Dear Editor,
Zygotic genome activation (ZGA) following fertilization is accomplished through a process termed the maternal-to-zygotic transition, during which the maternal RNAs and proteins are degraded and zygotic genome is transcriptionally activated.1 In mice, minor ZGA occurs from S phase of the zygote to G1 phase of the two-cell (2C) embryo, while major ZGA takes place during the middle-to-late 2C stage with a burst of transcription of totipotent cleavage stage-specific genes and retrotransposons.2 Dux has been recently identified and considered as a master inducer that regulates the ZGA process.3–5 Dux can directly bind and robustly activate 2C stage-specific ZGA transcripts and convert mouse embryonic stem cells (mESCs) to a 2C-like state with unique features that resembles the 2C embryos.4 Intriguingly, ~20% embryos with zygotic depletion of Dux unexpectedly reached morula or blastocyst stage even though defective ZGA program was detected.3
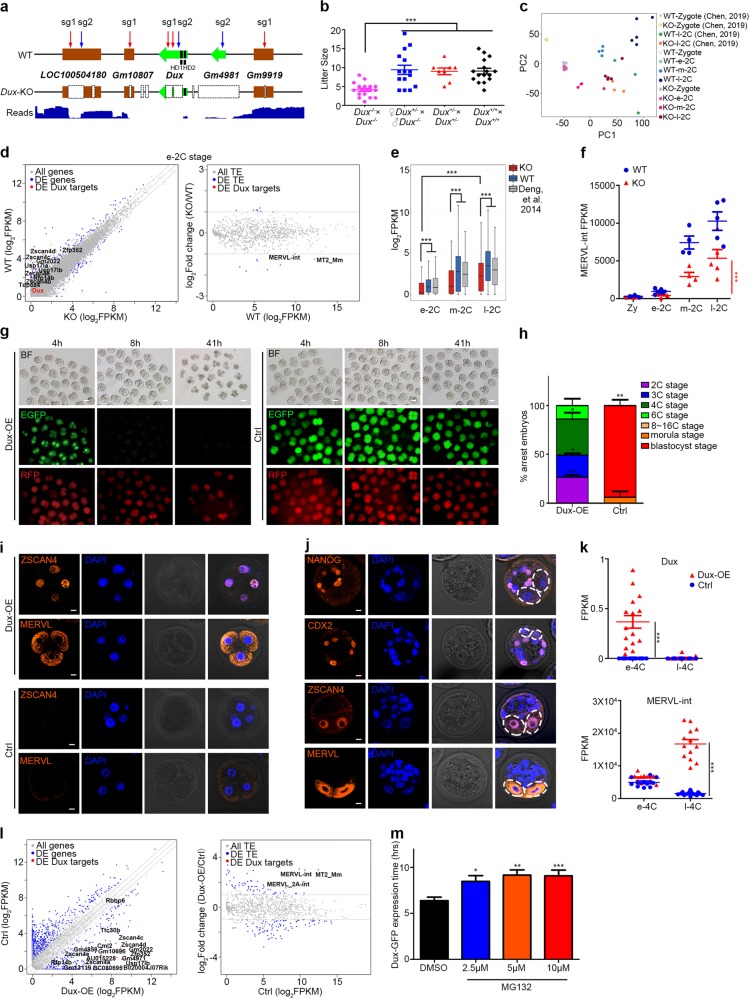
To determine whether Dux is a crucial factor for ZGA and embryo development, we generated a Dux-knockout (Dux-KO) mouse model using CRISPR/Cas9. Cas9 nuclease mRNA and two single guide RNAs (sgRNAs) targeting the macrosatellite tandem-array repeat region of Dux were injected into C57BL/6 zygotes. All injected embryos reached 2C stage and were transferred into oviduct of pseudo-pregnant recipients. F0 generation candidates with chimeric editing types were backcrossed with wild-type (WT) mice from the same genetic background. One of the F1 generation candidates carried large fragment deletion containing the second homeodomain of Dux (Fig. 1a). After backcrossing with WT individual again, the F1 candidate gave birth to heterozygous (Het) Dux+/− mice carrying the same mutation, indicating successful germline transmission. Homozygous Dux-KO mice were generated from heterozygous self-crossing, and the genotype was further confirmed by the third-generation long-read whole-genome sequencing, in which large fragment deletions in LOC100504180 (Duxf1), Dux (Duxf3) and Gm4981 (Duxf4) were observed. We also found short fragment deletions in Gm10807 (Duxf2) and Gm9919 (Duxf5) at the sgRNA targeting sites (Fig. 1a). Surprisingly, these homozygous Dux-KO mice could survive to adulthood, albeit they displayed a reduced frequency slightly deviated from a Mendelian distribution (Supplementary information, Fig. S1a). Notably, mating pairs of Dux-KO generated less offspring than WT pairs or Het pairs (Fig. 1b and Supplementary information, Fig. S1b). Thus, Dux depletion is not lethal, but it indeed leads to significantly decreased litter size. The phenotype, consistent with recent results,6 strongly suggests that Dux is important but not essential for in vivo embryo development.
Fig. 1.
a Schematic overview of Dux cluster locus in WT and Dux-KO mice, and distribution of third-generation long-read (30 kb) at the Dux cluster of Dux-KO mice. Red and blue arrowheads indicate targeting sites of sgRNAs. b Statistical analysis of litter size generated by WT, Dux-Het and Dux-KO mice. Each dot represents a single litter analyzed. ***P < 0.001; two-tailed Student’s t-test. Center lines and error bars indicate means and SD, respectively. c Principal component analysis of WT and Dux-KO embryos from zygotic stage to 2C stage. e-2C, early 2C stage; m-2C, middle 2C stage; l-2C, late 2C stage. d Scatter plots of gene expression (left panel) and transposable element (TE) expression (right panel) in WT and Dux-KO embryos at early 2C (e-2C) stage. FC > 2, FDR < 0.05. e Box plots of expression of 588 downregulated ZGA genes in WT and Dux-KO embryos from early to late 2C stages. e-2C, early 2C stage; m-2C, middle 2C stage; l-2C, late 2C stage. ***P < 0.001; Mann–Whitney–Wilcoxon two-sided test. The middle lines in the boxes represent medians. Box hinges indicate the twenty-fifth and seventy-fifth percentiles, and the whiskers indicate the hinge ± 1.5 × interquartile range. f Dot plots of MERVL-int expression in WT and Dux-KO embryos from zygotic to late 2C stages. Each dot represents an embryo. Zy, zygotic stage; e-2C, early 2C stage; m-2C, middle 2C stage; l-2C, late 2C stage. ***P < 0.001; two-tailed Student’s t-test. Center lines and error bars indicate means and SD, respectively. g Development progression of embryos 4 h, 8 h, and 41 h after mRNA injection in 2 blastomeres of 2C embryos. h hours, Dux-OE group with mRNA mixture of Dux-EGFP and RFP (left panels), Ctrl control group with mRNA mixture of EGFP and RFP (right panels). Scale bars, 50 μm. h Bar graph comparing the percentage of embryo at different developmental stages between Dux-OE and control groups 65 h after mRNA injection. *P < 0.05, **P < 0.01; two-tailed Student’s t-test. Error bars indicate SD. i Immunofluorescent staining of Dux-OE (upper panels) and control (lower panels) embryos with 2-blastomere injection at 3C or 4C stage. Scale bars, 10 μm. j Immunofluorescent staining of 1-blastomere-injected Dux-OE embryos at blastocyst stage. White dotted circles indicate the arrested blastomeres. Scale bars, 10 μm. k Dot plots showing expression of Dux (upper panel) and MERVL-int (bottom panel) in Dux-OE and control embryos at early and late 4C stages. Each dot represents an injected embryo. e-4C, early 4C stage; l-4C, late 4C stage. ***P < 0.001; two-tailed Student’s t-test. Center lines and error bars indicate means and SD, respectively. l Scatter plots of gene expression (left panel) and TE expression (right panel) in Dux-OE and control embryos at late 4C stage. FC > 2, FDR < 0.05. m Statistics of Dux-EGFP expression hours in MG132-treated and -untreated embryos. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed Student’s t-test. Error bars indicate SD
To further elucidate the effects of Dux depletion on embryos, maternal and zygotic (MZ) Dux-KO and WT embryos at zygotic, early 2C, middle 2C and late 2C stages were collected for single-cell RNA-seq (Smart-seq2). In agreement with published data7 (Supplementary information, Fig. S1c), Dux mRNA and reads were detected only in WT embryos at early 2C stage, while absent at Dux locus in KO embryos (Supplementary information, Fig. S1d, e). Moreover, Dux-KO embryos showed great similarity at transcriptional level to the KO embryos in recent report at late 2C stage6 (Fig. 1c), further validating the successful knockout of Dux.
As Dux is transiently expressed at early 2C stage, we mainly focused on its impact at 2C stage. Thus, we defined 6145 2C stage ZGA genes that were upregulated in early, middle or late 2C stage compared to zygotic stage in WT embryos (log2FC > 1.5 and FDR < 0.05, Supplementary information, Table S1). Dissecting different stages of 2C embryos revealed that Dux KO led to downregulation of MERVL and 129 ZGA genes (62.9% of downregulated genes) at early 2C stage, 213 ZGA genes (71.2%) at middle 2C stage, and 281 ZGA genes (78.1%) at late 2C stage, compared to WT 2C embryos (log2FC < –1.5 and FDR < 0.05, Fig. 1d; Supplementary information, Fig. S1f–h, Tables 2–4). Intriguingly, these ZGA transcripts showed robust upregulation at late 2C stage compared to early 2C stage in the absence of Dux, indicating that the activation of these ZGA transcripts, although delayed, still took place in Dux-KO embryos (Fig. 1e, f; Supplementary information, Fig. S1i–k). Thus, our single-cell RNA-seq results revealed that Dux-KO embryos at late 2C stage finally reached a state more similar to WT embryos at middle 2C stage than late 2C stage (Supplementary information, Fig. S1l). We speculated whether genetic compensation of Dux functional homologues triggered by Dux disruption as a mechanism expounded recently8 might be responsible for the activation of these ZGA transcripts and viability of Dux-KO mice. However, the expression of Gm4981 or Duxbl was not detected in Dux-KO embryos (Supplementary information, Fig. S1m).
Knockout of Dux resulted in delayed ZGA and decreased survivability of embryos. The phenotypic differences between Dux-KO individuals suggested that the potential compensatory factors may have to reach a threshold to support embryo development after Dux depletion as reported.9 MEVRL was found to generate chimeric transcripts with some 2C genes and its LTR (MT2) could promote the nearby 2C gene expression.10,11 Several studies also demonstrated that MERVL expression could be detected as early as zygotic stage2,7,10 (Fig. 1f), which is prior to endogenous expression of Dux. Remarkably, we found that MERVL was significantly upregulated at middle and late 2C stages in Dux-KO embryos, which is strongly correlated with the activation of some ZGA genes that generated chimeric transcripts with junctions to MERVL elements10 (Fig. 1f and Supplementary information, Fig. S1n). In sum, our data demonstrate that Dux is an important factor for ZGA by enhancing but not initiating the expression of ZGA transcripts such as Zscan4s, Tdpoz4, Usp17la, Zfp352 and MERVL.
To further determine the function of Dux in early embryo development, we injected the in vitro transcribed mRNA of Dux-EGFP (Dux-OE group) or EGFP (control group) into both blastomeres of 2C embryos (Fig. 1g and Supplementary information, Fig. S2a (left)) at late 2C stage when the endogenous Dux expression could not be detected already7 (Supplementary information, Fig. S1d, e). We also injected RFP mRNA simultaneously in both groups for labeling and tracing. Different from the control group in which both EGFP and RFP were robustly expressed in the cytoplasm until blastocyst stage, Dux-EGFP expression specifically occurred in the nuclei and lasted a few hours, and most of Dux-EGFP was dramatically diminished in 8 h after injection (Fig. 1g). More interestingly, the injected 2C embryos were arrested mainly at four-cell (4C) stage (Fig. 1g, h), similar to the phenomenon observed upon LINE1 knockdown that caused Dux upregulation as reported.12 Immunostaining showed that the arrested embryos robustly expressed Zscan4 and MERVL, the signatures of 2C embryo (Fig. 1i). To eliminate the artificial noise, we performed mRNA injection in only one blastomere of 2C embryos (Supplementary information, Fig. S2a (right)). The injected or non-injected blastomere showed no differences in developmental potential in the control group (Supplementary information, Fig. S2b, c). The blastomere injected with Dux-EGFP was mostly arrested after one cell cycle as monitored by the RFP signal and the other blastomere developed normally in Dux-OE group (Supplementary information, Fig. S2b, c). Immunostaining of such embryos at blastocyst stage showed that 2C-specific markers such as Zscan4 and MERVL were robustly expressed in the arrested blastomeres, while the markers for blastocyst such as Nanog and Cdx2 could only be detected in the blastocyst that developed from the non-injected blastomere (Fig. 1j). We could observe similar phenotype when injecting mRNA of Dux-EGFP or control EGFP into one blastomere of 4C embryos (Supplementary information, Fig. S2d, e). These results strongly suggest that prolonged expression of Dux in embryos significantly induced developmental arrest of blastomeres with 2C signatures.
To systematically study the transcriptional fluctuation caused by Dux expression in vivo, we collected the 2-blastomere-injected embryos of Dux-OE group and control group 6 h (early 4C stage) or 17 h (late 4C stage) after mRNA injection at late 2C stage. Transcriptional analysis revealed no significant differences between Dux-OE group and control group at early 4C stage (Supplementary information, Fig. S2f and Table S5). However, transcriptome changed violently in the Dux-OE embryos at late 4C stage (Supplementary information, Fig. S2g), with significant upregulation of Dux-related ZGA genes and retrotransposons (Fig. 1k, l; Supplementary information, Table S6). Collectively, these results demonstrate that Dux could promote the activation of some ZGA transcripts in vivo, while its prolonged expression would lead to developmental arrest and embryo death.
Both endogenous and exogenous expression of Dux last only a few hours in embryos. Especially when mRNA was injected into zygotes, Dux-EGFP was dramatically degraded within 3–5 h, showing no effect on development (Supplementary information, Fig. S3a–c and Table S7). This further demonstrated the importance of Dux degradation for embryo development. We speculated that there might be some mechanisms by which Dux protein is rapidly decomposed in vivo. Ubiquitin-proteasome system is a major intracellular protein degradation system during ZGA and early mouse embryo development.13 Treatment of proteasome inhibitor MG132 greatly delayed the attenuation of Dux-EGFP signal in zygote, thus resulting in developmental arrest (Fig. 1m and Supplementary information, Fig. S3d). Moreover, overexpression of Dux-EGFP in NIH3T3 cell line followed by immunoprecipitation-mass spectrometry revealed that Dux not only interacted with ZGA-related proteins including Zscan4 family, but also interacted with ubiquitin pathway-related proteins such as Uba52, Ubb, Ubc and Trim32, as well as Usp17la, Usp17le and Tdpoz5, which were also induced by Dux in 2C embryos (Supplementary information, Fig. S3e and Table S8).
Overall, our study demonstrated that Dux is important but not essential for early embryo development by enhancing rather than initiating ZGA. Albeit Dux-KO mice can survive to adulthood, knockout of Dux led to delayed ZGA and decreased development potential of embryos. On the other hand, Dux indeed triggered ZGA program in vivo; however, its expression should be silenced not only by LINE1 to recruit Nucleolin/Kap1 complex at DNA level,12 but also be degraded at both RNA and protein levels since prolonged activation of Dux severely impairs early embryo development. Exploring the mechanisms involved in Dux degradation may shed light on the deeper understanding of Facioscapulohumeral dystrophy (FSHD) caused by misexpression of DUX4,14 the human orthologue gene of Dux. Together, our data indicates that the activation of Dux is non-essential but the degradation of Dux is strictly necessary for embryo development in vivo.
Note: When we prepared the manuscript for submission, one study published by Dr Yi Zhang’s lab showed that Dux is not essential for ZGA.6 The major conclusion of their study is basically the same as our Dux KO part, but our study also demonstrates that the degradation of Dux is important for proper embryo development.
Supplementary information
Supplementary information, Figures and Methods
Acknowledgements
We thank our colleagues in the laboratory for their assistance with the experiments and comments on the manuscript. This work was supported by the Ministry of Science and Technology of China (2018YFA0108900 and 2016YFA0100400), the National Natural Science Foundation of China (31671530, 81630035, 81871164, 31871486 and 31741028), and the Rising-Star Program of Shanghai Science and Technology Commission (18QA1404400). In addition, M.G. wants to thank the support from Ni Wang over the past years.
Author contributions
P.Y. and Y.W. designed the experiments. M.G. and J.Z. performed the experiments; Y. Zhang analyzed data. Y.B., J.X. and C.X. assisted with generation of knockout mice; X.K., Y. Zhao and Y.L. performed embryo microinjection and manipulation; Z.T., K.L. and J.L assisted with generation of single cell RNA libraries. S.G., P.Y. and Y.W. conceived the project and provided mentoring. M.G. and Y.W. wrote the manuscript.
Data availability
All data sets that were generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE134832.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Mingyue Guo, Yanping Zhang.
Contributor Information
Peng Yang, Email: peng.yang@tongji.edu.cn.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
Yixuan Wang, Email: wangyixuan@tongji.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0238-4.
References
- 1.Schulz KN, Harrison MM. Nat. Rev. Genet. 2019;20:221–234. doi: 10.1038/s41576-018-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe KI, et al. Proc. Natl Acad. Sci. USA. 2018;115:E6780–E6788. doi: 10.1073/pnas.1804309115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Iaco A, et al. Nat. Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendrickson PG, et al. Nat. Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Nat. Genet. 2017;49:935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Zhang Y. Nat. Genet. 2019;51:947–951. doi: 10.1038/s41588-019-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Q, Ramskold D, Reinius B, Sandberg R. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 8.El-Brolosy MA, et al. Nature. 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macfarlan TS, et al. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, W. et al. Nucleic Acids Res. pii: gkz594. 10.1093/nar/gkz594 (2019).
- 12.Percharde M, et al. Cell. 2018;174:391–405. doi: 10.1016/j.cell.2018.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuchi C, et al. J. Reprod. Dev. 2018;64:65–74. doi: 10.1262/jrd.2017-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek A, Rahimov F, Jones PL, Kunkel LM. Trends Mol. Med. 2015;21:295–306. doi: 10.1016/j.molmed.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figures and Methods
Data Availability Statement
All data sets that were generated in this study have been deposited in the Gene Expression Omnibus under accession number GSE134832.