Abstract
The ‘non-linear’ genome, or the spatial proximity of non-contiguous sequences, emerges as an important regulatory layer for genome organization and function, including transcriptional regulation. Here, we review recent genome-scale chromosome conformation mappings (‘Hi-C’) in developing and adult human and mouse brain. Neural differentiation is associated with widespread remodeling of the chromosomal contact map, reflecting dynamic changes in cell-type-specific gene expression programs, with a massive (estimated 20–50%) net loss of chromosomal contacts that is specific for the neuronal lineage. Hi-C datasets provided an unexpected link between locus-specific abnormal expansion of repeat sequences positioned at the boundaries of self-associating topological chromatin domains and associated with monogenic neurodevelopmental and neurodegenerative disease. Furthermore, integrative analyses of cell-type-specific Hi-C and transcriptomes uncovered an expanded genomic risk space interacting with sequences conferring liability for schizophrenia and other cognitive disease. We predict that spatial genome exploration will deliver radically new insights into the brain’s nucleomes in health and disease.
General Introduction to the ‘Spatial Genome’
The 6Gb human diploid genome is spatially organized in the cell nucleus via myriads of chromosomal conformations, many of which bypass the ‘linear genome’ and thereby position stretches of non-contiguous stretches of intra- or, to a lesser degree, inter-chromosomal sequence into close physical proximity with each other. Orderly configuration of chromosomal conformations is thought to be an important prerequisite for proper genome organization and function. Recent articles have discussed the experimental and computational methods commonly used to explore the basic building blocks of mammalian ‘3-dimensional genomes’ (3DG) [1,2]. Here, we discuss some of the recent findings from 3DG studies that employ genome-wide (‘all sequences surveyed against all sequences’) Hi-C DNA-DNA proximity assays, involving chromatin restriction digestion followed by re-ligation, ideally within intact nuclei [2]. It should be noted that the actual proximity of non-contiguous DNA fragments ‘stitched together’ by DNA-ligase treatment—as the critical step in the Hi-C protocol following the chromatin digest—is thought to vary across 2–3 orders of magnitude on the nanometer scale [3] in the interphase nucleus. Importantly, Hi-C studies across a wide range of species and tissues revealed, for each chromosome, various degrees of ‘modular’ organization, or chromatin domains frequently referred to as topologically-associated domains (TADs), with smaller domains (‘subTADs’) nested into them. Each TAD and subTAD is comprised of long stretches of sequence, typically extending across hundreds of kilobases showing much higher chromosomal contact frequencies within as compared outside their domain boundaries, including the neighboring TADs [4]. Furthermore, TADs often assemble inside the nucleus with others TADs, preferentially those with of a similar chromatin architecture, ultimately giving rise to A/B (open/repressive) megabase-scale compartments, or chromosomal megadomains[5]. The function of TADs includes insulation by preventing spurious interactions between elements such as promoters and enhancers, which in turn could lead to aberrant levels of gene transcription, for instance of an oncogene [4,6]. Early reports indicated that TAD boundaries were enriched for a heterogenous set of genomic entities, such as housekeeping genes, transfer RNAs and short interspersed element (SINE) retrotransposons [7]. Furthermore, and somewhat surprisingly, the majority of TADs are considered to be conserved among cell types, and even species [7–10]. However, their potential for insulation and connections to other regions, both measured quantitatively, can be subject to cell-type-specific changes, primed perhaps by transcription factor binding at least during lineage specification [11]. Additionally, TAD insulation is highly correlated with transcription, such that novel TAD or subTAD borders could assemble around promoters of developmentally regulated genes [12].
Furthermore, the spatial genome also includes including tens of thousands of chromosomal loop formations, commonly defined as distinct pairwise contacts that, in Hi-C maps, sharply stand out from the surrounding ‘linear’ genome sequence, often interconnecting specific regulatory elements such as, for example, a gene promoter with a distal enhancer[2]. In contrast to TADs, a significant portion of chromosomal loopings is subject to cell type-specific regulation[13]. Note that the computational algorithms defining ‘loops’ in Hi-C datasets may differ between studies, with some authors reporting as many as 1×106 loop-like conformations in their Hi-C datasets [14]. Furthermore, higher order forms of promoter-enhancer loopings may exist, as exemplified by euchromatic enhancer islands ‘bundled’ together with odorant receptor genes via inter-chromosomal interactions[15]. Such type of olfactory receptor gene-specific chromosomal contacts apparently are positioned within, or at least in close proximity to constitutive and facultative heterochromatin that in olfactory sensory neurons—in contrast to many other cell types—includes a significant portion of central territories of the nuclear interior[16, 17]. Such types of higher order chromatin architectures are considered important for proper transcriptional regulation of the (single) active odorant receptor allele in an individual olfactory sensory neuron[18,19]. While compartments, TADs, and chromatin loops are important for genome organization including epigenetic and transcriptional regulation, the mechanisms governing the formation of such types of higher order chromatin are only partially understood. For instance, CTCF binding sites often are positioned in inward/convergent orientation, and to a lesser degree, tandem orientation, at the two contact sites of a particular loop or certain TADs [2,7,10,20]. Experimental inversion of CTCF binding sites at promoter-enhancer loops could in some instances alter normal patterns of gene expression [21]. Estimates of the proportion of loopings with convergent CTCF sites at loop anchor sequences range from 65% to 92% [2,20]. Of note, CTCF directionally recognizes binding sites via an 11 zinc finger array. Cohesin, as a multi-subunit protein complex, in turn, is assembled from the CTCF’s C-terminal end, resulting in loop-bound head-to-head CTCF configurations [21]. The orientation is purported to matter due to the biophysical “loop extrusion” model, whereby structural proteins such as cohesin continuously extrude chromatin until blocked by CTCF that is in an appropriately oriented site [22,23]. Cohesin removal, by deletion of cohesion-loading factor Nipbl or by auxin-induced degradation of RAD21 (a cohesin complex component), revealed a loss of CTCF loops and TADs but preservation of A/B compartment segregation, suggesting two independent mechanisms for 3DG and chromatin domain organization [24,25]. Similar to the decoupling of topological features impacted by cohesin depletion, depletion of CTCF also eradicated, in dose-dependent fashion, CTCF-bound chromosomal loops and TADs while compartments remained largely unaffected [26].
Neuronal differentiation is associated with genome-scale remodeling of the chromosomal contact map
Recent studies in developing mouse brain described developmentally regulated changes in 3DG folding in the context of multiple mechanisms including CTCF-dependent loop alterations, repressive chromatin remodeling and dynamic changes in cell- and lineage-specific transcription factor networks[12], and mobilization of genes into or out of heterochromatic environments such as the (nuclear) lamina-associated domains[27]. Similarly, during the course of isogenic differentiation of human neural precursors cells (NPCs), the genes bound to loopings that underwent pruning during the course of NPC-to-neuron-transition were significantly enriched for regulators of cell proliferation, morphogenesis and neurogenesis [13]. This is likely a reflection of the cells’ developmental dynamics including departure from precursor stage towards neuronal lineage commitment and neuronal differentiation [28]. Likewise, loops lost during NPC-to-glia-transition were significantly enriched for various neuron-specific functions, consistent with non-neuronal lineage commitment[28]. In addition, loss of many shorter range contacts and loopings during NPC-to-neuron transition was associated with concomitant increases in longer range (>100–200kb) contacts in both human and mouse in vitro model systems [12,13]. Of note, smaller subsets of short-range loops and contacts, including CTCF- and neuron-specific transcription factor occupied loop anchors, may indeed increase rather than decrease during the course of neuronal differentiation, reflecting functional dynamics in chromatin folding intimately linked to cell identity [13].
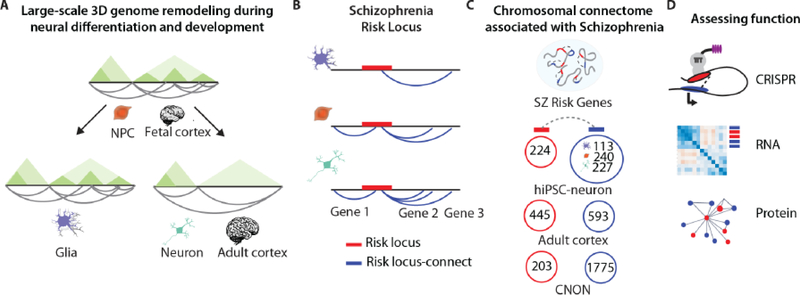
Such types of dynamic 3DG remodeling during the course of neural differentiation are unsurprising and in line with the overall association between chromosomal loopings and active gene expression. However, there is a somewhat unexpected, if not perplexing finding that emerged from the first wave of Hi-C studies in developing human and mouse brain: massive (estimated 25–50%) genome-wide net loss of chromosomal contacts in postmitotic neurons and adult cerebral cortex, as compared to neural progenitors and fetal cortex (Figure 2A). This large-scale net pruning of chromosomal contacts appears to be specific for neuronal differentiation[13]. The same phenomenon was independently reported in a study comparing Hi-C libraries in postmortem (human) fetal versus adult cortex[29]. Importantly, the two studies applied (i) different variations of the Hi-C protocol including different choices of enzymes for chromatin digest, and (ii) different biocomputing algorithms to count more broadly defined chromosomal contacts[29] versus chromosomal ‘loops’ as more conservatively defined distinct pair-wise contacts[13]. Furthermore, both studies [13,29] corroborated the findings from their Hi-C data by analyses of (previously published) additional Hi-C datasets from developing mouse and human brain [12,30]. In addition, genome-scale pruning of chromosomal loopings was specific for NPC-to-(glutamatergic) neuron generation and not observed in the isogenic parallel differentiation NPC-to-glia, and not associated with genome-wide shifts in the proportion of open chromatin [13]. Thus, based on the available evidence generated to date, one could summarize the above discussion that in mammalian brain, the process of neuronal differentiation involves a net loss of chromosomal contacts, apparently disproportionally affecting many ‘shorter range’ chromosomal contacts (<100–200Kb range) and accordingly, many of the smaller TADs, including nested subTADs[13,29] (Figure 2A). The biological significance of such developmental pruning of the chromosomal contact map in neurons is presently unclear and the field eagerly awaits additional studies that are to be expected to provide deeper insight into the massive 3DG remodeling in developing neurons.
Figure 2: Developmental remodeling of the spatial genome during the course of neural differentiation, with implications for the expanded genomic risk architecture of schizophrenia.
A) Independent Hi-C studies exploring (i) hiPSC-derived neural progenitor cells (NPCs) and neurons and glia derived from them by isogenic differentiation, and (ii) 3DG mappings in adult and fetal postmortem cortex, reveal large-scale changes in chromosomal conformation including pruning of loops and contacts, and widening of TADs across neuronal differentiation and development [13,29]. B) Cell-type-specific chromatin contacts anchored at schizophrenia risk loci identify genes from outside of the loci (“risk locus-connect”) interacting with common variants. Contacts defined in hiPSC NPCs/neurons/glia (n = 2 per cell type or 6 total, [13]) by binomial statistics, and in adult postmortem cortex (n = 3) [3+26], and neuronal cells derived from olfactory neuroepithelium (CNON) (n = 2) [53] by Fit Hi-C. Identification of risk locus-connect genes in multiple model systems expanding the known set of schizophrenia risk-associated genes. Rhie et al data [53]were re-assessed to consider vantage point from the latest 145 risk loci [49] used in Rajarajan et al [13] and Giusti-Rodriguez et al [29]as opposed to the older 132 used in the original study. Rajarajan et al [13] only considered cell-type-specific interactions. Giusti-Rodriguez et al [29] considered only interactions labeled as “promoter-promoter” or “promoter-enhancer” determined by open chromatin and histone modification signatures. Rhie et al [53] only considered interactions originating in bins containing high linkage disequilibrium regulatory (i.e., enhancer) variants as determined by histone modification and CTCF ChIP-seq. All three studies demonstrate the power of Hi-C to substantially increase the number of putative risk genes beyond location alone. Red circle = risk locus genes participating in Hi-C interactions; Blue circle = risk locus-connect genes participating in Hi-C interactions. D) Functionality of contacts of interest can be assessed through CRISPR (epi)genomic editing, transcriptome, and proteome studies.
Exploring mechanisms of neurological disease via spatial genome studies
References [31–34] provide an overview of early 3DG studies in the nervous system, which mostly explored candidate gene loci using polymerase chain reaction-read out of chromosome conformation capture (3C) assays and, by that approach, described phenomena such as activity-dependent regulation of chromosomal contacts in the context of gene expression changes during learning and memory[35] and in chronic neuropsychiatric disease[36–38)], or drug addiction[39]. In addition, findings from clinical genetics leave little doubt that proper 3DG regulation is critical for brain development and function, given that mutations and DNA structural variants impacting genes encoding chromosomal scaffolding proteins (including classical ‘loop organizers’ assembling as Cohesin-CTCF complex) are associated with neurological maladies including neurodevelopmental syndromes and—as exemplified by mutations of the nuclear lamina-associated protein LMNB1—also adult-onset demyelinating disease (reviewed in [31]).
Localized disruption of TAD architectures in neurological disease
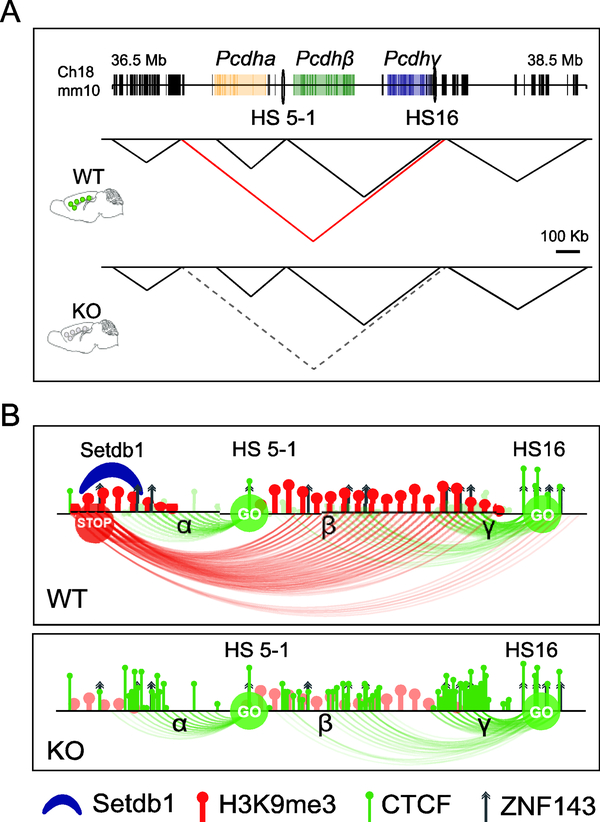
To date, very few studies have explored regulatory mechanisms governing structure and function of chromatin domains, or TADs, in the nervous system. A recent study in conditional mutant mice with neuron-specific ablation of the repressive histone H3-lysine 9 methyltransferase and neurodevelopmental risk gene, KMT1E (SETDB1)[40,41], reported that a small subset of megabase-spanning neuronal TADs (‘superTADs’) are dependent on SETDB1-mediated H3K9 methylation at the site of intra-TAD sequences flanking the TAD boundaries[42] (Figure 1A). Ablation of Setdb1 resulted in excessive CTCF occupancies, which together with the concomitant loss of H3K9 methylation is thought to disrupt repressive chromosomal conformation, including long-range contacts interconnecting the distal portions of the TADs [42] (Figure 1B). Such a disruption led to a partial disintegration of the superTADs, best exemplified by a large domain on mouse chromosome 18 that encompasses >70 genes, including the clustered Protocadherin genes[42]. This family of cell adhesion molecules is critically important for orderly development of neuronal connectivity[43], with the stochastic restraint and expression levels of individual Protocadherin genes in neurons regulated by a delicate balance of promoter-bound DNA methylation and sense transcription, CTCF promoter occupancy and specific promoter-enhancer loopings[42,44]. Importantly, localized disruptions of TAD architectures were associated with an abnormal expression of the affected Protocadherin genes in the adult mutant brains[42] or neural cell cultures [44], strongly suggesting that orderly formation or maintenance of TAD-bound chromosomal conformations associated with specific TADs provide a critical layer of transcription regulation in the affected neurons.
Figure 1: Selective loss of a large megabase TAD at the clustered Protocadherin gene locus after neuronal ablation of Kmt1e/Setdb1 methyltransferase.
(A) Schematic overview of TAD landscape of adult cortical neurons in 2Mb portion of chromosome 18, shown for wildtype (WT) and conditional Setdb1 knock-out (KO), as indicated. Dotted triangle in KO (corresponding to red triangle in WT) demarcates TAD that is massively weakened in KO neurons. Notice that subTADs and surrounding TAD landscape is minimally affected. (B) Chromatin structure and function at the clustered Protocadherin locus. Schematic presentation of 1 megabase TAD at site of Protocadherin genes (see A), illustrating repressive chromatin (‘STOP’) in TAD periphery connecting to intraTAD enhancer sequences (‘GO’). Top, WT, Bottom, KO. Loss of SETDB1 in neurons results in histone hypomethylation, massive excess of CTCF across the TAD, loss of repressive loopings at enhancers and zinc finger protein (incl. ZNF143) binding sites, triggering excessive transcription. See ref. [42] for details.
The potential importance of proper chromatin structures specifically at the site of TAD boundaries became dramatically clear when a recent study uncovered that a surprisingly large share, or 22 out of 27 neurological and medical conditions associated with abnormal expansion of short tandem repeat (STR) sequences had their disease-associated STR sequence located to the site of a TAD boundary[45]. These disease-associated STRs at TAD boundaries were defined by a very high CpG island density and included the specific STR sequences subject to an abnormal expansion, each associated with a monogenic neurodevelopmental or neurogenerative disease, including but not limited to FMR1 (fragile X syndrome), FXN (Friedreich’s ataxia), HTT (Huntington’s disease) and C9ORF72 (motor neuron disease) and ATXN1 (spinocerebellar ataxia 1)[45]. Unsurprisingly, given that TAD landscapes are largely invariant to cell type, the disease-relevant STR location at TAD boundaries was present in embryonic stem cell, blood, neural progenitor and postmortem brain tissue[45]. Interestingly, cell lines from FMR1 patients showed subtle changes in chromosomal conformations at the affected TAD boundaries including abnormal CTCF peak profiles within 100kb of the abnormally expanded STR[45]. However, it remains to be clarified whether the localized disruption of TAD architectures plays a role in the epigenetic dysregulation, including abnormal DNA methylation, and transcriptional shutdown of the FMR1 gene in fragile X cases through the positioning of disease-associated unstable DNA repeats at the site of TAD boundaries. The questions could be expected to be resolved soon.
3DG mappings uncover an expanded genomic risk space associated with schizophrenia
Schizophrenia is a common disorder, affecting 0.8% of the population world-wide[46], defined by core symptoms such as cognitive impairment and thought dysfunction, delusions and hallucinations, social withdrawal and a host of additional psychiatric symptoms[47]. The overwhelming majority of cases escape a mono- or oligogenic disease etiology thus far[48], while on the other hand common variants contributing to heritability risk are overwhelmingly positioned in non-coding DNA, with to date 145 loci identified by genome-wide association in the largest study involving 105,318 subjects [49]. Each of the 145 genomic loci linked to schizophrenia heritability harbors common variants, extending across 1bp to up to >1Mb of sequence in linkage disequilibrium (LD), making it exceedingly difficult to identify the causal variants[49]. Because the majority of functional elements in human non-coding DNA, including enhancers and repressors, are not bound to the nearest TSS but instead tethered via chromosomal contacts to genes located elsewhere on the chromosome[50], it is unsurprising therefore that, as discussed in[26], non-3DG based approaches such as, for example, gene expression quantitative trait loci (eQTL) and SNP prioritization algorithms had only limited success in assigning specific target genes to risk loci. In a pioneering Hi-C study, Won and colleagues integrated chromosomal conformations from fetal brain with schizophrenia GWAS noncoding variants and were able to highlight many candidate genes interacting with them, including those integrated in disease-relevant pathways such as cholinergic signaling and neurogenesis [30]. Another Hi-C study charting 3DG maps from fetal and adult human cortex reported that 1,197 (8.1%) of all brain-expressed protein coding genes are linked to a schizophrenia GWAS locus, with a majority of such genes being separated from the risk locus by hundreds of kilobases (median 305kb), and interact with a specific risk locus via chromosomal conformations bypassing the linear genome[29]. Furthermore, an integrative study by the PsychENCODE consortium[51], analyzed transcriptome and open chromatin landscape and transcriptional histone marks from altogether 2000 postmortem brains including hundreds of cases diagnosed with schizophrenia and integrating these profiles from the ‘linear genome’ with Hi-C data from fetal and adult reference brains[52]. The study mapped ~79,000 brain-active enhancers with their associated chromosomal contacts and TAD landscapes[52] and identified a vast number of eQTLs and gene regulatory networks and perhaps most importantly, applied deep machine learning algorithms that, for the first time, were able to predict presence or absence of disease (schizophrenia) based on a subject’s brain transcriptome and chromatin profiles[52]. The study approached disease prediction at probability level of 75%, reflecting a significant advancement over more conventional genomic approaches predicting disease only marginally above chance (50%) [52]. Likewise, Hi-C chromosomal contact mapping in iPSC-derived NPCs and their differentiated neurons and glia, increased the number of actively transcribed genes associated with a schizophrenia GWAS locus by approximately 2–3 fold (total N expressed genes interconnected with or located within a schizophrenia GWAS locus ranged from 201–386, depending on cell type (NPC, neuron, glia) [13] (Figure 2B). Because neurons, together with NPC, had the largest number of cell-type-specific chromosomal contacts anchored in a risk locus (as compared to glia and other non-neuronal cells), one could conclude that (as discussed above) while the overall spatial genome space contracts when NPC differentiate to neurons, the 3DG space associated with schizophrenia risk disproportionately increases as neurons differentiate[13]. Remarkably, the disease-related chromosomal connectome specific to NPC or neurons was associated with “clusters” of coordinated gene expression and protein interactions, with at least one cluster strongly enriched for regulators of neuronal connectivity and synaptic plasticity, and another cluster for chromatin-associated proteins, including transcriptional regulators[13]. Likewise, an expanded genome space involving higher order chromatin and chromosomal contacts anchored in schizophrenia risk loci has also been described for cultured primary sensory neurons from the olfactory neuroepithelium[53] (Figure 2B,C).
Outlook and future studies
Here, in this Current Opinions article, we report recent 3DG discoveries highly relevant for two very different categories of brain disorders. The first category includes rare monogenic neurological disorders associated with locus-specific abnormal expansion of short DNA repeats. The second disease category includes schizophrenia, a common psychiatric disease defined by an exceedingly complex genetic risk architecture. The fact that two such disparate disease categories, each afflicting the human brain in very different ways, were both newly informed via Hi-C-based approaches clearly speaks to the promise of spatial genome exploration in the fields of genomic medicine and neurobiology. We predict that 3DG studies, in cell-type-specific fashion, will provide new and critical insights into the genetic risk architectures of a broad range of neurological and psychiatric disorders and thereby providing a critical link between genome, epigenome and the ‘nucleome’ in normal and diseased brain. To mention just one example for work expected to be pursued in the near future, the next generation of schizophrenia-focused 3DG work could focus on the chromosomal risk connectome that showed a surprisingly strong correlation at the level of the transcriptome and, at least for a subset of expressed genes, also at the level of the proteome[13]. It is an open question currently whether or not the (schizophrenia) GWAS-bound genomic sequences converge on intra- and inter-chromosomal hubs enriched for specific transcription or splicing factors, in analogy to similar principles governing coordinated regulation of gene expression in sensory and peripheral systems [15,54,55]. To this end, it is encouraging that the three major functional categories associated with the genetic risk architecture of schizophrenia—neuronal connectivity, synaptic signaling and chromatin remodeling[56,57]—are also heavily represented within the developmentally regulated cell-type-specific chromosomal connectomes of cultured neurons and their precursors[13] and fetal brain tissue in vivo [30,58]. We have argued that cell-type-specific intersection of 3DG and genetic risk maps may further deepen understanding of the genomic underpinnings of normal and diseased cognition, and may lead to improved disease risk prediction as compared to cumulative schizophrenia risk allele burden estimates such as “polygenic risk score” (PRS) or “biologically-informed multilocus profile scores” (BIMPS), which currently do not take into account the spatial genome, a critical limitation that may explain why PRS and BIMPS are presently only minimally informative about disease risk [59].
Highlights.
Review of Hi-C genome-scale chromosome conformation (‘3D Genome’) mappings in brain
Discussion of developmental 3DG reorganization in differentiating neurons
Discussion of chromosomal contacts associated with schizophrenia risk sequences
Monogenic neurological disorders and DNA repeats at chromatin domain boundaries.
Acknowledgments
Funding: Work conducted in the authors’ laboratories is supported by NIH grants R01MH106056 and R01MH117790.
Footnotes
Conflicts of Interest: The authors report no conflict.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schmitt AD, Hu M, Ren B: Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol (2016) 17(12):743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL: A 3d map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell (2014) 159(7):1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giorgetti L, Heard E: Closing the loop: 3c versus DNA fish. Genome Biol (2016) 17(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowley MJ, Corces VG: Organizational principles of 3d genome architecture. Nature Reviews Genetics (2018) 19(12):789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibcus JH, Dekker J: The hierarchy of the 3d genome. Mol Cell (2013) 49(5):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, Reddy J et al. : Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science (New York, NY) (2016) 351(6280):1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B: Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012) 485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J et al. : Spatial partitioning of the regulatory landscape of the x-inactivation centre. Nature (2012) 485(7398):381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, Diao Y et al. : Chromatin architecture reorganization during stem cell differentiation. Nature (2015) 518(7539):331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S: Comparative hi-c reveals that ctcf underlies evolution of chromosomal domain architecture. Cell Rep (2015) 10(8):1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadhouders R, Vidal E, Serra F, Di Stefano B, Le Dily F, Quilez J, Gomez A, Collombet S, Berenguer C, Cuartero Y, Hecht J et al. : Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet (2018) 50(2):238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A, Cavalli G: Multiscale 3d genome rewiring during mouse neural development. Cell (2017) 171(3):557–572.e524.•• Bonev et al. generated a comprehensive genomics datasets for mouse neural differentiation in vivo and ex vivo, thereby linking repressive chromatin remodeling and transcription factor networks to dynamic changes in chromosomal conformations for specific brain cell types.
- 13.Rajarajan P, Borrman T, Liao W, Schrode N, Flaherty E, Casino C, Powell S, Yashaswini C, LaMarca EA, Kassim B, Javidfar B et al. : Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science (2018) 362(6420).• Rajarajan et al monitored developmentally regulated changes in the spatial genome of human neural precursor cells differentiating into neurons and glia and report massive pruning of the chromosomal contact map during the course of neuronal differentiation. The study also analyzed chromosomal contacts anchored in schizophrenia risk sequences, and examined co-regulation of gene expression, and (to a certain degree) proteomic interactions of such types of ‘risk-locus’ connected genes.
- 14.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B: A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature (2013) 503(7475):290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monahan K, Horta A, Lomvardas S: Lhx2- and ldb1-mediated trans interactions regulate olfactory receptor choice. Nature (2019) 565(7740):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armelin-Correa LM, Gutiyama LM, Brandt DY, Malnic B: Nuclear compartmentalization of odorant receptor genes. Proc Natl Acad Sci U S A (2014) 111(7):2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan L, Xing D, Daley N, Xie XS: Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat Struct Mol Biol (2019) 26(4):297–307. [DOI] [PubMed] [Google Scholar]
- 18.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R: Interchromosomal interactions and olfactory receptor choice. Cell (2006) 126(2):403–413. [DOI] [PubMed] [Google Scholar]
- 19.Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S: Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell (2012) 151(4):724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, Michalski P et al. : Ctcf-mediated human 3d genome architecture reveals chromatin topology for transcription. Cell (2015) 163(7):1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, Lu Y et al. : Crispr inversion of ctcf sites alters genome topology and enhancer/promoter function. Cell (2015) 162(4):900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP et al. : Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A (2015) 112(47):E6456–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA: Formation of chromosomal domains by loop extrusion. Cell Rep (2016) 15(9):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, LoeMie Y, Fonseca NA, Huber W, C HH, Mirny L, Spitz F: Two independent modes of chromatin organization revealed by cohesin removal. Nature (2017) 551(7678):51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, Huang X et al. : Cohesin loss eliminates all loop domains. Cell (2017) 171(2):305–320.e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG: Targeted degradation of ctcf decouples local insulation of chromosome domains from genomic compartmentalization. Cell (2017) 169(5):930–944 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, Reinders M et al. : Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell (2010) 38(4):603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Jenjaroenpun P, Bhinge A, Angarica VE, Del Sol A, Nookaew I, Kuznetsov VA, Stanton LW: Single-cell gene expression analysis reveals regulators of distinct cell subpopulations among developing human neurons. Genome Res (2017) 27(11):1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giusti-Rodriguez PM, Sullivan PF: Schizophrenia and a high-resolution map of the three-dimensional chromatin interactome of adult and fetal cortex. bioRxiv (2018) 406330. [Google Scholar]
- 30.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, Gandal MJ, Sutton GJ, Hormozdiari F, Lu D, Lee C et al. : Chromosome conformation elucidates regulatory relationships in developing human brain. Nature (2016) 538(7626):523–527.• Won et al were the first to study the 3D genome by Hi-C in human brain in the context of the genetic risk architecture of schizophrenia and other cognitive disease.
- 31.Rajarajan P, Gil SE, Brennand KJ, Akbarian S: Spatial genome organization and cognition. Nat Rev Neurosci (2016) 17(11):681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita Y, Yamashita T: Spatial organization of genome architecture in neuronal development and disease. Neurochem Int (2018) 119(49–56. [DOI] [PubMed] [Google Scholar]
- 33.Rajarajan P, Jiang Y, Kassim BS, Akbarian S: Chromosomal conformations and epigenomic regulation in schizophrenia. Prog Mol Biol Transl Sci (2018) 157(21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson LA, Tsai LH: In the loop: How chromatin topology links genome structure to function in mechanisms underlying learning and memory. Curr Opin Neurobiol (2017) 43(48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT et al. : Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell (2015) 161(7):1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bharadwaj R, Peter CJ, Jiang Y, Roussos P, Vogel-Ciernia A, Shen EY, Mitchell AC, Mao W, Whittle C, Dincer A, Jakovcevski M et al. : Conserved higher-order chromatin regulates nmda receptor gene expression and cognition. Neuron (2014) 84(5):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bharadwaj R, Jiang Y, Mao W, Jakovcevski M, Dincer A, Krueger W, Garbett K, Whittle C, Tushir JS, Liu J, Sequeira A et al. : Conserved chromosome 2q31 conformations are associated with transcriptional regulation of gad1 gaba synthesis enzyme and altered in prefrontal cortex of subjects with schizophrenia. J Neurosci (2013) 33(29):11839–11851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, Straubhaar J, Martin G, Akbarian S: Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the nmda receptor subunit nr2b. J Neurosci (2010) 30(21):7152–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engmann O, Labonte B, Mitchell A, Bashtrykov P, Calipari ES, Rosenbluh C, Loh YE, Walker DM, Burek D, Hamilton PJ, Issler O et al. : Cocaine-induced chromatin modifications associate with increased expression and three-dimensional looping of auts2. Biol Psychiatry (2017) 82(11):794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Q, Goldstein J, Wang P, Gadi IK, Labreche H, Rehder C, Wang WP, McConkie A, Xu X, Jiang YH: Chromosomal microarray analysis in clinical evaluation of neurodevelopmental disorders-reporting a novel deletion of setdb1 and illustration of counseling challenge. Pediatr Res (2016) 80(3):371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cukier HN, Lee JM, Ma D, Young JI, Mayo V, Butler BL, Ramsook SS, Rantus JA, Abrams AJ, Whitehead PL, Wright HH et al. : The expanding role of mbd genes in autism: Identification of a mecp2 duplication and novel alterations in mbd5, mbd6, and setdb1. Autism Res (2012) 5(6):385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y, Loh YE, Rajarajan P, Hirayama T, Liao W, Kassim BS, Javidfar B, Hartley BJ, Kleofas L, Park RB, Labonte B et al. : The methyltransferase setdb1 regulates a large neuron-specific topological chromatin domain. Nat Genet (2017) 49(8):1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirayama T, Yagi T: Regulation of clustered protocadherin genes in individual neurons. Semin Cell Dev Biol (2017) 69(122–130. [DOI] [PubMed] [Google Scholar]
- 44.Canzio D, Nwakeze CL, Horta A, Rajkumar SM, Coffey EL, Duffy EE, Duffie R, Monahan K, O’Keeffe S, Simon MD, Lomvardas S et al. : Antisense lncrna transcription mediates DNA demethylation to drive stochastic protocadherin alpha promoter choice. Cell (2019) 177(3):639–653 e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun JH, Zhou L, Emerson DJ, Phyo SA, Titus KR, Gong W, Gilgenast TG, Beagan JA, Davidson BL, Tassone F, Phillips-Cremins JE: Disease-associated short tandem repeats co-localize with chromatin domain boundaries. Cell (2018) 175(1):224–238 e215.• Sun et al analyzed human HiC datasets from a variety to cell types and tissues and show that many sequences associated with unstable DNA repeats (causing neurological disease once expanded beyond a certain treshold) each locate to the boundary of a specific chromatin domain, including TAD or subTAD. In case of the repeat expansion disorder, FMR1/Fragile X syndrome, the author are able to show disruption of the local TAD landscape at the site of the FMR1 gene affected by the abnormal repeat length.
- 46.Saha S, Chant D, Welham J, McGrath J: A systematic review of the prevalence of schizophrenia. PLoS Med (2005) 2(5):e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owen MJ, Sawa A, Mortensen PB: Schizophrenia. Lancet (2016) 388(10039):86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganna A, Satterstrom FK, Zekavat SM, Das I, Kurki MI, Churchhouse C, Alfoldi J, Martin AR, Havulinna AS, Byrnes A, Thompson WK et al. : Quantifying the impact of rare and ultra-rare coding variation across the phenotypic spectrum. Am J Hum Genet (2018) 102(6):1204–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pardinas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J et al. : Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet (2018) 50(3):381389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanyal A, Lajoie BR, Jain G, Dekker J: The long-range interaction landscape of gene promoters. Nature (2012) 489(7414):109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH et al. : The psychencode project. Nat Neurosci (2015) 18(12):1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, Xu M et al. : Comprehensive functional genomic resource and integrative model for the human brain. Science (New York, NY) (2018) 362(6420). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhie SK, Schreiner S, Witt H, Armoskus C, Lay FD, Camarena A, Spitsyna VN, Guo Y, Berman BP, Evgrafov OV, Knowles JA et al. : Using 3d epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation. Sci Adv (2018) 4(12):eaav8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V et al. : Higher-order inter-chromosomal hubs shape 3d genome organization in the nucleus. Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khanna N, Hu Y, Belmont AS: Hsp70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol (2014) 24(10):1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Network, Pathway Analysis Subgroup of Psychiatric Genomics C: Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci (2015) 18(2):199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D: Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci (2012) 15(12):1723–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, Geschwind DH: The dynamic landscape of open chromatin during human cortical neurogenesis. Cell (2018) 172(1–2):289–304 e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bogdan R, Baranger DAA, Agrawal A: Polygenic risk scores in clinical psychology: Bridging genomic risk to individual differences. Annu Rev Clin Psychol (2018) 14(119–157. [DOI] [PMC free article] [PubMed] [Google Scholar]