Abstract
Background:
Embryonic cells and cancer cells share various cellular characteristics important for their functions. It has been thus proposed that similar mechanisms of regulation may be present in these otherwise disparate cell types.
Results:
To explore how regulative embryonic cells are fundamentally different from cancerous cells, we report here that a fine balance of a tumor suppressor protein Retinoblastoma1 (Rb1) and a germline factor Vasa are important for proper cell proliferation and differentiation of the somatic cells during embryogenesis of the sea urchin. Rb1 knockdown blocked embryonic development and induced Vasa accumulation in the entire embryo, while its overexpression resulted in a smaller-sized embryo with differentiated body structures. These results suggest that a titrated level of Rb1 protein may be essential for a proper balance of cell proliferation and differentiation during development. Vasa knockdown or overexpression, on the other hand, reduced or increased Rb1 protein expression, respectively.
Conclusions:
Taken together, it appears that Vasa protein positively regulates Rb1 protein while Rb1 protein negatively regulates Vasa protein, balancing the act of these two antagonistic molecules in somatic cells. This mechanism may provide a fine control of cell proliferation and differentiation, which is essential for regulative embryonic development.
Keywords: Retinoblastoma1, cell differentiation, embryonic development, sea urchin
1. Introduction
Retinoblastoma (Rb1) is a pocket protein family conserved both in plants and animals (Dimaras and Corson, 2019; Kohno et al., 2016; Sage, 2012). It plays multiple roles, most notably, in cell cycle inhibition by serving as a check point factor (Moser et al., 2018) as well as in suppression of oncogene expression through its binding to E2F transcriptoin factor (Chang et al., 2018). In many human cancer cells, Rb1 is inactivated through its hyperphosphorylation or mutations, where Rb1 loses its binding to E2F. Free E2F then drives transcription of various cell cycle regulators and oncogenes, promoting cell proliferation and tumorigenesis. Other targets of Rb1/E2F pathway include cell fate determinants, of which include: key transcriptional regulators and members of the Notch, fibroblasts growth factor, Wnt, and TGF-beta signaling pathways (Julian et al., 2016). Overexpression of those factors under Rb1-inactivation has been proposed to contribute to plasticity of stem cells or cancer cells (Kohno et al., 2016). Furthermore, Rb1 is also reported to function independent of E2F transcription, and appears to regulate synchcrony of DNA synthesis and centrosome duplication as well as transcription of mitotic genes (Amato et al., 2009; Iovino et al., 2006). Rb1 is known to interact with over 100 proteins, have 15 different phosphrylation sites, and have multiple different functions in multiple types of cells and biological contexts (Morris and Dyson, 2001). Its dysregulation often contributes to turmorigenesis, which is why it is most well known as a tumor supressor.
More recently, as another potential function of Rb1, it has been proposed that Rb1 negatively regulates expression of germline factors in somatic cells. For example, when a malignant brain tumor was induced in Drosophila by inactivation of lethal (3) malignant brain tumor (l (3) mbt), a member of the Rb1 tumor suppressor complex, a quarter of the upregulated genes turned out to be factors required for the germline. Inhibition of each of those germline factors (e.g. vasa, piwi, aubergine, or nanos) halted tumor growth, suggesting that Rb1-mediated expression of these germline factors had an essential function in the somatic tumor (Janic et al., 2010). During embryonic development, on the other hand, Rb1 appears to play a critical role in promoting cell differentiation in several organisms such as C. elegans and Arabidopsis and its knockout drives overexpression of germline factors and prevents somatic differentiation (Dominado et al., 2016; Tu et al., 2018). Thus, Rb1 appears to play a critical role in pluripotency control through the regulation of germline factors in both cancer cells and embryonic cells. Based on these observations, we hypothesize that a fine balance of Rb1 (pluripotency suppressor) and germline factors (pluripotency activators) is essential for proper pluripotency regulation in the cells, and that its failure in some cases leads to cancers.
To test this hypothesis, we used the sea urchin, a close relative to chordates, as our model organism, Its embryo is transparent, fast developing in culture, well cell fate-mapped, easy to manipulate, and suitable for microscopy. Further, and most importantly, it is highly regulative: the sea urchin embryonic cells are known to remain highly multipotent and can change their gene expressions and cell fate regulations in response to environmental cues, which is critical for embryo’s survivability. They also express several germline factors both in the somatic lineage and the germline during embryonic development (Yajima et al., 2014; Yajima and Wessel, 2011 & 2015). The transcriptomic database suggests that several oncogenes and Rb1 are highly expressed during early embryogenesis (echinobase.org). These molecules often expressed in cancer cells function as essential developmental factors during embryogenesis, yet it is not entirely clear what mechanism allows this regulative but non-cancerous cell regulation in the embryo. In this study, we hypothesize a balanced act of a pluripotency suppressor (e.g. Rb1) and pluripotency activators (e.g. germline factors) is critical for controlled plasticity regulation in the embryo. We demonstrate that Rb1 has, indeed, critical roles in orchestrating a fine balance of cell proliferation and differentiation by regulating the protein level of Vasa, one of the germline factors in somatic lineages, controlling proper embryogenesis.
Results and Discussion
Sea urchin Rb1 and Rb1-like transcripts are uniformly expressed during early embryogenesis.
Rb1 is a member of the pocket protein family that consists of three proteins, Rb1, Rb1-like1/p107 and Rb1-like2/p130 in the human. Through database searches (echinobase.org), we identified that the sea urchin (Strongylocentrotus purpuratus) also has three Rb1 and Rb1-like gene orthologs, SpRb1 (SPU_011954), SpRb1-like1 (SPU_004292), and SpRb1-like2 (SPU_003798). The protein sequence alignment (Tables S1 & S2) followed by phylogenetic tree analysis (Fig. 1A) demonstrated that SpRb1 clusters with human/mouse Rb1 protein and SpRb1L2 clusters with human/mouse Rb1-like2 (p130), as predicted in the database. However, SpRb1L1 did not cluster well with human/mouse Rb1-like 1(p107) due to lower sequence similarity. SpRb1 protein was also identified as the most similar to human Rb1 protein with three conserved structural regions (Table S3): the N-terminus, the centrally located pocket domain, and the C-terminus. Each of these regions is known to contain multiple protein binding and phosphorylation sites (Hassler et al., 2007; Lamber et al., 2013). Based on these analyses, we determined SpRb1 (SPU_011954) as the sea urchin Rb1 ortholog as predicted in the database.
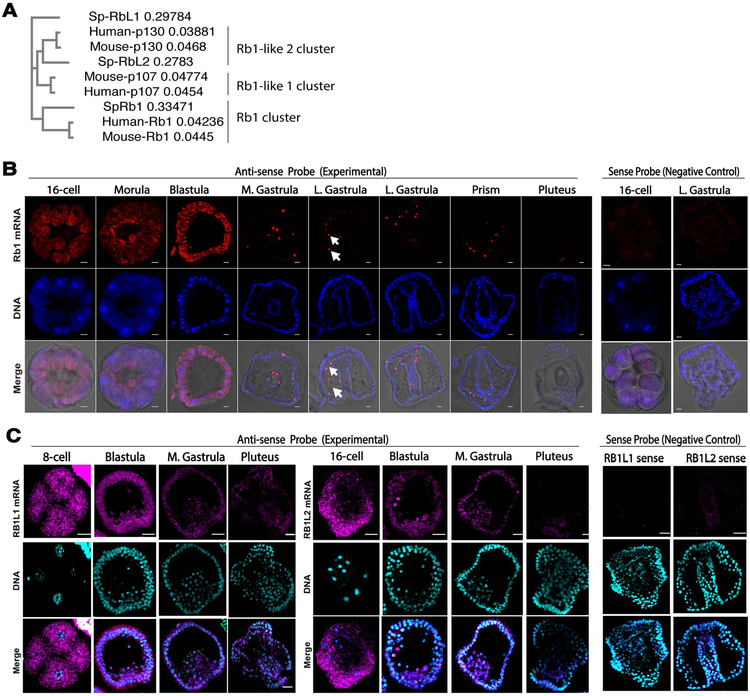
Fig.1. Rb1 transcript expression patterns during embryogenesis of the sea urchin.
A) A phylogenetic tree of sea urchin (Sp), human and mouse pocket protein family members. Rb1-like 1 is also called as p107 and Rb1-like2 as p130 in humans and mice. The tree is neighbor-joining constructed by Clustal omega. B) In situ hybridization images of SpRb1. SpRb1 mRNA (red) was uniformly expressed until blastula stage, and then became restricted into mesenchymal cells (arrows) after gastrula stage. DNA, blue. The merge is consisted of red, blue, and DIC channels. Images were taken by confocal laser microscopy. DNA, blue. Scale bars=10 μm. C) In situ hybridization images of SpRb1-like1 and –like2. SpRb1-like1 and –like2 mRNA (magenta) were both uniformly expressed until blastula stage and gradually decreased by pluteus stage. Images were taken by confocal laser microscopy. The merge is consisted of magenta and blue channels. DNA, blue. Scale bars=20 μm.
In several temporal gene expression databases (echinobase.org; http://www.spbase.org:3838/quantdev/; Wei et al., 2006), expression patterns of Rb1, Rb1-like1 and Rb1-like2 transcripts were all found to be similar, highly expressed during early embryogenesis, and decreased after the blastula stage at 15 hours post fertilization (hpf). Consistent with the temporal expression data, in situ hybridization (ISH) analysis of each of these genes identified abundant and uniform distribution of the transcript during early embryogenesis, and significant decrease after the gastrula or pluteus stage (Fig. 1B & C). The signal for Rb1-like2 was, in general, found much dimmer compared to that of Rb1 or Rb1-like1, consistent with the temporal expression data. Although the biological reason is not clear, Rb1 transcript was found enriched in the mesenchymal cells after blastula stage, which was less apparent for Rb1-like1 and Rb1-like2 transcripts (Figs. 1B, arrows). More specifically, those cells appeared to be a part of the secondary mesenchyme cells, like blastocoelar cells or pigment cells, based on their location in the embryo, yet we were unable to distinguish these two cell types due to a lack of pigmentation in the pigment cells at this early stage of the embryo in this study. The control embryos hybridized with each of the sense probes showed no detectable signal under the same experimental condition, suggesting specificity of the signal in the experimental groups (Fig. 1B & C, Negative Control). Since embryos undergo major cell differentiation at Day 2 to form endomesodermal tissues, Rb1 and Rb1-like1 & 2 genes may play more critical roles prior to these major cell differentiation events.
Rb1-like1 and Rb1-like2 are also reported to be involved in cell cycle regulation in humans. Rb1 is, however, the most well studied molecule among the Rb1-family proteins in other organisms because of its frequent mutation in cancerous cells as well as dominant function in tumor suppressive and pluripotency regulation (Kohno et al., 2016; Indovina et al., 2013). In this initial report, therefore, we focused on Rb1 and its functional contributions to regulative embryonic development in the sea urchin.
Rb1-GFP is uniformly distributed in the embryo during early embryogenesis.
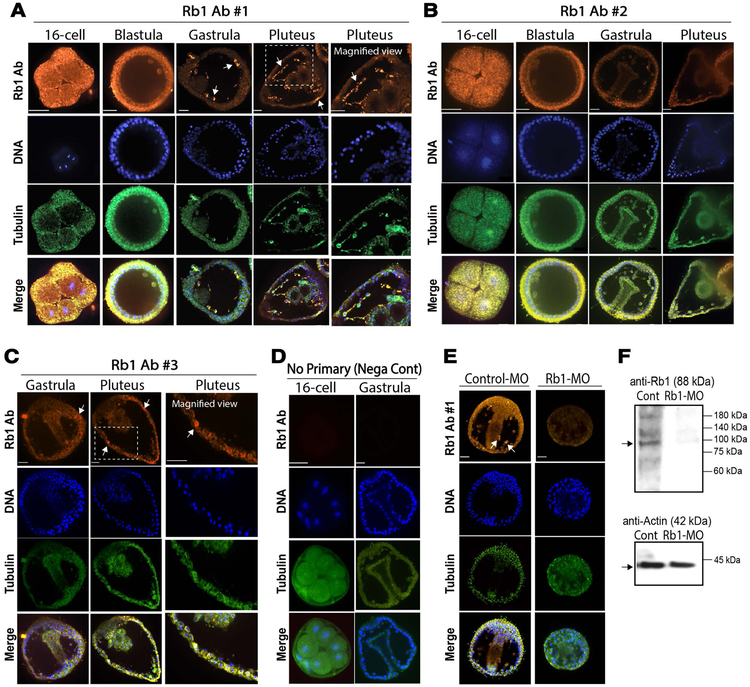
To identify Rb1 protein expression patterns during embryogenesis, we produced three peptide antibodies against SpRb1 (Rb1-Ab#1~3). Two of these antibodies were designed to detect the middle Rb1_A pocket domain and the third one was designed to detect the C-terminal Rb1_B domain (Table S3). Each antigen was designed within the amino acid sequence specific to Rb1 but not within the conserved region with other Rb1 family proteins such as Rb1-like1 or Rb1-like2. We first performed immunofluorescence using each of these antibodies. As a result, the signal was found uniformly distributed during early embryogenesis and then enriched into the mesenchymal cells after Day2 with Ab#1 and #3 (Fig. 2A & C), while the signal was rather uniform throughout development with Ab#2 (Fig. 2B). The negative control group with no primary antibody showed no detectable signal, suggesting the signal specificity of Rb1-Abs (Fig. 2D).
Fig.2. Rb1 protein distribution patterns during embryogenesis of the sea urchin.
A-D) Immunofluorescence results of SpRb1 antibodies #1~3 (A-C, orange channel) or negative control with no primary antibody (D) in normal embryos. Tubulin (green) and Hoechst/DNA (blue) were used as a counterstaining to visualize cell and embryo morphology. The antibody #1 showed the most prominent signal in mesenchymal cells (A, arrows). The antibody #3 also showed some enrichment in the mesenchymal cells (C, arrows), while the antibody #2 showed no signal enrichment in any specific cell types throughout development. E) Immunofluorescence results of control-MO (1.5mM stock of MVH-MO) and SpRb1-MO (1.5mM stock) embryos at Day 2 detected by SpRb1 antibody #1 (orange). The signal in the mesenchymal cells in the control group (arrows) was lost in the SpRb1-MO group. Images of A-E were taken by confocal laser microscopy with the same laser setting throughout of the samples. Orange, Rb1; Green, Tubulin; Blue, DNA. Scale bars=20 μm. F) Immunoblot results of control-MO (1.5mM stock of MVH-MO) and Rb1-MO (1.5mM stock) embryos at Day 2 detected by SpRb1 antibody #1. In the control group (left lane), SpRb1 antibody #1 detected a major band at the predicted size of 88 kDa. This signal was reduced by Rb1-MO (right lane). Actin (42 kDa) was used as a loading control.
The signal distribution of Rb1 Ab#1 appears to be more consistent with that of the SpRb1 transcript that showed specific enrichment in mesenchymal cells after Day 2. Therefore, we focused on the Rb1-Ab#1 and tested if morpholino antisense oligo against SpRb1 (SpRb1-MO) diminishes the signal in the mesenchymal cells. As a result, introduction of Rb1-MO, but not control-MO, reduced the Rb1-Ab#1 signal in mesenchymal cells, suggesting the specificity of the Rb1-MO as well as of the Rb1 Ab#1 (Fig. 2E). These similar results were obtained by immunoblot, yet multiple bands showed up even in the control group (Fig. 2F). This is consistent with other polyclonal Rb1 antibodies that were made for other organisms that are commercially available. These multiple bands may be due to modifications/breakdown of the Rb1 protein or cross-reactivity with other proteins, which need to be tested further in the future. Furthermore, the Rb1 protein signal in these embryos was found mostly in the cytoplasm rather in the nucleus throughout early development. Although Rb1 is known to be present both in the cytoplasm and in the nucleus of many organisms, it is expected to localize in the nucleus for its best-identified role in transcriptional repression of E2F or other transcription factors. Thus, the cytoplasmic Rb1 may be simply stored inactive and/or may have distinct functions from its roles in the nucleus in these embryos. A similar cytoplasmic Rb1 expression has been also reported in Drosophila embryo (Dominado et al., 2016), suggesting that cytoplasmic localization of Rb1 may be conserved among embryos of various organisms. Testing its possible cytoplasmic function during embryogenesis might be important in the future.
Rb1 functions in cell differentiation during embryogenesis.
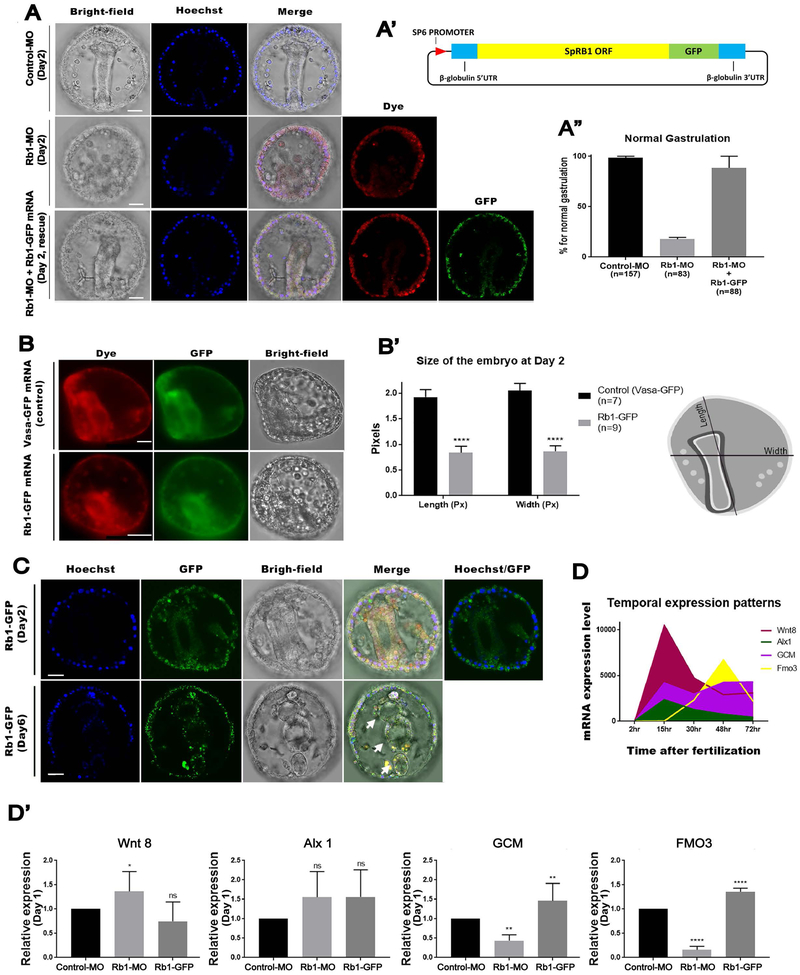
Next, we tested how Rb1 functions during embryogenesis of the sea urchin by introducing Rb1-MO into fertilized eggs. In these Rb1-knockdown (kd) embryos, 83.13 % of the embryos failed in gastrulation, whereas 100 % of the control embryos successfully gastrulated at Day 2 (Fig. 3A & A’). This Rb1-kd phenotype was rescued by co-introduction of Rb1-MO with a titrated level of Rb1-GFP mRNA (1.5 μg/uL stock) (Fig. 3A-A”, rescue). These results further suggest the specificity of Rb1-MO as well as the functionality of Rb1-GFP mRNA in the embryo. The genome editing technology (e.g. CRISPR/Cas9 system) was not used in this study as a knockdown approach because of the maternal load of Rb1 mRNA during early embryogenesis of the sea urchin (Lin and Su, 2015; Shevidi et al., 2018). This alternative approach will be, however, important to test these Rb1-knockdown phenotypes in the larval stage in the future. Overexpression (OE) of Rb1-GFP (2 μg/uL stock), on the other hand, caused smaller-sized embryos compared to the controls injected with Vasa-GFP mRNA, which is known to cause no phenotype (Gustafson et al., 2011a; Yajima and Wessel, 2011) at Day 2 (Fig. 3B & B’). These embryos constructed differentiated body parts such as mouth, esophagus, gut and anus (Fig. 3C, arrows) yet remained smaller in size even at Day 6~8, which may be due to smaller number of cells in the embryo. Taking all of these results together, Rb1-kd appears to compromise progression of development (e.g. gastrulation), resulting in embryos with not fully differentiated cells. On the contrary, Rb1-OE may progress tissue formation to some extent yet without sufficient numbers of cell divisions, resulting in the smaller size embryos.
Fig.3. Rb1 knockdown compromised development while Rb1 overexpression prematurely progressed development of the embryo.
A – A”) Rb1-MO inhibited gastrulation, and this phenotype was rescued by co-introduction of Rb1-GFP mRNA at 98.2% as shown in the graph (A”). () indicates the total number of gastrulated embryos counted for each group at Day 2. DNA (blue), Rb1-GFP protein (green), Dye (red) and Bright-field. Images were taken by confocal laser microscopy. (A’) indicates a construct design of Rb1-GFP. Scale bars=20 μm. B & B’) Rb1-GFP overexpression induced the smaller-sized embryos compared to the controls (Vasa-GFP embryos) at Day 2. The embryo size (Length and Width in pixels) was measured by Image J (n=7 for Vasa-GFP and n=9 for Rb1-GFP) as shown in the drawing on the right (A’). Images were taken by fluorescent microscopy. Scale bars=20 μm. C) The Rb1-GFP embryos at Day 2 and Day 6. Upper arrow indicates the differentiated body structure of fore-gut, the arrow in the middle indicates the middle-gut and the last arrow marks the hind-gut. Hoechst (blue), Rb1-GFP (green), and Bright-field. Images were taken by confocal laser microscopy. Scale bars=20 μm. D) Temporal mRNA expression pattern of each gene of interest is shown in the graph. The data was extracted from echinobase.org. X-axis indicates hours post fertilization. Images were taken by confocal laser microscopy. Scale bars=20 μm. D’) RT-qPCR results of Wnt8, Alx1, Glial Cell Missing (GCM), or Flavin-containing monooxygenase 3 (Fmo3) expression in the control, Rb1-MO and Rb1-GFP OE embryos at Day 1 (23~26 hours post fertilization). Copy number of each gene expression was normalized by that of Ubiquitin, a housekeeping gene, to obtain the Relative expression value. The relative value was then divided by that of the control to obtain the final relative value. Each graph was a combination of the four independent experimental results. Data are presented as mean ± s.e.m. *P<0.05.
To gain an insight if Rb1 indeed impacts expression of any fate determinants, we analyzed mRNA expression levels of several developmental markers at Day 1 (23~26 hours post fertilization) in Rb1-MO and Rb1-OE embryos, respectively. These markers include Wnt8, Alx1, Glial Cell Missing (GCM), and Flavin-containing monooxygenase3 (FMO3) (Fig. 3D). Wnt8 is highly expressed in the vegetal blastomeres during early embryogenesis for endomesoderm specification (Wikramanayake et al., 2004). Alx1 expresses the highest at Day1 in the mesoderm lineages and is essential for skeletogenic cell specification (Ettensohn et al., 2003). GCM (Ettensohn et al., 2003) and FMO3 (Calestani et al., 2003) both express the highest after Day 2 in the mesoderm lineages, and are important for secondary mesenchyme cell specification and pigment compound biosynthesis, respectively. We found that Wnt8 and Alx1 expressions were less impacted, yet that the pigment cell markers GCM and FMO3 were downregulated by Rb1-MO and upregulated by Rb1-OE, respectively (Fig. 3D’). Since both Rb1 transcript and protein appear to be enriched into mesenchymal cells after Day 1 (Figs. 1B & 2A), these results suggest that Rb1 may have specific functions in pigment cells.
Rb1 knockdown induced Vasa accumulation in the entire embryo.
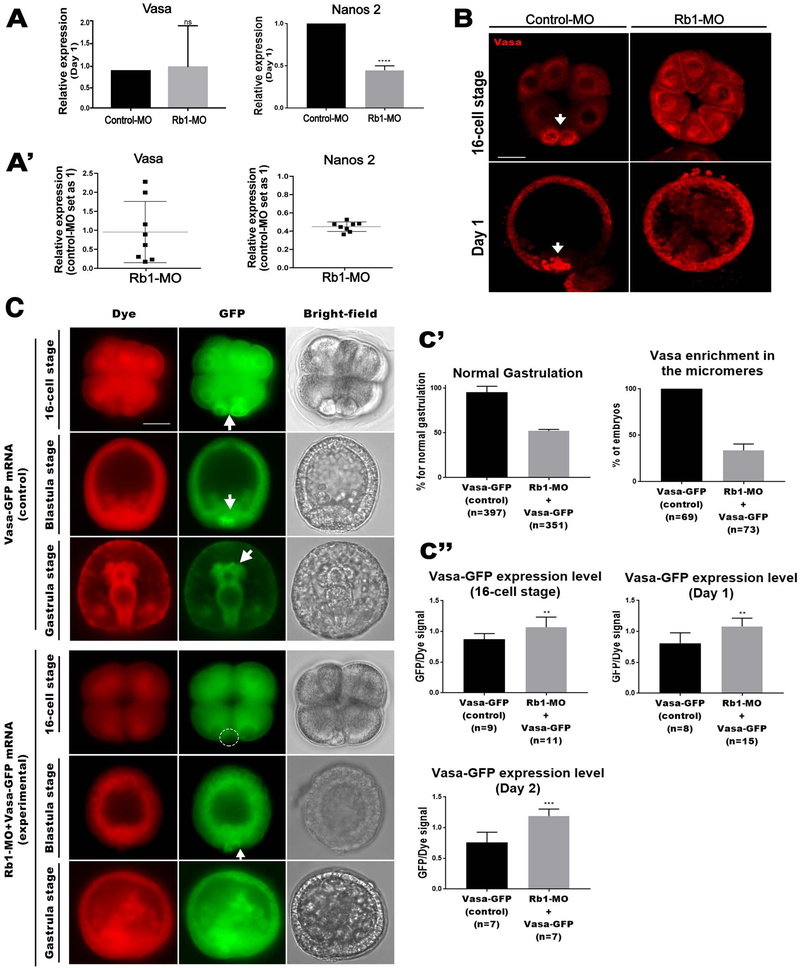
To test if the Rb1-kd drives overexpression of the germline factors as seen in other organisms (Dominado et al., 2016; Janic et al., 2010; Zhao et al., 2017), we analyzed the expression levels of Vasa and Nanos 2, representative germline markers present in various organisms at Day 1 (23~26 hours post fertilization). In the sea urchin embryo, Vasa mRNA is maternally present and becomes gradually restricted into the germline after blastula stage (Voronina et al., 2008), while Nanos2 mRNA expression is more transient and starts after 32-cell stage in the germline and decreases by Day 2. Both genes are considered to be important for germline specification and/or maintenance (Fresques et al., 2016; Voronina et al., 2008). As a result, however, Nanos2 mRNA level was found to be downregulated by Rb1-kd (Fig. 4A). One report suggests that Nanos transcription is directly upregulated by Rb1 inactivation in Drosophila (Miles et al., 2014), which is inconsistent with our result in this sea urchin embryo. Further investigation is thus needed in the future to reveal the underlying mechanisms of Rb1-mediated transcriptional regulation in this organism.
Fig.4. Rb1-knockdown induced Vasa accumulation in the entire embryo.
A-A’) RT-qPCR analyses of Vasa and Nanos2 in the control and Rb1-MO embryos at Day 1 (23~26 hours post fertilization). Copy number of each gene expression was normalized by that of Ubiquitin to obtain the Relative expression value. The relative value was then divided by that of the control to obtain the final relative value. Each graph was the combination for the five or more independent experimental results. (A’) shows the individual data points used in (A). Data are presented as mean ± s.e.m. *P<0.05. B) Vasa immunofluorescence of the Control-MO (16-cell stage, n=60; blastula stage, n=65) and Rb1-MO (16-cell stage n=50; blastula stage, n=72) embryos. Images were taken by confocal laser microscopy. Scale bars=20 μm. C) Live imaging of Vasa-GFP expressing embryos (control) and Vasa-GFP mRNA + Rb1-MO at 16-cell, blastula and gastrula stages. In the control group, the Vasa signal was enriched in the micromeres at the 16-cell stage, and in the germline at blastula and gastrula stages (Upper panels, arrows). This enrichment of Vasa was diminished in the experimental group (Lower panels). A dashed circle and an arrow indicate a lack of Vasa in the micromere and the germline, respectively. Images were taken by fluorescence microscopy. Dye (red), Rb1-GFP (green), and Bright-field. Images were taken by fluorescent microscopy. Scale bars= 20 μm. C’) Introduction of Vasa-GFP mRNA + Rb1-MO compromised gastrulation (Left graph) and Vasa enrichment in the micromeres (Right graph). () indicates the total number of embryos counted for each phenotype at Day 2 or at 16-cell stage, respectively. C”) The relative level of Vasa-GFP expression in the Vasa-GFP (control) and in Rb1-MO+Vasa-GFP (experimental) embryos at 16-cell, blastula and gastrula stages. The level of GFP signal was normalized by that of dye signal. () indicates the total number of the embryos measured for each group at 16-cell stage, Day 1, and Day 2, respectively.
Vasa mRNA level was, on the other hand, was unchanged by Rb1-kd (Fig. 4A), which is consistent with a previous report that showed Vasa upregulation not at the transcript level but only at the protein level in Drosophila brain tumor (Janic et al., 2010). We, therefore, tested if Rb1-kd increases Vasa protein expression level. In the sea urchin embryo, Vasa protein is uniformly present until 8-cell stage and then becomes restricted into the micromeres at the 16-cell stage and into the future germline (Fig. 4B, arrows) (Yajima and Wessel, 2011). In the Rb1-MO injected embryos, Vasa protein accumulated everywhere in the entire embryo at 16-cell the stage (Fig. 4B). This ectopic accumulation of Vasa continued, and the embryos appeared highly disorganized at blastula stage. These results suggest that Rb1 regulates Vasa at the protein level in the sea urchin embryo, as seen in Drosophila. The consistent observations in these distantly related organisms suggest that Vasa may not be a direct target of Rb1 for transcription, but rather functions in parallel as Rb1’s counterpart. Additionally, a significant fluctuation of Vasa mRNA expression was found in these Rb1-MO embryos among five different batches of animals we tested in this work (Fig. 4A’). This fluctuation was never seen in Nanos mRNA expression and may reflect a dynamic regulatory response against ectopic accumulation of Vasa protein in these Rb1-kd embryos. The detailed regulatory mechanism in this process is, however, unknown and needs to be tested in the future.
In the sea urchin embryo, Vasa protein restriction into the germline is regulated by its active protein degradation in the somatic lineages (Gustafson et al., 2011b). In normal development, simple Vasa mRNA introduction results in only more active degradation of the Vasa protein in the soma, leading to no extra Vasa accumulation or developmental defects (Yajima and Wessel, 2015, 2011). Therefore, Vasa protein accumulation in the Rb1-MO embryos was likely caused by a lack of Vasa protein clearance in the somatic lineage. If this is true, Vasa mRNA introduction under the Rb1-kd condition may cause over-accumulation of Vasa protein in the soma. To test this, we co-introduced Vasa-GFP mRNA and fluorescent red dye into the fertilized eggs in the presence or absence of Rb1-MO (Fig. 4C). In the presence of Rb1-MO, the resultant embryos showed compromised Vasa enrichment in the micromeres and gastrulation as expected (Fig. 4C & C’). In those embryos, the level of Vasa protein accumulation was measured by the GFP signal (green), while the injection amount of Vasa-GFP mRNA was measured by the dye signal (red). Vasa protein level was indeed found increased by Rb1-kd, compared to the control embryos without Rb1-MO introduction (Fig. 4C’). These results suggest that Rb1 is responsible for the clearance of Vasa protein in the somatic lineages during embryogenesis.
One remaining question is that the Rb1 mRNA, as well as its protein, appear to be uniformly present in the micromeres and the germline, yet Vasa is accumulated in this lineage in normal development. Therefore, another mechanism that inactivates Rb1 and/or that drives extra Vasa protein production in the germline might be present in this embryo. Since Rb1 is known to undergo extensive post-transcriptional and post-translation modifications as well as interact with over 100 different proteins for multiple different functions in different cell types (Morris and Dyson, 2001), it is highly possible that Rb1 is under different regulation in the germline.
Vasa positively regulates Rb1 protein expression.
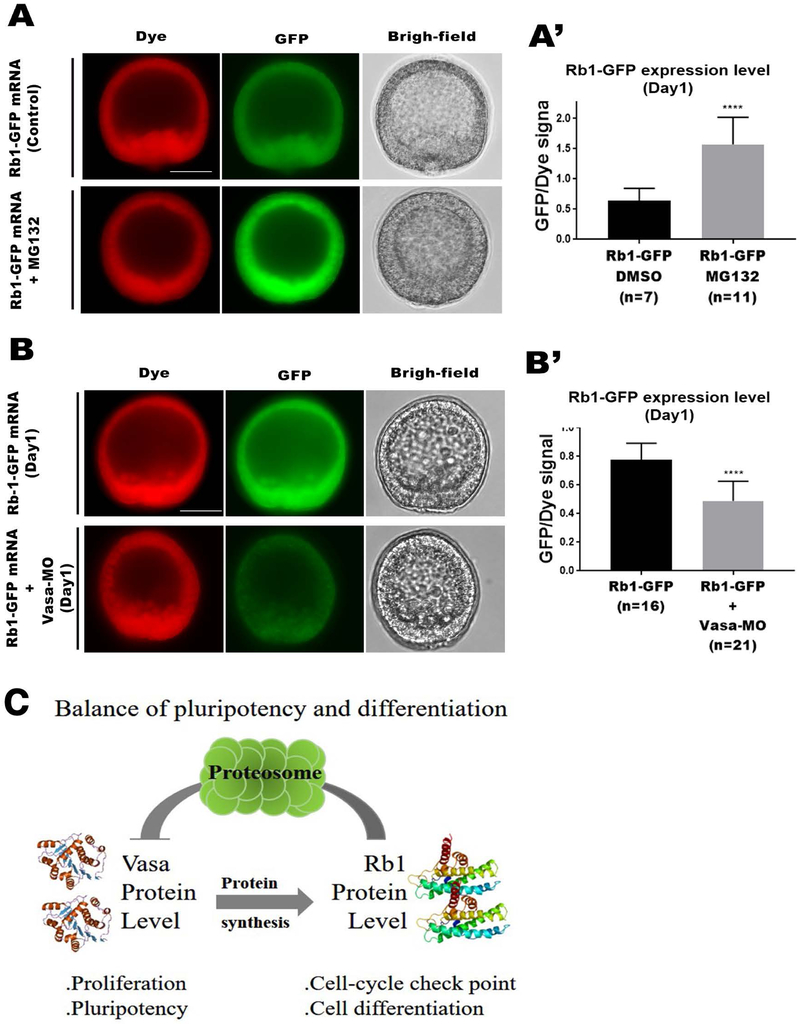
Vasa is an RNA-helicase and is implicated in translational regulation (Sengoku et al, 2006, Cell). In the sea urchin, other than its function in the germline, Vasa functions in cell cycle progression and is responsible for ~80% of general protein synthesis in the entire embryo (Yajima and Wessel, 2015). Therefore, we speculate that Rb1 may be also one of the Vasa’s targets for mRNA translation. To test this hypothesis, we overexpressed or knocked down the Vasa protein and analyzed if each of those manipulations upregulate or down-regulate Rb1 protein level, respectively (Fig. 5A & B). As mentioned above, Vasa protein level is controlled by its protein degradation. Therefore, we blocked Vasa protein degradation by treating the Rb1-GFP expressing embryos with MG132, a specific proteinase inhibitor previously demonstrated to induce Vasa accumulation in the sea urchin embryo (Gustafson et al., 2010). The level of Rb1-GFP protein expression (green) was then normalized to the amount of Rb1-GFP mRNA injected (red dye) to determine the relative value of Rb1 protein expression level in the resultant embryos. As a result, in the MG132-treated group, the relative Rb1 protein level was found to be increased compared to that of the DMSO-treated control group (Fig. 5A-A’). Similarly, the relative Rb1 protein expression level was measured in the presence or absence of Vasa-MO that was previously shown to specifically block endogenous Vasa protein synthesis (Yajima and Wessel, 2011). In Vasa-kd embryos, the Rb1 protein level was found to be reduced compared to that of the control embryos (Fig. 5B-B’). Although we could not exclude the possibility that both MG132 and Vasa-MO only indirectly affected Rb1 protein expression levels in this work, these results consistently suggest a possibility that Vasa positively regulates Rb1 protein expression as one of its targets.
Fig.5. Vasa positively regulates Rb1 protein expression.
A-B’) Live imaging of Rb1-GFP embryos treated with DMSO (control; Upper panels) or with MG132 (Lower panels) at Day1 (A & A’). Live imaging of Rb1-GFP (control; Upper panels) or Rb1-GFP+Vasa-MO (Lower panels) embryos at Day 1 (B & B’). The graphs indicate the relative Rb1-GFP expression level in each embryo group (A’ & B’). The total level of Rb1-GFP was normalized by that of the dye to obtain the relative Rb1-GFP expression level using Image J. () indicates the total number of embryos analyzed for each group at Day 1. Dye (red), Rb1-GFP (green), and Bright-field. Images were taken by fluorescent microscopy. Scale bars= 20 μm. Scale bars= 20 μm. C) A hypothetical model for Rb1 function in embryonic development. Rb1 facilitates cell differentiation by negatively regulating Vasa protein while Vasa positively regulates Rb1 protein, maintaining a fine balance of cell proliferation and differentiation in the somatic lineages during embryogenesis.
A fine balance of Rb1 and Vasa activities is critical for regulative but non-cancerous embryonic cell regulation.
In this report, we demonstrated that Rb1 is uniformly expressed and responsible for facilitating proper embryogenesis when the cells are still undifferentiated in the sea urchin. During this early time frame, a germline factor Vasa is also uniformly expressed and appears to be responsible for facilitating pluripotency in this embryo (Yajima and Wessel, 2015). Our results suggest that Vasa positively regulates Rb1 protein expression while Rb1 negatively regulates Vasa protein accumulation, and that expressions of both molecules decrease as development proceeds. These observations suggest that Vasa and Rb1 antagonistically regulate each other in the somatic lineage of the early embryo (Fig. 5C). Although this mechanism may not be applicable in the germline, in the somatic lineage, this antagonistic relationship of Rb1 and Vasa appears to be critical for controlling a fine balance and timing of cell proliferation and differentiation during early embryogenesis of the sea urchin. Indeed, in this study, Rb1-kd resulted in excessive Vasa protein accumulation in the entire embryo, mimicking tumorigenic cell development reported in other organisms (Dominado et al., 2016; Janic et al., 2010). Perhaps, during early embryonic cell specifications, Rb1 serves as pluripotency suppressor while the germline factors serve as pluripotency activators, and a failure of this fine balance causes developmental failure, or in some cases tumorigenesis. This concept may help us understand the mechanisms of controlled pluripotency/plasticity regulation in embryonic cells, which is distinct from uncontrolled plasticity of cancerous cells even though a similar set of molecules is involved in regulation of both cell types. The further mechanistic studies on how Rb1 and Vasa (and other germline factors) may directly interact with each other for embryonic cell regulation will be critical for understanding the fundamental mechanisms of controlled plasticity in embryonic cells.
Experimental Procedures
Animals and embryo cultures
Strongylocentrotus purpuratus were obtained from Pat Leahy, Kerchoff Marine Laboratories, California Institute of Technology or by Josh Ross, South Coast Bio-Marine LLC. Long Beach, California, USA. Females were shed by KCl (0.5 M) injection. Sea urchin eggs were de-jellied with the pH4 seawater for 10 minutes and prepared for fertilization. Eggs were fertilized in 1 mM 3-aminotriazol (Sigma, St. Louis, MO, USA) to prevent the cross-linking of fertilization envelopes. Fertilized eggs were cultured in seawater at 16 °C in petri dishes until desired developmental stage.
Generation of plasmid constructs and in vitro transcription.
All constructs were prepared in the pSP64 vector that was optimized for in vitro transcription. SpVasa-GFP fusion construct was previously made in Gustafson et al. (2010). Since we had a technical difficulty in cloning the entire fragment of SpRb1 (SPU_011954) through a single PCR of the S. purpuratus cDNA library, the open reading frame (ORF) of SpRb1 was amplified as two pieces using the following primer sets designed within the N-terminal half and the C-terminal half of SpRb1, respectively: SpRb1-N (F- ATGGACGAGAGTGTAGCCAATGTT, R- GAGGCTTGAACCCCTACCTCAC) and SpRb1-C (F- GAGGTAGGGGTTCAAGCCTCAGAG, R-AGAATGTACTTACCGAGATGCTC). Each of the amplified fragments was then sequenced and confirmed to be a match with the annotated SpRb1 sequence in the database. To make Rb1-GFP fusion construct, using the above sequencing results as well as the database, a gBlock fragment (IDT, USA) that contained the entire SpRb1 ORF was prepared and inserted at ApaI and EcoRV sites of the pSP64-GFP vector to construct SpRb1-GFP. Subcloning reaction was designed to be in frame with GFP and conducted by following the In-Fusion HD Cloning Kit (Clonetech) protocol. For in vitro transcription, all plasmids were linearized with SalI restriction enzyme and subject to in vitro mRNA synthesis by following the protocol of mMESSAGE mMACHINE™ SP6 kit (Thermo Fisher Scientific, #AM1340).
Morpholino Design and Injection
Morpholino was made by Gene Tools (Oregon, USA) and was designed in the 5’UTR region of SpRb1 (AAACATTGGCTACACTCTCGTCCAT). A control morpholino was designed against mouse Vasa (MVH, TAGCTTCAGGTTCCTCTCCGCTCCA) that should not bind to the sea urchin mRNAs. The stock concentration of 1.5 mM MO was found to be most effective in showing the gastrulation defect phenotype that was rescued by co-introduction of Rb1-GFP mRNA, and thus used in this study unless indicated separately. Injection of MO and/or mRNA was performed as described previously (Yajima et al., 2007). Approximately 6 pl of 1.5 mM stock MO and/or 1-2 μg/μl mRNA along with a fluorescent red dye (Dextran, tetramethylrhodamine, 3000 MW, anionic. Ref#D3307) were injected into fertilized eggs. The number of embryos expressing GFP was counted at 16-cell, blastula and/or gastrula stages. The total number of injected embryos was determined using injection dye as a marker. Phenotypes were counted by examining the morphology and/or GFP-expression of the embryos in live by fluorescence microscopy (AXIO Vert.A1, Zeiss).
RNA Whole Mount in situ Hybridization (ISH)
For fluorescent whole amount in situ hybridization (ISH), we followed the protocol outlined in Andrikou et al. (2013). The fluorescent signal was developed with fluorophore-conjugated TSA cyanine 3 (1:400 reagent diluents, Perkin Elmer, ref#NEL760001KT). Labeled probes were in vitro transcribed from linearized DNA with digoxigenin-11-UTP (Roche, USA). Since little overlap was seen among the nucleotide sequences of SpRb1, SpRb1-like1 and SpRb1-like2, the probe for each gene was designed to cover the majority of the ORF using the following primers: SpRb1 (F-CCACTTACTGCGTGCTTCAA, R-GAGGCTTGAACCCCTACCTC), SpRb1-Like1 (F- GACAAGCTGTGTGAGGTCCA, R- TGCAGGATAGCCTCTTCGAT) and SpRb1-Like2 (F- TCCGAGGAGATCGTGCTGACTAC, R- CGATCGTGATCGCCGTTTTTC). Template of the probe was sequenced prior to probe generation and cloned in the pGEM®-T Easy Vector (Promega, Madison, WI, USA) for subsequent in vitro transcription using either SP6 or T7 MEGAscript Transcription kit (Thermo Fisher Scientific, #AM1330 or AM1333). Hoechst 33342 Solution (ThermoFisher, ref#62249) was used as a counter-staining at a final concentration of 0.1 mg/ml. Samples were imaged with Olympus FV3000 confocal microscope.
SpRb1 Antibody production and Immunoblotting
Three affinity purified peptide antibodies against SpRb1 were made in rabbit by GenScript (Piscataway, NJ, USA) with the following antigen sequences: #1 – CMLIKEEKRLSTSDF; #2- CFVKNEPRLTEEARK; #3- CTYKNVYMGEGKESA. The exact location of each antigen sequence within SpRb1 is indicated in Table S3. Immunoblot was performed as described previously (Poon et al., 2019). Briefly, 2 μl each of embryonic lysate made from Day 2 embryos was run on a 10% polyacrylamide gel (Lonza, Rockland, ME) and transferred to nitrocellulose membranes for immunoblotting with rabbit polyclonal SpRb1 antibody #1~3 each or with mouse monoclonal β-Actin antibody (Cell Signaling Technology, #3700) at 1:2000 followed by HRP-conjugated anti-rabbit (Cell Signaling Technology, #7047) or –mouse secondary antibody (Cell Signaling Technology, #7076), respectively. The reacted proteins were detected by incubation in a chemiluminescence solution (1.25 mM luminol, 68μM coumeric acid, 0.0093% hydrogen peroxide and 0.1 M Tris pH 8.6) for 1-10 min, exposed to film and developed. Each experiment was performed at least two independent times.
Immunofluorescence
Immunofluorescence was performed as previously described in Yajima et al. (2007) and Yajima and Wessel, (2011). Embryos were fixed with 90% Methanol for 1 hour at −20°C, rinsed with PBS, incubated with the primary antibody overnight at 4°C and with secondary antibody for 3 hours at room temperature. Affinity purified primary antibodies were used at 1:100 for each of anti-SpRb1 antibodies #1~3 and 1:200 for anti-Vasa (Yajima and Wessel, 2011). Cy3 goat anti-rabbit immunoglobulin G (IgG) antibody (Life Technologies, ref#A10520) was used as a secondary antibody at 1:200. Fluorescent images were taken by confocal laser microscopy (Olympus FV3000) or by fluorescence microscopy (AXIO, Vert.A1, Zeiss).
Quantitative RT-PCR
RT-PCR was performed with the following primer sets: SpVasa (SPU_008908): F-TCAACTACGACCTCCCAAGC & R-TCTCGCAATGTTAGCATCCTT; SpFmo3 (SPU_017374): F-TTCATCCCTCCCATCCCCTA & R-CTGCCAGACGTTCATGATGG; SpGCM (SPU_006462): F-CACAGACACGAGAGGCCTAA & R- ACGGATATGTGAGCGGAGAG; SpWnt8 (SPU_020371): F-TGTCGTTCATTCAAGCCATC & R-TATCACTCGCCATTCGTTCA; SpNanos2 (SPU_003591): F-GCAAGAACAACGGAGAGAGC & R-CCGCATAATGGACAGGTGTA; SpAlx1 (SPU_025302): F-GCTTGATGAATCTGGCAATG & R-CTCCTCTTTGGCTTACTGAAGG; SpUbi-F (SPU_000014): F-CACAGGCAAGACCATCAC & R-GAGAGAGTGCGACCATCC. Embryos at the desired developmental stage were collected and subject to total RNA extraction with Arcturus ™ PicoPure ™, RNA Isolation Kit (Thermo Fisher Scientific). The cDNA library was made by Superscript ™IV (Thermo Fisher Scientific), and 0.5 μl each of cDNA was used for quantitative PCR (qPCR) reactions. qPCR was performed on the 7300 Real-Time PCR system (Applied Biosystems, Foster City, CA) with the Luna® Universal qPCR Master Mix (New England BioLabs Inc.). Data for each gene was normalized against ubiquitin levels.
MG132 treatment
The 12hpf morula stage embryos injected with 1.5 μg/μl Rb1-GFP mRNA were cultured with 10 μM MG132 (Z-Leu-Leu-Leu-CHO; Enzo Life Science International, Inc., Plymouth Meeting, PA) or with 0.5% Dimethyl sulfoxide (DMSO) as negative controls at 16°C for 8 hours.
Fluorescent intensity analysis
Fluorescent images were taken using Metamorph imaging software (Universal imaging Corporation, Downington, PA). The exact same acquisition setting with the fixed exposure time and gamma was used for each channel throughout all sample groups for each cycle of the experiments. The control and experimental groups were prepared and imaged at a time for each cycle of experiments to minimize technical and batch variations. The obtained images were then analyzed for average intensity measurement using Image J, by selecting a region of interest (e.g. the whole embryo) for each channel. The value of GFP fluorescence was then normalized by that of Dye fluorescence to obtain a relative value for each sample, and the average of those relative values was presented for each sample group in the graph.
Supplementary Material
Table S1. Protein sequence aligment of sea urchin (Sp), human and mouse pocket potein family members.
Table S2. Protein sequence aligment of SpRb1, SpRb-like1 & 2 and human Rb1.
Table S3. Protein sequence aligment of SpRb1and human Rb1. The conserved domains are highlighted: DUF3452 (Domain of unknown function) is functionally uncharacterized; RB_A (Retinoblastoma-associated protein A domain) has the cyclin fold; RB_B (Retinoblastoma-associated protein B domain) has the cyclin fold and the Rb pocket that is a conserved groove among RB1-related proteins. The antigen sequences for three SpRb1 peptide antibodies are underlined.
Highlights.
A tumor suppressor Rb1 is important for proper embryonic development of the sea urchin.
Rb1 overexpression incompletely progresses and its knockdown compromises embryonic development.
Rb1 knockdown results in excessive Vasa accumulation in the entire embryo.
A fine balance of Rb1 and Vasa is critical for regulative embryonic development.
Acknowledgements:
We thank members of the PRIMO at Brown University for active discussion.
Funding: This work was supported by the NIH (1R01GM126043-01) and American Heart Association Scientist Development Grant to MY (14SDG18350021).
References
- Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A. 2009. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol. 10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrikou C, Iovene E, Rizzo F, Oliveri P, Arnone MI. 2013. Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors. Evodevo 4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. 2003. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 130: 4587–4596. [DOI] [PubMed] [Google Scholar]
- Chang MM, Lai MS, Hong SY, Pan BS, Huang H, Yang SH, Wu CC, Sunny SH, Chuang JI, Wang CY, Huang BM. 2018. FGF9/FGFR2 increase cell proliferation by activating ERK1/2, Rb/E2F1 and cell cycle pathways in mouse Leydig tumor cells. Cancer Sci 109:3503–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaras H, Corson TW. 2019. Retinoblastoma, the visible CNS tumor: A review. J Neurosci Res 97:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominado N, La Marca JE, Siddall NA, Heaney J, Tran M, Cai Y, Yu F, Wang H, Somers WG, Quinn LM, Hime GR. 2016. Rbf Regulates Drosophila Spermatogenesis via Control of Somatic Stem and Progenitor Cell Fate in the Larval Testis. Stem Cell Reports 7:1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettensoshn CA, Illies MR, Oliveri P, De Jong DL. 2003. Alx1 , a member of the Cart1 / Alx3 / Alx4 subfamily of Paired-class homeodomain proteins , is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130:2917–2928. [DOI] [PubMed] [Google Scholar]
- Fresques T, Swartz SZ, Juliano C, Morino Y, Kikuchi M, Akasaka K, Wada H, Yajima M, Wessel GM. 2016. The diversity of nanos expression in echinoderm embryos supports different mechanisms in germ cell specification. Evol Dev 18:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, Wessel GM. 2011a. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol 349:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, Wessel GM. 2011b. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol 349:440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler M, Singh S, Yue WW, Luczynski M, Lakbir R, Sanchez-Sanchez F, Bader T, Pearl LH, Mittnacht S. 2007. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol Cell 28:371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indovina P, Marcelli E, Casini N, Rizzo V, Giordano A. 2013. Emerging roles of RB family: new defense mechanisms against tumor progression. J Cell Physiol 228:525–535. [DOI] [PubMed] [Google Scholar]
- Iovino F, Lentini L, Amato A, Di Leonardo A. 2006. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer 5:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. 2010. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330:1824–1827. [DOI] [PubMed] [Google Scholar]
- Julian LM, Liu Y, Pakenham CA, Dugal-Tessier D, Ruzhynsky V, Bae S, Tsai SY, Leone G, Slack RS, Blais A. 2016. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ 23:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno S, Kitajima S, Sasaki N, Takahashi C. 2016. Retinoblastoma tumor suppressor functions shared by stem cell and cancer cell strategies. World J Stem Cells 8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamber EP, Beuron F, Morris EP, Svergun DI, Mittnacht S. 2013. Structural insights into the mechanism of phosphoregulation of the retinoblastoma protein. PLoS One 8:e58463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Su YH. 2016. Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Dev Biol 409:420–428. [DOI] [PubMed] [Google Scholar]
- Miles WO, Korenjak M, Griffiths LM, Dyer MA, Provero P, Dyson NJ. 2014. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J 33:2201–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. 2001. Retinoblastoma protein partners. Adv Cancer Res 82:1–54. [DOI] [PubMed] [Google Scholar]
- Moser J, Miller I, Carter D, Spencer SL. 2018. Control of the Restriction Point by Rb and p21. Proc Natl Acad Sci 115:E8219–E8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon J, Fries A, Wessel GM, Yajima M. 2019. Evolutionary modification of AGS protein contributes to formation of micromeres in sea urchins. Nat Comm 10:3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, McClay DR. 2014. Sub-circuits of a gene regulatory network control a developmental epithelial-mesenchymal transition. Development 141:1503–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J 2012. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev 26:1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevidi S, Uchida A, Schudrowitz N, Wessel GM, Yajima M. 2017. Single nucleotide editing without DNA cleavage using CRISPR/Cas9-deaminase in the sea urchin embryo. Dev Dyn 246:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Huo Z, Liu M, Wang D, Xu A, Zhou R, Zhu D, Gingold J, Shen J, Zhao R, Lee DF. 2018. Generation of human embryonic stem cell line with heterozygous RB1 deletion by CRIPSR/Cas9 nickase. Stem Cell Res 28:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. 2008. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol 314:276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. 2006. A database of mRNA expression patterns for the sea urchin embryo. Dev Biol 300:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. 2004. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis 39:194–205. [DOI] [PubMed] [Google Scholar]
- Yajima M, Gustafson EA, Song JL, Wessel GM. 2014. Piwi regulates Vasa accumulation during embryogenesis in the sea urchin. Dev Dyn 243:451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Kiyomoto M, Akasaka K. 2007. Ars insulator protects transgenes from long-term silencing in sea urchin larva. Dev Genes Evol 217:331–336. [DOI] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. 2011. The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development 138: 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. 2015. Essential elements for translation: the germline factor Vasa functions broadly in somatic cells. Development 142:1960–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, Komaki S, Prusicki MA, Maruyama D, Forner J, Medzihradszky A, Wijnker E, Harashima H, Lu Y, Schmidt A, Guthörl D, Logroño RS, Guan Y, Pochon G, Grossniklaus U, Laux T, Higashiyama T, Lohmann JU, Nowack MK, Schnittger A. 2017. RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356:396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Protein sequence aligment of sea urchin (Sp), human and mouse pocket potein family members.
Table S2. Protein sequence aligment of SpRb1, SpRb-like1 & 2 and human Rb1.
Table S3. Protein sequence aligment of SpRb1and human Rb1. The conserved domains are highlighted: DUF3452 (Domain of unknown function) is functionally uncharacterized; RB_A (Retinoblastoma-associated protein A domain) has the cyclin fold; RB_B (Retinoblastoma-associated protein B domain) has the cyclin fold and the Rb pocket that is a conserved groove among RB1-related proteins. The antigen sequences for three SpRb1 peptide antibodies are underlined.