In the present study, Moeller et al. identify a previously unrecognized pathway through which intestinal epithelial cells expressing the novel chitin-binding receptor FIBCD1 can recognize and control intestinal fungal colonization, limit fungal dysbiosis, and dampen intestinal inflammation.
Abstract
Host–microbiota interactions are critical in regulating mammalian health and disease. In addition to bacteria, parasites, and viruses, beneficial communities of fungi (the mycobiome) are important modulators of immune- and tissue-homeostasis. Chitin is a major component of the fungal cell wall, and fibrinogen C containing domain 1 (FIBCD1) is a chitin-binding protein; however, the role of this molecule in influencing host–mycobiome interactions in vivo has never been examined. Here, we identify direct binding of FIBCD1 to intestinal-derived fungi and demonstrate that epithelial-specific expression of FIBCD1 results in significantly reduced fungal colonization and amelioration of fungal-driven intestinal inflammation. Collectively, these results identify FIBCD1 as a previously unrecognized microbial pattern recognition receptor through which intestinal epithelial cells can recognize and control fungal colonization, limit fungal dysbiosis, and dampen intestinal inflammation.
Introduction
Since diverging from plants over one billion years ago, fungi have coevolved with animals as an integral part of all ecosystems (Wainright et al., 1993; Peay et al., 2016). Like bacteria, fungi are a component of the microbiota at barrier surfaces in mammals, where emerging studies suggest they play an important role in shaping host immunity and tissue homeostasis (Belkaid and Harrison, 2017; Iliev and Leonardi, 2017). Alterations in intestinal fungal composition and dysbiosis have been associated with human inflammatory disorders, such as inflammatory bowel disease (IBD), colorectal cancer, and allergy (Sokol et al., 2017; Wypych and Marsland, 2018; Coker et al., 2019), in which mutations or deficiencies in fungal recognition receptors or downstream signaling pathways have been shown to substantially influence disease severity (Iliev et al., 2012; Wang et al., 2016, 2018; Li et al., 2018; Malik et al., 2018). Nevertheless, how fungi interact with the mammalian host and the molecular mechanisms mediating fungal recognition and resultant host responses remains incompletely defined.
Chitin, a linear polysaccharide of β-(1,4)-linked N-acetylglucosamine (GlcNAc), is one of the most abundant natural biopolymers, and is an essential structural component of crustaceans, insects, nematodes, protozoa, and fungi. Since it is absent in vertebrates, chitin is recognized as a pathogen-associated molecular pattern and targeted by the mammalian immune system (Brodaczewska et al., 2015; Elieh Ali Komi et al., 2018). Exposure to chitin can induce mammalian immune responses, often resembling responses associated with helminth infection and allergic inflammation (Reese et al., 2007; Satoh et al., 2010; Van Dyken et al., 2014). FIBCD1, a transmembrane protein tetramer that structurally resembles the ficolin family of pattern recognition receptors, binds chitin and other acetylated structures with high affinity in vitro (Schlosser et al., 2009; Thomsen et al., 2010). Unlike the ficolins, FIBCD1 does not bind to conventional microbe-associated molecular patterns, such as LPS, peptidoglycan, lipoteichoic acid, or β-glucan, yet it appears to bind chitin, recognize chitin-rich regions of Aspergillus fumigatus and modulate cellular responses to fungal cell wall components in a human lung epithelial cell line in vitro (Schlosser et al., 2009; Jepsen et al., 2018).
Results and discussion
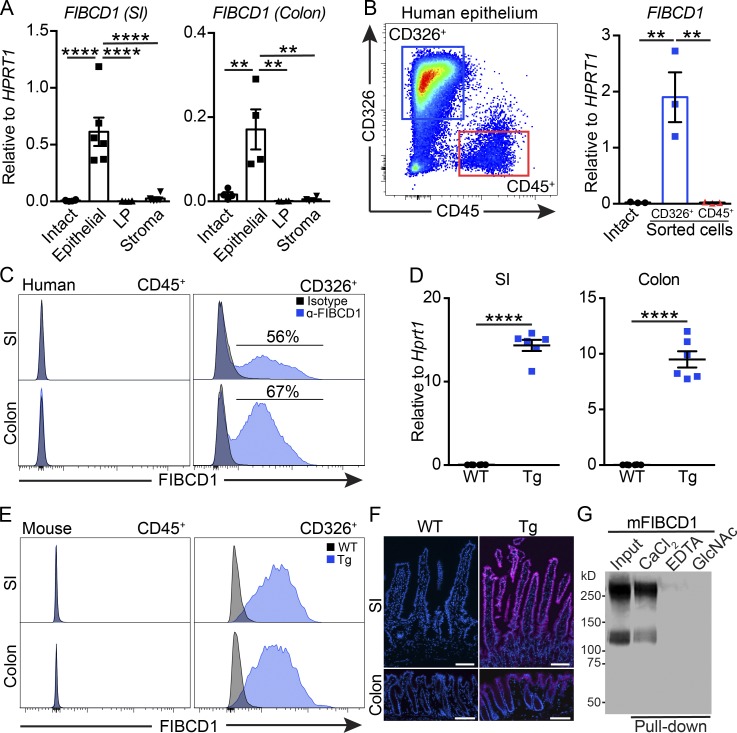
The biological functions of FIBCD1 in vivo are currently poorly understood; however, previous immunohistochemical studies in human tissues suggest that FIBCD1 expression is restricted to cells of the epithelial lineage, particularly at the intestinal barrier surface (von Huth et al., 2018). To examine this further, we isolated small intestine (SI) and colon from healthy human organ donors and assessed the expression of FIBCD1 in freshly isolated live human cells by quantitative RT-PCR and flow cytometry. In both human SI and colon, FIBCD1 expression was highest in the epithelial compartment compared with intact tissue, lamina propria (LP), or remaining stroma (Fig. 1 A). Moreover, FIBCD1 expression was restricted to sort-purified human CD45−CD326+ intestinal epithelial cells (IECs) when compared with the hematopoietic CD45+CD326− cell population (Fig. 1 B). Flow cytometry analysis further confirmed a highly specific surface expression of FIBCD1 on the CD45−CD326+ IEC population (Fig. 1 C). Together, these findings identify that FIBCD1 is a membrane protein specifically expressed on epithelial cells lining the human intestinal tract.
Figure 1.
FIBCD1 is expressed on the surface of human IECs, and this can be recapitulated in novel transgenic mice. (A) Analysis of FIBCD1 expression in fractionated SI and colon from healthy human donors (n = 4–6 samples per group). (B) Gating strategy and analysis of FIBCD1 expression in CD45+CD326− hematopoietic cells and CD45−CD326+ epithelial cells sorted from human colon (n = 3 samples per group). (C) Representative histograms of FIBCD1 expression in CD45+CD326− hematopoietic cells and CD45−CD326+ epithelial cells from human SI and colon. (D) Expression of Fibcd1 in SI and colon mouse tissues from WT and Fibcd1Tg (Tg) littermates (n = 6 samples per group). (E) Representative histograms depicting FIBCD1 expression in CD45+CD326− hematopoietic cells and CD45−CD326+ epithelial cells from SI and colon of WT and Tg littermates. (F) Representative immunofluorescence staining for FIBCD1 in SI and colon from WT and Tg littermates. Blue, DAPI; magenta, FIBCD1. Scale bars, 100 µm. (G) Representative pull-down experiment showing acetyl-group-specific and Ca2+-dependent binding of soluble recombinant mFIBCD1 ectodomain to chitin. Results in A and B are pooled data based on three to six independently collected samples from healthy human donors. Data in C–G are representative of at least three independently performed experiments with similar results. Statistics: Data are presented as mean ± SEM where dots represent individual mice. One-way ANOVA followed by Tukey post hoc test was used to analyze results in A and B, and unpaired Student’s t test was used to analyze data in D. **, P < 0.01 and ****, P < 0.0001.
In mice, expression of Fibcd1 mRNA is detectable in intestinal tissues, yet below the limit of detection in sorted cell populations (Fig. S1, A and B). Thus, to investigate the potential in vivo functional significance of FIBCD1 in intestinal tissues, we developed a gain-of-function transgenic mouse model (Fibcd1Tg) using the Villin promoter, previously shown to drive IEC-specific expression in mouse intestinal tissues (Pinto et al., 1999). Expression of Fibcd1 in SI and colon from Fibcd1Tg mice was initially verified by quantitative RT-PCR (Fig. 1 D), and further analyses by flow cytometry and immunofluorescence microscopy confirmed specific staining for FIBCD1 on CD45−CD326+ IECs lining the intestinal villi and crypts of Fibcd1Tg mice (Fig. 1, E and F). Murine FIBCD1 (mFIBCD1) is a 459–amino acid protein that by sequence alignment shares 90.6% identity with its human orthologue (Schlosser et al., 2009). As in humans, mFIBCD1 is predicted to be a type II transmembrane protein containing a C-terminal fibrinogen-related domain, which shares 97.2% identity with human FIBCD1 and includes a fully conserved S1 acetyl-group binding site previously shown to bind acetylated structures such as chitin (Schlosser et al., 2009; Thomsen et al., 2010; Shrive et al., 2014). We identified that soluble recombinant ectodomain of mFIBCD1 (mFIBCD1Ecto) assembled into homo-tetramers (Fig. S1 C) and bound chitin and other acetylated structures, in a Ca2+-dependent manner, through the conserved S1 acetyl-group binding site (Fig. 1 G and Fig. S1, D–G). Moreover, when expressed, full-length mFIBCD1 localized to the membrane and had the ability to internalize bound ligand (Fig. S1, H and I). At steady state, Fibcd1Tg mice displayed no overt phenotype compared with WT littermates when assessed for breeding potential, offspring frequencies, weight development, or intestinal barrier integrity (Fig. S1, J–M). Moreover, IEC-specific expression of FIBCD1 did not influence the immune cell composition in the LP of SI (Fig. S1, N–P) or colon (Fig. S1, Q–S). Overexpression of FIBCD1 in the A549 lung epithelial cell line in vitro has been shown to suppress genes associated with epithelial responses at steady state as well as upon stimulation with fungal cell wall components and a variety of TLR agonists (Jepsen et al., 2018). Yet, when sort-purified CD45−CD326+ IECs from Fibcd1Tg mice were assessed for the expression of a wide range of genes associated with mucus production (Muc2 and Muc5Ac), cell adhesion/tight junction formation (Cd326, Icam1, Zo1, Cldn1, and Cldn2), immune responses (Ccl2, Ccl20, Il-1b, Il-18, Il-25, Il-33, and Tslp) and production of anti-microbial peptides (Reg3a and Reg3g), no significant differences were observed compared with WT littermates (Fig. S1, T–W). Collectively, these data suggest that Fibcd1Tg mice are healthy with no aberrant phenotype, and although transgenic, they can serve as a suitable tool that recapitulates the in vivo expression of FIBCD1 in the human intestine.
To test the functional role of FIBCD1 expression on IECs in vivo, we employed the parasitic whipworm Trichuris muris, a widely used model of intestinal helminth infection that is transmitted through ingestion of chitin-containing embryonated T. muris eggs (Preston and Jenkins, 1985; Cliffe and Grencis, 2004; Klementowicz et al., 2012). We identified that FIBCD1 recognized and specifically bound the chitin-rich polar plugs of T. muris eggs (Fig. S2 A). We further identified T. muris excretory-secretory (E/S) antigen, consisting of a mixture of immune-modulatory proteins, lipids, and chitinous carbohydrates (Jenkins and Wakelin, 1983; Drake et al., 1994; Hasnain et al., 2012; Eichenberger et al., 2018), as potential ligands of FIBCD1 (Fig. S2 B). Immunity to T. muris is dependent on type 2 cytokine responses that promote IEC proliferation, goblet cell responses, and expulsion of the parasites (Cliffe and Grencis, 2004; Klementowicz et al., 2012). However, when we tested the role of FIBCD1 in immunity to T. muris infection, Fibcd1Tg mice exhibited equivalent type 2 cytokine responses and protective immunity as compared with WT littermates (Fig. S2, C–J). Similarly, Fibcd1Tg mice did not exhibit defects in type 2 immunity or parasite expulsion when infected with other chitin-containing helminth parasites, Heligmosomoides polygyrus or Nippostrongylus brasiliensis (Fig. S2, K–Q). Together, these data suggest that while FIBCD1 binds helminth-derived components that contain chitin in vitro, FIBCD1 is not required to modulate type 2 immunity following helminth infection.
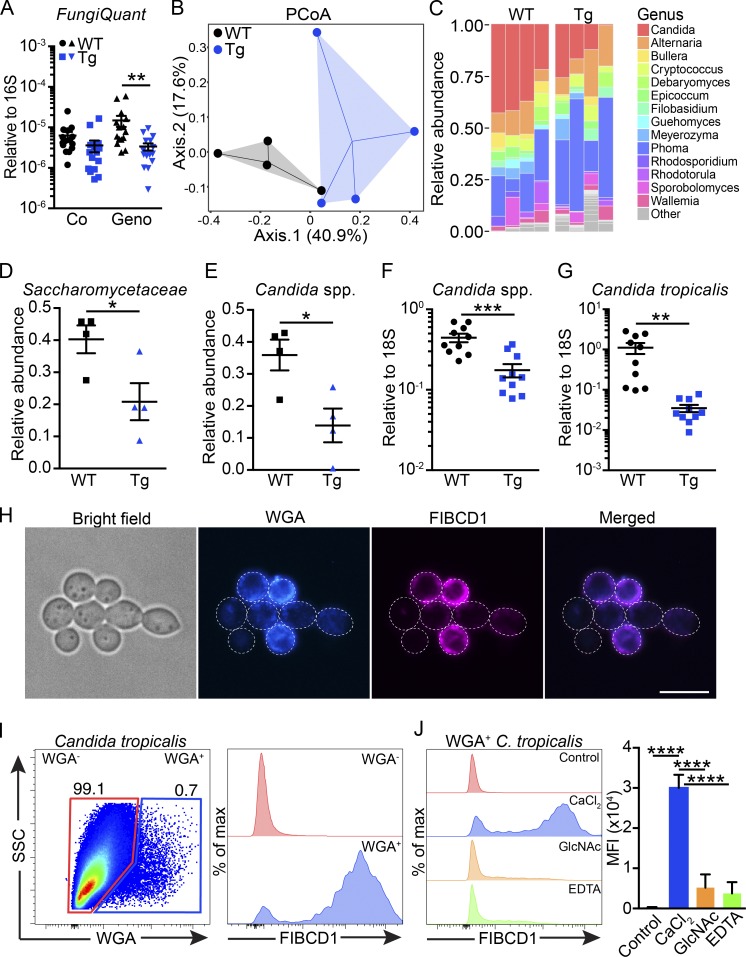
Since FIBCD1 is a novel receptor of chitin capable of binding chitin-rich regions of A. fumigatus (Jepsen et al., 2018) and recent studies have highlighted the importance of interactions between host and fungi present at barrier surfaces (Iliev et al., 2012; Wheeler et al., 2016; Leonardi et al., 2018), we tested whether IEC-restricted expression of FIBCD1 would influence the composition of the gut mycobiome. Initial characterization by quantitative PCR (qPCR) of the relative abundance of fungal 18S ribosomal DNA in WT and Fibcd1Tg littermates cohoused in mixed litters did not reveal any significant changes in fungal burden (Fig. 2 A). However, following separation and 2 wk of additional housing based on genotype, we observed a significant reduction in the abundance of fungal 18S ribosomal DNA in Fibcd1Tg mice compared with WT littermates (Fig. 2 A). To further characterize the impact of FIBCD1 on the composition of intestinal fungal communities, we performed high-throughput internal transcribed spacer 1 (ITS-1) sequencing analysis of fungal ribosomal DNA. ITS-1 sequencing revealed significant changes in the fungal composition in Fibcd1Tg mice compared with WT littermates, where the abundance of species belonging to the Saccharomycetaceae family, dominated by the prevalent Candida genus, was significantly reduced in Fibcd1Tg mice (Fig. 2, B–E). Like humans, mice are predominantly colonized with fungal species belonging to the Ascomycota phylum, particularly Candida spp., but mice exhibit a striking absence of Candida albicans and a prevalence of Candida tropicalis relative to humans (Iliev et al., 2012). In line with this and our ITS-1 sequencing data, we observed a significant reduction in the relative abundance of Candida spp. and specifically C. tropicalis in Fibcd1Tg mice compared with WT littermates when assessed by qPCR (Fig. 2, F and G). Since bacteria and fungi may interact in the intestine, we also evaluated the bacterial composition of Fibcd1Tg mice compared with WT littermates. Analysis by qPCR and sequencing of 16S bacterial ribosomal DNA did not identify FIBCD1-mediated changes to the composition of the bacterial microbiota (Fig. S2, R–V). Thus, we conclude that FIBCD1 selectively regulates the composition of the intestinal mycobiome.
Figure 2.
Expression of FIBCD1 by IECs modulates the intestinal mycobiome and FIBCD1 binds chitin-containing fungi. (A) Assessment of fungal burden (FungiQuant) relative to 16S in WT and Fibcd1Tg (Tg) littermates cohoused (Co) or housed for additional 2 wk based on genotype (Geno; n = 15 samples per group). (B) PCoA ordination based on Bray–Curtis dissimilarities in colonic fungal OTUs (n = 4 samples per group). (C) Fungal composition of the 14 major genera in WT and Tg littermates (n = 4 samples per group). (D and E) Relative abundance of Saccharomycetaceae (family) and the abundant Candida (genus) in WT and Tg littermates quantified by ITS-1 sequencing (n = 4 samples per group). (F and G) Relative abundance of Candida spp. and C. tropicalis normalized to FungiQuant in WT and Tg littermates quantified by qPCR (n = 10 samples per group). (H) Representative fluorescence imaging of recombinant FIBCD1FReD binding a proportion of live C. tropicalis. Blue, WGA; magenta, FIBCD1FReD. Scale bar, 10 µm. (I) Representative scatter plot and histogram of WGA+ C. tropicalis binding recombinant FIBCD1FReD. (J) Histograms and quantification of mean fluorescence intensity (MFI) demonstrating Ca2+-dependent and GlcNAc-specific binding of FIBCD1 to WGA+ C. tropicalis (n = 3 samples per group). Results shown in A–G are representative of two independently performed experiments with similar results. Data shown in H–J are representative of at least three independently performed experiments with similar results. Statistics: Data are presented as mean ± SEM where dots represent individual mice. Two-way ANOVA followed by Holm–Sidak post hoc test was used to analyze results in A, unpaired Student’s t test was used to analyze data in D–G, and one-way ANOVA followed by Tukey post hoc test was used to analyze results in J. *, P < 0.05, **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001. Max, maximum; SSC, side scatter.
As FIBCD1 binds chitin-rich regions of the opportunistic pathogen A. fumigatus and C. tropicalis along with most fungal species contains chitin as a structural component of the cell wall (Latgé, 2007), we next investigated the ability of recombinant FIBCD1 to bind C. tropicalis in vitro. By fluorescence microscopy and flow cytometry, we observed specific binding of the fibrinogen-related domain of FIBCD1 (FIBCD1FReD) to a fraction of exponentially growing C. tropicalis (Fig. 2, H–J). Binding was found to be restricted to cells staining positive for wheat germ agglutinin (WGA), a chitin-binding lectin recognizing exposed chitin in growing fungi (Rodrigues et al., 2008; Liedke et al., 2017). Binding was Ca2+ dependent and was abolished in the presence of GlcNAc, confirming specific binding through the conserved S1 binding site. Collectively, these results indicate that FIBCD1 potentially modulates the gut mycobiome through direct recognition of chitin on gut-resident fungi.
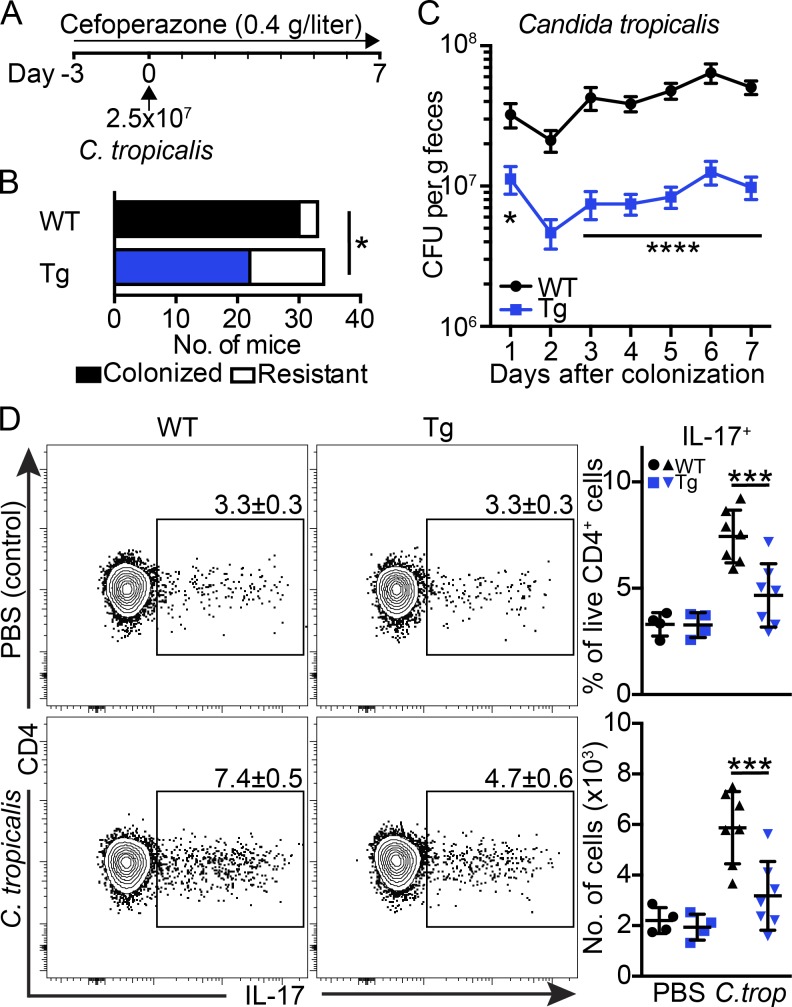
To directly assess the impact of IEC-restricted expression of FIBCD1 on fungal colonization, we colonized antibiotic-treated mice with C. tropicalis (Fig. 3 A). Antibiotics were added to decrease the potential effects of commensal bacteria and create a niche for fungal outgrowth (Leonardi et al., 2018). Colonization with a single-dose of C. tropicalis (2.5 × 107 cells) resulted in a robust 91% colonization efficiency in WT mice, whereas Fibcd1Tg littermates were significantly more resistant to colonization, displaying efficiencies <65% (Fig. 3 B). Further, in colonized mice, we observed a highly significant reduction in C. tropicalis CFUs in Fibcd1Tg mice compared with WT littermates, starting as early as 24 h after colonization (Fig. 3 C). Colonization with C. tropicalis has been shown to increase the number and ratio of IL-17–producing CD4+ T helper (Th) 17 cells in colonic LP (Leonardi et al., 2018). Consistent with heightened C. tropicalis burdens, WT mice exhibited elevated colonic Th17 responses in the LP compared with Fibcd1Tg littermates (Fig. 3 D). Taken together, these data indicate a rapid and specific role for FIBCD1 in limiting fungal colonization of the intestine independently of changes in the bacterial communities.
Figure 3.
Expression of FIBCD1 by IECs limits colonization with C. tropicalis and maintains immune homeostasis. (A–D) WT and Fibcd1Tg (Tg) littermates were treated with cefoperazone in drinking water from day −3 and throughout the experiment. At day 0, mice were colonized by oral gavage with 2.5 × 107 exponentially growing C. tropicalis (C. trop) or PBS as control. (A) Schematic representation of the colonization model. (B) Quantification of colonized vs. resistant mice after C. tropicalis colonization (WT n = 32; Tg n = 34). (C) Quantification of CFUs of C. tropicalis in feces from WT and Tg littermates during the course of the experiment (mean shown, n = 12 samples per group). (D) Scatter plots, frequencies, and absolute numbers of IL-17–producing CD4+ T cells in colonic LP at day 7 after colonization with C. tropicalis (n = 4–7 samples per group). Results shown in B are pooled from five independently performed experiments. Results shown in C are pooled from three independently performed experiments. Results shown in D are representative of three independently performed experiments with similar results. Statistics: Data are presented as mean ± SEM where dots represent individual mice. Fisher’s exact test was used to analyze results in B, and two-way ANOVA followed by Holm–Sidak post hoc test was used to analyze results in C and D. *, P < 0.05, ***, P < 0.001, and ****, P < 0.0001.
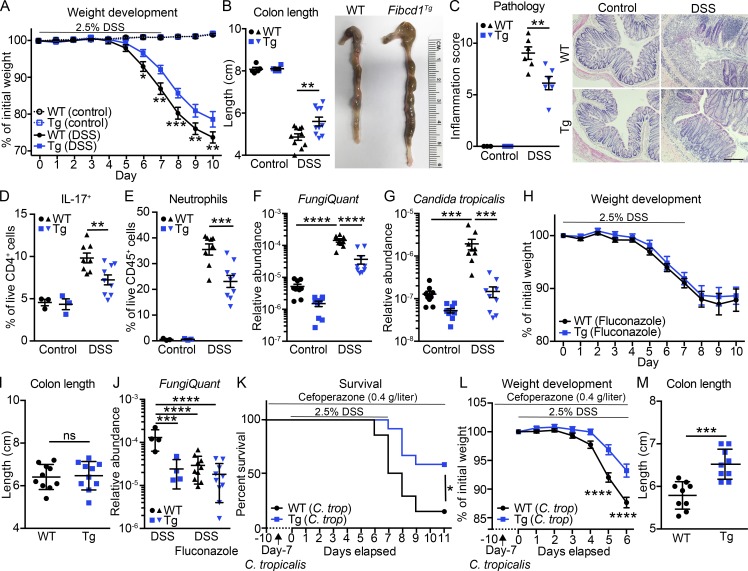
Since fungal dysbiosis has been associated with exacerbated inflammation in animal models of intestinal inflammation and in patients suffering from IBD (Iliev et al., 2012; Li et al., 2014; Lewis et al., 2015; Liguori et al., 2016; Wheeler et al., 2016; Sokol et al., 2017; Leonardi et al., 2018), we next investigated the potential role of IEC-restricted FIBCD1 expression in regulating intestinal inflammation using the dextran sodium sulfate (DSS)–induced model of intestinal damage and inflammation. When exposed to DSS in the drinking water, Fibcd1Tg mice exhibited significantly reduced weight loss compared with WT littermates (Fig. 4 A). At the time of sacrifice, Fibcd1Tg mice displayed significantly reduced colonic shortening and decreased intestinal inflammation compared with WT littermates (Fig. 4, B and C). Consistent with more severe disease, WT mice exhibited a significant increase in the absolute number and frequencies of Th17 cells, as well as infiltrating Ly6G+ neutrophils in colonic LP compared with Fibcd1Tg littermates (Fig. 4, D and E; and Fig. S3, A and B). Increased inflammation was also associated with a significant increase of Ly6chigh inflammatory monocytes in WT mice compared with Fibcd1Tg littermates, whereas the frequencies of regulatory T cells and innate lymphoid cells (ILCs) were unaffected by IEC-specific expression of FIBCD1 after DSS treatment (Fig. S3, C–E). Collectively, these data indicate that IEC-restricted expression of FIBCD1 reduces disease severity in DSS-induced intestinal inflammation.
Figure 4.
Expression of FIBCD1 by IECs ameliorates DSS-induced intestinal inflammation. (A–G) WT and Fibcd1Tg (Tg) littermates were exposed to 2.5% (wt/vol) DSS in the drinking water for 7 d followed by 3 d of regular water. (A) Weight development of mice during the course of the experiment (mean shown, n = 4–12 mice per group). (B) Assessment of colonic shortening at time of sacrifice (n = 4–10 samples per group). (C) Degree of inflammation examined by histopathology (n = 3–6 samples per group). Representative H&E-stained sections are depicted. Scale bar, 200 µm. (D) Frequencies of CD45+CD3+CD4+IL-17+ T cells in colonic LP (n = 3–9 samples per group). (E) Frequencies of infiltrating CD45+CD11b+Ly6G+ neutrophils in colonic LP (n = 3–9 samples per group). (F and G) Quantification of relative fungal abundance (FungiQuant) and specific burden of C. tropicalis before DSS exposure and at time of sacrifice (n = 9 samples per group). (H–J) WT and Tg littermates treated with fluconazole before being exposed to 2.5% (wt/vol) DSS in the drinking water for 7 d followed by 3 d of regular water. Fluconazole treatment (0.5 g/liter in drinking water) was initiated day −3 and maintained throughout the experiment. ns, not significant. (H) Weight development of mice during the course of the experiment (mean shown, n = 10 samples per group). (I) Assessment of colonic shortening at time of sacrifice (n = 10 samples per group). (J) Quantification of total fungal abundance before DSS exposure and at time of sacrifice (n = 4–10 samples per group). (K–M) WT and Tg littermates were colonized with C. tropicalis as described in Fig. 3 A before being exposed to 2.5% (wt/vol) DSS in the drinking water for 7 d followed by 4 d of regular water. Cefoperazone treatment was maintained throughout the experiment. (K) Kaplan–Meier survival curves of WT and Tg littermates colonized with C. tropicalis followed by DSS treatment (n = 9–14 samples per group). (L) Weight development of C. tropicalis–colonized mice during the first 6 d of DSS treatment (mean shown, n = 21–23 samples per group). (M) Assessment of colon length at time of sacrifice (n = 9 samples per group). Results shown in A, H, K, and L are pooled from three independently performed experiments. Results shown in F, G, I, J, and M are pooled from two independently performed experiments. Results shown in B–E are representative of at least three independently performed experiments with similar results. Statistics: Data are presented as mean ± SEM where dots represent individual mice. Two-way ANOVA followed by Holm–Sidak post hoc test was used to analyze results in A–H, J, and L, unpaired Student’s t test was used to analyze data in I and M, and log-rank Mantel–Cox test was used to analyze results in K. *, P < 0.05, **, P < 0.01, ***, P < 0.001, and ****, P < 0.0001.
Since outgrowth of Candida spp. has been reported in DSS-driven intestinal inflammation (Jawhara et al., 2008), we performed a quantitative assessment by qPCR of fungal burden, as well as the specific burden of prevalent C. tropicalis before and after DSS treatment. In WT mice, we observed a striking increase in general fungal abundance, as well as specific burden of C. tropicalis after DSS treatment, an increase that was significantly diminished in Fibcd1Tg littermates (Fig. 4, F and G). Taken together, these data suggest that fungal outgrowth and in particular outgrowth of C. tropicalis may contribute to the severity of intestinal inflammation.
To further examine the potential significance of fungal outgrowth on disease severity, we next subjected WT and Fibcd1Tg littermates to the antifungal drug fluconazole in combination with DSS exposure. Short-term administration of fluconazole, targeting most Candida spp. and other dimorphic fungi, has previously been shown to ameliorate DSS-induced inflammation in susceptible mice (Iliev et al., 2012; Wang et al., 2016), whereas long-term treatment has been reported to increase DSS-induced disease severity (Wheeler et al., 2016). Short-term treatment with fluconazole in mice exposed to DSS in the drinking water ameliorated disease severity in WT mice to levels comparable to Fibcd1Tg littermates when assessed for weight loss and colon length (Fig. 4, H and I). Moreover, the DSS-driven induction of Th17 responses and neutrophil recruitment in fluconazole-treated mice were found to be comparable in WT and Fibcd1Tg littermates (Fig. S3, F and G). Further, treatment with fluconazole resulted in a significant reduction of total fungal burden as well as specific burden of C. tropicalis in WT mice after exposure to DSS to levels comparable to Fibcd1Tg littermates (Fig. 4 J and Fig. S3 H).
To assess whether FIBCD1-mediated alterations in the composition of gut-resident mycobiota directly impact the severity of intestinal inflammation observed during DSS-induced colitis, we colonized antibiotic-treated WT and Fibcd1Tg littermates with C. tropicalis and subjected them to DSS treatment 7 d after colonization. WT mice colonized with C. tropicalis displayed a significantly increased mortality compared with colonized Fibcd1Tg littermates, as well as a significant increased weight loss and reduced colon length when assessed after 6 d of DSS exposure (Fig. 4, K–M). Notably, only comparable levels of infiltrating Ly6G+ neutrophils were observed during early exposure to DSS in WT and Fibcd1Tg littermates (Fig. S3 I). Yet, consistent with exacerbated disease, as well as increased burden of C. tropicalis, WT mice exhibited a significant increase in the frequency of Th17 cells in colonic LP compared with Fibcd1Tg littermates (Fig. S3 J). Collectively, these data highlight a central role for intestinal fungi in modulating intestinal inflammation and illustrate a functional role for FIBCD1 as a novel pattern recognition receptor that can limit fungal-driven intestinal inflammation.
Emerging data indicate that alterations in the composition of the fungal mycobiome, coupled with fungal outgrowth and translocation across the epithelial barrier into the intestinal mucosa, are associated with prolonged antibiotic treatment, immunodeficiencies, and chronic inflammatory diseases like IBD (Brown et al., 2012; Li et al., 2014; Lewis et al., 2015; Liguori et al., 2016; Wheeler et al., 2016; Sokol et al., 2017). In this report, we identify a novel molecular pathway through which IECs lining the intestinal tract can recognize and control the mycobiota via chitin-binding FIBCD1. Specifically, IEC-intrinsic FIBCD1 expression can limit fungal colonization and fungal dysbiosis and reduce fungal-driven exacerbation of intestinal inflammation. These findings indicate that in addition to C-type lectin receptors, TLRs, nucleotide oligomerization domain (NOD)–like receptors, and other microbial sensors, FIBCD1 is a previously unknown evolutionarily conserved pattern recognition receptor that regulates mammalian-fungal interactions at the intestinal barrier surfaces.
Materials and methods
Mice
Mice overexpressing FIBCD1 specifically in the intestinal epithelium (Fibcd1Tg) were generated using the Villin promoter to drive transcription of Fibcd1 following an approach described previously (Pinto et al., 1999). In brief, murine full-length Fibcd1 was amplified from cDNA using the primers 5′-GTGCGTACGATGGTCCACGAGCGGTG-3′ and 5′-GTGACGCGTCTAGCGGTCCTCACGGACC-3′, and inserted via BsiWI and MluI restriction sites into the pBS KS Villin MES vector kindly provided by S. Robine (Institute Curie, Paris, France). Following verification by sequencing, the prepared targeting vector was digested with SalI and purified before being injected into the pronuclei of fertilized oocytes of C57BL/6N mice (Taconic). Mice carrying the transgene were identified by PCR analysis of tail genomic DNA using the primers 5′-AAAGCCGGGTGGGCAGGGTA-3′ and 5′-GCTGCCCACGGTCTTCCACC-3′. Amplification was performed according to the following: 3 min at 95°C; 30 cycles of 95°C (30 s), 68°C (30 s), and 72°C (30 s); and a final 3 min elongation at 72°C. Stable Fibcd1Tg lines were established by backcrossing transgene-carrying founder mice with C57BL/6J mice (Jackson Laboratory). Fibcd1Tg littermates were identified among offspring and analyzed extensively by qPCR to ensure stable and singular integration of the transgene. All experiments in this study were conducted using Fibcd1Tg/- mice (heterozygote for the transgene) derived from one transgenic founder backcrossed onto the C57BL/6J background for a minimum of eight generations (N8-13). All experiments were performed using sex- and age-matched littermates that were cohoused in litters of mixed genotypes if not specifically noted. All mice were maintained under specific pathogen–free conditions and were used in accordance with the Institutional Animal Care and Use Committee guidelines at Weill Cornell Medicine.
Recombinant FIBCD1 variants
Recombinant variants of mouse FIBCD1 were produced by PCR using IMAGE clone (ID: 6848779) as template. Full-length mouse FIBCD1 (mFIBCD1FL) and the putative ectodomain of mouse FIBCD1 (mFIBCD1Ecto) were generated using the reverse primer 5′-TGGTCTTGCTGGAAGATGG-3′ and the forward primers 5′-CTAGCGGTCCTCACGGAC-3′ and 5′-AACCACACCCACACACCAG-3′, respectively. The PCR fragments were cloned into the pcDNA/FRT/V5-His vector (mFIBCD1FL) or pSecTag/FRT/V5-His vector (mFIBCD1Ecto), according to the manufacturer’s recommendations (Thermo Fisher Scientific). Selected point mutations were generated using the Quickchange XL Site-directed mutagenesis system according to the manufacturer’s instructions (Agilent Technologies). Stable integration and expression of the mouse FIBCD1 variants in the mammalian Flp-in cell lines HEK293 or CHO (Thermo Fisher Scientific), as well as the expression and purification of the FIBCD1FReD in insect cells were performed as described previously (Schlosser et al., 2009; Thomsen et al., 2010).
Characterization of recombinant variants of mouse FIBCD1
Characterization of acetyl-group binding specificity by ELISA-based approaches, flow cytometry, and fluorescence confocal microscopy were performed as described previously (Schlosser et al., 2009; Thomsen et al., 2010) using the following antibodies: polyclonal chicken antibody raised against recombinant mFIBCD1Ecto (Davids Biotechnology) was used for ELISA and fluorescence microscopy, and mouse anti-FIBCD1 monoclonal antibody raised against FIBCD1FReD (clone 11–14-10) was used for flow cytometry. Pull-down experiments and Western blotting were performed as described previously (Schlosser et al., 2009; Thomsen et al., 2010), using polyclonal chicken antibody raised against recombinant mFIBCD1Ecto (Davids Biotechnology) and alkaline-phosphatase conjugated anti-chicken IgG (Sigma-Aldrich).
Isolation of cells from mouse intestinal tissue and mesenteric LNs (MLNs)
Intestinal tissue was removed, cleaned from remaining fat tissue, and opened longitudinally before it was washed in ice-cold PBS (Corning). Peyer’s patches from SI were identified and eliminated. Dissociation of epithelial cells was performed by incubation on a shaker at 37°C in PBS containing 2% FBS, 5 mM EDTA, and 1 mM dithiothreitol (all from Thermo Fisher Scientific) twice for 15 min. After each step, samples were vortexed, and the epithelial cell fraction was passed through a 100 µm filter and collected. The remaining tissue was chopped into small pieces, and enzymatic digestion was performed on a shaker at 37°C for 45 min in RPMI 1640 medium (Corning) containing 5% FBS, 0.4 U/ml dispase (Thermo Fisher Scientific), 1 mg/ml collagenase III (Worthington), and 20 µg/ml DNaseI (Sigma-Aldrich). Dissociated leukocytes were passed through a 70 µm filter and collected. The remaining tissue (stromal fraction) was washed twice in ice-cold PBS and then homogenized in TRIzol. Collected epithelial cells and leukocytes were further enriched by a 20%/40% or 40%/80% Percoll gradient centrifugation (Sigma-Aldrich), respectively, before further processing.
MLNs were removed and cleaned from remaining fat tissue before being chopped into smaller pieces and incubated in RPMI medium containing 5% FBS, 20 µg/ml DNaseI and 0.3 mg/ml Liberase TL (Roche) for 20 min at 37°C. Following incubation specimens were mechanically dissociated and filtered through a 70 µm cell strainer.
Isolation of cells from human tissues
Human organ donor material was obtained through an approved research protocol and material transfer agreement with LiveOnNY as described before (Wojno et al., 2015). Donors, three males and six females (LiveOnN donor ID: 309, 310, 311, 313, 318, 320, 335, 336, and 337; average weight ± SD, 82 ± 22 kg) were tested to be HIV−, hepatitis B−, and hepatitis C−, and did not have chronic disease or cancer. This work does not qualify as “human subject” research as confirmed by the institutional review board of Columbia University. Tissues were collected after the donor organs were flushed with preservation solution. Isolation and preparation of compartmentalized cell fractions (IECs, LP leukocytes, and remaining stroma) were performed as described previously (Klose et al., 2017). Recovered IECs and LP leukocytes were stained with fluorescent-labeled antibodies and sort-purified. In addition, RNA was extracted from isolated IECs, LP leukocytes, remaining stroma, and whole unfractionated tissue using TRIzol.
Flow cytometry and cell sorting
Fc receptors were routinely saturated with anti-CD16/32-blocking antibody (mouse; BioLegend) or FcR Blocking Reagent (human; Miltenyi Biotec) before staining according to the manufacturer’s instructions. Dead cells were excluded with Fixable Aqua Dead Cell Stain or DAPI (both from Thermo Fisher Scientific). Single-cell suspensions were incubated on ice in PBS containing 2% FBS and 1 mM EDTA supplemented with antibodies. For surface staining of mouse specimens or exclusion of lineage-positive cells, the following antibodies against CD4 (GK1.5), CD45 (30F11), CD3ε (145-2C11), B220 (RA3-6B2), CD11b (M1/70), CD11c (N418), Ly6G (1A8), CD326 (G8.8), FIBCD1 (11–14-10), CD19 (eBio1D3), CD5 (53–7.5), CD64 (X54-5/7.1), FcεRI (MAR-1), Ly6c (HK1.4), MHCII (M5/114.15.2), CD90.2 (30-H12), and CD127 (A7R34) were used. Intracellular staining with anti–IL-5 (TRFK5), IL-13 (eBio13A), IL-17 (eBio17B7), and IFNγ (XMG1.2) antibodies was performed using Cytofix/Cytoperm Fixation/Permeabilization Solution and Perm/Wash buffer (both from BD Biosciences). Intra-nuclear staining for FoxP3 (FJK-16s), RoRγt (B2D), GATA3 (TWAJ), and Tbet (eBio4B10) was performed using the Foxp3 transcription factor staining buffer set (eBioscience). Human samples were surface stained using antibodies against CD45 (HI30), CD326 (9C2), and FIBCD1 (HG-Hyb-12-2) before analysis. All antibodies used in flow cytometry analyses except anti-FIBCD1 antibodies were purchased from eBioscience, BioLegend, or BD Biosciences. All flow cytometry experiments were acquired using a five-laser, 18-color custom configuration Fortessa flow cytometer and the FACS Diva software (BD Biosciences) or sort-purified by using a five-laser, 18-color custom configuration FACSAria cell sorter (BD Biosciences). Collected data were analyzed with FlowJo V10.4.1 software (TreeStar).
Quantitative RT-PCR and PCR
For transcriptional analyses, tissue samples and sorted cells were homogenized in TRIzol (Thermo Fisher Scientific) and stored at −80°C until processed. RNA was extracted with chloroform, and RNA concentration was determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific). Reverse transcription of total RNA was performed using the High Capacity cDNA Reverse Transcription kit according to the manufacturer’s recommendations (Thermo Fisher Scientific). Quantification of expression was detected on a QuantStudio 6 Flex Real-Time PCR (Thermo Fisher Scientific) using the following TaqMan Gene Expression Assays (Applied Biosystems): mouse Fibcd1 (Mm00619121_m1) and human FIBCD1 (Hs00262524_m1); the following SYBR Green primers: Muc2 (QT01060773), Muc5ac (QT01161104), Cd326 (QT00248276), Icam1 (QT00155078), Cldn1 (QT00159278), Cldn2 (QT01059212), Ccl2 (QT00167832), Ccl20 (QT02326394), Il-1b (QT01048355), Il-18 (Qt00171129), Il-25 (Qt00134645), Il-33 (Qt00135170), and Reg3g (QT00147455) supplied by Qiagen; and the following SYBR Green primers: Zo (6678355a1), Tslp (10946698a1), and Reg3a (6755308a) obtained from Primerbank (Wang et al., 2012). Gene expression was normalized as n-fold difference to mouse Hprt1 (Mm00446968_m1) or human HPRT1 (Hs02800695_m1) according to the cycling threshold. Analyses of the relative abundance of bacteria and fungi by qPCR was performed using the standard SYBR green protocol on the QuantStudio 6 Flex using the following primer pairs: 16S (5′-ACTCCTACGGGAGGCAGCAGT-3′/5′-ATTACCGCGGCTGCTGGC-3′; Barman et al., 2008), Bacteroides (5′-GGTTCTGAGAGGAGGTCCC-3′/5′-GCTGCCTCCCGTAGGAGT-3′; Barman et al., 2008), Firmicutes (5′-GGAGYATGTGGTTTAATTCGAAGCA-3′/5′-AGCTGACGACAACCATGCAC-3′; Guo et al., 2008), SFB (5′-AGGAGGAGTCTGCGGCACATTAGC-3′/5′-TCCCCACTGCTGCCTCCCGTAG-3′; Suzuki et al., 2004), 18S/FungiQuant (5′-GGRAAACTCACCAGGTCCAG-3′/5′-GSWCTATCCCCAKCACGA-3′; Liu et al., 2012), Candida spp. (5′-CCTGTTTGAGCGTCGTTT-3′/5′-TCCTCCGCTTATTGATAT-3′; Schabereiter-Gurtner et al., 2007), and C. tropicalis (5′-TTTGGTGGCGGGAGCAATCCT-3′/5′-CGATGCGAGAACCAAGAGATCCGT-3′; Iliev et al., 2012). Relative abundance was standardized to input DNA using 16S as reference. Since DSS has been identified as an inhibitor of PCR, all samples from DSS models were supplemented with 0.01 g/liter spermine (Sigma-Aldrich) before analysis and run in log-fold dilutions to ensure correct quantification (Krych et al., 2018).
Intestinal barrier integrity assay
Intestinal barrier integrity was assessed as previously described (An et al., 2007). In brief, 8–10-wk-old Fibcd1Tg and WT littermates were deprived of food and water for 4 h before being fed 200 µl of FITC-dextran (4 kD; Sigma-Aldrich) at 600 mg/kg by oral gavage. Blood was collected 4 h later by retro-orbital bleeding, and fluorescence intensity in the serum was measured (excitation, 490 nm; emission, 530 nm) using a Victor3 fluorometer (PerkinElmer). The serum concentration of FITC-dextran was subsequently determined from a standard curve of FITC-dextran serially diluted in mouse serum.
Helminth infections
T. muris was maintained and E/S antigens prepared as described in Zaph et al. (2006). Infection was established by administering 200 embryonated eggs by oral gavage, after which worm burden and immunological readouts were assessed at day 18 after infection. For H. polygyrus infection, 250 purified infective L3 larvae were administered by oral gavage, after which worm burden and immunological readouts were assessed at day 18 after infection. For N. brasiliensis infection, 500 purified infective L3 larvae were injected subcutaneously, after which worm burden and immunological readouts were accessed at day 7 after infection.
In vitro stimulation and ELISA
For cytokine staining, bulk MLNs or colonic LP leukocytes were incubated in RPMI containing 10% FBS, 10 mM Hepes (Gibco), and 100 U/ml penicillin/100 µg/ml streptomycin (Gibco) in 96-well microtiter plates for 4 h at 37°C in 5% CO2. Incubation was performed in the presence of 1 µg/ml PMA, 1 µg/ml ionomycin, and 10 µg/ml brefeldin A (all from Sigma-Aldrich). For T. muris antigen stimulation, isolated MLNs (2 × 106 cells) were cultured in 48-well microtiter plates for 48 h (37°C and 5% CO2) in 0.5 ml DMEM media (Corning) supplemented with 10% FBS, 10 mM Hepes, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were stimulated with 50 µg/ml T. muris E/S antigen prepared according to Zaph et al. (2006) or PBS serving as control. Detection of IL-4 (eBioscience), IL-5 (eBioscience), IL-13 (R&D Systems), and IFNγ (eBioscience) in the supernatants of cultured cells and total IgE in serum (eBioscience) was performed according to the manufacturer’s instructions. Quantification T. muris–specific IgG1 in serum was performed using microtiter plates coated with 5 µg/ml T. muris E/S antigens followed by detection using anti-IgG1 antibodies (BD Biosciences) according to the manufacturer’s recommendations.
Histology
Intestinal tissues were harvested and fixed in 4% paraformaldehyde (PFA; Bioworld) before paraffin embedding. From each specimen, two sections (spaced 500 µm apart) were obtained and used for the assessment. Periodic acid–Schiff (PAS) staining and H&E staining were performed by Idexx. Images were acquired using an inverted Nikon Eclipse Ti microscope. Quantification of goblet cell hyperplasia and inflammation was performed using ImageJ v1.49 software.
Fecal DNA isolation for ITS-1 and 16S sequencing
Fibcd1Tg and WT littermates were cohoused until 6 wk of age before being separated in cages based on genotype. After 2 wk of housing based on genotype, two or three fecal pellets from each mouse were collected, snap-frozen in liquid nitrogen, and stored at −80°C until processed. Fecal DNA was isolated following lyticase (Sigma-Aldrich) treatment, bead-beating, and processing using the QIAmp Fast stool DNA kit (Qiagen) as described previously (Tang et al., 2015).
16S rRNA bacterial sequencing
16S rRNA gene sequencing was performed using methods adapted from the National Institutes of Health Human Microbiome Project (Peterson et al., 2009). In brief, the 16S rDNA V4 region was amplified by PCR and sequenced on the MiSeq platform (Illumina) using a 2 × 250-bp paired-end protocol. The read pairs were demultiplexed based on unique molecular barcodes, and subsequently merged using USEARCH v7.0.1090 (Edgar, 2010), allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at the first base with Q5, after which a quality filter was applied discarding reads with >0.05 expected errors. Merged and filtered 16S sequences were clustered into operational taxonomic units (OTUs) using the UPARSE algorithm (Edgar, 2013) with a similarity cutoff value of 97%. OTUs were mapped to a modified version of the SILVA Database containing only the 16S V4 region to determine taxonomies. Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs. OTU tables were rarefied before calculation of Bray–Curtis dissimilarities, with rarefaction depth equal to either 10,000 reads or the minimum sample depth, whichever was largest. 16S sequencing data have been submitted to the National Center for Biotechnology Information Sequence Read Archive under Bioproject accession no. PRJNA560208.
ITS-1 fungal sequencing
Deep sequencing of fungal ITS-1 region was performed essentially as described previously (Leonardi et al., 2018). In brief, the fungal ITS-1 region was amplified by PCR and sequenced using a 2 × 300-bp paired-end protocol on the MiSeq platform (Illumina). All libraries were subjected to quality control using qPCR, DNA 1000 Bioanalyzer (Agilent), and Qubit (Life Technologies) to validate and quantitate library construction before preparing a Paired End flow cell. The generated raw FASTQ ITS-1 sequencing data were filtered to enrich for high-quality reads, removing the adapter sequence by cutadapt v1.4.1 or any reads that did not contain the proximal primer sequence (Tang et al., 2015). Sequence reads were then quality-trimmed by truncating reads not having an average quality score of 20 (Q20) over a 3-bp sliding window and removing reads shorter than 100 bp (Tang et al., 2015). These high-quality reads were then aligned to Targeted Host Fungi ITS-1 database, using BLAST v2.2.22 and the pick_otus.py pipeline in the QIIME v1.6 wrapper with an identity percentage ≥97% for OTU picking (Altschul et al., 1990). The alignment results were subsequently tabulated across all reads, using the accession identifier of the ITS reference sequences as surrogate OTUs and using a Perl script (Tang et al., 2015). The R packages Phyloseq (1.16.2) and Vegan (2.4–3) were used for determining community properties such as Bray–Curtis index, principal coordinate analysis (PCoA) scaling of Bray–Curtis dissimilarities, and relative abundance at various taxonomic levels. R version 3.3.1 was used. ITS-1 sequencing data have been submitted to the National Center for Biotechnology Information Sequence Read Archive under Bioproject accession no. PRJNA560213.
Recognition and binding of FIBCD1 to C. tropicalis
C. tropicalis (ATCC 750) was obtained from the American Type Culture Collection. C. tropicalis used for experiments was cultured in aerobic conditions at 37°C in Sabouraud Dextrose Broth (EMD Chemicals). Assessment of FIBCD1 binding was performed as follows: Overnight cultures of fungi were harvested, washed in PBS, and either fixed in 4% PFA for 1 h on ice or further processed alive. After PFA removal and washing, 2 × 107 cells were stained with 1 µg FIBCD1FReD (Thomsen et al., 2010) in Tris-buffered saline containing 1% BSA supplemented with 5 mM CaCl2, 5 mM EDTA, or 50 mM GlcNAc (all from Sigma-Aldrich) for 2 h on ice. After extensive washing in corresponding binding buffers, detection of bound FIBCD1FReD was performed in Tris-buffered saline (1% BSA and 5 mM CaCl2) using 1 µg/ml allophycocyanin-labeled anti-FIBCD1 antibody (clone 11–14-10) for 1 h on ice. Staining for exposed chitin was performed in parallel using 5 µg/ml Alexa Fluor 350–conjugated WGA (Thermo Fisher Scientific). Binding of FIBCD1 was visualized using a Nikon Eclipse Ti microscope (Nikon) or assessed by flow cytometry using a Fortessa flow cytometer and the FACS Diva software.
C. tropicalis colonization and fungal immune responses
Colonization with C. tropicalis was performed as described in Leonardi et al. (2018). In brief, 6–8-wk-old cohoused Fibcd1Tg and WT littermates were separated in cages based on genotype before the start of the experiment. 0.4 g/liter cefoperazone (Sigma-Aldrich) supplemented with 4 g/liter artificial sweetener (Sweet’n Low) was provided ad libitum in drinking water from day −3 precolonization and continued throughout the experiment. Mice were fed 2.5 × 107 C. tropicalis by gavage at day 0, after which fungal colonization was assessed by CFUs daily until termination at indicated time points. Mice with CFU counts below 104 colonies per gram feces were considered resistant to colonization. Candida-specific immune responses were assessed by flow cytometry in cells isolated from colon LP.
DSS-induced acute intestinal inflammation
Chemical induction of intestinal inflammation using DSS was performed using 8–10-wk-old sex-matched Fibcd1Tg and WT littermates. In brief, acute intestinal inflammation was induced by oral administration of 2.5% (wt/vol) DSS (36–50 kD; MP Biomedicals) in drinking water for 7 d, followed by ordinary tap water. During this time period, the mice were weighted daily and subsequently sacrificed at day 10. At the time of sacrifice, fecal specimens and colon were collected for immunological analyses by ELISA, histology, qPCR, and flow cytometry. In C. tropicalis–colonized mice as well as corresponding controls, treatment with 0.4 mg/ml cefoperazone supplemented with 4 g/liter artificial sweetener (Sweet’n Low) was continued throughout the experiments. DSS treatment was initiated at day 8 after colonization in these experiments. 0.5 g/liter fluconazole (Sigma-Aldrich) supplemented with 4 g/liter artificial sweetener (Sweet’n Low) was provided ad libitum in drinking water from 3 d before exposure to DSS and continued throughout the experiments.
Histological assessment of inflammation
Assessment of inflammation was performed essentially as described by Nguyen et al. (2011). In brief, colonic sections were H&E-stained and assessed for disease severity by trained personnel blinded to treatment and genotype. Scoring was based on three parameters; severity of inflammation (0–3), crypt damage (0–5), and ulceration (0–3). The combined values were added to give a histological score between 0 and 11. Images were acquired using an inverted Nikon Eclipse Ti microscope.
Monoclonal antibodies raised against FIBCD1
Mouse monoclonal anti-FIBCD1 antibody HG-HYB-12-2 was produced as previously described (Schlosser et al., 2009). Mouse monoclonal anti-FIBCD1FReD antibody clone 11–14-10 was produced by immunization of Fibcd1−/− C57BL/6N mice. The mice were immunized twice with 20 µg of recombinant FIBCD1FReD using GERBU as adjuvant (GERBU Biotechnik) with more than 2 wk between each immunization. The mice were boosted once by an intra-peritoneal injection with 20 µg recombinant FIBCD1FReD without adjuvant 3 d before isolation of B cells. Hybridoma cells were produced by fusion between isolated B cells and myeloma cells and adapted for serum-free media. Secreted antibodies were purified using a protein G column on a fast protein liquid chromatography apparatus (ÄKTA). Generation of antibodies was performed under license from the Danish National Animal Experiments Inspectorate (reference no. 2012-15-2934-00076). Antibodies used for flow cytometry were labeled with allophycocyanin according to the manufacturer’s recommendations (Thermo Fisher Scientific).
Statistical analysis
All statistical tests were performed with Prism v6.0 (GraphPad). Two-way ANOVA followed by Holm–Sidak post hoc test was used to compare experimental groups with two independent variables. One-way ANOVA followed by Tukey post hoc tests were applied to all experimental results with one independent variable in which more than two groups were compared. In analyses with only two groups and one independent variable, Student’s t test was applied. All weight curves were analyzed by two-way repeated measures ANOVA followed by post hoc testing corrected for multiple comparisons using the Tukey method. For comparisons of Kaplan–Meier survival curves, log-rank (Mantel–Cox) tests were used. For analysis of C. tropicalis colonization efficiency, two-tailed Fisher’s exact test was used. Error bars depict the SEM unless otherwise stated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows expression of Fibcd1 in mouse intestinal tissues and the initial characterization of transgenic mice specifically overexpressing FIBCD1 in the IECs. Fig. S2 shows that epithelial-specific expression of FIBCD1 does not influence immunity to helminth infection or changes the composition of gut bacteria. Fig. S3 expands upon the observed change to the immune cell composition in colon LP of WT and Fibcd1Tg littermates in models of DSS-driven intestinal inflammation.
Supplementary Material
Acknowledgments
We thank Artis laboratory members and Sonnenberg laboratory members for discussion and critical reading of the manuscript. We also thank Cure for IBD and the Rosanne H. Silberman Foundation for their kind support of the Jill Roberts Institute.
This work was supported by grants from the Novo Nordisk Foundation (grant 14052 to J.B. Moeller), Fonden til lægevidenskabens Fremme (grant 16-248 to J.B. Moeller), Hørslev-Fonden (grant 203866 to J.B. Moeller), Augustinus Fonden (grant 15-1374 to J.B. Moeller), Aage og Johanne Louis-Hansens Fond (grants L-153 and 18-2B-2769 to J.B. Moeller), Civilingeniør Frode V Nyegaard og Hustrus Fond (grant to J.B. Moeller), Dagmar Marshalls Fond (grant to J.B. Moeller), Knud Højgaards Fond (grant 48-780 to J.B. Moeller), Crohn’s and Colitis Foundation of America (grants to I. Leonardi and D. Artis), the Jill Roberts Institute (grant to G.G. Putzel), the National Institutes of Health (grants DK113136 and AI137157 to I.D. Iliev, and grants AI074878, AI095466, AI095608, and AI102942 to D. Artis), the Burroughs Wellcome Fund (grant to D. Artis), Cure for IBD (grant to D. Artis), and the Rosanne H. Silbermann Foundation (grant to D. Artis).
D. Artis has contributed to scientific advisory boards at MedImmune, Pfizer, Food Allergy Reseach and Education, and the Kenneth Rainin Foundation. All other authors declare no competing financial interests.
Author contributions: J.B. Moeller, I. Leonardi, A.-L. Flamar, N.J. Bessman, A. Schlosser, G.G. Putzel, T. Thomsen, M. Hammond, C.S. Jepsen, G.L. Sorensen, I.D. Iliev, U. Holmskov, and D. Artis designed and performed the research. J.B. Moeller, U. Holmskov, and D. Artis analyzed the data. K. Skjødt generated and provided monoclonal anti-FIBCD1 antibodies, E.-M. Füchtbauer provided the Fibcd1Tg mice and D.L. Farber provided the human intestinal samples through LiveOnNY. J.B. Moeller and D. Artis wrote the manuscript with input from the other authors.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- An G., Wei B., Xia B., McDaniel J.M., Ju T., Cummings R.D., Braun J., and Xia L.. 2007. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 204:1417–1429. 10.1084/jem.20061929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M., Unold D., Shifley K., Amir E., Hung K., Bos N., and Salzman N.. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76:907–915. 10.1128/IAI.01432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., and Harrison O.J.. 2017. Homeostatic Immunity and the Microbiota. Immunity. 46:562–576. 10.1016/j.immuni.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodaczewska K., Donskow-Łysoniewska K., and Doligalska M.. 2015. Chitin, a key factor in immune regulation: lesson from infection with fungi and chitin bearing parasites. Acta Parasitol. 60:337–344. 10.1515/ap-2015-0047 [DOI] [PubMed] [Google Scholar]
- Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., and White T.C.. 2012. Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Cliffe L.J., and Grencis R.K.. 2004. The Trichuris muris system: a paradigm of resistance and susceptibility to intestinal nematode infection. Adv. Parasitol. 57:255–307. 10.1016/S0065-308X(04)57004-5 [DOI] [PubMed] [Google Scholar]
- Coker O.O., Nakatsu G., Dai R.Z., Wu W.K.K., Wong S.H., Ng S.C., Chan F.K.L., Sung J.J.Y., and Yu J.. 2019. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut. 68:654–662. 10.1136/gutjnl-2018-317178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake L., Korchev Y., Bashford L., Djamgoz M., Wakelin D., Ashall F., and Bundy D.. 1994. The major secreted product of the whipworm, Trichuris, is a pore-forming protein. Proc. Biol. Sci. 257:255–261. 10.1098/rspb.1994.0123 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 10:996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Eichenberger R.M., Talukder M.H., Field M.A., Wangchuk P., Giacomin P., Loukas A., and Sotillo J.. 2018. Characterization of Trichuris muris secreted proteins and extracellular vesicles provides new insights into host-parasite communication. J. Extracell. Vesicles. 7:1428004 10.1080/20013078.2018.1428004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elieh Ali Komi D., Sharma L., and Dela Cruz C.S.. 2018. Chitin and Its Effects on Inflammatory and Immune Responses. Clin. Rev. Allergy Immunol. 54:213–223. 10.1007/s12016-017-8600-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Xia X., Tang R., Zhou J., Zhao H., and Wang K.. 2008. Development of a real-time PCR method for Firmicutes and Bacteroidetes in faeces and its application to quantify intestinal population of obese and lean pigs. Lett. Appl. Microbiol. 47:367–373. 10.1111/j.1472-765X.2008.02408.x [DOI] [PubMed] [Google Scholar]
- Hasnain S.Z., McGuckin M.A., Grencis R.K., and Thornton D.J.. 2012. Serine protease(s) secreted by the nematode Trichuris muris degrade the mucus barrier. PLoS Negl. Trop. Dis. 6:e1856 10.1371/journal.pntd.0001856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I.D., and Leonardi I.. 2017. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat. Rev. Immunol. 17:635–646. 10.1038/nri.2017.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I.D., Funari V.A., Taylor K.D., Nguyen Q., Reyes C.N., Strom S.P., Brown J., Becker C.A., Fleshner P.R., Dubinsky M., et al. . 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 336:1314–1317. 10.1126/science.1221789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhara S., Thuru X., Standaert-Vitse A., Jouault T., Mordon S., Sendid B., Desreumaux P., and Poulain D.. 2008. Colonization of mice by Candida albicans is promoted by chemically induced colitis and augments inflammatory responses through galectin-3. J. Infect. Dis. 197:972–980. 10.1086/528990 [DOI] [PubMed] [Google Scholar]
- Jenkins S.N., and Wakelin D.. 1983. Functional antigens of Trichuris muris released during in vitro maintenance: their immunogenicity and partial purification. Parasitology. 86:73–82. 10.1017/S0031182000057188 [DOI] [PubMed] [Google Scholar]
- Jepsen C.S., Dubey L.K., Colmorten K.B., Moeller J.B., Hammond M.A., Nielsen O., Schlosser A., Templeton S.P., Sorensen G.L., and Holmskov U.. 2018. FIBCD1 Binds Aspergillus fumigatus and Regulates Lung Epithelial Response to Cell Wall Components. Front. Immunol. 9:1967 10.3389/fimmu.2018.01967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klementowicz J.E., Travis M.A., and Grencis R.K.. 2012. Trichuris muris: a model of gastrointestinal parasite infection. Semin. Immunopathol. 34:815–828. 10.1007/s00281-012-0348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S.N., Mahlakõiv T., Moeller J.B., Rankin L.C., Flamar A.L., Kabata H., Monticelli L.A., Moriyama S., Putzel G.G., Rakhilin N., et al. . 2017. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature. 549:282–286. 10.1038/nature23676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krych Ł., Kot W., Bendtsen K.M.B., Hansen A.K., Vogensen F.K., and Nielsen D.S.. 2018. Have you tried spermine? A rapid and cost-effective method to eliminate dextran sodium sulfate inhibition of PCR and RT-PCR. J. Microbiol. Methods. 144:1–7. 10.1016/j.mimet.2017.10.015 [DOI] [PubMed] [Google Scholar]
- Latgé J.P. 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66:279–290. 10.1111/j.1365-2958.2007.05872.x [DOI] [PubMed] [Google Scholar]
- Leonardi I., Li X., Semon A., Li D., Doron I., Putzel G., Bar A., Prieto D., Rescigno M., McGovern D.P.B., et al. . 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science. 359:232–236. 10.1126/science.aao1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., et al. . 2015. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 18:489–500. 10.1016/j.chom.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang C., Tang C., He Q., Li N., and Li J.. 2014. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J. Clin. Gastroenterol. 48:513–523. 10.1097/MCG.0000000000000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Leonardi I., Semon A., Doron I., Gao I.H., Putzel G.G., Kim Y., Kabata H., Artis D., Fiers W.D., et al. . 2018. Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe. 24:847–856.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedke S.C., Miranda D.Z., Gomes K.X., Gonçalves J.L.S., Frases S., Nosanchuk J.D., Rodrigues M.L., Nimrichter L., Peralta J.M., and Guimarães A.J.. 2017. Characterization of the antifungal functions of a WGA-Fc (IgG2a) fusion protein binding to cell wall chitin oligomers. Sci. Rep. 7:12187 10.1038/s41598-017-12540-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori G., Lamas B., Richard M.L., Brandi G., da Costa G., Hoffmann T.W., Di Simone M.P., Calabrese C., Poggioli G., Langella P., et al. . 2016. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohn’s Colitis. 10:296–305. 10.1093/ecco-jcc/jjv209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.M., Kachur S., Dwan M.G., Abraham A.G., Aziz M., Hsueh P.R., Huang Y.T., Busch J.D., Lamit L.J., Gehring C.A., et al. . 2012. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 12:255 10.1186/1471-2180-12-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A., Sharma D., Malireddi R.K.S., Guy C.S., Chang T.C., Olsen S.R., Neale G., Vogel P., and Kanneganti T.D.. 2018. SYK-CARD9 Signaling Axis Promotes Gut Fungi-Mediated Inflammasome Activation to Restrict Colitis and Colon Cancer. Immunity. 49:515–530.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.T., Dalmasso G., Torkvist L., Halfvarson J., Yan Y., Laroui H., Shmerling D., Tallone T., D’Amato M., Sitaraman S.V., and Merlin D.. 2011. CD98 expression modulates intestinal homeostasis, inflammation, and colitis-associated cancer in mice. J. Clin. Invest. 121:1733–1747. 10.1172/JCI44631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peay K.G., Kennedy P.G., and Talbot J.M.. 2016. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 14:434–447. 10.1038/nrmicro.2016.59 [DOI] [PubMed] [Google Scholar]
- Peterson J., Garges S., Giovanni M., McInnes P., Wang L., Schloss J.A., Bonazzi V., McEwen J.E., Wetterstrand K.A., Deal C., et al. NIH HMP Working Group . 2009. The NIH Human Microbiome Project. Genome Res. 19:2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Robine S., Jaisser F., El Marjou F.E., and Louvard D.. 1999. Regulatory sequences of the mouse villin gene that efficiently drive transgenic expression in immature and differentiated epithelial cells of small and large intestines. J. Biol. Chem. 274:6476–6482. 10.1074/jbc.274.10.6476 [DOI] [PubMed] [Google Scholar]
- Preston C.M., and Jenkins T.. 1985. Trichuris muris: structure and formation of the egg polar plugs. Z. Parasitenkd. 71:373–381. 10.1007/BF00928339 [DOI] [PubMed] [Google Scholar]
- Reese T.A., Liang H.E., Tager A.M., Luster A.D., Van Rooijen N., Voehringer D., and Locksley R.M.. 2007. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 447:92–96. 10.1038/nature05746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues M.L., Alvarez M., Fonseca F.L., and Casadevall A.. 2008. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot. Cell. 7:602–609. 10.1128/EC.00307-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Takeuchi O., Vandenbon A., Yasuda K., Tanaka Y., Kumagai Y., Miyake T., Matsushita K., Okazaki T., Saitoh T., et al. . 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11:936–944. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Schabereiter-Gurtner C., Selitsch B., Rotter M.L., Hirschl A.M., and Willinger B.. 2007. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 45:906–914. 10.1128/JCM.01344-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser A., Thomsen T., Moeller J.B., Nielsen O., Tornøe I., Mollenhauer J., Moestrup S.K., and Holmskov U.. 2009. Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J. Immunol. 183:3800–3809. 10.4049/jimmunol.0901526 [DOI] [PubMed] [Google Scholar]
- Shrive A.K., Moeller J.B., Burns I., Paterson J.M., Shaw A.J., Schlosser A., Sorensen G.L., Greenhough T.J., and Holmskov U.. 2014. Crystal structure of the tetrameric fibrinogen-like recognition domain of fibrinogen C domain containing 1 (FIBCD1) protein. J. Biol. Chem. 289:2880–2887. 10.1074/jbc.M113.520577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C., Cohen D., Liguori G., Bourrier A., Nion-Larmurier I., et al. . 2017. Fungal microbiota dysbiosis in IBD. Gut. 66:1039–1048. 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Meek B., Doi Y., Muramatsu M., Chiba T., Honjo T., and Fagarasan S.. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA. 101:1981–1986. 10.1073/pnas.0307317101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Iliev I.D., Brown J., Underhill D.M., and Funari V.A.. 2015. Mycobiome: Approaches to analysis of intestinal fungi. J. Immunol. Methods. 421:112–121. 10.1016/j.jim.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen T., Moeller J.B., Schlosser A., Sorensen G.L., Moestrup S.K., Palaniyar N., Wallis R., Mollenhauer J., and Holmskov U.. 2010. The recognition unit of FIBCD1 organizes into a noncovalently linked tetrameric structure and uses a hydrophobic funnel (S1) for acetyl group recognition. J. Biol. Chem. 285:1229–1238. 10.1074/jbc.M109.061523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Mohapatra A., Nussbaum J.C., Molofsky A.B., Thornton E.E., Ziegler S.F., McKenzie A.N., Krummel M.F., Liang H.E., and Locksley R.M.. 2014. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity. 40:414–424. 10.1016/j.immuni.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Huth S., Moeller J.B., Schlosser A., Marcussen N., Nielsen O., Nielsen V., Sorensen G.L., and Holmskov U.. 2018. Immunohistochemical Localization of Fibrinogen C Domain Containing 1 on Epithelial and Mucosal Surfaces in Human Tissues. J. Histochem. Cytochem. 66:85–97. 10.1369/0022155417743694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright P.O., Hinkle G., Sogin M.L., and Stickel S.K.. 1993. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 260:340–342. 10.1126/science.8469985 [DOI] [PubMed] [Google Scholar]
- Wang T., Pan D., Zhou Z., You Y., Jiang C., Zhao X., and Lin X.. 2016. Dectin-3 Deficiency Promotes Colitis Development due to Impaired Antifungal Innate Immune Responses in the Gut. PLoS Pathog. 12:e1005662 10.1371/journal.ppat.1005662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Fan C., Yao A., Xu X., Zheng G., You Y., Jiang C., Zhao X., Hou Y., Hung M.C., and Lin X.. 2018. The Adaptor Protein CARD9 Protects against Colon Cancer by Restricting Mycobiota-Mediated Expansion of Myeloid-Derived Suppressor Cells. Immunity. 49:504–514.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Spandidos A., Wang H., and Seed B.. 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40(Database issue, D1):D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J., Brown J., Funari V.A., Wang H.L., Crother T.R., et al. . 2016. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 19:865–873. 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojno E.D., Monticelli L.A., Tran S.V., Alenghat T., Osborne L.C., Thome J.J., Willis C., Budelsky A., Farber D.L., and Artis D.. 2015. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8:1313–1323. 10.1038/mi.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypych T.P., and Marsland B.J.. 2018. Antibiotics as Instigators of Microbial Dysbiosis: Implications for Asthma and Allergy. Trends Immunol. 39:697–711. 10.1016/j.it.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Zaph C., Rook K.A., Goldschmidt M., Mohrs M., Scott P., and Artis D.. 2006. Persistence and function of central and effector memory CD4+ T cells following infection with a gastrointestinal helminth. J. Immunol. 177:511–518. 10.4049/jimmunol.177.1.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.