Lam et al. characterize a novel hematological/autoinflammatory disorder due to a de novo recurrent missense mutation of CDC42. The authors use in silico, in vitro, and in vivo analyses to correlate the molecular mechanisms altering CDC42 function to the observed phenotype.
Abstract
Hemophagocytic lymphohistiocytosis (HLH) is characterized by immune dysregulation due to inadequate restraint of overactivated immune cells and is associated with a variable clinical spectrum having overlap with more common pathophysiologies. HLH is difficult to diagnose and can be part of inflammatory syndromes. Here, we identify a novel hematological/autoinflammatory condition (NOCARH syndrome) in four unrelated patients with superimposable features, including neonatal-onset cytopenia with dyshematopoiesis, autoinflammation, rash, and HLH. Patients shared the same de novo CDC42 mutation (Chr1:22417990C>T, p.R186C) and altered hematopoietic compartment, immune dysregulation, and inflammation. CDC42 mutations had been associated with syndromic neurodevelopmental disorders. In vitro and in vivo assays documented unique effects of p.R186C on CDC42 localization and function, correlating with the distinctiveness of the trait. Emapalumab was critical to the survival of one patient, who underwent successful bone marrow transplantation. Early recognition of the disorder and establishment of treatment followed by bone marrow transplant are important to survival.
Graphical Abstract
Introduction
The diagnosis and delineation of novel genetic syndromes is often difficult given the limited availability of patients, genetic heterogeneity, and clinical variability. Hemophagocytic lymphohistiocytosis (HLH) has recently been characterized as a clinical syndrome with hyperinflammation driven by excessive activation and expansion of macrophages and CD8+ T lymphocytes (Jordan et al., 2011; Rosado and Kim, 2013). Typical features include persistent high fever, liver involvement, splenomegaly, intravascular activation of coagulation associated with pancytopenia, and usually an increase in ferritin. Although this syndrome is clinically unique, the mechanisms underlying this disorder are diverse (Sepulveda and de Saint Basile, 2017; Chinn et al., 2018). Mutations leading to defective cytotoxicity by natural killer (NK) and CD8+ T lymphocytes are the typical cause of monogenic HLH, typically termed primary HLH. However, a markedly higher number of patients present with HLH in the absence of genetically defective cytotoxicity in the context of infections, rheumatic inflammatory diseases and malignancy. HLH in its various forms, both in children and adults, is invariably fatal if untreated.
Cell division cycle 42 (CDC42) is a member of the Ras-homologous (Rho) GTPase family functioning as a signaling node controlling a number of cellular processes, including adhesion, migration, polarity, cell cycle, and proliferation (Zhou et al., 2013; Baschieri et al., 2014). CDC42 functions as a molecular switch by cycling between a guanosine 5′-triphosphate (GTP)–bound (active) and a guanosine diphosphate (GDP)–bound (inactive) state. Two CDC42 isoforms have been characterized. While isoform 1 is ubiquitously expressed, isoform 2 is primarily found in the brain. CDC42 function is controlled by three different classes of regulators: guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (Dvorsky and Ahmadian, 2004). The GTPase primarily acts through its spatial and temporal localized interaction with multiple downstream effectors, such as IQGAP1, p21-activated kinase (PAK), and Wiskott–Aldrich syndrome protein (WASP). Reversible localization of CDC42 at the cytoplasmic leaflet of the plasma membrane and other intracellular membranes is regulated by Rho GDP-dissociation inhibitor (RhoGDI) and IQGAP1. The former controls the dynamic membrane–cytoplasm shuttling of the GTPase (Gibson and Wilson-Delfosse, 2001; Gibson et al., 2004), while the latter promotes CDC42 translocation from the Golgi apparatus to the plasma membrane (Swart-Mataraza et al., 2002). These regulatory events play a crucial role in controlling CDC42 function, cytoskeletal rearrangement, cell polarity, and migration. Notably, altered binding of CDC42 to IQGAP1 induces multiple leading edge formation and aberrant multipolarized morphology (Fukata et al., 2002). Actin rearrangements and cell migration are also promoted by CDC42 interaction with its effector, WASP, a critical actin regulator and mediator of NK cell cytotoxicity (Orange et al., 2002; Ridley et al., 2003). Finally, CDC42 function requires posttranslational processing at the C-terminus, including prenylation at Cys188 (geranyl-geranylation, most commonly) followed by proteolytic cleavage of the last three residues and carboxyl-methylation (Aicart-Ramos et al., 2011).
We and others recently identified germline heterozygous mutations in CDC42 as the event underlying a remarkably heterogeneous collection of neurodevelopmental phenotypes (Takenouchi et al., 2015; Martinelli et al., 2018). Core clinical features of these traits include variable growth dysregulation; facial dysmorphism; intellectual disability; cardiac defects; immunological, hematological, and lymphatic abnormalities; and brain malformations. Mutations were found to variably disrupt CDC42 function by altering the switch between the active and inactive states of the GTPase and/or affecting its interaction with effectors (Martinelli et al., 2018). As a result, multiple cellular and developmental processes were differentially perturbed. Remarkably, the biochemical and functional characterization of mutations allowed the identification of genotype–phenotype relationships, suggesting a link between the specific impact of individual mutation class and its phenotypic expression (Martinelli et al., 2018). Mutations were documented to behave either as activating or inactivating, with the latter specifically associated with thrombocytopenia. Multisystem/organ involvement occurred in all groups.
Using whole-exome sequencing/whole-genome sequencing (WES/WGS) coupled to biochemical and functional validation, we describe a novel hematological and autoinflammatory phenotype in four unrelated patients caused by the same de novo missense mutation of CDC42 (NM_001791, c.556C>T, p.R186C). The disease differs considerably from those previously associated with CDC42 mutations and is characterized by neonatal onset of pancytopenia, autoinflammation, rash, and episodes of HLH (NOCARH). Through in silico, in vitro, and in vivo analyses, we describe the mechanism by which this specific amino acid change affects CDC42 function, intracellular signaling, and cellular and developmental processes. Finally, we provide a clinical delineation of this disorder and document the clinical response to IFN-γ and IL-1β neutralization and hematopoietic stem cell transplantation (HSCT).
Results
Clinical features of patients
The four unrelated patients included in the study showed a superimposable previously unrecognized multisystem disease characterized by neonatal onset of pancytopenia, persistent fever, skin rash, hepatosplenomegaly, and persistently elevated inflammatory markers in the absence of any evidence of neurodevelopmental involvement (Tables 1, S1, and S2; and Fig. 1 A). A suspicion of neonatal-onset multisystem inflammatory disease in three of the four patients led to treatment with IL-1 inhibitors with incomplete improvement of fever and rash, but not cytopenia. In all cases, high chronic doses of glucocorticoids were required to treat inflammation. All patients developed HLH, which was fatal in three subjects. Based on the shared features, we propose the term of NOCARH syndrome for this trait. A detailed description of the patients’ clinical history is provided in the Materials and methods.
Table 1. Features of patients sharing the de novo missense CDC42 c.556C>T (p.R186C) mutation.
| Pt 1 | Pt 2 | Pt 3 | Pt 4 | |
|---|---|---|---|---|
| Outcome and status | Alive, 6 yr | Dead, 6 mo | Dead, 1.5 yr | Dead, 4.5 mo |
| Fever | + | + | + | + |
| Skin rash | + | + | + | + |
| Facial dysmorphism | − | − | − | − |
| Failure to thrive | + | + | + | + |
| Hepatomegaly | + | + | + | + |
| Splenomegaly | + | + | + | − |
| CNS inflammatory diseasea | + | − | − | − |
| Gastrointestinal symptomsb | + | + | + | + |
| Cardiac abnormalities | − | − | − | − |
| HLHc | + | + | + | + |
| Anemiad | + | + | + | + |
| Thrombocytopeniad | + | + | + | + |
| Neutropeniae | + | + | + | + |
| Monocytopenia | + | + | N/A | N/A |
| Acute-phase response | + | + | + | + |
| BM dysplasia | + | + | + | + |
| Other notable features | +f | − | − | +g |
CNS, central nervous system; N/A, not applicable.
Pt 1, three episodes at age 2 yr with generalized seizures and white and gray matter lesions in MRI; Pt 3, increased CSF protein and MRI with leptomeningitis.
Pt 1, diarrhea, intestinal bleeding at age 11 mo, and intestinal infarction at age 5 yr; Pt 2, chronic diarrhea; Pt 3, small intestine inflammation; Pt 4, severe, unremitting enterocolitis from birth and diarrhea with intestinal bleedings and infarctions.
Pt 1, four episodes; Pt 2, a single episode leading to death; Pt 3, died of secondary HLH after transplant; Pt 4, four episodes.
Pt 1, constant transfusion requirement until age 2 yr; Pt 2, intermittent transfusion requirement during flares; Pt 3, transfusion dependent; Pt 4, transfusion dependent.
Pt 1, Pt 2, and Pt 3, profound neutropenia; Pt 4, mild neutropenia.
Suspected trigonocephaly.
Very small thymus; arthritis.
Figure 1.
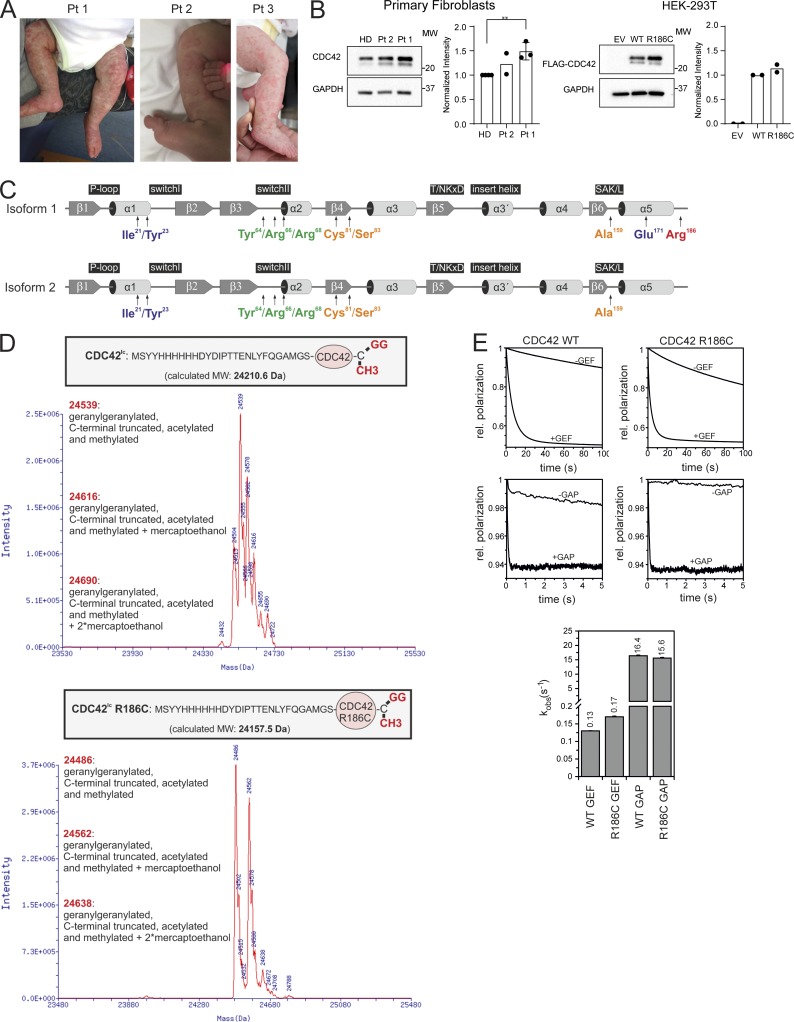
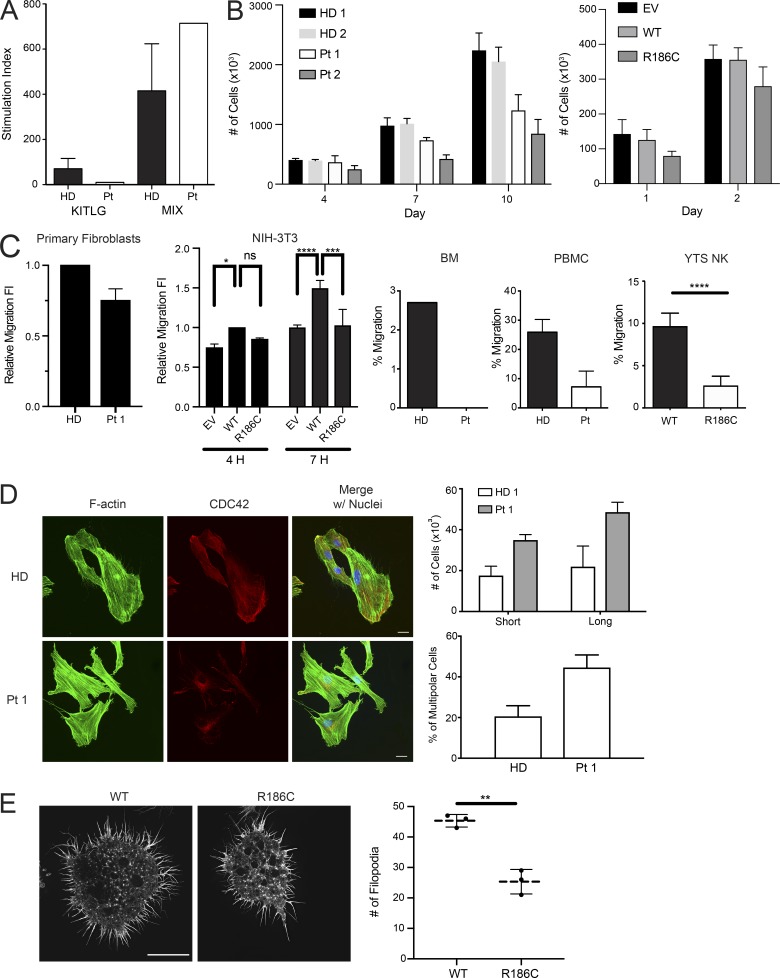
Clinical features of patients carrying the c.556C>T change (p.R186C) in CDC42, and biochemical profiling of the disease-causing mutant. (A) Skin findings of the affected subjects. (B) CDC42 levels in Pt 1 (n = 3, normalized intensity relative to HDs; **, P < 0.01, unpaired t test) and Pt 2 (n = 2) primary fibroblasts and HEK-293T cells (n = 2) expressing FLAG-tagged WT and mutant CDC42 or an empty vector (EV). (C) CDC42 domain organization indicating key functional regions and locations of residues affected by disease-causing variants. The two CDC42 isoforms are shown. Isoform 1 is ubiquitously expressed, while isoform 2 is brain restricted. The missense variants affecting E171 and R186 only affect isoform 1, while mutations affecting the other residues involve both isoforms. (D) p.R186C does not affect CDC42 posttranslational processing. MS analysis of WT and R186C CDC42 proteins shows a strong peak at 24,539 daltons and 24,486 daltons, respectively, indicating that CDC42R186C is properly processed at the C-terminus. Da, daltons. (E) p.R186C does not affect CDC42 GTPase activity and nucleotide exchange behavior. GTPase activity and nucleotide exchange reaction in CDC42R186C (right) are compared with WT CDC42 (left). kobs values (observed dissociation constant) are reported (bottom). Representative polarization curves are shown (n = 4–6). MW, molecular weight; SAK/L, serine-alanine-lysine/leucine motif.
Genetic studies
Two patients (patient 1 [Pt 1] and Pt 3) were enrolled in the Undiagnosed Disease Programs at the Ospedale Pediatrico Bambino Gesù and Baylor-Hopkins Center for Mendelian Genomics (CMG) at Baylor College of Medicine and the NK cell Evaluation and Research Program at Baylor and Columbia University. Using a trio-based WES strategy, a de novo missense CDC42 change (NM_001791, Chr1:22417990, c.556C>T, p.R186C) affecting an isoform 1–specific exon was identified as the only candidate variant underlying the disorder in both subjects (Table S3). Occurrence of mutations in genes known to be associated with HLH or other immunological and hematologic disorders was excluded. Similarly, there was no other rare substantively damaging variant shared by the two patients. The missense mutation had not previously been reported in the Exome Aggregation Consortium/Genome Aggregation Database (gnomAD) and was predicted to be damaging by in silico tools (Combined Annotation-Dependent Depletion [CADD] v1.3: 24.5, and Mendelian clinically applicable pathogenicity [M-CAP] v1.3: 0.0531). Based on this finding, a third patient (Pt 2) having clinical features resembling those of the two previously studied subjects underwent Sanger sequencing for the relevant CDC42 coding portion, revealing heterozygosity for the same c.556C>T transition, which was validated to occur as a de novo event. Finally, through networking, a fourth patient (Pt 4) sharing a similar clinical phenotype, and the same de novo CDC42 mutation was identified. In this case, the mutation was revealed by WGS using a trio-based strategy (Table S3), which excluded the occurrence of functionally and clinically relevant variants in HLH- and autoinflammation-related genes.
Biochemical and functional studies
To explore the functional consequence of the identified missense change, we initially assessed CDC42 levels in primary fibroblasts (Pt 1 and Pt 2) and HEK-293T cells transiently expressing the FLAG-tagged CDC42 mutant. In primary fibroblasts, the protein was expressed to similar levels without any apparent accelerated degradation (Fig. 1 B). Similarly, no reduction in the level of the mutant was observed in transfected cells. These data suggested that the disease-associated missense change does not significantly decrease CDC42 synthesis/stability, pointing to a specific perturbing effect on protein function.
Compared with the previously identified variants of CDC42 underlying neurodevelopmental disorders (Takenouchi et al., 2015; Martinelli et al., 2018), the p.R186C amino acid substitution occurs in a unique region at the C-terminus, within the hypervariable region (Fig. 1 C). Specifically, Arg186 lies in proximity of Cys188, which serves as substrate site for CDC42 prenylation. This irreversible posttranslational modification is required for proper CDC42 subcellular localization and function. To assess a possible effect of the p.R186C substitution on CDC42 prenylation, mass spectrometry (MS) analysis of the purified CDC42 mutant was performed, revealing proper C-terminal processing and lipid incorporation (Fig. 1 D), excluding the possibility of a defective or aberrant CDC42 posttranslational processing. Structural studies indicate that Arg186 is located far from the GDP/GTP–binding pocket and major regions controlling CDC42 interaction with positive and negative regulators (switch regions I and II; Phillips et al., 2008). Consistently, fluorescence-based cell-free assays showed that replacement of Arg186 by cysteine did not have a significant impact on either CDC42 basal and GEF-catalyzed GDP/GTP exchange activity or basal and GAP-stimulated GTPase activity of the protein (Fig. 1 E).
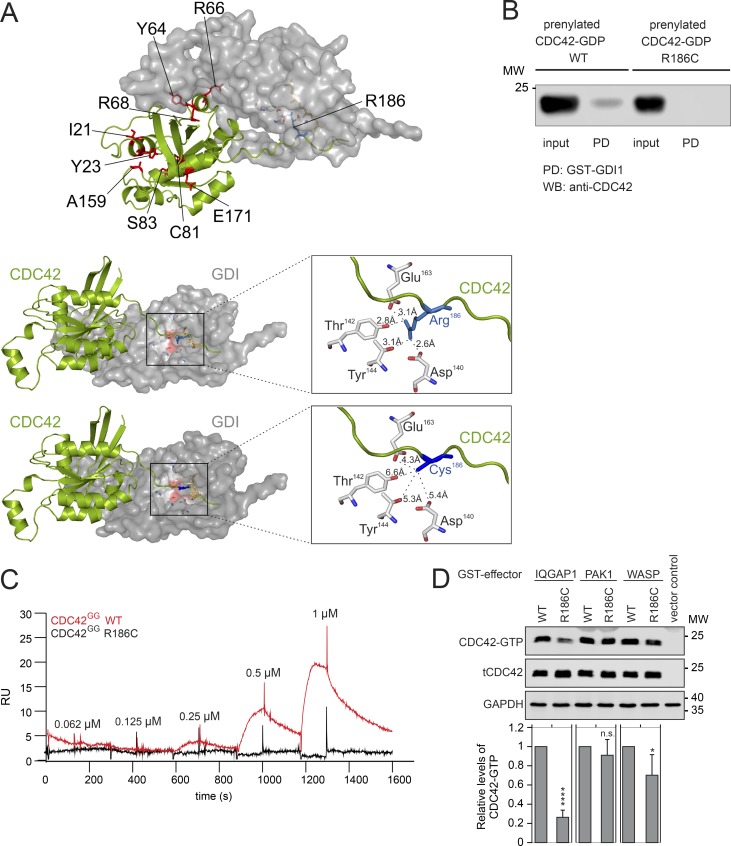
Arg186 is a solvent-exposed residue located at the surface of CDC42 in one of the two major sites of binding to RhoGDI (Hoffman et al., 2000; Fig. 2 A). The residue stabilizes the geranylgeranyl-mediated CDC42–RhoGDI interaction. This interaction has a key role in the control of CDC42 function, as it negatively regulates stable binding of the GTPase to the cytoplasmic leaflet of membranes and modulates CDC42 trafficking (Gibson and Wilson-Delfosse, 2001; Gibson et al., 2004). Based on the predicted disruptive effect of the Arg-to-Cys substitution on RhoGDI binding (Fig. 2 A), pull-down assays were performed to confirm defective interaction between the CDC42 mutant and RhoGDI (Fig. 2 B). The impaired binding of prenylated CDC42R186C protein to RhoGDI was also demonstrated by surface plasmon resonance (SPR) analysis (Fig. 2 C). These data strongly suggest a perturbation of proper subcellular localization and trafficking of CDC42 as a major event contributing to pathogenesis. Notably, since other residues previously reported to be mutated in CDC42-related neurodevelopmental disorders are located at the interacting surface of the second major site of the GTPase mediating binding to RhoGDI, with a subset of them directly involved in RhoGDI binding (Fig. 2 A), the defective binding of CDC42 to RhoGDI likely does not represent a distinctive impairment of the CDC42R186C mutant. This was confirmed by pull-down assays using a panel of bacterially expressed mutants to assess the impact of amino acid substitutions in regions far from the C-terminal tail on RhoGDI binding, which consistently with the structural data suggest that other missense changes variably impact CDC42 binding to RhoGDI (data not shown).
Figure 2.
p.R186C affects CDC42 binding to RhoGDI, IQGAP1, and WASP. (A) Structure of CDC42 complexed with RhoGDI. Visualization of residues affected in human disease (left). Arg186 has a unique localization within the hypervariable region. Arg186 is surrounded by Asp140, Thr142, Tyr144, and Glu163 of RhoGDI (within 4 Å; right middle panel). p.R186C is predicted to disrupt the interaction between the hypervariable region of CDC42 and RhoGDI (right lower panel). (B) Pull-down (PD) assays showing RhoGDI interaction of prenylated WT and mutant CDC42. Binding of GST-fused RhoGDI to CDC42 proteins was analyzed by WB documenting defective CDC42R186C binding. A representative image is shown (n = 2). (C) SPR analysis of the RhoGDI–CDC42 interaction. Immobilized GST-tagged RhoGDI was titrated with increasing concentrations of WT (red) and R186C (black) CDC42GG proteins. No binding for the mutant protein was observed. RU, response unit (n = 1). (D) Representative WB visualizing pull-down of overexpressed FLAG-tagged WT and R186C CDC42 from COS-7 cell lysates by GST-fused effector proteins IQGAP1, PAK1, and WASP. The same amount of cell lysate was used as a loading control (tCDC42). Bar charts indicate the relative levels of GTP-bound WT and R186C CDC42 normalized to the levels of total CDC42. A strongly reduced and decreased binding of CDC42R186C to IQGAP1 and WASP was documented, respectively (mean ± SD, n = 3 independent experiments; *, P < 0.05; ****, P < 0.0001, one-way ANOVA with Sidak’s multiple comparison test). n.s., not significant; MW, molecular weight.
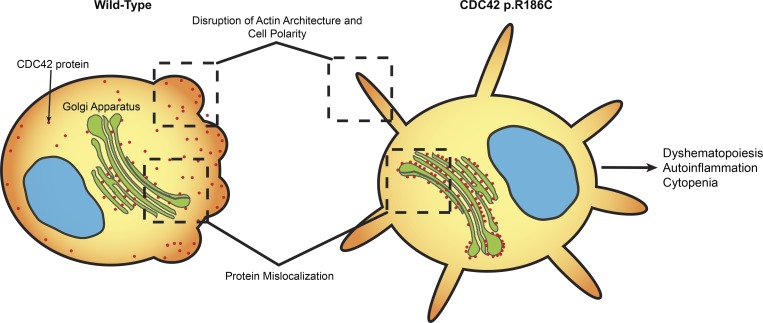
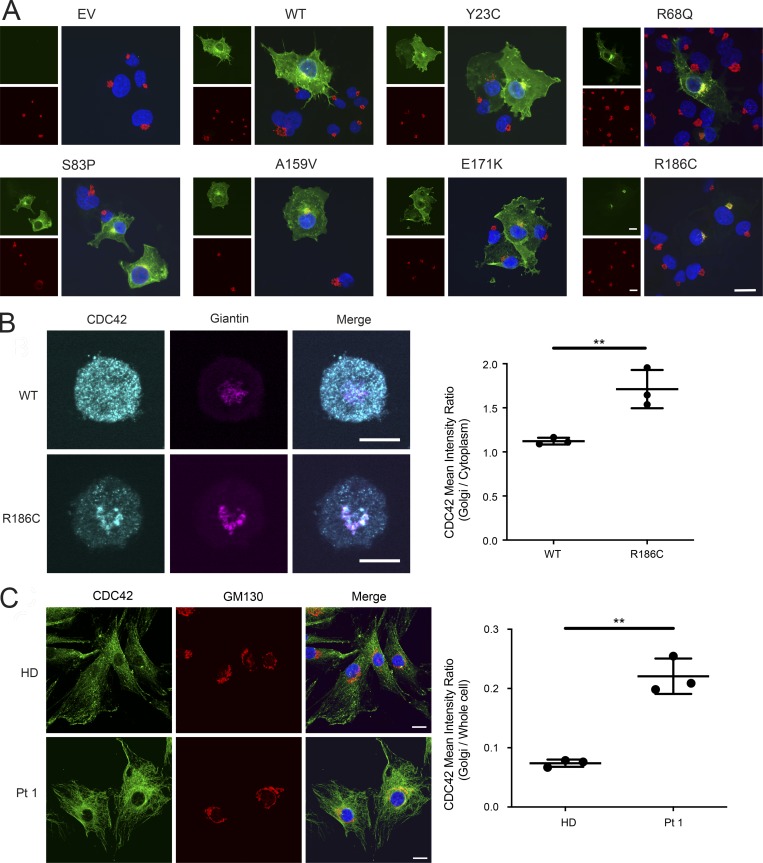
GTP-bound CDC42 activates multiple downstream signaling pathways by interacting with a variety of effectors (Dvorsky and Ahmadian, 2004; Nouri et al., 2017). We assessed the capability of the CDC42R186C mutant to transduce signals by measuring its direct binding to major effectors, including WASP, PAK1, and IQGAP1. Glutathione S-transferase (GST) pull-down assays coupled to immunoblotting demonstrated a decreased effector binding of the mutant protein compared with WT CDC42, with a significantly diminished binding to WASP and a dramatically reduced interaction with IQGAP1 (Fig. 2 D). Given the established role of RhoGDI and IQGAP1 in the control of the dynamic redistribution of CDC42 within cells, we analyzed the possible disruption of the subcellular localization of the CDC42R186C protein by using complementary cellular models. Depending on the cellular context and stimuli, CDC42 localizes to the cytoplasmic leaflet of multiple compartments, vesicles, and plasma membrane and partly the cytoplasm (Valdés-Mora and Lacal, 2011). Surprisingly, a constitutive cis/medial-Golgi–restricted localization of CDC42R186C was observed in the totality of COS-1 cells ectopically expressing the mutant protein (Fig. 3 A). On the other hand, a similar Golgi-specific targeting was not observed for a representative panel of previously identified disease-associated CDC42 mutants (e.g., p.Y23C, p.R68Q, p.S83P, p.A159V, and p.E171K; Martinelli et al., 2018), supporting the unique consequence of the p.R186C change on CDC42 subcellular distribution. This distinctive localization was observed in a disease-relevant cellular model, the YTS NK cell line, generated by CRISPR/Cas9 technology to express the mutant allele in the homozygous state (Fig. 3 B). Consistent with these findings, an enriched Golgi localization was also observed in the heterozygous primary fibroblasts obtained from Pt 1 (Fig. 3 C).
Figure 3.
p.R186C leads to aberrant subcellular localization of CDC42. (A) CDC42R186C shows a Golgi apparatus–restricted localization. Immunofluorescence staining of FLAG-tagged CDC42 proteins (green) and GM130 (red), a marker of cis/medial-Golgi apparatus, in COS1 cells transiently transfected with mock DNA (empty vector [EV]), WT CDC42, or mutant alleles carrying different disease-causing mutations (Y23C, R68Q, S83P, A159V, E171K, and R186C). Composite colocalization images are shown in the right panels with nuclei in blue. At least 200 FLAG-tagged CDC42-expressing cells were analyzed for each sample. Representative image of three independent repeats. Scale bar, 20 µm (applicable to all other images shown). (B) Subcellular localization of CDC42 in YTS NK cell lines homozygous for the p.R186C mutation. CDC42R186C was found to be predominantly localized to the Golgi apparatus with reduced cytoplasmic signal. CDC42 signal intensity within the Golgi apparatus was quantified using giantin as a Golgi-specific marker relative to the cytoplasm and presented as a ratio. Representative images of three independent repeats are shown. The graph shows the mean + SD of three independent experiments with n = 38 cells for WT and n = 35 cells for R186C; **, P ≤ 0.01, unpaired two-tailed t test. Scale bar, 10 µm (applicable to all images shown). (C) Immunofluorescence staining of CDC42 (green), GM130 (red), and nuclei (blue) in primary fibroblasts expressing WT CDC42 (HD) or heterozygous for p.R186C (Pt 1). Visual composite colocalization images are shown in the merge panels. Colocalization (orange/yellow overlay) of CDC42 and cis-Golgi is detected in mutant CDC42–expressing cells. Images refer to representative pictures of three independent experiments. Using GM130 as a mask for cis/medial-Golgi, CDC42 fluorescence intensity was quantified as the ratio of Golgi to whole-cell staining using ImageJ software (mean ± SD, n = 3; **, P < 0.01, unpaired two-tailed t test). Scale bar, 20 µm (applicable to all images shown).
We investigated the consequences of altered CDC42 localization and function on major cellular processes controlled by CDC42, including proliferation, polarized migration, and cytoskeletal rearrangement. In vitro proliferation of CD34+ cells from Pt 1 assessed in response to KIT ligand (KITLG) or a growth factor mixture (KITLG, FLT3L, thrombopoietin, and IL3) revealed a specific unresponsiveness to KITLG, suggesting a selective hematopoietic proliferation signaling defect (Fig. 4 A). Defective proliferation was also observed in fibroblasts derived from Pt 1 and Pt 2, and NIH-3T3 cells transiently transfected to overexpress the CDC42 mutant (Fig. 4 B). Notably, Pt 1 fibroblasts and NIH-3T3 cells overexpressing the mutant protein also showed reduced polarized migration (Fig. 4 C). Consistently, defective chemotaxis toward C-X-C motif chemokine 12 (CXCL12) was observed in Pt 1 purified bone marrow (BM) CD34+ cells, peripheral blood mononuclear cells (PBMCs), as well as in the homozygous CDC42R186C YTS NK cell line (Fig. 4 C). CDC42 is critical for cytoskeletal rearrangement primarily through the formation of filopodia, an F-actin–based structure contributing to cell adhesion, polarization, and migration (Etienne-Manneville, 2004; Mattila and Lappalainen, 2008; Jacquemet et al., 2015). Thus, imaging of filopodial pattern and cell polarization was performed in serum-stimulated fibroblasts from Pt 1. A higher number of cells with filopodia and an aberrantly polarized morphology compared with control cells was observed. In particular, aberrant polarization consisted of multiple lateral F-actin flat protruding edges (multipolar cells), in contrast with the front-rear canonical polarization (Fig. 4 D). Interestingly, in a cytolytic immunological synapse model on glass surface utilizing higher resolution imaging, a lower number of filopodia was detected in the mutant YTS cells, with a higher proportion of these cells retracting when activated on a CD18/28 surface (Fig. 4 E). Together, these data documented a variable perturbation of diverse cell-specific processes mediated by remodeling of the actin architecture, including cell polarity and migration.
Figure 4.
p.R186C is associated with defects in proliferation, migration, and formation of actin-based structures. (A) Proliferation of BM CD34+ cells from Pt 1 in response to stem cell factor (SCF/KITLG) or a cytokine mixture (MIX; n = 1). (B) Proliferation of primary fibroblasts from Pt 1 (n = 3) and Pt 2 (n = 3) at indicated time points of culture, and NIH-3T3 cells transiently expressing WT or mutant CDC42 or an empty vector (EV). (C) Migration assays of primary fibroblasts from Pt 1 (n = 3), transfected NIH-3T3 cells (n = 3; *, P < 0.05; **, P < 0.01; ****, P < 0.0001, two-way ANOVA with Sidak’s multiple comparisons), purified BM CD34+ sorted cells (n = 1), PBMCs (n = 2), and a YTS CRISPR/Cas9-modified cell line (n = 3; ****, P < 0.0001, unpaired two-tailed t test; mean ± SEM for CD34+/PBMCs and mean ± SD for NK cells, NIH-3T3 cell line, and primary fibroblasts). Migration was assayed using a wound-healing assay on primary fibroblasts and transfected NIH-3T3 cells and directed migration toward chemoattractant CXCL12 in BM CD34+ cells, PBMCs, and YTS NK cells. Decreased directed migration of all tested cell types was observed. FI, fold increase of migratory cells. (D) Cytoskeletal rearrangements of cells expressing CDC42R186C. Multipolarization and filopodia in primary fibroblasts from Pt 1 (n = 3) compared with fibroblasts from an HD. Immunofluorescence staining of CDC42 (red), F-actin (green), and nuclei (blue) in cells stimulated 3 h with 20% serum. In HD fibroblasts, CDC42 localizes to the front leading edge, whereas in Pt 1 fibroblasts, the protein mostly localizes to the perinuclear area. On the right of the panel, quantification of multipolar cells and number of cells with short and long filopodia length are shown. Cells bearing multiple F-actin flat protruding edges (butterfly shaped) were considered multipolar. While HD fibroblasts polarize showing a front (leading edge), a large proportion of Pt 1 fibroblasts show multiple protruding edges. Filopodia twofold longer than nuclei diameter were considered long filopodia. Filopodial length and number were notably increased in fibroblasts, suggesting a disruption of CDC42-dependent actin architecture. Scale bar, 20 µm (applicable to all images shown). (E) Filopodia dynamics of the YTS cells on an activating CD18/CD28 surface. Filopodia were imaged using SIM-TIRF microscopy with representative images showing decreased filopodia count (mean ± SD, n = 3; **, P < 0.01, unpaired two-tailed t test with Welch’s correction) in cells expressing CDC42R186C. Scale bar, 10 µm (applicable to all images shown). n.s., not significant.
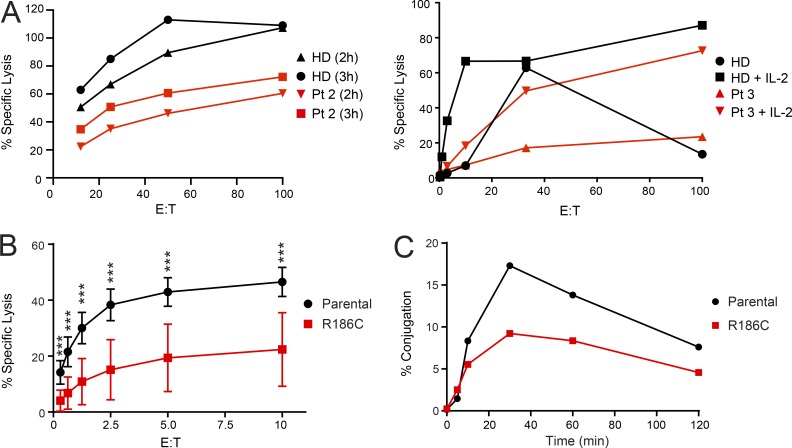
HLH occurring in infants and young children is generally associated with defects in immune regulation, including function of NK cells (Henter et al., 2007). We observed a slightly to markedly reduced NK cell activity in Pt 2 and Pt 3 through clinical assessments of immune cell function (Fig. 5 A). The homozygous CDC42R186C YTS cell line was used as an immune cell model to directly study the impact of the mutant on NK cytotoxicity. Chromium release assay against MHC class I–negative 721.221 target cells demonstrated reduced cytotoxic function in the mutant cell line through a cell-intrinsic defect (Fig. 5 B). The reduction in cytotoxic function was in part due to a reduction in the ability to form conjugates with target cells (Fig. 5 C) and impaired migration (Fig. 4 C). Consistent with the observed altered filopodia structure and number, these findings suggest that immune dysfunction in the patients is likely linked to actin dysregulation (Sinai et al., 2010).
Figure 5.
p.R186C is associated with impaired NK cell cytotoxic function. (A) Assessment of NK cell cytotoxicity in Pt 2 and 3. Using NK cell cytotoxicity assays, Pt 2 at diagnosis had a decreased cytotoxic function compared with an HD at the shown E/T ratios at time points 2 h and 3 h. For Pt 3 at diagnosis, NK cells were stimulated with IL-2, and cytotoxicity was also decreased. (B) Functional characterization of YTS NK cell model. Standard Cr-51 release assay of YTS NK cell lines against 721.221 target cells. The mutant cell line showed a significant decrease in cytotoxicity (pooled mean ± SD, n = 3 independent repeats each with triplicates; ***, P < 0.001, Mann–Whitney U test). (C) Co-culture conjugation assay of YTS NK cell lines against 721.221 target cells. Compared with parental YTS NK cells, a reduced ability of the YTS NK cells expressing the mutant allele to form conjugates with 721.221 target cells was also observed. A representative figure is shown (n = 4).
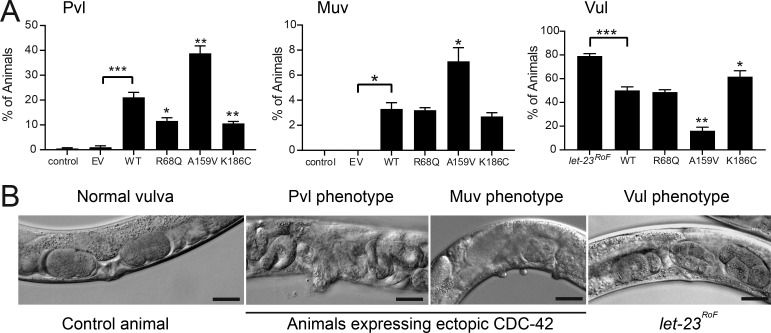
The unique behavior of the CDC42R186C mutant was also validated in vivo. In Caenorhabditis elegans, CDC-42 controls a number of developmental programs, including vulval development (Reiner and Lundquist, 2018). Our previous studies documented that CDC42 mutations differentially perturb vulval induction and morphogenesis, with some amino acid changes up-regulating multiple signaling pathways, including those mediated by LET-60/rat sarcoma (RAS), and others behaving as hypomorphic mutations on WSP-1/WASP-dependent signaling (Martinelli et al., 2018). In particular, overexpression of WT cdc-42 at early/mid larval stage 3 (L3) was shown to induce the formation of a single protrusion at the site of the vulva (protruding vulva [Pvl] phenotype) in a WSP-1–dependent manner. Ectopic expression of the WT protein also elicited a low penetrant multivulva (Muv) phenotype and partially rescued the vulvaless (Vul) phenotype of nematodes carrying a hypomorphic let-23/EGFR allele, demonstrating hyperactivation of signal flow through LET-60 and the MAPK cascade. Compared with WT CDC-42, the K186C (homologue of R186C) mutant induced a less penetrant Pvl phenotype and less efficiently rescued the Vul phenotype, indicating a hypomorphic effect of this mutation on both LET-60/RAS and WSP-1/WASP signaling pathways (Fig. 6, A and B; and Table S4).
Figure 6.
p.R186C affects C. elegans vulval development. (A) Hypomorphic effect of the mutation on pathways controlling C. elegans vulval development. Compared with WT CDC-42, the K186C mutant induces a less penetrant Pvl phenotype and less efficiently rescues the Vul phenotype of animals carrying a hypomorphic let-23/EGFR allele, indicating a hypomorphic behavior on multiple signaling pathways. The R68Q and A159V mutants, representative of group I (substitutions characterized by impaired binding to regulators and effectors) and group II (gain-of-function changes) mutations, respectively, are shown for comparison. Error bars indicate SEM of three independent experiments. Asterisks specify significant differences between animals expressing WT CDC-42 and those expressing the empty vector (EV) or the let-23(sy1) allele (*, P < 0.05; ***, P < 1.2e-6; two-tailed Fisher’s exact test), and between animals expressing WT and mutant CDC-42 (*, P < 0.05 [Muv] or P < 0.005 [Pvl and Vul]; **, P < 0.00002 [Pvl] or P < 3.2e-6 [Vul]). Number of animals are reported in Table S4. (B) Representative images of C. elegans phenotypes. Scale bars, 20 µm.
Collectively, the in vitro and in vivo biochemical and functional data consistently indicate a unique impact of the identified disease-causing p.R186C change on CDC42 subcellular localization and function, which likely underlies the distinctiveness of the trait.
Clinical profiling of NOCARH syndrome
The four unrelated patients sharing the same de novo missense CDC42 mutation (p.R186C) showed a similar multisystem inflammatory disease characterized by pancytopenia, fever, skin rash, hepatosplenomegaly, and persistently elevated inflammatory markers. Differently from what had previously been reported for other CDC42 mutations (Martinelli et al., 2018), no neurodevelopmental involvement was observed. In these patients, Wiskott–Aldrich syndrome, idiopathic myelofibrosis, dyskeratosis congenita, and classical immunodeficiency were rapidly excluded based on established clinical, biochemical, and genetic diagnostic criteria. Treatment with IL-1 inhibitors partially improved fever and rash, and high chronic doses of glucocorticoids were required to treat inflammation. Fatal HLH developed in three cases.
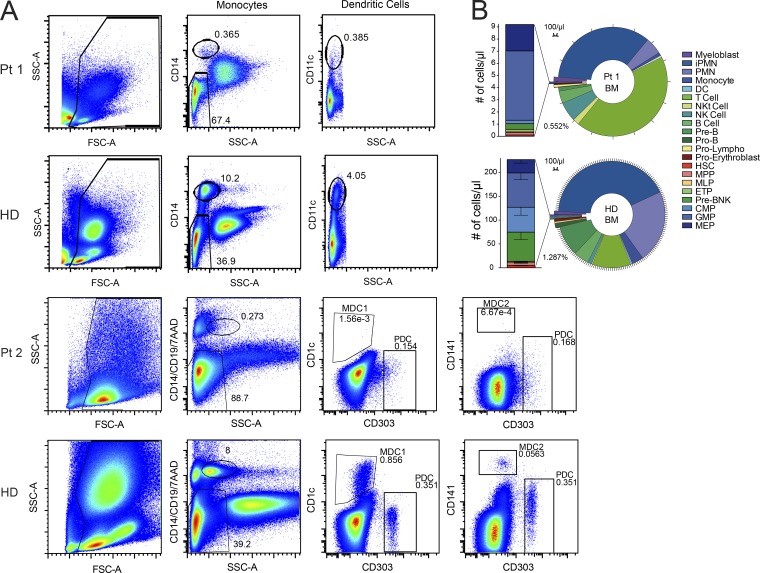
Trilineage dyshematopoiesis was observed in BM with prevalence of early differentiation elements, and decreased clonogenic progenitor content having altered clonogenicity was documented (Fig. 7 and Table S5). This indicates that the p.R186C substitution in CDC42 affects hematopoiesis, ultimately altering BM composition.
Figure 7.
p.R186C leads to disruption of certain hematopoietic compartments. (A) Flow cytometric analysis of monocyte and dendritic cell (DC) immunophenotype on whole lysed peripheral blood of Pt 1 (n = 1) and Pt 2 (n = 1) showing a severe reduction of monocytes and myeloid dendritic cells. Upper plots (Pt 1 and Pt 2) from left to right: after elimination of debris, we gated on CD14 or CD14, CD19, 7AAD SSClow cells to identify monocytes. In the CD14− SSClow population, we gated on CD11c+ cells for dendritic cell identification in Pt 1. In the CD14−CD19−7AAD− SSClow population, we gated on CD1c, CD141, or CD303 to distinct the dendritic cell major subtypes (MDC1, type 1 myeloid dendritic cell; MDC2, type 2 myeloid dendritic cell; PDC, plasmacytoid dendritic cell) in Pt 2. Numbers within the plots show the frequency of monocytes and dendritic cells in the total events displayed. SSC-A, SSC-area; FSC-A, FSC-area. (B) The ring chart shows absolute counts of distinct hematopoietic subpopulations on total CD45+ cells (indicated in the legend) in BM of pediatric HDs (n = 6) and Pt 1. The stacked bar graph on the left is a zoom on the absolute count of hematopoietic stem and progenitor cell subtypes within the LIN−CD34+ compartment. PreB/NK, B and NK cell progenitor; CMP, common myeloid progenitor; ETP, early T progenitor; GMP, granulocyte/monocyte progenitor; iPMN, immature polymorphonucleated cell; MEP, megakaryocyte/erythrocyte progenitor; MLP multi-lymphoid progenitor; MPP, multi-potent progenitor; NKt, NK T cell; PMN, polymorphonucleated cell.
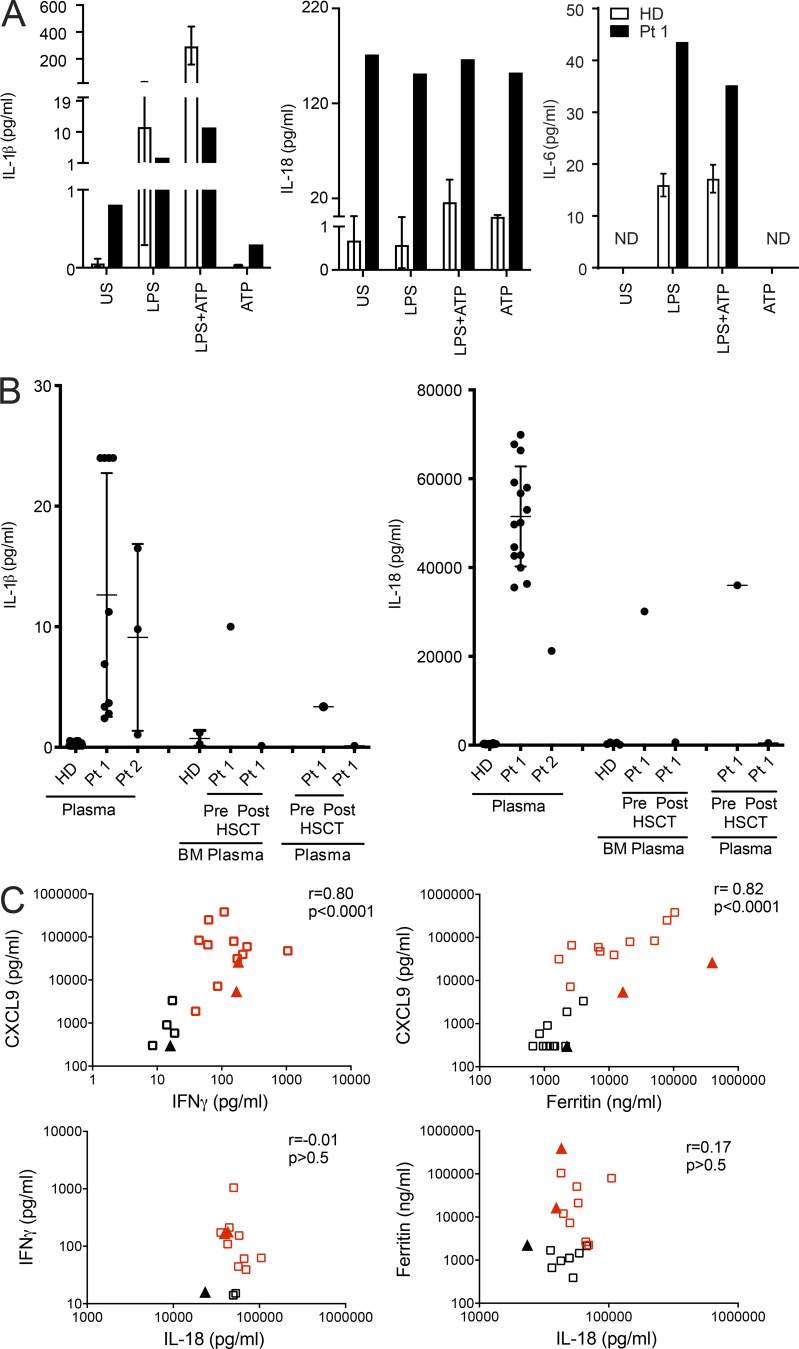
Since typical autoinflammatory features were present, we investigated IL-1β production. Increased IL-1β secretion was measured via ex vivo spontaneous IL-1β release from unstimulated BM mononuclear cells as well as high levels of IL-1β in BM supernatants and plasma (Fig. 8, A and B). This finding demonstrates that IL-1β overproduction is likely responsible for part of the phenotype, particularly the observed features typically associated with IL-1β–driven inflammasopathies (i.e., fever, rash and elevated acute phase proteins; Harapas et al., 2018). Remarkably, patients showed strikingly increased production of IL-18. This was demonstrated by a 300-fold higher ex vivo spontaneous IL-18 release from unstimulated BM mononuclear cells and by levels of IL-18 in BM supernatant and plasma that were ∼1,000-fold higher than those of healthy donors (HDs; Fig. 8, A and B). Stimulation of BM mononuclear cells with LPS or LPS plus ATP did not increase further IL-18 production as compared with unstimulated cells, suggesting the presence of constitutive maximal activation of IL-18 processing and release in the patient’s marrow. In contrast, no ex vivo spontaneous IL-6 release from unstimulated BM mononuclear cells was observed (Fig. 8 A), and circulating IL-6 levels in Pt 1 and Pt 2 were comparable to those observed in healthy subjects (data not shown). Importantly, IL-18 is a potent costimulator of IFN-γ production (Okamura et al., 1995), and IFN-γ appears to be a common mediator of all forms of HLH (Grom et al., 2016). Consistently, IFN-γ levels were increased in patients, particularly during HLH episodes. In multiple samples available from Pt 1, we observed that elevated IFN-γ levels were strictly correlated (r = 0.82) with elevated levels of CXCL9 (Fig. 8 C), a chemokine specifically induced by IFN-γ that has been proposed as biomarker of HLH (Bracaglia et al., 2017). CXCL9 levels were strictly correlated with levels of ferritin (r = 0.82), which is the most typical marker of HLH activity. In line with the finding that IL-18 alone is not sufficient to induce IFN-γ, we did not find a significant correlation between IL-18 and IFN-γ or ferritin levels (Fig. 8 C). Accordingly, in Pt 2, high IFN-γ levels were also associated with high CXCL9 and ferritin levels (Table S1). In addition, serum IL-6 levels were measured, and persistent elevation was not detected (Fig. 8 A). Notably, in Pt 1, the last severe HLH episode not responding to repeated glucocorticoid pulses, cyclosporine A, and high-dose anakinra (10 mg/kg/d) was successfully treated with emapalumab, a monoclonal antibody targeting IFN-γ. Altogether, these data show that the R186C change in CDC42 leads eventually to activation of the IFN-γ pathway through overproduction of IL-18, which has been demonstrated to be associated with secondary HLH development in the context of autoinflammatory diseases (Canna et al., 2014; Weiss et al., 2018).
Figure 8.
p.R186C is associated with high production of inflammasome-related cytokines IL-1β and IL-18 in vivo and ex vivo. (A) HD (n = 3) and Pt 1 BM mononuclear cells were left unstimulated (US) or stimulated with LPS (10 µg/ml) for 5 h, with or without ATP (1 mM) for an additional 1 h. Compared with unstimulated HD cells, unstimulated Pt 1 cells release high levels of IL-1β and IL-18. In LPS-primed HD cells, ATP addition caused a marked increase in IL-1β and IL-18 release, while this is not observed in LPS-primed Pt 1 cells. IL-6 levels released in supernatants were also measured; no spontaneous release was observed. ND, not detectable. Secreted cytokines were measured by ELISA, and results obtained were normalized by the absolute number of monocytes per milliliter BM. (B) IL-1β and IL-18 levels were measured in plasma samples collected from Pt 1 and Pt 2 at different time points over the course of the disease. IL-1β and IL-18 levels were also measured in BM, and plasma samples were obtained from Pt 1 30 d before and after HSCT. For HDs, plasma n = 24 and BM n = 5. For Pt 1 and Pt 2, n = 1. Mean ± SD is indicated. (C) Plasma levels of IFN-γ, CXCL9, and IL-18 were measured in Pt 1 (squares) and Pt 2 (triangles) and correlated with ferritin levels, a typical marker of HLH activity. IFN-γ and CXCL9 levels, but not IL-18 levels, are significantly correlated with ferritin levels. Red squares and triangles indicate samples collected when Pt 1 and Pt 2 fulfilled at least five criteria required for HLH diagnosis.
The surviving patient (Pt 1) underwent allogeneic HSCT from his HLA-haploidentical father after selective depletion of αβ T cells and CD19+ cells from the graft for preventing occurrence of graft-versus-host disease and Epstein-barr virus–related posttransplant lymphoproliferative disease, respectively (Locatelli et al., 2017). The conditioning regimen used treosulfan, thiotepa, and fludarabine. To avoid HLH flares, the patient was given emapalumab and anakinra until days +28 and +100 after HSCT, respectively. Neutrophil and platelet recovery occurred on days +18 and +12 after HSCT; successful engraftment of donor hematopoiesis was associated with normalization of IL-1β and IL-18 production (Fig. 8 B). No inflammatory or HLH flare has been observed up to day +210.
Discussion
Here, we characterize a previously unrecognized and distinctive hematological/autoinflammatory disorder due to a specific missense mutation of CDC42 (p.R186C). By using complementary biochemical and functional analyses, we provide evidence that the disease-causing amino acid substitution has unique consequences on CDC42 function, leading to a perturbation of hematopoiesis, immune function, and inflammatory response. We also provide a first characterization of the clinical profile of the associated phenotype and propose the acronym of NOCARH syndrome to highlight the major features of the condition, including neonatal onset of cytopenia, autoinflammation, rash, and episodes of HLH.
CDC42 codes for a small GTPase of the Rho family controlling multiple signaling pathways regulating cell polarity and migration, endocytosis, and cell cycle progression by cycling between an active (GTP-bound) and an inactive (GDP-bound) state (Etienne-Manneville and Hall, 2002; Etienne-Manneville, 2004). Recently, a number of missense mutations in this gene have been associated with developmental traits characterized by an unusually broad spectrum of anomalies, with core clinical features including defective growth, intellectual disability, brain malformations, facial dysmorphism, hearing/vision problems, cardiac malformations, and immune and hematologic and lymphatic defects (Takenouchi et al., 2015; Martinelli et al., 2018). Genotype–phenotype correlations were identified and linked to a variable functional impact of mutations, suggesting specific consequences of individual mutations on cellular processes controlling development. In contrast to those phenotypes, NOCARH syndrome is a hematological and autoinflammatory condition with no major involvement of other systems, which is in line with the documented peculiar functional consequences of the p.R186C change on protein function.
Our immunophenotype and clonogenic data are consistent with previous findings showing that controlled inactivation of CDC42 in BM cells results in altered frequency and distribution of hematopoietic stem cells, decreased abundance of long-term hematopoietic stem cells, and occurrence of myeloid and erythroid developmental defects, as well as functional defects in engraftment, migration, and BM retention (Yang et al., 2007a,b). Notably, present findings confirm the key role of the GTPase in controlling processes implicated in hematopoietic stem and progenitor cell fate and behavior.
Based on structural modeling and biochemical and cellular testing, we predicted and confirmed the mutated protein to have impaired interaction with known regulators and effectors, including RhoGDI, IQGAP1, and WASP, leading to aberrant subcellular localization, actin cytoskeleton rearrangement, and reduced migration. CDC42 mislocalization data are consistent with the physiological role played by RhoGDI and IQGAP1, key mediators of membrane cycling of CDC42 (Gibson and Wilson-Delfosse, 2001; Swart-Mataraza et al., 2002; Gibson et al., 2004). The observation that cells expressing the p.R186C variant undergo deep cytoskeletal rearrangements leading to multiple leading edges is congruous to findings by Fukata et al. (2002) showing that cells expressing IQGAP1 mutants defective in CDC42 binding display aberrant multipolar morphology. This supports the notion that the IQGAP1–CDC42 interaction is crucial to cell polarization and migration. Notably, the finding that CDC42 aberrant localization and trafficking is coupled to altered polarization and migration is in line with the relevant role played by CDC42 at the Golgi and the leading edge in the control of cell polarity (Baschieri et al., 2014). Compositional and functional assessment revealed impaired proliferation and polarized migration, further emphasizing the requirement of proper CDC42 function in these processes (Martinelli et al., 2018). Notably, although we were unable to identify a specific CDC42 functional perturbation associated with the p.R186C change as causally linked to the complex phenotype of NOCARH syndrome, this mutant seems unique in its mislocalization, which suggests this is a key factor in the pathogenesis for this uniquely presenting disease.
Our findings indicate that aberrant CDC42 function can involve components of hyperinflammation and immune impairment in NK cells, contributing to the HLH phenotype observed in NOCARH. The underlying pathogenetic mechanism of the p.R186C mutation in the context of anemia and thrombocytopenia has yet to be determined, suggesting a context-dependent effect on specific blood cell compartments. Future studies involving murine models carrying the heterozygous mutation would be needed to assess the effect on hematopoiesis in a similar context to that of our patients.
Several mutations in genes with a role in actin cytoskeleton remodeling of immune cells have been demonstrated to be involved in the pathogenesis of hematological/autoinflammatory diseases, underlining the importance of the actin cytoskeleton in modulating inflammatory responses. Indeed, mutations in the actin-binding protein cofilin (CFL1; Seeland et al., 2018), WAS (Li et al., 2017), DOCK8 (Dasouki et al., 2011), and ARPC1B (Brigida et al., 2018), as well as in RAC2, a Rho GTPase structurally and functionally related to CDC42 (Caye et al., 2015; Hsu et al., 2019), cause abnormal migration, proliferation, and/or differentiation of lymphoid and/or myeloid cells and are associated with features of autoinflammation. Moreover, aberrant actin depolymerization due to an inactivating mutation of the actin-depolymerizing cofactor Wdr1 has been demonstrated to cause in mice an autoinflammatory disease characterized by spontaneous autoinflammation, thrombocytopenia, and neutrophilia (Kim et al., 2015). In these mice, the autoinflammatory phenotype is IL-18, but not IL-1β, dependent and is driven by activation of the pyrin inflammasome. In our study, we could not investigate the exact molecular mechanism involved in the overproduction of IL-18 and the role of CDC42 in inflammasome regulation. However, the spontaneous release of IL-18 by BM mononuclear cells, associated with high IL-18 circulating levels, has been exclusively described in patients carrying mutations in the NLRC4 protein (Canna et al., 2014) who present with recurrent episodes of HLH. Moreover, it is well known that inactivation of RhoA GTPases by many bacterial toxins is sensed by the inflammasome (Jamilloux et al., 2018). Interestingly, IQGAP1 has also been described as a novel regulator of caspase-1 in macrophages infected with Yersinia pestis (Chung et al., 2014). Based on the above-mentioned observations regarding the connection between the actin cytoskeleton and inflammation, we propose the term “inflammatory actinopathies” to group these diseases, including NOCARH, differently from actinopathies with neurological and myologic features.
In conclusion, NOCARH is an autoinflammatory disease at high risk for HLH, similarly to what has been observed for the disorder caused by NLRC4 gain-of-function mutations (Canna et al., 2014; Romberg et al., 2014). Given the observed autoinflammation associated with markedly elevated IL-18 levels, the mechanism is most likely associated with the role of IL-18 as a costimulus for IFN-γ production. The characteristic inflammation in two patients in conjunction with elevated IL-18 levels support IL-18 as a disease biomarker for diagnosing NOCARH and potentially related syndromes of dysregulated CDC42 function that involve inflammation. One patient survived severe HLH, and this appears to have been dependent on timely therapeutic neutralization of IL-1 and IFN-γ. Early recognition and establishment of treatment for NOCARH, based on the unique association of early-onset trilinear cytopenia and autoinflammation, are crucial to prevent multiorgan failure and subsequent death.
Materials and methods
Detailed clinical history of patients
Pt 1
Pt 1 is a 6-yr-old Caucasian girl born from healthy unrelated parents with no family history of genetic disease. At birth, she presented with persistent high fever; erythematous skin lesions of the face, trunk, and limbs; a large abdomen because of hepatosplenomegaly (spleen diameter, 6.4 cm); and suspected trigonocephaly (Table 1). Laboratory tests showed an increase in inflammatory markers and a severe trilinear cytopenia requiring frequent red blood cell and platelet transfusions during the first year of life. BM biopsy at disease onset revealed fibrosis with trilineage dyshematopoiesis (prevalence of early differentiation elements in erythroid and myeloid lineage, dysmorphic and reduced megakaryocytes, increased lymphocytes, and abundant CD68+ histiocytes with focal or absent phagocytosis; Tables S1 and S2). Given the partial clinical overlap with chronic infantile neurological cutaneous articular syndrome–neonatal-onset multisystem inflammatory disease (CINCA-NOMID), she was started on glucocorticoids and daily therapy with anakinra, with improvement of fever and rash, but no effect on cytopenia. Failure to thrive was present. Treatment with G-CSF was started with partial response. Recurrence of inflammatory symptoms following tapering and/or discontinuation of glucocorticoids was observed. At age of 11 mo, she presented with recurrent episodes of intestinal bleeding, associated with persistent/chronic diarrhea. Meckel’s diverticulum was excluded. At 2 yr and 6 mo, she presented with three episodes of generalized seizures followed by hypotonia, hypersomnia, and transient hemiparesis requiring in one case admission to an intensive care unit. Magnetic resonance imaging (MRI) performed during the first episode showed lesions suggestive of inflammatory central nervous system involvement; during the third episode, the MRI showed one additional lesion of the brainstem. CSF was sterile, CSF white blood cells were 18/mmc, and no hemophagocytic cells were detected. All the episodes were treated with a high dose of i.v. glucocorticoid with good response; therapy with levetiracetam was started. At the age of 5 yr, because of incomplete control of the inflammatory state, anakinra was switched to canakinumab. Starting from 5 yr of age, she developed four episodes of full-blown HLH, fulfilling the HLH-2004 diagnostic criteria. All episodes were treated with high dose of i.v. glucocorticoids and cyclosporine-A with resolution in all cases except one. The last episode was severe and associated with massive intestinal ischemia and necrosis requiring surgical resection and consequent ileocolostomy. This episode was unresponsive to high-dose glucocorticoids and IL-1 inhibition. Treatment with emapalumab, an anti–IFN-γ antibody, was added with rapid resolution of the episode. At age of 6 yr, the patient underwent allogeneic HSCT from her HLA-haploidentical father after a selective depletion of αβ T cells and CD19+ cells from the graft. The conditioning regimen consisted of a combination of treosulfan, thiotepa, and fludarabine. To avoid flares of HLH, the patient was given emapalumab and anakinra until days +28 and +100 after the allograft, respectively. Neutrophil and platelet recovery occurred on days +18 and +12 after HSCT. Normalization of the acute phase response and no inflammatory or HLH flare have been observed up to day +210.
Pt 2
Pt 2 was a male born from Caucasian healthy unrelated parents. Since the first day of life, he presented with persistent fever, skin rash, mild hepatosplenomegaly, failure to thrive, increase in inflammatory markers, and trilinear pancytopenia that required red blood cell and platelet transfusions. Extensive microbiological screening revealed a positive IgM titer for parvovirus B19, with B19 parvovirus PCR negative on whole blood. At age 57 d, he was first seen at one of our centers transferred from another hospital. He presented with failure to thrive (height and weight below the fifth percentile), persistent fever, skin rash, mild hepatosplenomegaly, and chronic diarrhea and positive blood stool. Inflammatory bowel disease was suspected, and temporary parenteral nutrition started. Laboratory tests showed elevated acute-phase reactants and severe trilinear cytopenia requiring recurrent red blood cells and platelet transfusions. BM biopsy revealed dyshematopoiesis and some lymphohistiocytic aggregates without significant hemophagocytosis. The disease course was characterized by a persistent inflammatory state despite several treatment attempts with glucocorticoids, antibiotics, high doses of i.v. immunoglobulins, and cyclosporine-A. Suspecting an autoinflammatory condition, treatment with anakinra was also started with only partial improvement of the clinical and laboratory parameters. At age 7 mo, he developed severe HLH with multiorgan failure that rapidly progressed to death. Extensive immunological workup showed severe monocytopenia and reduction of plasmacytoid, myeloid dendritic cells, neutropenia, increased central memory CD4+ cells with reduced naive and recent thymic emigrant CD31+ cells, increased regulatory T cells with high memory phenotype, and increased switched B cell memory cells, plasma cells, and autoreactive B cells associated with reduced transitional B cells (Table S2).
Pt 3
Pt 3 was a female born from nonconsanguineous Caucasian parents with no family history of inherited diseases. At birth, she presented with generalized rash, intermittent cytopenia (leukopenia, thrombocytopenia, and anemia requiring transfusions), and generalized lymphadenopathy. Hepatosplenomegaly, nutritional problems, and stagnant growth were also noted within the first few months after birth. The anemia was profound and required several red blood cell transfusions. At age 3 mo, the condition was thought to be CINCA-NOMID, and treatment with anakinra was started, which was temporarily effective for rash and overall inflammation, but not cytopenia. Treatment was stopped due to respiratory infections and no clear effect of the treatment. 6 mo later, she started treatment per modified HLH-2004 protocol with steroids and weekly etoposide-phosphate over 8 wk. No neurological deficits were noted, but the patient declined over time, requiring increasing erythrocyte and thrombocyte transfusions. She was then transferred to another hospital, and for the first few weeks, she was stable, but she developed signs of an HLH relapse with increased hepatosplenomegaly, high ferritin, high triglycerides, bicytopenia, and a florid rash. Histologically, hemophagocytosis was not detected. She also developed breathing difficulties of unknown origin. She was commenced on a modified HLH-2004 protocol with the addition of antithymocyte globulin, infliximab, and anakinra. There was some clinical response on this treatment and a decision to opt for a stem cell transplant was made. While she was being investigated and prepared for transplant, she continuously deteriorated; she was treated with two courses of alemtuzumab in this time period. She received a CD3/CD19-depleted haploidentical transplant and died 4 d later.
Pt 4
Pt 4 was a boy born in week 34 to nonconsanguineous parents from a Christian minority originating from the Middle East with a history of thalassemia minor and familial cardiomyopathy. The patient was small for gestational age and presented at birth with bloody diarrhea, cholestasis, ascites, and a generalized rash. He had an unusually small thymus for his age. The patient also rapidly developed additional symptoms, including stiff/contracted joints, high fever, highly elevated inflammatory markers, intermittent cytopenia (thrombocytopenia and anemia requiring transfusions), hepatomegaly, and diatheses (bleeding from the mucosas; Table 1). At 2 wk of age, an extensive workup was initiated. Investigations excluded known gut, liver, and metabolic diseases. Immunological investigations slightly lowered number of T cells with prevalence of naive T cells. Results of functional tests of NK/T cells and degranulation ability were normal. Serum IL-6 and IL-18 was remarkably high on repeated occasions. An autoinflammatory condition in combination with a primary immunodeficiency was suspected. The patient was treated with high dose glucocorticoids and anakinra. Because of insufficient response tocilizumab was also added. This intensive antiinflammatory treatment was effective for some of the rash, ascites, cholestasis, as well as intermittently for overall inflammation and cytopenia. The patient needed supportive treatment in the intensive care unit on six separate occasions, four of which were due to macrophage activation syndrome–HLH episodes, likely triggered by infection.
Severe gastrointestinal symptoms and unexplained mucosal bleedings persisted. The patient continued to have watery diarrhea. Gastroduodenal endoscopy showed severe inflammation with ongoing bleeding. Biopsies of the duodenum showed acute and chronic inflammation with ulcers. In connection with colonoscopy, the patient suffered a perforation and needed surgery. Since no matched related SCT donor was available, a search for an unrelated matched donor was initiated; however, his conditions were too unstable with no signs of remission, and HSCT was considered too risky. Because of the symptoms of agonizing pain and high levels of anxiety, and in the presence of no response to treatments, the antiinflammatory therapies were gradually stopped, and the patient was transferred to another hospital for palliative care, where he died at 4.5 mo of age.
Mutation analyses
This study was approved by the Ospedale Pediatrico Bambino Gesù and Baylor College of Medicine Ethical Committees. Clinical data and DNA specimens from the subjects included in the study were collected following procedures in accordance with the ethical standards of the declaration of Helsinki protocols and approved by the review boards of all involved institutions, with signed informed consents from the participating subjects/families (study protocol for Pt 1 and 3: “Genome Sequencing to Elucidate the Causes and Mechanisms of Mendelian Genetic Disorders” [H-29697] of Baylor College of Medicine; study protocol for Pt 1 and 2: 1702_OPBG_2018).
Pt 1
Exome capture was performed using SureSelect AllExon v5+UTR (Agilent) and sequenced on a HiSeq platform (Illumina). WES data were processed and analyzed using an in-house–implemented pipeline as previously described (Flex et al., 2016; Stray-Pedersen et al., 2017; Bauer et al., 2018). Briefly, reads were aligned to human genome build GRCh37/UCSC hg19, and variants were quality filtered according to the Genome Analysis Toolkit’s 2016 best practices, annotated, and filtered against public (dbSNP150 and gnomAD V.2.0.1) and in-house (>1,300 population-matched exomes) databases to retain private and rare (unknown frequency or minor allele frequency <0.1%) variants located in exons with any effect on the coding sequence (CDS) and within splice site regions. Functional annotation of variants was performed using SnpEff v.4.3 and dbNSFP V.2.9, and functional impact was analyzed by CADD v.1.3 and M-CAP v.1.0.(Cingolani et al., 2012; Liu et al., 2013; Kircher et al., 2014; Jagadeesh et al., 2016). Average coverage was 91× and 20× for ≥86% of the target. Among 127,405 high-quality variants, 14,224 affect either the CDS or splice sites, while 338 have low or unknown frequencies according to the aforementioned frequency thresholds. For sequencing statistics, see Table S3. Relevant variants and the de novo origin of the c.556C>T change were validated by Sanger sequencing.
Pt 2
Mutation analysis was performed by Sanger sequencing using DNA obtained from circulating leukocytes. Primer pairs are available upon request.
Pt 3
Initial mutation analysis was performed by Sanger sequencing (and Baylor College of Medicine CMA v9.1), which was negative for NLRP3, IL1RN, LPIN2, NOD2, MEFV, TNFRSF1A, PSTPIP1, MVK, and PSMB8. The patient was identified in a shared CMG database at Baylor College of Medicine by searching for rare, likely damaging variants (using the Exome Aggregation Consortium and CADD) in CDC42 in a cohort of unsolved immunodeficiency probands. On further study, patient had underwent trio-based WES in Norway, revealing the presence of the de novo c.556C>T change in CDC42, and proband-only WES was repeated at Baylor College of Medicine. Sanger sequencing of proband and parents was then performed, confirming the presence of a de novo variant (Table S3).
Pt 4
Blood samples from the patient and his parents were obtained with informed consent according to the Declaration of Helsinki. The study was approved by the Regional Ethics Review Board in Stockholm, Sweden. DNA samples were subjected to trio-based WGS with a TruSeq DNA PCR-free protocol followed by sequencing on an Illumina HiSeq X machine with an average coverage of 34×. Sequencing reads were analyzed with the pipeline SpeedSeq. In brief, reads were mapped to the human genome build GRCh37 with Burrows–Wheeler aligner, while calling of single-nucleotide and structural variants was performed, respectively, with FreeBayes and Lumpy. No mutations were identified in HLH/autoinflammation–related genes. The genomic region spanning CDC42 was visualized with the Integrative Genomics Viewer, indicating a de novo c.556C>T mutation, which was validated by Sanger sequencing (Table S3).
MS
WT CDC42 from TNAO38 insect cells were dissolved in 50% (vol/vol) acetonitrile and 0.2% (vol/vol) formic acid at a final concentration of 2 mg/ml. Proteins were subjected to a C4 HPLC column (MassPrep Online Desalting Cartridge, dimensions 2.1 × 10 mm; Waters) equilibrated with 20% (vol/vol) acetonitrile and 0.1% (vol/vol) formic acid. For HPLC separation, the following conditions were used: HPLC-system U300 series (Agilent Technology), a flow rate of 500 µl/min; eluent A, 0.1% (vol/vol) formic acid in water; eluent B, 0.1% (vol/vol) formic acid in acetonitrile; gradient conditions, 20% B for 0.5 min, linear gradient up to 60% B in 1.5 min, linear gradient up to 90% B in 0.5 min, 90% B for 0.5 min, and reequilibration of the column. The HPLC system was coupled on-line to an ion trap mass spectrometer (VelosPro; ThermoFisher Scientific) equipped with an electrospray ionization source. Full spectra were acquired using a mass-to-charge range of 700–2,000. Obtained spectra were deconvoluted using the program package Promass (ThermoFisher Scientific). Masses obtained from the respective spectra are described with respect to calculated molecular weights (CH3, methyl group; Ic, insect cells; GG, geranylgeranyl moiety).
Constructs and proteins for biochemical analysis
pGEX vectors were used for bacterial overexpression of CDC42WT, CDC42R186C, RhoGDI-1, p50GAP, ITSN1, WASP (GTPase-binding domain), PAK1 (GTPase-binding domain), and IQGAP1 (863–1,657). All proteins were isolated as GST fusion proteins in Escherichia coli BL21 (DE3) purified after cleavage of the GST tag via gel filtration (Superdex 75 or 200; Pharmacia). Nucleotide-free and fluorescent nucleotide-bound CDC42 were prepared using alkaline phosphatase (Roche) and phosphodiesterase (Sigma-Aldrich) at 4°C as described. Fluorescent nucleotide was methylanthraniloyl (mant-) GppNHp (guanosine 5′-β,γ-imidotriphosphate), a nonhydrolysable GTP analogue. All proteins were analyzed by SDS-PAGE and stored at −80°C. The GTPase assay and nucleotide exchange reaction were performed with a Hi-Tech Scientific (SF-61) stopped-flow instrument. The release of fluorescently labeled GDP (mantGDP) was measured basally and with the catalytic GEF domain of ITSN1. Hydrolysis of tamraGTP was measured basally and following stimulation with the catalytic GAP domain of ARHGAP1 (p50GAP). The excitation wavelengths were 543 nm and 362 nm for tamraGTP and mantGDP, respectively. Fluorescence experiments were performed in a Fluoromax 4 fluorimeter in polarization mode. Human WT and mutant CDC42 were subcloned into pFastBacHTB vector (Invitrogen) and fused with an N-terminal hexa-histidine (6xHis) tag and transformed into DH10BAC strain for insect cell purification.
Fluorescence assays
The GTPase assay and nucleotide exchange reaction were performed with a Hi-Tech Scientific (SF-61) stopped-flow instrument. The excitation wavelengths were 543 nm and 362 nm for tamraGTP (Jena Bioscience) and mantGDP (Jena Bioscience), respectively. For the GTPase assay, equal volumes (600 µl) of 0.2 µM CDC42-tamraGTP and 10 µM of p50GAP in 30 mM Tris/HCl, pH 7.5, 10 mM K2HPO4/KH2PO4, 5 mM MgCl2, and 3 mM dithiothreitol at 25°C were rapidly mixed and transferred to a fluorescence detection cell within seconds. For the nucleotide exchange reaction, 0.2 µM CDC42-mantGDP and 40 µM GDP + 10 µM ITSN1 were used.
Structural analysis
To elucidate the impact of substituted arginine to cysteine at residue 186, the protein–protein interaction between CDC42 and RhoGDI-1 (PDB: 1DOA) was analyzed. Residues in vicinity ≤4 Å were considered as part of binding interface of Arg186. Analysis and illustrations were made using the PyMOL molecular viewer.
SPR
A Biacore X100 instrument (GE Healthcare) was used in a single-cycle mode to analyze the kinetics of interaction between WT or R186C CDC42GG with RhoGDI. SPR experiments were performed at 25°C in HBS-P+ running buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 0.05% [vol/vol] surfactant P20; GE Healthcare). GST capturing method was used to immobilized GST antibody on the surface of CM5 chip. 4 µM GST-GDI (5 µl/min) in HBS-P buffer injects for the period of 180 s in order to immobilized GST-GDI over the chip surface. Regeneration of the chip performed using 10 mM glycine, pH 3.0, at the end of each experiment. WT and R186C CDC42GG were injected in 0.06–1 µM concentrations with twofold increase in each injection step. Final curve was fitted in a 1:1 binding model.
Cell lines and cultures
Dermal mutant fibroblasts were obtained from patients, and HEK-293T, COS-1, and NIH-3T3 cell lines and primary control dermal fibroblasts were obtained from American Type Culture Collection and grown in high-glucose DMEM supplemented with 10% FCS, 2 mM L-glutamine, and 10 U/ml penicillin/streptomycin (all from Sigma-Aldrich). YTS cells, a subclone of the YT cell line, were cultured in R10 composed of RPMI-1640 supplemented with 10% FBS (Premium; Atlanta Biologicals), 2 mM L-glutamine (ThermoFisher Scientific), 1 mM sodium pyruvate (ThermoFisher Scientific), 1% MEM nonessential amino acid (100×; ThermoFisher Scientific), 10 mM Hepes (ThermoFisher Scientific), and 1% penicillin/streptomycin (ThermoFisher Scientific).
Plasmids for CDC42 expression in HEK-293T, COS-1, and NIH-3T3 cell lines
WT human CDC42 isoform 1 (NP_001782.1) was cloned into a pcDNA3-FLAG backbone using PCR and standard cloning methods via BamHI and EcoRI digestion. Mutant constructs carrying the R186C change were generated by site-directed mutagenesis using the QuikChange XL kit (Agilent Technologies) and verified by direct sequencing. Each of the FLAG-tagged CDC42 protein expression constructs or the empty vector was transfected using Fugene 6 (Roche). 24 h after transfection, cells were assayed for cell growth or processed for wound healing assays, immunofluorescence, or immunoprecipitation analysis. Transfection efficiency was verified by Western blot (WB) analysis of the protein lysate.
Pull-down of CDC42 variants from COS-7 cell lysates
COS-7 cells were transfected with pcDNA3.1-Flag plasmids containing WT and R186C CDC42 and transiently overexpressed for 48 h. 10 µg of each plasmid was used for transfection of the COS-7 cells (900,000 cells per 10-cm Petri dish). Empty vector was used as control. 20 µl TurboFect transfection reagent (ThermoFisher Scientific) was added to the plasmids and incubated for 20 min at room temperature before adding it to the cells. Medium was changed after 4 h, and cells were incubated for 48 h. Cells were lysed in an ice-cold buffer containing 50 mM Tris/HCl, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 1% Igepal Ca-630, 10% glycerol, 20 mM β-glycerolphosphate, 1 mM Na3VO4, and one tablet EDTA-free protease inhibitor (Roche). Cell lysates were incubated with glutathione beads coupled to GST-effector proteins for 30 min at 4°C. Beads were washed five times, heated in 1× Laemmli buffer at 95°C for 10 min, and subjected to SDS-PAGE for immunoblot analysis. WB was performed using an anti-FLAG antibody (F3165, M2; Sigma-Aldrich) and anti-GAPDH antibody (21185, clone 14C10; Cell Signaling Technology).
Polarized migration
Polarized migration of primary fibroblasts and NIH-3T3 cells on fibronectin-coated wells (10 μg/ml; Sigma-Aldrich) were evaluated by wound-healing assays. Monolayers of cells were scratched with a 200-µl micropipette tip and incubated in low-serum medium (2% FBS in DMEM high glucose without Phenol red, 2 mM L-glutamine, and 1% penicillin/streptomycin; all purchased from Sigma-Aldrich) in the presence of thymidine (10 mM, T1895; Sigma-Aldrich) to inhibit cell proliferation. Images were acquired at different time points using a Nikon Eclipse TS100 microscope, a Nikon Plan Fluor 10×/0.13 objective, and a Nikon Coolpix 990 digital camera. Cells that had migrated in the wounded area were counted in four fields per well, and the fold increase of migratory cells was evaluated after scratch at the time points indicated.
Proliferation and viability
Proliferation and viability of transiently transfected NIH-3T3 cells and primary fibroblasts were quantified by manual counting using a Neubauer hemocytometer. Cell viability was detected by the exclusion of trypan blue dye (5 mg/ml in PBS, ECB4004L; Euroclone). Cells were counted at different time points by phase contrast using a Leitz Ortholux II microscope and a Leitz 10×/0.13 objective.
Immunofluorescence analysis of primary fibroblasts and COS-1 cells
Cells were washed in PBS, fixed (4% paraformaldehyde [PFA], 20 min, room temperature), washed twice, treated for free aldehydes quenching (50 mM NH4Cl, 10 min at room temperature), and washed again. After 5-min permeabilization with 0.5% Triton X-100 in PBS, cells were blocked for 30 min with 2% FBS and 1% BSA in PBS. Then, cells were sequentially incubated with primary and secondary antibodies 1.5 h at 37°C, 1:500 mouse anti-FLAG (F-1804; Sigma-Aldrich) or 1:50 mouse anti-CDC42 (SC8401; Santa Cruz); 30 min at room temperature, 1:100 goat anti mouse Alexa Fluor 488 (A11017; ThermoFisher Scientific); 30 min room temperature, 1:50 rabbit anti-GM130 (PA1-077; ThermoFisher Scientific); and 30 min at room temperature, 1:100 goat anti-rabbit Alexa Fluor 568 (A11011; ThermoFisher Scientific). All antibodies were diluted in 0.5% BSA and 0.1% saponin in PBS. Unbound antibodies were removed after each incubation by three washes with washing buffer (0.2% BSA in PBS). After a 30-min incubation with phalloidin-Alexa Fluor 647 (Life Technologies), coverslips were washed and then mounted on glass slides with 5 µg/ml Hoechst33342 (Life Technologies) nuclear dye in antifade reagent (ThermoFisher Scientific) and analyzed. Imaging was performed on an Olympus FV1000 using excitation spectral laser lines at 405, 488, and 633 nm. Signals from different fluorescent probes were taken in sequential scanning mode. Images were captured using Olympus FluoView Viewer Software. Golgi localization of CDC42 in primary fibroblasts was quantitated using GM130 as a mask for cis/medial-Golgi, and CDC42 fluorescence intensity was quantified as the ratio of Golgi to whole-cell staining by using ImageJ software. Multipolar and filopodia-bearing cells were determined (blinded to sample information) by manually counting in FluoView Viewer ≥200 cells from ≥10 randomly chosen images. Cells having multiple F-actin flat protruding edges (butterfly shaped) were considered multipolar, while cells showing a front (leading edge) and a rear were considered unipolar. To calculate the average filopodial length, the distance from the cell base to the tip was determined. Actin protrusions <1 µm were not considered as filopodia. Filopodia measured as twofold longer than nuclei diameter were considered long filopodia, while the remaining filopodia were considered short.
CRISPR cell line production
The immortalized YTS human NK cell line was used as the model for immune function. In collaboration with the Cell Based Assay Screening Service at Baylor College of Medicine, a homozygous knock-in of the c.556C>T/p.R186C mutation was placed into chromosome 1, isoform 1 of CDC42. Briefly, guide RNAs were screened in 293T cells for editing efficiency. Each guide RNA was incorporated into an all-in-one vector encoding Cas9 and eGFP.
A single-stranded donor oligonucleotide template (antisense; 5′-GAATGTATTTGACGAAGCAATATTGGCTGCCCTGGAGCCTCCAGAACCGAAAAAGTCCTGCAGGTGTGTGCTGCTATGAACATCTCTCCAGAGCCCTTTCTGCACAGCTGGTGTCGGC-3′) was synthesized incorporating the desired mutation, a silent mutation encoding for the SbfI restriction site, and another mutation to reduce Cas9 recutting. Constructs were transfected into the parental YTS cell line (Neon electroporator, 1,400 V/10 ms/three pulses, 2.5e6 YTS cells). Cells were sorted based on eGFP 48 h later and plated by limiting dilution. Clones were screened based on genomic extraction and SbfI digestion and then sequenced using Sanger sequencing of the targeted region for verification. Clones were expanded in R10 medium and then underwent repeat Sanger sequencing for the presence of the CDC42 mutation and for the lack of off-site mutagenesis at GBA3, a known pseudogene of CDC42. A single clone was used for all YTS experiments presented.
YTS fixed-cell immunofluorescence confocal microscopy
YTS cells were washed once with complete R10 and resuspended to a final concentration of 1e6/ml in complete R10. 200,000 cells were plated onto a poly-lysine–coated slide and incubated at 37°C for 20 min for adherence. After incubation, cells were gently washed three times with PBS and fixed and permeabilized with BD cytofix/cytoperm (554655; BD Biosciences) for 20 min at room temperature. All subsequent washes and antibody incubations are done with PBS-S (PBS + 1% BSA + 0.1% saponin). Cells were incubated with anti-CDC42 (1:300, 187643; Abcam) for 1 h at room temperature followed by goat anti-rabbit Alexa Fluor 568 (1:300, A11011; ThermoFisher Scientific) for 1 h. Lastly, cells were incubated with phalloidin-Alexa Fluor 647 (1:100, A22287; ThermoFisher Scientific) and anti-giantin Alexa Fluor 488 (1:200, #908701/Poly19087; BioLegend) for 1 h. After washing, cells were mounted with Prolong Diamond (P36970; ThermoFisher Scientific) to a #1.5 coverslip and allowed to cure for 24 h before sealing with nail varnish. Imaging was done using Metamorph on a Zeiss Axio Observer CSU-X confocal microscope with a 63×/1.4 NA objective. Colocalization was conducted in ImageJ software where a segmentation mask was created around the Golgi apparatus (using giantin thresholding) and cell membrane (using CDC42 thresholding) and average intensity signal within the Golgi and the cytoplasm were measured. The ratios between the signal within the Golgi and the cytoplasm were calculated.
SIM-TIRF microscopy and filopodial analysis
A Lab-Tek II 8-well chambered #1.5 coverglass (#155409; Nunc) was coated with 5 μg/ml anti-CD18 (IB4, single batch) and 5 μg/ml anti-CD28.2 (LEAF anti-CD28.2, 302923; BioLegend) and incubated for ≥30 min at 37°C. YTS cells were resuspended in R10 without FBS and phenol red (imaging media) and resuspended in imaging media at ∼50,000 cells/150 μl (0.33e6/ml). 50,000 YTS cells plated in 150 μl imaging media and incubated at 37°C for exactly 20 min. After, cells were immediately fixed with 4% PFA for 20 min at room temperature. Cell were then washed once with PBS and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. After three additional PBS washes, cells were stained with phalloidin-Alexa Fluor 568 (1:50, A12380; ThermoFisher Scientific) for ≥1 h and cells were imaged thereafter without buffer exchange. Imaging was done on a GE DeltaVision OMX-SR microscope under structured illumination microscopy–total internal reflection fluorescence (SIM-TIRF) mode using a 60×/1.42 PlanApoN objective and a pco.edge 4.2 camera. Images were then reconstructed in GE Softworx using experimentally measured OTF files. Filopodia were manually counted for cells that spread onto the surface on ImageJ. Cells with retraction fibers and not filopodia were omitted from calculation.
Immune cytotoxicity assay for NK cell lines and Pt 2 and Pt 3
For the NK cell line, a chromium-based assay was used. For Pt 2, the DELFIA cytotoxicity assay was used as described below. For Pt 3, a flow cytometric–based cytotoxicity assay was performed similarly to that previously described (Bryceson et al., 2009; Valiathan et al., 2012). Target cells (721.221) were incubated with 100 μCi of 51Cr (sodium chromate in normal saline, NEZ030005MC; PerkinElmer) for 1 h at 37°C, washed three times in complete R10, and resuspended in complete R10 at a final concentration of 105 cells/ml. Unlabeled effector YTS cells were washed once in complete R10 and resuspended to a final concentration of 106/ml. 105 (200 μl) effector cells were plated into the first well of a 96-well, round-bottomed polystyrene tissue culture–treated plate and serially diluted 1:2, except for the spontaneous and total lysis control wells, which contain only target cells. 104 51Cr signal from the LumaPlate was read using a TopCount NXT (PerkinElmer) at the conditions indicated by the manufacturer. Percent lysis is calculated as: [(TopCount measured cpm − spontaneously released cpm)/(total cpm − spontaneously released cpm)] × 100. Total cpm was derived from a target containing well that was lysed with 1% IGEPAL (I3021; Sigma-Aldrich) in water.
The DELFIA cytotoxicity assay is based on loading cells with an acetoxymethyl ester of a fluorescence enhancing ligand. After the ligand has penetrated the cell membrane the ester bonds are hydrolyzed within the cell to form a hydrophilic ligand, which no longer passes through the membrane. After cytolysis, the released ligand is introduced to a europium solution to form a fluorescent chelate. The measured signal correlates directly with the amount of lysed cells. 100 µl of loaded target cells (5,000 cells) is pipetted into a round-bottomed sterile plate, and 100 µl effector cells of varying cell concentration is added. An E/T ratio from 6:1 to 100:1 is commonly used for NK cells. Additional wells containing only target cells are plated for detection of background, spontaneous release, and maximum release. Cells are then incubated 2 and 3 h in a humidified 5% CO2 atmosphere at 37°C. After incubation, the plate is centrifuged for 5 min at 500 ×g. 20 µl the supernatant is transferred to a flat-bottomed plate with 200 µl Eu solution, the plate is shaken at 250 rpm for 15 min, and fluorescence is measured. Percent lysis is calculated as: [(TopCount measured cpm − spontaneously released cpm)/(total cpm − spontaneously released cpm)] × 100. To measure spontaneous release, incubate the target cells (100 µl) with 100 µl of medium instead of effector cells. After centrifugation, transfer 20 µl of the supernatant to the flat-bottomed plate and add 200 µl Eu solution. Shake for 15 min and measure. To measure maximum release, incubate the target cells (100 µl) with 100 µl of medium supplemented with 10 µl of lysis buffer. After centrifugation, transfer 20 µl of the supernatant to the flat-bottomed plate and add 200 µl Eu solution. Shake for 15 min and measure.
Transwell migration assay for immune cells
A standard Boyden chamber assay was used. Corning transwell polycarbonate membrane cell culture inserts (6.5 mm, 5-µm pore polycarbonate, 3421; Corning) were coated overnight with 10 µg/ml fibronectin in PBS at 4°C. YTS cells were washed once using complete R10 and resuspended to a final concentration of 2–4e6 cells/ml. 600 µl of complete R10 supplemented with 100 ng/ml SDF1a/CXCL12 (300-28A; Peprotech) chemoattractant was plated into a 24-well tissue culture–treated plate. Fibronectin was removed from the transwell, and the transwell was placed into the wells containing CXCL12-supplemented media. 100 µl (200,000–400,000 cells) YTS cells were plated onto the upper chamber of the transwell. The cells were incubated at 37°C for 4 h. After incubation, the cells were counted by measuring the volume in the lower chamber and counting the number of events at medium speed for 1 min on an LSR Fortessa Flow Cytometer. The live events were gated for based on forward scatter (FSC) and side scatter (SSC) and used for the migration measurement. The percent migration was calculated as the number of cells that migrated into the lower chamber divided by the number of cells in a control well (input cell count) containing the same number of cells but no transwell.
Immune conjugation assay
YTS NK cells and 721.221 target cells were washed once with complete R10. YTS cells were then resuspended in PBS at 1e6/ml and incubated with efluor 450 (#65-0842-85; 1:1,000, 10 min, and 37°C; eBioscience). 721.221 cells were resuspended in PBS at 1e6/ml and incubated with Cell Trace CFSE (#C34554; 1:2,000, 10 min, and 37°C). Both labeled cells were then washed once with complete R10 and counted. Labeled YTS cells were resuspended to 1e6/ml and labeled 721.221 cells resuspended to 2e6/ml in complete R10. Cells were then combined at an E/T ratio of 1:2 (100,000 YTS cells:200,000 721.221 cells) and incubated at 37°C for 0, 5, 10, 30, and 60 min. The conjugation reaction was stopped by vortexing for 3 s and fixed with 4% PFA in PBS for ≥10 min before acquisition. Cells were then counted on a BD LSR Fortessa flow cytometer and gated based on negative and single-positive control cells. At least 20,000 live cells were counted based on FSC/SSC gating. Percent conjugation was calculated as: (double-positive cells)/(single-positive eFluor 450/YTS cells + double-positive cells) × 100.
Peripheral blood immunophenotype
All flow cytometric analyses were performed on EDTA blood samples within 24 h of venipuncture. After red blood cell lysis with ammonium chloride, lymphocytes were washed, resuspended in PBS, and stained with the following mouse anti-human antibodies to identify T and B cell subsets: CD45RA APC-H7 (clone T6D11; Miltenyi Biotec), CD3 PerCP (clone BW264/56; Miltenyi Biotec), CCR7 PE (clone 3D12; eBioscience), CD4 APC (clone OKT4; Becton Dickinson), CD8 PE-Cy7 (clone RPA-T8; Becton Dickinson), CD19 PE-CY7 (clone SJ25C1; Becton Dickinson), CD16 PE (clone 3G8), CD56 PE, CD27 FITC (clone M-T271, Becton Dickinson), TCR α-beta APC (clone T10B9; Becton Dickinson), TCR gamma-delta FITC (11F3; Miltenyi Biotec), CD21 PE (clone B-ly4; Becton Dickinson), CD24 PE (clone ML5; Becton Dickinson), IgD FITC (clone IA6-2; Becton Dickinson), Goat F(ab)2 anti-Human IgM (µ)-Alexa Fluor 647 (Jackson ImmunoResearch), and CD38 FITC (clone HIT2; Becton Dickinson). Cells were incubated with the appropriate antibody cocktail for 30 min at 4°C and then washed with PBS and resuspended in PBS for flow cytometric acquisition. At least 50,000 events were acquired within the lymphogate. Data were acquired on a FACSCanto II (Becton Dickinson) and analyzed with FlowJo software (Tree Star).
Dendritic cell flow cytometric analysis
Dendritic cell analysis was performed with the Blood Dendritic cell enumeration kit (#130-091-086; Miltenyi Biotec). This assay is based on dendritic cell–specific surface antigens CD303 (BDCA-2), CD141 (BDCA-3), and CD1c (BDCA-1). Three distinct dendritic cell subsets were identified in whole blood: plasmacytoid dendritic cells as CD303+ (FITC clone: AC144, BDCA-2), type 1 myeloid dendritic cells (MDC1s) as CD1c+ (PE clone: AD5-8E7, BDCA-1), and type 2 myeloid dendritic cells (MDC2s) as CD141+ (APC clone: Ad4-14H12, BDCA-3) surface expression. Gate exclusion for CD19, CD14, and dead cells was performed. After 10 min of incubation in ice with the mix of antibodies, red blood cells were lysed. Cells were washed with PBS, fixed with 1% PFA for 10 min, acquired with a FACSCanto II (Becton Dickinson), and analyzed with FlowJo software (Tree Star Inc.; version 8.8.6). At least 106 total events were acquired.
Clinical flow cytometry for BM analysis/composition
The detailed protocol for whole BM staining has been previously reported (Basso-Ricci et al., 2017). In brief, Precision Count beads (BioLegend) were added to 100 μl of P1 BM sample to allow absolute quantification of hematopoietic cell subsets, and red blood cell lysis was performed. The lysed sample was labeled with fluorescent antibodies for CD3-BV605, CD56-PE-Cy5, CD14-BV510, CD61/41-PE-Cy7, CD135-PE, CD34-BV421, CD45RA-APC-Cy7 (BioLegend), CD33-BB515, CD66b-BB515, CD38-BUV737, CD45-BUV395, CD90-APC, CD10-BV786, CD11c-BV650, CD19-APCR700, CD7-PE-Cy5.5, and CD71-BV711 (BD Biosciences). Titration assays were performed to assess the best antibody concentration. After surface marking, the cells were incubated with PI (BioLegend) to stain dead cells. All samples were acquired using a BD LSR Fortessa (BD Bioscience) flow cytometer after calibration with SPHERO rainbow calibration particles (Spherotech), and raw data were collected using BD FACSDIVA software. The data were subsequently analyzed with FlowJo software, and the graphical output was automatically generated using GraphPad Prism.
Functional characterization of proliferation and migration of CD34+ cells
Purification of CD34+ cells was performed by two steps of immunomagnetic selection (CD34+ microbeads, 130–097-047; Miltenyi Biotec). For proliferation, 104 BM CD34+ cells were stimulated with stem cell factor (SCF; 100 ng/ml) or a growth factor mixture (SCF 300 ng/ml, thrombopoietin 100 ng/ml, IL3 60 ng/ml). Proliferation was evaluated at day 5 of stimulation by 16 h liquid scintillation counting of 3H-thymidine (Amersham Biosciences). The stimulation index was calculated as the ratio between stimulated and nonstimulated cells. For the transwell migration assay, 0.5 × 106 peripheral blood lymphocytes or 0.5 × 104 BM CD34+ cells were seeded on a transwell chambers in presence of SDF1-α/CXCL12 (100 ng/ml; Peprotech). Migration was assessed after 3 h. To evaluate clonogenic potential of CDC42− cells, 2 × 105 BM mononuclear cells as well as 103 BM CD34+ cells were plated in Methocult medium (H4434; Stemcell Technologies), and the number of colony-forming units per cell was scored at day 14 to determine number and type of colonies.
BM mononuclear cells stimulation and cytokine measurements
BM mononuclear cells were isolated from HD (n = 3) and Pt BM and were left unstimulated or stimulated with LPS (10 µg/ml) for 5 h, with or without the addition of ATP (1 mM) for an additional hour. Secreted cytokines were measured by in conditioned media. Plasma and supernatant levels of IFNγ, CXCL9, and IL-6 were assessed using enzyme-linked immunosorbent assays with human Duoset ELISA kits, and IL-1β plasma levels were measured using the high-sensitivity quantikine ELISA kit (all purchased from R&D Systems). Plasma IL-18 levels were measured using an ELISA kit obtained from MBL.
C. elegans studies
WT cdc-42 cDNA (open reading frame clone R07G3.1; ThermoFisher Scientific) was cloned into the pPD49.83 heat shock–inducible vector (a kind gift of Andrew Fire, Stanford University, Stanford, CA). The c.556_558AAG>TGT trinucleotide substitution (p.K186C corresponding to human p.R186C) was introduced by site-directed mutagenesis (Stratagene). Germline transformation and genetic crosses were performed using standard techniques. Constructs were injected at a concentration of 100 ng/ml. The pJM67 plasmid (pelt-2::NLS::GFP; 30 ng/ml) was used as coinjection marker. To explore vulval defects, synchronized animals from three independent lines for each construct were grown at 20°C and heat shocked (33°C, 90 min followed by 30°C, 30 min) in parallel at the early (Muv and Vul) or mid (Pvl) L3 larval stages. Adults were scored blindly at a Leica MZ10F dissecting microscope to check for the presence of Pvl phenotype, multiple ectopic pseudovulvae (Muv phenotype) and lack of a vulva (Vul phenotype). After each cross, the genotype was confirmed by Sanger sequencing. Isogenic worms that had lost the transgene were cloned separately and used as controls. Microscopy observations were performed with a Nikon Eclipse 80i instrument equipped with Nomarski differential interference contrast optics on live animals mounted on 2% agarose pads containing 10 mM sodium azide as anesthetic. The N2 (Bristol) and PS21 (let-23(sy1), let-23/EGFR hypomorphic allele) strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Statistical analyses
Data are represented as means ± SD or SEM where indicated. Statistical analyses were conducted in GraphPad Prism. WB of patient CDC42 expression intensity measurements and protein localization intensity measurements were analyzed with the Student’s t test. GTPase function assays were analyzed using a Student’s t test. For variance analysis in pull-down assays, an ordinary one-way ANOVA was performed using the Sidak’s multiple comparison test. Proliferation assays were analyzed using either a two-way ANOVA with Dunnett’s multiple comparisons for patient fibroblasts. Migration assays were assessed using a Student’s t test (for YTS NK cell and primary fibroblasts) or a two-way ANOVA with Tukey’s multiple comparisons (for NIH-3T3 cells). Multipolarity cell counts, filopodia number, and filopodia length were assessed using a Welch’s t test. NK cell cytotoxicity was assessed using the Mann–Whitney U test of three pooled replicates. NK cell conjugation was assessed using a Welch’s t test. For C. elegans studies, P values were calculated using two-tailed Fisher’s exact test. All remaining assays were performed with n of ≤2. Statistical significance is indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) for all tests except two-tailed Fisher’s exact test (*, P < 0.005; **, P < 0.00002, ***, P < 1.2e−6).
Online supplemental material
Table S1 reports clinical laboratory data for Pt 1 and Pt 2. Table S2 shows the hematological and immunological profiles of Pt 1–3. Table S3 provide WES/WGS metrics and data output. Table S4 reports on the vulval phenotypes in transgenic C. elegans expressing WT CDC-42 or the K186C, R68Q, and A159V disease-causing mutants. Table S5 reports on the clonogenic assay of BM cells from Pt 1.
Supplementary Material
Acknowledgments
We would like to thank the Baylor College of Medicine Medical Scientist Training Program for training support.
This work was supported by grants from Fondazione Bambino Gesù (Vite Coraggiose to M. Tartaglia), Italian Ministry of Health (Ricerca Corrente to F. Locatelli, C. Cancrini, F. De Benedetti and M. Tartaglia; Ricerca Finalizzata NET-2011-02350069 to C. Cancrini and A. Aiuti), Associazione Italiana per la Ricerca sul Cancro (IG21614 to M. Tartaglia), National Institutes of Health (R01AI120989-05 and R01AI067946 to J.S. Orange), National Human Genome Research Institute/National Heart, Lung, and Blood Institute (grant UM1HG006542 to the Baylor-Hopkins Center for Mendelian Genomics), German Research Foundation (AH 92/8-1 to M.R. Ahmadian), German Federal Ministry of Education and Research (01GM1902C to M.R. Ahmadian), German Research Foundation (International Research Training Group 1902 Intra- and Interorgan Communication of the Cardiovascular System, IRTG 1902-P6, to M.R. Ahmadian), and E-Rare (NSEuroNet to M.R. Ahmadian and M. Tartaglia; and EUROCID to C. Cancrini and A. Aiuti).
The authors declare no competing financial interests.
Author contributions: M.T. Lam and S. Coppola performed the in vitro studies and contributed to manuscript drafting; O.H.F. Krumbach, M. Akbarzadeh, A. Pastore, S. Levi Mortera, S. Camerini, L. Farina, M. Buchholzer, R. Dvorsky, L. Pannone, and P. Janning performed the in silico, biochemical, and MS analyses; G. Prencipe, C. Cifaldi, I. Brigida, S. Scala, S. Di Cesare, A. Pascarella, F. Conti, P. Merli, L. Farina, L. Basso-Ricci, M. Chiriaco, R. Carsetti, Y.T. Bryceson, L. Torralba-Raga, K. Ramme, V. Rosti, C. Bracaglia, and V. Messia performed the hematological and immunological studies; E. Zara, P. Netter, A.F. Carisey, M. Diehl, and E.M. Mace performed the in vitro studies and assisted with experimental design and data analysis; A. Insalaco, A. Stray-Pedersen, H.C. Erichsen, A.C. Horne, K. Ramme, P. Palma, A. Finocchi, F. Locatelli, C. Cancrini, and A. Aiuti provided the clinical data and biological material; S. Martinelli, L. Pannone, and M. Di Rocco performed the C. elegans studies; M. Niceta, F. Pantaleoni, A. Ciolfi, T.N. Cao, Z.H. Coban-Akdemir, S.N. Jhangiani, D.M. Muzny, R.A. Gibbs, A.C. Horne, Y.T. Bryceson, L. Torralba-Raga, I.K. Chinn, J.R. Lupski, and E.M. Mace contributed to the genomic analyses; and M.R. Ahmadian, J.S. Orange, F. De Benedetti, and M. Tartaglia coordinated the study, conceived and designed the experiments, analyzed the data, and contributed to manuscript drafting.
References
- Aicart-Ramos C., Valero R.A., and Rodriguez-Crespo I.. 2011. Protein palmitoylation and subcellular trafficking. Biochimica et Biophysica Acta (BBA) -. Biomembranes. 1808:2981–2994. 10.1016/j.bbamem.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Baschieri F., Confalonieri S., Bertalot G., Di Fiore P.P., Dietmaier W., Leist M., Crespo P., Macara I.G., and Farhan H.. 2014. Spatial control of Cdc42 signalling by a GM130-RasGRF complex regulates polarity and tumorigenesis. Nat. Commun. 5:4839 10.1038/ncomms5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso-Ricci L., Scala S., Milani R., Migliavacca M., Rovelli A., Bernardo M.E., Ciceri F., Aiuti A., and Biasco L.. 2017. Multiparametric Whole Blood Dissection: A one-shot comprehensive picture of the human hematopoietic system. Cytometry A. 91:952–965. 10.1002/cyto.a.23148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C.K., Calligari P., Radio F.C., Caputo V., Dentici M.L., Falah N., High F., Pantaleoni F., Barresi S., Ciolfi A., et al. . 2018. Mutations in KCNK4 that Affect Gating Cause a Recognizable Neurodevelopmental Syndrome. Am. J. Hum. Genet. 103:621–630. 10.1016/j.ajhg.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracaglia C., de Graaf K., Pires Marafon D., Guilhot F., Ferlin W., Prencipe G., Caiello I., Davì S., Schulert G., Ravelli A., et al. . 2017. Elevated circulating levels of interferon-γ and interferon-γ-induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 76:166–172. 10.1136/annrheumdis-2015-209020 [DOI] [PubMed] [Google Scholar]
- Brigida I., Zoccolillo M., Cicalese M.P., Pfajfer L., Barzaghi F., Scala S., Oleaga-Quintas C., Álvarez-Álvarez J.A., Sereni L., Giannelli S., et al. . 2018. T-cell defects in patients with ARPC1B germline mutations account for combined immunodeficiency. Blood. 132:2362–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y.T., Fauriat C., Nunes J.M., Wood S.M., Björkström N.K., Long E.O., and Ljunggren H.-G.. 2009. Natural Killer Cell Protocols, Cellular and Molecular Methods. Methods Mol. Biol. 612:335–352. 10.1007/978-1-60761-362-6_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S.W., de Jesus A.A., Gouni S., Brooks S.R., Marrero B., Liu Y., DiMattia M.A., Zaal K.J.M., Sanchez G.A.M., Kim H., et al. . 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140–1146. 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caye A., Strullu M., Guidez F., Cassinat B., Gazal S., Fenneteau O., Lainey E., Nouri K., Nakhaei-Rad S., Dvorsky R., et al. . 2015. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat. Genet. 47:1334–1340. 10.1038/ng.3420 [DOI] [PubMed] [Google Scholar]
- Chinn I.K., Eckstein O.S., Peckham-Gregory E.C., Goldberg B.R., Forbes L.R., Nicholas S.K., Mace E.M., Vogel T.P., Abhyankar H.A., Diaz M.I., et al. . 2018. Genetic and mechanistic diversity in pediatric hemophagocytic lymphohistiocytosis. Blood. 132:89–100. 10.1182/blood-2017-11-814244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L.K., Philip N.H., Schmidt V.A., Koller A., Strowig T., Flavell R.A., Brodsky I.E., and Bliska J.B.. 2014. IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM. MBio. 5:e01402–e01414. 10.1128/mBio.01402-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., and Ruden D.M.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasouki M., Okonkwo K.C., Ray A., Folmsbeel C.K., Gozales D., Keles S., Puck J.M., and Chatila T.. 2011. Deficient T Cell Receptor Excision Circles (TRECs) in autosomal recessive hyper IgE syndrome caused by DOCK8 mutation: implications for pathogenesis and potential detection by newborn screening. Clin. Immunol. 141:128–132. 10.1016/j.clim.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorsky R., and Ahmadian M.R.. 2004. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 5:1130–1136. 10.1038/sj.embor.7400293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2004. Cdc42--the centre of polarity. J. Cell Sci. 117:1291–1300. 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2002. Rho GTPases in cell biology. Nature. 420:629–635. 10.1038/nature01148 [DOI] [PubMed] [Google Scholar]
- Flex E., Niceta M., Cecchetti S., Thiffault I., Au M.G., Capuano A., Piermarini E., Ivanova A.A., Francis J.W., Chillemi G., et al. . 2016. Biallelic Mutations in TBCD, Encoding the Tubulin Folding Cofactor D, Perturb Microtubule Dynamics and Cause Early-Onset Encephalopathy. Am. J. Hum. Genet. 99:962–973. 10.1016/j.ajhg.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., and Kaibuchi K.. 2002. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell. 109:873–885. 10.1016/S0092-8674(02)00800-0 [DOI] [PubMed] [Google Scholar]
- Gibson R.M., and Wilson-Delfosse A.L.. 2001. RhoGDI-binding-defective mutant of Cdc42Hs targets to membranes and activates filopodia formation but does not cycle with the cytosol of mammalian cells. Biochem. J. 359:285–294. 10.1042/bj3590285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R.M., Gandhi P.N., Tong X., Miyoshi J., Takai Y., Konieczkowski M., Sedor J.R., and Wilson-Delfosse A.L.. 2004. An activating mutant of Cdc42 that fails to interact with Rho GDP-dissociation inhibitor localizes to the plasma membrane and mediates actin reorganization. Exp. Cell Res. 301:211–222. 10.1016/j.yexcr.2004.07.033 [DOI] [PubMed] [Google Scholar]
- Grom A.A., Horne A., and De Benedetti F.. 2016. Macrophage activation syndrome in the era of biologic therapy. Nat. Rev. Rheumatol. 12:259–268. 10.1038/nrrheum.2015.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapas C.R., Steiner A., Davidson S., and Masters S.L.. 2018. An Update on Autoinflammatory Diseases: Inflammasomopathies. Curr. Rheumatol. Rep. 20:40 10.1007/s11926-018-0750-4 [DOI] [PubMed] [Google Scholar]
- Henter J.I., Horne A., Aricó M., Egeler R.M., Filipovich A.H., Imashuku S., Ladisch S., McClain K., Webb D., Winiarski J., and Janka G.. 2007. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer. 48:124–131. 10.1002/pbc.21039 [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., and Cerione R.A.. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell. 100:345–356. 10.1016/S0092-8674(00)80670-4 [DOI] [PubMed] [Google Scholar]
- Hsu A.P., Donkó A., Arrington M.E., Swamydas M., Fink D., Das A., Escobedo O., Bonagura V., Szabolcs P., Steinberg H.N., et al. . 2019. Dominant activating RAC2 mutation with lymphopenia, immunodeficiency, and cytoskeletal defects. Blood. 133:1977–1988. 10.1182/blood-2018-11-886028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Hamidi H., and Ivaska J.. 2015. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 36:23–31. 10.1016/j.ceb.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., and Bejerano G.. 2016. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 48:1581–1586. 10.1038/ng.3703 [DOI] [PubMed] [Google Scholar]
- Jamilloux Y., Magnotti F., Belot A., and Henry T.. 2018. The pyrin inflammasome: from sensing RhoA GTPases-inhibiting toxins to triggering autoinflammatory syndromes. Pathog. Dis. 76:fty020 10.1093/femspd/fty020 [DOI] [PubMed] [Google Scholar]
- Jordan M.B., Allen C.E., Weitzman S., Filipovich A.H., and McClain K.L.. 2011. How I treat hemophagocytic lymphohistiocytosis. Blood. 118:4041–4052. 10.1182/blood-2011-03-278127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.L., Chae J.J., Park Y.H., De Nardo D., Stirzaker R.A., Ko H.-J., Tye H., Cengia L., DiRago L., Metcalf D., et al. . 2015. Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J. Exp. Med. 212:927–938. 10.1084/jem.20142384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sun X., Wang J., Zhao Q., Dai R., Wang Y., Zhou L., Westerberg L., Ding Y., Zhao X., and Liu C.. 2017. Defective thymic output in WAS patients is associated with abnormal actin organization. Sci. Rep. 7:11978 10.1038/s41598-017-12345-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jian X., and Boerwinkle E.. 2013. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34:E2393–E2402. 10.1002/humu.22376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli F., Merli P., Pagliara D., Li Pira G., Falco M., Pende D., Rondelli R., Lucarelli B., Brescia L.P., Masetti R., et al. . 2017. Outcome of children with acute leukemia given HLA-haploidentical HSCT after αβ T-cell and B-cell depletion. Blood. 130:677–685. 10.1182/blood-2017-04-779769 [DOI] [PubMed] [Google Scholar]
- Martinelli S., Krumbach O.H.F., Pantaleoni F., Coppola S., Amin E., Pannone L., Nouri K., Farina L., Dvorsky R., Lepri F., et al. University of Washington Center for Mendelian Genomics . 2018. Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am. J. Hum. Genet. 102:309–320. 10.1016/j.ajhg.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- Nouri K., Timson D.J., and Ahmadian M.R.. 2017. New model for the interaction of IQGAP1 with CDC42 and RAC1. Small GTPases.:1–7. 10.1080/21541248.2017.1321169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H., Tsutsi H., Komatsu T., Yutsudo M., Hakura A., Tanimoto T., Torigoe K., Okura T., Nukada Y., Hattori K., et al. . 1995. Cloning of a new cytokine that induces IFN-γ production by T cells. Nature. 378:88–91. 10.1038/378088a0 [DOI] [PubMed] [Google Scholar]
- Orange J.S., Ramesh N., Remold-O’Donnell E., Sasahara Y., Koopman L., Byrne M., Bonilla F.A., Rosen F.S., Geha R.S., and Strominger J.L.. 2002. Wiskott-Aldrich syndrome protein is required for NK cell cytotoxicity and colocalizes with actin to NK cell-activating immunologic synapses. Proc. Natl. Acad. Sci. USA. 99:11351–11356. 10.1073/pnas.162376099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.J., Calero G., Chan B., Ramachandran S., and Cerione R.A.. 2008. Effector proteins exert an important influence on the signaling-active state of the small GTPase Cdc42. J. Biol. Chem. 283:14153–14164. 10.1074/jbc.M706271200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner D.J., and Lundquist E.A.. 2018. Small GTPases. WormBook. 2018:1–65. 10.1895/wormbook.1.67.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Romberg N., Al Moussawi K., Nelson-Williams C., Stiegler A.L., Loring E., Choi M., Overton J., Meffre E., Khokha M.K., Huttner A.J., et al. . 2014. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46:1135–1139. 10.1038/ng.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado F.G.N., and Kim A.S.. 2013. Hemophagocytic lymphohistiocytosis: an update on diagnosis and pathogenesis. Am. J. Clin. Pathol. 139:713–727. 10.1309/AJCP4ZDKJ4ICOUAT [DOI] [PubMed] [Google Scholar]
- Seeland I., Xiong Y., Orlik C., Deibel D., Prokosch S., Küblbeck G., Jahraus B., De Stefano D., Moos S., Kurschus F.C., et al. . 2018. The actin remodeling protein cofilin is crucial for thymic αβ but not γδ T-cell development. PLoS Biol. 16:e2005380 10.1371/journal.pbio.2005380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda F.E., and de Saint Basile G.. 2017. Hemophagocytic syndrome: primary forms and predisposing conditions. Curr. Opin. Immunol. 49:20–26. 10.1016/j.coi.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Sinai P., Nguyen C., Schatzle J.D., and Wülfing C.. 2010. Transience in polarization of cytolytic effectors is required for efficient killing and controlled by Cdc42. Proc. Natl. Acad. Sci. USA. 107:11912–11917. 10.1073/pnas.0913422107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray-Pedersen A., Sorte H.S., Samarakoon P., Gambin T., Chinn I.K., Coban Akdemir Z.H., Erichsen H.C., Forbes L.R., Gu S., Yuan B., et al. . 2017. Primary immunodeficiency diseases: Genomic approaches delineate heterogeneous Mendelian disorders. J. Allergy Clin. Immunol. 139:232–245. 10.1016/j.jaci.2016.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart-Mataraza J.M., Li Z., and Sacks D.B.. 2002. IQGAP1 is a component of Cdc42 signaling to the cytoskeleton. J. Biol. Chem. 277:24753–24763. 10.1074/jbc.M111165200 [DOI] [PubMed] [Google Scholar]
- Takenouchi T., Kosaki R., Niizuma T., Hata K., and Kosaki K.. 2015. Macrothrombocytopenia and developmental delay with a de novo CDC42 mutation: Yet another locus for thrombocytopenia and developmental delay. Am. J. Med. Genet. A. 167A:2822–2825. 10.1002/ajmg.a.37275 [DOI] [PubMed] [Google Scholar]
- Valdés-Mora F., and Lacal J.C.. 2011. CDC42 (cell division cycle 42 (GTP binding protein, 25kDa)). Atlas Genet. Cytogenet. Oncol. Haematol. (11). 10.4267/2042/44606 [DOI] [Google Scholar]
- Valiathan R., Lewis J.E., Melillo A.B., Leonard S., Ali K.H., and Asthana D.. 2012. Evaluation of a flow cytometry-based assay for natural killer cell activity in clinical settings. Scand. J. Immunol. 75:455–462. 10.1111/j.1365-3083.2011.02667.x [DOI] [PubMed] [Google Scholar]
- Weiss E.S., Girard-Guyonvarc’h C., Holzinger D., de Jesus A.A., Tariq Z., Picarsic J., Schiffrin E.J., Foell D., Grom A.A., Ammann S., et al. . 2018. Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood. 131:1442–1455. 10.1182/blood-2017-12-820852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang L., Geiger H., Cancelas J.A., Mo J., and Zheng Y.. 2007a Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc. Natl. Acad. Sci. USA. 104:5091–5096. 10.1073/pnas.0610819104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wang L., Kalfa T.A., Cancelas J.A., Shang X., Pushkaran S., Mo J., Williams D.A., and Zheng Y.. 2007b Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood. 110:3853–3861. 10.1182/blood-2007-03-079582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Johnson J.L., Cerione R.A., and Erickson J.W.. 2013. Prenylation and membrane localization of Cdc42 are essential for activation by DOCK7. Biochemistry. 52:4354–4363. 10.1021/bi301688g [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.