Abstract
Field cancerization is a premalignant process marked by clones of oncogenic mutations spreading through the epithelium. The timescales of intestinal field cancerization can be variable and the mechanisms driving the rapid spread of oncogenic clones are unknown. Here we use a Cancer rainbow (Crainbow) modelling system for fluorescently barcoding somatic mutations and directly visualizing the clonal expansion and spread of oncogenes. Crainbow shows that mutations of ß-catenin (Ctnnb1) within the intestinal stem cell results in widespread expansion of oncogenes during perinatal development but not in adults. In contrast, mutations that extrinsically disrupt the stem cell microenvironment can spread in adult intestine without delay. We observe the rapid spread of premalignant clones in Crainbow mice expressing oncogenic Rspondin-3 (RSPO3), which occurs by increasing crypt fission and inhibiting crypt fixation. Crainbow modelling provides insight into how somatic mutations rapidly spread and a plausible mechanism for predetermining the intratumor heterogeneity found in colon cancers.
Subject terms: Functional genomics, Cancer genetics, Mechanisms of disease
Pre-malignant cells harbouring oncogenic mutations can populate and spread throughout a tissue. Here, using a rainbow mouse system, the authors explore how clonal expansion in the mouse intestine might explain high levels of intra-tumoural heterogeneity observed in the disease.
Introduction
The process of field cancerization displaces normal cells with fields of premalignant cells harboring mutations to proto-oncogenes and tumor suppressors. Field cancers are operationally defined by Graham and colleagues as “a group of cells that are considered to be further along an evolutionary path toward cancer”1. Fields can present with or without morphologic pathology. The initiation and spread of field cancers increase the risk of a tumorigenic event setting the stage for sporadic cancers1–3. Determining how field cancerization occurs may lead to new strategies for cancer surveillance and treatment. The extent of field cancerization in the intestine varies from a few centimeters adjacent to a tumor or, in other circumstances, can include nearly the entire colonic epithelium4,5. The current model of intestinal field cancerization begins with the acquisition of a somatic mutation in an intestinal stem cell (ISC) within a crypt. Somatic mutations increase the competitive fitness of the ISC, resulting in the replacement of neighboring wild-type (WT) ISCs and the fixation of the mutation in a single crypt6. Premalignant crypts then duplicate by crypt fission to outcompete and displace adjacent normal crypts7–9 (Fig. 1a). The entire process is thought to occur slowly over several decades7,10.
Fig. 1.
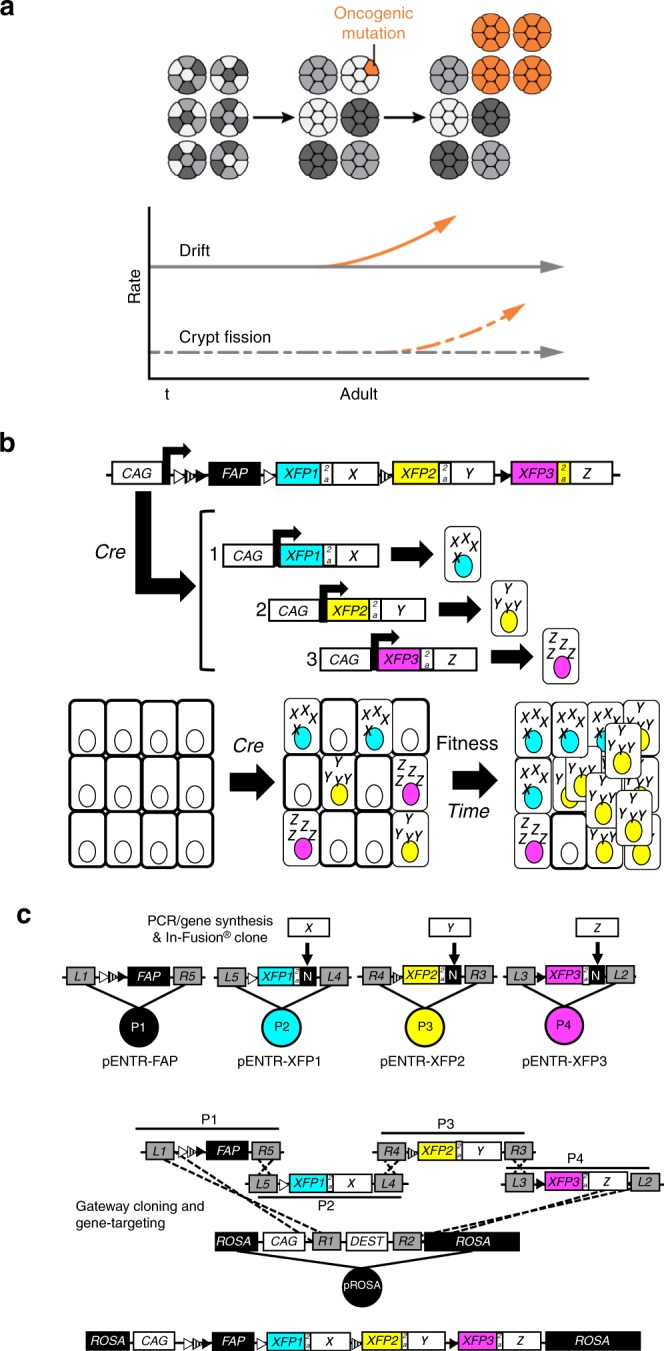
Engineering cancer rainbow mice. a The current two-step model of intestinal field cancerization. Somatic mutations increase stem cell fitness and lead to the fixation of premalignant clones within crypts (orange). Crypt fission results in duplication of premalignant crypts and the lateral propagation of somatic fields. b The Crainbow expression vector incorporates 4-tandem cassettes downstream of the ubiquitous chicken-β-actin promoter (CAG). The construct confers ubiquitous expression of a membrane-targeted and chemically inducible near-infrared fluorogen-activating peptide (FAP-Mars1) as a control. Cell type specific activation of Cre recombinase mediates recombination at one of three positions through inclusion of three pairs of orthogonal lox sites (LoxN: white triangle, Lox2272: hatched triangle, and LoxP: filled triangle). Single-copy transgene insertion provides a single outcome per cell and barcoding of each genetic fate by fluorescent imaging. c Candidate tumor driver genes are synthesized/PCR amplified and then In-Fusion® cloned into a bicistronic expression cassette. Multi-site Gateway™ cloning is used to directionally clone each pENTR plasmid into a Gateway™-compatible ROSA-targeting vector in a single step21.
One reason for the proposed slow progression of field cancers is the paradoxical finding that premalignant ISCs with somatic mutations, such as KrasG12D, APC-loss, or mutant TP53, can be stochastically replaced by neighboring WT ISCs. Therefore, field cancers can be prematurely extinguished by the healthy intestine10. A second reason for the proposed slow progression of field cancers is that healthy adult intestinal crypts infrequently duplicate—a process termed crypt fission. Less than 2% of crypts are undergoing fission in adults. Each crypt may only undergo one fission event every 30–40 years in the healthy intestine9,11. Therefore, the spread of field cancers is also severely limited. Crypt fission can be increased by somatic mutations. However, in familial adenomatous polyposis (FAP) patients and in mouse models of APC inactivation, the rate of increase is modest and variable8,9.
Growing evidence suggests that rapid field cancerization can occur in the intestine as a result of changes to the crypt microenvironment, epithelial injury, and age. First, perturbations to the microenvironment can lead to the selective loss of WT ISCs and their rapid replacement by more fit premalignant ISCs. The increase in ISC replacement results in the accelerated fixation of somatic mutations within intestinal crypts and the efficient initiation of a field cancer12. Second, chronic epithelial injury induces crypt fission and can spread field cancers throughout the entire colonic epithelium in less than 4 years4,13. Third, rapid field cancerization can also occur if somatic mutations are acquired during intestinal development when more than 20% of the crypts are actively undergoing crypt fission14,15. However, somatic mutations that overcome the constraints of intestinal homeostasis and drive rapid field cancerization in otherwise healthy adult intestine have still not been found.
Rspondin-3 (RSPO3) is a microenvironmental factor that regulates ISCs and when overexpressed induces crypt hyperplasia16. Oncogenic fusions of RSPO3 with the protein tyrosine phosphatase receptor type K (PTPRK) are also known drivers of colon cancer17,18. RSPO3 and its oncogenic fusions are compelling candidates that could drive the rapid spread of intestinal field cancers. Current mouse models lack the resolution to easily investigate the cellular and molecular roles of RSPO3 in field cancerization. Convenient solutions also do not exist for expressing and directly comparing multiple RSPO3 mutations within a single isogenic mouse. Coincidentally, mouse models for broadly investigating the functional genomics of field cancerization are also needed. Therefore, we have developed a cancer rainbow (Crainbow) mouse modelling platform that combines the desirable features of “Brainbow”19,20 based lineage tracing with functional genomics screening into one seamless and interchangeable platform. Crainbow provides a means to induce multiple somatic mutations and visualize two essential attributes of field cancerization—ISC competition and clone spreading.
Crainbow modeling directly demonstrates that somatic mutations in the neonatal intestine clonally spread throughout the intestine during a critical period of intestinal growth and development15. In addition, RSPO3 and its fusion isoforms are identified as a class of oncogenes that extrinsically transforms ISC behavior resulting in the widespread expansion of oncogenes throughout the adult epithelium in only a few weeks. Crainbow modelling is a transformative modelling technology and is a broadly applicable tool for visualizing the cellular and molecular dynamics of the early events that drive cancer.
Results
Engineering and validating cancer rainbow mouse models
Crainbow is a genetic model system for labelling and visualizing individual cells that express somatic mutations. Included in the Crainbow transgene are four positions that either express an inert fluorescent protein (position 0) or three spectrally resolvable fluorescent proteins paired with an oncogenic mutation of choice (positions 1–3). In addition, these candidate driver genes are fused to unique epitopes to ensure that their resultant protein products can be immunolocalized in tissue. In this manner, simple activation by Cre recombinase can induce spatiotemporal expression of fluorescently barcoded tumor driver genes and single-cell visualization of cell fitness, cell signalling, and the clonal spread of oncogenic mutations (Fig. 1b). In this report, several adaptations were made to overcome previous limitations in construct engineering and imaging21. First, a seamless and efficient cloning approach for building Crainbow targeting vectors was utilized21–24 (Fig. 1c). Second, the fluorescent protein (XFP) palette for imaging in vivo and ex vivo was optimized. This optimization included the use of a chemically inducible near-infrared fluorescent protein (FAP-Mars121) for mitigating an overabundant control signal20, targeting XFPs to the nucleus for cell counting, and delivery of two separate vector systems with different XFP palettes (Supplementary Figs. 1–3). Third, transgene expression was increased by including the woodchuck hepatitis posttranscriptional regulatory element (WPRE)25. Fourth, positional bias inherent to Cre/LoxP systems was empirically determined in a control mouse. Three Crainbow mouse models are developed in this study and are summarized in Table 1.
Table 1.
Overview of cancer rainbow (Crainbow) mouse lines.
| Symbol | Name | Oncogenea | Key observations | Map |
|---|---|---|---|---|
| NCAT | N-terminal truncated ßcatenin |
Cre Off: Mars1 FAP Cre On: 1. ΔNßcat 2. ΔNßcat 3. ΔNßcat |
The positional recombination efficiency of Crainbow is biased 2.3-fold at position 1, 0.36-fold at position 2, and 0.32-fold at position 3. | Supplementary Fig. 4, Supplementary Data 1 |
| MCAT | Multiple isoforms of ßcatenin |
Cre Off: Mars1 FAP Cre On: 1. ΔNßcat 2. Ccat/Lef1 3. ΔNßcatΔC |
Clone expansion occurs rapidly during perinatal development but is constrained by intestinal epithelial homeostasis in adult. | Supplementary Fig. 5, Supplementary Data 2 |
| ROBO | RSPO3 Crainbow |
Cre Off: Mars1 FAP Cre On: 1. RSPO3 2. PTPRKe1:RSPO3e2–5 3. PTPRKe1–7:RSPO3e2–5 |
RSPO3 oncogenes expand the crypt microenvironment to induce crypt fission and promote rapid oncogenic clone spreading in the adult epithelium. | Supplementary Fig. 7, Supplementary Data 3 |
aDescription of oncogenes and Crainbow lines. (NCAT) ΔNßcat (aa. 80–781) is based upon the previously described N-terminal truncation mutant and escapes degradation to increase Wnt-signaling26. NCAT mice express ΔNßcat in positions 1–3 and are used to calculate positional bias in Crainbow transgenes. (MCAT) MCAT mice express three isoforms of ßcat that act as oncogene prototypes. ΔNßcat was directly compared with ßcat isoforms that possess an increased Wnt-signaling potential (Ccat/Lef1: aa.693-781/aa.57-398)33,34 or a decreased Wnt-signaling potential (ΔNßcatΔC, aa.80-693)33–35. MCAT mice are used to test the comparative ability of Crainbow modelling and measure oncogenic clone competition of three oncogenes. (ROBO) RSPO3 is expressed by the crypt microenvironment and controls stem cell homeostasis50. The recently described oncogenic fusions of PTPRKe1:RSPO3e2–5 and PTPRKe1–7:RSPO3e2–5 are known drivers of colorectal cancer16–18. ROBO mice are used to assess how oncogenic activation of microenvironmental cues could also be potent drivers of the rapid spread of premalignant clones in the adult intestinal epithelium. Oncogenes are pseudocolored to match the fluorescent protein barcode
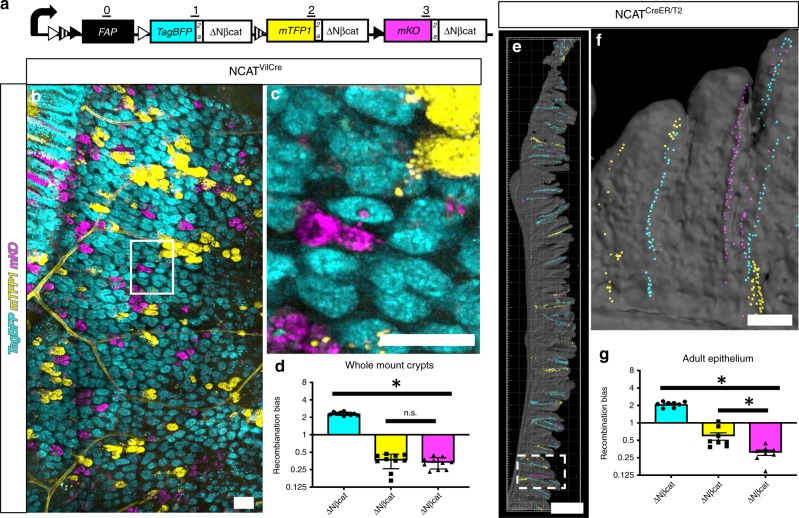
A Crainbow control mouse was developed to validate transgene expression and calculate positional bias. The NCAT Crainbow control mouse model (N-terminally truncated allele of ßcatenin) encodes the same N-terminally truncated and oncogenic form of ßcatenin (ΔNßcat) at each position26. Each ΔNßcat is coexpressed with TagBFP, mTFP1, or mKO (Fig. 2a, Supplementary Fig. 4). Simple confocal imaging for each fluorescent protein lineage marker can be used to assess recombination induced positional bias and the clonal spread of oncogenic mutations. NCAT Crainbow mice were bred to mice that express Villin-Cre recombinase to generate NCAT(+/−):VilCre(+/−) mice (NCATVilCre) and thereby induce recombination of Crainbow in the developing intestinal epithelium (e12.5)27. NCATVilCre mice were sacrificed between postnatal days (PND) 9–20 and the intestine was imaged by confocal microscopy. Since TagBFP quenched after fixation, imaging was performed on freshly isolated tissue when possible. Wholemount preparations of NCATVilCre intestine allowed confocal imaging through the serosa and into the intestinal crypt (Fig. 2b). Intestinal crypts harbor a population of ISCs that symmetrically divide and stochastically differentiate. This results in neutral drift behavior of the ISC and crypts that are populated by a single clone of stem cells—a process also known as crypt fixation28,29. Confocal imaging of intestinal crypts revealed that crypt fixation had already occurred in most crypts by the time of analysis, as evidenced by monoclonal crypts that expressed TagBFP (cyan), mTFP1 (yellow), or mKO (magenta) (Fig. 2b, c). Unbiased recombination of Crainbow should result in a 1/3 chance of activating positions 1, 2, or 3. Therefore, XFP crypts were counted and normalized to this expected probability of 0.33 to calculate recombination bias. Values equal to 1 reflect no bias, whereas >1 are positively biased and <1 are negatively biased. Crainbow activation has a 2.3-fold positive bias for position 1 and a concomitant negative bias that was similar for positions 2 and 3 (Fig. 2d). NCAT mice were also bred to ROSA-CreER/T2 mice to generate NCAT(+/−):ROSACreER/T2(+/−) mice (NCATCreER/T2) and treated with tamoxifen to assess positional bias in adult mice. NCATCreER/T2 were sacrificed 12 days post tamoxifen injection to allow for ISC labelling and clonal replacement of the differentiated cell lineage. As expected, stripes of TagBFP, mTFP1, and mKO recombined epithelial cells emanating from the crypt base were observed by confocal microscopy of vibratome sliced small intestine (Fig. 2e, f). Quantification of recombination bias revealed a positive bias for position 1 and negative bias for positions 2–3. This bias was similar to the bias observed in developmental NCATVilCre studies (Fig. 2g). The NCAT studies provided a basis for visualizing the clonal spreading of oncogenes in the intestine using Crainbow mouse models.
Fig. 2.
Validating Crainbow functionality. a Diagram of NCAT-Crainbow mice. See also Table 1 and Supplementary Fig. 1. b NCATVilCre small intestine (N = 10 mice, PND9–PND20) was prepared as a whole-mount and confocal imaged for XFP expression in the intestinal crypts. c Inset in “b” at higher magnification. d Crypts were color segmented and counted. The experimentally observed value was normalized to the predicted stochastic outcome (0.33/XFP). The asterisk denotes statistical significance by one-way ANOVA (TagBFP vs. mTFP1: p < 1e–6, TagBFP vs. mKO: p < 1e–6, mTFP1 vs. mKO: not significant). NCATCreER/T2 mice (N = 8, 19–22 weeks of age) were injected with 200 mg/kg tamoxifen. Mice were sacrificed 12 days post tamoxifen injection, and the small intestine was vibratome sectioned and antibody stained to recover the quenched TagBFP signal. e Sections were imaged by confocal microscopy for XFP expression (TagBFP: cyan, EYFP: mTFP1, mKO: magenta; segmented nuclear masks shown and overlaid with surface rendered tissue outline). f Rotated inset in “e” at higher power. g Nuclei were segmented, counted, and normalized to the predicted stochastic outcome (0.33/XFP). The asterisk denotes statistical significance by one-way ANOVA (TagBFP vs. mTFP1: p < 1e–6, TagBFP vs. mKO: p < 1e–6, mTFP1 vs. mKO: 8.4e–4). (SEM included for each graph). Scale Bars = 100 µm in b, c, f and 2 mm in e. Source data are provided as a “Source Data file”.
Intestinal growth accelerates the clonal spread of oncogenes
Genetic inactivation of adenomatous polyposis coli (APC) potentiates Wnt signalling and is a known driver of colon cancer30–32. ßcatenin is a downstream signaling effector and a prime candidate for increasing stem cell fitness and initiating field cancers. Several isoforms of ßcatenin have been used previously to study Wnt-signaling. ΔNßcat is already described above in NCAT mice and potentiates Wnt-signaling by escaping degradation. The Ccat/Lef1 isoform is a fusion of the C-terminal transactivation domain of ßcat to the transcription factor Lef1. Ccat/Lef1 potentiates Wnt signalling but cannot be sequestered by epithelial cadherin (CDH1)33,34. ∆Nßcat∆C lacks the N-terminal domain of ßcat and escapes degradation but also lacks the C-terminal transactivation domain and is thereby unable to transduce nuclear Wnt signalling33–35. These somatic mutations vary in their Wnt-signaling aptitude and are excellent prototypes for validating Crainbow as a robust system that quantifies the clonal expansion of oncogenes in vivo. Therefore, an MCAT-Crainbow mouse model (Multiple forms of ßcatenin, Fig. 3a and Supplementary Figs. 3, 5) was engineered. MCAT also used mTFP1, EYFP, and mKO palette for more favorable confocal imaging.
Fig. 3.
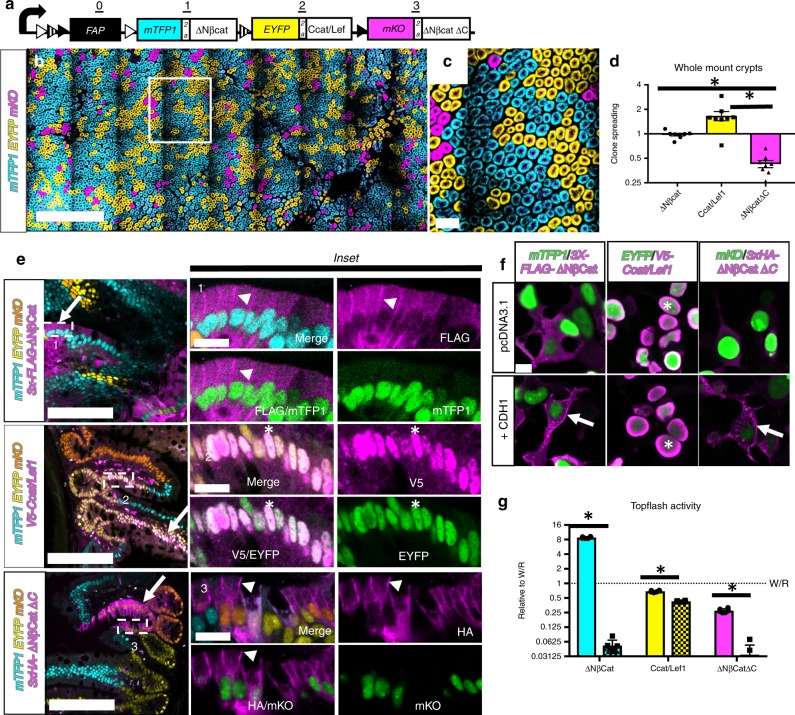
Widespread expansion of oncogenic clones during perinatal development. a Diagram of MCAT-Crainbow mice. See also Table 1 and Supplementary Fig. 5. b MCATVilCre small intestine (N = 10 mice, 3–6 weeks of age) prepared as a wholemount and confocal imaged. c Inset in “b” at higher magnification. d MCATVilCre Crypts were color segmented, counted and normalized to the positional bias calculated in NCATVilCre mice. Asterisk denotes statistical significance by one-way ANOVA (mTFP1 vs. EYFP: p = 0.003, mTFP1 vs. mKO: p = 0.016, EYFP vs. mKO = 3e–6). e Immunostaining for FLAG, V5, or HA epitopes (magenta) specific to each ßcat isoform in MCATVilCre small intestine vibratome slices and merged with fluorescent lineage markers (mTFP1: cyan, EYFP: yellow, and mKO: orange). Arrows denote isoform expression with cognate lineage reporter (FLAG and mTFP1, V5 and EYFP, and HA and mKO). Corresponding insets depict higher magnification images. Arrowheads denote membrane-localized ßcat, whereas asterisk denotes nuclear-localized ßcat. Epitope stains (magenta) are also presented as merged and as a single-channel image with its cognate fluorescent lineage reporter (green). f HEK cells were transiently transfected with MCAT isoforms, fixed, stained, and imaged for the indicated epitope (magenta) and fluorescent reporter (green). Cells were also cotransfected with epithelial cadherin (CDH1) as indicated. Arrows denote sequestration of ßcat at the plasma membrane, and the asterisk denotes nuclear ßcat. g Wnt signalling activity for each oncogene in the absence of CDH1 (solid bar) or in the presence of overexpressed CDH1 (hatched bar) (N = 6 wells per condition and independently repeated in four experiments). TOP FLASH activity was normalized to WNT/RSPO-stimulated control cells (dashed line). Asterisk denotes statistical significance by two-way ANOVA and Bonferroni’s multiple comparisons test (cyan < 1e–6, yellow = 0.01, magenta = 0.02). (SEM included for each graph). Scale Bars = 1 mm in b, 100 µm in c/e, 15 µm in e: insets 1–3, and 10 µm in f. Source data are provided as a “Source Data file”.
MCAT Crainbow mice were bred to mice that express Villin-Cre recombinase to generate MCAT(+/−):VilCre(+/−) mice (MCATVilCre) and thereby induce recombination of Crainbow in the developing intestinal epithelium (e12.5)27. Whole-mount confocal imaging of 3–6-week-old MCATVilCre small intestine revealed that crypt fixation had already occurred, as was evident by a single color of epithelial cells in each crypt (Fig. 3b, c). The spread of oncogenic clones through the intestine was also studied in MCAT mice. The same ΔNßcat allele was used in Position 1 of NCAT and MCAT mice. Therefore, a relative clone spreading potential was calculated by normalizing the MCAT crypt frequency to the positional recombination bias calculated in NCATVilCre mice. Ccat/Lef1 expression induced a relative 1.6-fold increase in clone spreading potential. In contrast, ΔNßCatΔC induced a respective 0.4-fold reduction in clone spreading potential and ΔNßcat was unchanged (Fig. 3d). The proximal to distal axis of the intestine differs in Wnt/ßcatenin signalling potential36. ΔNßCatΔC expression resulted in a similar decrease in clone spreading potential in the colon and the effects of Ccat/Lef1 were mitigated and more similar to ΔNßcat (Supplementary Fig. 6a, b).
Epithelial cadherin (CDH1) sequesters ßcat at the lateral plasma membrane acting as a molecular “sink” for excess ßcat that ultimately inhibits Wnt-signalling37,38. We hypothesized that the increased spread of Ccat/Lef1 clones was due to the loss of CDH1 sequestration activity. Antibody staining for the epitope tags fused to each ßcat isoform revealed that each isoform was correctly expressed with its cognate XFP (Fig. 3e, arrows). FLAG immunostaining for ΔNßcat and HA immunostaining for ΔNßCatΔC showed plasma membrane sequestration of ßcat with no detectable nuclear signal (Fig. 3e, arrowheads). In contrast, V5 immunostaining for Ccat/Lef1 revealed nuclear localization with no detectable expression at the plasma membrane (Fig. 3e, asterisks). A similar effect was observed in a complementary in vitro transfection system (Fig. 3f). In addition, a TOP-FLASH luminescent assay for Wnt signalling39 demonstrated that only Ccat/Lef1 could effectively transduce Wnt/ßcat signalling in the presence of CDH1 (Fig. 3g). These results confirmed that Crainbow modelling could be used for inducing and directly comparing multiple premalignant epithelial clones harboring oncogenic mutations and that these clones effectively spread during perinatal intestinal development.
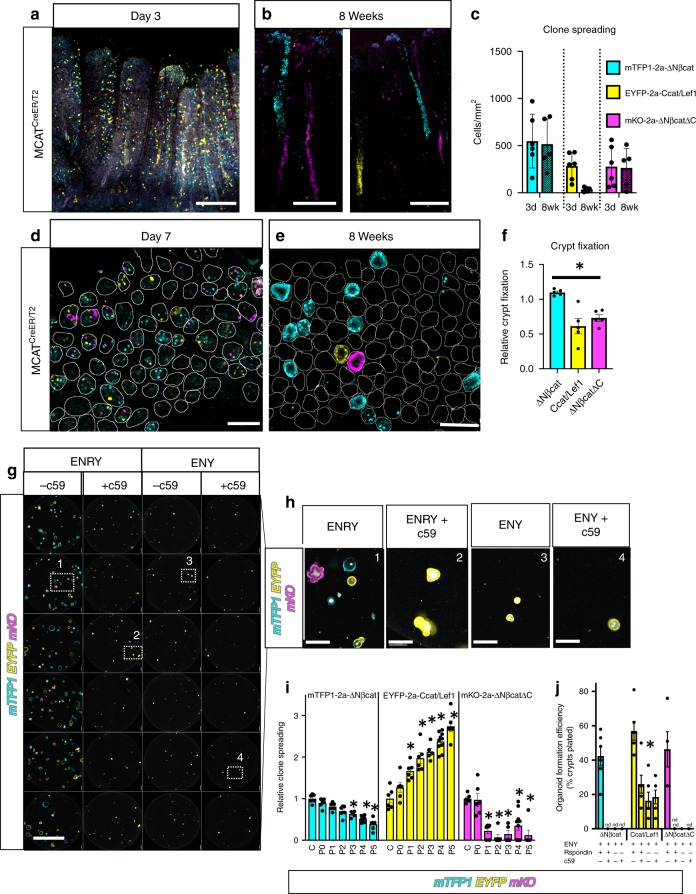
MCAT mice were also used to study the evolution of oncogenic clones in adult intestine. MCAT mice were bred to ROSA-CreER/T2 mice to generate MCAT(+/−):ROSACreER/T2(+/−) mice (MCATCreER/T2). Adult MCATCreER/T2 mice were treated with 200 mg/kg tamoxifen and chased for 3 days or 8 weeks. Sparse recombination was observed throughout the villus and crypt epithelium at day 3 (Fig. 4a). Prior lineage tracing studies of the intestinal epithelium revealed that recombined villus epithelial cells are lost within a few days and are replaced by clones of epithelial cells emanating from recombined ISCs29. Clones of MCAT recombined cells could be similarly observed 8 weeks after tamoxifen induced recombination (Fig. 4b). Clones were restricted to narrow stripes of recombined epithelial cells within each villus and were typically only a few cells wide. The widespread recombination of an entire villus was not observed. An estimate for clone spread was determined by counting the total recombined cells at day 3 versus 8 weeks. The total number of recombined cells did not increase from day 3 to 8 weeks for any of the oncogenic ßcat isoforms. An apparent but statistically insignificant decrease in total recombined Ccat/Lef1 cells was also observed (Fig. 4c). Therefore, clone spreading does not significantly occur in adult MCAT intestine.
Fig. 4.
The crypt microenvironment impairs the spread of oncogenic clones in adults. a, b MCATCreER/T2 mice (>6 weeks of age) were I.P. injected with 200 mg/kg of tamoxifen and sacrificed 3 days (N = 8) or 8 weeks later (N = 5). The small intestine was vibratome sectioned and imaged. c Nuclei were segmented, counted, and normalized to the total volume of tissue imaged. Clone spread was determined by comparing the change in total recombined nuclei for each fate at day 3 and 8 weeks. Statistical significance by mixed effects modelling and Sidak multicomparison correction (∆Nßcat: p = 0.99, Ccat/Lef: p = 0.14 ∆Nßcat∆C: p = 0.99). d, e MCATCreER/T2 (>6 weeks of age) were injected three times (Day 1, 3, 5) with 200 mg/kg tamoxifen and sacrificed at Day 7 (N = 4) or at 8 weeks (N = 5). The small intestine was whole mount imaged by confocal microscopy. Representative crypts are outlined and f the relative fraction of crypt fixation events for each ßcat isoform was measured. Statistical significance by one-way ANOVA and Holm-Sidak’s multiple comparison test (∆Nßcat vs. Ccat/Lef1: p = 0.011 (∆Nßcat vs. ∆Nßcat∆C: p = 0.0062, Ccat/Lef1 vs. ∆Nßcat∆C p = 0.259). g MCATVilCre crypts were isolated and cultured as in the indicated media (+/−c59 inhibitor at 10 nM) and then imaged by confocal microscopy. h High-magnification view of wells (N = 6) from each condition in “g” and rotated corresponding insets. i MCATVilCre organoids were subcultured (P0–P5, and N = 5–10 wells per subculture) and the entire well imaged by confocal microscopy and organoids of each lineage were counted. Relative clone spreading for each organoid fate was calculated by normalization to the crypt number (C) at isolation (see also Supplementary Fig. 6c). The asterisk denotes statistical significance by mixed effects modelling and Dunnett post hoc relative to Crypt control (C) (∆Nßcat: p = 0.03, p = 0.002, p < 0.0001 and Ccat/Lef1: p < 0.0001 for each asterisk and ∆Nßcat∆C p < 0.0001 for each asterisk). j Organoid formation efficiency for MCAT crypts cultured in “g–h.” The asterisk denotes statistical significance by two-way ANOVA and Tukey’s multiple comparisons test. ENRY conditions: not significant between ßcat isoforms, whereas Ccat/Lef1 was significantly different from ∆Nßcat and ∆Nßcat∆C for each treatment (ENRY + c59: p = 0.0003, ENY: p = 0.03, and ENY + c59: p = 0.01). (SEM included for each graph). Scale bars = 200 µm in a, b, 100 µm in d, e, 2 mm in g, and 200 µm in h. Source data are provided as a “Source Data file”.
The fixation of somatic mutations within a crypt is an initiating event in cancer field formation. A previous report documented that ISCs with potent somatic mutations can still be replaced by neighboring stem cells within the crypt10. Therefore, crypt fixation rate was measured in MCAT mice. MCATCreER/T2 mice were treated three times with 200 mg/kg tamoxifen (Days 1, 3, and 5) to abundantly label multiple ISCs within the crypts. Mice were sacrificed at Day 7 or at 8 weeks. Whole-mount confocal imaging of intestinal crypts at day 7 revealed that 42% of crypts were labelled. Multiple examples of monoclonal, biclonal, or triclonal crypts were evident (Fig. 4d). Prior lineage tracing studies demonstrated that crypt fixation occurs within 4–8 weeks29. Whole-mount confocal imaging of MCAT intestinal crypts 8 weeks after recombination revealed that crypt fixation had occurred in 15% of the crypts counted (Fig. 4e). The relative fraction of fixed crypts for each MCAT lineage was calculated at 8 weeks and normalized for positional bias in order to calculate the relative crypt fixation potential of each ßcat isoform. The results indicated that the relative fixation potential of mTFP1:ΔNßcat ISCs was modestly increased compared to the reduced fixation potential of EYFP:Ccat/Lef1 and mKO:ΔNßCatΔC ISCs (Fig. 4f). The direct measurement of clone spreading and crypt fixation within MCAT mice indicates that the normal adult intestine is resistant to the clonal expansion of oncogenic mutations.
Interestingly, efficient and widespread field cancerization during development has previously been attributed to the plasticity of crypts during development—including crypt fission and nascent crypt formation14. Intestinal organoid cultures effectively model crypt plasticity40. Therefore, MCAT organoids were used as an ex vivo correlate of the clonal expansion of oncogenes observed during development. ΔNßcat−, Ccat/Lef1−, ΔNßCatΔC−expressing crypts were isolated from MCATVilCre mice and similarly established viable organoids in full media (Fig. 4g, h, Panel 1). MCAT organoids were passaged so that each lineage could be longitudinally studied for alterations in competitive fitness over time. Interestingly, CCat/Lef1 expressing organoids rapidly expanded after multiple rounds of subculturing, whereas ΔNßcat and ΔNßCatΔC expressing organoids did not (Fig. 4i and Supplementary Fig. 6c). Paneth cells are the source of Wnts in organoid culture and are required for organoid growth41. Organoids were treated with c59 to prevent Wnt acylation and thereby inhibit Wnt secretion42. Only Ccat/Lef1 expressing organoids grew in the absence of Wnt (Fig. 4g, h, j, Panel 2). Similarly, organoids require exogenous Rspondins (RSPO)s for their growth43. Only Ccat/Lef1 organoids could grow without Rspondin (Fig. 4g, h, j, Panel 3). Ccat/Lef1 organoids could also grow in the absence of Wnts and RSPO (Fig. 4g, h, j, Panel 4). Therefore, the oncogenic activation of Wnt/ßcat signaling endows ISCs with increased fitness that results in their preferential expansion during a permissive period of intestinal development but not in healthy adult intestine.
Rapid spreading of oncogenes in the adult intestine by microenvironmental oncogenesis
The abundant crypt fission and formation of nascent crypts in organoids is likely due in part to the artificial reconstitution of the microenvironment, in which the cells are grown, including important niche factors like (RSPO)s41,43. RSPO3 is a critical regulator of the intestinal microenvironment16 and in vivo its expression is normally limited to the subepithelial mesenchyme of WT intestine (Fig. 5a). RSPO3 fusions with PTPRK have been found in colon cancer patients17 and are protumorigenic in mice18. Therefore, a Crainbow RSPO3 model was generated to test the hypothesis that misexpression of RSPO3 and its fusion products perturb the microenvironment of the crypt and induce the spread of oncogenic clones by increasing crypt fission. ROBO Crainbow mice (RSPO3-Crainbow) encode RSPO3, PTPRKe1:RSPO3e2–5, and PTPRKe1–7:RSPO3e2–5 at positions 1–3 of Crainbow and are coexpressed with the mTFP1, EYFP, and mKO fluorescent protein barcode (Fig. 5b and Supplementary Fig. 7).
Fig. 5.
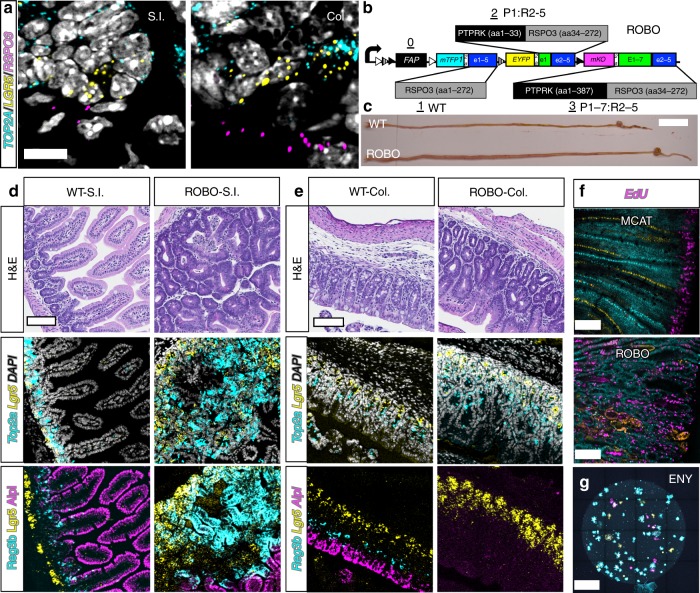
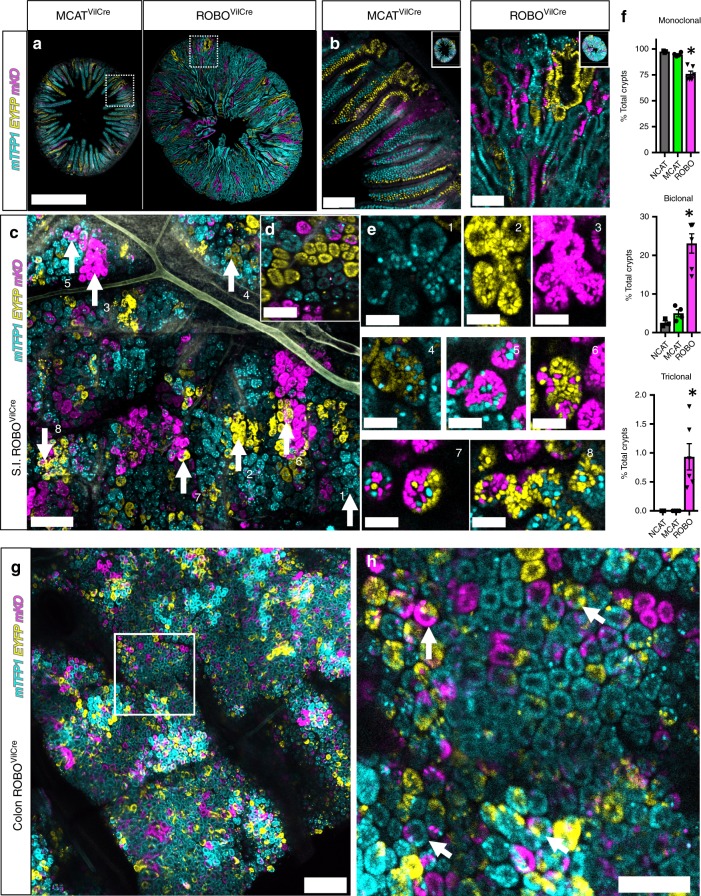
Ectopic expansion of the intestinal stem cell compartment by expression of oncogenic RSPO3. a Multiprobe RNA FISH and confocal microscopy for Top2a (cyan), Lgr5 (yellow), and Rspo3 (magenta) in paraffin-embedded sections of the small intestine (SI) and colon (Col). b ROBOVilCre mice expressing oncogenic human RSPO3. Position 0: control and membrane-targeted FAP-Mars1, Position 1: mTFP1 (cyan) coexpressed with 3×FLAG-RSPO3, EYFP (yellow) coexpressed with V5-PTPRKe1:RSPO3e2–5, and Position 3: mKO (magenta) coexpressed with 3×HA-PTPRKe1–7:RSPO3e2–5 (see also Supplementary Fig. 7). c Gross examination of the ROBOVilCre mice and gastrointestinal tracts at PND16 compared with WT littermate controls. WT and ROBO d small intestines (SI) and e colons (Col) were paraffin embedded and sectioned for H&E pathology and coregistry of indicated cell type markers by multiprobe RNA FISH. Images are insets from whole-slide imaged Swiss rolls (see Supplementary Figs. 8–13). f MCATVilCre and ROBOVilCre mice were injected with EdU (magenta) and coimaged for each lineage reporter by confocal microscopy (mTFP1: cyan, EYFP: yellow, mKO: orange). g ROBOVilCre organoids can grow without exogenous RSPO3 (ENY) and were imaged by confocal microscopy for each lineage reporter. Scale bars = 10 µm in a, 2 cm in c, 100 µm in d, e, 50 µm in f, and 1 mm in g. Source data are provided as a “Source Data file”.
ROBO Crainbow mice were bred to mice that express Villin-Cre recombinase to generate ROBO(+/−):VilCre(+/−) mice (ROBOVilCre) and thereby induce recombination of Crainbow in the developing intestinal epithelium (e12.5)27. The resultant ROBOVilCre pups developed distended abdomens and were moribund by the time of weaning. Gross anatomical examination revealed dilated and elongated small and large intestines compared with WT mice (Fig. 5c). Whole-slide imaging of H&E stained Swiss-roll WT and ROBOVilCre small and large intestines revealed an expansion of the crypt zone in the small intestine, basal crypt hyperplasia, architectural distortion, and occasional luminal crypt abscesses (Fig. 5d, Supplementary Figs. 8–13). Similarly, the colonic crypts were expanded by basal crypt hyperplasia and architectural distortion including crypt branching and budding (Fig. 5e, Supplementary Figs. 8–13). RNA FISH was used to molecularly map the stem cell compartment in WT and ROBOVilCre intestine. A potent expansion of the stem cell compartment in ROBOVilCre mice was observed and included an overabundance of Lgr5-positve stem cells and Top2a-positive transiently amplifying (TA) cells44 (Fig. 5d, e, Supplementary Figs. 8–13). Conversely, a marked reduction in differentiated Alpi-positive enterocytes45 was observed in ROBOVilCre intestine. Reg3b, a recently described marker of enterocytes adjacent to the stem cell crypt45,46, was found in scattered cells interposing the TA zone of the crypt and more mature enterocytes of the villus. Reg3b expressing cells were also expanded in ROBOVilCre small intestine when compared with WT. In the colon, Reg3b expressing cells were less frequent in both WT and ROBOVilCre mice (Fig. 5d, e, Supplementary Figs. 8–13). ROBOVilCre intestine also had an expanded proliferative zone and was in contrast to the normal proliferative zone found in MCATVilCre intestinal crypts (Fig. 5f). ROBOVilCre organoids also grew in the absence of exogenous RSPOs (Fig. 5g). These data confirm that ectopic overexpression of Rspondins can restructure the microenvironment of the intestine leading to crypt hyperplasia and the ectopic expansion of proliferative stem cells throughout the epithelium16,47. We have termed this process microenvironmental oncogenesis.
Previous reports showed that RSPO3 and RSPO3:PTPRK fusions are protumorigenic in mice but the lack of lineage tracing prevented full understanding of how each form of the protein affects stem cell dynamics18. Therefore, we assessed stem cell clonality and lineage development in ROBOVilCre small intestine and compared this to MCATVilCre small intestine. Confocal imaging revealed that ROBOVilCre intestine was significantly larger than age-matched MCATVilCre intestine (Fig. 6a). At higher magnification, MCATVilCre intestine displayed the typical pattern of single-color clones in the villus that emanate from the base of the intestinal crypt. In contrast, ROBOVilCre mice had fewer large single-color clones and instead were characterized by smaller clones, in which each lineage was heterogeneously mixed throughout the villus and crypt (Fig. 6b). This suggested that crypt fixation had been inhibited in ROBOVilCre mice. Whole-mount confocal imaging crypts revealed that the ROBOVilCre crypts were markedly polyclonal (Fig. 6c). This was in direct contrast to the monoclonality and crypt fixation evident in control MCATVilCre crypts (Fig. 6d). Instances of monoclonality (Fig. 6e, insets 1–3), biclonality (Fig. 6e, insets 4–6), and triclonality (Fig. 6e, insets 7, 8) were observed in the ROBOVilCre crypts. In addition, crypt fission and crypt budding could be observed in ROBOVilCre crypts by confocal microscopy and confirmed the histopathology assessment (Fig. 6e, insets 1–3). Crypt fixation was scored in ROBOVilCre and compared with MCATVilCre and the control NCATVilCre mice. This analysis revealed that 95% and 97% of crypts were monoclonal in MCATVilCre and NCATVilCre mice, respectively. In contrast, only 76% of crypts were monoclonal or fixed in ROBOVilCre mice (Fig. 6f). Approximately 23% of crypts were biclonal in ROBOVilCre mice, whereas only 5% and 3% of crypts were biclonal in MCATVilCre and NCATVilCre mice, respectively. Triclonal crypts were found in 1% of the ROBOVilCre mice but were not observed in MCATVilCre or NCATVilCre mice. Crypt fixation was also inhibited in the colon (Fig. 6g, h). These data demonstrated that microenvironmental oncogenesis inhibits the neutral drift behavior of ISCs and attenuates the rate of crypt fixation.
Fig. 6.
Microenvironmental oncogenesis attenuates crypt fixation. a Vibratome cross-sections of MCATVilCre and ROBOVilCre small intestines were confocal imaged and tiled for each lineage reporter (PND17). b Higher magnification of areas indicated in “a”. c Whole-mount confocal imaging of small intestines for each fluorescent lineage reporter in ROBOVilCre mice. d Whole-mount confocal imaging of MCATVilCre small intestines. e Indicated callouts (1–8) from “c” at higher magnification. f The percentage of each crypts that are monoclonal (crypt fixation), biclonal, or triclonal was calculated for NCATVilCre mice (N = 3 and 938 crypts), MCATVilCre mice (N = 9, and 6006 crypts analyzed at PND17) and ROBOVilCre mice (N = 6 and 3557 crypts analyzed). (SEM included for each graph). g Wholemount confocal imaging of ROBOVilCre colon at PND18. h Region of interest in “g” at higher magnification. Arrows show examples of crypts where fixation has not occurred. Scale bars = 1 mm in a, 100 µm in b/d, g, h, 200 µm in c, and 40 µm in e. Source data are provided as a “Source Data file”.
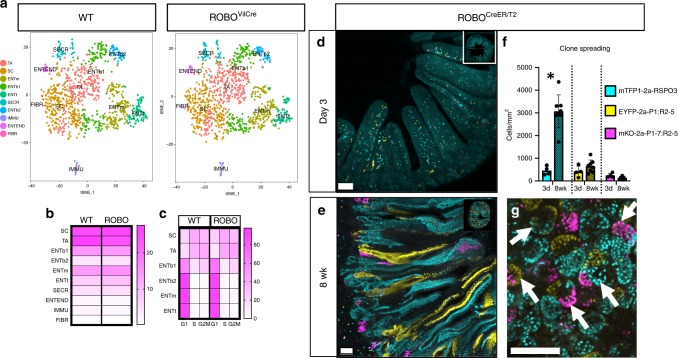
Next, lineage dynamics and signalling were assessed by single-cell RNA sequencing (scRNAseq) of crypt enriched tissue isolated from WT and ROBOVilCre small intestine. t-SNE clustering and visualization of scRNAseq data revealed ten cell types. Two of these cell types were fibroblasts and immune cells. The remaining eight include stem cells, transiently amplifying cells, secretory Paneth and goblet cells, enteroendocrine cells, and three populations of enterocytes that could be qualitatively associated with distinct spatial zones within the villus46 (Fig. 7a, Supplementary Fig. 14). Surprisingly, the overall fractional contribution of each cell type and the cell-cycle state did not change significantly between WT and ROBO crypts (Fig. 7b, c). Signalling was also largely unaffected as only 25 genes (23 genes in TA cells and two genes in ENTb1) were differentially expressed (Supplementary Data 4). The scRNAseq data suggest that microenvironmental oncogenesis extrinsically modifies the stem cell compartment.
Fig. 7.
Microenvironmental oncogenesis drives heterogenous premalignant clones in adults. a Single cells were isolated from PND16 WT and ROBOVilCre by fractionation to enrich for crypt epithelium. scRNAseq and tSNE visualization was performed for WT and ROBOVilCre cells. (SC: stem cell, TA: transiently amplifying cell, ENT: enterocytes that included bottom (b1: S/G2M phase and b2: G1), middle (m), and top (t), and were recently described45, SECR: secretory Paneth and Goblet cells, IMMU: immune, ENTEND: enteroendocrine, FIBR: fibroblast). For cell type markers, see also Supplementary Fig. 14. b Heatmap visualization of cell types present in WT and ROBOVilCre isolates as a percentage of total cells. c Heat map visualization of cell cycle phase as a percentage of each cell type. d–g ROBOCreER/T2 (N = 9 mice, 6–15 weeks of age) were I.P. injected with 200 mg/kg of tamoxifen and sacrificed 3 days (N = 4) or 8 weeks later (N = 5), and the small intestine was vibratome sectioned and imaged by tiling confocal microscopy. Confocal imaged vibratome sections of ROBOCreER/T2 small intestines at d 3 days or e 8 weeks post tamoxifen injection. f Nuclei were segmented and counted and normalized to the total volume imaged. Clone spread was determined by comparing the total recombined nuclei for each fate at 8 weeks relative to day 3. The asterisk denotes statistical significance by mixed effects modelling and Sidak multicomparison correction (RSPO3: <0.000001, PTPRKe1:RSPO3e2–5 (P1:R2-5): p = 0.31 and PTPRKe1–7:RSPO3e2–5 (P1-7:R2-5): p = 0.94). g Evidence for attenuated crypt fixation 8 weeks post tamoxifen injection as revealed by chimeric crypts in whole-mount confocal imaging. (SEM included for each graph). Scale bars = 100 µm in d, e, g. Source data are provided as a “Source Data file”.
Last, we hypothesized that microenvironmental oncogenesis can promote the expansion of oncogenic clones in adult intestine. To test this, ROBO mice were bred to ROSA-CreER/T2 mice to generate ROBO(+/−):ROSACreER/T2(+/−) mice. The resultant ROBOCreER/T2 mice were injected with a single dose of tamoxifen and chased for either 3 days or 8 weeks. Both small and large intestinal mucosa showed crypts with dysplasia of the ISC compartment at 8 weeks (Supplementary Fig. 15). Confocal imaging of each lineage demonstrated that there was sporadic recombination throughout the crypt and villus epithelium at day 3 (Fig. 7d). We found that at 8 weeks recombined cells had spread throughout the ROBOCreER/T2 intestinal epithelium and were therefore no longer constrained to a local clone within single crypts (Fig. 7e). This was in clear contrast to similarly performed experiments in MCATCreER/T2 mice (Fig. 4a–c). We quantified clone spreading by counting the total number of recombined cells at day 3 and 8 weeks. The results indicated oncogenic clones had significantly spread in ROBOCreER/T2 mice. Specifically, WT RSPO3 clones increased by 6.7-fold, and PTPRKe1:RSPO3e2–5 clones increased by 1.55-fold whereas PTPRKe1–7:RSPO32–5 clones decreased by 0.55-fold (Fig. 7f). In addition, the resultant crypts and clones were heterogenous, confirming that neutral drift and crypt fixation was also impaired in adults (Fig. 7g). Together, our data illustrate that microenvironmental oncogenesis can induce and rapidly spread heterogenous oncogenic clones in the adult intestine by increasing crypt fission, expanding the stem cell compartment, and inhibiting crypt fixation.
Discussion
The current study developed a mouse platform useful for modeling the oncogenic and cellular determinants of ISC competition and the clonal spread of somatic mutations throughout the intestine. Crainbow mouse models were used to induce somatic activations and then visualize the evolution and competition of premalignant epithelial clones by fluorescent imaging. The spatiotemporal activation of somatic mutations was used to identify the time and microenvironmental constraints present within the intestine that inhibit the clonal expansion of premalignant epithelium. Crainbow modelling revealed that the microenvironment inherently restricts this process to a susceptible period of intestinal development. However, in the adult, oncogenic activation of microenvironmental factors expands the stem cell compartment and leads to the rapid spread of oncogenic clones.
Crainbow lineage tracing experiments demonstrated that oncogenic activation of ßcat (MCAT-Crainbow) results in widespread expansion of premalignant clones during a critical period of perinatal development but not when activated in adult intestine (Fig. 8a). This illustrates how early APC inactivation can lead to increased tumor burden in adults14. The ISC compartment of mice is established during perinatal growth of the intestine and is initiated by the nascent formation of intestinal crypts48. The nascent crypts then multiply throughout the intestine by undergoing several rounds of crypt fission until a mature intestine is formed, at which time the rate of crypt fission slows15. MCAT Crainbow studies show that somatic activations can hijack the normal mechanics of intestinal growth and efficiently spread premalignant clones before adulthood. The growth of the human intestine occurs throughout the first 2 years of life and is also characterized by a sustained increase in crypt fission that markedly attenuates when the intestine reaches maturity11. Therefore, the infant’s environmental burdens, diet, and injury should be closely monitored to guard against the spread of oncogenic clones. Future work should identify additional genetic markers that could be used as noninvasive measures of premalignant clones to identify children with increased risk of colon cancer in adulthood.
Fig. 8.
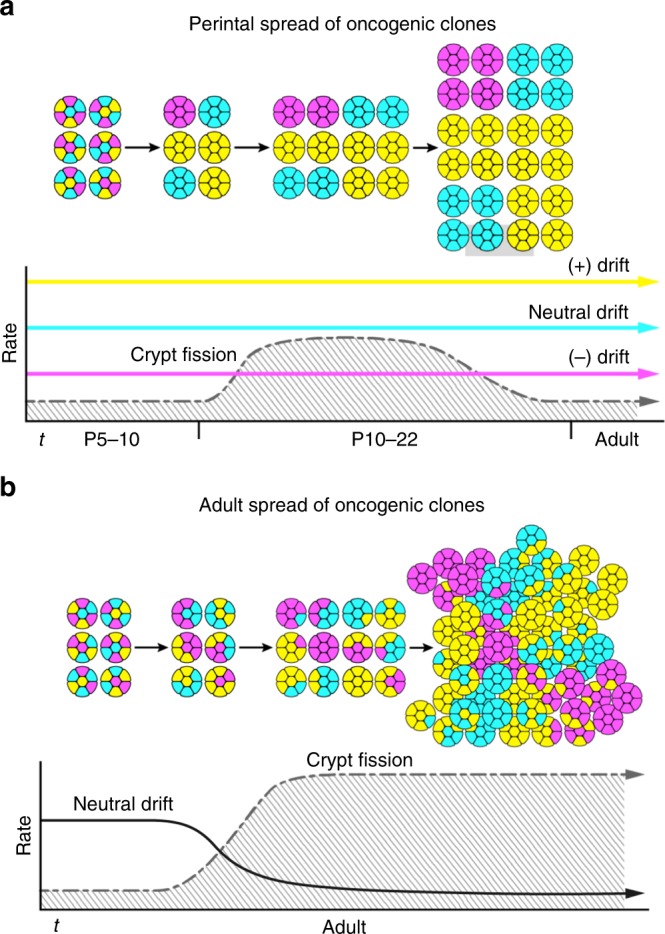
Models for the initiation and spread of oncogenic clones. a Early acquisition of somatic mutations during a critical period drives efficient spread of oncogenic clones before adulthood. Intestinal development selects for ISCs with increased fitness. This could bias crypt fixation (i.e., neutral drift dynamics) or crypt formation during development. Premalignant clones can then spread throughout the intestinal epithelium due to the high rate of crypt fission that is observed in mice (postnatal days 5–22, P5–22)15 and humans (0–2 years of age)11. b Microenvironmental oncogenesis induces crypt fission and inhibits crypt fixation (i.e., neutral drift dynamics) in the adult intestine. This results in an expanded stem cell compartment and the rapid spread of heterogeneous premalignant clones in the adult intestine.
The lack of adenomas in ßcat Crainbow mice (NCAT or MCAT) was surprising—especially since Harada et al.26 have shown previously that extensive intestinal polyps form when the N-terminal domain of endogenous ßcatenin is deleted. Our in vitro signaling assays confirmed ΔNßcat and Ccat/Lef1 isoforms were oncogenic as measured by Wnt/Rspondin independent TOP-FLASH activity. Interestingly, epithelial cadherin expression robustly inhibited the TOP-FLASH activity of ΔNßcat but not Ccat/Lef1. The in vivo subcellular localization of ΔNßcat and Ccat/Lef1 confirmed this finding by showing Ccat/Lef1 was localized to the nucleus whereas ΔNßcat was found at the lateral adherens junction. These data support the ßcatenin:Cadherin “sink” model previously proposed37. We conclude that the overexpression of ßcat in either MCAT or NCAT mice is unable to overcome the inhibition of Cdh1, further confirming the importance of Cdh1 as a tumor suppressor38. Additional complexity may also be due to the compensatory and/or competitive roles of the endogenous ßcat allele in the NCAT/MCAT Crainbow models.
MCAT organoid experiments provided additional insight into the spread of oncogenic clones by showing that Ccat/Lef1 expressing organoids outcompete other ßcat isoforms and grow in the absence of microenvironmental factors. Wnt deprivation in vivo was similarly shown in a recent study to lead to accelerated crypt fixation rates of pre-malignant ISCs12. Therefore, selective pressure, like injury or microenvironmental perturbations, could result in an evolutionary bottleneck, the loss of less-fit ISCs, and the dramatic expansion of a premalignant ISC population. The crypt microenvironment also maintains ISC homeostasis and is reorganized during crypt morphogenesis41,48. Therefore, oncogenes that restructure the crypt microenvironment can also spread rapidly in adults. RSPO3 was an obvious candidate due to its role as a secreted microenvironmental factor that controls crypt homeostasis and the recent discovery of its fusions with PTPRK in colon cancer16,17,49. Crainbow lineage tracing revealed that the oncogenic activation of RSPO3 (ROBO-Crainbow) expands the stem cell microenvironment. This leads to rapid spread of oncogenic clones in the adult intestine—a process we term microenvironmental oncogenesis (Fig. 8b). The altered crypt microenvironment results in ISC hyperplasia, increased crypt fission, and a decreased rate of crypt fixation. The reduction in crypt fixation demonstrates that crypt fixation is not required for the optimal spread of clones. Importantly, a reduced rate of crypt fixation also resulted in heterogeneous clones that spread throughout the intestine. This phenomenon begins to explain the genetic mechanisms underlying intratumor heterogeneity3.
Crainbow modelling was also used to compare gain-of-function activity for the PTPRK:RSPO3 fusion isoforms found in colon cancer17. Endogenous PTPRK is expressed throughout the intestinal epithelium (Supplementary Fig. 14), whereas the expression of endogenous RSPO3 is restricted to stromal cells residing adjacent to the crypt50 (Fig. 5a). The PTPRK:RPSO3 fusion results in juxtaposition of RPSO3 into the open reading frame of PTPRK and should therefore drive ectopic expression of RSPO3 and expand the microenvironment. However, each isoform may not be functionally equivalent. WT RSPO3 and the PTPRKe1:RSPO3e2−7 isoform both increased clone spreading potential, albeit at different rates. We were surprised to find an almost twofold decrease in clone spreading potential for the PTPRKe1:RSPO3e2−7 isoform. PTPRK is a transmembrane protein tyrosine phosphatase and through homophilic interactions on adjacent cells stabilizes cell adhesion and reduces epithelial invasiveness51. The PTPRKe1–7:RSPO32–5 fusion retains the Ig-like domain and the fibronectin-III repeat necessary for homophilic binding. Therefore, the loss of clone spreading potential for the PTPRKe1–7:RSPO32–5 fusion could be due to potential binding with endogenous PTPRK and a reduction in epithelial invasiveness. RSPO3 is also a secreted protein and may have autocrine and paracrine effects on adjacent stem cells and the microenvironment.
In recent work, breast cancer evolution and functional genomics was studied using a compound cross of five mouse lines52. The inability to resolve multiple mutations within the same tissue and obligate laborious breeding strategies limit the utility of such an approach. The major technical benefit of our work is to directly compare oncogenic mutations in the same model system. Current models do not easily support such experiments as they are unwieldy and not appropriately controlled. For instance, the comparison of multiple transgenic models is not recommended—transgene variegation, background strain, immunological variation, and genetic drift are just a few variables confounding the analysis. Crainbow is a transformative solution to this problem by enabling in vivo functional genomics at single cell resolution. Several imaging modalities and experimental workflows are compatible with Crainbow and together enable the direct visualization of stem cell fitness and premalignant clone competition within the intestine—together two key attributes of field cancerization. Field cancerization is a defining event of many cancers that include cancers of the breast, skin, and lung1. Crainbow modelling is easily adaptable and can be used to visualize the molecular and cellular attributes of field cancerization in any organ.
Several decades ago, Hubel and Wiesel demonstrated the importance of environmentally sensitive critical periods in the developing brain53. We broaden this paradigm to include critical periods of development sensitive to the acquisition and propagation of somatic mutations. The oncogenic activation of RSPO3 in adult intestine reactivates the developmental critical period and facilitates the widespread expansion of premalignant clones. The accelerated spread of oncogenic clones in the adult intestine provides a cellular and molecular mechanism for driving the rapid formation of colon cancers.
Methods
Crainbow mouse engineering
Infusion® (Takara, 638910) and Multisite Gateway™ Cloning (Life Technologies, 12537-100) techniques were used to engineer four customizable Gateway™ pENTR™ plasmids. All cloning steps were followed from supplier provided protocols. gBlocks® (Integrated DNA Technologies) or PCR products were used to construct positions 2–4 pENTRs with tripartite nuclear localization sequences immediately followed by a unique fluorescent protein terminated by a P2A54 cleavage sequence and a WPRE expression enhancing element25,55. Position one pENTR™ was engineered using a geneblock with Mars:Sci1 FAP56,57 followed by WPRE. Each gBlock® or PCR product was flanked by unique attB recombination sites for BP cloning to Gateway pDONR221 vectors specific to each position 1–4. The individual reactions were transformed to One Shot® Mach1 Competent cells (ThermoFisher C862003), and minipreps were Sanger sequenced to validate insert presence and sequence fidelity. Each construct after BP cloning became a pENTR™ vector with attL sites ready for LR cloning into any expression or targeting vector that contains the DEST recombination cassette. All pENTR™ vectors were whole plasmid sequenced at Massachusetts General Hospital Center for Computational & Integrative Biology DNA Core (https://dnacore.mgh.harvard.edu/new-cgi-bin/listing.action) and aligned in SnapGene (GSL Biotech LLC). Each pENTR™ was designed with a multiple cloning site containing a unique SnaBI restricition site for infusion cloning a gene of interest to pENTRs 2–4 between the P2A sequence and the WPRE element. Genes were PCR amplified or synthesized with SnaBI (ThermoFisher FD0404) compatible ends for Infusion cloning to SnaBI linearized pENTR™ vectors following supplier protocol. LR Clonase II® (Life Technologies, 11791-100) was used in an overnight reaction for Multisite Gateway™ Cloning into a ROSA26 mouse targeting vector22,23 that we previously adapted for Gateway cloning21. Max Efficiency® Stbl2™ competent cells were transformed (ThermoFisher Scientific 10268-019) and grown at 30 °C to reduce spontaneous recombination. Minipreps were isolated and restriction digestion strategies were used to map potentially positive clones. Maxipreps were grown from these miniprep cultures under the same conditions, and whole plasmid sequencing was performed. ROSA-Crainbow targeting vectors were linearized by restriction digestion and then purified by Phenol:Choloroform:Isoamyl Alcohol (ThermoFisher Scientific 15593031) extraction and Ethanol precipitation. Linearized DNA was resuspended in Tris-EDTA Buffer (1×, Sigma T9285) at 1 ug/uL. Mouse embryonic stem cell gene targeting and morulae injections were performed by the Duke Transgenic Mouse facility21.
Mouse husbandry and genotyping
Experiments performed in this study were conducted in accordance with an approved protocol for the ethical use of animals in research (Duke Institutional Animal Care and Use Committee protocols: A109-16-05, A079-16-04, and A034-19-02). Mice were housed in an AALAC certified specific pathogen free facility and given food and water ad libitum. Crainbow lines were either genotyped by PCR and by Mars1-SCi1 staining of toes (Supplementary Fig. 16). In addition to Crainbow lines, the following mouse lines were purchased from the Jackson Laboratory. ROSA FLPe—Jax-129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J (Stock No: 003946)58, Villin Cre—Jax-B6.Cg-Tg(Vil1-cre)997Gum/J (Stock No: 004586)27, ROSACreER—JAX-B6;129-Gt(ROSA)26Sortm1(cre/ERT)Nat/J (Stock No: 004847)59. Crainbow models and plasmids are available from the corresponding author upon reasonable request. Tamoxifen (Sigma, T5648) was resuspended in corn oil and delivered by intraperitoneal injection at 200 mg/kg.
Mouse organoid culturing
Mouse intestinal enteroids (organoids) were made according to established protocols60–62. Briefly, mice were sacrificed according to approved protocol and the entire intestine removed. The small intestine was cut at the intestine and cecum junction then cut in half to separate the proximal from the distal small intestine. The upper and proximal half (i.e., duodenum/jejunum) of the small intestine was flushed with 10–20 ml ice-cold PBS (Genesee 25–507) using a blunted syringe. The tissue was cut longitudinally and then cut into 2 cm pieces and washed with ice-cold PBS. Intestinal pieces were in 5 ml ice-cold 5 mM EDTA (Sigma E6758) while rotating slowly in a cold room for 30 min and then placed on ice for an additional 30 min. Tissue pieces were moved to a 15 ml tube (Supplier) containing 5 ml ice cold PBS. The tube was vigorously inverted 20–30 times, the tissue was collected and then transferred to the next 15 mL tube containing 5 ml ice-cold PBS. This process was repeated a total of five times. Ten microliters samples from each tube were assessed on a light microscope to find the fraction containing the highest density of crypts. The fraction(s) with the most crypts were filtered through a 70 µm filter (Fisher 22363548) into a BSA (10 mg/mL, Roche 10735086001)-coated conical. Isolated crypts were centrifuged at 400 g for 4 min and resuspended in ice-cold Matrigel (Corning 356237) at a concentration of 80 crypts/10 µl Matrigel. 10 µl crypt/matrigel suspension was plated per well on a 48 well cell culture plate (Corning 3548) that was prewarmed at 37 °C. The crypt/matrigel patty was polymerized by moving the plate to a 37 °C incubator for 30 min and then 200 uL base media (ENRY: EGF, Noggin, Rspondin-1, and Y-27632) was added per well. Briefly, base ENRY media was made in DMEM/F12 (Gibco 11330-032) with 1× Antibiotic/Antimycotic (Sigma A5955), 1× N-2 Supplement (Gibco 17502-048), 1× B-27 Supplement (Gibco 12587-010), 50 ng/mL EGF (Peprotech AF-100-15-100UG), 100 ng/mL Noggin (R&D Systems 1967-NG-025), 500 ng/mL Rspondin-1 (R&D Systems 4645-RS-025), and 10 uM Y-27632 (Cayman Chemical 10005583). Media was made and stored for up to one at 4 °C and changed every 2–3 days. Weekly subculture was performed by removing growth media, adding cold Gentle Dissociation Reagent (StemCell Technologies 07174), and pipetting until matrigel was dissociated from the plastic and then pipetted again until organoids were broken open to release dead cells from the organoid lumen. Dissociated organoids were centrifuged at 400 g for 4 min to pellet cells and washed once with ice cold 0.1% BSA in PBS. Organoids were pelleted by centrifugation at 400 g for 4 min and then resuspended in ice-cold matrigel. For experiments using the PORCN inhibitor Wnt-C59 (Cayman Chemical 16644), a stock solution was made at 1 mM in DMSO and then diluted to a final 10 nM in ENRY or ENY media.
Multiprobe RNA FISH
Advanced Cell Diagnostics RNAscope® Multiplex Fluorescent kit (323100) was used to identify mRNA targets in FFPE tissues sections from Crainbow mice. We followed the steps outlined in the user manual for FFPE sample preparation and pretreatment as well as for probe hybridization and signal development. In addition, we used dyes from the Opal® 7-color Kit (PerkinElmer, Inc NEL801001KT) for probe labelling as per company specifications. Confocal imaging was performed as described below. The following probes were used in this study: Mm-Alpi 436781, Mm-Rspo3 402011, mKO-fp 540071, EYFP-C2 312131-C2, mTFP1-C3 500271-C3, Mm-Top2a-C4 491221-C4, Mm-Lgr5-C3 312171-C3, and Mm-Reg3b-C2 478091-C2.
Single-cell RNAseq
We used the BioRad ddSEQTM single-cell isolator (BioRad 12004336) for single cell capture. Briefly, intestinal crypts were isolated according to standard organoid culturing protocol. Next, isolated crypts were incubated for approximately 1 h in 0.25% Trypsin (ThermoFisher Scientific 15090046) diluted in PBS (Genesee Scientific 25–507) with 1 mg/mL Collagenase/Dispase (Sigma 10269638001), 4 uL/mL DNase I solution (ThermoFisher Scientific 90083), and 1 uM Q-VD-OPH (Cayman Chemical 15260) with pipetting every 10 min to further dissociate cell clumps. Solution was neutralized by adding three volumes of 20% FBS diluted in PBS with Ca+ and Mg+ (Sigma D8662) and passed through a 20 um mesh filter. Cells were centrifuged 400 g for 4 min and resuspended in 1 mL 0.1% BSA + 1:250 anticlumping agent (ThermoFisher Scientific 01-0057AE). Viahance™ (Biopal, Inc CP-50VQ02) reagent was used to remove dead cells and debris by adding 100 uL to cell suspension and incubating first at room temperature for 5 min then on a magnet for 10 min. The cell suspension was removed and washed in 10 ml of PBS, centrifuged, and resuspended in 0.1% BSA for counting using Trypan Blue dye and BioRad TC20™ Automated Cell Counter. Single cells were brought to a concentration of 3000/µl and libraries generated using SureCell WTA 3′ Library Prep Kit (Illumina, San Diego, CA) according to company specification. Sequencing was performed according to company specification on an Illumina NextSeq 500 in hi-output mode. Data were preprocessed in BaseSpace Sequence Hub to build gene matrix files (Illumina, San Diego CA). The gene matrix files consisting of raw counts at the gene level for each cell which was analyzed using Rstudio and Seurat(ver.2)63. Briefly, the gene counts for all the cells in the different conditions were combined into one matrix, normalized, and adjusted for cell cycle effects. The cell cycle phase for each cell was also determined. Unsupervised clustering was done to separate out the cell types and markers for the cell types were identified using differential gene expression. These markers were then used for identifying the cell subpopulations such as stem cells and enterocytes.
Luminescent signaling assays
Experiments were performed using the HEK 293 STF stable cell line kindly provided by Dr. Jeremy Nathans and described in Xu Q, et al.39 (PMID 15035989). STF cells were cultured in DMEM (Corning 10-013-CV) supplemented with 10% FBS (Sigma F2442) and 1% antibiotic/antimycotic (Sigma A5995) and passaged to a Poly-D-Lysine coated 96 well plate at 75,000 cells per well. We used 3 ul of Genecellin (Bulldog Bio GC1000) per 1 ug of cDNA for transfection and added the mixture during cell plating. We used 100 ng/well Renilla Luciferase (RLuc) cDNA, 10 ng/well ßcatenin Crainbow cDNAs, 90 ng/well E-cadherin (CDH1, Addgene 45769), and used empty pcDNA3.1+ to equalize total amount of cDNA across different conditions. The media was changed 24 h after transfection to MEM (ThermoFisher Scientific 51200-038) supplemented with 10 mM HEPES (ThermoFisher Scientific 15630080) and 1% GlutaMAX (ThermoFisher Scientific 35050061). For stimulation 10% L Cell (ATCC CRL-2648) or L Cell Wnt-3A (ATCC CRL-2647) conditioned media was added to MEM with or without R spondin-3 (R&D Systems 3500-RS) to a final concentration of 83.3 ng/ml. Plates were assayed 24 h later using Luc-Pair™ Duo-Luciferase HS Assay Kit (GeneCopeia LF005) and read on a DLR compatible luminometer.
ßcatenin in vitro sequestration assays
STF cells were plated on Fibronectin (Millipore FC010) coated 35 mm glass bottom dishes (MatTek P35G-0-10-C) at 1 × 106 cells per dish and transfected 1 µg of indicated ßcatenin Crainbow plasmid +/− E-cadherin (CDH1) and 6 µl Genecellin as above. The next day media was changed to MEM with 10% L cell conditioned media. Cells were fixed at room temperature for 15 min in 4% PFA, washed once with PBS, and permeabilized/blocked in FISHX (0.25% Triton-X diluted in 1% Fish Gelatin(Rockland MB-067-0100)) for 20 min at room temperature. Primary antibodies (Anti-HA: Abcam ab9111, Anti-V5: ThermoFisher R960-25, and Anti-Flag M2: Sigma F1804) were diluted in FISHX at 1:500 and incubated at room temperature for 1 h. Samples were washed 3 times in PBS and incubated with appropriate antibody Alexafluor™-633 (ThermoFisher Goat anti-Chicken A-21103 or anti-Mouse A-21050) diluted 1:1000 FISHX for 1 h at room temperature.
Tissue isolation and histopathology
For whole-mount imaging, tissues were fixed in 10% NBF overnight. A 1 cm piece of intestine was cut open lengthwise and placed villus down on a slide with Vectashield mounting medium (Vector Laboratories Cat. No: H-1000). The slide (VWR Micro Superfrost Plus, Cat. No. 48311-703) had a 22 × 22 mm coverslip (VWR micro cover glass, 22 × 22 mm, Cat.No. 48366-227) taped on both ends so a large coverslip (VWR micro cover glass, 22 × 50 mm, Cat.No. 48393-195) could span over the tissue, compressing it enough to have a flat imaging plane. NCAT whole-mount imaging was performed on unfixed and freshly isolated tissue because fixation quenched the TagBFP signal. For cross-sections (70–100 um) intestine were sectioned on a compresstome® VF-300-0Z (Precisionary). For histology, tissues were fixed in 10% NBF overnight and embedded in paraffin at the UC Davis Center for Genomic Pathology Laboratory. Tissue sections (4 μm) were stained with Mayer’s hematoxylin and eosin (H&E) and whole slide imaging performed on an AT2 Scanscope (Leica Biosystems, Inc.) and stored in the Spectrum database (Leica). Images were viewed using Imagescope software (Leica).
Tissue staining
Antibody staining of vibratome slices was also performed for TagBFP and epitope tags as described below. For HA and V5 epitope tags, tissues were permeabilized with FISHX for 1 h, washed, and then blocked with Fc block (Innovex, NB309-5). Rb-anti-HA (Cell Signaling, Clone C29F4, Cat#3724) or Rb-anti-V5 (Cell Signaling, Clone D3H8Q, Cat#13202 s) were diluted 1:500 in FISHX and incubated overnight at 4 °C overnight, washed, and then blocked in FISHX with 5% Normal Goat Serum (NGS, Thermofisher 1610064) for 30 min. Tissues were then stained for 1.5 h with Alexa Fluor™-633 goat anti-rabbit (Thermofisher A21070) diluted 1:500 in FISHX/5%NGS. For FLAG immunostaining, Ms-IgG1-anti-FLAG (Sigma, F1804) was used at 1:250 and the protocol was similar to HA and V5 immunostaining but with the inclusion of a mouse-on-mouse blocking reagent (Jackson Immuno Research Laboratories, AffiniPure Fab Fragment Donkey Anti-Mouse IgG, 715-007-003) during tissue permeabilization and appropriate secondary antibody (Alexa Fluor™-633 goat anti-mouse IgG(g1), ThermoFisher A21126). TagBFP immunostaining was performed similar to HA and V5 staining but with a cross-reactive GuineaPig (GP) anti-TagRFP antibody (Kerafast EMU107) diluted 1:500 in FISHX and appropriate secondary antibody (Alexa Fluor™-633 goat-anti-guinea pig IgG(H + L), Thermofisher A21105). Stained tissue sections were mounted with Vectashield mounting medium (Vector Laboratories Cat. No: H-1000and imaged by confocal microscopy) on glass slides (VWR Micro Superfrost Plus, Cat. No. 48311-703) and appropriate coverslip (VWR micro cover glass, 22 × 22 mm, Cat.No. 48366-227) or (VWR micro cover glass, 22 × 50 mm, Cat.No. 48393-195). For in situ assessment of proliferation mice were I.P. injected with EdU (Sigma Cat. 900584, 50 mg/kg) 2 h before sacrifice. EdU staining was performed according to company protocol using a Click-iT™ Plus kit to prevent quenching of XFPs (Click-iT™ EdU Alexa Fluor™ 647 imaging kit, C10340).
Confocal microscopy and image analysis
Imaging was performed on a Zeiss LSM 880 confocal microscope equipped with seven lasers (405, 458, 488, 514, 561, 594, and 633 nm), 34 spectral array detectors (32 GaAsP and two PMTs). All lenses used Zeiss Plan-Apochromat, and they included 10×/0.45 NA, 20×/0.80 NA, 40×/1.20 NA Imm Corr, and 63×/1.40 NA Oil DIC. Dichroic beam splitters included: 458/514/561/633. Excitation laser and appropriate detector ranges were used for multispectral acquisition of XFPs and fluorescent stains. Images were processed and analyzed with Imaris (Bitplane), FIJI64, and the scikit-image65 Python package. Imaris was used to view images in 3D, adjust images for figures, and find and count spots, and in some cases count crypts. FIJI and the scikit-image package were used to build analysis pipelines for preprocessing data. Briefly, images were first preprocessed by subtracting channels where necessary to remove noise, followed by background subtraction, and gaussian or median filtering. Wholemount images were z-projected into one plane and either segmented by hand (crypt chimerism) or by thresholding and using a watershed morphological segmentation plugin (MorphoLibJ, IJBP-plugins66) in FIJI). Nuclei were segmented by intensity thresholding using the Spot module in IMARIS and organoids (Fig. 3c–e) were manually counted in IMARIS.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank Dr. Brigid Hogan for excellent discussions, editing, and review of this paper. We also thank Dr. David Virshup for thoughtful review of this paper. The authors also thank Dr. Alan S. Waggoner for the generous gift of SCi1 fluorogen. This work was supported by NCI K12 Training grant 512-CA100639-10, NCI 1K22CA212058, Sage Biosciences IACM Supplement 3U24CA209923-01S1, Duke Surgery Start-up funds to J.C.S. and the Duke Surgery Gardner Award to J.C.S. and H.K.L. J.D.G. is supported by an NICHD 5T32HD040372. This work was also supported by NCI Grants R21CA173245 and 1R33CA191198 to L.S.B, H.K.L, and M.G.C. This work was also supported by DOD grant W81XWH-12-1-0447 to H.K.L.
Source data
Author contributions
P.G.B. designed experiments, collected data, and analyzed data. L.K.R. designed experiments, collected data, and analyzed data. V.L. collected data and analyzed data. W.L.R. maintained the mouse colony. P.J.N. provided analysis support and instrumentation support. C.B. and M.F. conducted ES gene targeting. R.J.F. provided expertise for organoid growth. J.D.G analyzed data and interpreted data. B.R.S. designed experiments and discussed data. P.A. performed bioinformatics and scRNAseq data analysis. A.D.B. and R.D.C. performed histopathology. L.S.B., M.G.C., H.K.L., and J.C.S. conceptualized Crainbow and designed experiments. J.C.S. performed experiments, analyzed data, and wrote the paper.
Data availability
The scRNAseq datasets analyzed in the current study are available at GEO, accession GSE132402. The source data underlying Figs. 2d, g, 3d, g, 4c, f, i, j, 6f, and 7b are provided as Source Data file. The data supporting this study are available in the Article, Supplementary Information, Source Data and available from the authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peter G. Boone, Lauren K. Rochelle.
These authors jointly supervised this work: Larry S. Barak, Marc G. Caron, H. Kim Lyerly, Joshua C. Snyder.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-019-13330-y.
References
- 1.Curtius K, Wright NA, Graham TA. An evolutionary perspective on field cancerization. Nat. Rev. Cancer. 2018;18:19–32. doi: 10.1038/nrc.2017.102. [DOI] [PubMed] [Google Scholar]
- 2.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 3.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galandiuk S, et al. Field cancerization in the intestinal epithelium of patients with Crohn’s ileocolitis. Gastroenterology. 2012;142:855–864 e858. doi: 10.1053/j.gastro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jothy S, et al. Field effect of human colon carcinoma on normal mucosa: relevance of carcinoembryonic antigen expression. Tumour Biol. 1996;17:58–64. doi: 10.1159/000217967. [DOI] [PubMed] [Google Scholar]
- 6.Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014;15:62–69. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicholson AM, et al. Fixation and spread of somatic mutations in adult human colonic epithelium. Cell Stem Cell. 2018;22:909–918 e908. doi: 10.1016/j.stem.2018.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasan HS, et al. APC in the regulation of intestinal crypt fission. J. Pathol. 1998;185:246–255. doi: 10.1002/(SICI)1096-9896(199807)185:3<246::AID-PATH90>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 9.Baker AM, et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014;8:940–947. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 11.Cummins AG, et al. Crypt fission peaks early during infancy and crypt hyperplasia broadly peaks during infancy and childhood in the small intestine of humans. J. Pediatr. Gastroenterol. Nutr. 2008;47:153–157. doi: 10.1097/MPG.0b013e3181604d27. [DOI] [PubMed] [Google Scholar]
- 12.Huels DJ, et al. Wnt ligands influence tumour initiation by controlling the number of intestinal stem cells. Nat. Commun. 2018;9:1132. doi: 10.1038/s41467-018-03426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, Bjerknes M, Amar J, Gardiner G. Crypt production in normal and diseased human colonic epithelium. Anat. Rec. 1986;216:44–48. doi: 10.1002/ar.1092160108. [DOI] [PubMed] [Google Scholar]
- 14.Fischer JM, Schepers AG, Clevers H, Shibata D, Liskay RM. Occult progression by Apc-deficient intestinal crypts as a target for chemoprevention. Carcinogenesis. 2014;35:237–246. doi: 10.1093/carcin/bgt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dehmer JJ, et al. Expansion of intestinal epithelial stem cells during murine development. PLoS One. 2011;6:e27070. doi: 10.1371/journal.pone.0027070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilkens J, et al. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut. 2017;66:1095–1105. doi: 10.1136/gutjnl-2016-311606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han T, et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 2017;8:15945. doi: 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 20.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat. Methods. 2013;10:540–547. doi: 10.1038/nmeth.2450. [DOI] [PubMed] [Google Scholar]
- 21.Snyder Joshua C., Rochelle Lauren K., Ray Caroline, Pack Thomas F., Bock Cheryl B., Lubkov Veronica, Lyerly H. Kim, Waggoner Alan S., Barak Larry S., Caron Marc G. Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled receptor-5 diminishes cell fitness. Journal of Biological Chemistry. 2017;292(17):7208–7222. doi: 10.1074/jbc.M116.756635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 24.Cheo DL, et al. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada N, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 29.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 31.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 32.Su LK, Vogelstein B, Kinzler KW. Association of the APC tumor suppressor protein with catenins. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 33.Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 1998;18:4807–4818. doi: 10.1128/MCB.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vleminckx K, Kemler R, Hecht A. The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis. Mech. Dev. 1999;81:65–74. doi: 10.1016/S0925-4773(98)00225-1. [DOI] [PubMed] [Google Scholar]
- 35.Cong F, Schweizer L, Chamorro M, Varmus H. Requirement for a nuclear function of beta-catenin in Wnt signaling. Mol. Cell. Biol. 2003;23:8462–8470. doi: 10.1128/MCB.23.23.8462-8470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theodosiou NA, Tabin CJ. Wnt signaling during development of the gastrointestinal tract. Dev. Biol. 2003;259:258–271. doi: 10.1016/S0012-1606(03)00185-4. [DOI] [PubMed] [Google Scholar]
- 37.Fagotto F, Funayama N, Gluck U, Gumbiner BM. Binding to cadherins antagonizes the signaling activity of beta-catenin during axis formation in Xenopus. J. Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Q, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/S0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 40.Langlands AJ, et al. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLoS Biol. 2016;14:e1002491. doi: 10.1371/journal.pbio.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabiri Z, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 43.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 44.Yan KS, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moor AE, et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell. 2018;175:1156–1167 e1115. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 46.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KA, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 48.Guiu Jordi, Hannezo Edouard, Yui Shiro, Demharter Samuel, Ulyanchenko Svetlana, Maimets Martti, Jørgensen Anne, Perlman Signe, Lundvall Lene, Mamsen Linn Salto, Larsen Agnete, Olesen Rasmus H., Andersen Claus Yding, Thuesen Lea Langhoff, Hare Kristine Juul, Pers Tune H., Khodosevich Konstantin, Simons Benjamin D., Jensen Kim B. Tracing the origin of adult intestinal stem cells. Nature. 2019;570(7759):107–111. doi: 10.1038/s41586-019-1212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 50.Greicius G, et al. PDGFRalpha(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA. 2018;115:E3173–E3181. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fearnley, G. W. et al. The homophilic receptor PTPRK selectively dephosphorylates multiple junctional regulators to promote cell-cell adhesion. Elife8, 10.7554/eLife.44597 (2019). [DOI] [PMC free article] [PubMed]
- 52.Rios AC, et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D imaging. Cancer Cell. 2019;35:618–632 e616. doi: 10.1016/j.ccell.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 53.Hubel DH, Wiesel TN. The period of susceptibility to the physiological effects of unilateral eye closure in kittens. J. Physiol. 1970;206:419–436. doi: 10.1113/jphysiol.1970.sp009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mastroyiannopoulos NP, Feldman ML, Uney JB, Mahadevan MS, Phylactou LA. Woodchuck post-transcriptional element induces nuclear export of myotonic dystrophy 3′ untranslated region transcripts. EMBO Rep. 2005;6:458–463. doi: 10.1038/sj.embor.7400390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snyder JC, et al. A rapid and affordable screening platform for membrane protein trafficking. BMC Biol. 2015;13:107. doi: 10.1186/s12915-015-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Ming, Chakraborty Subhasish K., Sampath Padma, Rojas Juan J., Hou Weizhou, Saurabh Saumya, Thorne Steve H., Bruchez Marcel P., Waggoner Alan S. Fluoromodule-based reporter/probes designed for in vivo fluorescence imaging. Journal of Clinical Investigation. 2015;125(10):3915–3927. doi: 10.1172/JCI81086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. doi: 10.1002/1526-968X(200011/12)28:3/4<106::AID-GENE30>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 59.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- 61.Seiler KM, et al. Tissue underlying the intestinal epithelium elicits proliferation of intestinal stem cells following cytotoxic damage. Cell Tissue Res. 2015;361:427–438. doi: 10.1007/s00441-015-2111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoenborn Alexi A., von Furstenberg Richard J., Valsaraj Smrithi, Hussain Farah S., Stein Molly, Shanahan Michael T., Henning Susan J., Gulati Ajay S. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes. 2018;10(1):45–58. doi: 10.1080/19490976.2018.1474321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Walt S, et al. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The scRNAseq datasets analyzed in the current study are available at GEO, accession GSE132402. The source data underlying Figs. 2d, g, 3d, g, 4c, f, i, j, 6f, and 7b are provided as Source Data file. The data supporting this study are available in the Article, Supplementary Information, Source Data and available from the authors upon reasonable request.