Abstract
The last two decades have witnessed explosive advances in our understanding as to how the organization of chromatin, the association of DNA with histones and vast numbers of non-histone regulatory proteins, controls the expression of specific genes in brain. Prominent among such regulatory mechanisms are modifications of histones, along with the “writers,” “erasers,” and “readers” of these modifications. Much of the work delineating these mechanisms has contributed to the idea that a ‘histone code’ may be a central determinant of a gene’s activity and its potential to be activated or repressed in response to environmental perturbations (both beneficial and aberrant). Indeed, increasing evidence has demonstrated the significance of histone regulation in neurological plasticity and disease, although we are still at the earliest stages of examining all of the many potential chromatin changes involved. In this short review, we provide an emerging perspective on putative roles for histones, and their combinatorial readouts, in the context of neural plasticity, and we provide a conceptual framework for future mechanistic studies aimed at uncovering causal links between the neural ‘histone code’ and brain function/disease.
Introduction
While DNA provides a blueprint for our growth, development and survival, our DNA alone cannot fully account for the broad spectrum of phenotypic variance observed. In all eukaryotic organisms, DNA is tightly packaged into chromatin, where it is wrapped around a core octamer of histone proteins, containing two copies each of H3, H4, H2A and H2B, or variants thereof (e.g., H3.3, H2A.Z, etc.). Chromatin serves as a signal integration platform, where it provides cells with appropriate instructions for RNA transcription and protein synthesis. Within each cell, the readout of our DNA is finely tuned so that specific genetic loci can be temporally and/or spatially regulated in response to environmental cues to promote phenotypic outcomes. This level of control is dictated by a vast array of molecular processes, collectively referred to as “epigenetics.” Transitions toward different activity states of DNA are modulated by the accessibility of chromatin to various general and non-general transcription factors, with heterochromatic organization linked to gene repression, and euchromatic conformations associated with permissive gene expression1,2. The transition between these different states is mediated by numerous factors that impact DNA interactions with histones themselves. These factors are comprised of a large number of enzymes (e.g., “writers” and “erasers”) and associated protein effector complexes (i.e., “readers”), which are responsible for depositing/removing/interacting with particular chemical modifications to influence the structure of chromatin and, in turn, regulate gene transcription3. Accurate deposition and removal of these modifications is critical for both normal neural development and adulthood brain plasticity. Given that neurons are generally post-mitotic, thereby remaining in a mature state for the entirety of an organism’s life, it is not surprising that histone regulation (which additionally includes nucleosome remodeling, histone variant exchange, histone turnover, etc.) in the central nervous system (CNS) plays a governing role in neuronal plasticity. While a multitude of data exists highlighting the importance of histones in these processes, we will attempt to illuminate their involvement using select examples associated with brain homeostasis and their regulation in the context of neurodevelopmental disease. Furthermore, we will discuss a critical need for the field to begin moving towards a more mechanistic/causal investigatory framework in order to begin understanding potential contributions of chromatin regulation to neural plasticity.
The combinatorial nature of the ‘histone code’ and its relevance to brain plasticity
Histone modifications, along with their respective contributions to the regulation of chromatin structure, induce long-lasting influences on the development and plasticity of post-mitotic neurons and have been demonstrated to function in a combinatorial manner. Just as histone modifications can be described as the ‘alphabet’ of the so-called ‘histone code,’ varied combinations of these ‘letters’ can result in a phenotypic syntax. As such, pre-existing modifications on a given histone/nucleosome may directly or indirectly impede/enhance the ability of specific “reader” protein complexes and/or chromatin modifiers to recognize, and thereby influence additional substrate sites on the same or adjacent transcriptional unit. Such pattern recognition is ultimately critical for transcriptional homeostasis. Following groundbreaking work in 1999, which identified the first histone modification “reader” complexes (e.g., histone lysine acetylation recognition by bromodomain containing proteins4), the ‘histone code’ hypothesis5 was articulated, proposing that different combinations of histone modifications will result in distinct influences on gene expression. This is mediated in part, by the manner in which downstream effector proteins decipher these combinations (in trans) in conjunction with alterations in electrostatic interactions between histones and DNA (in cis). Since that time, a plethora of additional histone modification binding proteins/domains and their structures have been resolved–including chromodomains, Tudor domains, plant homeodomains (PHD) and others–further extending the breadth of combinatorial readout possibilities. While discussions of the ‘histone code’ as a discrete form of molecular language has led to a healthy debate in the scientific community regarding what actually constitutes a ‘code,’ most researchers remain in agreement that such chromatin related processes serve a crucial role in the mediation of cellular phenotypes.
Disruptions in histone regulatory proteins contribute to neurodevelopmental dysplasticity
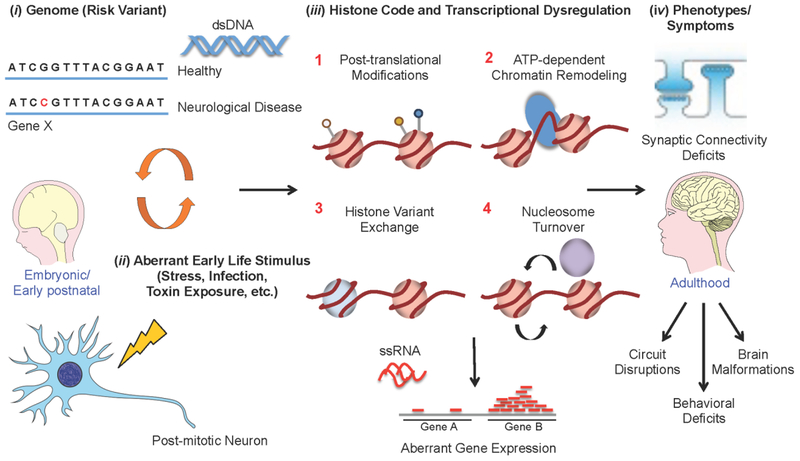
Neurodevelopmental plasticity is a complex and dynamic process that is heavily influenced by gene x environment interactions, whereby alterations in gene expression influence critical processes, such as axon guidance and synaptic connectivity. It is well understood that ‘peaks’ of plasticity landmark critical stages of neurodevelopment (both embryonic and early postnatal), allowing the brain to adapt in size as diverse neuronal circuits are formed. These circuits, which are constantly fine-tuned during ‘peak’ periods, remain highly malleable in order to encompass the capacity to be altered in their relative strengths during times of activity-/experience-dependent plasticity. Such flexibility remains critical into adulthood, as constant integration of cues and exposures from the environment is required to promote positive physiological and behavioral adaptations (e.g., the development of new memories, proper stress responsivity, etc.). Therefore it is not surprising that experiences of aberrant environmental stimuli during critical periods can exert negative consequences on gene expression, leading to deleterious phenotypes (Fig 1). Tight regulation of gene expression is controlled in an activity-dependent manner, a process that is ultimately orchestrated by the localization and/or expression of chromatin regulatory machinery6. Thus, distinct patterns of histone modifications, which act in unison with DNA methylation, expression of non-coding RNAs (ncRNAs) and recruitment of transcription factors, need to be properly maintained and appropriately responsive to environmental cues to achieve normal levels of plasticity3,5,7,8.
Fig. 1: Histone dysregulation in neurological disease.
Aberrant alterations of histone PTMs and chromatin structures are thought to result in disrupted patterns of neurodevelopment and subsequent synaptic/behavioral abnormalities in adulthood. These disorders are precipitated by a combination of (i) genetic risk variants (i.e., DNA mutations) and (ii) environmental risk factors (e.g., early life stress, exposure to toxins, infections, etc.), the latter of which can result in (iii) profound alterations in gene expression through aberrant regulation of histones (e.g., 1–PTMs, 2–chromatin remodeling, 3–histone variant exchange and 4–nucleosome turnover) and other “epigenetic” phenomena. (iv) These molecular processes may then be responsible for many of the physiological and behavioral phenotypes observed in neurological disease during adulthood. Much work is still needed, however, to truly be able to causally link such “epigenetic” mechanisms to neurological illness, a process that will require both the development of novel methodologies (e.g., in vivo protein trans-splicing, etc.) and a concerted effort by the field to assess molecular outcomes within a brain specific framework.
Several neurological disorders involve mutations in genes that encode proteins associated with chromatin remodeling complexes, as well as histone PTM “writers,” “erasers” and “readers”2,9. For example, X-linked mental retardation syndrome and α-thalassaemia result from mutations in the gene encoding ATRX, a chromatin-remodeling factor. In addition, mutations in the CREB binding protein (CBP), the histone H3K4me3-demethylase, JARID1C, and the histone H3K9me1/2-methyltransferase complex, G9a/GLP (EHMT2/1), result in Rubinstein-Taybi syndrome, epilepsy X-linked mental retardation and Kleefstra syndrome, respectively10–12. More recent studies have also identified de novo mutations in genes encoding chromatin modifiers, such as KMT2D, SMARCA2, SIRT1 and KAT6A in neonatal brain arterio-venous malformations13. Additional data now exist demonstrating that mutations in histones themselves can result in human disease. Independent mutations in genes encoding H4 (K9fs) and H3 (R130C) have been reported14, providing direct links between histone mutations and mental retardation/intellectual disabilities. Additionally, following a mutational screen in zebrafish, the H3.3 variant protein was directly implicated in processes associated with neuronal differentiation, whereby a missense mutation (D123N) was demonstrated to result in abnormal development of the cranial neural crest.
Down syndrome (DS): an example of a classic ‘genetic’ disorder likely influenced by “epigenetic” phenomena
It is becoming increasingly clear that numerous additional disorders that have long been thought to be purely genetic in nature, may also have epigenetic components. DS is one such likely example. DS is the most common genetic cause of intellectual disability worldwide resulting from triplication of chromosome 21 (HSA21) in humans. Despite much progress in understanding the genetics of DS, the genes encoded on HSA21 that directly contribute to intellectual disability and their associated molecular mechanisms, are not fully understood. Since the completion of the Human Genome Project in 2000, which provided a complete DNA sequence for chromosome 21 (HSA21)15, many labs throughout the world have focused their attention toward gaining a better understanding of the contributions of HSA21 protein-coding genes, some of which are overexpressed in DS, to processes associated with learning and memory. However, variability in cognitive deficits (ranging from mild to moderate) remains difficult to explain16,17. For example, if one were to reductionistically assume a 1:1 relationship between gene abundance and transcription, trisomy 21 would theoretically be expected to result in a 1.5 fold increase in the expression of genes being triplicated; however, numerous gene expression studies in both human brain/cells18,19 and in rodent models of DS20–22 have shown this to be incorrect. For example, one recent study in human cells from DS cases demonstrated that > 50% of analyzed genes on HSA21 displayed expression levels significantly higher or lower than expected23. Furthermore, RNA-seq analyses of postmortem brain tissues have demonstrated that many genes across all chromosomes are altered in their expression in DS (both up- and downregulated), and that progressive increases in differential gene expression can be observed across development in trisomy 2119.
Such incongruous findings between genotype and gene expression in DS heavily suggest that epigenetic mechanisms may play an important role in the disease. Limited studies of epigenetic mechanisms in DS are further surprising given that numerous putative chromatin regulators (e.g., DNMT3L, BRWD1, CHAF1B, etc.) are themselves encoded within Down Syndrome Critical Regions (DSCRs) on HSA21 and have been found to be dysregulated in DS tissues24. While definitive links between epigenetic dysregulation and DS have not yet been fully substantiated, findings such as those described above are certainly worth consideration in future investigations, as well as in studies of additional disorders currently thought to be purely genetic in cause.
Towards a conceptual framework for future mechanistic studies of neuroepigenetic phenomena
Until recently, little attention has been paid to exploring chromatin related mechanistic differences that may exist between brain/neurons vs. proliferating cells and tissues. This is to say that neuroepigenetics researchers have oftentimes relied heavily on mechanistic and biochemical information about chromatin related phenomena gleaned from mitotic cellular systems to inform their hypotheses as to how such events in brain may relate to observed phenotypic responses. On the one hand, it is reasonable to assume that some, if not many, of these basic mechanisms may be shared between brain and non-neural tissues (e.g., the euchromatic histone PTM, H3K4me3, is most certainly a permissive mark in both dividing and non-dividing cells). However, some of these previously defined mechanisms may not hold weight when examined in the context of the CNS. As such, it will likely be necessary to begin shifting away from the more descriptive and phenomenological examinations of histone PTMs, and their associated proteomic machinery, that have dominated the neuroepigenetics field over the last 15 years. For example, while it is tempting to attribute ‘established’ chromatin mechanisms to brain specific phenomena, it is important to consider that numerous brain specific and/or enriched epigenetic processes may also exist (e.g. brain enriched histone PTMs, etc.). Exploring such mechanisms will require better integration of basic techniques and approaches that can be borrowed from the field of chromatin biochemistry. Below, we discuss two examples that highlight how moving the field towards a more mechanistic investigatory framework may help to yield greater insights into the many potentially unexplored aspects of chromatin biology that govern brain development and plasticity.
The dangers in evaluating neuroepigenetic phenomena in the context of mechanistic studies in proliferating cells: the case of the variant histone H3.3
Histone variants, which contain amino acid sequence substitutions that distinguish them from their canonical counterparts (e.g., canonical H2A vs. variant H2A.Z), are proteins that can replace canonical histones in a locus specific manner, oftentimes resulting in differential structural and transcriptional responses. Disruptions in histone variant biology can result in deleterious phenotypes resulting in human disease. Of particular importance is the question as to whether histone variants, which are typically expressed and deposited into chromatin in a cell cycle dependent manner, might function differently in post-mitotic cells. Since many canonical histones cannot be actively incorporated into chromatin in post-replicative systems, it was hypothesized that neuronal differentiation may result in an imbalance between variant and non-variant histones.
The histone variant H3.3 is one such example that had been observed to progressively accumulate over the course of development in rodent brain, with concomitant reductions in canonical H3.1 and H3.2 observed25. Such a phenomenon begs the question as to what, if any, specific roles H3.3 may play in the developing and adult brain. Previous assessments in dividing cells suggested that H3.3 may preferentially be marked throughout the genome by specific modification ‘signatures’ that are typically associated with active gene expression26. This work contributed to the so-called genomic “barcode” hypothesis27,28, which posited that specific histone variants may then function to influence chromatin states during cellular differentiation. Such mechanisms of modification enrichment, however, have been poorly described in post-mitotic systems, such as neurons, which rely exclusively on a single histone H3 protein (i.e., the variant H3.3) for global replacement during cellular development. If true, such phenomena might directly challenge the notion of histone “barcoding” in brain. Consistent with this hypothesis, one study recently demonstrated a direct role for calcium dependent Daxx (an H3.3 chaperone) phosphorylation in the regulation of H3.3 deposition in response to neural activity, specifically at regulatory elements known to mediate immediate early gene (IEG) expression29. While these data indicated H3.3 as a potentially dynamic protein in neurons during periods of cellular plasticity, it stopped short of addressing more global roles for H3.3 variant exchange and accumulation in brain during neurodevelopment and into adulthood.
Extending upon this work, we and others uncovered additional roles for histone variant dynamics (e.g., histone incorporation/eviction, nucleosome sliding and disassembly) during activity-dependent transcription in brain29–32. Specifically, we found that nucleosome turnover, which requires active incorporation of replication-independent H3.3 is required for neural (both neuronal and glial) specific gene expression patterns, synaptic development and connectivity, dendritic spinogenesis and cognition30. More recently, it was also found that these phenomena are disrupted in specific pathophysiological states (e.g., major depressive disorder) associated with abnormal brain plasticity. Taken together, these data suggest that histone turnover itself may serve as a fundamental mediator of transcriptional plasticity within brain; more work, however, is needed to fully delineate the contributions of these phenomena to neurological function and disease.
Novel chromatin phenomena in brain: the case of histone serotonylation
Since its earliest days, the chromatin field has continually recognized that our understanding of the ‘histone code’ remains incomplete, owing primarily to the fact that new histone PTMs are constantly being discovered across a large variety of cellular systems. Few attempts to investigate whether neural cells may also exhibit previously unexplored modifications have been undertaken.
Monoaminergic systems in brain play critical roles in the regulation of a wide variety of neurodevelopmental and adult cognitive/reward processes. Furthermore, many drugs used to treat individuals with neurodegenerative disease (e.g., Parkinson’s, Huntington’s), as well as psychiatric syndromes ranging from major depressive disorder (MDD) to drug addiction and schizophrenia target monoaminergic systems33. In addition to its presence in vesicles, past and recent data have demonstrated the presence of extravesicular monoamines in the nucleus and soma of monoamine producing neurons34,35. The presence of extravesicular monoamines has been suggested to be critically important during periods of activity-dependent neurotransmission due their ability to rapidly replace depleted vesicularized monoaminergic pools at the synapse following somatic release34; it had remained unclear, however, whether these monoamines might play additional roles independent of neurotransmisson.
Serotonin, as well as other monoamines, had previously been shown to form covalent bonds with certain cytoplasmic and/or matrix associated proteins via transamidation by the tissue Transglutaminase 2 (TGM2) enzyme, a modification that alters the signaling properties of monoaminylated substrates36–39. We identified histone H3 as a direct substrate for monoaminylation, and later identified H3 glutamine 5 (H3Q5) as the exclusive reactive amino acid for this mark (e.g., H3Q5serotonyl)40. Given H3Q5ser’s proximity to lysine 4 on H3, we examined the impact of pre-establishing nucleosomal H3K4me3 on TGM2 activity in vitro. In doing so, we found that TGM2 activity is not perturbed by the presence of H3K4me3, indicating that these marks can likely co-exist in vivo (e.g., H3K4me3Q5ser). The existence of this modified histone protein (both in the presence and absence of H3K4me3) was unambiguously confirmed in serotonergic cells, peripheral organs (e.g., colon, blood) and in brain, indicating that the dual mark acts to enhance binding of adjacent H3K4me3 interacting proteins [e.g., “readers,” such as the general transcription factor complex TFIID41], rendering serotonylated nucleosomes transcriptionally permissive. While much work still needs to be done to fully delineate the entirety of mechanisms guiding H3 serotonylation’s role in promoting active transcription, we believe that this work nicely highlights the existence of a previously unknown brain enriched modification that seemingly acts to fine tune the neural histone code in order to achieve normal patterns of neuronal transcription during development. In future studies, explorations into whether additional monoamines (e.g., dopamine, histamine, etc.) may similarly be covalently modified to histones, as well as what roles such marks may play in neuroplasticity and disease, promise to shed new light on previously undiscovered epigenetic mechanisms that have eluded chromatin researchers working in other biological systems.
Emerging methodologies for mechanistically defining causal links between histone PTMs and brain function
Over the past 15 years, neuroepigeneticists have made great strides in furthering our understanding as to how histone regulation may contribute to aspects of neurodevelopment, plasticity and behavior. However, it is our opinion that the field has only begun to scratch the surface of deciphering how, or if, these complicated patterns of chromatin regulation directly contribute to disruptions in neural plasticity. Further attempts to describe the precise transcriptional mechanisms involved in these complex processes will necessitate a more mechanistic, and unbiased, combination of chromatin biochemical, proteomic and molecular approaches in order to begin integrating such mechanistic insights with more typical phenomenological outputs.
For example, it is common in the field for researchers to assign chromatin-associated functions to alterations in the expression/activity of chromatin regulatory enzymes and remodeling complexes. This emphasis is logical given that adequate tools do not yet exist to directly manipulate histone PTMs in brain without altering the enzymatic machinery. While it is true that numerous chromatin-regulatory proteins (e.g., HATs, HDACs, HMTs, HDMs, “readers,” etc.) have been shown to display altered patterns of regulation in neurons, it is becoming apparent that most, if not all, of these proteins are capable of modifying/binding to additional non-histone substrates (both in the nucleus and cytoplasm). Such processes complicate current interpretations of most genetic studies examining histone related proteins. While changes in enzyme activity/”reader” associations may indeed contribute to the mediation of chromatin landscapes, it remains difficult to assign causality. Many groups are beginning to address this dilemma through mutational assessments, whereby it is possible to directly manipulate catalytic activities and/or binding outcomes of chromatin regulatory proteins (e.g., acetyl bromodomains, methyl PHD fingers, etc.), allowing for more direct examinations of the epigenetic machinery in vivo. However, this issue is further complicated by the fact that, until recently, neuroepigenetics researchers have tended to study histone marks in isolation, often ignoring their combinational functionality, an issue that is incredibly challenging to address without the development of novel approaches that will allow for direct manipulations of histone PTMs themselves in the absence of perturbing associated modifying enzymes and/or effector complexes.
Advanced technologies that allow researchers to directly modify the epigenomic landscape, sometimes even in a locus specific manner, are gaining traction. For example, via the use of zinc finger proteins (ZFPs) and/or CRISPR/dCas9 fused to DNA sequence recognition domains42, it is now possible to deliver conjugated domains of enzymatic activators or repressors of gene expression (e.g., VP64, p65, G9a) to targeted loci in order to assess their impact on gene expression through the artificial establishment of endogenous chromatin modifications. While these approaches allow for locus-specific establishment/removal of histone PTMs, which is potentially useful for investigating roles for PTMs on a gene by gene basis, the ability to alter global levels marks in a cell-type specific manner–in a single or combinational fashion, is also emerging in the form of a new methodology referred to as protein trans-splicing. Tom Muir, Yael David and colleagues43 have recently employed ultra-fast trans-splicing split inteins to incorporate non-native histone PTMs into chromatin in living cells. Protein splicing utilizes intervening polypeptides, known as inteins (i.e., “protein introns”), which cleave themselves from host proteins (extein) to generate novel spliced protein products44. In their landmark Nature Chemistry paper, David and colleagues genetically fused IntN to a truncated histone H2B protein, expressed it by transfection in 293T cells, and then delivered an IntC carrying synthetic cargo (complementary to the truncated H2B), using a cell-penetrating peptide. With this new method, it was possible to synthesize H2B (and presumably other histones as well, such as H3) carrying an array of PTMs in vivo. Thus, approaches like these carry with them the exciting possibility of being able to one day directly assess the contributions of specific histone marks in brain to neuronal patterns of transcription, development and behavior, both in normal and pathophysiological states.
Conclusion
Dysregulation of histone PTMs and related chromatin structures are believed to result in aberrant patterns of neurodevelopment and subsequent behavioral abnormalities later in life. With an ever growing interest into how “epigenetic” landscapes may be perturbed in neurological disorders, it is now more urgent than ever that neuroepigeneticists begin approaching related research questions with interpretive caution and mechanistic awareness. As such, we posit that more faithful examinations of chromatin regulatory phenomena specifically within the context of the CNS–in part, using more targeted molecular approaches, such as those described above–promise to improve our basic understanding of “epigenetic” regulation in brain plasticity and may further aid in the development of future therapeutics that can more accurately target these processes to alleviate disease.
Highlights.
Histone regulation in brain is a central determinant of neural development and plasticity.
Many neurological diseases are linked to aberrant histone regulation in the CNS.
New methods for investigating histones may help to causally link them to biological function.
Acknowledgements
We thank the National Institutes of Health (DP1DA042078, R01MH116900, R21DA044767 to I.M.), as well as the Brain and Behavior Research Foundation (NARSAD Young Investigator Award to L.A.F.), for their gracious support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Maze I, Noh KM & Allis CD Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacology 38, 3–22, doi: 10.1038/npp.2012.124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borrelli E, Nestler EJ, Allis CD & Sassone-Corsi P Decoding the epigenetic language of neuronal plasticity. Neuron 60, 961–974, doi:S0896–6273(08)00885–4 [pii] 10.1016/j.neuron.2008.10.012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T & Allis CD Translating the histone code. Science 293, 1074–1080, doi: 10.1126/science.1063127 293/5532/1074 [pii] (2001). [DOI] [PubMed] [Google Scholar]
- 4.Dhalluin C et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496, doi: 10.1038/20974 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45, doi: 10.1038/47412 (2000). [DOI] [PubMed] [Google Scholar]
- 6.McClung CA & Nestler EJ Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology 33, 3–17, doi: 10.1038/sj.npp.1301544 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R & Bird A Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics 33 Suppl, 245–254, doi: 10.1038/ng1089 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Spitale RC, Tsai MC & Chang HY RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics: official journal of the DNA Methylation Society 6, 539–543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levenson JM & Sweatt JD Epigenetic mechanisms in memory formation. Nat Rev Neurosci 6, 108–118, doi:nrn1604 [pii] 10.1038/nrn1604 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Alarcon JM et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42, 947–959, doi: 10.1016/j.neuron.2004.05.021 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Tahiliani M et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447, 601–605, doi:nature05823 [pii] 10.1038/nature05823 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Gibbons RJ, Picketts DJ, Villard L & Higgs DR Mutations in a putative global transcriptional regulator cause X-linked mental retardation with CE±-thalassemia (ATR-X syndrome). Cell 80, 837–845, doi:Cited By (since 1996) 281 Export Date 23 February 2012 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Duran D et al. Mutations in Chromatin Modifier and Ephrin Signaling Genes in Vein of Galen Malformation. Neuron 101, 429–443 e424, doi: 10.1016/j.neuron.2018.11.041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najmabadi H et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63, doi: 10.1038/nature10423 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Hattori M et al. The DNA sequence of human chromosome 21. Nature 405, 311–319, doi: 10.1038/35012518 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Jiang J et al. Translating dosage compensation to trisomy 21. Nature 500, 296–300, doi: 10.1038/nature12394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prandini P et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet 81, 252–263, doi: 10.1086/519248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weick JP et al. Deficits in human trisomy 21 iPSCs and neurons. Proc Natl Acad Sci U S A 110, 9962–9967, doi: 10.1073/pnas.1216575110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olmos-Serrano JL et al. Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination. Neuron 89, 1208–1222, doi: 10.1016/j.neuron.2016.01.042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A & Deutsch S Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet 5, 725–738, doi: 10.1038/nrg1448 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Kahlem P et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res 14, 1258–1267, doi: 10.1101/gr.1951304 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S & Antonarakis SE Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res 14, 1268–1274, doi: 10.1101/gr.2090904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ait Yahya-Graison E et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet 81, 475–491, doi: 10.1086/520000 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dekker AD, De Deyn PP & Rots MG Epigenetics: the neglected key to minimize learning and memory deficits in Down syndrome. Neurosci Biobehav Rev 45, 72–84, doi: 10.1016/j.neubiorev.2014.05.004 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Pina B & Suau P Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Developmental Biology 123, 51–58 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Hake SB et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. The Journal of biological chemistry 281, 559–568, doi: 10.1074/jbc.M509266200 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Hake SB & Allis CD Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proceedings of the National Academy of Sciences of the United States of America 103, 6428–6435, doi: 10.1073/pnas.0600803103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein E & Hake SB The nucleosome: a little variation goes a long way. Biochemistry and cell biology = Biochimie et biologie cellulaire 84, 505–517, doi: 10.1139/o06-085 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Michod D et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron 74, 122–135, doi: 10.1016/j.neuron.2012.02.021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maze I et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 87, 77–94, doi: 10.1016/j.neuron.2015.06.014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun H et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med 21, 1146–1153, doi: 10.1038/nm.3939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM & Sweatt JD Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–586, doi: 10.1038/nature13707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charney DS Monoamine dysfunction and the pathophysiology and treatment of depression. J Clin Psychiatry 59 Suppl 14, 11–14 (1998). [PubMed] [Google Scholar]
- 34.Colgan LA, Putzier I & Levitan ES Activity-dependent vesicular monoamine transporter-mediated depletion of the nucleus supports somatic release by serotonin neurons. J Neurosci 29, 15878–15887, doi: 10.1523/JNEUROSCI.4210-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young AB, Pert CD, Brown DG, Taylor KM & Snyder SH Nuclear localization of histamine in neonatal rat brain. Science 173, 247–249 (1971). [DOI] [PubMed] [Google Scholar]
- 36.Walther DJ et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet alpha-granule release. Cell 115, 851–862 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Watts SW, Priestley JR & Thompson JM Serotonylation of vascular proteins important to contraction. PLoS One 4, e5682, doi: 10.1371/journal.pone.0005682 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hummerich R & Schloss P Serotonin--more than a neurotransmitter: transglutaminase-mediated serotonylation of C6 glioma cells and fibronectin. Neurochem Int 57, 67–75, doi: 10.1016/j.neuint.2010.04.020 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Hummerich R, Thumfart JO, Findeisen P, Bartsch D & Schloss P Transglutaminase-mediated transamidation of serotonin, dopamine and noradrenaline to fibronectin: evidence for a general mechanism of monoaminylation. FEBS Lett 586, 3421–3428, doi: 10.1016/j.febslet.2012.07.062 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Farrelly LA et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539, doi: 10.1038/s41586-019-1024-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauberth SM et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036, doi: 10.1016/j.cell.2013.01.052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton PJ, Lim CJ, Nestler EJ & Heller EA Neuroepigenetic Editing. Methods Mol Biol 1767, 113–136, doi: 10.1007/978-1-4939-7774-1_5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David Y, Vila-Perello M, Verma S & Muir TW Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat Chem 7, 394–402, doi: 10.1038/nchem.2224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eryilmaz E, Shah NH, Muir TW & Cowburn D Structural and dynamical features of inteins and implications on protein splicing. J Biol Chem 289, 14506–14511, doi: 10.1074/jbc.R113.540302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
References to Annotate:
- Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45, doi: 10.1038/47412 (2000).- [DOI] [PubMed] [Google Scholar]; This highly influential hypothesis piece discussed the network of post-translational modifications that decorate histone tails, and posited that such signals can be “read” by specific chromatin interacting proteins–in a combinatorial manner–to regulation gene expression (i.e., the ‘histone code’).
- David Y, Vila-Perello M, Verma S & Muir TW Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat Chem 7, 394–402, doi: 10.1038/nchem.2224 (2015).- [DOI] [PMC free article] [PubMed] [Google Scholar]; This study described the incorporation of non-native histone PTMs in living cells using a novel protein trans-splicing technology.
- Hamilton PJ, Lim CJ, Nestler EJ & Heller EA Neuroepigenetic Editing. Methods Mol Biol 1767, 113–136, doi: 10.1007/978-1-4939-7774-1_5 (2018).- [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses current advances in neuroepigenetic editing approaches, further highlighting methodological considerations critical to neuroepigenetics research.
- Farrelly LA et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539, doi: 10.1038/s41586-019-1024-7 (2019).- [DOI] [PMC free article] [PubMed] [Google Scholar]; This study revealed a previously unidentified role for serotonin, an important neurotransmitter in the CNS, as a novel histone post-translational modification that influences permissive gene expression in brain.
- Maze I et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 87, 77–94, doi: 10.1016/j.neuron.2015.06.014 (2015).- [DOI] [PMC free article] [PubMed] [Google Scholar]; This article demonstrated that nucleosomal histones, specifically those containing the variant H3.3–long believed to be ‘static’ in post-mitotic chromatin–are dynamically turned over during both neurodevelopment and in adult brain to control fundamental aspects of neuronal transcriptional and phenotypic plasticity.