Abstract
Recent advances in molecular diagnostics have shown that lesions affecting both copies of the gene for tumor suppressor protein 53 (TP53) count among the most powerful predictors for high-risk disease in multiple myeloma (MM). However, the functional relevance and potential therapeutic implications of single hits to TP53 remain less well understood. Here, we have for the first time approximated the different constellations of mono- and bi-allelic TP53 lesions observed in MM patients within the frame of a single MM cell line model and assessed their potential to disrupt p53 system functionality and to impart drug resistance. Both types of common first hit: point mutation with expression of mutant p53 protein or complete loss of contribution from one of two wildtype alleles strongly impaired p53 system functionality and increased resistance to melphalan. Second hits abolished remaining p53 activity and increased resistance to genotoxic drugs even further. These results fit well with the clinical drive to TP53 single- and double-hit disease in MM patients, provide a rationale for the most commonly observed double-hit constellation (del17p+ TP53 point mutation), and underscore the potential increases in MM cell malignancy associated with any type of initial TP53 lesion.
Subject terms: Transposition, Myeloma
Introduction
The past 15 years have seen rapid and ongoing changes in the treatment of multiple myeloma (MM), a plasma cell neoplasm which accounts for about 13% of hematologic cancers1,2. These advances have already led to substantial improvements in the numbers for progression-free and overall survival, with ever more therapeutic options available for combinatorial use and promising efficacy even in relapse settings3. About 15% of newly-diagnosed MM is classified as “high-risk disease” and expected to show an unfavourable course of treatment with (much) shorter than average survival. This classification is based on retrospective correlation of cytogenetic and clinical parameters with outcome, and is being updated and refined as more molecular information becomes available4–6. One of the most constant features in high-risk myeloma are lesions that affect the TP53 gene, which encodes the tumor suppressor protein p53. Typically, such hits have been associated with deletions of the short arm of chromosome 17 (del17p) and determined cytogenetically by fluorescence in situ hybridization7. The possibility of involvement of genes other than TP53 in del17p-associated oncogenicity has still not fully been discounted8, but presence of TP53 in the minimally deleted region and the demonstration that TP53 hemizygosity entails p53 haploinsufficiency strongly support its cardinal role8,9. Additionally, advances in tumor cell sequencing have revealed the existence of point mutations affecting the p53 coding sequence in MM patients. Such mutations are most often observed in association with del17p, thus marking out MM cells with hits to both TP53 alleles (“double-hit disease”), although combinations of wildtype + mutant can also be found10–13. The TP53 double-hit constellation represents one of the most dire prognosis groups in newly diagnosed MM, whereas hardly any4 or only a moderate effect14 have been reported for TP53 single-lesion disease.
Although MM with TP53 lesions appears to be per se responsive to therapies with novel agents15–17, the acquisition of additional oncogenic driver events, often in combination with outgrowth of a TP53 double-hit clone, appears to underlie the fast progress into intractable and fatal disease18. However, little is known about whether and to what extent the different types and constellations of TP53 lesions, i.e. a deletion-first (haploinsufficiency) vs. a mutation-first (potential dominant negativity) vs. a double-hit scenario (most often high expression of just mutant p53 protein) may affect p53 system functionality and drug responsiveness in MM, and if such knowledge could inform therapeutic decisions. Additionally, depending on the actual mutation present, gain-of-function activities of p53 are also a possibility19. Here we have used the TP53wt/wt MM cell line AMO-1 in a functional approach that emulates the types of single-hit and double-hit lesions to TP53 observed in MM patients, and which provides a means to compare their effect within the frame of an isogenic cell line model.
Results
Generation of mono-and bi-allelic TP53 lesions in MM cells and analysis of p53 system functionality in AMO-1 clones
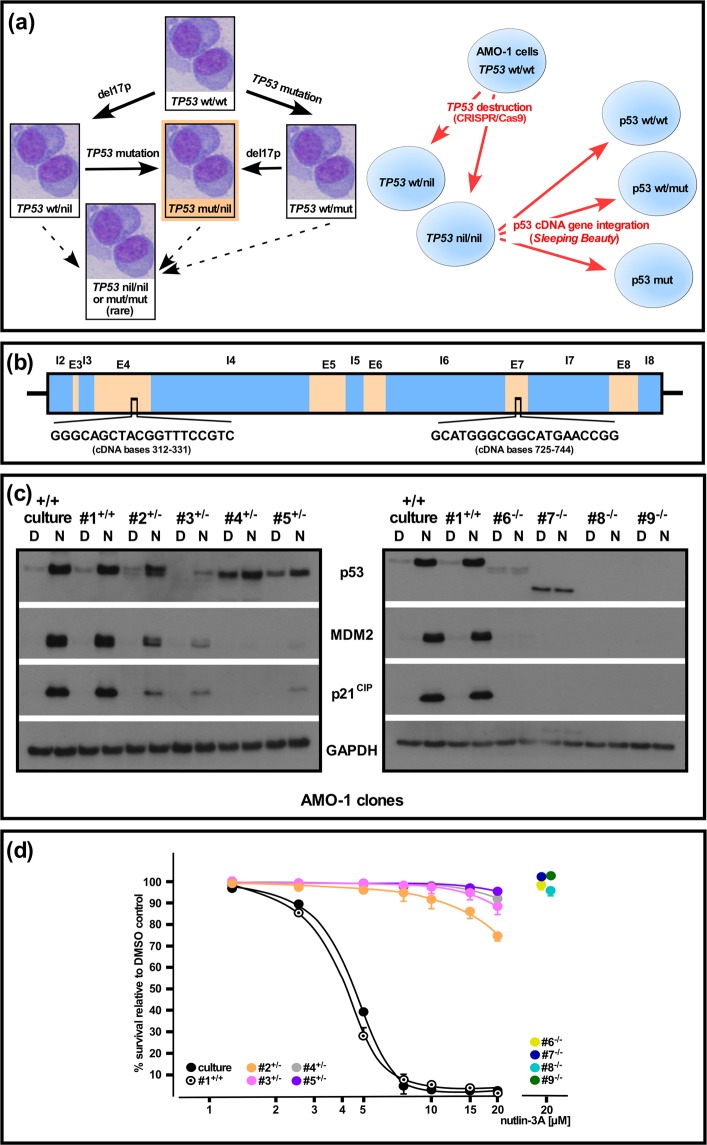
In order to get better insights into the functional consequences of the different types of TP53 lesions in MM we decided to try to emulate the different single- and double-hit constellations within a single MM cell line model. The two basic steps involved were an initial destruction of one or both TP53 alleles in TP53-wildtype MM cells via the CRISPR/Cas9 method, followed by concerted introduction of cDNA-expression cassettes for wildtype + mutant p53 via the Sleeping Beauty transposon system (Fig. 1a). Two different target sequences for CRISPR/Cas9-mediated TP53 disruption were tested (Fig. 1b) and the respective guide-RNA expression vectors were co-electroporated with an expression plasmid for EGFP to permit manual selection of the most efficiently transfected cells for further clonal upgrowth. Of the four TP53 wildtype MM cell lines initially tested (AMO-1, MM.1s, MOLP-8, NCI-H929) only AMO-1 yielded sufficient numbers of clones to permit further analysis, probably due to its favourable combination of relatively high electroporation efficiency for plasmids and fast growth rate. A total of 85 clones were checked for TP53 defects by PCR/sequence analysis off genomic DNA covering the respective CRISPR/Cas9 target sites and/or Western blotting for p53 and its downstream targets MDM2 and p21CIP after overnight treatment with the MDM2 inhibitor nutlin 3A (Fig. 1c). Clones showing TP53 defects on sequencing (i.e. a clean sequence turning to unreadability once the sequences between the two alleles diverge) were further characterized by cloning of PCR products into vector pGEM-T Easy to assess whether one or both TP53 alleles were affected and to identify the precise molecular defects. One TP53wt/wt clone (clone #1) and four clones each with either TP53wt/− or TP53−/− status were selected for further analysis (Fig. 1c,d and See Supplementary Fig. 1 for an overview of the specific changes to TP53 and the predicted effects on the translated p53 protein). As expected, the wildtype clone was indistinguishable in its reactions to treatment with nutlin 3A from the parental cell line, whereas clones with double disruption of TP53 showed no activity of the p53 system at all, as evidenced by absence of upregulation of p53/MDM2/p21CIP in Western blotting and of nutlin 3a-induced cell death (Fig. 1c,d). Clones “hemizygous” for TP53 (in the sense that they harbored one wildtype allele and one affected by a CRISPR/Cas9-mediated indel with presumed lack of contribution to p53 levels from the affected allele) displayed some variability in their ability to mount a nutlin-induced p53 response, but these effects were in all cases strongly impaired compared to parental AMO-1 cells with two wildtype copies of TP53, and barely affected their survival when challenged with the MDM2 inhibitor (Fig. 1c,d).
Figure 1.
Generation and functional analysis of homo- and heterozygous TP53 lesions in an AMO-1 MM cell line model system. (a) Pathways to TP53 lesions. Left: Primary MM cells will either initially lose part of chromosome 17 (del17p) or acquire an inactivating TP53 point mutation as “first-hit”. Both will then normally give rise to a del17p/point mutation “double hit” constellation (highlighted in orange), which is also the most common arrangement found in MM cell lines. Right: TP53 destruction in AMO-1 cells by CRISPR/Cas9 followed by establishment of p53 cDNA gene expression in TP53 nil/nil clones via Sleeping Beauty to generate p53 wt/wt, p53 wt/mut and p53 nil/mut scenarios. (b) Schematic intron (I)/exon (E) structure of parts of the human TP53 gene, indicating the two target sites for CRISPR/Cas9-mediated disruption. (c) Characterization of p53 system functionality in p53 wt AMO-1 cells (culture, clone #1), AMO-1 clones with a single disrupted TP53 allele (clones 2–5) or with two disrupted TP53 alleles (clones 6–9). Cells were treated overnight with 10 µM nutlin-3A (N) or the applicable amount of solvent (DMSO, D) and analyzed for p53 system components by Western blotting. Staining for GAPDH served as loading control. See Supplementary Fig. 4 for full representation of the data. (d) Kill curves for 3-day-treatment with nutlin-3A for TP53+/+, TP53+/− and TP53−/− AMO-1 clones. Only the highest drug concentration tested is depicted for the fully-resistant TP53−/− clones (#6–9). Kill curves were calculated from 2 independent experiments. Error bars depict s.e.m.
Functional analysis of the effect of p53 point mutation in the AMO-1 MM cell line model system
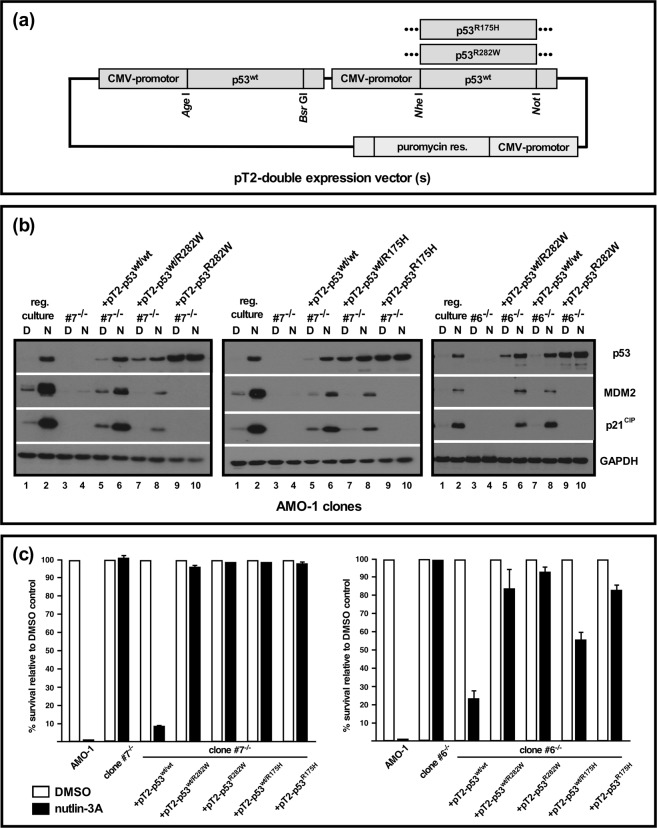
We next tested whether stable integration of vectors for constitutive re-expression of p53 (i.e. of cDNA copies) into AMO-1 TP53−/− clones was feasible and sufficient to re-instate a functional p53 system. A modified Sleeping Beauty transposon vector (Fig. 2a and Supplementary Fig. 2) was used to concomitantly introduce either two identical CMV-promotor driven expression cassettes for wildtype p53, or for combinations of wildtype p53 plus point mutated p53, thus emulating a “mutation first” scenario. Two different point mutants, corresponding to known mutation hotspots (R175H and R282W) were used and transposed into TP53−/− clones. Initial transient transfection experiments and reverse transcription cDNA analysis with p53/vector-specific primers confirmed that both alleles were about equally well expressed from such double-cassette vectors (Supplementary Fig. 2). Following electroporation and puromycin selection transposed cultures were ready for use in experiments within 12–14 days. The polyclonal cultures that resulted for TP53−/−/p53wt/wt cells showed only slightly elevated steady state levels of p53 protein when compared to regular AMO-1 cells by Western blotting (Fig. 2b: left and middle: lanes 5 vs. lanes 1, right: lane 7 vs. lane 1). Levels of p53 were strongly enhanced after overnight treatment with nutlin-3A and roughly on a par with the increase observed in parental TP53 wildtype cells (Fig. 2b: left and middle: lanes 6 vs. lanes 2, right: lane 8 vs. lane 2), showing that the introduction of two constitutively expressed wildtype p53 cDNA-gene copies under CMV-promotor control could still functionally be integrated into the MDM2/p53 degradation/transcription cycle that governs the levels of p53 in AMO-1 cells. Likewise, MDM2 inhibitor treatment led to increased levels of the p53 downstream targets MDM2 and p21CIP, although not necessarily to levels as high as in parental AMO-1 cells (Fig. 2b). However, functional reinstatement of the p53 system in knockout clones also largely restored the p53-mediated cell death response (Fig. 2c), showing that this cellular model system indeed displayed all basic features required to analyze p53 function. We therefore tested next the stable transposition of TP53−/− cells with Sleeping Beauty vectors expressing combinations of wildtype p53 and either the mutant R282W (Fig. 2b, left, clone #7; right, clone #6) or the mutant R175H (Fig. 2b, clone 7, middle). In addition to higher steady-state levels of p53 (Fig. 2b: left and middle: lanes 7 vs. lanes 1; right: lane 5 vs. lane 1) these cells also showed attenuated nutlin-3A-mediated p53 responses with modest increases in total levels of p53 and smallish increases in the levels of MDM2 and p21CIP (Fig. 2b). Accordingly, these changes were not sufficient to trigger substantial cell death responses (Fig. 2c), underpinning the dominant negative functional effect that these p53 mutants are supposed to exert on the tetrameric transcription factor. As expected, introduction of just a single mutant p53 cDNA gene led to high steady-state levels of p53 that remained unchanged by treatment with nutlin-3A (Fig. 2b: lanes 9,10), and no functional responses of the p53 system (Fig. 2b,c) – a situation mimicking that encountered in many MM cell lines which display a TP53−/mut phenotype20.
Figure 2.
Functional analysis of single p53 point mutations with or without concomitant expression of wildtype p53 in AMO1-TP53−/− cells. (a) Schematic representation of Sleeping Beauty vectors with two individually addressable CMV promotor-driven expression cassettes to establish stably transfected polyclonal cultures with p53wt/wt, p53wt/R282W or p53wt/R175H cDNA gene expression. (b) Sleeping Beauty-mediated introduction into AMO-1 TP53−/− cells (clone #7) of either two CMV promotor-driven cDNA genes for wildtype p53 (pT2-p53wt/wt), of one wildtype plus one mutant cDNA gene (pT2-p53wt/R282W (left, in clone #7; right, in clone #6)) or pT2-p53wt/R175H (middle, in clone #7), or of single mutant p53 cDNA genes, to emulate “mutation first” and “deletion plus mutation” scenarios. Western blotting for p53 system components after overnight challenge with 10 µM nutlin-3A. See Supplementary Fig. 5 for full representation of the data. (c) Survival analysis (annexin V/PI-staining) for two AMO-1 TP53−/− clones expressing different constellations of wildtype and mutant p53 after 3 days incubation with nutlin-3A or an applicable amount of solvent (DMSO). Kill rates calculated from 2 experiments. Error bars depict s.e.m.
Impact of TP53/p53 status on the sensitivity for genotoxic drugs and proteasome inhibitors in the AMO-1 MM cell line model system
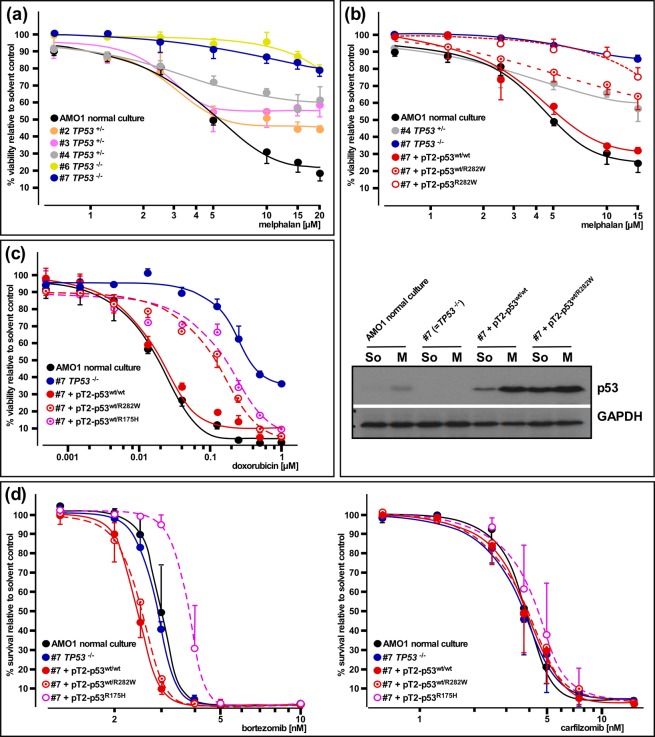
We next tested the consequences of TP53 single or double deletion for the sensitivity of AMO-1 cells against the clinically employed alkylans melphalan. AMO-1 clones that had either one or both alleles of TP53 destroyed by CRISPR/Cas9-mediated disruption were treated with the drug and viability curves (alamarBlue metabolic assay) measured after three days (Fig. 3a). The effect of the drug at higher concentrations was severely blunted by hemizygosity for TP53 (Fig. 3a: clones #2, #3, #4), and became very small in TP53−/− clones (Fig. 3a: clones #6, #7). We then tested the effects of melphalan treatment on AMO1 TP53−/− cells (Fig. 3b: clone #7, Supplementary Fig. 3a: clone #6, 3b: clone #7) after Sleeping Beauty-mediated stable introduction of two cDNA genes for either wildtype p53 or for combinations of wildtype and mutant p53 (Fig. 3b and Supplementary Fig. 3). Whereas re-instatement of a wildtype p53 status led to near-complete resensitization against melphalan, expression of the wildtype + mutant combinations produced an attenuated response very similar in size to that seen in the CRISPR/Cas9-generated clones hemizygous for TP53, again underpinning a dominant negative effect of acquisition of a TP53 mutation in a “mutation first” scenario (Fig. 3b and Supplementary Fig. 3). Accordingly, when comparing the effects of melphalan treatment on the p53 levels in TP53−/− cells either transposed with the p53wt/wt or the p53wt/R282W combination, the latter showed a decidedly smaller relative increase in p53 levels (Fig. 3b; absolute levels are higher in these cells because of the different steady-state level due to the presence of mutant p53, see also Fig. 2b). As expected, expression of mutant p53 in the TP53−/− cells did not alter the already unresponsive behaviour of such clones in any way (Fig. 3b). Treatment of AMO-1 cells with doxorubicin, another genotoxic compound that exerts at least part of its anti-myeloma effects via the p53 pathway, also showed a strong influence of the p53 status (Fig. 3c). Again, both settings emulating a “mutation-first” scenario (introduction of p53wt/R175H or p53wt/R272W into TP53−/− cells) led to levels of resistance decidedly higher than normal AMO-1 TP53 wildtype cells, or those in which two copies of p53 wildtype cDNA genes were expressed, but did not quite attain the insensitivity imparted by full p53 loss (Fig. 3c). Lastly, we tested the influence of the p53 status on the sensitivity of AMO-1 cells for proteasome inhibitors. No differences in the reaction to carfilzomib was found for any of the different TP53/p53 constellations (Fig. 3d, right), and at best slight shifts of the kill curves were found with bortezomib treatment. These were, if significant at all, perhaps a reflection of the slightly higher levels of wildtype p53 protein in the CMV-promotor driven contexts (resulting in slight sensibilisation) and of strong overexpression of mutant p53 in the mutant only expression scenario (resulting in slight desensibilisation). No difference was found between AMO1 wildtype cells and the TP53−/− clone (Fig. 3d, left), confirming that functionally p53 appears not to be significantly involved in the proteasome inhibitor-mediated apoptosis of these cells.
Figure 3.
Analysis of drug efficacy as a function of the TP53/p53 status. (a) Response to melphalan in AMO-1 wildtype cells and various clones with either one or both TP53 alleles disrupted. Viability assays (alamarBlue) after 3-day drug treatment. (b) Top: Response to melphalan in AMO-1 TP53−/− cells without (blue curve) or with (red curves) stable expression of combinations of wildtype and mutant p53 cDNA genes via Sleeping Beauty. Wildtype (black curve) and TP53 hemizygous AMO-1 clone #4 (grey curve) also included for comparison. Viability assays (alamarBlue) after 3-day drug treatment. Bottom: Western blot showing melphalan treatment-induced increases in p53 in AMO-1 cells with at least one (cDNA-) gene for p53. See Supplementary Fig. 5 for full representation of the data. (c) Response to doxorubicin in AMO-1 TP53−/− cells without (blue curve) or with (reddish curves) stable expression of combinations of wildtype and mutant p53 cDNA genes via Sleeping Beauty. Two different p53 mutant proteins (R175H, R282W) tested. Wildtype (black curve) AMO-1 cells also included for comparison. Viability assays (alamarBlue) after 3-day drug treatment. (d) Response to proteasome inhibitors in AMO-1 TP53−/− cells without (blue curve) or with (reddish curves) stable expression of combinations of wildtype and/or mutant p53 cDNA genes via Sleeping Beauty. Wildtype (black curve) AMO-1 cells also included for comparison. Survival assays (annexin V/propidiumiodide) after 3-day drug treatment. Kill curves calculated from 3 experiments. Error bars depict s.e.m.
Discussion
The presence of genetic lesions affecting both alleles of the tumor suppressor gene TP53, most commonly achieved via a combination of deletion (del17p) and point mutation, has now firmly been established as the major indicator for unfavourable prognosis in newly diagnosed multiple myeloma4,14,21. Additionally, a sizeable subgroup of MM patients will acquire de novo first hits to TP53, and/or progresses from single-hit to double-hit constellations during the course of their disease under treatment, thus in effect genetically switching into a class with high-risk features11–13,22,23. These observations also suggest that, whereas clones that harbour a single hit to TP53 do have a selection advantage, acquisition of a second hit still provides a significant, and probably often decisive step towards intractable malignancy. However, whereas it is now possible to monitor the presence, emergence or rise of subclones with TP53 deficiencies at consecutive stages of the disease, it remains unclear if, when and how such knowledge could potentially be used to inform therapeutic decisions.
Here, we have for the first time approximated the main types of TP53 lesion observed in MM patients within the frame of a single isogenic MM cell line model, and analyzed their impact on the functionality of the p53 system and their potential role for resistance against anti-MM drugs. Importantly, these models also comprise the “single-hit” constellations, i.e. situations in which either p53 contribution from one of two TP53 alleles is lost (approximating the del17p case), or in which only one of two expressed TP53 alleles is point mutated. The first situation has previously been associated with haploinsufficiency10, whereas the second situation (normally associated with dominant negativity) is encountered in MM patients, but is not represented by any MM cell line.
With a number of different haploinsufficient clones and two different p53 hotspot mutations tested, both types of single-hit models strongly impaired the functionality of the p53 system, and attenuated the response of AMO-1 cells to genotoxic drugs to similar extent. Furthermore, acquisition of a second hit to TP53 (here approximated by different models that either lead to strong expression of only mutant p53 protein or no expression of any p53 protein at all) again produced identical effects inasmuch as even the last vestiges of p53 system functionality were abolished and resistance to genotoxic drugs was still further enhanced. These results are important for three reasons: First, these models functionally underscore the strong gains in fitness and drug resistance that MM cells can potentially acquire from any type of single TP53 lesion, and which supports their rise to subclonal prominence. Second, the observation that the second hit to TP53 still further increases the survival potential of single-hit AMO-1 cells corresponds well with the clinically observed drive towards “double-hit” disease. Full TP53 inactivation in combination with genetic events that boost proliferation probably provides one of the fastest routes to outgrowth of intractable MM subclones. Third, these data lend support for the notion that it is indeed the hit to TP53 – and possibly to TP53 only – that represents the oncogenic element in the del17p lesion, and that it is the associated loss of p53 system function (and not any gain-of-function mechanism) that generally drives the spread of TP53 lesions across the MM holotumor. If, as implied by our experiments, double-hit disease is the favoured outcome, and in addition the functional consequences of single TP53 deletion and single TP53 mutation are essentially the same, then the selection of the generally observed constellation of bi-allelic TP53 lesions: del17p+ mutation, makes perfect sense. If a deletion is the easiest way to lose a functional TP53 copy, but a second deletion poses a problem because it normally also entails bi-allelic loss of other genes that might be important for MM cell fitness (but which are not necessarily related to malignancy), then the second event is most likely a precise hit to TP53, i.e. an inactivating mutation. Conversely, if that precise hit happened to be the initial lesion, it should most often be followed by del17p, providing an MM cell with all the gains of removing the remaining wildtype TP53 allele at no or little collateral costs.
Even though no single cell line model is likely to reflect all aspects of the consequences of TP53 lesions in MM patients, the AMO-1 system represented here has proven a robust and coherent model system for functional and pharmacological analysis of the consequences of single- and double-hits to TP53. Importantly, it implies that the different types of single-hit clones may already acquire considerable resilience against genotoxic drugs, but that this does not represent a primary resistance mechanism against other therapeutic approaches, such as proteasome inhibitors. The clinical need for treatment of TP53 double-hit MM patients is imminent, but unfortunately there are no proven drug approaches that target this lesion, as efforts to pharmacologically re-activate mutant p53 have not yet led to specific therapies24. Moreover, since MM cells are adept at developing resistance also against therapies that do not rely on functional p53, it is possible that even with the best efforts, and including the use of novel approaches such as immunotherapies, MM patients with double-hits to TP53 may always represent the clinically most disfavoured group. From a longer-term perspective it may therefore be rewarding to identify therapy approaches with the declared aim to keep the number of MM patients that develop TP53 double-hit disease as low as feasible. Increasingly precise molecular tumor diagnostics, including MRD molecular status assessments, can now be applied before and along the course of treatment. The presence or rise of established or novel MM subclones with single or double TP53 deficiencies could thus be detected and the information potentially be used for therapeutic decisions as well as for retrospective analyses that promote our understanding of the precise genetic features that underlie the unfavourable clinical courses of MM with TP53 lesions. Based on the results presented here, it would for example be important to know if patients who present with MM subclones that display single hits to TP53 should rather not be treated with genotoxic drugs in order to minimize the risk of acquisition of the second hit. Likewise, it would be important to know if patients with MM subclones that display single TP53 lesions should preferentially be treated with therapies that are known to work on MM cells with TP53 defects, and if such genetic constellations are an indication for use of powerful combination approaches, perhaps with inclusion of immunotherapies, in an attempt to nip emergent (pre-) high risk clones in the bud. Over time, such information will automatically accrue as part of the collective treatment histories of the global MM patient population, but it will only be interpretable if all aspects regarding the (change of) status of TP53 lesion are precisely documented.
Materials and Methods
Cell culture
Cell line AMO-1 was bought from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) and immediately expanded into stock and working banks. The cell culture was reinstated from working bank-aliquots every 3–4 months. Standard MM cell culture was performed in RPMI-1640 medium supplemented with 10% FBS, 1 mM sodium pyruvate, 2 mM glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. Cells were kept at 5% CO2 and 37 °C. Cultures were regularly checked for mycoplasma negativity25.
CRISPR/Cas9 constructs
Vectors for CRISPR/Cas9-mediated TP53 disruption were based on the GeneArt CRISPR nuclease: CD4 enrichment kit (Invitrogen, Carlsbad, USA; A21175). The following oligonucleotides were used to create dsDNA fragments for cloning and generation of functional guide-RNA expression vectors (nucleotide sequence corresponding to the human TP53 gene in bold): construct 1: TP53-1-forward: 5′-GGGCAGCTACGGTTTCCGTCGTTTT-3′, TP53-1-reverse: 5′-GACGGAAACCGTAGCTGCCCCGGTG-3′; construct 2: TP53-2-forward: 5′-GCATGGGCGGCATGAACCGGGTTTT-3′, TP53-2-reverse: 5′ CCGGTTCATGCCGCCCATGCCGGTG-3′.
Determination of the TP53 status of MM cell line clones
Clones were initially screened for p53 system functionality while still in 96-well format culture, by exposing about 10.000 cells overnight to either DMSO or 10 µM nutlin-3A, followed by Western analysis for p53 system induction. Additionally, genomic DNA was prepared for all clones and parts of the TP53 gene which included the CRISPR target site(s) amplified by conventional PCR using primers 5′-TGAAGACCCAGGTCCAGATG and 5′-CACTGACAGGAAGCCTAAGG (site 1) and/or 5′-TTGCCACAGGTCTCCCCAAG and 5′-TGAGTGGGAGCAGTAAGGAG (site 2), respectively. PCR products were Sanger sequenced to either confirm wildtype status (clean correct reads) or presence of single/double lesions (clean reads turning unreadable from the base at which both alleles diverge, or in one case (clone #8) a clean read containing an insertion). The latter clones were further characterized by cloning of PCR products into vector pGEM-T Easy and Sanger sequencing of minipreparations to (i) reveal the exact lesion(s) present, (ii) confirm the presence of wild-type TP53 in single-lesion clones (Supplementary Fig. 1).
Construction of p53 cDNA expression vectors
Wild-type p53 cDNA was amplified from AMO-1 first-strand cDNA using primers that flank the coding region either with NheI/NotI or with BsrGI/AgeI restriction sites and cloned into suitably modified pBluescript II vectors. Single base changes to generate p53 hotspot mutants R175H (selected for its presence and previous confirmation of p53 system loss of function in MM cell line OPM220) and R282W (selected for its presence in a primary MM sample showing additional loss of the initially remaining wildtype TP53 gene in a later biopsy) were introduced according to the QuikChange protocol. A pT2 (Sleeping Beauty) transposon vector conferring puromycin resistance was modified to contain either a single or two near identical CMV promotor-driven expression cassettes. In the latter case, the only difference between these cassettes was that they were individually selectable for subcloning using either the NheI/NotI or the BsrGI/AgeI combinations. Wildtype and/or mutant p53 cDNAs were finally subcloned from the respective pBluescript II vectors into Sleeping Beauty to generate vectors for p53wt/wt and p53wt/mut cDNA-gene expression, or for expression of single p53 cDNA-gene copies.
Electroporation and selection of AMO-1 clones or transposed cultures
The protocol for MM cell electroporation is described in detail in ref. 26. TP53 guide-RNA expression vectors were used in electroporation mixtures at concentrations of 25 µg/ml when used alone, or at 20 µg/ml each when used together. Additionally, an expression plasmid for EGFP was co-transfected, and after 3 days single bright green cells were manually selected from the transfection culture and transferred into 96-well plates for clonal upgrowth and characterization of TP53 defects.
Electroporation of Sleeping Beauty vectors into AMO-1 cells was performed with 20 µg/ml of Sleeping Beauty plasmid, 30 µg/ml of pTX100 transposase expression plasmid and 5 µg/ml of EGFP expression plasmid for easy assessment of transfection efficiency. After overnight recovery the transfected cultures were subjected to puromycin selection (1 µg/ml, 10–14 days) to establish polyclonal cultures of transposed cells.
Drug treatment of MM cells
MM cells were seeded into 96 well plates (10.000 cells per well), incubated for 3 days with the respective compounds and subjected to alamarBlue or annexin V/propidiumiodide viability/cell death measurements. Drug solutions were always freshly prepared from concentrated stocks (solvents for stock solutions: acidic ethanol for melphalan, DMSO for all other compounds). Drug treatment of cells intended for use in Western blotting was performed overnight.
Western blotting and antibodies
Western blotting was performed exactly as described27. The primary antibodies used were: anti-β-actin (Sigma, Deisenhofen, Germany; A5316), anti-MDM2 (Santa Cruz, Heidelberg, Germany (sc-965)), anti-p21Cip/Waf (Santa Cruz; sc-397), anti-p53 (Santa Cruz; sc-126), and anti-GAPDH (Sigma; G9545). Secondary antibody F(ab′)2 fragments coupled to horseradish peroxidase and specific for rabbit-IgG (No. 111-036-045) or mouse-IgG (No. 115-036-072) were obtained from Jackson ImmunoResearch, Newmarket, UK. A freshly made solution of luminol (2.5 mM), p-coumaric acid (0.2 mM) and H2O2 (0.01%) in 100 mM Tris-HCl (pH 8.8) was used as reagent for chemiluminescent detection.
Other reagents
Annexin V was prepared following a published protocol28, and coupled to either fluoresceinisothiocyanate (Sigma; F7250) or to PromoFluor 647 using its N-hydroxysuccinimidyl ester (PromoCell, Heidelberg, Germany; PK-PF647-1). Drugs used in this study were obtained from Sigma-Aldrich (melphalan; M2011), Selleck Chemicals, München, Germany (bortezomib; S1013, carfilzomib; S2853, nutlin-3A; S8059), and Merck, Darmstadt, Germany (doxorubicin; 324380). A solution of Na-resazurin (Sigma; R7017) in PBS was used as reagent in viability assays (alamarBlue).
Flow cytometry
Cells were washed with PBS, pelleted and resuspended in 200 µl of cold annexin V binding buffer (10 mM HEPES/NaOH, 140 mM NaCl, 2.5 mM CaCl2; pH7.4) containing 1 µl of annexin V-PromoFluor 647 solution (see Reagents) and 1 µg/ml propidium iodide. Flow cytometry was performed with a FACS Calibur (BD Biosciences, Heidelberg, Germany). Datafiles were analysed with FlowJo version 8.8.7 (Tree Star, Inc., Ashland, USA).
Data analysis
Dose-effect curves were calculated from at least two independent experiments by non-linear regression analysis (sigmoidal shape, variable slope setting) using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA). Regression analysis was not performed for data sets when it was obvious that a dose-effect relationship did not exist. The figures depict functional effects calculated with respect to the individual solvent treated controls of the respective clones. Data permitting appraisal of the magnitude of differences (generally slight) between the clones or Sleeping Beauty-transposed polyclonal cultures and the parental cell line are presented in Supplementary Figs. 1c and 3c.
All data generated or analysed during this study are included in this published article (and its Supplementary Information Files). The described materials are freely available for scientific purposes.
Supplementary information
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (KFO 216). U.M. was supported by a grant from the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg.
Author contributions
U.M., M.R., D.S. and T.S. performed research; S.B., H.W., R.C.B. and T.S. analysed data; K.M.K. and T.S. edited/wrote manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Umair Munawar and Markus Roth.
Supplementary information
is available for this paper at 10.1038/s41598-019-54407-4.
References
- 1.Palumbo A, Anderson K. Multiple myeloma. N. Engl. J. Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Landgren O, Rajkumar SV. New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin. Cancer Res. 2017;22:5428–5433. doi: 10.1158/1078-0432.CCR-16-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar SK, et al. Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: a multicenter IMWG study. Leukemia. 2017;31:2443–2448. doi: 10.1038/leu.2017.138. [DOI] [PubMed] [Google Scholar]
- 4.Walker BA, et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33:159–170. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palumbo A, et al. Revised international staging system for multiple myeloma: a report from International Myeloma Working Group. J. Clin. Oncol. 2015;33:2863–2869. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolli N, et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia. 2018;32:2604–2616. doi: 10.1038/s41375-018-0037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merz M, et al. Baseline characteristics, chromosomal alterations, and treatment affecting prognosis of deletion 17p in newly diagnosed myeloma. Am. J. Hematol. 2016;91:E473–E477. doi: 10.1002/ajh.24533. [DOI] [PubMed] [Google Scholar]
- 8.Perrot, A., Corre, J. & Avet-Loiseau, H. Risk stratification and targets in multiple myeloma: from genomics to bedside. ASCO Educational Book, 675–680 (2018). [DOI] [PubMed]
- 9.Teoh PJ, et al. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia. 2014;28:2066–2074. doi: 10.1038/leu.2014.102. [DOI] [PubMed] [Google Scholar]
- 10.Lodé L, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010;95:1973–1976. doi: 10.3324/haematol.2010.023697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kortüm KM, et al. Targeted sequencing using a 47 gene multiple myeloma panel (M3P) in −17p high risk disease. Br. J. Haematol. 2015;168:507–510. doi: 10.1111/bjh.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lionetti M, et al. Molecular spectrum of TP53 mutations in plasma cell dyscrasias by next generation sequencing: an Italian cohort study and overview of the literature. Oncotarget. 2016;7:21353–21361. doi: 10.18632/oncotarget.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin M, et al. Prevalence and timing of TP53 mutations in del(17p) myeloma and effect on survival. Blood Cancer J. 2017;7:e610. doi: 10.1038/bcj.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanendrarajan S, et al. The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica. 2017;102:e364–e367. doi: 10.3324/haematol.2017.168872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen YC, et al. Treatment patterns and clinical outcomes in high-risk newly diagnosed multiple myeloma carrying the 17p deletion: an observational multi-center retrospective study. Am. J. Hematol. 2018;93:810–815. doi: 10.1002/ajh.25098. [DOI] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, et al. Carfilzomib significantly improves the progression-free survival of high-risk patients in multiple myeloma. Blood. 2016;128:1174–1180. doi: 10.1182/blood-2016-03-707596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonneveld P, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127:2955–2962. doi: 10.1182/blood-2016-01-631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker BA, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2014;132:587–597. doi: 10.1182/blood-2018-03-840132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller PAJ, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stühmer T, et al. Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood. 2005;106:3609–3617. doi: 10.1182/blood-2005-04-1489. [DOI] [PubMed] [Google Scholar]
- 21.Weinhold N, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128:1735–1744. doi: 10.1182/blood-2016-06-723007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chavan SS, et al. Bi-allelic inactivation is more prevalent at relapse in multiple myeloma, identifying RB1 as an independent prognostic marker. Blood Cancer J. 2017;7:e535. doi: 10.1038/bcj.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryland GL, et al. Novel genomic findings in multiple myeloma identified through routine diagnostic sequencing. J. Clin. Pathol. 2018;71:895–899. doi: 10.1136/jclinpath-2018-205195. [DOI] [PubMed] [Google Scholar]
- 24.Perdrix A, et al. PRIMA-1 and PRIMA-1Met (APR-246): from mutant/wild type p53 reactivation to unexpected mechanisms underlying their potent anti-tumor effect in combinatorial therapies. Cancers. 2017;9:172. doi: 10.3390/cancers9120172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uphoff CC, Drexler HG. Detecting mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol. Med. 2004;88:319–326. doi: 10.1385/1-59259-406-9:319. [DOI] [PubMed] [Google Scholar]
- 26.Steinbrunn T, Chatterjee M, Bargou RC, Stühmer T. Efficient transient transfection of human multiple myeloma cells by electroporation - an appraisal. Plos One. 2014;9:e97443. doi: 10.1371/journal.pone.0097443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hausmann S, et al. Loss of serum and glucocorticoid-regulated kinase 3 (SGK3) does not affect proliferation and survival of multiple myeloma cell lines. Plos One. 2015;10:e0122689. doi: 10.1371/journal.pone.0122689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Logue SE, Elgendy M, Martin SJ. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat. Protoc. 2009;4:1383–1395. doi: 10.1038/nprot.2009.143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.