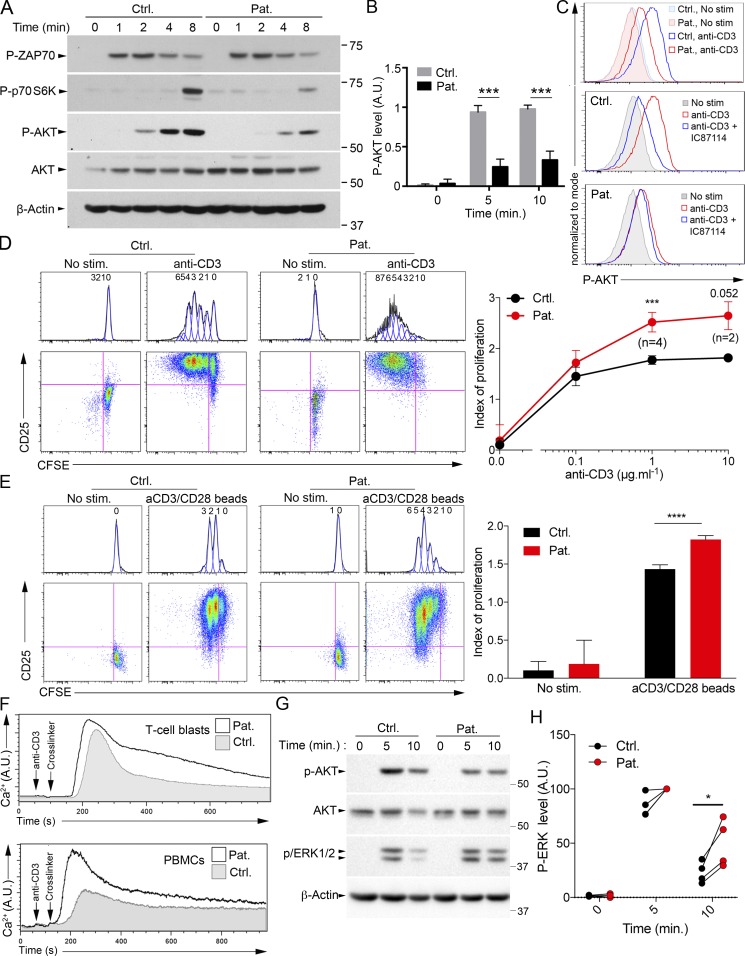
The R821H mutation in PIK3CD is associated with diminished activation of AKT and increased T cell proliferation and calcium flux. (A) Immunoblots of phosphorylated AKT (P-AKT), p70 S6K (P-p70 S6K), and ZAP70 (P-ZAP70) in T cell blasts from a control (Ctrl.) and the patient (Pat.) stimulated with anti-CD3 antibody for 0, 1, 2, 4, and 8 min. Total AKT and ACTIN were used as loading controls. Molecular weight markers are shown on the right. (B) Densitometry quantifications of immunoblots of P-AKT of T cells stimulated as in A for 0, 5, and 10 min. (C) FACS histograms of intracellular P-AKT in activated T cells not stimulated (no stim.) or stimulated with anti-CD3 in the presence or not of the p110δ pharmacological inhibitor IC87114 (IC). (D and E) FACS dot-plots and corresponding histograms of proliferation assays (after gating on CD3+ cells) assessed with the CFSE dye. T cell blasts (D) or PBMCs (E) stimulated or not with immobilized 1 µg ml−1 anti-CD3 (D, left), different doses of anti-CD3 (D, right) or anti-CD3/CD28 coated beads (E). Staining with anti-CD25 antibody as an activation marker. Numbers ahead each peak correspond to the number of divisions. Calculated proliferation indexes from FACS data are shown on the right. (F) Intracellular Ca2+ levels by real time flow cytometry in T cell blasts or PBMCs of Ctrl. (gray) or Pat. (black line) after stimulation with anti-CD3 and a cross-linker. (G) Same as A, with immunoblots of phosphorylated ERK (P-ERK). (H) Normalized densitometry quantifications of P-ERK immunoblots. Stimulation of patient cells at 5 min was used as 100%. *, P < 0.1; ***, P < 0.001; ****, P < 0.0001 with unpaired (B) or paired (D, E, and H) Student’s t test from data of five (B), two (0.1 and 10 µg ml−1; D), or four (0 and 1 µg ml−1; D) or four (E and H) independent experiments with mean and SD. In D, at 10 µg ml−1, P = 0.052 as indicated. Data are representative of three independent experiments for A, C, and F. A.U., arbitrary units.
Figure 7.
Increased proliferation of activated T cells from the patient is not observed in his sister. (A and B) FACS histograms from plots of proliferation assays of T cell blasts (A) or PBMCs (B) from a control (Ctrl.), the patient (Pat.), or his sister assessed with the CellTrace Violet dye. Numbers ahead each peak correspond to the number of divisions. Cells were (A) stimulated or not (0) with immobilized 1 or 10 µg ml−1 anti-CD3 antibody or anti-CD3/CD28–coated beads or (B) stimulated by coculture or not (no stim.) with LCLs expressing (LCL-CD70+) or not CD70 (LCL-CD70−) after preincubation with anti-CD3 antibody. (C) FACS dot-plots of CD27 and CD3 of PBMCs. Numbers represent the percentage of CD27+ CD3+ cells in the gates. (D) Intracellular Ca2+ levels analyzed by real-time flow cytometry in T cell blasts from the control (black line), the patient (red line), or his sister (blue line) after stimulation with anti-CD3 and a cross-linker. Data are from a single experiment (in which the cells of the patient and his sister were compared) but representative of several independent experiments in which the patient and his sister were tested separately and compared with controls (see Fig. 6, D and E; and Fig. S4 G).
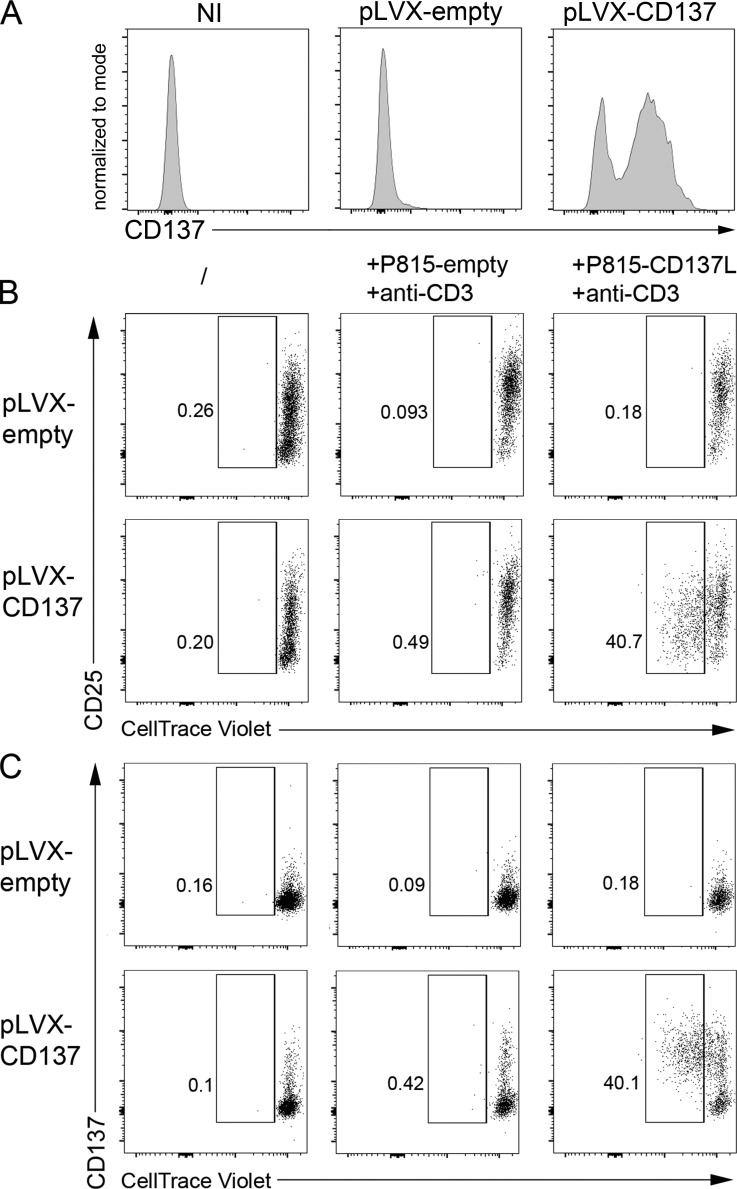
To formally prove that the premature stop codon p.G57fsX91 in CD137 was responsible for the defect of CD137L-triggered proliferation, CD137 expression was restored in T cells of the sister by transfecting PBMCs with a CD137-containing lentiviral vector or an empty vector (Figs. 3 and S2). CD137 expression was achieved in >40% of CD8+ T cells of the sister (Fig. 3 A). Restored CD137 expression by both CD8+ and CD4+ T cells was associated with their ability to proliferate and to express CD25 when cocultured with P815-CD137L, but not with P815-empty (Figs. 3 B and C and S2 and data not shown). Overexpression of CD137 by control CD8+ and CD4+ T cells only resulted in a slight increase of proliferating CD137+ T cells. These results demonstrate that the mutation in TNFRSF9 identified in the patient and his sister behaves as an amorphic mutation leading to impaired T cell expansion toward cells expressing CD137L. Thus, the loss of costimulatory function of CD137 in activated T cells of the patient and his sister could impair clearance of EBV-infected cells resulting from defective expansion of EBV-specific T cells.
Figure 3.
Correction of CD137 expression in CD137-deficient T cells restores their capacity to proliferate in response to CD137L-expressing cells. (A) FACS histograms of CD137 expression of T cells from the sister infected or not (NI) with a lentiviral vector containing or not (pLVX empty) a cDNA for CD137 (pLVX-CD137). (B and C) FACS dot-plots of proliferation assays from T cells shown in A (same as cells shown in Fig. 2 E). Cells were stained with an anti-CD25 antibody (B) as an activation marker or with an anti-CD137 antibody (C) and CellTrace Violet dilution analyzed by flow cytometry after gating on CD8+ T cells. Numbers correspond the percentage of proliferating cells in gates. Experiments in A–C were done in duplicate with the same results. One replicate is shown.
Identification of a homozygous missense LOF mutation in PIK3CD in the patient, but not in his sister
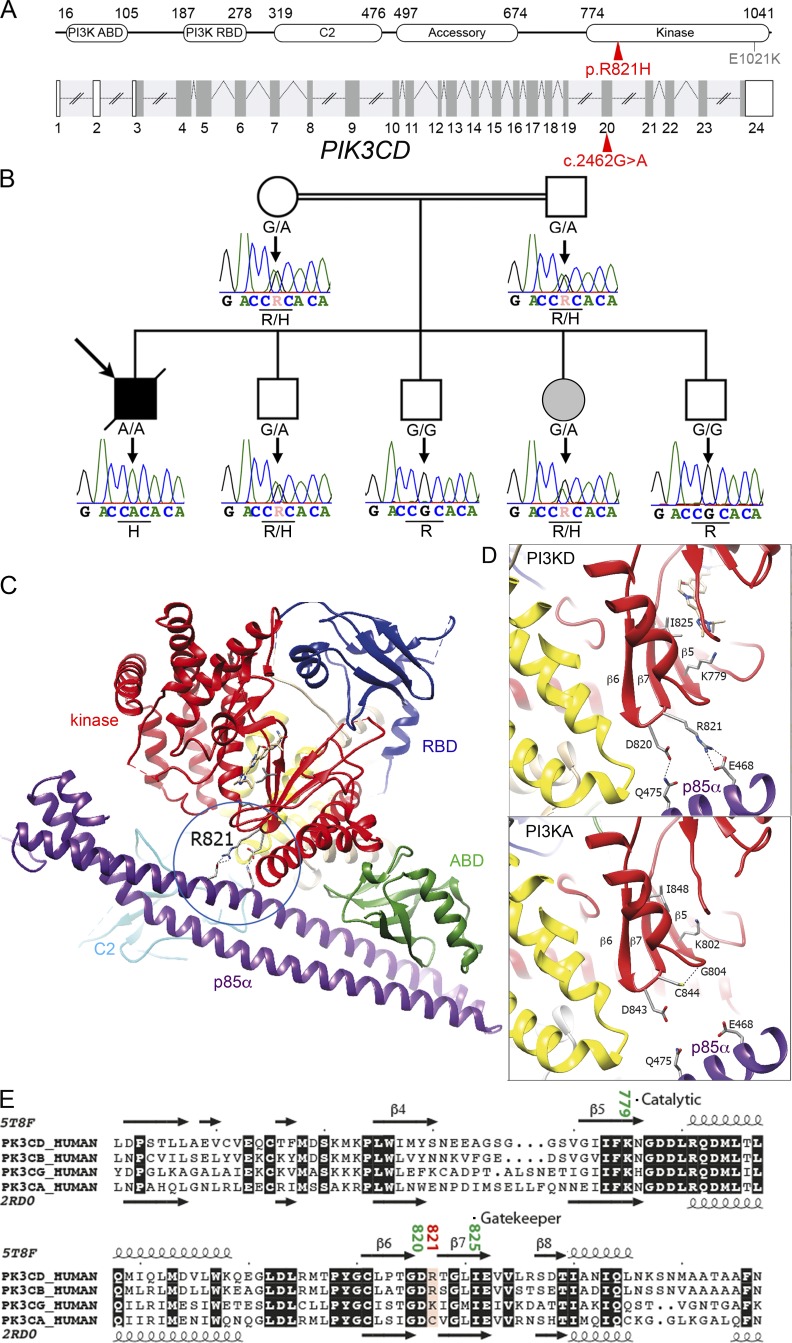
Because the patient’s sister has not developed any clinical manifestation despite the persistence of EBV viremia and circulating EBV-infected T cells, we postulated that the patient could be a carrier of an additional genetic factor that might account for the overt clinical phenotype. First, we looked for an allelic reversion event of the TNFRSF9 mutation in the patient’s sister that could have explained why she has no clinical signs. We failed to detect any reversion event in PBMCs and PHA-activated T cells from the sister (as well as from the patient) using Sanger and next-generation sequencing (Fig. S3). Therefore, WES results were reanalyzed considering the sister as not affected. One significant missense biallelic mutation in PIK3CD was identified in the patient at position 9783218 (rs573872848) on chromosome 1, (NM_005026.3 c.2462G>A) in exon 20 that led to a R821H substitution (Fig. 4 A). In public WES and whole-genome sequencing databases and own databases, the mutation was only found as heterozygous with a frequency of 3.8 × 10−5. The mutation was further verified by Sanger sequencing confirming the autosomal recessive inheritance in the kindred (Fig. 4 B). Both parents were heterozygous carriers of the mutation as well as the sister and one brother, while the other siblings were not mutated. The patient was thus the only homozygous carrier of the mutation. PIK3CD encodes p110δ, the catalytic subunit of the phosphoinositide 3-kinase (PI3K). PIK3CD deficiency was recently reported to cause immunodeficiency in four patients, but the functional consequences of the PIK3CD defect were not studied (Sogkas et al., 2018; Cohen et al., 2019; Swan et al., 2019). p110δ is highly expressed in lymphocytes and is the main active isoform of the PI3K catalytic subunit in human T cells (Okkenhaug et al., 2007; Okkenhaug and Fruman, 2010). p110δ associates with the regulatory p85α subunit which is also shared with other isoforms of PI3K catalytic subunit, including the p110γ and p110α (Engelman et al., 2006).
Figure 4.
Identification in the patient of a homozygous mutation in PIK3CD affecting the catalytic site. (A) Schematic representation of PIK3CD coding the p110δ. Intron–exon organization (coding regions in dark gray) and corresponding protein structure. ABD, adaptor-binding domain; RBD, Ras-binding domain. The mutation is indicated in red, and the previously GOF mutation E1021K in gray. (B) Family pedigree with DNA electropherograms and the PIK3CD c.2462 G>A genotype of each individual (also see Fig. 1 A). (C) Ribbon representation of the p110δ–p85α complex (PDB: 5T8F), highlighting possible bonds between the p110δ kinase domain and p85α, in addition to contacts made by C2 and ABD. (D) Comparison of the experimental three-dimensional structures of PIK3CD (PI3KD; PDB: 5T8F; Castanedo et al., 2017) and PIK3CA (PI3KA; PDB: 2RD0; Huang et al., 2007) in complex with p85α. The catalytic residues K779 (PI3KD) and K802 (PI3KA) are shown and specific contacts with p85α (in purple) are highlighted. (E) Alignment of isoforms of p110 within the kinase domain. The observed secondary structures of p110δ/PIK3CD (PDB: 5T8F) and p110α/PIK3CA (PDB: 2RD0) are reported above and below the alignment, respectively (UniProtKB accession numbers PIK3CD_HUMAN: O00329, PK3CB_HUMAN: P42338, PK3CG_HUMAN: P48736, and PK3CA_HUMAN: P42336).
Reduced kinase activity of the mutant PIK3CD/p110δ R821H
The R821H mutation is located in a highly conserved region of the p110δ catalytic domain. To characterize the impact of this mutation, analysis of the three-dimensional structures of p110δ–p85α complexes was undertaken. p110δ associates with its p85α subunit through interactions outside the kinase domain and involves the adaptor-binding domain (ABD) and the C2 domains (Huang et al., 2007; Vadas et al., 2011; Burke, 2018; Fig. 4 C). The R821 is located in the β6-β7 loop of the kinase domain in the vicinity of the active site that includes the gatekeeper I825 and catalytic lysine K779 (Fig. 4 D, upper panel). R821 and D820 are the only amino acids in the kinase domain able to establish bonds, after side-chain rotamer change, with p85α Q475 (H-bond) and E468 (salt-bridges). Thus, these interactions provide a specific contact point between the p110δ kinase domain and p85α, in addition to the well-recognized contacts made between p85α and the ABD and the C2 domain, which are shared by all isoforms. The former specific contact is lost in the p110δ R821H mutant protein. Such an interaction does not exist between p110α and p85α (Huang et al., 2007), where the equivalent amino acid to R821 in p110δ is a cysteine (C844), which makes an endogenous H-bond with the G802 residue, thereby contributing to the structure of the catalytic site (Fig. 4 D, lower panel; and Fig. 4 E). Similarly, the R821 likely contributes to the structure of the catalytic site through a salt bridge formed with the E468 of p85α. When this specific conformation is disrupted, kinase activity is likely impaired.
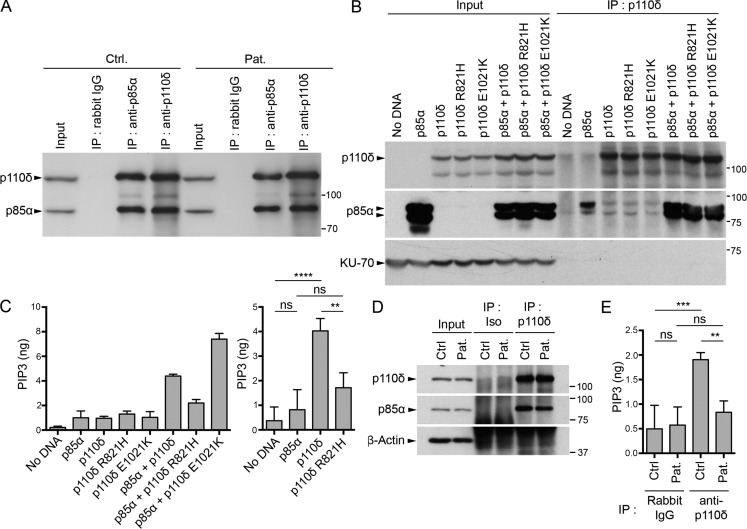
We therefore tested the respective consequences of the R821H substitution on both p110δ enzymatic activity and binding to p85α (Fig. 5). First, whole-cell extracts lysates from control and patient cells showed comparable levels of p110δ and p85α, indicating that the mutation had no impact on p110δ expression (Fig. 5 A). Similarly, immunoprecipitations (IPs) of p85α or p110δ recovered from both donor and patient cell extracts showed comparable amounts of p110δ and p85α, showing that the R821H mutation in p110δ did not disrupt association with p85α. Consequences of the R821H mutation on p110δ kinase activity was next examined in HEK293T cells that transiently expressed WT p110δ, the R821H mutant, or an overactive E1021K p110δ mutant and were cotransfected or not with p85α. Total cell extracts showed comparable expression of both WT and mutated forms of p110δ (Fig. 5 B). In IP experiments, recovery of WT and mutated p110δ was similar and comparable amounts of p85α were found to be associated with p110δ. Immunoprecipitates were then tested for PI3K activity. No production of PIP3 was detected in cells expressing p110δ alone in the absence of p85α, highlighting the need of the p85α subunit for actual kinase activity, as previously reported (Yu et al., 1998). Cotransfection of p85α and WT p110δ led to a significant production of PIP3 that was significantly diminished when p85α was coexpressed with the R821H p110δ (Fig. 5 C). As expected, cotransfection of p85α with the overactive E1021K p110δ further increased PIP3 production. Similar diminution in PIP3 production was observed from T cell blast protein extracts of the patient when compared with protein extracts of control cells (Fig. 5, D and E). These data demonstrate that the p110δ R821H mutation behaves as an LOF hypomorphic mutation resulting in a profound defect in PI3K activity.
Figure 5.
The R821H mutation in PIK3CD impairs p110δ kinase activity, but not binding to p85α. (A and B) Analysis of the p85α–p110δ association. (A) T cell lysates from the patient (Pat.) or a control (Ctrl.) after IP with control IgG (Rabbit IgG) or with antibody to p85α or p110δ followed by WB with anti-p85α or anti-p110δ. Total lysates (input) are shown on the left; molecular weight markers are shown on the right. (B) Lysates of HEK293T cells ectopically expressing or not p85α and/or WT, R821H, or E1021K p110δ, followed or not (input controls) by IP with anti-p110δ antibody (IP: p110δ) and WB with anti-p110δ, -p85α, and -KU-70 antibody as loading control. HEK293T expressed small amount of p85α that is not detected in the input but detectable in the IP of the p110δ, p110δ R821H, and p110δ E1021K (without ectopic p85α expression). (C) In vitro PI3K assays after IP, as in B, followed by quantification of produced PIP3 (left). Statistical analyses are shown on the right. (D) Same as A, except that total lysates are shown (input) and WB with anti–β-actin was used as a loading control. (E) In vitro PI3K assay of IPs obtained as in D followed by quantification of produced PIP3. Data are representative of two independent experiments (A, B, and C, left, and D) or are mean and SD of two independent experiments with three (C, right) or two (E) kinase assay replicates per group. ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA with post hoc Bonferroni t test).
Reduced PI3K-dependent AKT/S6K signaling in patient T cells
Activation of the AKT and p70-S6K kinase pathway is dependent on PI3K activity in T cells (Okkenhaug et al., 2007). Increased phosphorylation of AKT and p70-S6K in T cells is considered a hallmark of patients carrying PIK3CD GOF mutations associated with activated PI3K delta syndrome type 1 (Angulo et al., 2013; Lucas et al., 2014). Phosphorylation of p70-S6K and AKT on serine 473 and threonine 308 (not shown), respectively, were found to be significantly reduced in T cell blasts of the patient in response to TCR/CD3 stimulation in comparison to control cells (Fig. 6, A–C). However, tyrosine phosphorylation of ZAP-70 (Fig. 6 A) and whole tyrosine phosphorylation signals (data not shown) that are independent of PI3K activity were equivalent between patient and control cells. Residual AKT phosphorylation in patient cells is likely explained by the hypomorphic nature of the mutation and/or by the activity of other PI3K catalytic subunit isoforms. Of note, the p110δ-specific pharmacological inhibitor IC87114 reduced TCR/CD3–induced AKT phosphorylation in control cells to a level comparable to that seen in cells of the patient (Fig. 6 C). These results show that decreased kinase activity of the mutant R821H p110δ leads to reduced AKT and p70-S6K activation in patient T cells.
Increased proliferation of patient T cells associated with an elevated Ca2+ influx
PI3K-dependent signaling is known to regulate many lymphocyte functions, including cell survival and proliferation. Hence, we assessed T cell proliferation using a vital dye tracer. It was striking to observe that patient T cells directly tested from PBMCs or T cell blasts in culture exhibited a significantly increased proliferation in comparison to control cells in response to TCR/CD3 stimulation (Fig. 6, D and E). However, T cell blasts from the patient exhibited normal degranulation and activation-induced cell death upon anti-CD3 stimulation as well as normal apoptosis upon anti-FAS stimulation, indicating that not all T cell functions were increased (Fig. S4, A–C). We also examined intracellular Ca2+ mobilization and the MAPK/ERK pathway in response to TCR activation, which are known to be downstream and dependent of phosphatidylinositol-4,5-bisphosphate (PIP2) metabolism in T cells. Ca2+ flux and phospho-ERK to anti-CD3 stimulation were found to be increased in patient T cells when compared with control cells (Fig. 6, F–H). Consistent with increased Ca2+ flux, phospho-PLCγ1 was found to be enhanced in T cells of the patient upon anti-CD3 stimulation (Fig. S4 D).
T cells of the sister, who is heterozygous for the PIK3CD mutation, were also evaluated, showing an intermediate decrease of TCR-dependent AKT phosphorylation and Ca2+ flux upon CD3 stimulation when compared with the patient (Fig. 7 D; and Fig. S4, E and F), while T cell proliferation upon stimulation with CD3 alone or CD3/CD28–coated beads was not significantly different from that of controls cells (Figs. 7 A and S4 G). Taken together, these results indicate that the LOF of R821H mutation in PIK3CD independently of the mutation in TNFRSF9 is associated with increased TCR-dependent proliferation and calcium mobilization in T cells.
We previously reported that CD27, another costimulatory molecule of the TNFR superfamily expressed by T cells, is required for expansion of EBV-specific T cells toward EBV-infected B lymphoblastoid cell lines (LCLs) expressing CD70, the ligand of CD27 (Izawa et al., 2017). CD27-dependent T cell proliferation toward LCL-expressing CD70 (LCL-CD70+) or CD70-negative LCL (LCL-CD70−) was analyzed with T cells from the patient and his sister (Fig. 7 B). LCL-CD70+, but not LCL-CD70−, triggered proliferation of T cells from the sister that was normal or slightly increased compared with control cells. Proliferation of T cells from the patient was increased in the same conditions. CD27 was found normally expressed on T cells from the sister and the patient (Fig. 7 C). The increased CD27-dependent proliferation in the patient might suggest a compensatory mechanism for the absence of the CD137 pathway and/or related to the increased proliferation rate of activated T cells associated with the R821H mutation in PIK3CD.
Imbalanced phospholipase C gamma 1 (PLCγ1)–dependent signals in PIK3CD-deficient T cells
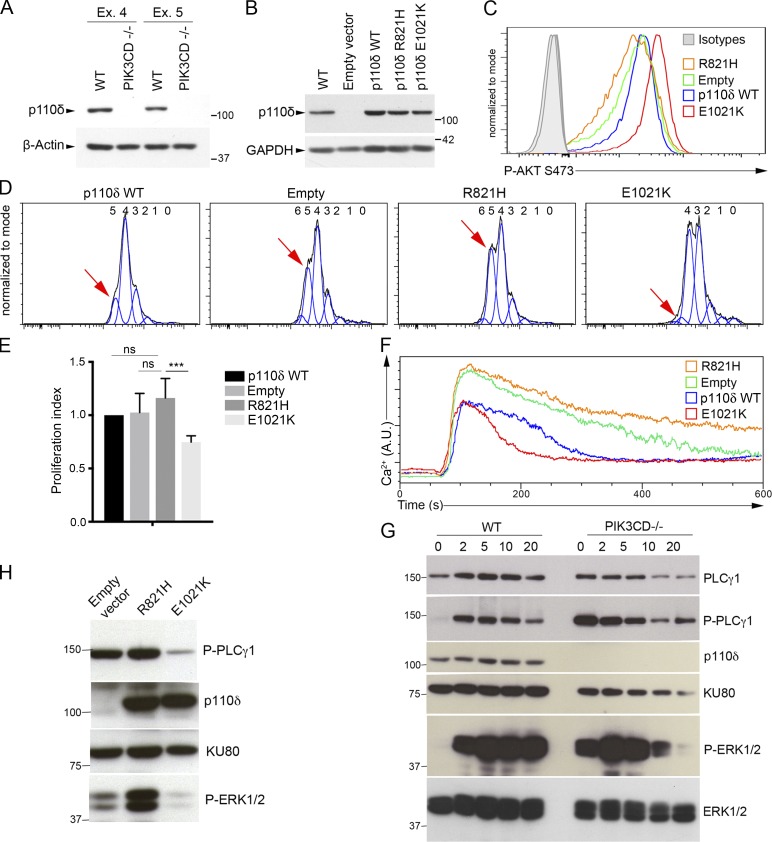
Because of the lack of available material from the patient and in order to further demonstrate the effect of the R821H p110δ mutation on T cell responses, two Jurkat T cell lines deficient for p110δ (PIK3CD−/−) were generated by the CRISPR-Cas9 technology using RNA guides targeting respectively exon 4 or exon 5 of PIK3CD (referred as CRISPR-Ex4 and CRISPR-Ex5). They were then reconstituted with the R821H mutant, the GOF E1021K mutant, or WT p110δ/PIK3CD (Figs. 8 and S5). Absence of p110δ expression in CRISPR-Ex4 and CRISPR-Ex5 cell lines was confirmed by Western blot (WB) analysis (Fig. 8 A). CRISPR-Ex4 and CRISPR-Ex5 cell lines transduced with WT, R821H p110δ, or E1021K p110δ coding lentiviruses showed comparable levels of p110δ (Figs. 8 B and S5). Constitutive AKT phosphorylation was examined and found to be increased in cells reconstituted with the activating E1021K mutant and reduced in cells expressing the inactive R821H mutant or in cells transduced with the empty vector (Figs. 8 C and S5). Of note, in Jurkat cells, AKT phosphorylation/activation and proliferation are constitutive and uncoupled to the TCR activation, in contrast to Ca2+ mobilization, which still requires TCR-CD3 stimulation (Shan et al., 2000; Astoul et al., 2001; Abraham and Weiss, 2004). When compared with E1021K p110δ–expressing cells, proliferation of p110δ-deficient cells and R821H p110δ–expressing cells was significantly increased (Fig. 8, E and D; and Fig. S5). Anti-CD3-induced calcium flux was also found to be increased (Figs. 8 F and S5). The abnormal T lymphocyte phenotypes of the patient were thus recapitulated in these Jurkat cell lines. Taken together, these results demonstrate that impaired PI3K activity leads to an increased calcium flux and cell proliferation, while increased PI3K activity results in the opposite phenotypes.
Figure 8.
Imbalanced PLCγ1–dependent signals in PIK3CD-deficient Jurkat T cells. (A and B) Immunoblots of p110δ expression (A) of PIK3CD-deficient (PIK3CD−/−) Jurkat cell lines obtained from CRISPR-Cas9 targeting of exon 4 (Ex. 4) or exon 5 (Ex. 5) of PIK3CD or nontargeted (WT) Jurkat cell lines (B) of WT or exon 4–targeted PIK3CD-deficient Jurkat cells (CRISPR-Ex4-PIK3CD−/−) reconstituted with an empty vector or a vector coding for WT (p110δ WT), R821H, or E1021K p110δ. (C) FACS histograms of intracellular phosphorylated AKT at serine 473 (Ser473 P-AKT) in the different exon 4 PIK3CD−/− Jurkat cell lines shown in B. Isotype controls are in gray. (D) FACS histograms from plots of proliferation assays of the different CRISPR–exon 4–PIK3CD−/− Jurkat cell lines shown in B assessed with the CellTrace Violet dye. Numbers ahead each peak correspond to the number of divisions. The red arrow indicates the peak of the fifth division in each histogram. (E) Indexes of proliferation calculated from D. (F) Intracellular Ca2+ levels analyzed by real-time flow cytometry in the different CRISPR–exon 4–PIK3CD−/− Jurkat cell lines shown in B stimulated with anti-CD3 antibody. (G) Immunoblots of phosphorylated PLCγ1 (P- PLCγ1) and ERK1/2 (P-ERK1/2) in WT Jurkat (WT) or CRISP–exon 4–PIK3CD−/− Jurkat (PIK3CD−/−) stimulated with anti-CD3 for 0, 1, 2, 5, 10, and 20 min. Total amounts of PLCγ1, PIK3CD (p110δ), KU80, and ERK1/2 are shown. (H) Same as G in nonstimulated PIK3CD−/− Jurkat (exon 4) reconstituted cells shown in B. (G and H) One representative experiment of two is shown. Similar experiments done with cells PIK3CD−/− cells obtained by targeting exon 5 showing the same results (see Fig. S4). Molecular weight markers are shown on the right (A and B) and left (G and H). Data are representative of three (A and B) or two (C, D, and F) independent experiments with three (D) or one (C and F) biological replicate per group in each. Two-way ANOVA test in E with mean and SD of two independent experiments with three and two replicates. ns, not significant; ***, P < 0.001, n = 5.
Calcium entry and ERK1/2 activation are initiated by PLCγ1, which shares with p110δ the same PIP2 substrate (Rhee, 2001; Engelman et al., 2006; Okkenhaug et al., 2007). TCR-dependent activation of PLCγ1 results in the cleavage of PIP2 into inositol-1,4,5-triphosphate (IP3) and diacylglycerol. IP3 and diacylglycerol are second messengers that activate Ca2+ mobilization and the MAPK pathway, respectively. Thus, our findings strongly suggest the existence of a balance between PLCγ1–dependent and PI3K-dependent signaling pathways that could compete for PIP2 availability. To test this hypothesis, we examined PLCγ1 and MAPK activation in response to TCR-CD3 stimulation in WT Jurkat cells compared with PIK3CD-deficient Jurkat cell lines (Figs. 8 G and S5). Phosphorylation of PLC-γ1 and ERK1/2 kinases, which are known to reflect PLCγ1 and MAPK activation, was tested in response to anti-CD3 stimulation. Strikingly, a constitutive phosphorylation of PLCγ1 and ERK1/2 was strongly detectable in PIK3CD-deficient cell lines, but not in WT cells. In PIK3CD-deficient cells, TCR-CD3 activation led to a further increased ERK1/2 phosphorylation that was more intense, fast, and blunted more rapidly compared with WT cells. Activation did not further increase PLCγ1 phosphorylation, which was already very high in unstimulated cells. In p110δ-deficient cells reconstituted with R821H, a marked constitutive phosphorylation of PLCγ1 and ERK1/2 was detectable, while in cells reconstituted with the GOF E1021K mutant, constitutive PLCγ1 and ERK1/2 phosphorylation was low (Figs. 8 H and S5). Therefore, these data show that in the absence of p110δ or impaired p110δ activity, PLCγ1 is more active, leading to increased calcium flux and activation of the downstream MAPK pathway. This may likely result in increased proliferation.
Discussion
We have identified herein an immunodeficient patient carrier of two homozygous LOF mutations in PIK3CD and TNFRSF9 with recurrent infections and an uncontrolled EBV-induced T cell proliferative disorder with features of CAEBV. His asymptomatic sister, who poorly controlled EBV replication, shared the same homozygous TNFRSF9 LOF mutation. These differential genotypes contributed to dissect the potential roles of each gene alteration.
Recently, four patients with severe infections and inflammatory colitis associated with hypogammaglobulinemia and loss of B cells have been reported to be carriers of homozygous frameshift mutations in PIK3CD resulting in premature stop codons leading to absence of protein or removing the catalytic domain (Sogkas et al., 2018; Cohen et al., 2019; Swan et al., 2019). These patients had normal T cell subsets and counts, and T cell proliferation in response to various stimuli was reported to be normal or enhanced (like in our patient). No EBV infection was reported in these patients, but it was not known whether these patients had encountered EBV. However, the clinical phenotype of our patient partially differs from those patients. There are several explanations for this difference. First, the R821H mutation of our patient is hypomorphic and associated with a normal expression and residual PIK3CD enzymatic activity, in contrast to the mutations in PIK3CD-deficient patients, which are either null or lead to a truncated protein. Thus, this may explain that in our patient, the B cell phenotype is attenuated. Second, our patient did not develop colitis. CD137 deficiency may impair T cell responses avoiding the occurrence of colitis in our patient. Indeed, the inflammatory colitis in one patient was examined and found to be associated with a substantial increase in activated effector CD8+ T compartments in the lamina propria, suggesting an abnormal expansion of effector CD8+ T cells (Swan et al., 2019). Of note, CD137 costimulation is well characterized in humans to induce CD8 T cell expansion with effector functions (see below), thus supporting that in our patient, such an expansion of CD8+ T cells in the lamina propria would be impaired. Interestingly, two patients with dual PIK3CD and small kinetochore-associated protein deficiency have been reported (Sharfe et al., 2018). Notably, these patients did not develop inflammatory colitis and exhibited strong T cell defects, including proliferation, in contrast to the other patients with PIK3CD deficiency. These T cell defects are likely caused by the small kinetochore-associated protein deficiency that impaired kinetochore/spindle microtubule dynamics during mitosis and might also explain why these two patients did not develop inflammatory colitis.
Heterozygous activating GOF mutations in PIK3CD have also been reported. They are responsible for activated PI3K delta syndrome type 1 syndrome, a complex immunodeficiency associated with variable clinical phenotypes (Angulo et al., 2013; Lucas et al., 2014; Coulter et al., 2017). To our knowledge, EBV-associated T/NK-cell lymphoproliferative disorders have not been reported in patients with GOF (and LOF) mutations in PIK3CD. Functional features associated with PIK3CD GOF mutations are increased activation-induced T cell death, low T cell proliferation, and increased phosphorylation of AKT and p70-S6K kinase (Angulo et al., 2013; Lucas et al., 2014; Carpier and Lucas, 2018). The T cell abnormalities detected herein in the patient are the opposite, consisting of increased T cell proliferation, decreased phosphorylation of AKT and p70-S6K kinase, normal senescent CD57+ T cells, and preserved activation-induced cell death. Therefore, these functional abnormalities (associated with the PIK3CDR821H mutation) correspond to dysfunctions that would have been expected to be caused by PIK3CDLOF mutations (in opposition to those associated with PIK3CD GOF mutations).
The observed association of the PI3KCDR821H LOF mutation with increased proliferation, ERK1/2 activation, and calcium flux comes as a relative surprise given the known role of the PI3K–AKT pathway in cell survival. However, an identical phenotype was reproduced in p110δ-deficient Jurkat cell lines obtained by CRISPR-Cas9, demonstrating that these abnormalities are not the consequence of unrelated genetic variations in the patient and/or indirect consequence of EBV infection of T cells. These results suggest that there is a physiological balance between PLCγ1 and PI3K-dependent pathways in T cells. Indeed, PLCγ1 and PI3Kδ (p110δ) activities share the same substrate, PIP2, and hence are competing to access to it (Okkenhaug et al., 2007). We propose that the skewed signaling toward PLC-γ1 resulting from PIK3CD deficiency may provide a permissive cellular background in T cells to EBV infection or/and for the persistence and expansion of infected T cells because of the related proliferation advantage. The mechanisms involved in the entry and maintenance of EBV in T cells are not known (Fujiwara et al., 2014). However, a recent study identified somatic cancer driver mutations in EBV-positive infected T cells of CAEBV patients (Okuno et al., 2019). These mutations accumulated when the disease progressed and were associated with a shorter overall survival. The homozygous germline PIK3CDR821H found in the patient might be viewed as a driver mutation allowing EBV-infected T cells to persist and acquire additional somatic mutations, leading to uncontrolled proliferation. Thus, a growth selective advantage could be a very significant contributing factor to the onset of CAEBV originating from germline (the present report) or somatic mutations.
The persistent EBV replication and the presence of circulating EBV-infected T cells in the healthy sister led us to the discovery of a homozygous deleterious mutation in TNFRSF9 shared by the patient and his sister. It is unlikely that the CD137 deficiency plays a role in the functional abnormalities detected in T cells of the patient, as these functional phenotypes were reproduced in p110δ-deficient Jurkat cells. Rather, it is likely that CD137 deficiency accounts for the impaired immune control of EBV-infected T cells. CD137 is a highly potent costimulatory molecule of T cell responses during viral infections (Wortzman et al., 2013). It has been shown that CD137 ligation sustains antigen-specific CD8+ T cell responses (Shuford et al., 1997; DeBenedette et al., 1999; Takahashi et al., 1999; Lee et al., 2002; Maus et al., 2002). Furthermore, targeting CD137 with agonistic antibodies also demonstrated potent effects on antitumoral and viral responses (Chester et al., 2018). Chimeric antigen receptor (CAR) T cell therapy using CARs containing the intracytoplasmic domain of CD137 was also shown to be the most efficient to sustained CAR T cell responses (Li et al., 2018).
CD27 is a well-known costimulatory molecule belonging to the TNFR super family, like CD137 (Watts, 2005). In contrast to CD137, CD27 is highly expressed on both resting and activated T cells. Importantly, primary immunodeficiencies caused by mutations in CD27 or CD70, the ligand of CD27, have been reported in patients with a high susceptibility to develop EBV-driven B cell lymphoproliferative disorders (van Montfrans et al., 2012; Salzer et al., 2013; Alkhairy et al., 2015; Abolhassani et al., 2017; Izawa et al., 2017). We showed that CD70 expression on EBV-infected B cells is required for expansion of EBV-specific T cells via cosignals deliver by CD27 (Izawa et al., 2017). Similarly, CD137-CD137 ligand (CD137L) interactions with antigen presenting cells possibly including EBV-infected T cells or B cells, may play an important similar nonredundant role in the expansion of EBV-specific T cells notably required to target EBV-infected T cells. Supporting this assumption, CD137L is expressed by T cells and was shown to induce T cell trans-costimulation resulting in potent tumor rejection (Stephan et al., 2007). Furthermore, EBV-unrelated cutaneous T cell lymphomas have been shown to express significant amount of CD137L (Kamijo et al., 2018). The recent description of EBV-driven B cell lymphomas in two patients with CD137 deficiency is strongly in favor of a key role of CD137 in immunity to EBV (Alosaimi et al., 2019), although the clinical asymptomatic feature of the sister (in our study) suggests incomplete penetrance. In this study, expansion and function of antigen-specific CD137-deficient CD8+ T lymphocytes in coculture with B-LCLs were impaired. With our observations, this supports that CD137 delivers to EBV-specific T cells cosignals required for their expansion.
In conclusion, we report the first genetically well-documented case of EBV-driven T/NK cell lymphoproliferative disorder associated with homozygous mutations in PIK3CD and TNFRSF9, highlighting the potential role of these pathways in the immunity to EBV when EBV infects T cells. Additional genetic and/or environmental factors may also participate to this susceptibility. These results provide the first evidence of the multifactorial genetic inheritance underlying EBV-associated T/NK cell lymphoproliferative disorders, that can be now considered as the result in part of a primary immune defect. This observation also constitutes one of the first examples of a recessive immunodeficiency resulting from two gene defects with mechanisms likely to independently contribute (and even synergize) to cause the disease.
Materials and methods
Ethics
Written informed consent was obtained from all humans in this study in accordance with the Helsinki declaration and local legislation and ethical guidelines from the Comité de Protection des Personnes de l’Ile de France II, Hôpital Necker-Enfants Malade, Paris. Blood from healthy donors was obtained at Etablissement français du sang under approved protocols (convention 15/EFS/012).
WES
Genomic DNA was extracted from whole blood according to standard methods, and WES and data analysis were performed as previously described (Martin et al., 2014; Izawa et al., 2017; Winter et al., 2018). Genomic DNA regions flanking PIK3CD and TNFRSF9 mutations were amplified using the forward primer 5′-CTTGACCATGCCATTTGC-3′ and reverse primer 5′-CTTGGACTTCAGCCAGTTG-3′ (for PIK3CD) or the forward primer 5′-CGTGTACCACTATTGTCTGCC-3′ and the reverse primer 5′-GAACTCATACCTTTACACTGCC-3′ (for TNFRSF9) with Platinum Taq DNA Polymerase (Invitrogen) according to manufacturer’s recommendations, gel purified with the High Pure PCR Product Purification Kit (Roche), sequenced with the BigDye terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), and analyzed on a 3500xL Genetic Analyzer (Applied Biosystems). All collected sequences were analyzed using DNADynamo (BlueTractorSoftware). To search for reversion events, amplicons of TNFRSF9 obtained by RT-PCR were also sequenced by WES methods to analyze single reads.
Cell cultures
PBMCs were isolated by Ficoll-Paque (Lymphoprep; Proteogenix) density-gradient centrifugation and washed and resuspended at a density of 106 cells/ml in complete Panserin 401 medium (Pan Biotech) containing 5% human male AB serum (BioWest), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco). T cell blasts were expanded by incubating PBMCs for 72 h with 2.5 µg ml−1 PHA (Sigma-Aldrich); dead cells were then removed by Ficoll-Paque density-gradient centrifugation (Lymphoprep), and T cell blasts were cultured in complete Panserin 401 medium culture supplemented with 100 IU/ml recombinant human IL-2 (R&D Systems). Murine mastocytoma P815 cell lines, EBV-infected B-LCLs, and Jurkat T cell lines were cultured in complete RPMI 1640 GlutaMAX medium (Invitrogen) containing 10% heat-inactivated fetal calf serum (Gibco), 100 U/ml penicillin, and 100 µg ml−1 streptomycin (Gibco). HEK293T cells were cultured in complete DMEM, high glucose, GlutaMAX supplement, pyruvate medium (Gibco) containing 10% heat-inactivated fetal calf serum (Gibco), 100 U/ml penicillin, and 100 µg ml−1 streptomycin (Gibco).
Constructs and RT-PCR for TNFRSF9
WT PIK3CD and TNFRSF9 coding sequences were obtained by RT-PCR from control T cell blasts with the forward primer 5′-CACCATGCCCCCTGGGGTGGACTG-3′ and the reverse primer 5′-GGGAGGAGCCACTACTGCCTGTTG-3′ for PIK3CD and with the forward primer 5′-CACCATGGGAAACAGCTGTTACAA-3′ and the reverse primer 5′-TCACAGTTCACATCCTCCTTCTTC-3′ for TNFRSF9. The PIK3CD and TNFRSF9 cDNAs were inserted into the plasmid pcDNA3.1 directional TOPO (Invitrogen) according to manufacturer’s instructions. The c.2462G>A (p.R821H) and c.3061G>A (p.E1021K) mutants were obtained with a Q5 site-directed mutagenesis kit (Promega) with the following primers: 2462GA-F, 5′-ACCGGGGACCACACAGGCCTC-3′; 2462GA-R, 5′-GGGGAGGCAGCCATAGGG-3′; 3061GA-F, 5′-GAAGTTTAACAAAGCCCTCCGTG-3′; and 3061GA-R, 5′-ACTCGGAAGTGCTTCAGTG-3′. Coding sequences were confirmed by Sanger sequencing and subcloned into the bicistronic lentiviral plasmid (pLenti7.3/V5 TOPO; Invitrogen) or modified by replacing puromycin coding sequences by those of GFP as the reporter gene (pLenti7.3-GFP). p85α coding expression plasmid (p3XFLAG-CMV-p85α) was previously reported (Deau et al., 2014).
CD137/TNFRSF9 transcript expression was analyzed by semiquantitativePCR with the forward primer 5′-ATGGGAAACAGCTGTTACAA-3′ and the reverse primer 5′-TCACAGTTCACATCCTCCTTCTTC-3′ for TNFRSF9 and with the forward primer 5′-ATGCCATCACTGCCACCCAG-3′ and the reverse primer 5′-CCTGCTTCACCACCTTCTTG-3′ for GAPDH as a control for amplification and normalization.
CRISPR-Cas9 genome editing
The pSpCas9(BB)-2A-GFP (pX458) plasmid was a gift from Feng Zhang (Massachusetts Institute of Technology, Boston, MA; plasmid 48138; Addgene). All sgRNAs were designed using the Massachusetts Institute of Technology CRISPR Design Tool (http://crispr.mit.edu). 24-bp oligonucleotides containing the target sequences in exon 4 and exon 5 of PIK3CD were synthesized (Eurogentec) with a 4-bp overhang (later shown in lowercase) to enable cloning into the BbsI site. sgRNA sequences are as follows: sgEx4F, 5′-caccgAGGTGAACACATAGGCCTCG-3′; sgEx4R, 5′aaacCGAGGCCTATGTGTTCACCTc-3′; sgEx5F, 5′-caccgGTACCCTGCGGCTCCCGAAC-3′; sgEx5R, 5′-aaacGTTCGGGAGCCGCAGGGTACc-3′. Pairs of synthetized oligos were annealed, phosphorylated, ligated to linearized vector, and transformed into Stbl3 bacteria (Life Technologies). sgRNA insertion was confirmed by Sanger sequencing using sequencing primer (5′-TTTCTTGGGTAGTTTGCAGTTTT-3′). The plasmids were serially transfected three times into Jurkat T cells with Nepa21 electroporator (Nepagene). Cells were subcloned by limit dilution and cultured for 3 wk. DNA was extracted, and regions flanking the targeted site were amplified and sequenced using the following primers: Ex4F, 5′-ACTGTCCCTCCAGCTGCTGT-3′; Ex4R, 5′-GAGCTACCTTTGCCGATGAGGAGG; Ex5F, 5′-CCGCAGGCCTCCACGAGTTT-3′; Ex5R, 5′-CTCACCTCGCTGCCCTCAAACT. Two clones presenting biallelic frameshift mutations in exon 4 (g.1:9715598delG, c.199delG, p.A67fsX42) or exon 5 (g.1:9716036_046, c.558_568del, p.P186PfsX) were selected.
Lentiviral gene transfer
PIK3CD-deficient Jurkat cell lines with empty pLenti-7.3-GFP or pLenti-7.3-GFP encoding WT, p.R821H, and p.E1021K p110δ were performed by lentivirus-mediated gene transfer. HEK293T cells were cotransfected with the lentiviral plasmid together with the plasmids pVSVG and pGag-pol at a molar ratio of 1:1:1 with Lipofectamine 2000 (Invitrogen) according to supplier’s instructions. Supernatants containing virus particles were harvested 36 h after transfection and freshly used for infection of Jurkat cells. All Jurkat cell lines expressed comparable levels of CD3 and CD28. Transductions of CD137-deficient T cells or control T cells with the pLenti-7.3-CD137 as previously described (Martin et al., 2014).
Immunoblotting
Stimulation of cells, protein extraction, and immunoblotting protocols have been described previously (Hauck et al., 2012; Martin et al., 2014; Winter et al., 2018). Briefly, cells (5 × 106 cells per ml) were stimulated by anti-CD3 antibody (1 µg/ml, clone OKT3) cross-linked with a rabbit anti-mouse IgG (2 µg/ml) for the different time periods. Cells were washed in PBS, and proteins were extracted with cell lysis buffer (1% NP-40 [NP-40 alternative; Calbiochem], 50 mM Tris, pH 8, 150 mM NaCl, 20 mM EDTA, 1 mM Na3VO4, 1 mM NaF, complete protease inhibitor cocktail [Roche], and phosphatase inhibitor cocktails 2 and 3 [Sigma]). Proteins were denatured by boiling 10 min with sample buffer (125 mM Tris, pH 6.8, 3% SDS, 10% glycerol, 5% 2β-mercaptoethanol, and 0.01% bromophenol blue), separated by SDS-PAGE, and transferred on polyvinylidene fluoride membrane (Millipore). Membranes were blocked with milk or BSA-based buffer before incubation with antibodies. The following antibodies were used for immunoblotting: anti-AKT (11E7, #4685S), anti-AKT phosphorylated on T308 (244F9, #4056S), anti-AKT phosphorylated on S473 (193H12, #4058S), anti-Erk1/2 (p44/42 MAPK, 137F5, #4695S), anti-phosphorylated Erk1/2 (phospho-p44/42 MAPK, 20G11, #4376S), anti-Ku70 (D35, #4103S), anti-p110α (C73F8, #4249S), anti-p85α(#4292), anti-p70 S6K (#9202S), anti-phosphorylated p70 S6K (108D2, #9234S), anti-PLC-γ1 (#2822S), anti-phosphorylated PLCγ1 (D6M9S, #140008S), anti-PTEN (138G6, #9559S), anti-phosphorylated tyrosine (P-Tyr-100, #9411S), anti-ZAP-70 (99F2, #2705S), and anti-phosphorylated ZAP-70 (#2704S; all from Cell Signaling Technology), anti-PI3K p110δ (H-219, sc-7176; Santa Cruz), and anti–β-actin (A2066, Sigma-Aldrich). Membranes were then washed and incubated with secondary anti-mouse (GE-Healthcare) or anti-rabbit (Cell Signaling Technology) HRP-linked antibodies. Pierce ECL WB substrate and Bio-Rad Clarity ECL substrate were used for revelation. For reprobing, membranes were incubated with Restore WB stripping buffer (Thermo Scientific) before blocking. Densitometry analyses were performed using ImageJ software and normalized to loading WB controls.
Immunochemistry
Staining and in situ hybridization were performed on an automated stainer (Bond Max; Leica Biosystems). The presence of EBV was demonstrated by in situ hybridization for the small RNA–encoding regions 1 and 2 (EBER). Antibodies used and dilution were anti-CD3 (polyclonal anti-CD3, 1/200; DAKO), anti-CD20 (polyclonal anti-CD3, 1/200; DAKO), and anti-CD8 (clone C8/144B; 1/200; DAKO).
PI3 kinase assay and IPs
HEK293T cells were transfected with the indicated expression plasmids using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. T cell blasts or transfected (HEK293T cells 48 h after transfection) were lysed as described above. 500 mg protein was incubated with 2 µg polyclonal rabbit anti-anti-p110δ from Santa-Cruz (clone H-219) or normal rabbit IgG from Cell Signaling Technology (#2729) for 1 h, and immune complexes were recovered with Protein A Sepharose beads (GE Healthcare) at 4°C. The beads were then washed three times with 20 mM Tris-HCl, pH 7.4, 137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM sodium orthovanadate; three times with 0.1 M Tris-HCl, pH 7.4, 5 mM LiCl, and 1 mM sodium orthovanadate; and two times with 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA. For immunoblot, proteins were eluted from beads by boiling with SDS sample buffer for 10 min and were processed as described above. Kinase assay was performed with PI3K Activity ELISA:pico kit (Echelon) according to supplier recommendations. Briefly, immunoprecipitates were incubated with PI(4–5)P2 substrate for 6 h, and enzyme reaction and PIP3 standards were then incubated with a PIP3-binding protein. The mixture was transferred on a PIP3-coated microplate for competitive binding. Afterwards, a peroxidase-linked detector was added, allowing colorimetric detection of the amount of PIP3 produced by comparison with the standard curve.
Flow cytometry
Cell membrane staining and the flow cytometry–based phenotypic analyses were performed according to standard flow cytometry methods. The following validated antibodies were used: anti-CD3 (UCHT1), anti-CD4 (OKT4), anti-CD8 (RPA-T8), anti-CD14 (M5E2), anti-CD19 (HIB19), anti-CD25 (BC96), anti-CD27 (O323), anti-CD28 (CD28.2), anti-CD56 (HCD56), anti-CD57 (NK.1), anti-CD107a/b (H4A3/HAB4), anti-CD137 (4B4-1), anti-CD161 (HP-3G10), anti-TCR Vα7.2 (3C10), and anti-IgM (MHM88; all purchased from BioLegend); anti-CD16 (3G8), anti-CD45RA (HI100), anti-CD45RO (UCHL1), anti-CD197/CCR7 (3D12), anti-Perforin (dG9), anti-TCRαβ (OT31), and anti-IgD (IA6-2; BD Biosciences); anti-CD21 (BL13), anti-TCRγδ (IMMY510), anti-TCR Vα24 (C15), and anti-TCR Vβ11 (X21; Beckman Coulter). These antibodies were conjugated to FITC, PE, phycoerythrin-cyanin5 (PE-Cy5), PE-Cy5.5, PE-Cy7, peridinin-chlorophyll, peridinin-chlorophyll-Cy5.5, allophycocyanin, allophycocyanin-Cy7, allophycocyanin-Vio7, Brilliant Violet 421 (BV421), BV510, BV605, BV650, BV711, or BV785. For phosflow staining, 106 cells were incubated in 1 ml PBS with 1 µg/ml anti-CD3, pelleted, resuspended in 100 µl hot PBS containing 2 µg/ml rabbit anti-mouse cross-linker, incubated at 37°C for 5 min, fixed with 1 ml preheated phosflow fix buffer 1 (BD Biosciences), and then permeabilized with phosflow perm buffer 3 (BD Biosciences) according to manufacturer’s instructions. Cells were then incubated with Alexa Fluor 647–conjugated anti-AKT phosphorylated on S473 antibody (D9E; Cell Signaling) or Alexa Fluor 647–conjugated anti-AKT phosphorylated on T308 antibody (C31E5E; Cell Signaling). All data were collected on an LSRFortessa X-20 cytometer (BD Biosciences).
Proliferation assays
T cell blasts were washed and cultured without IL-2 for 72 h to synchronize the cells. T cell blasts or PBMCs were labeled with CellTrace Violet dye (Invitrogen) or CFSE (Invitrogen) according to the manufacturer’s instructions. Cells were then cultured for 4 to 7 d in complete Panserin 401 medium alone or in the presence of 0.1, 1, or 10 µg/ml immobilized anti-CD3 antibody (clone OKT3; eBioscience), dynabeads Human T-Activator CD3/CD28 (Invitrogen), 100 IU/ml IL-2, 2.5 µg ml−1 PHA, or 10−5 M ionomycin (Sigma-Aldrich) plus 10−7 M PMA (Sigma-Aldrich). Cells were surface stained for CD3, CD4, CD8, and CD25 detection and analyzed by flow cytometry (LSRFortessa X-20; BD Biosciences). Proliferation assays with coculture of T cells with the P815 cells or EBV B cells have been previously described elsewhere (Wen et al., 2002; Izawa et al., 2017). Briefly, irradiated P815 expressing or not CD137L or irradiated EBV B cell lines (LCLs) expressing CD70 were preincubated with soluble 0.25 µg ml−1 anti-CD3 antibody, washed, and cocultured with PBMCs or T cell blasts labeled with CellTrace Violet dye. The proliferation index (corresponding to the total number of divisions divided by the number of cells that went into division) was calculated using FlowJo software (TreeStar).
Calcium flux analysis
Ca2+ influx was assessed by real-time flow cytometry, as previously described (Martin et al., 2014). Briefly, cells were loaded with 5 µM Indo-1 AM (Molecular Probes) in presence of 2.5 mM of probenecid (PowerLoad; Molecular Probes), washed, and surface stained for CD4 and CD8 detection. Cells were analyzed in real time with a FACS ARIA II flow cytometer (BD Biosciences). During acquisition, 1 µg ml−1 anti-CD3 antibody was added to the cells, followed by 10 µg ml−1 of F(ab′)2 rabbit–anti-mouse IgG cross-linker (Jackson ImmunoResearch) and finally incubated with a calcium ionophore (1 mM ionomycin; Sigma). Changes in the intracellular calcium concentration are quantified by a shift in the indo-1 emission peak from 485 nm (indo-blue) for unbound dye to 405 nm (indo-violet) when the indo-1 molecule is bound to calcium. Data were analyzed using kinetic tool of FlowJo software (TreeStar). Intracellular Ca2+ levels correspond to the normalized ratio of 405 nm/485 nm indo-1 emission peaks.
Apoptosis assay
T cell blasts were left unstimulated or stimulated for 12 h with 0.01, 0.1, 1, and 10 µg ml−1 immobilized anti-CD3 (clone OKT3) or cross-linked anti-FAS antibody (clone Apo1.3) as previously described (Rigaud et al., 2006). Cells were then washed and stained for viability (viaprobe; BD Biosciences); surface expression of CD3, CD4, and CD8; and surface localization of phosphatidylserines using PE-conjugated Annexin-V (BD Biosciences) and analyzed by flow cytometry. Apoptotic cells corresponding to Annexin V+/viaprobe− cells.
Degranulation assay
T cell blasts were stimulated for 4 h with 0.3, 3, and 30 µg ml−1 of immobilized anti-CD3 in the presence of PE-conjugated anti-LAMP-1/2 (H4A3, H4B4; BD Biosciences) as previously described (Martin et al., 2014). Cells were then washed and stained for surface expression of CD3 and CD8 and analyzed by flow cytometry.
Statistical analyses
Data were analyzed using paired and unpaired Student’s t test or one-way ANOVA with a post hoc Bonferroni t test using PRISM software (GraphPad).
Online supplemental material
Fig. S1 shows the infiltrate of EBV-infected CD3+CD8+ T cells in hydroa vacciniforme–like lesions from a skin biopsy of the patient. Fig. S2 shows that correction of CD137 expression in CD4+ CD137-deficient T cells from the sister restores their capacity to proliferate in response to CD137L-expressing cells. Fig. S3 shows that sequencing of the mutation by Sanger or NGS does not reveal any reversion genetic event in lymphocytes from the patient and his sister. Fig. S4 shows additional functional analyses of T cells from the patient and his sister, including apoptosis, degranulation, phospho-PLC-γ1, phospho-AKT, calcium flux, and proliferation. Fig. S5 shows signaling analyses of PIK3CD/p110δ-deficient Jurkat T cells obtained by targeting PIK3CD exon 5 by CRISPR-Cas9 and reconstituted or not with WT, R821H, or E1021K p110δ.
Supplementary Material
Acknowledgments
We acknowledge the patient, his family, and the healthy donors for cooperation and blood gifts. S. Latour is a senior scientist at the Centre National de la Recherche Scientifique (France).
R. Rodriguez was supported by the Ministère de la Recherche (France) and Ligue Contre le Cancer (France), D. Boutboul by the Agence Nationale de la Recherche (France), S. Winter and D. Jorge Cordeiro by the Imagine Institut PhD program funded by the Fondation Bettencourt Schueller, and B. Fournier by the Fondation pour la Recherche Médicale (FDM20170638301). This work was supported by grants from the Ligue Contre le Cancer Equipe Labelisée (France; S. Latour), Institut National de la Santé et de la Recherche Médicale (France), Rare Diseases Fondation (France; S. Latour), the Agence Nationale de la Recherche (ANR-14-CE14-0028-01 and ANR-18-CE15-0025-01 [S. Latour] and ANR-10-IAHU-01 [Imagine Institut]), and the European Research Council (ERC-2009-AdG_20090506 n°FP7-249816; A. Fischer).
The authors declare no competing financial interests.
Author contributions: R. Rodriguez, D. Jorge Cordeiro, S. Winter, S. Latour, and B. Fournier designed the research and performed experiments and analyzed the data. C. Picard, D. Boutboul, K. Izawa, C. Lenoir, E. Martin, J. Bruneau, S. Fraitag, and I. Callebaut performed experiments and analyzed the data. B. Neven identified the patients and provided clinical data. B. Neven, S. Latour, and A. Fischer analyzed clinical data. S. Kracker and T.H. Watts provide key reagents. A. Fischer and R. Rodriguez participated in the writing. S. Latour wrote the manuscript and supervised the research.
References
- Abolhassani, H., Edwards E.S., Ikinciogullari A., Jing H., Borte S., Buggert M., Du L., Matsuda-Lennikov M., Romano R., Caridha R., et al. 2017. Combined immunodeficiency and Epstein-Barr virus-induced B cell malignancy in humans with inherited CD70 deficiency. J. Exp. Med. 214:91–106. 10.1084/jem.20160849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, R.T., and Weiss A.. 2004. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 4:301–308. 10.1038/nri1330 [DOI] [PubMed] [Google Scholar]
- Alkhairy, O.K., Perez-Becker R., Driessen G.J., Abolhassani H., van Montfrans J., Borte S., Choo S., Wang N., Tesselaar K., Fang M., et al. 2015. Novel mutations in TNFRSF7/CD27: Clinical, immunologic, and genetic characterization of human CD27 deficiency. J. Allergy Clin. Immunol. 136:703–712.e10. 10.1016/j.jaci.2015.02.022 [DOI] [PubMed] [Google Scholar]
- Alosaimi, M.F., Hoenig M., Jaber F., Platt C.D., Jones J., Wallace J., Debatin K.M., Schulz A., Jacobsen E., Möller P., et al. 2019. Immunodeficiency and EBV-induced lymphoproliferation caused by 4-1BB deficiency. J. Allergy Clin. Immunol. 144:574–583.e5. 10.1016/j.jaci.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo, I., Vadas O., Garçon F., Banham-Hall E., Plagnol V., Leahy T.R., Baxendale H., Coulter T., Curtis J., Wu C., et al. 2013. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 342:866–871. 10.1126/science.1243292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astoul, E., Edmunds C., Cantrell D.A., and Ward S.G.. 2001. PI 3-K and T-cell activation: limitations of T-leukemic cell lines as signaling models. Trends Immunol. 22:490–496. 10.1016/S1471-4906(01)01973-1 [DOI] [PubMed] [Google Scholar]
- Bukczynski, J., Wen T., Ellefsen K., Gauldie J., and Watts T.H.. 2004. Costimulatory ligand 4-1BBL (CD137L) as an efficient adjuvant for human antiviral cytotoxic T cell responses. Proc. Natl. Acad. Sci. USA. 101:1291–1296. 10.1073/pnas.0306567101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J.E. 2018. Structural Basis for Regulation of Phosphoinositide Kinases and Their Involvement in Human Disease. Mol. Cell. 71:653–673. 10.1016/j.molcel.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Carpier, J.M., and Lucas C.L.. 2018. Epstein-Barr Virus Susceptibility in Activated PI3Kδ Syndrome (APDS) Immunodeficiency. Front. Immunol. 8:2005. 10.3389/fimmu.2017.02005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanedo, G.M., Blaquiere N., Beresini M., Bravo B., Brightbill H., Chen J., Cui H.F., Eigenbrot C., Everett C., Feng J., et al. 2017. Structure-Based Design of Tricyclic NF-κB Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K). J. Med. Chem. 60:627–640. 10.1021/acs.jmedchem.6b01363 [DOI] [PubMed] [Google Scholar]
- Chester, C., Sanmamed M.F., Wang J., and Melero I.. 2018. Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood. 131:49–57. [DOI] [PubMed] [Google Scholar]
- Cohen, J.I. 2015. Primary Immunodeficiencies Associated with EBV Disease. Curr. Top. Microbiol. Immunol. 390:241–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S.B.W., Bainter W., Johnson J.L., Lin T.Y., Wong J.C.Y., Wallace J.G., Jones J., Qureshi S., Mir F., Qamar F., et al. 2019. Human primary immunodeficiency caused by expression of a kinase-dead p110δ mutant. J. Allergy Clin. Immunol. 143:797–799.e2. 10.1016/j.jaci.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter, T.I., Chandra A., Bacon C.M., Babar J., Curtis J., Screaton N., Goodlad J.R., Farmer G., Steele C.L., Leahy T.R., et al. 2017. Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: A large patient cohort study. J. Allergy Clin. Immunol. 139:597–606.e4. 10.1016/j.jaci.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deau, M.C., Heurtier L., Frange P., Suarez F., Bole-Feysot C., Nitschke P., Cavazzana M., Picard C., Durandy A., Fischer A., and Kracker S.. 2014. A human immunodeficiency caused by mutations in the PIK3R1 gene. J. Clin. Invest. 124:3923–3928. 10.1172/JCI75746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBenedette, M.A., Shahinian A., Mak T.W., and Watts T.H.. 1997. Costimulation of CD28- T lymphocytes by 4-1BB ligand. J. Immunol. 158:551–559. [PubMed] [Google Scholar]
- DeBenedette, M.A., Wen T., Bachmann M.F., Ohashi P.S., Barber B.H., Stocking K.L., Peschon J.J., and Watts T.H.. 1999. Analysis of 4-1BB ligand (4-1BBL)-deficient mice and of mice lacking both 4-1BBL and CD28 reveals a role for 4-1BBL in skin allograft rejection and in the cytotoxic T cell response to influenza virus. J. Immunol. 163:4833–4841. [PubMed] [Google Scholar]
- Edwards, E.S.J., Bier J., Cole T.S., Wong M., Hsu P., Berglund L.J., Boztug K., Lau A., Gostick E., Price D.A., et al. 2019. Activating PIK3CD mutations impair human cytotoxic lymphocyte differentiation and function and EBV immunity. J. Allergy Clin. Immunol. 143:276–291.e6. 10.1016/j.jaci.2018.04.030 [DOI] [PubMed] [Google Scholar]
- Engelman, J.A., Luo J., and Cantley L.C.. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7:606–619. 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- Fujiwara, S., Kimura H., Imadome K., Arai A., Kodama E., Morio T., Shimizu N., and Wakiguchi H.. 2014. Current research on chronic active Epstein-Barr virus infection in Japan. Pediatr. Int. 56:159–166. 10.1111/ped.12314 [DOI] [PubMed] [Google Scholar]
- Hauck, F., Randriamampita C., Martin E., Gerart S., Lambert N., Lim A., Soulier J., Maciorowski Z., Touzot F., Moshous D., et al. 2012. Primary T-cell immunodeficiency with immunodysregulation caused by autosomal recessive LCK deficiency. J. Allergy Clin. Immunol. 130:1144–1152.e11. 10.1016/j.jaci.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Huang, C.H., Mandelker D., Schmidt-Kittler O., Samuels Y., Velculescu V.E., Kinzler K.W., Vogelstein B., Gabelli S.B., and Amzel L.M.. 2007. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 318:1744–1748. 10.1126/science.1150799 [DOI] [PubMed] [Google Scholar]
- Izawa, K., Martin E., Soudais C., Bruneau J., Boutboul D., Rodriguez R., Lenoir C., Hislop A.D., Besson C., Touzot F., et al. 2017. Inherited CD70 deficiency in humans reveals a critical role for the CD70-CD27 pathway in immunity to Epstein-Barr virus infection. J. Exp. Med. 214:73–89. 10.1084/jem.20160784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo, H., Miyagaki T., Shishido-Takahashi N., Nakajima R., Oka T., Suga H., Sugaya M., and Sato S.. 2018. Aberrant CD137 ligand expression induced by GATA6 overexpression promotes tumor progression in cutaneous T-cell lymphoma. Blood. 132:1922–1935. 10.1182/blood-2018-04-845834 [DOI] [PubMed] [Google Scholar]
- Kimura, H., and Cohen J.I.. 2017. Chronic Active Epstein-Barr Virus Disease. Front. Immunol. 8:1867. 10.3389/fimmu.2017.01867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., Hoshino Y., Kanegane H., Tsuge I., Okamura T., Kawa K., and Morishima T.. 2001. Clinical and virologic characteristics of chronic active Epstein-Barr virus infection. Blood. 98:280–286. 10.1182/blood.V98.2.280 [DOI] [PubMed] [Google Scholar]
- Kimura, H., Ito Y., Kawabe S., Gotoh K., Takahashi Y., Kojima S., Naoe T., Esaki S., Kikuta A., Sawada A., et al. 2012. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood. 119:673–686. 10.1182/blood-2011-10-381921 [DOI] [PubMed] [Google Scholar]
- Latour, S., and Winter S.. 2018. Inherited Immunodeficiencies With High Predisposition to Epstein-Barr Virus-Driven Lymphoproliferative Diseases. Front. Immunol. 9:1103. 10.3389/fimmu.2018.01103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W., Park S.J., Choi B.K., Kim H.H., Nam K.O., and Kwon B.S.. 2002. 4-1BB promotes the survival of CD8+ T lymphocytes by increasing expression of Bcl-xL and Bfl-1. J. Immunol. 169:4882–4888. 10.4049/jimmunol.169.9.4882 [DOI] [PubMed] [Google Scholar]
- Li, G., Boucher J.C., Kotani H., Park K., Zhang Y., Shrestha B., Wang X., Guan L., Beatty N., Abate-Daga D., and Davila M.L.. 2018. 4-1BB enhancement of CAR T function requires NF-κB and TRAFs. JCI Insight. 3:e121322. 10.1172/jci.insight.121322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, C.L., Kuehn H.S., Zhao F., Niemela J.E., Deenick E.K., Palendira U., Avery D.T., Moens L., Cannons J.L., Biancalana M., et al. 2014. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat. Immunol. 15:88–97. 10.1038/ni.2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E., Palmic N., Sanquer S., Lenoir C., Hauck F., Mongellaz C., Fabrega S., Nitschké P., Esposti M.D., Schwartzentruber J., et al. 2014. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature. 510:288–292. 10.1038/nature13386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus, M.V., Thomas A.K., Leonard D.G., Allman D., Addya K., Schlienger K., Riley J.L., and June C.H.. 2002. Ex vivo expansion of polyclonal and antigen-specific cytotoxic T lymphocytes by artificial APCs expressing ligands for the T-cell receptor, CD28 and 4-1BB. Nat. Biotechnol. 20:143–148. 10.1038/nbt0202-143 [DOI] [PubMed] [Google Scholar]
- Okano, M., Kawa K., Kimura H., Yachie A., Wakiguchi H., Maeda A., Imai S., Ohga S., Kanegane H., Tsuchiya S., et al. 2005. Proposed guidelines for diagnosing chronic active Epstein-Barr virus infection. Am. J. Hematol. 80:64–69. 10.1002/ajh.20398 [DOI] [PubMed] [Google Scholar]
- Okkenhaug, K., and Fruman D.A.. 2010. PI3Ks in lymphocyte signaling and development. Curr. Top. Microbiol. Immunol. 346:57–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug, K., Ali K., and Vanhaesebroeck B.. 2007. Antigen receptor signalling: a distinctive role for the p110delta isoform of PI3K. Trends Immunol. 28:80–87. 10.1016/j.it.2006.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno, Y., Murata T., Sato Y., Muramatsu H., Ito Y., Watanabe T., Okuno T., Murakami N., Yoshida K., Sawada A., et al. 2019. Defective Epstein-Barr virus in chronic active infection and haematological malignancy. Nat. Microbiol. 4:404–413. 10.1038/s41564-018-0334-0 [DOI] [PubMed] [Google Scholar]
- Park, S., and Ko Y.H.. 2014. Epstein-Barr virus-associated T/natural killer-cell lymphoproliferative disorders. J. Dermatol. 41:29–39. 10.1111/1346-8138.12322 [DOI] [PubMed] [Google Scholar]
- Quintanilla-Martinez, L., Ridaura C., Nagl F., Sáez-de-Ocariz M., Durán-McKinster C., Ruiz-Maldonado R., Alderete G., Grube P., Lome-Maldonado C., Bonzheim I., and Fend F.. 2013. Hydroa vacciniforme-like lymphoma: a chronic EBV+ lymphoproliferative disorder with risk to develop a systemic lymphoma. Blood. 122:3101–3110. 10.1182/blood-2013-05-502203 [DOI] [PubMed] [Google Scholar]
- Rhee, S.G. 2001. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 70:281–312. 10.1146/annurev.biochem.70.1.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud, S., Fondanèche M.C., Lambert N., Pasquier B., Mateo V., Soulas P., Galicier L., Le Deist F., Rieux-Laucat F., Revy P., et al. 2006. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 444:110–114. 10.1038/nature05257 [DOI] [PubMed] [Google Scholar]
- Salzer, E., Daschkey S., Choo S., Gombert M., Santos-Valente E., Ginzel S., Schwendinger M., Haas O.A., Fritsch G., Pickl W.F., et al. 2013. Combined immunodeficiency with life-threatening EBV-associated lymphoproliferative disorder in patients lacking functional CD27. Haematologica. 98:473–478. 10.3324/haematol.2012.068791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, X., Czar M.J., Bunnell S.C., Liu P., Liu Y., Schwartzberg P.L., and Wange R.L.. 2000. Deficiency of PTEN in Jurkat T cells causes constitutive localization of Itk to the plasma membrane and hyperresponsiveness to CD3 stimulation. Mol. Cell. Biol. 20:6945–6957. 10.1128/MCB.20.18.6945-6957.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharfe, N., Karanxha A., Dadi H., Merico D., Chitayat D., Herbrick J.A., Freeman S., Grinstein S., and Roifman C.M.. 2018. Dual loss of p110δ PI3-kinase and SKAP (KNSTRN) expression leads to combined immunodeficiency and multisystem syndromic features. J. Allergy Clin. Immunol. 142:618–629. 10.1016/j.jaci.2017.10.033 [DOI] [PubMed] [Google Scholar]
- Shuford, W.W., Klussman K., Tritchler D.D., Loo D.T., Chalupny J., Siadak A.W., Brown T.J., Emswiler J., Raecho H., Larsen C.P., et al. 1997. 4-1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J. Exp. Med. 186:47–55. 10.1084/jem.186.1.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogkas, G., Fedchenko M., Dhingra A., Jablonka A., Schmidt R.E., and Atschekzei F.. 2018. Primary immunodeficiency disorder caused by phosphoinositide 3-kinase δ deficiency. J. Allergy Clin. Immunol. 142:1650–1653.e2. 10.1016/j.jaci.2018.06.039 [DOI] [PubMed] [Google Scholar]
- Stephan, M.T., Ponomarev V., Brentjens R.J., Chang A.H., Dobrenkov K.V., Heller G., and Sadelain M.. 2007. T cell-encoded CD80 and 4-1BBL induce auto- and transcostimulation, resulting in potent tumor rejection. Nat. Med. 13:1440–1449. 10.1038/nm1676 [DOI] [PubMed] [Google Scholar]
- Swan, D.J.D., Aschenbrenner D., Lamb C.A., Chakraborty K., Clark J., Pandey S., Engelhardt K.R., Chen R., Cavounidis A., Ding Y., et al. 2019. Immunodeficiency, autoimmune thrombocytopenia and enterocolitis caused by autosomal recessive deficiency of PIK3CD-encoded phosphoinositide 3-kinase δ. Haematologica.:haematol.2018.208397. 10.3324/haematol.2018.208397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, C., Mittler R.S., and Vella A.T.. 1999. Cutting edge: 4-1BB is a bona fide CD8 T cell survival signal. J. Immunol. 162:5037–5040. [PubMed] [Google Scholar]
- Tangye, S.G., Palendira U., and Edwards E.S.. 2017. Human immunity against EBV-lessons from the clinic. J. Exp. Med. 214:269–283. 10.1084/jem.20161846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.S., Long H.M., Brooks J.M., Rickinson A.B., and Hislop A.D.. 2015. The immunology of Epstein-Barr virus-induced disease. Annu. Rev. Immunol. 33:787–821. 10.1146/annurev-immunol-032414-112326 [DOI] [PubMed] [Google Scholar]
- Vadas, O., Burke J.E., Zhang X., Berndt A., and Williams R.L.. 2011. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 4:re2. 10.1126/scisignal.2002165 [DOI] [PubMed] [Google Scholar]
- van Montfrans, J.M., Hoepelman A.I., Otto S., van Gijn M., van de Corput L., de Weger R.A., Monaco-Shawver L., Banerjee P.P., Sanders E.A., Jol-van der Zijde C.M., et al. 2012. CD27 deficiency is associated with combined immunodeficiency and persistent symptomatic EBV viremia. J. Allergy Clin. Immunol. 129:787–793.e6. 10.1016/j.jaci.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, T.H. 2005. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. 23:23–68. 10.1146/annurev.immunol.23.021704.115839 [DOI] [PubMed] [Google Scholar]
- Wen, T., Bukczynski J., and Watts T.H.. 2002. 4-1BB ligand-mediated costimulation of human T cells induces CD4 and CD8 T cell expansion, cytokine production, and the development of cytolytic effector function. J. Immunol. 168:4897–4906. 10.4049/jimmunol.168.10.4897 [DOI] [PubMed] [Google Scholar]
- Winter, S., Martin E., Boutboul D., Lenoir C., Boudjemaa S., Petit A., Picard C., Fischer A., Leverger G., and Latour S.. 2018. Loss of RASGRP1 in humans impairs T-cell expansion leading to Epstein-Barr virus susceptibility. EMBO Mol. Med. 10:188–199. 10.15252/emmm.201708292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzman, M.E., Clouthier D.L., McPherson A.J., Lin G.H., and Watts T.H.. 2013. The contextual role of TNFR family members in CD8(+) T-cell control of viral infections. Immunol. Rev. 255:125–148. 10.1111/imr.12086 [DOI] [PubMed] [Google Scholar]
- Yu, J., Wjasow C., and Backer J.M.. 1998. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J. Biol. Chem. 273:30199–30203. 10.1074/jbc.273.46.30199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.