Abstract
As an individual becomes addicted to a drug of abuse, nerve cells within the brain’s reward circuitry adapt at the epigenetic level during the course of repeated drug exposure. These drug-induced epigenetic adaptations mediate enduring changes in brain function which contribute to life-long, drug-related behavioral abnormalities that define addiction. Targeting these epigenetic alterations will enhance our understanding of the biological basis of addiction and might even yield more effective anti-addiction therapies. However, the complexity of the neuroepigenetic landscape makes it difficult to determine which drug-induced epigenetic changes causally contribute to the pathogenic mechanisms of drug addiction. In this review, we highlight the evidence that epigenetic modifications, specifically histone modifications, within key brain reward regions are correlated with addiction. We then discuss the emerging field of locus-specific neuroepigenetic editing, which is a promising method for determining the causal epigenetic molecular mechanisms that drive an addicted state. Such approaches will substantially increase the field’s ability to establish the precise epigenetic mechanisms underlying drug addiction, and could lead to novel treatments for addictive disorders.
Epigenetic modifications in addiction
The neuroepigenetic landscape
The term ‘epigenetics’ is commonly used to describe the dynamic molecular modifications deposited upon chromatin within a cell’s nucleus, which has the functional consequence of regulating DNA-related processes, such as DNA repair, chromatin organization, and RNA transcription and splicing, among other functions. Drug addiction researchers have become interested in studying epigenetics due to the fact that an individual’s experience, specifically volitional, repeated drug consumption, alters the chromatin landscape within the brain in a region- and cell type-specific manner1. It is widely hypothesized that, by regulating DNA-related processes, these drug-induced epigenetic alterations contribute to aberrant cellular function that drives drug addiction pathogenesis. There may, therefore, be therapeutic potential in targeting key drug-induced epigenetic modifications within the brain as a way of combating an individual’s spiral into an addictive state. However, to achieve this goal, it is vital to first untangle the remarkable complexity of addiction-related epigenetics to determine which specific, drug-induced epigenetic modifications casually contribute to distinct aspects of the pathophysiological maladaptations underlying drug addiction. The burgeoning field of locus-specific neuroepigenetic editing is uniquely well suited to pursue this goal and is the focus of this review.
To contextualize the complexity of the neuroepigenetic state, it is important to understand the diversity of epigenetic modifications. For simplicity, we concentrate here on histone post-translational modifications (PTMs), but other epigenetic regulatory events include DNA modifications and the actions of non-coding RNAs2. Chromatin is the macromolecular complex consisting of DNA wrapped tightly around histone protein octamers to form nucleosomes. Chromatin enables the dense packing of nucleosomes to fit within the cell’s nucleus, and provides an instructive scaffold that is responsive to external cues. Histones are highly basic and enriched for lysine and arginine residues. PTMs of these and other residues on histone N-terminal tails, which protrude from the nucleosome core, alter the steric properties and charge distribution of chromatin, controlling DNA-related processes. Histone subunits can be modified by numerous PTMs including acetylation, methylation, phosphorylation, ADP ribosylation, ubiquitylation, and sumoylation, among a growing list of newly discovered modifications, which occur on >50 distinct sites2–4. The diversity of modifications, and the innumerable sites multiplied across the ~2 meters of linear DNA within a given cell, underscore the vast complexity of the epigenetic landscape.
Histone PTMs are reversible: they are dynamically deposited by “writer’ enzymes, recognized by “reader” proteins which mediate the cellular response, and removed by “eraser” enzymes2. It is the delicate balance between the function of writers and erasers that dictates the global epigenetic state and downstream functions within a given brain cell. The expression and function of numerous writers, erasers, and readers have been observed to be altered both in addicted humans and in animal models of addiction1,5,6. Restoring normal function to these proteins through the use of small molecules represents a novel area for anti-addiction pharmacotherapies7, yet understanding the proteins to target, and how their regulation would affect the broader addictive phenotype, remains inadequately explored.
Drug experience alters the neuroepigenetic landscape
Drug addiction is characterized by the compulsive seeking and taking of drug despite adverse consequences8. Vulnerability to addiction emerges through approximately equal parts of genetic predisposition and environmental risk, strongly suggesting an important role for epigenetic mechanisms. All abused drugs target the mesolimbic dopamine circuitry9, which serves the evolutionary purpose of reinforcing activities important for an individual’s survival and reproduction, such as seeking palatable food and sex10. The mesolimbic circuitry is composed of dopaminergic neurons in the midbrain ventral tegmental area (VTA) and their innervation of medium spiny neurons (MSNs), the predominant cell type within the nucleus accumbens (NAc)11. All natural rewards and abused drugs share the property of acutely increasing dopaminergic neurotransmission in NAc12. Chronic drug use induces long-lasting structural, electrophysiological, and transcriptional changes in this region, which are considered the sustained biological substrates of addiction (Figure 1). For this reason, researchers are examining epigenetic maladaptations within NAc cell types as driving addiction pathogenesis13.
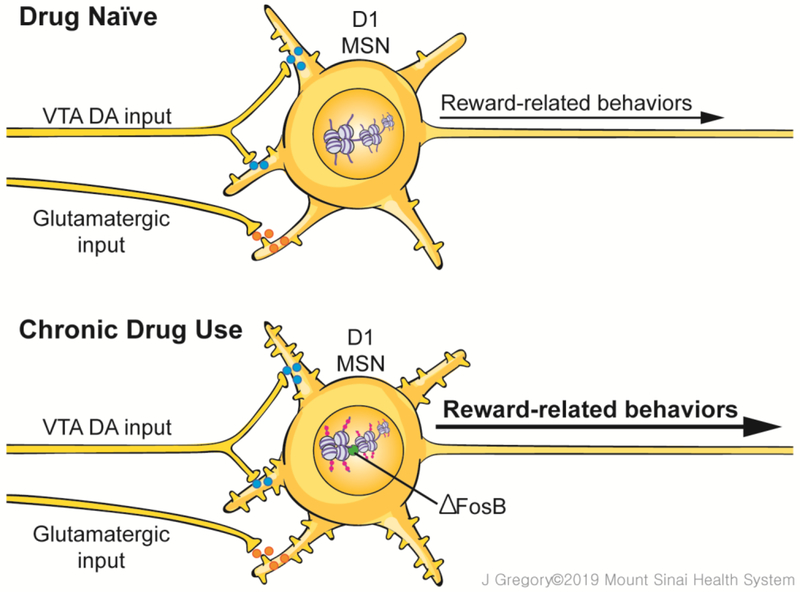
Figure 1. Chronic drug use alters the epigenome in reward processing neurons.
Top: In drug naïve conditions, medium spiny neurons (MSNs) within the nucleus accumbens (NAc) receive dopaminergic inputs from the ventral tegmental area (VTA) and glutamatergic inputs from several cortical and thalamic brain regions. These MSNs receive and integrate reward-related signals, and the homeostasis in nuclear epigenome writer and eraser enzymes within these MSNs enable normal reward-processing required for organismal survival. The NAc contains two types of MSNs, D1- and D2-type, named for the dopamine receptor they predominantly express. Only a D1-type MSN is shown. Bottom: Chronic drug use perturbs the balance in writer and eraser enzymes, resulting in a multitude of epigenetic adaptations at specific loci within the MSN nucleus. These adaptations, in conjunction with drug induction of certain transcription factors (e.g., ΔFosB). mediate transcriptional changes at many genes, including those that encode neurotransmitter receptors, cytoskeletal proteins, and ion channels, among many others. The cumulative consequence of these transcriptional adaptations is altered MSN morphology (e.g., increased dendritic spine density is shown) and physiological function in relation to reward processing which underlies behavioral maladaptations that define addiction.
The functional consequence of histone PTMs within NAc is well illustrated by histone acetylation. This PTM occurs on all core histone subunits, is catalyzed by histone acetyltransferases (HATs), and is reversed by histone deacetylases (HDACs). Histone acetylation is typically associated with an ‘opening’ of higher order chromatin structure14—that is, increased spacing between nucleosomes—which facilitates transcription activation by making the surrounding DNA more accessible to transcription factors and the transcriptional machinery. In support of the importance of histone acetylation in drug addiction, acute or chronic exposure to psychostimulants, opiates, alcohol, or nicotine all serve to increase total cellular levels of acetylation of histones H3 and H4 in NAc15–20. This increase in total acetylation levels is the net result of drug-induced alterations to the balance of HAT and HDAC function. cAMP response elementbinding protein (CREB)-binding protein (CBP), a HAT critical to learning and memory21, is required for cocaine-induced increases in histone acetylation in the NAc22. Conversely, chronic cocaine23 or alcohol24 use reduces HDAC enzymatic activity and disrupts sub-cellular HDAC localization25. Further, while chronic cocaine exposure promotes an increase in NAc expression of sirtuins26, class III HDACs, the genomic accessibility, particularly within gene promoters, is reduced27. This limits the capacity of this enzyme to reduce histone acetylation, specifically at key gene sites.
Indeed, the deposition of histone acetylation that occurs in NAc in addiction models is highly locus specific. For instance, in response to acute psychostimulant exposure, H4 acetylation occurs within the promoters of immediate early genes like c-Fos and Fosb, which associates with their rapid expression in response to drug consumption15,28. For Fosb, this increase in histone acetylation requires CBP29, indicating gene-specific actions of these chromatin modifiers upon drug-experience.
The biological relevance of chromatin modifiers to drug-related behavior is further strengthened by NAc-targeted viral or genetic manipulations. Viral over-expression of the transcription factor CREB reduces the rewarding properties of cocaine30 and morphine31, indicating that CREB activation may be a general mechanism for reward tolerance. Conversely, viral over-expression of ΔFosB promotes the rewarding properties of cocaine32 and morphine33, revealing how transcriptional regulators can oppositely impact drug-related behaviors. Further, genetic deletion of the HAT CBP impairs cocaine sensitivity and cocaine-related memory22, and a sizeable body of work demonstrates that direct manipulations to HDACs alter behaviors related to multiple classes of abused substances23,28,34,35. Manipulation of enzymes that control histone or DNA methylation also control behavioral responses to drugs of abuse36,37. As well, viral delivery of accessory subunits to chromatin remodeling proteins alters drug-related behaviors38. Taken together, these direct manipulations to chromatin modifiers indicate that the actions of these factors contribute to addiction pathogenesis. However, it remains unclear whether these and many other chromatin modifications are required, causal events in driving key transcriptional adaptations, or if they are simply correlated with such regulation. This has remained a challenging question, since all pharmacological, viral, and genetic tools manipulate histone PTMs genome-wide. The advent of locus-specific neuroepigenetic editing within specific brain cell types has enabled researchers to make crucial advances in establishing the casual consequences of gene-specific PTMs within the context of drug addiction.
Determining causality with locus-specific neuroepigenetic editing
The introduction of easy-to-use, artificial fusion proteins has launched the new field of in vivo neuroepigenetic editing in brain, making it possible for the first time to explore the causal epigenetic mechanisms driving drug addiction. The long-term goal of this research is to distill which drug-induced epigenetic marks contribute most prominently to the deleterious pathogenic mechanisms of addiction, and to use this knowledge to guide the design of novel therapies to correct these maladaptations with minimal off-target effects. In theory, this approach could yield superior addiction treatments with the capacity to halt or even reverse the pathogenesis of an addicted state.
Neuroepigenetic editing refers to the targeted rewriting of the epigenome at a single genomic locus within a given neuronal or other cell type in a given brain region. The ability to accomplish this novel approach is built upon the fields of gene editing and epigenome engineering, which have enjoyed a renaissance within the past eight years, but have been applied almost exclusively in vitro39–43. The much more recent ability to adapt these technologies in awake, behaving animals has allowed for key advances in studying the causal epigenetic mechanisms underlying neuropsychiatric syndromes44–46.
DNA-binding domains
Neuroepigenetic editing is accomplished through the expression of bi-functional constructs consisting of a programmable, DNA-binding domain that binds a desired genomic locus with high affinity and specificity, and an effector moiety, which mediates an epigenetic modification confined exclusively to the chromatin landscape proximal to DNA-binding (Figure 2). The first used DNA-binding domains were zinc finger proteins (ZFPs)47,48, derived from eukaryotic transcription factors, or transcription activation-like effectors (TALEs)49,50, derived from plant pathogenic prokaryotes. Fusing either of these DNA-binding domains to effector moieties allowed researchers to demonstrate the feasibility and utility of epigenetic editing for a variety of neuroscience research-related approaches51–56. However, as ZFPs and TALEs utilize protein-DNA interactions to mediate DNA-targeting, the synthesis and validation of these tools are time-consuming, expensive, and technically challenging, which has limited their application.
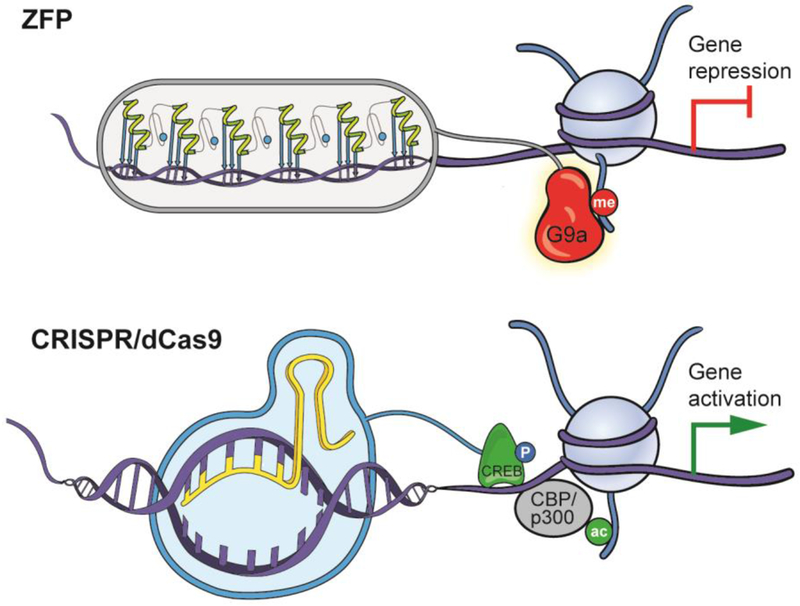
Figure 2. Neuroepigenetic editing tools.
Such tools are bi-functional constructs that consist of a DNA-binding domain that is designed to target a desired sequence of genomic DNA with high affinity and specificity and an effector moiety that mediates an epigenetic modification proximal to the region of DNA-binding. This results in epigenetic editing restricted to a single genomic locus, the specificity of which must be validated extensively to ensure minimal off-target binding. The most widely used DNA-binding domains are zinc finger proteins (ZFPs) and, more recently, RNA-guided CRISPR/dCas9. Here, we highlight a ZFP-G9a fusion that our group has generated for targeted gene repression in brain52 (top), as well as a dCas9-CREB fusion for in vivo gene-targeted activation81 (bottom). Figure modified with permission from reference 44: Hamilton, P. J., Lim, C. J., Nestler, E. J. & Heller, E. A. Neuroepigenetic Editing. Methods in molecular biology 1767, 113-136, doi:10.1007/978-1-4939-7774-1_5 (2018).
The more recent development of CRISPR/dCas9-based eukaryotic genome targeting has radically simplified the design and synthesis of DNA-targeting domains57. In contrast to ZFP- and TALE-based platforms, CRISPR/dCas9 DNA-targeting is mediated by programming a 20 bp sequence within a single guide RNA (sgRNA) (see Figure 2). The sgRNA complexes with nuclease-deficient dCas9 and targets the complex to the genomic region where the guide sequence binds its complementary DNA sequence58. Tethering an effector moiety to the sgRNA or, more commonly, to dCas9 allows for DNA-targeted localization of a desired effector. The ease and cost-effectiveness of design and synthesis of the CRISPR/dCas9 platform has helped usher in a new era of epigenome engineering research.
Effector moieties
Shortly after applying programmable DNA-binding domains for genome editing, researchers began fusing to transcriptional activator domains as a means of inducing expression of targeted endogenous genes. One of the first, and most effective, effector moieties was VP64, derived from herpes simplex virus (HSV), which directly recruits RNA polymerase II for gene-targeted transcriptional activation59. VP64 does not directly mediate epigenetic reprogramming, although permissive chromatin remodeling is associated with its function60,61; rather, VP64 is a scaffold for recruiting gene-activating transcriptional machinery62. By targeting VP64 to the promoter region of a single gene with ZFP-63, TALE-60,64, or CRISPR/dCas9-60,65 DNA-binding domains, laboratories have consistently reported the ability to induce transcription of targeted genes in vitro and even in vivo61,66.
Many laboratories have pursued improvements to the magnitude of gene activation, which is accomplished by concentrating effector moieties at a single genomic region to induce a proportional increase in gene activation. One approach was termed SunTag, a peptide scaffold containing repeating binding sites for effector domains like VP6467. Another strategy was termed CRISPR-SAM, wherein the sgRNA is re-engineered at protruding RNA loops with RNA aptamers to serve as a scaffold for recruitment of activation domains42. A third strategy to concentrate effector domains is to target multiple DNA-binding sites within a given promoter region simultaneously57. These approaches are useful for in vitro assays where large magnitude effects are often desired, yet can be prohibitively complex when attempting to deliver the multiple components to targeted cells within the brain.
To achieve bi-directional control of gene expression, repression of an endogenous gene is most universally accomplished with Kruppel-associated box (KRAB) effector moieties. KRAB, a transcriptional repression domain found in human zinc finger transcription factors, recruits heterochromatin-forming complexes, which in turn deposit repressive H3K9me3 marks and represses transcription68. Like VP64, the KRAB domain does not possess enzymatic activity, but instead recruits secondary factors to edit the epigenome. Promoter- or enhancer-targeted KRAB has been utilized in cell culture and in brain69,70.
While these methods of bi-directional regulation are more physiologically-relevant than conventional transgenic overexpression or knockout approaches, directly regulating genes in this manner lacks the construct validity of replicating the actual aberrant molecular mechanisms present within the addicted state. Indeed, decades of research have revealed the specific transcription factors and epigenetic writers, readers, and erasers implicated in the pathogenesis of drug addiction. In the past few years, there has been an explosion in the use of effector moieties for neuroepigenetic editing. These include: DNA-modifying enzymes like DNMT3a (which methylates CpG)71,72 and TET1 (which catalyzes hydroxymethyl-CpG)73,74; proteins that control histone PTMs including: NFκB subunit p65 (which recruits HATs to acetylate core histones)52–54, histone methyltransferase G9a (which catalyzes H3K9me2)52–54, p300 HAT (which acetylates all four core histones)43, SID (which recruits HDACs)75,76, LSD1 (which demethylates H3K4 and H3K9)77, PRDM9 (which methylates H3K4 and H3K36)78, and DOT1L (which methylates H3K79)79,80; and even transcription factors like CREB—thus mimicking a known mechanism of transcriptional regulation81. The modularity of designing neuroepigenetic editing constructs presents the ability to apply a wide range of effector domains for epigenome editing purposes that more closely models the molecular mechanisms underlying drug addiction.
Utility in studying drug addiction
Neuroepigenetic editing approaches present several advantages over conventional transgene overexpression or knockout approaches, including controlling the expression of the targeted gene from its endogenous promoter (potentially via physiologically relevant mechanisms depending on the effector moiety utilized), as well as controlling the magnitude of gene expression levels within physiological ranges. These advantages justify the more widespread use of these approaches, particularly in researching neuropsychiatric syndromes like drug addiction. However, the major hurdle for applying neuroepigenetic editing to studies of addiction is the challenge in delivering these tools to the brain of awake, behaving animals. Delivery of such constructs to intact tissues is an active area of investigation for both research and therapeutic purposes82. To date, scientists have applied several approaches for in vivo brain delivery, most involving injection of viral vectors via stereotaxic surgery (Figure 3).
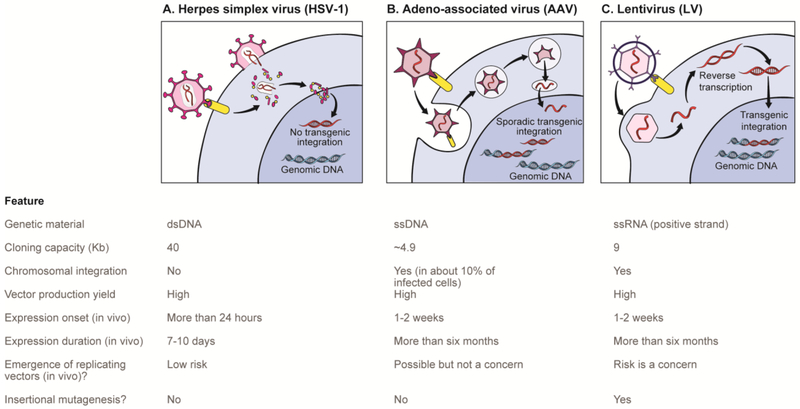
Figure 3: In vivo delivery methods for neurocpigcnctic editing tools.
A summary of the properties of routinely used viral vectors available for delivery of neuroepigenetic editing tools to the brain of awake, behaving animals. While other approaches exist, the vast majority of published work on neuroepigenetic editing in brain cells utilize these viral delivery strategies coupled with stereotaxic surgery. Figure reproduced with permission from reference 44: Hamilton, P. J., Lim, C. J., Nestler, E. J. & Heller, E. A. Neuroepigenetic Editing. Methods in molecular biology 1767, 113-136, doi:10.1007/978-1-4939-7774-1_5 (2018).
Viruses like adeno-associated virus (AAV) are challenging to use for neuroepigenetic editing owing to their limited packaging capacity (~4.5 kb). The most commonly utilized Cas9, derived from Streptococcus pyogenes, is ~4.1 kb83. Fusing even the smallest effector moiety pushes it above the AAV packaging limit. While other Cas9 species are ~1 kb shorter (e.g., Staphylococcus aureus)84, most researchers have elected to utilize viruses with larger packaging capacities. The most widely applied viruses are HSVs, which mediate short-lived transgene expression (up to 7 days) with a large packaging capacity (~14 kb), or lentivirus, which mediates long-term transgene expression of >6 months with a packaging capacity of 8-10 kb. Figure 3 compares the features of these viral delivery methods. Importantly, incorporation of loxP sites within these vectors, combined with the use of Cre-driver lines, makes it possible to target neuroepigenetic editing constructs to specific types of neurons or other cell types within a brain region of interest54,55.
There are now several published examples of utilizing locus-specific neuroepigenetic editing to demonstrate that a single type of histone PTM (i.e., histone acetylation or methylation) at a single gene promoter in a single brain region or even cell type within that region alters expression levels of that gene and induces downstream behavioral effects in drug addiction and related models52–56. The first demonstration came in 2014 when Heller et al.52 designed a suite of ZFPs that bound the promoter region of the FosB gene and were fused to either G9a or p65. These constructs were packaged in HSVs and delivered to the NAc of mice via stereotaxic surgery. Depending on promoter localization and the effector moiety, these FosB-ZFFs deposited the specific histone PTM (methylation with G9a, acetylation with p65) proximal to their DNA-binding site and consequently bi-directionally regulated NAc levels of mRNA and protein expression of FosB, and the splice variant ΔFosB—whose accumulation in this brain region promotes behavioral responses to cocaine and other drugs of abuse, as mentioned earlier. The authors demonstrated further that such deposition of PTMs and regulation of FosB expression occurred solely at the FosB locus, and that FosB-G9a decreased, whereas FosB-p65 increased, the locomotor behaviors associated with repeated cocaine exposure. This provided an authoritative demonstration that the epigenetic state at the FosB locus and subsequent NAc accumulation of ΔFosB is necessary and sufficient to modulate cocaine-evoked behaviors. Heller, Hamilton et al. 201653 built on these approaches by directing ZFP tools to Cdk5, a locus epigenetically regulated by chronic cocaine exposure, and demonstrating that both the rewarding and locomotor properties of cocaine were dependent on the epigenetic state of Cdk5. These studies reveal that epigenetic regulation at the FosB and Cdk5 loci are not merely correlative, but actually casual molecular adaptations that drive the pathogenesis of addictive behaviors, and establish more generally that manipulation of a single type of PTM at a single locus in a single brain region controls the expression level of the targeted gene and downstream behavioral outcomes. While not yet applied to study addiction, CRISPR-based neuroepigenetic editing approaches have been used in targeted brain regions and have proved fruitful for exploring other brain illnesses73,74,81,85, supporting their utility to study addiction. These findings establish that epigenetic modifications at a given locus can in and of themselves mediate transcriptional regulation in fully differentiated adult neurons in vivo, and these approaches enable a form of in vivo manipulation that was previously unattainable.
Conclusions and future directions
The ability to deposit or remove specific epigenetic marks at restricted gene sites within targeted cell types in key brain regions provides researchers with the unparalleled ability to experimentally interrogate the causal contribution of these epigenetic molecular mechanisms to neuropsychiatric syndromes, like addiction. The modularity and flexibility of neuroepigenetic editing approaches make them ideal and adaptable tools to untangle in vivo epigenetic complexity. It is theoretically possible, for example, to use CRISPR/dCas9 to target multiple genes within the same cells and thereby experimentally manipulate networks of drug-regulated genes. By combining these approaches with cutting-edge animal models of addiction, like drug self-administration, researchers will be able to isolate the specific epigenetic maladaptations that drive the most damaging elements of drug addiction, like binging, craving, and relapse. These novel insights will guide the development of targeted, and likely more effective, pharmacotherapies in an effort to alleviate the human suffering associated with drug addiction.
Highlights:
Chronic drug use alters the epigenetic state of neurons and other cell types within brain reward regions.
Epigenetic adaptations are correlated with addiction, but their causal contributions are incompletely characterized.
Neuroepigenetic editing enables locus-targeted epigenetic manipulations in specific brain cells.
Such locus-directed epigenome editing reveals specific causal contributions to addiction pathogenesis.
Acknowledgments
Funding Sources:
The preparation of this review was supported in part by funding from National Institute on Drug Abuse: K99 DA045795 to P.J.H and R01 DA07359 to E.J.N.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
References of interest:
Of interest (*): 13,52,56,73
Of special interest (**): 54,61,66,81
- 1.Robison AJ & Nestler EJ Transcriptional and epigenetic mechanisms of addiction. Nature reviews Neuroscience 12, 623–637, doi: 10.1038/nrn3111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nature reviews. Genetics 17, 487–500, doi: 10.1038/nrg.2016.59 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Bowman GD & Poirier MG Post-translational modifications of histones that influence nucleosome dynamics. Chemical reviews 115, 2274–2295, doi: 10.1021/cr500350x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ & Kouzarides T Regulation of chromatin by histone modifications. Cell research 21, 381–395, doi: 10.1038/cr.2011.22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogge GA & Wood MA The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 38, 94–110, doi: 10.1038/npp.2012.154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egervari G, Ciccocioppo R, Jentsch JD & Hurd YL Shaping vulnerability to addiction - the contribution of behavior, neural circuits and molecular mechanisms. Neuroscience and biobehavioral reviews 85, 117–125, doi: 10.1016/j.neubiorev.2017.05.019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kennedy PJ & Harvey E Histone Deacetylases as Potential Targets for Cocaine Addiction. CNS & neurological disorders drug targets 14, 764–772 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Koob GF & Volkow ND Neurobiology of addiction: a neurocircuitry analysis. The lancet. Psychiatry 3, 760–773, doi: 10.1016/S2215-0366(16)00104-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koob G & Kreek MJ Stress, dysregulation of drug reward pathways, and the transition to drug dependence. The American journal of psychiatry 164, 1149–1159, doi: 10.1176/appi.ajp.2007.05030503 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley AE & Berridge KC The neuroscience of natural rewards: relevance to addictive drugs. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 3306–3311, doi:20026361 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyman SE, Malenka RC & Nestler EJ Neural mechanisms of addiction: the role of reward-related learning and memory. Annual review of neuroscience 29, 565–598, doi: 10.1146/annurev.neuro.29.051605.113009 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Diana M The dopamine hypothesis of drug addiction and its potential therapeutic value. Frontiers in psychiatry 2, 64, doi: 10.3389/fpsyt.2011.00064 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Feng J et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome biology 15, R65, doi: 10.1186/gb-2014-15-4-r65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shogren-Knaak M et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847, doi: 10.1126/science.1124000 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Kumar A et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48, 303–314, doi: 10.1016/j.neuron.2005.09.023 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Shen HY et al. Additive effects of histone deacetylase inhibitors and amphetamine on histone H4 acetylation, cAMP responsive element binding protein phosphorylation and DeltaFosB expression in the striatum and locomotor sensitization in mice. Neuroscience 157, 644–655, doi: 10.1016/j.neuroscience.2008.09.019 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Schroeder FA et al. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 33, 2981–2992, doi: 10.1038/npp.2008.15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renthal W et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62, 335–348, doi: 10.1016/j.neuron.2009.03.026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine A et al. Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. Science translational medicine 3, 107ra109, doi: 10.1126/scitranslmed.3003062 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botia B, Legastelois R, Alaux-Cantin S & Naassila M Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice. PloS one 7, e47527, doi: 10.1371/journal.pone.0047527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett RM & Wood MA Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learning & memory 15, 460–467, doi: 10.1101/lm.917508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malvaez M, Mhillaj E, Matheos DP, Palmery M & Wood MA CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 16941–16948, doi: 10.1523/JNEUROSCI.2747-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renthal W et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron 56, 517–529, doi: 10.1016/j.neuron.2007.09.032 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Pandey SC, Ugale R, Zhang H, Tang L & Prakash A Brain chromatin remodeling: a novel mechanism of alcoholism. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 3729–3737, doi: 10.1523/JNEUROSCI.5731-07.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi M et al. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron 73, 108–120, doi: 10.1016/j.neuron.2011.10.032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson D et al. Essential role of SIRT1 signaling in the nucleus accumbens in cocaine and morphine action. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 16088–16098, doi: 10.1523/JNEUROSCI.1284-13.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson D et al. SIRT1-FOXO3a regulate cocaine actions in the nucleus accumbens. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 3100–3111, doi: 10.1523/JNEUROSCI.4012-14.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renthal W et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 7344–7349, doi: 10.1523/JNEUROSCI.1043-08.2008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine AA et al. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proceedings of the National Academy of Sciences of the United States of America 102, 19186–19191, doi: 10.1073/pnas.0509735102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlezon WA Jr. et al. Regulation of cocaine reward by CREB. Science 282, 2272–2275 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Barrot M et al. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proceedings of the National Academy of Sciences of the United States of America 99, 11435–11440, doi: 10.1073/pnas.172091899 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grueter BA, Robison AJ, Neve RL, Nestler EJ & Malenka RC FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proceedings of the National Academy of Sciences of the United States of America 110, 1923–1928, doi: 10.1073/pnas.1221742110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zachariou V et al. An essential role for DeltaFosB in the nucleus accumbens in morphine action. Nature neuroscience 9, 205–211, doi: 10.1038/nn1636 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Li X et al. Role of Dorsal Striatum Histone Deacetylase 5 in Incubation of Methamphetamine Craving. Biological psychiatry 84, 213–222, doi: 10.1016/j.biopsych.2017.12.008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L et al. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 35, 913–928, doi: 10.1038/npp.2009.193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maze I et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216, doi: 10.1126/science.1179438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaPlant Q et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nature neuroscience 13, 1137–1143, doi: 10.1038/nn.2619 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H et al. BAZ1B in Nucleus Accumbens Regulates Reward-Related Behaviors in Response to Distinct Emotional Stimuli. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 3954–3961, doi: 10.1523/JNEUROSCI.3254-15.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823, doi: 10.1126/science.1231143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821, doi: 10.1126/science.1225829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451, doi: 10.1016/j.cell.2013.06.044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588, doi: 10.1038/nature14136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology 33, 510–517, doi: 10.1038/nbt.3199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamilton PJ, Lim CJ, Nestler EJ & Heller EA Neuroepigenetic Editing. Methods in molecular biology 1767, 113–136, doi: 10.1007/978-1-4939-7774-1_5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang MG & West AE Editing the Neuronal Genome: a CRISPR View of Chromatin Regulation in Neuronal Development, Function, and Plasticity. The Yale journal of biology and medicine 89, 457–470 (2016). [PMC free article] [PubMed] [Google Scholar]
- 46.Savell KE & Day JJ Applications of CRISPR/Cas9 in the Mammalian Central Nervous System. The Yale journal of biology and medicine 90, 567–581 (2017). [PMC free article] [PubMed] [Google Scholar]
- 47.Klug A The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual review of biochemistry 79, 213–231, doi: 10.1146/annurev-biochem-010909-095056 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Gersbach CA, Gaj T & Barbas CF 3rd. Synthetic zinc finger proteins: the advent of targeted gene regulation and genome modification technologies. Accounts of chemical research 47, 2309–2318, doi: 10.1021/ar500039w (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boch J et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512, doi: 10.1126/science.1178811 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Moscou MJ & Bogdanove AJ A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501, doi: 10.1126/science.1178817 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Snowden AW, Gregory PD, Case CC & Pabo CO Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Current biology : CB 12, 2159–2166 (2002). [DOI] [PubMed] [Google Scholar]
- 52*.Heller EA et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nature neuroscience 17, 1720–1727, doi: 10.1038/nn.3871 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heller EA et al. Targeted Epigenetic Remodeling of the Cdk5 Gene in Nucleus Accumbens Regulates Cocaine- and Stress-Evoked Behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 4690–4697, doi: 10.1523/JNEUROSCI.0013-16.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Hamilton PJ et al. Cell-Type-Specific Epigenetic Editing at the Fosb Gene Controls Susceptibility to Social Defeat Stress. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 43, 272–284, doi: 10.1038/npp.2017.88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aleyasin H et al. Cell-Type-Specific Role of DeltaFosB in Nucleus Accumbens In Modulating Intermale Aggression. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 5913–5924, doi: 10.1523/JNEUROSCI.0296-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Sase AS et al. Sex-Specific Regulation of Fear Memory by Targeted Epigenetic Editing of Cdk5. Biological psychiatry, doi: 10.1016/j.biopsych.2018.11.022 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183, doi: 10.1016/j.cell.2013.02.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sander JD & Joung JK CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology 32, 347–355, doi: 10.1038/nbt.2842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall DB & Struhl K The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein cross-linking in vivo. The Journal of biological chemistry 277, 46043–46050, doi: 10.1074/jbc.M208911200 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Polstein LR et al. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome research 25, 1158–1169, doi: 10.1101/gr.179044.114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Liao HK et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507 e1415, doi: 10.1016/j.cell.2017.10.025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ingles CJ, Shales M, Cress WD, Triezenberg SJ & Greenblatt J Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351, 588–590, doi: 10.1038/351588a0 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Stege JT, Guan X, Ho T, Beachy RN & Barbas CF 3rd. Controlling gene expression in plants using synthetic zinc finger transcription factors. The Plant journal : for cell and molecular biology 32, 1077–1086 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Crocker J & Stern DL TALE-mediated modulation of transcriptional enhancers in vivo. Nature methods 10, 762–767, doi: 10.1038/nmeth.2543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Black JB et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell stem cell 19, 406–414, doi: 10.1016/j.stem.2016.07.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.Zhou H et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nature neuroscience 21, 440–446, doi: 10.1038/s41593-017-0060-6 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646, doi: 10.1016/j.cell.2014.09.039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SS et al. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proceedings of the National Academy of Sciences of the United States of America 93, 15299–15304 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Groner AC et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS genetics 6, e1000869, doi: 10.1371/journal.pgen.1000869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng Y et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nature neuroscience 21, 447–454, doi: 10.1038/s41593-018-0077-5 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Vojta A et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic acids research 44, 5615–5628, doi: 10.1093/nar/gkw159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stepper P et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic acids research 45, 1703–1713, doi: 10.1093/nar/gkw1112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73*.Liu XS et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247 e217, doi: 10.1016/j.cell.2016.08.056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu XS et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–992 e976, doi: 10.1016/j.cell.2018.01.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cong L, Zhou R, Kuo YC, Cunniff M & Zhang F Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nature communications 3, 968, doi: 10.1038/ncomms1962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476, doi: 10.1038/nature12466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kearns NA et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nature methods 12, 401–403, doi: 10.1038/nmeth.3325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cano-Rodriguez D et al. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nature communications 7, 12284, doi: 10.1038/ncomms12284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lei Y et al. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nature communications 8, 16026, doi: 10.1038/ncomms16026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwon DY, Zhao YT, Lamonica JM & Zhou Z Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nature communications 8, 15315, doi: 10.1038/ncomms15315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81**.Lorsch ZS et al. Zfp189 Mediates Stress Resilience Through a CREB-Regulated Transcriptional Network in Prefrontal Cortex. bioRxiv, 403733, doi: 10.1101/403733 (2018). [DOI] [Google Scholar]
- 82.Lino CA, Harper JC, Carney JP & Timlin JA Delivering CRISPR: a review of the challenges and approaches. Drug delivery 25, 1234–1257, doi: 10.1080/10717544.2018.1474964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826, doi: 10.1126/science.1232033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ran FA et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191, doi: 10.1038/nature14299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen LF et al. Enhancer Histone Acetylation Modulates Transcriptional Bursting Dynamics of Neuronal Activity-Inducible Genes. Cell reports 26, 1174–1188 e1175, doi: 10.1016/j.celrep.2019.01.032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]