Knipfer et al. identify a critical role for the type 2–related chemokine receptor CCR8 on ILC2 functions during type 2 immune responses. CCR8 supports ILC2 survival by autocrine secretion of the ligand CCL1. The authors demonstrate that CCL1/CCR8 signaling protects against helminth infections.
Abstract
Group 2 innate lymphoid cells (ILC2s) possess indispensable roles during type 2–mediated inflammatory diseases. Although their physiological and detrimental immune functions seem to depend on the anatomical compartment they reside, their tissue tropism and the molecular and immunological processes regulating the self-renewal of the local pool of ILC2s in the context of inflammation or infection are incompletely understood. Here, we analyzed the role of the CC-chemokine receptor CCR8 for the biological functions of ILC2s. In vitro and in vivo experiments indicated that CCR8 is in comparison to the related molecule CCR4 less important for migration of these cells. However, we found that activated mouse and human ILC2s produce the CCR8 ligand CCL1 and are a major source of CCL1 in vivo. CCL1 signaling to ILC2s regulates their proliferation and supports their capacity to protect against helminthic infections. In summary, we identify a novel chemokine receptor–dependent mechanism by which ILC2s are regulated during type 2 responses.
Introduction
Innate lymphoid cells (ILCs) embody a heterogeneous group of developmentally related cells that arise from shared lymphoid precursor populations, including the common lymphoid progenitor, but typically lack recombination-activating gene (Rag)–dependent rearranged antigen receptors (Diefenbach et al., 2014; Vivier et al., 2018). ILC2s share important developmental and transcriptional signatures with T helper type 2 cells (Th2 cells) and rapidly produce large amounts of cytokines, mainly IL-5, IL-9, and IL-13, upon exposure to alarmin-like factors such as IL-33, IL-25, and thymic stromal lymphopoietin (Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Wilhelm et al., 2011). Due to their strategic location and relative abundance in tissues harboring barrier functions such as gut, lung, and skin, they have been shown to vitally contribute to early phases of host protection against helminthic and viral infections and other environmental triggers (Monticelli et al., 2011; Klose and Artis, 2016; Kindermann et al., 2018). On the other hand, dysregulated ILC2 activation has been shown to be an important trigger of tissue remodeling and type 2–mediated immunopathologies, including atopic dermatitis and allergic asthma (Barlow and McKenzie, 2014; Lambrecht and Hammad, 2015; Ealey et al., 2017).
ILC2s arise from fetal progenitors and colonize mucosal tissues in the perinatal phase. Data from parabiosis experiments clearly indicated that they are in adult tissues rather long-lived and tissue resident, while local pools or precursor populations are supposed to be responsible for ILC2 replenishment (Gasteiger et al., 2015; Moro et al., 2016). However, some hematogenous trafficking of ILC2s to the lung was described in settings of allergic lung inflammation or prolonged parasitic infections (Gasteiger et al., 2015; Karta et al., 2018; Stier et al., 2018). Furthermore, intestinal ILC2 populations activated specifically via the cytokine IL-25 were described as precursors of inflammatory ILC2s (iILC2s) in lungs and livers of mice (Huang et al., 2018). In the context of human ILC2s, the lipid mediators cysteinyl leukotriene E4 and prostaglandin D2 were demonstated to have chemoattractive capacity (Salimi et al., 2017).
Chemokine receptors belong to the class A rhodopsin-like family of G-protein–coupled receptors and play fundamental roles in normal physiology as well as in inflammatory and infectious diseases. Besides their well-established roles in orchestrating immune cell migration, diverse other regulatory functions implicated in growth, survival, or cytokine production of cells became evident. Within ILCs, distinct sets of chemokine receptors are expressed by individual subsets that correlate with their tissue distribution (Kim et al., 2016). It has been demonstrated that ILC2s express CCR9, CXCR4, and CXCR6, which appear to be functionally involved in their tissue distribution (Roediger et al., 2013; Kim et al., 2015; Stier et al., 2018). Interestingly, data from global transcriptome analyses (Robinette et al., 2015) showed that the closely related Th2-associated CC-type chemokine receptors CCR8 and CCR4 are selectively expressed in ILC2s compared with other ILCs, including natural killer cells. While CCR4 was also found on human ILC2s (Salimi et al., 2013), the expression of CCR8 on human ILC2s remains unknown. Several studies correlated an upregulation of CCR4, CCR8, and their cognate chemokines (CCR4: CCL17 and CCL22; CCR8: CCL1 and CCL8 [mouse]/CCL18 [human]) to asthma and atopic dermatitis (Kakinuma et al., 2001; Panina-Bordignon et al., 2001; Gombert et al., 2005; Vijayanand et al., 2010). Moreover, CCR8 and CCR4 signaling was described as an important skin-homing mechanism for Th2 and regulatory T (T reg) cells (Reiss et al., 2001; Schaerli et al., 2004; Sather et al., 2007), and the CCR8 ligands CCL1 and CCL8 were shown to induce chemotaxis of Th2 cells (Zingoni et al., 1998). In line, Ccr8−/− mice are protected from allergic skin inflammation due to impaired CCL8-mediated Th2 cell recruitment (Islam et al., 2011). Similarly, type 2–challenged Ccr8-deficient mice exhibited impaired eosinophil recruitment to lungs and attenuated type 2 cytokine levels (Chensue et al., 2001). However, the chemotactic role for CCR8 on T cells is controversial, as in models of allergic lung inflammation, no impairment in lung infiltration of Th2 cells was observed in Ccr8−/− mice (Chung et al., 2003; Mikhak et al., 2009) or in the case of antibody-mediated CCL1 blockade (Bishop and Lloyd, 2003). Whether CCR8 expression plays a similar role in ILC2s as in T cells remains to be elucidated.
Here, we demonstrate that CCR8, different from CCR4, seems to be dispensable for systemic ILC2 migration. Conversely, our in vitro and in vivo findings clearly implicate CCL1 signaling to CCR8 as significant pathway supporting the accumulation and biological functions of ILC2s. Moreover, our study identified mouse and human activated ILC2s as a source of CCL1 protein, suggesting that the CCL1/CCR8 axis supports the tissue-specific ILC2 functions in an auto-/paracrine manner.
Results
CCR8 is expressed on ILC2s but does not mediate their migration
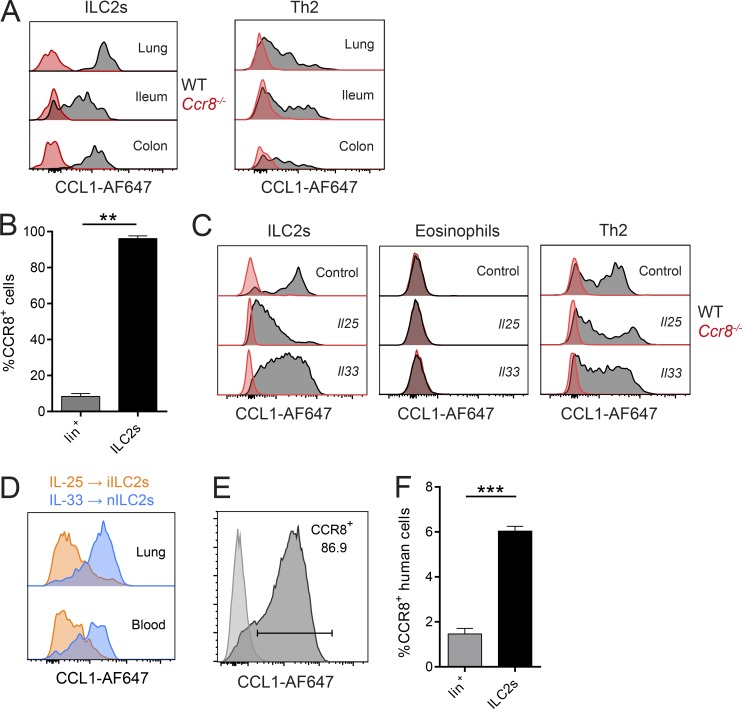
Consistent with previous global transcriptome analyses (Robinette et al., 2015), we observed that CCR8 is highly and specifically expressed on sorted ILC2s compared with ILC3s (not shown) on the transcriptional level. On the protein level, this chemokine receptor was widely present on the surface of ILC2s isolated from lungs and intestinal tissues of naive C57BL/6 mice, as evidenced by flow cytometry (Fig. 1 A). Similar results were obtained when lung ILC2s isolated from mice infected with the helminthic parasite Nippostrongylus brasiliensis (Fig. 1 B) or systemically treated with the alarmins IL-25 and IL-33 (Fig. 1 C) were analyzed. Notably, we found no expression of CCR8 on eosinophils, while our data confirm expression on T cells (Fig. 1, A and C). Interestingly, lung ILC2s obtained from IL-25–treated mice seemingly display slightly reduced expression of CCR8 compared with ILC2s isolated from naive mice or IL-33–treated mice. Because IL-25 and IL-33 were previously implicated in the differential accumulation of iILC2s and “natural” ILC2s (nILC2s; Huang et al., 2015), we next directly compared the expression of CCR8 on iILC2s and nILC2s. These experiments revealed that IL-25–induced iILC2s have lower CCR8 surface expression than IL-33–induced nILC2s (Fig. 1 D and Fig. S1, A and B). We also found that ILC2s maintain CCR8 surface expression after culture and expansion in vitro (Fig. 1 E). In line with these findings in mice, CCR8 was present on a subset of Lin−CD127+CD160+CRTH2+ ILC2s obtained from human peripheral blood (Fig. 1 F), and such human ILC2s also maintained CCR8 expression during in vitro expansion (Fig. S1 C).
Figure 1.
Analysis of CCR8 expression on ILC2s. (A–F) Flow cytometric analyses of CCR8 surface expression using fluorophore-coupled recombinant CCL1 (CCL1-AF647) or a human anti-CCR8 antibody (F). (A) ILC2s (Lin−Thy1+KLRG1+ICOS+ST2+) or Th2 (CD3+CD4+ST2+) cells in lung or the ileal/colonic lamina propria of naive WT and Ccr8−/− mice were analyzed by flow cytometry. (B) WT mice were infected with N. brasiliensis, and lung lineage+ lymphocytes (lin+) and ILC2s (Lin−Thy1+KLRG1+ST2+) were analyzed after 9 d. A Mann–Whitney U test was applied. (C) Lung cells of WT and Ccr8−/− mice challenged for 3 d with DNA vectors encoding Il25 or Il33 or empty vectors (control) were analyzed for CCR8 on ILC2s (Lin−Thy1+KLRG1+ICOS+), eosinophils (CD11b+SiglecF+CD11c−SSChi), and Th2 cells (Lin+Thy1+ST2+). (D) To investigate CCR8 expression on ILC2 subsets, WT mice were treated with Il25 (to induce iILC2s) or Il33 (to induce nILC2s) vectors for 5 d, and iILC2s (Lin−CD127+KLRG1hiST2−) or nILC2s (Lin−CD127+KLRG1intST2+) from lungs and blood were analyzed and compared by flow cytometry. (E) Murine sorted and in vitro–expanded ILC2s were analyzed and compared with a fluorescence minus one (FMO) control. (F) ILC2s (Lin−CD127+CD161+CRTH2+) and lineage+ (Lin+) lymphocytes in freshly isolated human PBMCs were analyzed. An unpaired t test was applied. All results are representative of two or more independent experiments. A and C–E show representative histograms of one animal of two to four animals in total per experimental group (A, C, and D) or four independent sorting experiments (E). Bar graphs represent five mice (B) or three donors (F). Data represent mean ± SEM. **, P ≤ 0.01; ***, P ≤ 0.001 by Mann–Whitney U tests or unpaired t test.
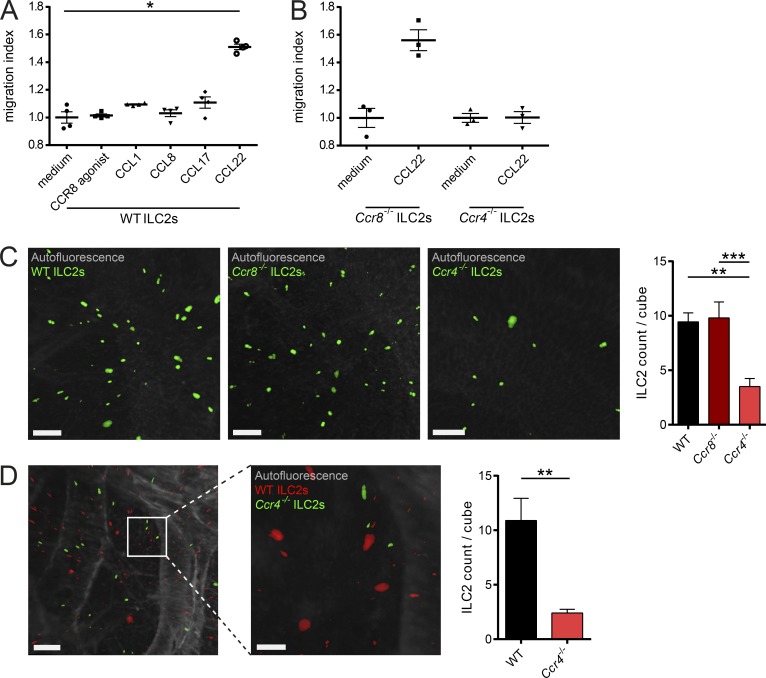
CCR8+ memory T helper cells have been shown to migrate in response to the CCR8 ligand CCL1 (Soler et al., 2006). In the next series of experiments, we therefore aimed in analyzing the functional role of CCR8 for ligand-induced cell migration of ILC2s. In transwell chemotaxis assays, CCL1 and CCL8, the known natural CCR8 agonists in mice, as well as the selective synthetic agonist ZK756326 showed no chemotactic activity for ILC2s. By contrast, ligands for the related chemokine receptor CCR4, CCL17 and CCL22, prompted significant migration of WT and Ccr8−/− ILC2s (Fig. 2, A and B; and Fig. S2 A). To confirm these data in an in vivo setting, we employed a procedure that we recently developed for the analysis of lung homing of lymphocytes using light-sheet microscopy (Schulz-Kuhnt et al., 2019; Fig. S2, B and C). In this model, C57BL/6 recipient mice were intranasally challenged with the protein allergen papain and adoptively transferred with fluorescence-labeled WT, Ccr4−/−, and Ccr8−/− ILC2s to analyze their homing capacity to the lung in vivo. Light-sheet microscopy revealed equal spread of cells of both WT and Ccr8−/− ILC2s groups to the inflamed lungs. Conversely, transferred Ccr4−/− ILC2s failed to migrate efficiently into lungs (Fig. 2 C). Additional competitive homing experiments with WT and Ccr4−/− ILC2s further supported the notion that efficient lung ILC2 migration in this adoptive transfer model depends on Ccr4 expression (Fig. 2 D and Video 1). Along this line, Ccr4−/− mice accumulated fewer iILC2s in their lungs compared with WT mice in the context of systemic IL-25 abundance (Fig. S2 D).
Figure 2.
CCR4, but not CCR8, mediates ILC2 migration. (A and B) To assess the migratory behavior of ILC2s, in vitro–expanded WT (A) and Ccr8−/− or Ccr4−/− (B) ILC2s were used. Chemotaxis assays were performed using transwell inserts against gradients of the chemokines CCL1, CCL8, CCL17, or CCL22 (100 ng/ml each) as well as an activating CCR8 agonist (5 µM). (C and D) To investigate ILC2 lung homing, WT mice were pretreated with papain, and 106 fluorescent-labeled WT, Ccr4−/−, or Ccr8−/− ILC2s (C) or a 50:50 mix (D) were adoptively transferred by i.v. injection. 24 h later, lungs were collected, processed, and analyzed by light-sheet microscopy. (C) One-way ANOVA was applied. Scale bars are 100 µm. Images were acquired with a 10× zoom factor. (D) The pictures show snapshots of Video 1. Scale bars are 200 µm (left) or 100 µm (right). Images were acquired with a 25× zoom factor. (A and B) Each dot represents a technical replicate. One representative experiment out of two independently performed experiments is shown. (C and D) Data represent one out of two independent sorting experiments with one or two mice per group. Data represent mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by Mann-Whitney U tests or, if indicated, one-way ANOVA.
Collectively, these data indicate that CCR4 receptor activation by CCL22 may drive ILC2 migration, while CCR8 signaling is seemingly dispensable for ILC2 migration in the different settings we analyzed.
CCL1 is an autocrine ILC2-stimulating factor
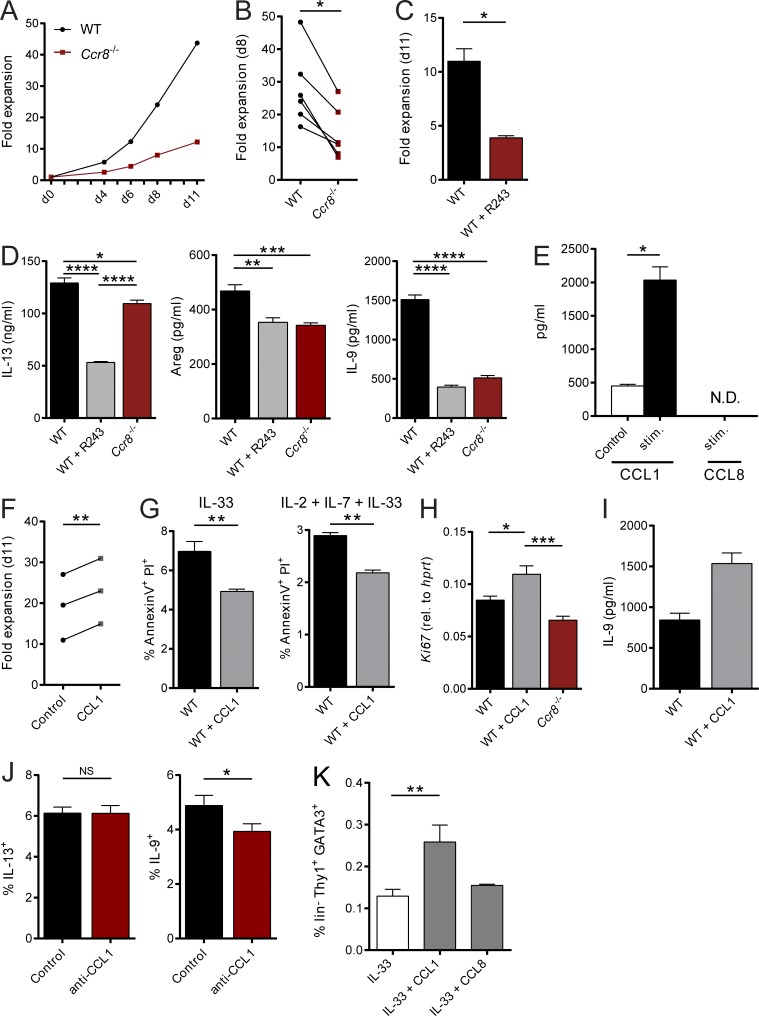
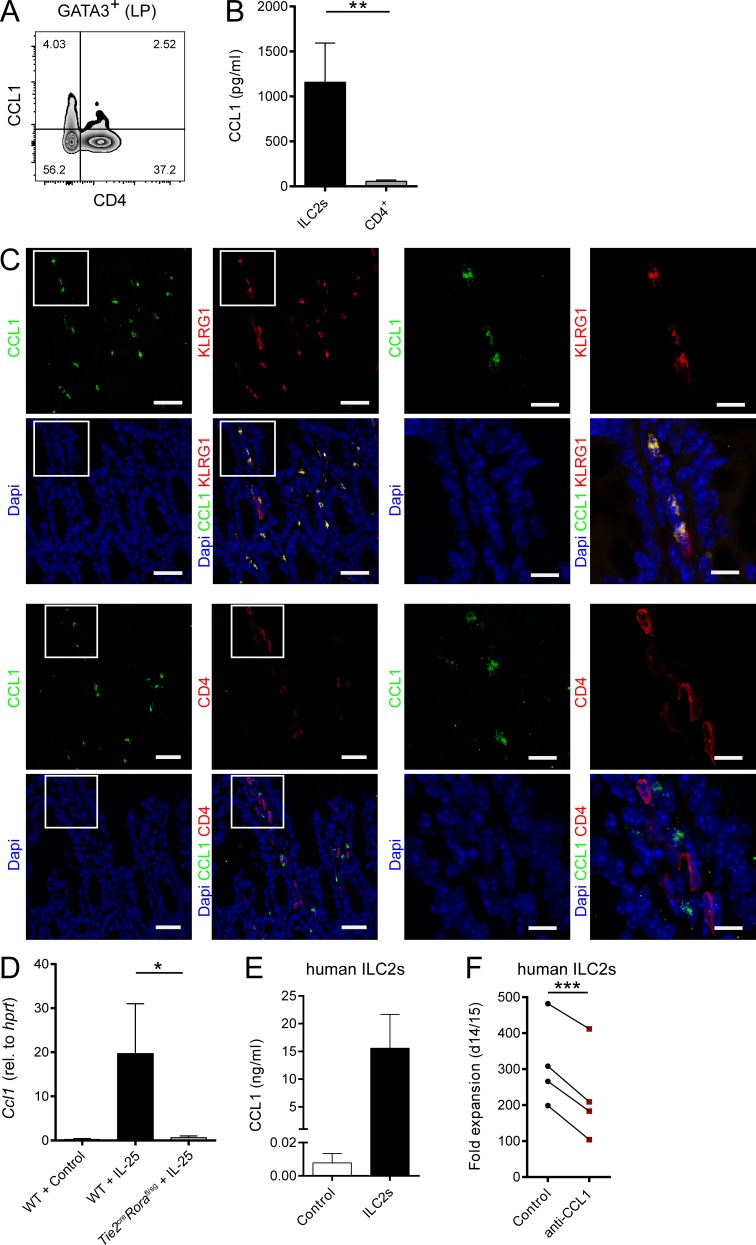
Beyond their functions in the context of migration and homing, several chemokines, including CCL1, have been shown to directly control the growth and effector functions of their target cells (Iellem et al., 2000; Hoshino et al., 2007). Given that our in vitro and in vivo data were not indicative of a role of CCR8 in ILC2 migration, we next analyzed, whether CCR8 signaling regulates such ILC2 functions in vitro. Interestingly, highly sort-purified murine Ccr8−/− ILC2s expanded significantly less than control ILC2s when cultured in vitro in the presence of activating cytokines (Fig. 3, A and B). Similarly, the in vitro growth of WT ILC2s was moderated by treatment of expansion cultures with the CCR8 antagonist R243 (Oshio et al., 2014; Fig. 3 C). Moreover, we found significantly lower concentrations of the cytokines IL-13, Amphiregulin (Areg), and in particular IL-9 in supernatants of WT ILC2s treated with R243 or Ccr8−/− ILC2s compared with controls (Fig. 3 D). Because CCL1 was shown to mediate an autocrine antiapoptotic loop in T cell leukemia cells (Ruckes et al., 2001) and the cytokine cocktail we used for ILC2 expansion did not contain CCR8 ligands, we next assessed the capacity of ILC2s to secrete CCR8 ligands. Specific ELISA analysis showed that CCL1 protein secretion was detectable in resting ILC2s and markedly increased upon stimulation with IL-2, IL-7, and IL-33 cytokines, while no CCL8 protein was present in ILC2 supernatants (Fig. 3 E). To ascertain the effects of CCL1 on ILC2s directly, we next added rCCL1 to ILC2 cultures and monitored their expansion over time. Thereby, we recovered more ILC2s in the presence of CCL1 (Fig. 3 F). Moreover, reduced numbers of late apoptotic/necrotic cells were present in cultures of resting ILC2s stimulated with IL-33 alone or in combination with IL-2 and IL-7 as analyzed by Annexin V/propidium iodide (PI) staining (Fig. 3 G). Besides, CCL1 supplementation led to increased expression of the cell proliferation marker Ki67 (Fig. 3 H) and accumulation of IL-9 protein (Fig. 3 I), while the addition of neutralizing anti-CCL1 antibodies reduced the numbers of IL-9–producing, but not IL-13–producing, ILC2s in cultures (Fig. 3 J). Noteworthy, none of these effects were induced by addition of CCL8 (not shown). Similarly, CCL1, but not CCL8, overexpression increased the alarmin-mediated accumulation of ILC2s in vivo (Fig. 3 K). In additional experiments, we found by intracellular flow cytometry that a proportion of Gata3+CD4− cells in the intestinal lamina propria of naive mice produced CCL1 (Fig. 4 A). Moreover, intestinal sort-purified ILC2s of such mice secreted significantly more CCL1 protein than CD4+ T cells (Fig. 4 B). In the context of IL-33 overexpression, the majority of CCL1+ cells coexpress KLRG1, while no coexpression with CD4 was detected, further supporting the notion of ILC2s as significant CCL1 producers (Fig. 4 C). Accordingly, in vivo ILC2 accumulation by IL-25 overexpression strongly induced Ccl1 transcripts in WT mice. By contrast, ILC2-deficient Tie2creRorafl/sg mice (Omata et al., 2018) did not display Ccl1 mRNA induction upon IL-25 treatment (Fig. 4 D).
Figure 3.
The CCR8 ligand CCL1 is an autocrine survival factor for ILC2s. (A–C and F) Sorted mouse ILC2s were cultured as described in Materials and methods and cell numbers determined at the indicated days to calculate fold expansion. (A and B) WT and Ccr8−/− ILC2 numbers were compared at indicated time points. (A) One representative experiment is shown. (B) Each replicate represents one independent sorting experiment. An unpaired t test was applied. (C) WT ILC2s were expanded in the presence of the CCR8 inhibitor R243. (D) In vitro–expanded WT ILC2s, WT ILC2s plus R243, and Ccr8−/− ILC2s were restimulated with IL-2, IL-7, and IL-33 (20 ng/ml each). Production of IL-13, Amphiregulin, and IL-9 was measured in the supernatants after 24 h. A one-way ANOVA was applied. (E) To measure secretion of CCL1 and CCL8 by in vitro–expanded murine WT ILC2s, cells were stimulated (IL-2, IL-7, and IL-33; 20 ng/ml each) under addition of PMA/ionomycin, and supernatants were collected after 24 h. (F) WT ILC2s were expanded in the presence of CCL1 (50 ng/ml). Each replicate represents one independent sorting experiment, and a paired t test was applied. (G) To measure cell death, WT ILC2s expanded with or without CCL1 were restimulated for 24 h with IL-33 or IL-2, IL-7, and IL-33 (20 ng/ml each), and Annexin V/PI stainings were performed. (H) WT ILC2s expanded with or without CCL1 or Ccr8−/− ILC2s were restimulated for 24 h with IL-2, IL-7, and IL-33 (20 ng/ml each) and Ki67 expression determined by qPCR. A one-way ANOVA was applied. (I) IL-9 was measured in the supernatant of WT ILC2s expanded with or without CCL1 for 11 d. P = 0.0571. (J) Flow cytometry of WT ILC2s restimulated with IL-2, IL-7, and IL-33 (20 ng/ml each) and a neutralizing anti-CCL1 antibody (10 µg/ml) for 24 h. Monensin was added to the culture for the last 4 h of stimulation. (K) WT mice were challenged with DNA vectors for IL-33, CCL1, or CCL8 as indicated. After 5 d, liver single-cell suspensions were generated and analyzed by flow cytometry. The graphs show pooled data of two independent experiments with at least four mice per group. A one-way ANOVA was applied. (C–E and G–J) Data represent technical replicates of one out of at least two independent experiments. Data represent mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001 by Mann-Whitney U tests or, if indicated, paired t test, unpaired t test, or one-way ANOVA. N.D., not detected.
Figure 4.
ILC2s are a major in vivo source of CCL1. (A) Flow cytometric analysis of gut lamina propria (LP) cells. Lymphocytes were gated on GATA3+ cells and analyzed for CD4 and CCL1 expression. (B) ILC2s and CD4+ T cells were sorted from intestinal lamina propria cells of naive RoracreRosa26-tdtomatofl/fl mice. Cells were cultivated in the presence of PMA/ionomycin, and supernatants were collected after 48 h for CCL1-specific ELISA analysis. (C) Small intestinal tissue sections of C57BL/6 WT mice overexpressing IL-33 were stained with DAPI (blue), anti-CCL1 (green), and anti-KLRG1 (red, top) or anti-CD4 (red, bottom) antibodies and analyzed by confocal microscopy. Scale bars are 30 µm (left) or 10 µm (right). Pictures show results of one representative animal. (D) WT and Tie2creRorafl/sg mice were challenged with DNA vectors for systemic release of IL-25 or empty vectors (control). Livers were collected after 5 d, and the presence of Ccl1 transcripts was investigated by qPCR. (E) In vitro–expanded human ILC2s were restimulated with IL-2, IL-25, and IL-33, and supernatants were collected after 3 d for CCL1 measurements. CCL1 levels were compared with supernatants of feeder cells only. Each replicate represents one independent sorting experiment. (F) Sorted human ILC2s were expanded with or without neutralizing anti-CCL1 antibodies and cell number determined to investigate fold expansion. Each dot represents one independent sorting experiment. Paired t test was applied. All graphs show data of one representative experiment out of at least two independent experiments. Experimental groups consisted of at least three mice or donors. Data represent mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 by Mann–Whitney U tests or, if indicated, a paired t test.
Importantly, these findings with murine ILC2s translate to human cells, as high CCL1 levels were present in culture supernatants of ILC2s sorted from peripheral blood, and treatment of human ILC2 cultures with neutralizing anti-CCL1 antibodies significantly inhibited their in vitro expansion (Fig. 4, E and F).
Taken together, these data indicate that a CCL1/CCR8-dependent autocrine loop may support alarmin-induced ILC2 growth and survival.
Ccr8 expression on ILC2s drives antihelminthic immunity
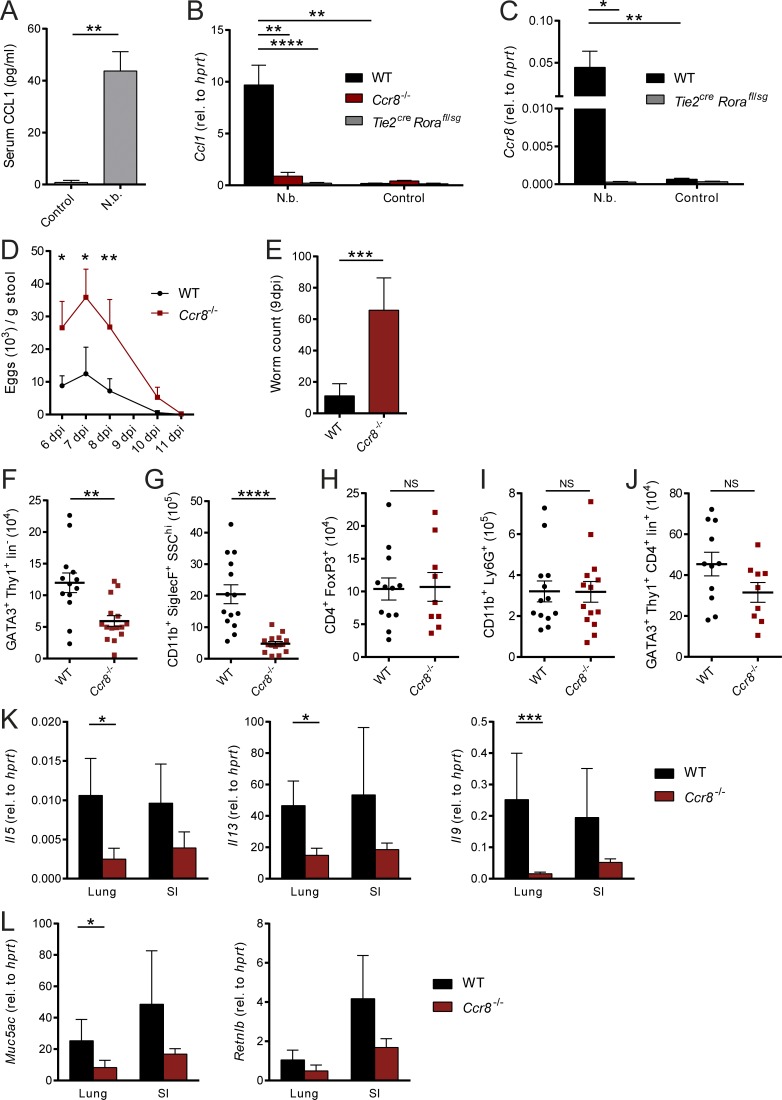
Multiple in vivo studies suggest that ILC2s considerably contribute to protective immunity to parasitic infections (Kindermann et al., 2018). Given that our in vitro data strongly related a CCL1/CCR8-mediated feed-forward mechanism to the biological functions of ILC2s and CCL1, we next aimed in addressing the functional role of the CCL1/CCR8 axis in an infection model with the prototypical type 2 immunity–inducing helminthic parasite N. brasiliensis. In this model, CCL1 serum concentrations were highly elevated in infected C57BL/6 mice compared with controls (Fig. 5 A). In line, pulmonary Ccl1 transcripts were increased upon infection. However, we observed no strong induction of Ccl1 expression in infected Ccr8−/− mice similar to ILC2-deficient Tie2creRorafl/sg mice (Fig. 5 B), while T and B cell deficiency did not affect Ccl1 expression (Fig. S3 A). These data were supported by staining of the small intestine, as the majority of CCL1+ cells coexpressed KLRG1, but not CD4 (Fig. S3 E). In addition, Ccr8 transcripts were almost absent in infected Tie2creRorafl/sg mice, confirming ILC2s as a critical Ccr8-expressing cell type in this model (Fig. 5 C). Importantly, starting at day 6 after infection with 500 L3 larvae, after adult worms have populated the small bowel and matured, Ccr8−/− mice exhibited increased egg production compared with the WT control mice (Fig. 5 D). While we found in Ccr8-sufficient mice only a few worms in their small intestines at 9 d post-infection (dpi), significantly increased worm numbers were present in the absence of CCR8 at this time point (Fig. 5 E). We observed no changes in the proportions of ILC2s, myeloid cell subsets, and T cells in lungs of Ccr8−/− mice at steady state (Fig. S3 B). However, considerably fewer ILC2s (Figs. 5 F and S3 F) and eosinophils (Fig. 5 G) accumulated in the lungs of Ccr8−/− mice in response to N. brasiliensis infection, while FoxP3+ T reg cell (Fig. 5 H), neutrophil (Fig. 5 I), and Th2 cell numbers (Figs. 5 J and S3 F) were not significantly affected. Consistently, this impairment in type 2–related immune cell infiltration in infected Ccr8−/− mice is reflected by the decreased presence of transcripts of the prototypical type 2 cytokines IL-5, IL-13, and IL-9 in both lungs and small intestines (Fig. 5 K). Moreover, transcripts of the Muc5ac and Retnlb genes, which encode for proteins, related to goblet cell activation, mucus hypersecretion, and worm expulsion, were decreased in tissue lysates generated from lungs or guts of these mice (Fig. 5 L).
Figure 5.
Ccr8−/−mice are impaired in N. brasiliensis defense due to defective type 2 immune responses. (A–L) Mice of indicated genotypes were infected with 500 L3 N. brasiliensis (N.b.) larvae. (A) Serum concentrations of CCL1 in WT mice were measured in response to N. brasiliensis infection at 9 dpi. (B and C) Expression of Ccl1 and Ccr8 was determined in naive (control) and N. brasiliensis–infected WT, Ccr8−/−, and Tie2creRorafl/sg lungs by qPCR. One-way ANOVA was applied. (D) Parasite eggs in stool samples of WT and Ccr8−/− mice were counted 6 dpi to 11 dpi. (E) Adult worm counts in small intestinal tissues were determined 9 dpi. (F–J) ILC2 (GATA3+Thy1+Lin−), eosinophil (CD11b+SiglecF+SSChi), T reg cell (CD4+FoxP3+), neutrophil (CD11b+Ly6G+), and Th2 cell (GATA3+Thy1+CD4+Lin+) numbers per lung were determined by flow cytometry 9 dpi. (K and L) Expression of the type 2–related effector cytokines Il5, Il13, and Il9 (K) as well as the mucins Muc5ac and Retnlb (L) were determined in lung and small intestinal (SI) tissues by qPCR 9 dpi. All graphs show pooled data of two to four representative experiments with at least four mice in each experimental group. Each dot represents one animal. Data represent mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; Mann–Whitney U tests or, if indicated, one-way ANOVA.
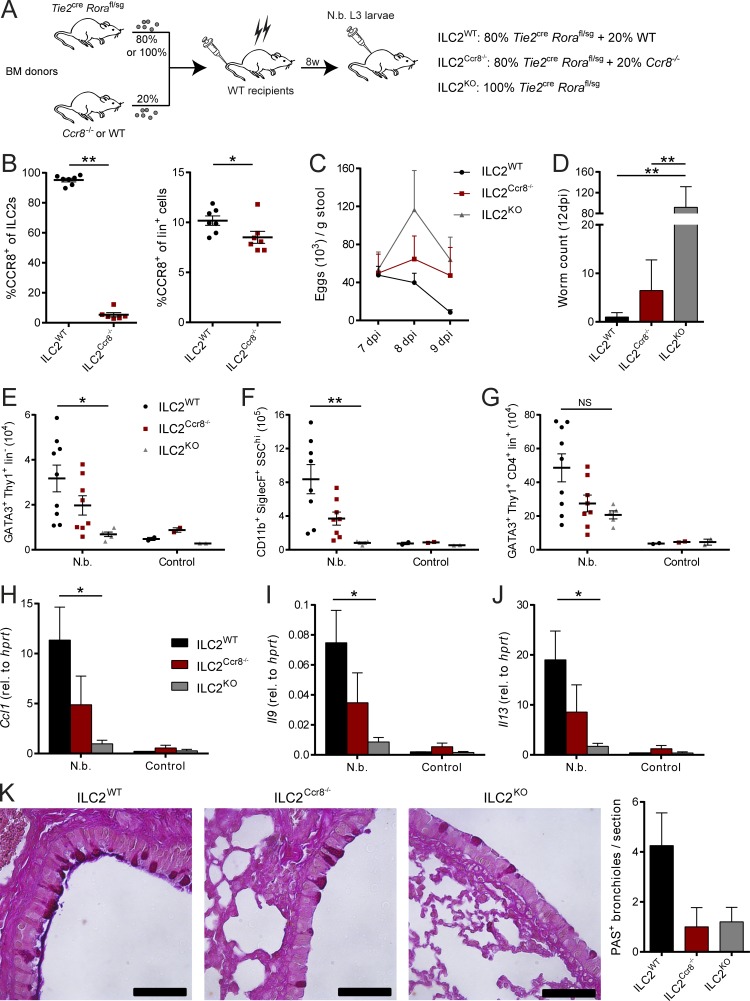
Besides ILC2s, different immune cell subsets, including myeloid cell and T cell subsets, have been described to express CCR8 (Heymann et al., 2012; Barsheshet et al., 2017; Sokol et al., 2018). Having shown the importance of CCR8 for efficient antihelminthic immunity, we next addressed the importance of CCR8 expression on ILC2s more specifically by generation of mixed bone marrow chimeric mice in which Ccr8 deficiency was limited to ILC2s. To this end, WT C57BL/6 mice were lethally irradiated and subsequently adoptively transferred with bone marrow cells from ILC2-deficient (Fig. S3 C) Tie2creRorafl/sg mice and Ccr8−/− mice mixed at a ratio of 80:20 (Ballesteros-Tato et al., 2016). Control chimeras received 20% WT bone marrow in place (Fig. 6, A and B). A further control group was transferred with 100% bone marrow of Tie2creRorafl/sg mice, a strain with high susceptibility to N. brasiliensis infection (Fig. S3 D). In this setting, mice receiving Ccr8−/− bone marrow displayed attenuated worm clearance, as indicated by increased egg counts in stools (Fig. 6 C) and a delayed capacity for parasite expulsion (Fig. 6 D). Furthermore, the reduction in parasite-directed immunity in these mice was accompanied by reduced numbers of lung ILC2s (Fig. 6 E), eosinophils (Fig. 6 F), and Th2 cells (Fig. 6 G) compared with control mice. We also observed impaired lung expression of Ccl1 (Fig. 6 H), Il9 (Fig. 6 I), and Il13 (Fig. 6 J) as well as decreased numbers of periodic acid–Schiff (PAS+) airway epithelial cells in mice that received Ccr8−/− bone marrow (Fig. 6 K), supporting the concept that CCR8 expression on ILC2s is required for mounting efficient antihelminthic immune responses in our model.
Figure 6.
ILC2-specific deletion of CCR8 results in impaired immunity against N. brasiliensis infection. (A–K) Mixed bone marrow (BM) chimeras with 80% Tie2creRorafl/sg and 20% WT (ILC2WT) or Ccr8−/− (ILC2Ccr8−/−) bone marrow were generated. Additionally, chimeras with 100% Tie2creRorafl/sg (ILC2KO) bone marrow were created. After 8 wk, mice were infected with 500 L3 N. brasiliensis larvae or left untreated (control). (B) To confirm the absence of CCR8 on ILC2s in ILC2Ccr8−/− mice, flow cytometry of lung ILC2s (Lin−Thy1+KLRG1+ST2+) and lineage+ lymphocytes was performed. CCR8 surface expression was detected using fluorophore-coupled recombinant CCL1 (CCL1-AF647). (C) Parasite egg counts were determined in stool samples. (D) Adult worm counts in small intestinal tissues were determined 12 dpi. (E–J) ILC2 (E; GATA3+Thy1+Lin−), eosinophil (F; CD11b+SiglecF+SSChi), and Th2 (G; GATA3+Thy1+CD4+Lin+) numbers per lung were analyzed by flow cytometry 12 dpi. Ccl1 (H), Il9 (I), and Il13 (J) expression in lung tissues was determined by qPCR 12 dpi. (K) AB-PAS staining was performed from lung paraffin sections. Scale bars are 50 µm. Pictures show results of one representative animal and quantification of one representative experiment with three to five animals in one experimental group. Graphs in B–J show pooled data of two representative experiments out of three independent experiments in total. Each dot represents one animal. Data represent mean ± SEM. *, P ≤ 0.05; **, P ≤ 0.01; Mann–Whitney U tests (B) or one-way ANOVA (D–J).
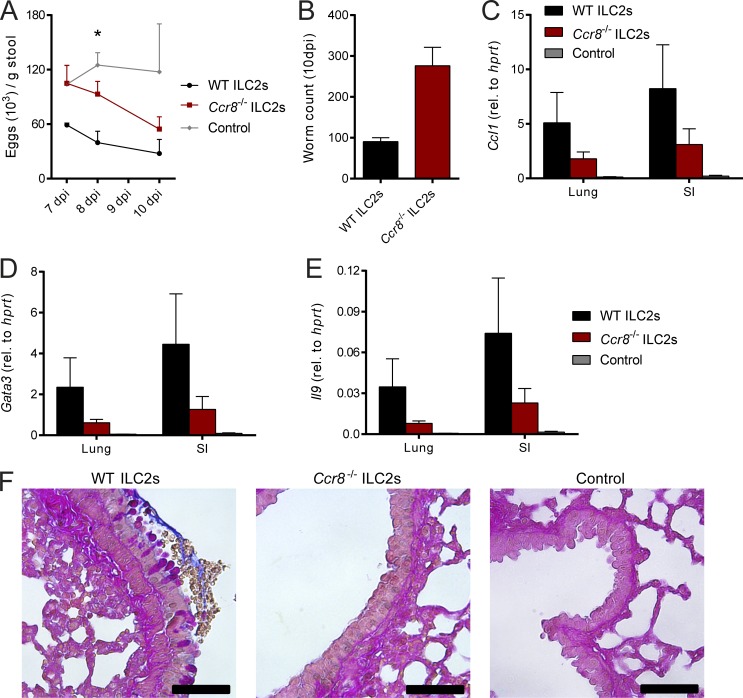
To further ascertain the functional importance of ILC2 intrinsic functions of CCR8 in the immune response to parasitic infection, we adoptively transferred sort-purified WT or Ccr8−/− ILC2s into alymphoid host mice. These mice possess severe immunodeficiencies, including a complete lack of the T cell and ILC compartment, leading to the inability to defend against N. brasiliensis infection (Halim et al., 2012). Although adoptive transfer of WT ILC2s into Rag2−/−il2rg−/− mice partially restored host defense against N. brasiliensis, Ccr8−/− ILC2 chimeras tended to have increased infection burdens, as indicated by higher egg counts (Fig. 7 A) during the course of infection and also showed a trend to higher worm numbers at 10 dpi (Fig. 7 B). In line with this, mice receiving Ccr8-deficient ILC2s tended to have decreased pulmonary expression of Ccl1 (Fig. 7 C), the transcription factor Gata3 (Fig. 7 D), and Il9 (Fig. 7 E) compared with control mice. PAS staining of lung tissues also showed a trend toward reduced mucus secretion in the context of CCR8 deficiency on ILC2s (Fig. 7 F).
Figure 7.
CCR8 on ILC2s is critical for efficient type 2 immunity against N. brasiliensis infection upon adoptive cell transfer. (A–F) Sorted WT or Ccr8−/− ILC2s were adoptively transferred into Rag2−/−il2rg−/− mice or received no ILC2s (control) and infected with N. brasiliensis. (A) Parasite eggs were counted in stool samples. (B) Mice were sacrificed after 10 d, and adult worm counts in small intestinal tissues were determined. (C–E) Ccl1, Gata3, and Il9 expression was determined in lung and small intestinal tissues by qPCR. (F) AB-PAS staining was performed from lung paraffin sections. Scale bars are 50 µm. Pictures show results of one representative animal. Graphs in A–E show pooled data of two independent experiments with two to five mice in each experimental group. Data represent mean ± SEM. *, P ≤ 0.05 by one-way ANOVA.
Discussion
In summary, our findings indicate a critical role for CCR8 expression on ILC2s during inflammatory type 2 responses and provide clear evidence that ILC2-intrinsic CCL1 production potentiates lung-resident ILC2 responses via an autocrine mechanism. Unique secretion patterns of chemokines are central for specific and fine-tuned functioning of the immune network. CCR8 has been associated with atopic diseases and is expressed on subsets of Th2 cells, T reg cells, and myeloid cells (Heymann et al., 2012). While in some models, Ccr8−/− mice showed decreased type 2 immune responses, these defects were mostly thought to be related to defective CCR8 expression by Th2 cells. Recent data also suggest an important role of CCR8 for the local migration of CD301b+ dermal dendritic cells during cutaneous type 2 immune responses (Sokol et al., 2018). In this study, production of the CCR8 ligand CCL8 by macrophages promoted dermal dendritic cell attraction to draining lymph nodes, while lymph node CCL1 production was not induced by Th2 cell skewing immunization. In addition, CCL8 was required for migration of Th2 cells to the skin in a model of atopic dermatitis (Islam et al., 2011). In the lung, however, we observed induction of CCL1 expression upon IL-25 injection, papain treatment, or worm infection, indicating that tissue-specific expression patterns of ligands define the role of CCR8 signaling in different type 2–mediated diseases. CCL1 did not provoke ILC2 migration but rather supported their maintenance after alarmin-mediated activation. A similar self-feeding mechanism was recently described for T reg cells, as autocrine CCL1 potentiates the proliferation of T reg cells in mouse model of multiple sclerosis (Barsheshet et al., 2017). Consistent with the notion of CCL1 as regulator of cell survival, it was previously associated with antiapoptotic activity on T-lymphoma cells (Spinetti et al., 2003) potentially through regulation of the RAS/MAPK pathway (Louahed et al., 2003). Whether similar pathways are functional in CCL1-stimulated ILC2s remains to be further addressed. While our data indicate that CCL1 stimulation supports ILC2 survival and/or proliferation, we did not observe profound direct effects on the production of the signature cytokines IL-13 and IL-5. However, effects on the production of IL-9, an important autocrine growth factor of ILC2s (Turner et al., 2013), were stronger, indicating that the IL-9 pathway may represent a target of the ILC2 regulatory capacity of CCL1/CCR8.
Although we did not find that CCL8 triggers mouse ILC2 migration across a wide range of chemokine concentrations in vitro, and Ccr8−/− ILC2s displayed no lung homing defects in an in vivo adoptive transfer model, a very recent study found that this chemokine exhibited in mice amoeboid-like movement of ILC2s, thereby regulating their local distribution in the peribronchial and perivascular space during type 2–associated lung inflammation (Puttur et al., 2019). These data indicate that critical factors in the local tissue of the lung seemingly regulate the CCR8-dependent tropism of tissue-resident ILC2s or potentially precursor populations. Conversely, our data indicate that this chemokine receptor in contrast to CCR4 is not important for migration of other ILC2 subtypes such as iILC2s. Altogether, the study by Puttur et al. (2019) and the data provided in this study indicate that both CCL1 and CCL8 synergistically support type 2–mediated immunity by differentially regulating ILC2s. Notably, it remains unclear how CCL1 and CCL8 differentially regulate the fate of ILC2s and other CCR8-expressing cells. However, such functional selectivity has been observed for other important chemokine receptor ligands (Anderson et al., 2016) and may in the case of CCR8 be exploited for targeted therapy of type 2–mediated diseases.
Synchronized cell recruitment and maintenance of Th2 cell–related populations is required for protective immunity to helminthic infections. In consequence, hosts have developed highly coordinated chemokine responses to combat helminths and deal with the tissue destruction induced by them. Although our findings clearly show that CCR8 signaling is pivotal for optimal worm expulsion, some important outstanding questions remain. For example, further studies will have to address the individual in vivo roles of both mouse CCR8 ligands for parasite clearance. Noteworthy, CCL8 is in humans not a ligand for CCR8 (Islam et al., 2011). Thus, it will be interesting to elucidate whether CCL18, the functional human analogue of murine CCL8, triggers ILC2 migration or other potential functions in ILC2s. Besides ILC2s, CCR8 is known to be expressed on several other immune cell types, and in particular, CCL1 regulation of T cell subsets such as T reg and Th2 cells has been implicated in inflammatory reactions. As we have identified ILC2s as important in vivo source of this chemokine during parasitic infections, CCL1 secretion by ILC2s may also influence their crosstalk with other cells, including T cells. Indeed, our data in bone marrow chimeric mice lacking CCR8 on ILC2s indicate that besides ILC2s, other CCR8-expressing cells are potentially important for optimal N. brasiliensis clearance. The availability of conditional alleles for CCR8 and CCL1 will certainly be of critical importance to further characterize the role of autocrine CCL1/CCR8 signaling for regulation of ILC2s in vivo and to identify exactly the cell type–specific roles of these molecules in the complex immune cell network that regulates type 2–mediated inflammation. Although current evidence supports the notion that ILC2s are primarily tissue-resident and not subject to extensive migration, we observed a clear chemotactic response to ligands of CCR4, which has been shown to be expressed by mouse and human ILC2s (Salimi et al., 2013; Moro et al., 2016). Hence, it will be interesting to study whether this pathway supports in vivo scenarios where ILC2 migration has been described, such as the IL-33–dependent egress of ILC2s from the bone marrow or IL-25–dependent trafficking of iILC2s out of the intestinal mucosa.
Together, our data support the concept that the CCL1/CCR8 axis may support the maintenance of the local pool of ILC2s in type 2–mediated inflammatory settings such as parasitic infections.
Materials and methods
Animals and mouse protocols
Ccr8−/− mice (Chensue et al., 2001) were kindly provided by F. Tacke (Department of Medicine III, University Hospital Center Aachen, Aachen, Germany), and Ccr4−/− mice (Chvatchko et al., 2000) were received from D. Anz (Department of Medicine II, University Hospital Center Munich, Munich, Germany). Roracre mice (Chou et al., 2013) were kindly provided by D. O’Leary (The Salk Institute for Biological Studies, La Jolla, CA). Rag1−/−, Rag2−/−il2rg−/−, C57BL/6, Rosa26Stop-tdtomatofl/fl, and Tie2cre mice were originally obtained from The Jackson Laboratory and bred in-house. Rorafl/sg mice were generated from a strain provided by the European Conditional Mouse Mutagenesis Program. Tie2creRorafl/sg, RoracreRosa26Stop-tdtomatofl/fl, Ccr4−/−, and Ccr8−/− mice were backcrossed to the C57BL/6 background for 10 generations. In experiments with Tie2creRorafl/sg and Ccr4−/− mice, littermate controls were used. In experiments with Rag1−/− and Ccr8−/− mice, C57BL/6 mice were used as controls. Here, cohoused mice were used if possible; otherwise, exchange of bedding for 4 wk was performed to normalize the microbiota. We used 8–12-wk-old background-, age-, and sex-matched mice for experiments. For helminth infection, mice were subcutaneously injected with 500 viable L3 stage larvae of N. brasiliensis (kindly provided by D. Voehringer, Department of Infection Biology, University Hospital Center Erlangen, Erlangen, Germany). Mice were housed with neomycin sulfate (2 g/liter) and polymyxin B (100 mg/liter) in the drinking water for 5 d. Feces were collected from each individual at the indicated time points, and eggs were counted under an Axiophot microscope (Zeiss). After 9–12 d, mice were sacrificed, and lungs and small intestines were collected and further analyzed as previously described (Mchedlidze et al., 2016). Worm burdens in small intestines were enumerated in dissected tissue under a stereo microscope (SZX7; Olympus). In vivo expression of cytokines using minicircle-based vectors was performed as previously described (McHedlidze et al., 2013). Animal experiments were approved by the local animal ethical committee of the government of Unterfranken, Würzburg, Germany.
Bone marrow chimeras
For generation of mixed bone marrow chimeras, 107 donor bone marrow cells obtained from femurs and tibias were injected i.v. into lethally irradiated (10 grays) C57BL/6 recipient mice. To create ILC2-specific Ccr8-deficient mice, mixed donor bone marrow was transferred, containing 80% Tie2creRorafl/sg and 20% WT or Ccr8−/− bone marrow cells. 100% Tie2creRorafl/sg bone marrow was used to create control mice. After 8 wk, N. brasiliensis infection was performed.
Adoptive transfer of ILC2s
To transfer ILC2s into alymphoid mice, 2 × 106 in vitro–expanded and rested WT or Ccr8−/− ILC2s were injected i.v. into Rag2−/−il2rg−/− mice. Subsequently, mice were infected with N. brasiliensis and analyzed after 10 d.
Flow cytometry
To obtain lung, liver, and lamina propria single-cell suspensions, organs were removed and dissociated using the gentleMACS Octo dissociator (Miltenyi Biotec) according to manufacturer’s instructions. Because experiments with Ccr8−/− mice revealed that binding of several commercially available anti-mouse CCR8 mAbs is seemingly unspecific, we used custom-made fluorochrome-labeled CCL1 proteins for the detection of CCR8 surface expression. Briefly, tissue cell suspensions were incubated with murine CCL1-AF647 (5 nM; Almac) for 30 min at 37°C and washed before labeling with antibodies. For intracellular cytokine measurements, cells were stimulated with Cell Stimulation Cocktail plus protein transport inhibitors (eBioscience) or Monensin Solution (eBioscience) according to manufacturer’s instructions before antibody staining. Antibodies for flow cytometry were purchased from Miltenyi Biotech, unless specified otherwise. For ILC2 staining, a premixed lineage cocktail containing biotinylated anti-CD3, anti-CD5, anti-CD11b, anti-B220, anti-NK1.1, anti-Ter-119, anti-Gr1, and anti-SiglecF mAbs was used. Streptavidin conjugated to Brilliant Violet 421 (BioLegend) or VioBright FITC was applied in a secondary staining. The following fluorochrome-tagged antibodies were used for surface staining: anti-Thy1.2 (30-H12), anti-KLRG1 (2F1), anti-inducible T cell costimulator (anti-ICOS; 7E.17G9), anti-ST2 (DJ8; MD Biosciences), anti-CD127 (A7R34; BioLegend), anti-CD4 (GK1.5; eBioscience), anti-CD11b (M1/70; Invitrogen), anti-sialic acid–binding Ig-type lectin F (anti-SiglecF; ES22-10D8), anti-CD11c (N418; Invitrogen), and anti-CCR4 (2G12; BioLegend). Depending on the experiment, cells were subsequently fixed and permeabilized using the Foxp3 Transcription Factor Staining Buffer Kit (Invitrogen) according to the manufacturer’s instructions followed by intracellular staining with fluorochrome-coupled anti-GATA3 (REA174), anti-FoxP3 (FJK-16s; eBioscience), anti-IL-9 (RM9A4; BioLegend), anti–IL-13 (eBio13A; Invitrogen), or anti-CCL1 (148113; R&D Systems; labeled in-house with the Mix-n-Stain CF 647 Kit [Sigma-Aldrich] according to the manufacturer’s instructions) antibodies.
Cell death was investigated using the Annexin V Kit with PI (Miltenyi Biotec) according to the manufacturer’s instructions.
Samples were measured on an LSRFortessa cell analyzer (BD Biosciences). Gating strategies for analysis of murine ILC2s and eosinophils and in vitro–expanded ILC2s are provided in Fig. S4, A–C.
Mouse ILC2 isolation and culture
For potent ILC2 induction in vivo, mice were hydrodynamically injected with an IL-25 vector and sacrificed after 3 d. Subsequently, single-cell suspensions from spleen and mesenteric LNs were prepared using the gentleMACS Octo device (Miltenyi Biotec) according to the manufacturer’s instructions. ILC2s were identified using the following sort panel: ICOS VioBlue+ (7E.17G9; Miltenyi Biotec), KLRG1 PE+ (REA1016; Miltenyi Biotec), CD5 FITC− (REA421; Miltenyi Biotec), CD3 FITC− (17A2; BioLegend), CD45R FITC− (REA755; Miltenyi Biotec), NK1.1 PE-Vio770− (PK136; Miltenyi Biotec), CD49b PE-Vio770− (REA981; Miltenyi Biotec), CD11b APC-Vio770− (REA592; Miltenyi Biotec), CD11c APC-Vio770− (N418; Miltenyi Biotec), and FcεR1a PE-Cy7− (MAR-1; Invitrogen). FACS purification was achieved by using a MoFlo Astrios EQ device (Beckman Coulter) in the Core Unit Cell Sorting Erlangen.
ILC2s were expanded for several days in DMEM GlutaMAX medium (Gibco) supplemented with 10% FBS (Gibco), 1× MEM nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 20 mM Hepes (Carl Roth), 50 µM 2-mercaptoethanol (Sigma-Aldrich), 1% penicillin-streptomycin (Sigma-Aldrich), and recombinant IL-7, IL-25, IL-33 (50 ng/ml each; Immunotools), IL-2 (50 ng/ml; BioLegend), and thymic stromal lymphopoietin (20 ng/ml; Invitrogen). In some experiments, additional CCL1 (50 ng/ml; BioLegend), the CCR8 inhibitor R243 (5 µM; Aobious), or an anti-CCL1 antibody (10 µg/ml, 148113; R&D Systems) was added. To transfer ILC2s to a quiescent state for further experiments, cells were maintained in medium with IL-2 and IL-7 (10 ng/ml) for ≥48 h before restimulation as indicated. Depending on the experiment, additional stimulation with PMA (50 ng/ml) and ionomycin (500 ng/ml) was performed.
In another experimental setting, ILC2s and T cells were sorted from the colon of naive RoracreRosa26Stop-tdtomatofl/fl mice. Intestinal lamina propria cells were isolated using the gentleMACS Octo device (Miltenyi Biotec) according to the manufacturer’s instructions. ILC2s were identified using the following sort panel: tdTomato+, KLRG1 APC+ (2F1; eBioscience), CD5 FITC− (REA421; Miltenyi Biotec), CD3 FITC− (17A2; BioLegend), CD45R FITC− (REA755; Miltenyi Biotec), NK1.1 PE-Vio770− (PK136; Miltenyi Biotec), NKp46 PE-Cy7− (29A1.4; eBioscience), CD11b APC-Vio770− (REA592; Miltenyi Biotec), and CD11c APC-Vio770− (N418; Miltenyi Biotec). In the same staining, T cells were identified as CD4 Brilliant Violet 421+ (GK1.5; BioLegend) and FITC+. Cells were directly stimulated with PMA and ionomycin and supernatants collected for ELISA.
Characterization, purification, and ex vivo culture of human ILC2s
All studies with human material were approved by the institutional review board and ethics committee of the University Hospital of Erlangen (approval number 40_60B). Written informed consent was obtained from all subjects. Human peripheral blood mononuclear cells (PBMCs) were isolated from EDTA-treated peripheral blood of healthy donors via density gradient centrifugation using Lymphocyte Separation Media (Anprotec). For characterization and isolation of human ILC2s (Lin−CD127+CD161+CRTH2+ lymphoid cells), cells were stained for 20 min at 4°C using a Human Hematopoietic Lineage Antibody Cocktail (eBioscience) combined with the following fluorescence-labeled antibodies: anti-CCR8 (L263G8; BioLegend), anti-CD11c (MJ4-27G12; Miltenyi Biotec), anti-CD127 (REA614; Miltenyi Biotec), anti-CD161 (191B8; Miltenyi Biotec), anti-CRTH2 (BM16; BioLegend), and respective isotype controls. For flow cytometric characterization, stained samples were fixed with BD CellFIX (BD Biosciences) and acquired with a MACSQuant Analyzer 10 (Miltenyi Biotec). Sort purification of human ILC2s was performed with a FACS Aria II SORT (BD Biosciences).
Sort-purified human blood ILC2s were co-cultured with γ-irradiated (45 grays) allogenic PBMCs pooled from three different donors in Yssels T cell medium with 1% human serum antibody and 1% Penicillin-Streptomycin in 96-well round-bottom plates at a density of 102 ILC2s and 4 × 105 feeder PBMCs per well. ILC2 expansion was induced by the following stimuli: PHA (1 µg/ml; Sigma-Aldrich), recombinant human (rh) IL-2 (100 IU/ml; Miltenyi Biotech), rh IL-25 (50 ng/ml; eBioscience), and rh IL-33 (50 ng/ml; BioLegend). Medium and stimulation were replenished every 3–4 d. On days 13–15, expanded human ILC2s were rested in rh IL-2 (20 IU/ml) for 2 d. Afterward cells were seeded at 105 cells per well and restimulated with rh IL-2, rh IL-25, and rh IL-33 for 3 d. The effect of the monoclonal anti-CCL1 antibody (500 ng/ml, 35305; Invitrogen) on ILC2 expansion was determined after 14–15 d by flow cytometry. The gating strategy for analysis of human ILC2s is provided in Fig. S4 D.
Chemotaxis experiments
Cell migration was investigated using 5-µm transwell inserts for 24-well cell culture plates (Sarstedt). Briefly, 2 × 105 in vitro–expanded ILC2s in ILC2 medium were placed in the upper compartment. The well below was filled with ILC2 medium containing the recombinant chemokine CCL1, CCL8, CCL17, or CCL22 (all from BioLegend) at the indicated concentration or 5 µM of the activating CCR8 agonist ZK756326 (Selleckchem). After incubation for 2.5 h in a 37°C incubator, the migrated cells in the lower compartment were counted using a Neubauer chamber. The chemotaxis index was defined as the ratio of migrated cells to the respective stimulus and migrated cells without chemokines.
Cytokine and chemokine measurements
For determination of murine CCL1, CCL8, and Amphiregulin concentrations in sera or cell culture supernatants, Duoset ELISA Kits from R&D Systems were used according to the manufacturer’s instructions. To detect IL-9, the mouse IL-9 ELISA MAX Deluxe Kits (BioLegend) was used. For measurement of IL-13 concentrations, a mouse IL-13 ELISA Kit (Invitrogen) was employed. Secretion of CCL1 by human ILC2s was detected via Human ELISA Kits (Invitrogen) in ILC2 supernatants collected after restimulation.
Gene expression analysis
RNA was isolated from snap-frozen tissues or cultured cells using the peqGOLD Micro Spin Total RNA Kits (Peqlab) according to the manufacturer’s instructions. cDNA was synthesized with the Script RT-PCR kit from Jena Bioscience. Quantitative PCR (qPCR) analyses were performed using predesigned QuantiTect Primer assays (Qiagen) in a CFX Connect system (Bio-Rad). Relative expression of the indicated genes was calculated using hypoxanthine phosphoribosyltransferase 1 (hprt) as reference gene.
Histology and immune fluorescence
Lung samples were fixed in buffered formalin (Roti-Histofix; Carl Roth) at 4°C for 24 h, dehydrated, and embedded in liquid paraffin. 4-µm sections were cut using a microtome and stained with Alcian blue and PAS reagents (AB-PAS; Carl Roth) according to standard laboratory procedures. Pictures were acquired on a DMI4000B microscope (Leica Microsystems) at a 50× zoom factor. Equal lung lobes were used and tissues cut at similar positions to ensure comparable surface area. The number of PAS+ bronchioles per section was counted and compared.
For immune fluorescence, the following antibodies were used: anti-CD4 AF647 (GK1.5; BioLegend), anti-KLRG1 AF647 (2F1; Invitrogen), and anti-CCL1 (R&D Systems; labeled in-house with the Mix-n-Stain CF 555 Kit according to the manufacturer’s instructions). Pictures were acquired on a Leica SP5 confocal microscope.
Light-sheet fluorescence microscopy
For induction of airway inflammation, three doses of intranasal papain (100 µg each) were given daily before adoptive ILC2 transfer. Subsequently, 106 WT, Ccr8−/−, Ccr4−/− ILC2s, or a 50:50 mix of WT and Ccr4−/− ILC2s were stained with the CellTrace Yellow or Cell Proliferation Dye eFluor670 (Invitrogen) according to the manufacturer’s instructions and injected i.v. into recipient mice.
To prepare samples for light-sheet microscopy, mice were sacrificed at indicated time points after ILC2 transfer after transcardial perfusion with PBS and 5 mM EDTA followed by 4% paraformaldehyde solution (Klingberg et al., 2017). Lungs were reconstructed to physiological shape by intratracheal insertion of 0.75% low-gelling-temperature agarose (Biozym). Afterwards, lungs were removed and postfixed in 4% paraformaldehyde for 2 h. For dehydration, samples were incubated in increasing ethanol series of 50%, 70%, and 100% for 4 h each, repeating the latter step twice. Sample clearing was achieved by transferring the lungs into ethyl cinnamate (Sigma-Aldrich). Whole lungs were imaged with a LaVision Ultramicroscope II (BioTec) with an Olympus MVX10 Zoom Microscope Body (Olympus), a LaVision BioTec Laser Module, and an Andor Neo sCMOS Camera using the ImSpector Pro software (Abberior Instruments). Zoom factors of 10× or 25× were set, and Z-step size was 5 µm. Ethyl cinnamate was used as imaging medium.
Software and statistical analysis
Results are displayed as means with error bars showing ± SEM. Mann–Whitney U tests were applied using Prism 6.07 (GraphPad) software. Here, P values < 0.05 were considered significant indicated by asterisks, if not indicated elsewhere. Flow cytometry data were analyzed with FlowJo 10 software (Tree Star). Imaris 8.1.2 software (Bitplane) was used for visualization and three-dimensional reconstruction of light-sheet microscopy data.
Quantification of light-sheet data was achieved by randomly defining at least nine areas of similar volume (500 µm × 500 µm × 500/1,000 µm) dispersed over the whole sample (Schulz-Kuhnt et al., 2019). Fluorescent cells were counted manually using Imaris software.
Online supplemental material
Fig. S1 shows flow cytometric analyses of CCR8 on iILC2s, nILC2s, and in vitro–expanded human ILC2s. Fig. S2 shows chemotaxis assays of ILC2s and analysis of their in vivo migratory behavior. Fig. S3 demonstrates flow cytometric analyses of immune cell populations in naive and N. brasiliensis–infected WT, Ccr8−/−, and Tie2creRorafl/sg mice as well as worm counts in small intestinal tissues of WT and Tie2creRorafl/sg; CCL1 expression analyses by qPCR and immunohistochemistry are also shown. Fig. S4 shows flow cytometry gating strategies of murine lung ILC2s and in vitro–expanded ILC2s, human ILC2s, and murine lung eosinophils. Video 1 visualizes in vivo lung homing of WT and Ccr4−/− ILC2s upon adoptive transfer of labeled cells.
Supplementary Material
Acknowledgments
We thank A. Grüneboom and the Optical Imaging Center Erlangen of the Friedrich–Alexander University Erlangen for supporting light-sheet fluorescence microscopy imaging.
This work has received funding from the German Research Foundation within the TRR241 (A03), GK1660, CRC1181 (A08, A02), and FOR2886 (TP01). Further support was given by the Interdisciplinary Center for Clinical Research of the University Erlangen-Nuremberg (A75). Cell sorting was supported by the FACS Core Unit at the Nikolaus Fiebiger Center, Erlangen.
The authors declare no competing financial interests.
Author contributions: L. Knipfer, A. Schulz-Kuhnt, M. Kindermann, and V. Greif performed experiments and analyzed data. D. Voehringer, C. Symowski, M.F. Neurath, and I. Atreya provided material and commented on the manuscript. L. Knipfer and S. Wirtz conceptualized the project and wrote the paper.
References
- Anderson C.A., Solari R., and Pease J.E.. 2016. Biased agonism at chemokine receptors: obstacles or opportunities for drug discovery? J. Leukoc. Biol. 99:901–909. 10.1189/jlb.2MR0815-392R [DOI] [PubMed] [Google Scholar]
- Ballesteros-Tato A., Randall T.D., Lund F.E., Spolski R., Leonard W.J., and León B.. 2016. T Follicular Helper Cell Plasticity Shapes Pathogenic T Helper 2 Cell-Mediated Immunity to Inhaled House Dust Mite. Immunity. 44:259–273. 10.1016/j.immuni.2015.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.L., and McKenzie A.N.. 2014. Type-2 innate lymphoid cells in human allergic disease. Curr. Opin. Allergy Clin. Immunol. 14:397–403. 10.1097/ACI.0000000000000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsheshet Y., Wildbaum G., Levy E., Vitenshtein A., Akinseye C., Griggs J., Lira S.A., and Karin N.. 2017. CCR8+FOXp3+ Treg cells as master drivers of immune regulation. Proc. Natl. Acad. Sci. USA. 114:6086–6091. 10.1073/pnas.1621280114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop B., and Lloyd C.M.. 2003. CC chemokine ligand 1 promotes recruitment of eosinophils but not Th2 cells during the development of allergic airways disease. J. Immunol. 170:4810–4817. 10.4049/jimmunol.170.9.4810 [DOI] [PubMed] [Google Scholar]
- Chensue S.W., Lukacs N.W., Yang T.Y., Shang X., Frait K.A., Kunkel S.L., Kung T., Wiekowski M.T., Hedrick J.A., Cook D.N., et al. 2001. Aberrant in vivo T helper type 2 cell response and impaired eosinophil recruitment in CC chemokine receptor 8 knockout mice. J. Exp. Med. 193:573–584. 10.1084/jem.193.5.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S.J., Babot Z., Leingärtner A., Studer M., Nakagawa Y., and O’Leary D.D.. 2013. Geniculocortical input drives genetic distinctions between primary and higher-order visual areas. Science. 340:1239–1242. 10.1126/science.1232806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.D., Kuo F., Kumer J., Motani A.S., Lawrence C.E., Henderson W.R. Jr., and Venkataraman C.. 2003. CCR8 is not essential for the development of inflammation in a mouse model of allergic airway disease. J. Immunol. 170:581–587. 10.4049/jimmunol.170.1.581 [DOI] [PubMed] [Google Scholar]
- Chvatchko Y., Hoogewerf A.J., Meyer A., Alouani S., Juillard P., Buser R., Conquet F., Proudfoot A.E., Wells T.N., and Power C.A.. 2000. A key role for CC chemokine receptor 4 in lipopolysaccharide-induced endotoxic shock. J. Exp. Med. 191:1755–1764. 10.1084/jem.191.10.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Koyasu S.. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 41:354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealey K.N., Moro K., and Koyasu S.. 2017. Are ILC2s Jekyll and Hyde in airway inflammation? Immunol. Rev. 278:207–218. 10.1111/imr.12547 [DOI] [PubMed] [Google Scholar]
- Gasteiger G., Fan X., Dikiy S., Lee S.Y., and Rudensky A.Y.. 2015. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science. 350:981–985. 10.1126/science.aac9593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombert M., Dieu-Nosjean M.C., Winterberg F., Bünemann E., Kubitza R.C., Da Cunha L., Haahtela A., Lehtimäki S., Müller A., Rieker J., et al. 2005. CCL1-CCR8 interactions: an axis mediating the recruitment of T cells and Langerhans-type dendritic cells to sites of atopic skin inflammation. J. Immunol. 174:5082–5091. 10.4049/jimmunol.174.8.5082 [DOI] [PubMed] [Google Scholar]
- Halim T.Y., MacLaren A., Romanish M.T., Gold M.J., McNagny K.M., and Takei F.. 2012. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 37:463–474. 10.1016/j.immuni.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Heymann F., Hammerich L., Storch D., Bartneck M., Huss S., Rüsseler V., Gassler N., Lira S.A., Luedde T., Trautwein C., and Tacke F.. 2012. Hepatic macrophage migration and differentiation critical for liver fibrosis is mediated by the chemokine receptor C-C motif chemokine receptor 8 in mice. Hepatology. 55:898–909. 10.1002/hep.24764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Kawamura Y.I., Yasuhara M., Toyama-Sorimachi N., Yamamoto K., Matsukawa A., Lira S.A., and Dohi T.. 2007. Inhibition of CCL1-CCR8 interaction prevents aggregation of macrophages and development of peritoneal adhesions. J. Immunol. 178:5296–5304. 10.4049/jimmunol.178.8.5296 [DOI] [PubMed] [Google Scholar]
- Huang Y., Guo L., Qiu J., Chen X., Hu-Li J., Siebenlist U., Williamson P.R., Urban J.F. Jr., and Paul W.E.. 2015. IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16:161–169. 10.1038/ni.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Mao K., Chen X., Sun M.A., Kawabe T., Li W., Usher N., Zhu J., Urban J.F. Jr., Paul W.E., and Germain R.N.. 2018. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 359:114–119. 10.1126/science.aam5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iellem A., Colantonio L., Bhakta S., Sozzani S., Mantovani A., Sinigaglia F., and D’Ambrosio D.. 2000. Inhibition by IL-12 and IFN-alpha of I-309 and macrophage-derived chemokine production upon TCR triggering of human Th1 cells. Eur. J. Immunol. 30:1030–1039. [DOI] [PubMed] [Google Scholar]
- Islam S.A., Chang D.S., Colvin R.A., Byrne M.H., McCully M.L., Moser B., Lira S.A., Charo I.F., and Luster A.D.. 2011. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat. Immunol. 12:167–177. 10.1038/ni.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma T., Nakamura K., Wakugawa M., Mitsui H., Tada Y., Saeki H., Torii H., Asahina A., Onai N., Matsushima K., and Tamaki K.. 2001. Thymus and activation-regulated chemokine in atopic dermatitis: Serum thymus and activation-regulated chemokine level is closely related with disease activity. J. Allergy Clin. Immunol. 107:535–541. 10.1067/mai.2001.113237 [DOI] [PubMed] [Google Scholar]
- Karta M.R., Rosenthal P.S., Beppu A., Vuong C.Y., Miller M., Das S., Kurten R.C., Doherty T.A., and Broide D.H.. 2018. β2 integrins rather than β1 integrins mediate Alternaria-induced group 2 innate lymphoid cell trafficking to the lung. J. Allergy Clin. Immunol. 141:329–338.e12. 10.1016/j.jaci.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Hashimoto-Hill S., and Kim M.. 2016. Migration and Tissue Tropism of Innate Lymphoid Cells. Trends Immunol. 37:68–79. 10.1016/j.it.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Taparowsky E.J., and Kim C.H.. 2015. Retinoic Acid Differentially Regulates the Migration of Innate Lymphoid Cell Subsets to the Gut. Immunity. 43:107–119. 10.1016/j.immuni.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindermann M., Knipfer L., Atreya I., and Wirtz S.. 2018. ILC2s in infectious diseases and organ-specific fibrosis. Semin. Immunopathol. 40:379–392. 10.1007/s00281-018-0677-x [DOI] [PubMed] [Google Scholar]
- Klingberg A., Hasenberg A., Ludwig-Portugall I., Medyukhina A., Männ L., Brenzel A., Engel D.R., Figge M.T., Kurts C., and Gunzer M.. 2017. Fully Automated Evaluation of Total Glomerular Number and Capillary Tuft Size in Nephritic Kidneys Using Lightsheet Microscopy. J. Am. Soc. Nephrol. 28:452–459. 10.1681/ASN.2016020232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S., and Artis D.. 2016. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 17:765–774. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2015. The immunology of asthma. Nat. Immunol. 16:45–56. 10.1038/ni.3049 [DOI] [PubMed] [Google Scholar]
- Louahed J., Struyf S., Demoulin J.B., Parmentier M., Van Snick J., Van Damme J., and Renauld J.C.. 2003. CCR8-dependent activation of the RAS/MAPK pathway mediates anti-apoptotic activity of I-309/ CCL1 and vMIP-I. Eur. J. Immunol. 33:494–501. 10.1002/immu.200310025 [DOI] [PubMed] [Google Scholar]
- McHedlidze T., Waldner M., Zopf S., Walker J., Rankin A.L., Schuchmann M., Voehringer D., McKenzie A.N., Neurath M.F., Pflanz S., and Wirtz S.. 2013. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity. 39:357–371. 10.1016/j.immuni.2013.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchedlidze T., Kindermann M., Neves A.T., Voehringer D., Neurath M.F., and Wirtz S.. 2016. IL-27 suppresses type 2 immune responses in vivo via direct effects on group 2 innate lymphoid cells. Mucosal Immunol. 9:1384–1394. 10.1038/mi.2016.20 [DOI] [PubMed] [Google Scholar]
- Mikhak Z., Fukui M., Farsidjani A., Medoff B.D., Tager A.M., and Luster A.D.. 2009. Contribution of CCR4 and CCR8 to antigen-specific T(H)2 cell trafficking in allergic pulmonary inflammation. J. Allergy Clin. Immunol. 123:67–73.e3. 10.1016/j.jaci.2008.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., and Koyasu S.. 2010. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 463:540–544. 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]
- Moro K., Kabata H., Tanabe M., Koga S., Takeno N., Mochizuki M., Fukunaga K., Asano K., Betsuyaku T., and Koyasu S.. 2016. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17:76–86. 10.1038/ni.3309 [DOI] [PubMed] [Google Scholar]
- Neill D.R., Wong S.H., Bellosi A., Flynn R.J., Daly M., Langford T.K., Bucks C., Kane C.M., Fallon P.G., Pannell R., et al. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 464:1367–1370. 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata Y., Frech M., Primbs T., Lucas S., Andreev D., Scholtysek C., Sarter K., Kindermann M., Yeremenko N., Baeten D.L., et al. 2018. Group 2 Innate Lymphoid Cells Attenuate Inflammatory Arthritis and Protect from Bone Destruction in Mice. Cell Reports. 24:169–180. 10.1016/j.celrep.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Oshio T., Kawashima R., Kawamura Y.I., Hagiwara T., Mizutani N., Okada T., Otsubo T., Inagaki-Ohara K., Matsukawa A., Haga T., et al. 2014. Chemokine receptor CCR8 is required for lipopolysaccharide-triggered cytokine production in mouse peritoneal macrophages. PLoS One. 9:e94445 10.1371/journal.pone.0094445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina-Bordignon P., Papi A., Mariani M., Di Lucia P., Casoni G., Bellettato C., Buonsanti C., Miotto D., Mapp C., Villa A., et al. 2001. The C-C chemokine receptors CCR4 and CCR8 identify airway T cells of allergen-challenged atopic asthmatics. J. Clin. Invest. 107:1357–1364. 10.1172/JCI12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.E., Liang H.E., Sullivan B.M., Reinhardt R.L., Eisley C.J., Erle D.J., and Locksley R.M.. 2010. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA. 107:11489–11494. 10.1073/pnas.1003988107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttur F., Denney L., Gregory L.G., Vuononvirta J., Oliver R., Entwistle L.J., Walker S.A., Headley M.B., McGhee E.J., Pease J.E., et al. 2019. Pulmonary environmental cues drive group 2 innate lymphoid cell dynamics in mice and humans. Sci. Immunol. 4:eaav7638 10.1126/sciimmunol.aav7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y., Proudfoot A.E., Power C.A., Campbell J.J., and Butcher E.C.. 2001. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J. Exp. Med. 194:1541–1547. 10.1084/jem.194.10.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.. Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S., Mitchell A.J., Tay S.S., Jain R., Forbes-Blom E., et al. 2013. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 14:564–573. 10.1038/ni.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckes T., Saul D., Van Snick J., Hermine O., and Grassmann R.. 2001. Autocrine antiapoptotic stimulation of cultured adult T-cell leukemia cells by overexpression of the chemokine I-309. Blood. 98:1150–1159. 10.1182/blood.V98.4.1150 [DOI] [PubMed] [Google Scholar]
- Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.C., Johnson D., Scanlon S.T., McKenzie A.N., et al. 2013. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210:2939–2950. 10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Stöger L., Liu W., Go S., Pavord I., Klenerman P., Ogg G., and Xue L.. 2017. Cysteinyl leukotriene E4 activates human group 2 innate lymphoid cells and enhances the effect of prostaglandin D2 and epithelial cytokines. J. Allergy Clin. Immunol. 140:1090–1100.e11. 10.1016/j.jaci.2016.12.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather B.D., Treuting P., Perdue N., Miazgowicz M., Fontenot J.D., Rudensky A.Y., and Campbell D.J.. 2007. Altering the distribution of Foxp3(+) regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 204:1335–1347. 10.1084/jem.20070081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli P., Ebert L., Willimann K., Blaser A., Roos R.S., Loetscher P., and Moser B.. 2004. A skin-selective homing mechanism for human immune surveillance T cells. J. Exp. Med. 199:1265–1275. 10.1084/jem.20032177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz-Kuhnt A., Zundler S., Grüneboom A., Neufert C., Wirtz S., Neurath M.F., and Atreya I.. 2019. Advanced Imaging of Lung Homing Human Lymphocytes in an Experimental In Vivo Model of Allergic Inflammation Based on Light-sheet Microscopy. J. Vis. Exp. (146). 10.3791/59043 [DOI] [PubMed] [Google Scholar]
- Sokol C.L., Camire R.B., Jones M.C., and Luster A.D.. 2018. The Chemokine Receptor CCR8 Promotes the Migration of Dendritic Cells into the Lymph Node Parenchyma to Initiate the Allergic Immune Response. Immunity. 49:449–463.e6. 10.1016/j.immuni.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler D., Chapman T.R., Poisson L.R., Wang L., Cote-Sierra J., Ryan M., McDonald A., Badola S., Fedyk E., Coyle A.J., et al. 2006. CCR8 expression identifies CD4 memory T cells enriched for FOXP3+ regulatory and Th2 effector lymphocytes. J. Immunol. 177:6940–6951. 10.4049/jimmunol.177.10.6940 [DOI] [PubMed] [Google Scholar]
- Spinetti G., Bernardini G., Camarda G., Mangoni A., Santoni A., Capogrossi M.C., and Napolitano M.. 2003. The chemokine receptor CCR8 mediates rescue from dexamethasone-induced apoptosis via an ERK-dependent pathway. J. Leukoc. Biol. 73:201–207. 10.1189/jlb.0302105 [DOI] [PubMed] [Google Scholar]
- Stier M.T., Zhang J., Goleniewska K., Cephus J.Y., Rusznak M., Wu L., Van Kaer L., Zhou B., Newcomb D.C., and Peebles R.S. Jr. 2018. IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J. Exp. Med. 215:263–281. 10.1084/jem.20170449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.E., Morrison P.J., Wilhelm C., Wilson M., Ahlfors H., Renauld J.C., Panzer U., Helmby H., and Stockinger B.. 2013. IL-9-mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J. Exp. Med. 210:2951–2965. 10.1084/jem.20130071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayanand P., Durkin K., Hartmann G., Morjaria J., Seumois G., Staples K.J., Hall D., Bessant C., Bartholomew M., Howarth P.H., et al. 2010. Chemokine receptor 4 plays a key role in T cell recruitment into the airways of asthmatic patients. J. Immunol. 184:4568–4574. 10.4049/jimmunol.0901342 [DOI] [PubMed] [Google Scholar]
- Vivier E., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N.J., Mebius R.E., et al. 2018. Innate Lymphoid Cells: 10 Years On. Cell. 174:1054–1066. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- Wilhelm C., Hirota K., Stieglitz B., Van Snick J., Tolaini M., Lahl K., Sparwasser T., Helmby H., and Stockinger B.. 2011. An IL-9 fate reporter demonstrates the induction of an innate IL-9 response in lung inflammation. Nat. Immunol. 12:1071–1077. 10.1038/ni.2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingoni A., Soto H., Hedrick J.A., Stoppacciaro A., Storlazzi C.T., Sinigaglia F., D’Ambrosio D., O’Garra A., Robinson D., Rocchi M., et al. 1998. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J. Immunol. 161:547–551. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.