Abstract
Pathological healing characterized by abnormal angiogenesis presents a serious burden to patients’ quality of life requiring innovative treatment strategies. Glycosaminoglycans (GAG) are important regulators of angiogenic processes. This experimental and computational study revealed how sulfated GAG derivatives (sGAG) influence the interplay of vascular endothelial growth factor (VEGF)165 and its heparin-binding domain (HBD) with the signaling receptor VEGFR-2 up to atomic detail. There was profound evidence for a HBD-GAG-HBD stacking configuration. Here, the sGAG act as a “molecular glue” leading to recognition modes in which sGAG interact with two VEGF165-HBDs. A 3D angiogenesis model demonstrated the dual regulatory role of high-sulfated derivatives on the biological activity of endothelial cells. While GAG alone promote sprouting, they downregulate VEGF165-mediated signaling and, thereby, elicit VEGF165-independent and -dependent effects. These findings provide novel insights into the modulatory potential of sGAG derivatives on angiogenic processes and point towards their prospective application in treating abnormal angiogenesis.
Subject terms: Molecular biophysics, Growth factor signalling, Computational models
Introduction
Sulfated glycosaminoglycans (sGAG) are linear, negatively charged polysaccharides consisting of repetitive disaccharide units (D.U.), which interact with a variety of mediator proteins and, thereby, modulating their biological activity1,2. Native GAG differ in their sulfation content and pattern as well as their monosaccharide composition, which results in a high complexity of GAG-mediated biological functions3–7.
Vascular endothelial growth factor (VEGF)-A is a 45 kDa homodimeric glycoprotein and the most potent and specific regulator of physiological and pathological angiogenesis8. It is required for the chemotaxis and differentiation of endothelial precursor cells (angioblasts), endothelial cell (EC) proliferation, the direct assembly of ECs into vascular structures (vasculogenesis) and angiogenic remodeling9. Alternative splicing of VEGF-A gives rise to at least eight isoforms10,11. VEGF165 is the predominant isoform in human tissues and contains a heparin (Hep) binding domain (HBD)10. VEGF165 exerts its biological effects through binding to the high affinity receptor tyrosine kinases VEGF receptor-1 (VEGFR-1) and -2 (VEGFR-2), as well as to the co-receptor neuropilin-1 (NRP-1), all of which are predominantly expressed on ECs10. However, the major receptor for the mitogenic, angiogenic and vascular permeability enhancing effects of VEGF is VEGFR-212. Upon ligand binding, VEGFR-2 undergoes dimerization and strong autophosphorylation of the cytoplasmic domains on specific tyrosine residues resulting in a mitogenic, chemotactic and prosurvival signal8,12,13. GAG such as Hep and heparan sulfate (HS) have profound effects on VEGF165 function, not only by binding VEGF165 directly, but also by interacting with VEGF receptors and NRP-114–16. In vitro studies demonstrated that cell surface HS interaction with VEGF165 enhance VEGF165-induced phosphorylation of VEGFR-2 and increase mitogenic activity as well as endothelial tube formation17–20. In addition, binding of VEGF165 to VEGFR-2 was affected by the size, degree of sulfation (DS), sugar ring stereochemistry and conformation of Hep21–23.
Currently, in a constantly aging population with increasing number of multimorbid patients24, controlling angiogenic factors represents a very important goal for regenerative medicine and tissue engineering in terms of improving healing processes, particularly in injured vascularized tissues such as bone and skin. Innovative biomaterials containing GAG derivatives with defined sulfation degree and pattern are promising tools for selectively influencing their molecular recognition by target mediator proteins such as growth factors and, thereby, modulating their biological activity. In previous studies, a regulatory effect of hyaluronic acid (HA) derivatives on angiogenic processes was revealed. On the one hand sulfated HA (sHA) interfered with the TIMP-3-mediated inhibition of VEGF-A mediated signaling. On the other hand, sHA-containing HA/collagen-based hydrogels were found to directly stimulate the proliferation of a porcine EC line25. However, these findings were limited to selected HA derivatives restricting a detailed and comprehensive understanding of the potential dual action of sulfated HA on angiogenic processes. Against this background, in the present study, the interactions between VEGF165 or its HBD domain and a broad range of HA and chondroitin sulfate (CS) derivatives with defined sulfation degrees and patterns were analyzed in comparison to native GAG using surface plasmon resonance (SPR) and computer-based molecular modeling techniques. Furthermore, the consequences of these interactions on VEGF165/VEGFR-2 complexation and the biological function of VEGF165 were evaluated in silico up to the atomic detail and in 2D in vitro cell culture experiments using human umbilical vein endothelial cells (HUVEC). The impact of different GAG was verified in a 3D in vitro angiogenesis assay by analyzing sprouting of HUVECs in the absence and presence of VEGF165. This allows for an in-depth understanding of the dual GAG activities and reveals whether it translates into a pro- or anti-angiogenic effect on ECs in a complex system.
Materials and Methods
Materials
Hyaluronan (HA) (from Streptococcus, MW = 1.1 × 106 g mol−1) was obtained from Aqua Biochem (Dessau, Germany). Sulfur trioxide/dimethylformamide complex (SO3–DMF, purum, 97%, active SO3 48%) as well as sulfur trioxide/pyridine complex (SO3–pyridine, pract.; 45% SO3) were acquired from Fluka Chemie, (Buchs, Switzerland). Hep extracted from porcine intestinal mucosa and the specific VEGFR-2 inhibitor SU1498 were available from Sigma-Aldrich (Schnelldorf, Germany). Hep hexasaccharide (dp 6) was obtained from Iduron (Manchester, UK). Recombinant human VEGF165 (293-VE-010/CF) and neutralizing VEGFR-2 antibody (MAB3572-100) were obtained from R&D Systems (Wiesbaden-Nordenstadt, Germany). For SPR measurements, the Series S Sensor Chips C1, CM5 and CM3, the Amine Coupling Kit and HBS-EP (10x) from GE Healthcare Europe GmbH (Freiburg, Germany) were used. The VEGF165 HBD was purified as previously described26.
Preparation of polymeric and oligomeric GAG derivatives
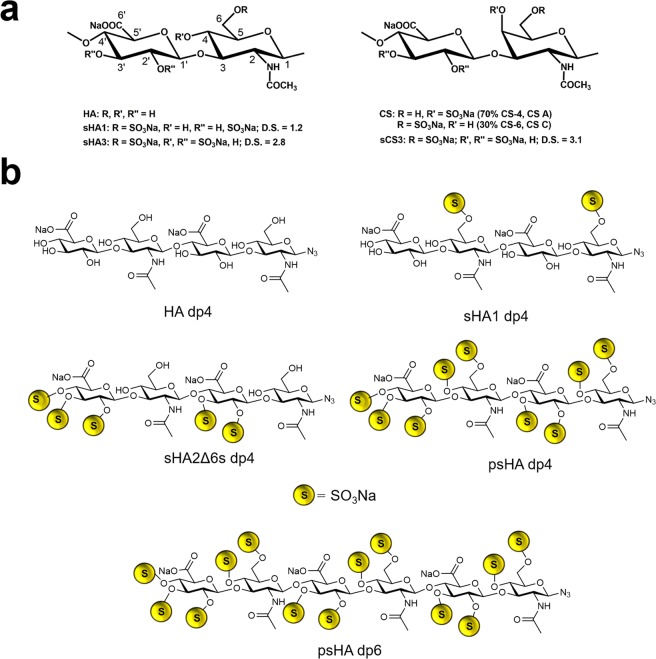
The polymeric HA and CS derivatives were synthesized and characterized according to previous protocols27–29. Analytical data of the used polymeric GAG derivatives (Fig. 1a) are summarized in Table 1. Preparation and characterization of oligomeric HA derivatives (Fig. 1b) was performed as previously described30–32.
Figure 1.
Structural characteristics of polymeric and oligomeric GAG.
Table 1.
Characteristics of polymeric GAG derivatives.
| Sample | HA | sHA1 | sHA3 | CS | sCS3 | Hep |
|---|---|---|---|---|---|---|
| D.S.a | — | 1.2 | 2.8 | 0.8 | 3.1 | 2.2 |
| Mw [g/mol]b | 48 300 | 27 300 | 28 700 | 20 700 | 19 900 | 18 000 |
| Sulfate group distributionc | — | 6, 2′, 3′ |
462′, 463′, 62′3′ |
4, 6 |
462′, 463′, 62′3′ |
2′6, 2N, 6 |
aDegree of sulfation (D.S.) as determined by elemental analysis, bweight-average (Mw) values as revealed with laser light scattering (LLS) detection and cthe sulfate group distribution as detected by nuclear magnetic resonance.
Surface plasmon resonance analyses
Immobilization of VEGF165, HBD of VEGF165 and VEGFR-2 to sensor chip surfaces
For SPR analysis, a BIACORE T100 instrument (GE Healthcare) was used. VEGF165 was immobilized to Series S Sensor Chip CM3 at 25 °C in HBS-EP running buffer (0.01 M HEPES, (pH 7.4), 0.15 M NaCl, 3 mM EDTA, 0.055 surfactant P20) using the amine coupling reaction according to the manufacturer’s protocol (GE Healthcare). 5–10 µg/ml growth factors diluted in sodium acetate buffer, pH 5.5 were injected at 5 µl/min until an immobilization level of approximately 1200 RU was achieved33 A surface without immobilizing one of the growth factors was used as a reference. The HBD of VEGF165 (VEGF165-HBD) was covalently bound to a Series S Sensor Chip CM5 at 25 °C in HBS-EP by amine coupling. Therefore, 30 µg/ml HBD dissolved in sodium acetate puffer (pH 5.5) was streamed for 45 s over an activated sensor chip surface (flow: 10 µl/min) resulting in an immobilization level of about 900 RU. 50 µg/ml VEGFR-2 dissolved in sodium acetate buffer (pH 4.5) was immobilized to a Series S Sensor Chip C1 at 5 µl/min to an immobilization level of approximately 220 RU.
SPR analysis of GAG binding to immobilized growth factors
Interaction studies were performed as described previously33. Briefly, GAG (diluted in HBS-EP) were injected for 300 s at 30 µl/min (at 37 °C in HBS-EP) and binding levels were recorded 10 s before injection stop. The injection was followed by a 10 min dissociation phase in running buffer at a flow rate of 30 µl/min. The sensor chip surface was regenerated after each sample injection with 5 M NaCl in 40 mM NaOH or 5 M NaCl in 10 mM NaOH in case of the VEGF165 HBD for 60 s at a flow rate of 30 µl/min. The baseline was allowed to stabilize for 1000 s with running buffer prior to injection of the next sample. Binding parameters were evaluated using the BIACORE T100 evaluation software 2.03. Data were double referenced by the response of the reference surface and the response of HBS-EP buffer alone relative to a baseline report point. Furthermore, binding levels were corrected for the respective molecular weight of the GAG derivatives to consider the fact that the SPR binding response is related to a mass increase at the sensor chip surface.
Isothermal titration calorimetry
Titrations of 60 µM psHA dp4 and 50 µM psHA dp6 to 15 µM VEGF165-HBD were carried out using a Microcal PEAQ-ITC calorimeter (Microcal, Malvern Panalytical, Malvern, GB) at 25 °C. VEGF165-HBD was dialyzed against 8 mM phosphate buffer (8 mM NaH2PO4∙2H2O, 2 mM KCl, pH 7.5) without DTT with 10 wt-% DMSO to reduce noise before measurements. To evaluate the influence of the salt concentration on binding of sulfated oligohyaluronans to VEGF165-HBD, additional measurements were performed in phosphate buffered saline, PBS (10 mM NaH2PO4∙2H2O, 2 mM KH2PO4, 3 mM KCl, 140 mM NaCl, pH 7.4) without DTT with 10 wt-% DMSO. The synthesized oligosaccharides psHA dp4 and psHA dp6 were dissolved in the same buffer to ensure a reasonable baseline. For each titration 19 to 25 injections of 1.5 and 1 µl of titrant, respectively, were conducted at 180 s intervals, while stirring at 750 rpm. Three different control experiments were performed: buffer in buffer titration, a 60 µM solution of psHA dp4 titrated to buffer and buffer titrated to a 15 µM VEGF165-HBD solution. Raw data were integrated, normalized and the titration curve fitted using Microcal PEAQ-ITC analysis software provided by Microcal. Binding curves were fitted to the single site model.
Molecular modeling and molecular dynamics simulation
Modeling of VEGF165 dimer and VEGF165/VEGFR-2 ternary complex
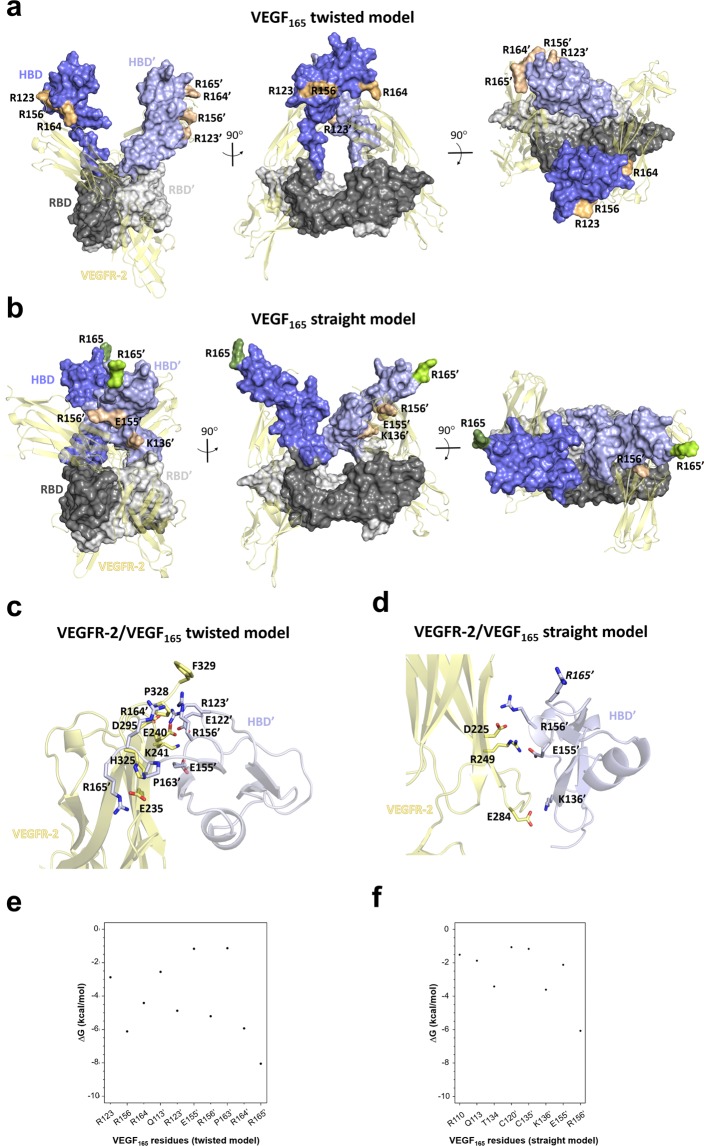
The full three-dimensional (3D) VEGF165 dimer structure was built taking into account the following experimentally available 3D crystal structures from the Brookhaven protein data bank (PDB) as templates: (i) the high-resolution crystal structures of the N-terminal region of VEGF corresponding to its receptor binding domain (VEGF-RBD), which is a dimer structure (PDB ID 2VPF, 1.9 Å)34, (ii) the NMR structure of the C-terminal region of VEGF corresponding to its heparin binding domain (VEGF-HBD; PDB ID 1VGH)35, and (iii) the structure of the complex of VEGFR-2 with the VEGF receptor binding domain (RBD) (complex 1:1, PDB ID 3V2A, 3.2 Å)36. Unfortunately, the full structure of VEGF165 dimer as such is not experimentally available. The amino acid Arg110 acting as a link between the N- and C- terminal domains is not resolved in any of the available experimentally solved structures. Therefore, in order to model the 3D spatial disposition of VEGF’s N- and C-terminal domains (i.e. RBD and HBD, respectively) with respect to each other in the dimer form of VEGF and with respect to the VEGFR-2, we did the following. First of all, based on literature about the stoichiometry of the VEFG-VEGFR-2 signaling complex from which it is known that the dimer VEGF signals through complexing with 2 receptor molecules13, we generated the complete structure of the dimer VEGF-RBD/VEGFR-2 complex (1:2) by applying the corresponding symmetry operations in PDB ID 3V2A. In the resulting structure, we modeled the missing residue Arg110 in extended conformation at the C-term on one of the VEGF-RBD (RBD-Arg110). We connected RBD-Arg110 to HBD-Ala111 in each of the 20 structures comprised in the HBD NMR ensemble (PDB ID 1VGH), and we examined the resulting configurations of the HBD domain with respect to the two receptor molecules and the two RBD by taking into consideration available information on the potential role of VEGF165-HBDs in the regulation of VEGFR-2 phosphorylation suggesting that they could also interact with the receptor17. This analysis allowed us to be able to exclude those configurations of HBD presenting steric clashes either with the receptor or with the RBD molecules. The best resulting model fulfilling the above-mentioned requirements resulted from the structure number 4 from the HBD NMR ensemble. As a next step, we performed a Arg110-Ala111 ϕ and φ dihedral scan and looked for interfacial electrostatics and shape compatibility of HBD with the receptor molecules and avoiding any steric hindrances. This resulted in two representative models of VEGF165 monomer consisting of two well-differentiated orientations of HBD with respect to VEGFR-2: i) HBD interacting with the close by VEGFR-2 and ii) HBD interacting with the farther VEGFR-2 (Fig. S1). In order to finally build the VEGF165 dimer complexed with two receptor molecules, we applied a twofold symmetry operation to the previously modeled VEGF165 monomer based on the symmetry exhibited by the VEGF-RBD/VEGFR-2 crystallographic structure (PDB ID 3V2A). The intermolecular disulfide bonds between the VEGF165-RBD monomers were correspondingly modeled. Next, and because receptor residues Ser264 to Lys271 and Lys278 to Gly282 are not resolved in the experimentally available X-ray VEGFR-2 structure, in order to complete the full complex structure those missing residues were modeled taking as template the VEGFR-2 structure in complex with VEGF-C (PDB ID 2X1W, 2.7 Å)37. As a last step, the two VEGF models were energetically refined in complex with the two receptor molecules by applying molecular dynamics (MD). Modeller as implemented in Discovery Studio (Accelrys Software) was used for the molecular modeling38, and AMBER14 was used for the MD simulations (40 ns; for details see MD simulations section)39. After refinement of the two models, it was observed that the HBD were disposing themselves with respect to the receptor molecules in two distinguishable arrangements: one, which we named “twisted” in which the RBD of one VEGF165 monomer and the HBD of the other VEGF165 monomer interact with the same VEGFR-2 molecule, and another one, which we named “straight” allowing the interaction of the HBD and RBD of a VEGF165 monomer with the same VEGFR-2 molecule (see Fig. 2 for details). For each model (twisted and straight), the lowest energy structure was selected for further docking studies with the GAG derivatives.
Figure 2.
Molecular modeling of the 3D structure of VEGF165 dimer and energy contribution analysis. Two distinct conformations of the VEGF165 dimer in complex with VEGFR-2 refined by MD simulations are obtained: “twisted” (a) and “straight” (b). VEGF165 models (twisted and straight) and VEGFR-2 are depicted in molecular surface and cartoon representation, respectively. The RBD is shown in gray (dark and light representing each monomer), the HBD in blue (dark and light representing each monomer) and VEGFR-2 in yellow. Charged residues at the HBD of VEGF165 interacting with VEGFR-2 are highlighted in pale (dark and light for each monomer) and numbered (VEGF165 monomers are distinguished by a comma). Residue R165, which is essential for NRP-1 recognition, is not interacting in the “straight” model and is shown in green (dark and light for each monomer). Close-up of the MD refined complex of VEGFR-2 with VEGF165-HBD twisted (c) and straight (d) models with relevant interacting residues in sticks, colored by atom type and labeled. The no interacting residue R165 in the VEGF165 straight model is labeled in italic. Per-residue energy analysis (calculated with MM-GBSA from MD simulations) of most contributing residues of VEGF165-HBD twisted (e) and straight (f) models in binding to VEGFR-2.
Modeling of GAG derivatives
The following GAG derivatives were modeled in AMBER1439 and MOE40 as previously described32,41: Hyaluronan (HA), sulfated hyaluronan (sHA1, sulfated either at position C4 or C6 of the disaccharide unit), high-sulfated hyaluronan (sHA3, sulfated at positions C4, C6 and C3′ in each disaccharide unit), chondroitin sulfate (CS, sulfated either at position C4 or C6 of the disaccharide unit), high-sulfated chondroitin sulfate (sCS3, sulfated at positions C4, C6 and C3′ in each disaccharide unit), tetrameric (dp4) hyaluronan azide derivatives HA, sHA1, sHA2Δ6s, psHA and psHA dp6 (Fig. 1). Based on previous work42, the hexamer GAG length (dp6) was considered as representative of polymeric GAG for our docking studies.
Molecular docking
Computer-based binding was performed by docking calculations32,41 of the GAG derivatives with each of the VEGF165 dimer refined models and with the respective receptor VEGFR-2. For this, we used Autodock 343. Autogrid3 was used to calculate the atomic potential of each structure covering the full surface with a grid box and spacing grid of 126 Å × 126 Å × 126 Å and 0.710 Å for VEGF165 and 126 Å × 126 Å × 126 Å and 0.580 Å for VEGFR-2. The GAG molecules were treated completely flexible, and the previously refined protein structures were considered rigid. The Lamarckian genetic algorithm with an initial population size of 300 and a termination condition of 10000 generations and 9995 × 105 energy evaluations was used. A total of 1000 independent runs were carried out. Spatial clustering of the top 50 docking solutions was performed with the DBSCAN algorithm44 as previously described45. From each of the clusters obtained, a representative GAG pose was selected for further refinement of the corresponding GAG-protein complex.
Molecular dynamics simulations
The GAG-protein complexes selected as representative from the docking studies were further refined by MD simulations in AMBER1439 as previously described32,41. Charges were taken from the GLYCAM 06-j force field46 for the different sulfated hyaluronan units and from the literature for sulfate groups47. AMM1-BCC charges were used for the azide group48. Parameters for the GAG part were taken from the GLYCAM-06j force field46, and for the proteins from the ff14SB force field39. Missing parameters of the azide group were taken from the General Amber Force Field (GAFF)49. Each GAG-protein complex was solvated in a truncated octahedral box of TIP3P water molecules and neutralized with Na+ or Cl− counterions. MD simulations were preceded by two energy-minimization steps: (i) only the solvent and ions were relaxed with position restraints for the solute (500 kcal/mol·Å2) using 1000 steps of steepest descendent minimization followed by 500 steps of conjugate gradient minimization; (ii) the entire system was minimized without restraints applying 3000 cycles of steepest descendent and 3000 steps of conjugate gradient equilibration. Then the system was heated up from 200 K to 300 K in 20 ps with weak position restraints (10 kcal/mol·Å2). Langevin temperature coupling with a collision frequency γ = 1 ps−1 was used at this step. The system was equilibrated under constant pressure of 1 atm using periodic boundary conditions (NPT conditions) at 300 K for 50 ps. A total of 20 ns MD simulation was carried out at 300 K NPT conditions for each complex. The SHAKE algorithm was used to constrain all bonds involving hydrogen atoms. A time step of 2 fs was used during SHAKE algorithm. A cutoff of 8 Å was applied to treat the non-bonded interactions, and the Particle Mesh Ewald (PME) method was used to treat long-range electrostatic interactions. MD trajectories were recorded every 10 ps. The pyranose rings in the GAG molecules were harmonically restrained. In the case of the complexes between VEGF165 and the oligohyaluronan derivatives, a 40 ns MD simulation was carried out. Trajectories were visualized with VMD50 and evaluated in terms of intermolecular H-bonds by using the CPPTRAJ module implemented in AMBER. At least 10% of hydrogen bond occupancy was taken as criterion for dynamic hydrogen bond formation. Energy decomposition per residue as well as binding free energy post-processing analysis of the last 300 frames from the MD simulations were performed in implicit solvent using the MM-GBSA method51,52 as implemented in AMBER14. Data analysis was carried out with the R-package53. Figures were created with PyMOL54.
Cell Culture
Cultivation of HUVECs
HUVECs (VEGF pre-screened; PromoCell, Heidelberg, Germany) were maintained in EC medium (PromoCell Endothelial Basal Medium, EBM) supplemented with 2% (v/v) fetal calf serum (FCS), 1% penicillin/streptomycin (Pen/Strep), 5 ng/ml rhEGF, 10 ng/ml rhbFGF, 20 ng/ml R3 IGF-1, 0.5 ng/ml rhVEGF, 1 µg/ml ascorbic acid, 22.5 µg/ml heparin and 0.2 µg/ml hydrocortisone at 37 °C in a humidified atmosphere with 5% CO2. Cells were grown to 80% confluence before splitting or stimulation. Passages of HUVECs used for experiments were not higher than four.
Phosphorylation of VEGFR-2
Phosphorylation of VEGFR-2 was determined as described previously33. HUVECs were seeded in 6-well plates at a density of 10,000 cells/cm2 in complete EC medium for 24 h at 37 °C. Prior to stimulation HUVECs were serum-starved for 24 h in EBM containing 0.1% BSA, 0.2% FCS and 1% Pen/Strep. Cells were treated for 10 min with 2.6 nM VEGF165 and 200 µM D.U. GAG, pre-incubated for 10 min at 37 °C. Activation of VEGFR-2 in HUVEC cells stimulated with VEGF165/GAG complexes was determined using two sandwich ELISA kits (Biotechne) detecting phosphorylated VEGFR-2 (P-VEGFR-2) and total VEGFR-2, respectively, according to the manufacturer’s protocol. As a control, VEGFR-2 phosphorylation was blocked by 100 nM inhibitor SU1498.
3D in vitro Angiogenesis Assay
The protocol used was adapted from PromoCell (Heidelberg, Germany) and Korff et al. (Korff T et al., Exp Cell Res 297:415–23, 2004). The modified angiogenesis assay was performed as described in33. HUVECs were trypsinized and resuspended in EBM containing 2% FCS and 10% (v/v) methocel (Sigma-Aldrich) stock solution. 1 × 103 cells/well were seeded in 96-well round-bottom well plates to generate spheroids within 18–24 h at 37 °C, 5% CO2. Spheroids were harvested (5 min, 1000 × g, RT) and embedded in gels of 3 mg/ml collagen type I and 1x M199 cell culture medium (Sigma-Aldrich), adjusted to a neutral pH using 0.2 M NaOH. Here, 600 µl of the pH-adjusted collagen I-M199 solution were added to 100 spheroids in methocel-FCS medium (1:1) and mixed carefully. 400 µl/well of the methocel-collagen solution containing the spheroids was added to 48-well plate and incubated at 37 °C for 30 min. Each well contained 50 spheroids, which were stimulated afterwards according to the respective experiment using EBM containing 0.5% FCS, 1% P/S, 25 ng/ml VEGF and 100 µM D.U. GAG. Growth factor and GAG were incubated for 10 min at 37 °C to form complexes and then added to the samples. For neutralization experiments 10 µg/ml anti-VEGFR-2 antibody (VEGFR-2 Ab) were used. Spheroids were treated with 100 µl stimulating solution per well for 24 h. All components (growth factor, GAG, neutralizing antibody) were added as 5x stock of the final concentration according to the final volume of 500 µl in the wells. 10 spheroids per condition were randomly selected and evaluated regarding the individual sprout length, the calculation of the cumulative sprout length and the number of sprouts analyzed using ImageJ.
Statistical analysis
All experiments were performed at least in triplicate, and results are presented as means ± standard deviation (SD). One-way ANOVA or two-way ANOVA with Tukey or Bonferroni post-test, respectively, were applied. P values < 0.05 were considered statistically significant.
Results
Molecular models of VEGF165 and VEGF165/VEGFR-2
The complete structure of the VEGF165 dimer has not been yet experimentally resolved. The structures of the VEGF165-RBD at the N-terminus and its VEGF165-HBD at the C-terminus are available separately in the Protein Data Bank (PDB), but no full VEGF165 structure is available containing both domains connected. In order to build the VEGF165 dimer structure, we have first made use of relevant biological information referred to the fact that the signaling complex requires the interaction of one VEGF165 dimer with two VEGFR-2 molecules (see Methods section for details) and, furthermore, that the VEGF165-HBDs could be implicated in the regulation of VEGFR-2 phosphorylation17 and, therefore, they could also interact with the receptor. Thus, the experimentally not resolved connecting residue Arg110 was modeled by attaching the HBD and the RBD of one monomer crystallographic structure taking into account the criteria of avoiding steric clashes (see section 2.5.1 for details). After performing a Phi and Psi dihedral angle scan on the linked residues Arg110-Ala111, two representative and distinguishable orientations with respect to the VEGFR-2 molecules were selected. The application of a twofold symmetry operation as observed in the RBD structure to the resulting VEGF165 monomer structure along the y-axis (see Fig. S1) led two VEGF165 dimer models that were further MD refined in complex with two VEGFR-2 molecules (see section 2.5.1 for details). Each VEGF165 dimer conformation was named according to the orientation of the HBDs and the VEGFR-2 molecules. The first one consists of a “twisted” conformation, in which the RBD of one VEGF165 monomer and the HBD of the second monomer interact with the same VEGFR-2 monomer molecule. The second one shows a “straight” conformation, which allows the interaction of the HBD and RBD of the same VEGF165 monomer with the same VEGFR-2 monomer molecule (Fig. 2).
Binding of the VEGF165 dimer to two VEGFR-2 molecules was more favorable for the twisted than for the straight model (ΔGVEGF165twisted/VEGFR-2 = −217.6 ± 16.3, ΔGVEGF165straight/VEGFR-2 = −149.7 ± 9.7). In the twisted model, residues Arg123, Arg156, Arg164 and Arg165 belonging to the HBD domains are involved in the recognition of VEGFR-2, whereas in the case of the straight model, those correspond to residues Lys136, Glu155 and Arg156. Thus, Arg156 constitutes a common recognition residue in both models (Fig. 2). Both VEGF165 conformations could potentially co-exist in their receptor-free state. However, taking into account previous studies on the interaction of VEGF with other molecules such as the NRP-1 co-receptors10,55, the VEGF165 straight model would be the most biologically relevant one as it would allow simultaneous interactions with VEGF receptors and co-receptors (Fig. 2).
SPR analysis of GAG binding to VEGF165 and molecular recognition of GAG by VEGF165
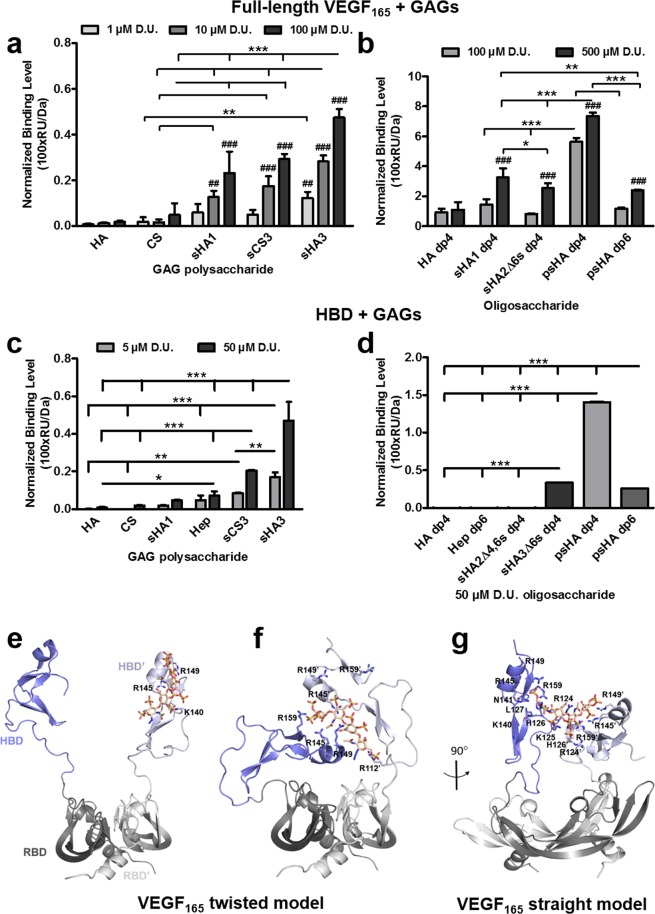
The interaction of solute GAG derivatives with immobilized VEGF165 was analyzed by SPR. While HA showed only a weak interaction with VEGF165, a higher sulfation and concentration of the polymeric GAG derivatives led to stronger binding, with sHA3 showing the highest binding strength (Fig. 3a). The ranking of binding strength for the polymeric GAG was as follows: HA ≤ CS < sHA1 < sCS3 < sHA3. Interestingly, sHA derivatives revealed higher binding compared to CS derivatives with a different sugar backbone but a comparable DS. The ranking of binding strength for the oligomeric derivatives to VEGF165 revealed the following: HA dp4 < sHA2Δ6s dp4, psHA dp6 < sHA1 dp4 < psHA dp4 (Fig. 3b), highlighting the importance of sulfation at the C6 position of the N-acetylglucosamine (GlcNAc) unit as well as the polymerization degree. Additional SPR measurements (Fig. 3c,d) using the VEGF165-HBD clearly demonstrated the direct binding of GAG derivatives to the HBD and support the GAG rankings for full-length VEGF165. It is of note that, while polymeric Hep bound to HBD, no binding could be detected for Hep dp6.
Figure 3.
Interaction of immobilized VEGF165 and HBD with solute GAG derivatives as determined by SPR. Binding levels for the interaction of (a) polymeric and (b) oligomeric GAG to VEGF165 and HBD (c,d) are displayed [adapted and modified from33]. All values represent the mean ± SD of n = 3 and are given as relative to baseline response and corrected for the respective molecular weight of GAG derivatives. Two-way ANOVA: *p < 0.05; **p < 0.01; ***p < 0.001 vs. respective treatment; ##p < 0.01; ###p < 0.001 vs. (a) HA or (b) HA dp4. Predicted recognition of psHA dp4 by VEGF165 dimer. (e) Initial complex VEGF165 twisted/psHA dp4. (f) Refined VEGF165 twisted/psHA dp4 (40 ns). (g) Refined VEGF165 straight/psHA dp4 (40 ns). VEGF165 is depicted in cartoon style, and relevant interacting residues are shown in sticks and colored by atom type. The RBD is shown in gray (dark and light representing each monomer), and the HBD is shown in blue (dark and light representing each monomer). psHA dp4 is shown in stick representation and colored by atom type.
Molecular docking was used to predict GAG recognition by the VEGF165 dimer. The resulting GAG-protein complexes were energy refined by MD. For VEGF165, the twisted and straight models resulting from the MD refinement in the presence of VEGFR-2 were used. For both VEGF165 conformations, all studied GAG were predicted to bind to the VEGF165-HBD, which is further supported by experimental studies using the HBD and GAG derivatives (Fig. 3c,d). In the case of the twisted model, GAG were predicted to bind in two different modes: parallel and/or perpendicular orientation with respect to the HBDs. For the straight model, two distinct binding modes were observed depending on the GAG type. A parallel binding mode to HBD was predicted for HA (both, polymeric and oligomer), sHA1 and sHA2Δ6s. In case of all sulfated polymeric GAG derivatives and persulfated oligohyaluronan derivatives, a parallel-curved binding mode was observed in which the GAG was bridging the two HBDs (Fig. S2a–e). This could be feasible considering that the separation between both HBDs is in the same range as the GAG dp6 lengths (ca. 30 Å).
Independently of the protein conformation, we observed that all studied sulfated polymers initially bound to one VEGF165-HBD were acting as a kind of “molecular glue” originating molecular recognition modes in which the GAG molecule sticks on the two VEGF165-HBDs along the MD energy refinement. This HBD-GAG-HBD stacking configuration appeared around the first 10 ns and was preserved through the rest of the MD simulation. For most of the tetrameric oligohyaluronan derivatives, a comparable stacking configuration was observed at about 20 ns. Therefore, to further explore and fully refine the obtained stacking configuration, for the oligohyaluronan derivatives the MD simulation was elongated to a total of 40 ns, in which such configuration remained stable (Fig. S3). In the case of the non-sulfated HAs the stacking configuration was not observed.
The binding energies computed with MM-GBSA51,52 for the polymeric GAG indicated that binding to VEGF165 was, in general, more favorable for the straight model. Noticeably, independently of the VEGF165 model used for our calculations, the binding obtained increased with the DS of the GAG (Table 2), which is in agreement with our experimental observations (Fig. 3a). In the case of the VEGF165 twisted model, binding was more favorable for HA than for CS derivatives of the same DS. Moreover, binding was more favorable for sHA1 with GlcNAc sulfated at C6 position than for the other investigated monosulfated GAG derivatives, and similar than sCS3 (Table 2, Fig. S2f). In contrast, in the straight model the predicted binding was similar for mono- as well as high-sulfated HA and CS. The binding energies obtained for the oligohyaluronans in the VEGF165 twisted model resulted in the following ranking: HA dp4 < sHA2Δ6s dp4 < sHA1 dp4 < psHA dp4 < psHA dp6 (Table 2) and, as observed for the polymeric analogues, GlcNAc of sHA1 showed more favorable binding energies than those from sHA2Δ6s (Fig. S2g). Interestingly, the binding energy ranking obtained for the straight model agreed well with the binding data obtained from the SPR experiments. The psHA dp4 showed the most favorable binding energy to the VEGF165 straight model among all oligohyaluronan azides (Table 2).
Table 2.
MM-GBSA binding free energies obtained for GAG in complex with VEGF165a.
| GAG | ΔGGAG-VEGF165twisted (kcal/mol) | ΔGGAG-VEGF165straight (kcal/mol) |
|---|---|---|
| HA dp4 | −24.9 ± 3.3 | −16.7 ± 3.4 |
| HA dp6 | −13.4 ± 6.1 | −38.8 ± 5.8 |
| sHA1 dp4 | −61.5 ± 6.3 | −55.5 ± 5.8 |
| sHA1 dp6 (sulfated at C4) | −35.8 ± 4.5 | −59.2 ± 7.4 |
| sHA1 dp6 (sulfated at C6) | −66.8 ± 8.3 | −55.3 ± 9.5 |
| sHA2Δ6s dp4 | −50.0 ± 10.6 | −60.7 ± 10.3 |
| sHA3 dp6 (sulfated at C4,C6,C3′) | −72.9 ± 12.7 | −106.7 ± 10.8 |
| CS4 dp6 (sulfated at C4) | −34.0 ± 4.8 | −65.2 ± 8.8 |
| CS6 dp6 (sulfated at C6) | −46.4 ± 7.3 | −52.6 ± 8.0 |
| sCS3 dp6 (sulfated at C4,C6,C3′) | −67.6 ± 9.9 | −108.6 ± 9.5 |
| psHA dp4 | −82.0 ± 9.0 | −153.7 ± 9.5 |
| psHA dp6 | −111.3 ± 7.6 | −121.9 ± 11.3 |
aShown for the energetically most favored binding modes.
The detailed analysis of the interactions established in the refined VEGF165 straight model indicated that the HBD-GAG-HBD stacking complex was stabilized by favorable contacts between psHA dp4 and residues Arg123, Arg124, Lys125, His126, Leu127, Lys140, Arg145, Arg149 and Arg159 of both HBDs, and Asn141 for one of the HBDs. Furthermore, the MD-based per-residue energy calculations showed a major number of favorable interacting residues of VEGF165 with psHA dp4 in comparison to psHA dp6. In particular, interactions of Arg124, His126, Leu127 and Arg159 with psHA dp4 were more favorable than with psHA dp6, while psHA dp6 led to interactions that were more favorable with Lys140 and, in lesser extent, with Lys125 in comparison to the tetramer analogue (Fig. S4). In the twisted model, the HBD-GAG-HBD stacking structure obtained for psHA dp4 exhibited a lower number of interactions, which involved some of the residues observed in the straight model (Arg145, Arg149 and Arg159 for both VEGF165 HBDs, and Arg110, Ala111 and Arg112 for one HBD) (Fig. 3f,g). The differences in binding energies obtained for psHA dp6 and dp4 with VEGF165 twisted model were due to the more favorable interactions of Lys140, Lys147, Lys162 and Arg164 with the hexamer analogue in comparison to the tetramer derivative, in which the major contributors to the binding energies were Arg110, Ala111, Arg112 and Arg145 (Fig. S4). In general, independently of the VEGF165 model, residues Arg145, Arg149 and Arg159 appear to be crucial and common for the recognition of all investigated sGAG (Fig. S4). Interestingly, previous VEGF165 mutagenesis studies indicated that simultaneous mutations involving Arg123, Arg124, Lys140, Arg145, Arg156 and Arg159 could almost abolish Hep binding to the protein56,57, which supports our molecular models. Likewise, the straight model was the one matching maximum number of significant mutagenesis interactions (Fig. S4). In conclusion, our findings suggest that GAG recognition by VEGF165 occurs through a HBD-GAG-HBD stacking complex involving both HBDs. Although in the absence of receptor the twisted and straight conformations of VEGF165 may be possible and both support a stacking conformation when recognizing a GAG molecule, in the case of the signaling complex (VEGF165 dimer and the VEGFR-2 dimer), and based on our models and the experimental indications on NRP-1 recognition10,55–57 as well as previous mutagenesis studies, the straight conformation appears to be the one being functionally relevant.
Isothermal titration calorimetry (ITC) was performed to validate the stoichiometry of the predicted HBD-GAG-HBD stacking conformation. The titration of a psHA dp4 and psHA dp6 to VEGF165-HBD in independent measurements (Figs. S5, S6 and S7) indicated a mean binding stoichiometry of N = 0.503 and N = 0.545 respectively, corresponding to one oligohyaluronan binding two VEGF165-HBDs. Binding of the psHA dp4 was observed to be driven by a negative binding enthalpy of −5 kJ mol-1 and by a strong positive entropic contribution of +32 kJ mol-1 (for TΔS) resulting in the dissociation constant KD of 364 ± 48 nM (Table 3, Fig. S6). The mean binding stoichiometry and the dissociation constant of sulfated oligohyaluronans binding to VEGF165-HBD were unaffected by the salt content of the buffer (ITC titrations done in phosphate buffer: N = 0.493, KD = 378 ± 21 nM, Fig. S5; ITC titrations done in PBS buffer: N = 0.503, KD = 364 ± 48 nM, Fig. S6). The higher sulfated, oligomeric hexahyaluronan psHA dp6 revealed three times stronger binding compared to psHA dp4 (psHA dp4: KD = 364 ± 48 nM, psHA dp6, KD = 124 ± 29 nM, Fig. S7) in agreement with the predicted ranking from our MM-GBSA calculations for the proposed VEGF165 twisted model (Table 2). The differences in binding between the tetra- and hexahyaluronan derivatives obtained through SPR and ITC could be explained considering the fact that ITC experiments were performed in solution, allowing the simultaneous complex formation of two HBD molecules with one GAG, which may be the reason for higher affinity for dp6 compared to dp4. In contrast, SPR experiments were performed with an immobilized HBD, which does not allow the formation of equivalent complexes that are possible in the ITC experiments. Furthermore, our theoretical models predicted a more favorable binding energy for psHA dp6 when considering the VEGF165 twisted model in comparison to the straight (Table 2). It should be considered that in our theoretical models two HBDs can simultaneously interact with the GAG, but they have some conformational restriction because of their covalent attachment to the RBDs (in ITC the HBD have more conformational freedom due to the fact that they do not have any kind of immobilization). The observed differences in binding could simply be a consequence of the accessibility of the HBD in different conditions.
Table 3.
Results of the ITC measurements for VEGF165-HBD and psHA dp4.
| compound | caprotein | caligand | Nb | ΔGc | ΔHd | TΔSe | KDf | (KD)g |
|---|---|---|---|---|---|---|---|---|
| psHA dp4h | 15 | 60 | 0.505 | −36.8 | −17.3 | +19.5 | 357 ± 21 | 378 ± 21 |
| 0.481 | −36.5 | −17.1 | +19.4 | 398 ± 10 | ||||
| psHA dp4i | 15 | 60 | 0.523 | −36.4 | −3.7 | +32.7 | 431 ± 245 | 364 ± 48 |
| 0.557 | −37.0 | −7.2 | +29.7 | 341 ± 73 | ||||
| 0.429 | −37.1 | −4.6 | +32.5 | 320 ± 200 | ||||
| psHA dp6i | 15 | 50 | 0.628 | −39.0 | −4.8 | +34.2 | 149 ± 80 | 124 ± 29 |
| 0.409 | −40.5 | −5.7 | +34.8 | 83 ± 30 | ||||
| 0.599 | −39.2 | −5.0 | +34.2 | 140 ± 65 |
aConcentration in µM, bmolar binding ratio of the ligand-protein interaction (stoichiometry), cbinding free energy in kJ∙mol−1, dbinding enthalpy in kJ∙mol−1, dentropic contribution term in kJ∙mol−1, fdissociation constant in nM, gmean of the calculated KD values, hITC measurements were performed in 8 mM phosphate buffer with 2 mM NaCl, iITC measurement were performed in 10 mM PBS buffer with 140 mM NaCl.
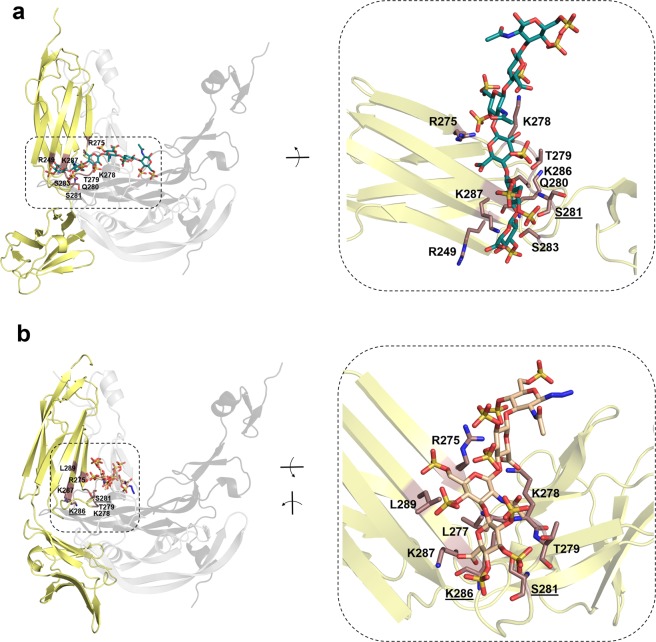
To investigate the effect of GAG binding to VEGF165 on the recognition of its receptor VEGFR-2, the VEGF165/VEGFR-2 interface was analyzed and compared to the GAG/VEGF165 recognition sites (Figs. 2 and S4). In case of the VEGF165 twisted model, residue Arg164, which is involved in binding of sGAG, is also engaged in VEGFR-2 recognition. In contrast, in the VEGF165 straight model GAG and VEGFR-2 do not compete for the same recognition site. Nevertheless, taking into account that sGAG promote HBD-GAG-HBD stacking complex formation with VEGF165, it could be assumed that the binding strength of the HBD toward VEGFR-2 would decrease with the consequent weakening of VEGF165/VEGFR-2 complex formation. To shed light onto this possible mechanism, the recognition of sHA3 dp6 by the ternary complex (i.e. two VEGFR-2 receptors and a dimeric VEGF165 molecule) was further investigated computationally. A HBD-GAG-HBD stacking configuration between VEGF165/sHA3 was also obtained in the presence of the two receptors. Indeed, for both conformations, twisted and straight, sHA3 stabilized the complex (ΔGVEGF165twisted-sHA3/VEGFR-2 = −252.6 ± 11.5, ΔGVEGF165straight-sHA3/VEGFR-2 = −172.1 ± 11.1), which resembles previous experimental observations with Hep in a length- and concentration-dependent-manner21,22. Interestingly, some of the interactions between the receptors and the VEGF165-HBDs were lost upon sHA3 binding and formation of the HBD-GAG-HBD stacking configuration (Fig. S8). In particular, Arg156 of VEGF165 constitutes a common missing interactive residue in both VEGF165 conformations, while, Asp63 and Glu64 in the VEGF165-RBD increase their interactions with VEGFR-2. The recognition of GAG by VEGFR-2 was also investigated (Fig. 4, Table S1). Here, high-sulfated HA derivatives did not show strong binding toward the receptor but, nevertheless, they established specific interactions with Ser281 and Lys286, which were also involved in the recognition of VEGF165.
Figure 4.
Molecular modeling of the interaction of VEGFR-2 with sGAG derivatives. Representative snapshots from 20 ns MD simulations of most favorable binding sites and modes of (a) sHA3 dp6 (ΔGVEGFR-2/sHA3 = −22.8 ± 3.2 kcal/mol) and (b) psHA dp4 (ΔGVEGFR-2/psHA dp4 = −43.0 ± 4.4 kcal/mol) in complex with VEGFR-2. VEGFR-2 is depicted in yellow cartoon representation with relevant interacting residues colored brown and labeled. Receptor residues recognizing GAG and also VEGF165 are shown in sticks colored by atom type and are labeled (underlined). For illustrative purposes, VEGF165 (not taken into account for calculations) is shown in grey cartoon transparency (dark and light representing each monomer) in the left panels. GAG derivatives are shown in stick and colored by atom type.
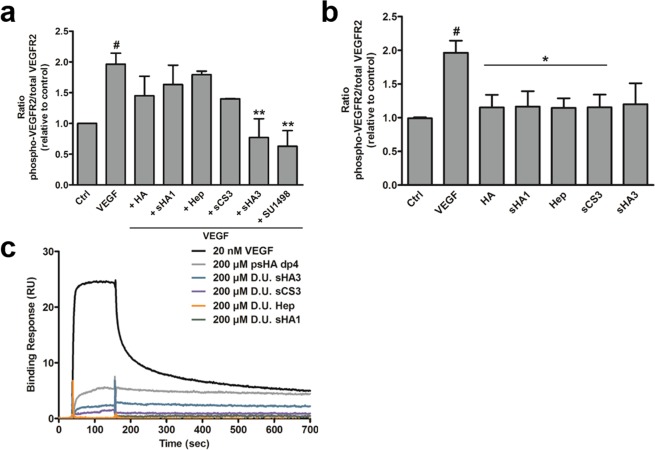
Phosphorylation of VEGFR-2 in the presence of GAG
In order to determine the direct effect of solute GAG on VEGFR-2 activation, the receptor phosphorylation in the absence or presence of VEGF165 was studied via ELISA. The presence of HA, sHA1, sCS3 and Hep had no significant influence on VEGF165-mediated VEGFR-2 phosphorylation in comparison to VEGF165 alone (Fig. 5a). In contrast, sHA3 displayed a significant inhibitory effect on VEGFR-2 phosphorylation comparable to the VEGFR-2 inhibitor SU1498. In the presence of sCS3, VEGFR-2 phosphorylation was reduced as well compared to VEGF165 alone, but not significantly and, therefore, not comparable to SU1498. In the absence of VEGF165, no effect of GAG alone on VEGFR-2 phosphorylation was observed (Fig. 5b). Nevertheless, SPR binding data indicated a weak interaction between sGAG and VEGFR-2 (Fig. 5c).
Figure 5.
Influence of GAG derivatives on VEGF165-mediated phosphorylation of VEGFR-2 (a,b) and direct interaction of solute GAG derivatives with immobilized VEGFR-2 (c). (a) HUVEC cells were stimulated with GAG for 10 min. Cells were lysed afterwards and lysates were applied to a specific phospho- and a total VEGFR-2 sandwich ELISA. The amount of phospho-VEGFR-2 relative to total VEGFR-2 is shown. Values represent the mean ± SD of n = 2. One-way ANOVA: #p < 0.05 vs. Ctrl; *p < 0.05 vs. VEGF165. (b) HUVEC cells were stimulated with VEGF165 alone or pre-formed VEGF/GAG complexes for 10 min. Cell lysates were applied to a specific phospho- and a total VEGFR-2 sandwich ELISA. The amount of phospho-VEGFR-2 relative to total VEGFR-2 is shown. SU1498, a specific VEGFR-2 inhibitor was used as a reference for reduced phosphorylation. Values represent the mean ± SD of n = 3. One-way ANOVA: #p < 0.05 vs Ctrl; **p < 0.01 vs. VEGF165 alone. (c) One representative sensorgram out of three independent measurements is shown [adapted and modified from33].
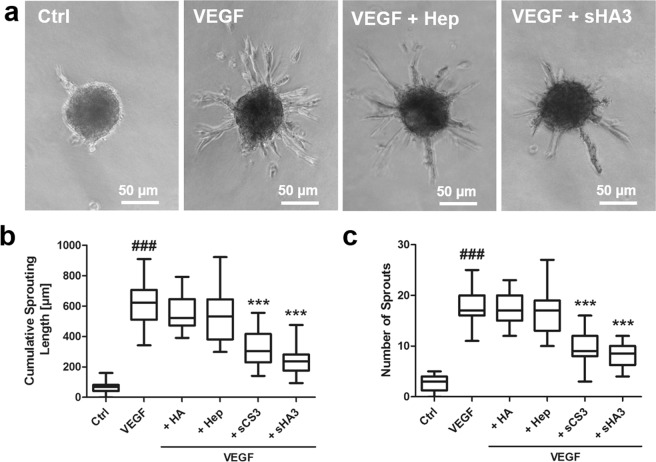
Influence of GAG derivatives on VEGF165-mediated sprouting of HUVEC spheroids
The biological consequences of VEGF165/GAG interaction were further evaluated in a 3D in vitro angiogenesis assay. HUVEC spheroids were treated with growth factor alone or pre-formed growth factor/GAG complexes. Treatment with VEGF165 led to the formation of numerous sprouts compared to the untreated control (Fig. 6). In the presence of HA and Hep no change in sprouting behavior of spheroids compared to VEGF165 treatment was determined. In contrast, sCS3 and sHA3 significantly inhibited VEGF165-mediated spheroid sprouting displayed by a reduced cumulative sprouting length and a reduced number of sprouts (Fig. 6b,c).
Figure 6.
Biological consequences of VEGF165/GAG interaction on VEGF165-mediated sprouting of HUVEC spheroids. 1 × 103 cells were seeded in medium containing 10% methyl cellulose to form spheroids, which were embedded in methyl cellulose/collagen-I gels and treated with VEGF165 alone or pre-formed VEGF165/GAG complexes. (a) Sprouting of HUVEC spheroids determined by light microscopy. (b) Cumulative sprouting length calculated using ImageJ and (c) number of sprouts. For (a) one representative microscope picture is shown and for (b) and (c) values represent the mean ± SD of n = 3 with 10 spheroids evaluated for each independent experiment. One-way ANOVA***p < 0.001 vs. VEGF165 alone; ###p < 0.001 vs. Ctrl. [adapted and modified from33].
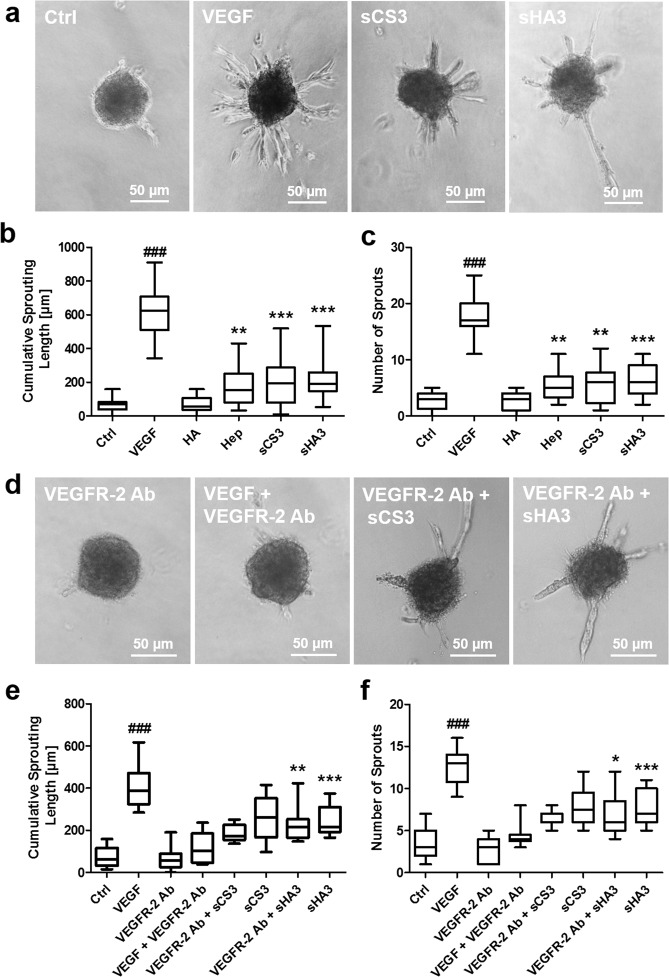
Growth factor-independent effects of GAG on spheroid sprouting
Treatment of HUVEC spheroids with GAG in the 3D in vitro angiogenesis assay revealed no induction of sprouting for HA itself (Fig. 7). In contrast, sGAG stimulated sprouting in a sulfation-dependent manner. Cumulative sprouting length and sprout number were comparably increased by sCS3 and sHA3 (Fig. 7b,c) indicating that these GAG were exhibiting VEGF-independent effects. To evaluate whether VEGFR-2 recognition by sGAG (Figs. 4 and 5c) initiates the pro-angiogenic effect of GAG observed in the 3D in vitro angiogenesis assays, additional experiments with VEGFR-2-blocking antibody were performed. VEGF-dependent sprouting of HUVEC spheroids was suppressed in the presence of 10 µg/ml anti-VEGFR-2 (Fig. 7d–f). However, the anti-VEGFR-2 antibody was unable to abrogate the effect of sCS3 and sHA3 on HUVEC spheroids, and these experiments revealed a comparable sprouting intensity as observed for GAG alone (Fig. 7e,f). For the neutralizing VEGFR-2 Ab used in the present study, no interaction with sHA3 and sCS3 was observed in SPR binding studies.
Figure 7.
Impact of GAG derivatives and role of VEGFR-2 for the pro-angiogenic effect of GAG derivatives on sprouting of HUVEC spheroids. VEGF165 was used as a positive control for spheroid sprouting in a 3D in vitro angiogenesis assay. (a) Sprouting of HUVEC spheroids determined by light microscopy. (b) Cumulative sprouting length calculated using ImageJ and (c) number of sprouts. For (a) one representative microscope picture is shown and for (b) and (c) values represent the mean ± SD of n = 3. One-way ANOVA: **p < 0.01; ***p < 0.001 vs. Ctrl. (d-f) Cells were treated with a VEGFR-2 neutralizing antibody (VEGFR-2 Ab) and GAG. (d) Sprouting of HUVEC spheroids determined by light microscopy. (e) Cumulative sprouting length calculated using ImageJ and (f) number of sprouts. For (d) one representative microscope picture is shown and for (e) and (f) values represent the mean ± SD of n = 3 with 10 spheroids evaluated for each independent experiment. One-way ANOVA: *p < 0.05; **p < 0.01; ***p < 0.001 vs. VEGF + VEGFR-2 Ab; ###p < 0.001 vs. Ctrl. [adapted and modified from33].
Discussion
Chemically defined sHA derivatives are promising to functionalize biomaterials since their sulfate groups modulate binding and biological activity of growth factors, like VEGF165 thereby influencing healing processes30,42,58,59. Pro-angiogenic and anti-angiogenic effects of GAG were previously described regarding their impact on VEGF biological activity, although often restricted to heparin17,19,60–64, which is known for its heterogeneity regarding the carbohydrate backbone, sulfation degree and pattern1.
Thus, in the present study, the potential dual action of sGAG was investigated with a broad range of HA and chondroitin sulfate (CS) derivatives with defined sulfation degrees and patterns in comparison to native GAG. First, the interaction between sGAG and VEGF165 or its HBD domain was analyzed using SPR and molecular modeling techniques. Second, the consequences of these interactions on VEGF165/VEGFR-2 complexation and the biological function of VEGF165 were evaluated in silico up to the atomic detail and in 2D in vitro cell culture experiments using HUVECs. Finally, the impact of different GAG was verified in a 3D in vitro angiogenesis assay by analyzing sprouting of HUVECs in the absence and presence of VEGF165. GAG displayed a concentration- and sulfation-dependent interaction with full-length VEGF165 and with its HBD. Interestingly, a preferred binding of sHA compared to CS derivatives with a comparable DS was observed, suggesting that the carbohydrate backbone of the GAG significantly influences the interaction. This is in line with previous findings for TGF-β1 and BMP-227,65. The reason for this might be that the molecular geometries in the carbohydrate backbones of sCS (GalNAc) and sHA (GlcNAc) render the respective sulfated groups to interact differently. Furthermore, the differences in the sulfation extent of specific positions within the D.U. might be another explanation. While sHA derivatives were completely sulfated at C6 of GlcNAc, and to a lesser extent at C2′ and C3′ of GlcA and C4 of GlcNAc, native CS contained 70% CS-4, sulfated at C4 of GalNAc before additional complete sulfation at C6 of GalNAc leading to sCS328.
The establishment of two distinct VEGF165 dimer molecular models, straight and twisted, which could co-exist in solution allowed us to investigate the mechanisms for GAG-VEGF165 recognition at atomic detail. In line with SPR, GAG were predicted to bind to the HBD at the C-terminus of VEGF165. In addition, SPR and free energy calculations demonstrated that GAG binding to HBD occurs in a sulfation-dependent manner. However, when comparing sulfated HA and CS derivatives, the differences in binding obtained from the theoretical models were not as apparent as with SPR analysis. For most of the analyzed GAG, the binding energy obtained was more favorable for the straight conformation. Our studies pointed toward a HBD-GAG-HBD stacking complex structure with the GAG embedded between the two HBD domains of the VEGF165 dimer. Furthermore, our theoretical models revealed that VEGF165 residues Arg145, Arg149 and Arg159 are crucial for sGAG recognition, which agree with previous mutagenesis studies56,57. Moreover, our analysis indicated that the VEGF165 straight model exhibited maximum number of significant GAG interactions matching the mutagenesis data (Fig. S4) shedding light on its biological relevance. In line with these findings, the fact that the VEGF165 straight model allows simultaneous interactions with VEGF receptors and co-receptors55 additionally supports its functional relevance.
In order to gain experimental evidence on the predicted molecular recognition of sGAG by VEGF165, the binding of psHA dp4 and dp6 to VEGF165-HBD was assessed by ITC. Calorimetric investigation revealed a mean stoichiometry close to that of a ternary complex suggesting that one psHA dp4/dp6 molecule binds to two VEGF165-HBD, which validates the predicted HBD-GAG-HBD stacking recognition mode. In these experiments, the physiological salt concentration did not affect the stoichiometry and KD values obtained for psHA dp4. Formation of the ternary complex was primarily driven by a positive entropic contribution. These thermodynamic data suggest that binding of two VEGF165-HBD by the sGAG is impelled - at least in part - by the desolvation of hydrophobic protein surfaces that are exposed to solvent in the GAG-free protein dimer and are joined together by sGAG binding. Interestingly, the formation of ternary complexes of psHA dp4 with other proteins such as IL-8 and IL-10 was driven by a strongly negative enthalpic contribution with a small negative entropic effect instead32.
SPR interaction analysis revealed that a tetrasaccharide is a sufficient length for sHA to interact with VEGF165, and a sulfation-dependent binding, as observed for polymeric sHA, was confirmed. Furthermore, the C6 sulfation of GlcNAc was shown to play an important role, as the binding strength for the higher sulfated sHA2Δ6s dp4, non-sulfated at C6, was lower compared to C6-sulfated sHA1. Our calculations supported this for the VEGF165 twisted model, while small differences in binding energies were obtained for the straight model. Interestingly, a higher dp does not necessarily enhance binding strength, as binding of psHA dp4 to full-length VEGF165 and VEGF165-HBD was stronger compared to dp6. This suggests that tetrasaccharides could bind to additional regions of VEGF165, as previously described for TIMP-366.
ITC results revealed a stronger binding of psHA dp6 to VEGF165-HBD than psHA dp4 (Table 3, Figs. S5, S6 and S7), which is in line with the theoretical binding energies obtained through the VEGF165 twisted model. The discrepancy between SPR and ITC psHA dp4 and dp6 binding data could be understood by considering the different experimental conditions in which the corresponding experiments are performed in each case. VEGF165-HBD is immobilized in SPR experiments, which may preclude certain complex formation accessible in solution phase in the ITC experiments. Our theoretical models which consider VEGF165 dimer showed how two HBDs can simultaneously interact with one GAG. Furthermore, the covalent attachment of the HBDs to the RBDs makes their conformational freedom lower than in the case of the free VEGF165-HBD in solution (ITC).
Considering that VEGF is assumed to act as a single entity, it can form a continuous binding surface for the interaction with large GAG67. Hence, the two identical HBDs of a dimer can be occupied by the same GAG chain. Robinson et al. showed that Hep dp7 is sufficient to fully occupy the Hep binding cleft of VEGF68. However, in case of smaller GAG it can be assumed that the two sites of the dimer could interact with the GAG independently of each other. Therefore, more than one molecule of psHA dp4 might interact with the VEGF165 dimer. In contrast to Hep oligosaccharides, psHA dp6 might be sufficient to stick on both HBDs, which in our models appear separated from each other by a distance equivalent to that of the GAG length. Our theoretical models suggested, nevertheless, a different molecular mechanism for the recognition of short sHA oligosaccharides. In the case of the VEGF165 twisted model, psHA dp4 and dp6 were initially located along one HBD or the two HBDs, while the recognition of the VEGF165 straight model by such HA derivatives resulted initially in a simultaneous binding to both VEGF-HBDs. Interestingly, after MD refinement, both models offered an equilibrated HBD-GAG-HBD stacking structure; mechanism which was indeed substantiated by ITC measurements as stated above. Our calculations showed more favorable interactions for psHA dp4 than dp6 toward the straight model (Fig. S4f, Table 2), which could explain the strikingly strong binding of psHA dp4 among the oligohyaluronans found by SPR. Interestingly, as stated above, this VEGF165 straight model is supported by previous studies to be the most biologically relevant form because it allows simultaneous interactions with VEGF receptors and co-receptors55. On the other hand, our predictions indicated more favorable binding for psHA dp6 than dp4 toward the VEGF165 twisted model, which is in line with the obtained KD data through ITC experiments. Differences in GAG accessibility to the HBDs may be responsible of the distinct binding obtained in different conditions. In conclusion, the binding strength to VEGF165 depends on the specific structural features of the oligosaccharides including their DS, sugar ring stereochemistry and conformation.
The interaction of VEGF165 with sHA3 led to a decreased VEGFR-2 mediated biological activity displayed by an impaired VEGFR-2 phosphorylation and a reduced HUVEC spheroid sprouting. However, EC sprouting was not completely blocked. First evidence regarding the inhibitory effect of sHA3 on the pro-angiogenic activity of VEGF165 was given by Rother et al.25. However, experiments were carried out only in a 2D model using a porcine EC line. Furthermore, there has been no comparison to other chemically and native, sulfated GAG. In the present study, a trend for reduced phosphorylation was observed also in the presence of sCS3 together with a significantly reduced sprouting of EC spheroids. Both high-sulfated GAG, therefore, exerted anti-angiogenic effects regarding the activity of VEGF165. This could be explained by previous findings30 revealing an impaired interaction of VEGF with VEGFR-2 in the presence of sGAG. Likewise, our atomic detailed models revealed that sGAG and VEGFR-2 were competing for Arg164 at the binding surface of VEGF165 in the twisted model. Furthermore, such molecular recognition mechanism would impair further interactions with other co-receptors such as NRP-1, which could negatively influence signaling. According to our predictions, and regardless of the possible coexistence of distinguishable twisted and straight conformations, the recognition of sGAG by VEGF165 points toward the formation of a HBD-GAG-HBD stacking complex structure in which the VEGF165-HBDs would lose contacts toward VEGFR-2, especially with Arg156 (Fig. S8), and consequently weakening the strength for HBD-receptor binding. This molecular mechanism could offer a plausible explanation to the experimental results, which is in accordance with previous observations showing the potential regulatory role of VEGF165-HBDs on VEGFR-2 phosphorylation17.
Still, sHA1 did not lead to a reduced phosphorylation of VEGFR-2, which is in accordance with a modest inhibitory effect on VEGF/VEGFR-2 interaction as previously shown30. It is in agreement with its weaker binding to full-length VEGF165 and VEGF165-HBD revealed by SPR and computer-based analysis. Here, free VEGF165 might still be able to interact with the receptor leading to an activation and subsequent signal transduction. Also for Hep, no significant effect on receptor activation and HUVEC spheroid sprouting was observed, even though a pronounced blocking effect on VEGFR-2 binding was reported30. These opposing results could be explained taking into account that in our cell culture experiments Hep might also interact with other proteins present, in contrast to SPR experiments where VEGF165 and VEGFR-2 are the only possible GAG interaction partners. Beneficial effects for Hep on several growth factor/receptor interactions are described, but differences between Hep sources can lead to high structural variability, resulting in different outcomes of experiments17,21,22.
In summary, high-sulfated GAG derivatives impair the biological activity of VEGF165 by hindering receptor activation and subsequent downstream signaling, which might be due to an impaired receptor binding. These findings suggest a potential local application of these particular GAG derivatives as components of biomaterials, e.g. functional wound dressings, to rebalance excessive angiogenesis associated with VEGF165/VEGFR-2 signaling, found in conditions like rheumatoid arthritis or diabetic retinopathy69,70.
This study revealed that GAG derivatives compete with VEGFR-2 leading to a reduced biological activity of angiogenic growth factors and, therefore, to an anti-angiogenic effect. In line with this, soluble Hep mimetics like PI-8871–73 and glycol-split Heps74 sequester angiogenic growth factors like VEGF165 thereby competing with endogenous cell surface HS and preventing ternary complex formation on the cell surface and receptor signaling. sGAG derivatives might exert anti-angiogenic effects through both, competition with cell-associated HS as well as growth factor receptors and, thus, be a promising option in therapeutic strategies aiming to inhibit angiogenesis.
In addition to the influence of GAG on growth factor activity, effects independent from these mediators were observed. SPR binding studies revealed an interaction of sGAG with VEGFR-2 (Fig. 5c). Xu et al. also found an in situ interaction of HS with VEGFR-2 using a proximity ligation assay75. Furthermore, Hep binding to VEGFR-2 was demonstrated in vitro in several studies14,18,22, while direct interaction was not observed in others64. Our theoretical models suggested that GAG binding to VEGFR-2 would preclude VEGF recognition.
In our SPR experiments, the binding strength of GAG toward VEGFR-2 was considerably lower compared to the binding of VEGF165. Such weak binding strength between GAG and VEGFR-2 did not lead to receptor activation, as the VEGFR-2 phosphorylation level in the presence of sGAG was comparable to the control without GAG and stimulating factors (Fig. 5). Interestingly, sGAG alone (Hep, sHA3, sCS3) were found to induce EC sprouting. This is in line with an enhanced EC proliferation in 3D GAG-containing hydrogels25. However, only low-sulfated GAGs that were crosslinked within the gel were included in these experiments, while the present study further revealed a positive effect of solute, high-sulfated GAG derivatives on ECs.
Therefore, Hep and both high-sulfated GAG derivatives might exert opposing effects on HUVECs. On one hand, they distinctly inhibited receptor binding of VEGF165 by interaction with this growth factor. On the other hand, they had a positive effect on HUVEC sprouting, independently from VEGF165. In case of Hep, there is probably a balance between these two effects, leading to no detectable effects on the biological activity of VEGF165 in the EC sprouting assay. This is further supported by the weaker interaction of Hep with the growth factors compared to sCS3 and sHA3. Therefore, only a portion of GAG is bound to the growth factor, while the rest might remain free and is able to activate EC sprouting. In contrast, sCS3 and sHA3, which strongly bind to VEGF165, remain predominantly growth factor-bound and, therefore, exerting a negative effect on VEGF165-mediated EC sprouting due to interference with growth factor receptor binding.
To understand the underlying mechanisms of the growth factor-independent effects of GAG, receptor-specific Abs were used to prevent GAG binding. Even though VEGFR-2 was blocked by the Ab, the added sGAG exerted a pro-angiogenic effect suggesting that VEGFR-2 is not involved. This is in line with the results on VEGFR-2 phosphorylation in the presence of sGAG alone (Fig. 5) and also with the low binding strength in SPR studies. We suggest that GAG modulate cell functions at several levels by upregulating proteins associated for instance with cell adhesion, cell signaling, matrix remodeling and endocytosis as reported for human mesenchymal stromal cells (hMSCs)76, which might be due to GAG internalization as shown previously77. In summary, sGAG do not only exert anti-angiogenic effects regarding the biological activity of VEGF165, but they are also pro-angiogenic in the absence of the exogenous growth factor. For the chemically high-sulfated GAG derivatives the effects were more pronounced compared to the native CS and Hep.
In addition, GAG derivatives have been shown to block inhibitors of angiogenesis, like TIMP-330, indicating an additional pro-angiogenic effect. These pro- and anti-angiogenic effects could be translated into GAG-containing biomaterials to improve the healing process by tuning their composition and selected GAG type. In case of low-sulfated GAG, we expect predominantly pro-angiogenic effects on EC behavior due to the rather weak GAG-growth factor interaction leaving a considerable amount of GAG available to interact with the cells directly. This is supported by previous findings with sHA1-containing HA/collagen hydrogels25. For high-sulfated GAG, the strong scavenging effect on angiogenic growth factors might dominate the EC-stimulating effects.
Conclusion
The aim of the present study was to elucidate the structure-function relationship of defined sGAG derivatives in their interplay with the angiogenic growth factor VEGF165 and to reveal the molecular mechanisms of the dual activity of GAG on angiogenic processes. The results obtained show that the interaction strength depends on GAG concentration, DS as well as sulfation pattern. In particular, sulfation at position C6 of GlcNAc plays an important role for these interactions. The established theoretical models in conjunction with experimental SPR and ITC results add novel insights into the mechanisms of GAG recognition by VEGF165 and its implication for receptor binding and, therefore, biological function. The carbohydrate backbone of the GAG had an additional influence on the interaction with VEGF165. GAG derivatives were found to exert growth factor-dependent but also independent effects. On one hand, GAG, upon binding to VEGF165, impair the interaction of the growth factor with its cognate receptor, thereby preventing downstream signaling. On the other hand, GAG alone exert pro-angiogenic effects on EC sprouting. Overall, the results obtained in this multidisciplinary approach contribute to a better understanding of the modulatory effects of GAG derivatives on angiogenic processes, which might be crucial to foster the rational design of functional biomaterials including GAG derivatives that specifically modulate angiogenesis and thereby healing processes.
Supplementary information
Acknowledgements
This project was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - Project number 59307082 - TRR67 [subprojects A3, A7, A8 and Z3]. The publication contains results and modified figures from the doctoral thesis “Interaction of glycosaminoglycans with growth factors and their receptors – implications for biological activity” of Dr. Linda Köhler (TU Dresden, 2018). We are thankful to Mario Hirt for technical support. The authors acknowledge support by the Open Access Publication Funds of the SLUB/TU Dresden.
Author contributions
L.K., G.R.-G., V.H., M.T.P. and J.R. designed the experiments. L.K., G.R.-G., K.B., S.R. and J.F. performed experiments. S.M., J.F. and S.K. synthesized the GAG derivatives under the direction of M.S. and J.R. L.K., G.R.-G., K.B., S.R., J.F., J.R., M.T.P. and V.H. analyzed the data and wrote the manuscript. S.D. provided the VEGF-HBD and reviewed the respective data. V.H., M.T.P., D.S. and J.R. supervised the project. All authors reviewed and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Linda Koehler and Gloria Ruiz-Gómez.
Contributor Information
M. Teresa Pisabarro, Email: Maria_Teresa.Pisabarro@tu-dresden.de.
Vera Hintze, Email: Vera.Hintze@tu-dresden.de.
Supplementary information
is available for this paper at 10.1038/s41598-019-54211-0.
References
- 1.Imberty A, Lortat-Jacob H, Perez S. Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 3.Gama CI, Hsieh-Wilson LC. Chemical approaches to deciphering the glycosaminoglycan code. Curr. Opin. Chem. Biol. 2005;9:609–619. doi: 10.1016/j.cbpa.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Gama CI, et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 5.Habuchi H, Habuchi O, Kimata K. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj. J. 2004;21:47–52. doi: 10.1023/B:GLYC.0000043747.87325.5e. [DOI] [PubMed] [Google Scholar]
- 6.Martinez P, et al. Macrophage polarization alters the expression and sulfation pattern of glycosaminoglycans. Glycobiology. 2015;25:502–513. doi: 10.1093/glycob/cwu137. [DOI] [PubMed] [Google Scholar]
- 7.Raman K, Mencio C, Desai UR, Kuberan B. Sulfation patterns determine cellular internalization of heparin-like polysaccharides. Mol. Pharm. 2013;10:1442–1449. doi: 10.1021/mp300679a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr. Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 9.Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- 10.Eming SA, Krieg T. Molecular mechanisms of VEGF-A action during tissue repair. J. Investig. Dermatol. Symp. Proc. 2006;11:79–86. doi: 10.1038/sj.jidsymp.5650016. [DOI] [PubMed] [Google Scholar]
- 11.Tischer E, et al. The human gene for vascular endothelial growth factor. Multiple protein forms are encoded through alternative exon splicing. J. Biol. Chem. 1991;266:11947–11954. [PubMed] [Google Scholar]
- 12.Muñoz-Chápuli R, Quesada AR, Angel Medina M. Angiogenesis and signal transduction in endothelial cells. Cell. Mol. Life Sci. 2004;61:2224–2243. doi: 10.1007/s00018-004-4070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koch S, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Cold Spring Harb. Perspect. Med. 2012;2:a006502. doi: 10.1101/cshperspect.a006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dougher AM, et al. Identification of a heparin binding peptide on the extracellular domain of the KDR VEGF receptor. Growth Factors. 1997;14:257–268. doi: 10.3109/08977199709021524. [DOI] [PubMed] [Google Scholar]
- 15.Mamluk R, et al. Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J. Biol. Chem. 2002;277:24818–24825. doi: 10.1074/jbc.M200730200. [DOI] [PubMed] [Google Scholar]
- 16.Park M, Lee ST. The fourth immunoglobulin-like loop in the extracellular domain of FLT-1, a VEGF receptor, includes a major heparin-binding site. Biochem. Biophys. Res. Commun. 1999;264:730–734. doi: 10.1006/bbrc.1999.1580. [DOI] [PubMed] [Google Scholar]
- 17.Ashikari-Hada S, Habuchi H, Kariya Y, Kimata K. Heparin regulates vascular endothelial growth factor165-dependent mitogenic activity, tube formation, and its receptor phosphorylation of human endothelial cells. Comparison of the effects of heparin and modified heparins. J. Biol. Chem. 2005;280:31508–31515. doi: 10.1074/jbc.M414581200. [DOI] [PubMed] [Google Scholar]
- 18.Gitay-Goren H, Soker S, Vlodavsky I, Neufeld G. The binding of vascular endothelial growth factor to its receptors is dependent on cell surface-associated heparin-like molecules. J. Biol. Chem. 1992;267:6093–6098. [PubMed] [Google Scholar]
- 19.Ono K, Hattori H, Takeshita S, Kurita A, Ishihara M. Structural features in heparin that interact with VEGF165 and modulate its biological activity. Glycobiology. 1999;9:705–711. doi: 10.1093/glycob/9.7.705. [DOI] [PubMed] [Google Scholar]
- 20.Terman B, et al. VEGF receptor subtypes KDR and FLT1 show different sensitivities to heparin and placenta growth factor. Growth Factors. 1994;11:187–195. doi: 10.3109/08977199409046916. [DOI] [PubMed] [Google Scholar]
- 21.Soker S, et al. Variations in the size and sulfation of heparin modulate the effect of heparin on the binding of VEGF165 to its receptors. Biochem. Biophys. Res. Commun. 1994;203:1339–1347. doi: 10.1006/bbrc.1994.2329. [DOI] [PubMed] [Google Scholar]
- 22.Wijelath E, et al. Multiple mechanisms for exogenous heparin modulation of vascular endothelial growth factor activity. J. Cell. Biochem. 2010;111:461–468. doi: 10.1002/jcb.22727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W, McCallum SA, Xiao Z, Zhang F, Linhardt RJ. Binding affinities of vascular endothelial growth factor (VEGF) for heparin-derived oligosaccharides. Biosci. Rep. 2012;32:71–81. doi: 10.1042/BSR20110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martino MM, et al. Extracellular matrix and growth factor engineering for controlled angiogenesis in regenerative medicine. Front. Bioeng. Biotechnol. 2015;3:45. doi: 10.3389/fbioe.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rother Sandra, Galiazzo Vanessa D., Kilian David, Fiebig Karen M., Becher Jana, Moeller Stephanie, Hempel Ute, Schnabelrauch Matthias, Waltenberger Johannes, Scharnweber Dieter, Hintze Vera. Hyaluronan/Collagen Hydrogels with Sulfated Hyaluronan for Improved Repair of Vascularized Tissue Tune the Binding of Proteins and Promote Endothelial Cell Growth. Macromolecular Bioscience. 2017;17(11):1700154. doi: 10.1002/mabi.201700154. [DOI] [PubMed] [Google Scholar]
- 26.Seyedarabi A, Cheng L, Zachary I, Djordjevic S. Production of soluble human vascular endothelial growth factor VEGF-A165-heparin binding domain in Escherichia coli. PLoS One. 2013;8:e55690. doi: 10.1371/journal.pone.0055690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hintze V, et al. Sulfated hyaluronan and chondroitin sulfate derivatives interact differently with human transforming growth factor-β1 (TGF-β1) Acta Biomater. 2012;8:2144–2152. doi: 10.1016/j.actbio.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 28.Hintze V, et al. Modifications of hyaluronan influence the interaction with human bone morphogenetic protein-4 (hBMP-4) Biomacromolecules. 2009;10:3290–3297. doi: 10.1021/bm9008827. [DOI] [PubMed] [Google Scholar]
- 29.van der Smissen A, et al. Growth promoting substrates for human dermal fibroblasts provided by artificial extracellular matrices composed of collagen I and sulfated glycosaminoglycans. Biomaterials. 2011;32:8938–8946. doi: 10.1016/j.biomaterials.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Rother S, et al. Sulfated Hyaluronan Alters Endothelial Cell Activation in Vitro by Controlling the Biological Activity of the Angiogenic Factors Vascular Endothelial Growth Factor-A and Tissue Inhibitor of Metalloproteinase-3. ACS Appl. Mater. Interfaces. 2017;9:9539–9550. doi: 10.1021/acsami.7b01300. [DOI] [PubMed] [Google Scholar]
- 31.Köhling Sebastian, Künze Georg, Lemmnitzer Katharina, Bermudez Marcel, Wolber Gerhard, Schiller Jürgen, Huster Daniel, Rademann Jörg. Chemoenzymatic Synthesis of Nonasulfated Tetrahyaluronan with a Paramagnetic Tag for Studying Its Complex with Interleukin-10. Chemistry - A European Journal. 2016;22(16):5563–5574. doi: 10.1002/chem.201504459. [DOI] [PubMed] [Google Scholar]
- 32.Köhling S, et al. Syntheses of defined sulfated oligohyaluronans reveal structural effects, diversity and thermodynamics of GAG-protein binding. Chem. Sci. 2019;10:866–878. doi: 10.1039/C8SC03649G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler, L. Interaction of glycosaminoglycans with growth factors and their receptors – implications for biological activity. Dissertation, TU Dresden, https://nbn-resolving.org/urn:nbn:de:bsz:14-qucosa2-325219, (2018).
- 34.Muller YA, Christinger HW, Keyt BA, de Vos AM. The crystal structure of vascular endothelial growth factor (VEGF) refined to 1.93 A resolution: multiple copy flexibility and receptor binding. Structure. 1997;5:1325–1338. doi: 10.1016/S0969-2126(97)00284-0. [DOI] [PubMed] [Google Scholar]
- 35.Fairbrother WJ, Champe MA, Christinger HW, Keyt BA, Starovasnik MA. Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure. 1998;6:637–648. doi: 10.1016/S0969-2126(98)00065-3. [DOI] [PubMed] [Google Scholar]
- 36.Brozzo MS, et al. Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood. 2012;119:1781–1788. doi: 10.1182/blood-2011-11-390922. [DOI] [PubMed] [Google Scholar]
- 37.Leppanen VM, et al. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc. Natl. Acad. Sci. USA. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Accelrys Software Inc., Discovery Studio Modeling Environment, Release 3.5, San Diego. (2012).
- 39.Case, D. A. et al. AMBER 2014, University of California, San Francisco (2014).
- 40.Molecular Operating Environment (MOE), version 2016; Chemical Computing Group Inc.: Montreal, QC, Canada (2016).
- 41.Thönes S, et al. Hyaluronan/collagen hydrogels containing sulfated hyaluronan improve wound healing by sustained release of heparin-binding EGF-like growth factor. Acta Biomater. 2019;86:135–147. doi: 10.1016/j.actbio.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 42.van der Smissen A, et al. Artificial extracellular matrix composed of collagen I and highly sulfated hyaluronan interferes with TGFbeta(1) signaling and prevents TGFbeta(1)-induced myofibroblast differentiation. Acta Biomater. 2013;9:7775–7786. doi: 10.1016/j.actbio.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 43.Morris Garrett M., Goodsell David S., Halliday Robert S., Huey Ruth, Hart William E., Belew Richard K., Olson Arthur J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry. 1998;19(14):1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 44.Ester, M., Kriegel, H.-P., Sanders, J. & Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. Proc. 2nd Int. Knowl. Discov. Data Min. (KKD96), 226–231, 10.1.1.121.9220 (1996).
- 45.Gehrcke JP, Pisabarro MT. Identification and characterization of a glycosaminoglycan binding site on interleukin-10 via molecular simulation methods. J. Mol. Graph. Model. 2015;62:97–104. doi: 10.1016/j.jmgm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Kirschner KN, et al. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huige CJM, Altona C. Force-Field Parameters for Sulfates and Sulfamates Based on Ab-Initio Calculations - Extensions of AMBER and CHARMm Fields. J. Comput. Chem. 1995;16:56–79. doi: 10.1002/jcc.540160106. [DOI] [Google Scholar]
- 48.Walker RC, Crowley MF, Case DA. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comput. Chem. 2008;29:1019–1031. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 49.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Wang JM, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 52.Miller BR, III, et al. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 53.R: a language and environment for statistical computing. R-package Development Core Team, http://www.r-project.org (2015).
- 54.Schrödinger, L. L. C., The PyMOL Molecular Graphics System, Version 1.8 (2009–2015).
- 55.Djordjevic S, Driscoll PC. Targeting VEGF signalling via the neuropilin co-receptor. Drug Discov. Today. 2013;18:447–455. doi: 10.1016/j.drudis.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Stammers R, Robinson CJ, Forster MJ, Mulloy B, Rafferty B. Vascular endothelial growth factor (VEGF): modulation of heparin-binding activity and bioactivity by site-directed mutagenesis. Endocrine Abstracts. 2005;10:P27. [Google Scholar]
- 57.Krilleke D, Ng YS, Shima DT. The heparin-binding domain confers diverse functions of VEGF-A in development and disease: a structure-function study. Biochem. Soc. Trans. 2009;37:1201–1206. doi: 10.1042/BST0371201. [DOI] [PubMed] [Google Scholar]
- 58.Clark RA, Ghosh K, Tonnesen MG. Tissue engineering for cutaneous wounds. J. Invest. Dermatol. 2007;127:1018–1029. doi: 10.1038/sj.jid.5700715. [DOI] [PubMed] [Google Scholar]
- 59.Koehler L, et al. Sulfated Hyaluronan Derivatives Modulate TGF-beta1:Receptor Complex Formation: Possible Consequences for TGF-beta1 Signaling. Sci. Rep. 2017;7:1210. doi: 10.1038/s41598-017-01264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiodelli P, Bugatti A, Urbinati C, Rusnati M. Heparin/Heparan sulfate proteoglycans glycomic interactome in angiogenesis: biological implications and therapeutical use. Molecules. 2015;20:6342–6388. doi: 10.3390/molecules20046342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Benedetto M, Starzec A, Vassy R, Perret GY, Crepin M. Distinct heparin binding sites on VEGF165 and its receptors revealed by their interaction with a non sulfated glycoaminoglycan (NaPaC) Biochim. Biophys. Acta. 2008;1780:723–732. doi: 10.1016/j.bbagen.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 62.Le Jan S, et al. Functional overlap between chondroitin and heparan sulfate proteoglycans during VEGF-induced sprouting angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2012;32:1255–1263. doi: 10.1161/ATVBAHA.111.240622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pike DB, et al. Heparin-regulated release of growth factors in vitro and angiogenic response in vivo to implanted hyaluronan hydrogels containing VEGF and bFGF. Biomaterials. 2006;27:5242–5251. doi: 10.1016/j.biomaterials.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 64.Teran M, Nugent MA. Synergistic Binding of Vascular Endothelial Growth Factor-A and Its Receptors to Heparin Selectively Modulates Complex Affinity. J. Biol. Chem. 2015;290:16451–16462. doi: 10.1074/jbc.M114.627372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hintze V, et al. Sulfated Glycosaminoglycans Exploit the Conformational Plasticity of Bone Morphogenetic Protein-2 (BMP-2) and Alter the Interaction Profile with Its Receptor. Biomacromolecules. 2014;15:3083–3092. doi: 10.1021/bm5006855. [DOI] [PubMed] [Google Scholar]
- 66.Rother S, et al. Structural and functional insights into the interaction of sulfated glycosaminoglycans with tissue inhibitor of metalloproteinase-3 - A possible regulatory role on extracellular matrix homeostasis. Acta Biomater. 2016;45:143–154. doi: 10.1016/j.actbio.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 67.Cochran S, et al. Probing the interactions of phosphosulfomannans with angiogenic growth factors by surface plasmon resonance. J. Med. Chem. 2003;46:4601–4608. doi: 10.1021/jm030180y. [DOI] [PubMed] [Google Scholar]
- 68.Robinson CJ, Mulloy B, Gallagher JT, Stringer SE. VEGF165-binding sites within heparan sulfate encompass two highly sulfated domains and can be liberated by K5 lyase. J. Biol. Chem. 2006;281:1731–1740. doi: 10.1074/jbc.M510760200. [DOI] [PubMed] [Google Scholar]
- 69.Hamilton JL, et al. Targeting VEGF and Its Receptors for the Treatment of Osteoarthritis and Associated Pain. J. Bone Miner. Res. 2016;31:911–924. doi: 10.1002/jbmr.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiselyov A, Balakin KV, Tkachenko SE. VEGF/VEGFR signalling as a target for inhibiting angiogenesis. Expert Opin. Investig. Drugs. 2007;16:83–107. doi: 10.1517/13543784.16.1.83. [DOI] [PubMed] [Google Scholar]
- 71.Ferro V, et al. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Semin. Thromb. Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 72.Hazel SJ. A novel early chorioallantoic membrane assay demonstrates quantitative and qualitative changes caused by antiangiogenic substances. J. Lab. Clin. Med. 2003;141:217–228. doi: 10.1067/mlc.2003.19. [DOI] [PubMed] [Google Scholar]
- 73.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 74.Casu B, et al. Short heparin sequences spaced by glycol-split uronate residues are antagonists of fibroblast growth factor 2 and angiogenesis inhibitors. Biochemistry. 2002;41:10519–10528. doi: 10.1021/bi020118n. [DOI] [PubMed] [Google Scholar]
- 75.Xu D, Fuster MM, Lawrence R, Esko JD. Heparan sulfate regulates VEGF165- and VEGF121-mediated vascular hyperpermeability. J. Biol. Chem. 2011;286:737–745. doi: 10.1074/jbc.M110.177006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kliemt S, et al. Sulfated Hyaluronan Containing Collagen Matrices Enhance Cell-Matrix-Interaction, Endocytosis, and Osteogenic Differentiation of Human Mesenchymal Stromal Cells. J. Proteome Res. 2013;12:378–389. doi: 10.1021/pr300640h. [DOI] [PubMed] [Google Scholar]
- 77.Salbach-Hirsch J, et al. Sulfated Glycosaminoglycans Support Osteoblast Functions and Concurrently Suppress Osteoclasts. J. Cell. Biochem. 2014;115:1101–1111. doi: 10.1002/jcb.24750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.