Abstract
Background
V600EBRAF mutated metastatic colorectal cancer (mCRC) is a subtype (10%) with overall poor prognosis, but the clinical experience suggests a great heterogeneity in survival. It is still unexplored the real distribution of traditional and innovative biomarkers among V600EBRAF mutated mCRC and which is their role in the improvement of clinical prediction of survival outcomes.
Methods
Data and tissue specimens from 155 V600EBRAF mutated mCRC patients treated at eight Italian Units of Oncology were collected. Specimens were analysed by means of immunohistochemistry profiling performed on tissue microarrays. Primary endpoint was overall survival (OS).
Results
CDX2 loss conferred worse OS (HR = 1.72, 95%CI 1.03–2.86, p = 0.036), as well as high CK7 expression (HR = 2.17, 95%CI 1.10–4.29, p = 0.026). According to Consensus Molecular Subtypes (CMS), CMS1 patients had better OS compared to CMS2-3/CMS4 (HR = 0.37, 95%CI 0.19–0.71, p = 0.003). Samples showing less TILs had worse OS (HR = 1.72, 95%CI 1.16–2.56, p = 0.007). Progression-free survival analyses led to similar results. At multivariate analysis, CK7 and CMS subgrouping retained their significant correlation with OS.
Conclusion
The present study provides new evidence on how several well-established biomarkers perform in a homogenousV600EBRAF mutated mCRC population, with important and independent information added to standard clinical prognosticators. These data could be useful to inform further translational research, for patients’ stratification in clinical trials and in routine clinical practice to better estimate patients’ prognosis.
Subject terms: Colorectal cancer, Prognostic markers
Background
V600EBRAF mutation is detected in 8–12% of colorectal cancer (CRC) patients, accounting for more than 90% of CRC BRAF mutations.1 It is an independent negative prognostic factor in CRC across all stages.2,3 Furthermore, a recent consensus work identified BRAF mutational status as one of the top five fundamental stratification characteristics in the initial evaluation of metastatic CRC (mCRC) patients, together with RAS mutations, patients’ performance status, primary tumour sidedness and presence of liver-limited disease.4
Despite the evidence of its prognostic significance, great heterogeneity in survival outcome is evident among V600EBRAF mutated mCRC.5 Indeed, some patients with V600EBRAF mutated mCRC may experience prolonged survival and durable response to therapies, while other patients develop rapid resistance.6 These observations led to the hypothesis that a better stratification based on clinical and molecular features should be explored when considering V600EBRAF mCRC as a separate disease. To achieve this goal, correct methodology, homogeneous patients’ cohorts and adequate sample size are of crucial importance. We recently proposed a clinical risk score prognostic calculator based on ECOG PS, tumour grading, presence of liver metastases, presence of lung metastases and presence of nodal involvement, CA19.9, CEA, LDH levels and neutrophils/lymphocytes ratio.7 Moreover, a ‘simplified’ version based only on the first five covariates was subsequently developed as functional and reliable tool for multivariate modelling of translational analyses.
Caudal type homeobox 2 (CDX2) is a gene encoding a protein involved in cell differentiation, adhesion and polarity. It has been hypothesised that V600EBRAF mutation and loss of CDX2, which are significantly associated, might cooperate in promoting CRC tumorigenesis.8,9 Dalerba et al. demonstrated that loss of CDX2 expression may confer poor prognosis to stage II-III CRC patients.10 Notwithstanding, no or limited information regarding the relative impact of BRAF mutations and other prognostic features were available.11
CRC has been classically associated to a CK20-positive and CK7-negative profile.12,13 Literature data suggest that among V600EBRAF mutated CRC, a higher prevalence of CK20-negative tumours may be found.14,15
Most importantly, up to 30% of V600EBRAF mutated cases show microsatellite instability (MSI). In early stages, V600EBRAF mutated microsatellite stable (MSS) CRC have a poorer prognosis16–18;conversely, the prognostic impact of microsatellite instability-high (MSI-H) status in V600EBRAF mutated patients is still debated. Venderbosch et al.2 retrospectively analysed a large cohort of 3063 patients from four different studies aiming to describe mutual influence on prognosis of microsatellite instability in BRAF mutated stage IV CRC and vice-versa. The prognostic influence of V600EBRAF mutation in MSS was confirmed, but other definitive conclusions were limited by excessive subgrouping. Another retrospective study including only 14 V600EBRAF mutated patients out of 55 MSI-H cases suggested a negative impact of V600EBRAF mutation in MSI-H patients, but again small sample size limited any reliable consideration on the prognostic impact of microsatellite instability among V600EBRAF mutated patients.19
A remarkable step forward in the description of CRC heterogeneity has been made by the Consensus Molecular Subtypes (CMS).20 The majority of V600EBRAF (up to 70%) are classified into CMS1 subgroup, while 7 and 17% are grouped in CMS2-3 and CMS4, respectively. This heterogeneous distribution supports the rationale for exploring the prognostic relevance of CMS subgrouping among V600EBRAF mCRC.
Another active field of interest in the definition of mCRC prognosis is the presence of tumour infiltrating lymphocytes (TILs). So far, no specific studies are available in literature concerning their role in V600EBRAF mutated tumours.
Finally, Barras et al. categorised V600EBRAF mutated CRC into two groups based on gene expression signatures: BM1 patients, accounting for approximately one third of cases, show activation of KRAS/mTOR/AKT/4EBP1 pathway, while BM2 group is characterised by dysregulation in the cell-cycle.5
Given the above reported assumptions, the aim of our work is to investigate the prognostic role of the most important and biologically sound prognostic markers in a large set of V600EBRAF mutated mCRC patients, in order to better explain the wide inter-patient heterogeneity observed in routine clinical practice.
Methods
Clinical and molecular data of V600EBRAF mutated mCRC patients referred to eight Italian Oncology Units between January 2005 and December 2016 were collected. In particular, for each patient data on demographic, tumour characteristics, 1st line systemic treatment, locally assessed RECIST1.1 response and survival were retrieved. The study received Ethics approval from the Coordinating centre, i.e. Istituto Oncologico Veneto IOV, Principal Investigator Dr Fotios Loupakis and was subsequently approved by each centre according to Italian national regulations code 2017/34. Cases were deemed eligible if clinical data and archival tissue either of primary tumour and/or metastases were available.
Available primary and/or metastatic formalin-fixed paraffin-embedded (FFPE) surgical samples were processed using the Galileo CK3500 Arrayer (www.isenet.it), a semiautomatic and computer-assisted Tissue microarray (TMA) platform. Tissue cores (3 cores per sample; 1 mm in diameter) were obtained from each primary and metastatic lesion, respectively. Small biopsy samples were processed separately. Immunohistochemical stainings were automatically performed using the Bond Polymer Refine Detection kit (Leica Biosystems, Newcastle Upon Tyne, UK) in the BOND-MAX system (Leica Biosystems) on 4 μm-thick sections. Primary antibodies, dilutions and scoring evaluation are available upon request. Specific methods and scoring systems for each marker are reported below.
CDX2
CDX2 expressions values were defined according to H-score, defined as the aggregate of total percentage of tumour cells expressing CDX2 at each particular intensity level from 0, +1 (weak intensity), +2 (moderate intensity) or +3 (strong intensity). In brief, the H-score was defined as: (Percent of CDX2 1 + tumour cells multiplied by intensity of 1) + (Percent of CDX2 2 + tumour cells multiplied by intensity of 2) + (Percent of CDX2 3 + tumour cells multiplied by intensity of 3). Thus, this composite score can range from 0 (a tumour which is completely negative) to a maximum of 300 (a tumour in which all the cells feature a 3+ staining). CDX2 results were split in tertiles as follows: 0–24 (low expression), 25–120 (intermediate expression), 121–300 (high expression).
Cytokeratins
Cytokeratin expression pattern was evaluated by CK7 and CK20 expression. CK7 expression was categorised in low (values 0–1) and high (values 2–3), whereas CK20 expression was indicated as negative (no expression) or positive (values 1, 2 or 3), according to staining intensity in more than 10% of cancer cells.
MSI-H status was defined in the absence of nuclear immunostaining for one of the couples MLH1/PMS2 or MSH2/MSH6 in tumour cell. The diagnostic performance of immunohistochemistry in identifying MSI-H cases was tested by microsatellite analysis (Titano kit, Diatech Pharmacogenetics) in a series of 20 MMRd and 20 MMRp tumours.21
Consensus molecular subtypes
CMS were assigned by assessing four IHC markers (FRMD6, ZEB1, HTR2B, CDX2) in combination with pan-cytokeratin (KER) to normalise results as reported in literature.22 Primary tumours and/or metastasis were then categorised into the 3 CMS classes (CMS1, CMS2/3 or CMS4) using the online classification tool (https://crcclassifier.shinyapps.io/appTesting). As previously described, MSI status was first used to define patients which belong to the CMS1 subtype.22,23
Tumour infiltrating lymphocytes
Presence of TILs was evaluated on haematoxylin and eosin (H&E) stained slides and dichotomised by using a cut-off of 2.0: low number of TILs for tumours showing an average number of TILs <2.0, high number of TILs for tumours with ≥2.0 TILs.24
BM1 and BM2 subgroups
To stratify tumours according to Barras et al.5 in BM1 and BM2 groups, we exploratively categorised each tumour based on the presence/absence of these five markers: CDK1, ATM, Phospho-Akt (Ser473), Cyclin D1 and Phospho-4E-BP1 (Thr70). Since BM1 is characterised by activation of PI3K/mTOR/AKT pathway, while BM2 of cell cycle pathway we assigned samples to BM1 or BM2 based on the coherence of the following parameters. Overexpression of Phospho-Akt, Phospho-4E-BP1, ATM and Cyclin D1 and downregulation of CDK1 were consistent with a BM1 profile. On the other hand, BM2 cases were characterised by overexpression of CDK1 and downregulation of the remaining markers. A tumour was considered positive in ATM if >10% of tumour cells were positive for nuclear ATM staining. The activation of the AKT/4E-BP1 cascade was defined in the presence of high expression levels of the phosphorylated forms of AKT and/or 4E-BP1. High levels of Cyclin D1 and CDK1 expression were defined in the presence of at least 50% of cancer cells positive (Cyclin D1 in the nucleus, CDK1 both in the nucleus and cytoplasm). Samples with 4 or 5 coherent parameters were defined as BM1 or BM2, whereas tumours in which 3 out of 5 parameters were coherent with the hypothesis were defined as borderline BM1 or BM2. Tumours with only 1 or 2 parameters coherent with the original classification were defined as not evaluable.
Clinical score
Simplified score for estimating the prognostic impact of major clinical and pathological was calculated considering 5 parameters as previously described7: grading, ECOG PS at diagnosis of metastatic disease and sites of metastases at diagnosis (liver, lung, nodes). To calculate the score the following criteria were applied: ECOG PS 0 = 0 points; ECOG PS 1 = 2 points; ECOG PS 2–3 = 3 points. Tumour grading 1 or 2 = 0 points, tumour grading 3 or 4 = 1 point. Presence of liver metastases = 1 point; presence of lung metastases = 2 points, presence of distant nodes metastases = 2 points. The score is calculated as the total sum of points. Patients were classified as ‘low-risk’ if they had a score ranging from 0 to 2; they were classified as ‘intermediate-risk’ if the score was 3 or 4; they were classified as ‘high-risk’ if their score ranged from 5 to 9.
Statistical analysis
The primary endpoint of the present analysis was Overall Survival (OS) for each variable analysed. OS was defined as time from metastatic disease diagnosis to death due to any cause. Secondary endpoints included: progression free survival (PFS) for each variable (PFS was defined as the time from 1st line treatment start date to 1st progression); the reproducibility of BM1/BM2 subgrouping and CMS distribution as assigned by means of IHC/TMA. For each determinant, comparison between samples from primary tumour and metastatic lesions was performed in order to explore their concordance.
Both OS and PFS and 95%CI were calculated using Kaplan–Meier method. Cox proportional Hazard model was adopted in the multivariate analysis including all covariates significantly correlated with survival in the univariate analysis.
PFS and OS were calculated in univariate analysis for the following molecular factors: CDX2, CK7, CK20 expression, CMS groups, BM1/BM2 groups and presence of TILs. Factors found significant at univariate analysis were included into the multivariate analysis for both OS and PFS including clinical score data as covariate.
Results
A total of 155 patients were included. Males and females were equally represented (50.3%/49.7%, respectively). As expected, frequent features were: right-sidedness of primary tumour (74.2%), presence of synchronous metastases (65.8%), hepatic or nodal involvement (53 and 38% respectively), with 63% of patients having a single metastatic site at the time of stage IV disease diagnosis. A large proportion of patients had previous primary tumour resection (87.1%). Baseline characteristics and major clinical parameters are summarised in Table 1.
Table 1.
Baselinea characteristics and major clinical parameters
| Characteristic | TOT = 155 |
|---|---|
| N (%) | |
| Sex | |
| Female | 77 (49.7%) |
| Male | 78 (50.3%) |
| Age | |
| Median (range) | 66 (28–85) |
| Age | |
| >70 | 55 (35.5%) |
| ≤70 | 100 (64.5%) |
| Baseline ECOG PS | |
| 0 | 112 (72.2%) |
| 1 | 37 (23.9%) |
| ≥2 | 6 (3.9%) |
| Primary tumour resected | |
| Yes | 135 (87.1%) |
| No | 20 (12.9%) |
| Primary tumour location | |
| Right | 115 (74.2%) |
| Left | 31 (20.0%) |
| Rectal | 9 (6.8%) |
| Presentation of metastases | |
| Synchronous | 102 (65.8%) |
| Metachronous | 53 (34.2%) |
| Number of metastatic sites | |
| Single | 97 (63%) |
| Multiple | 57 (37%) |
| Missing | 1 |
| Sites of metastases at diagnosis | |
| Liver | 82 (53%) |
| Lung | 27 (17.4%) |
| Distant nodes | 59 (38%) |
| Other | 28 (18.1%) |
| Missing | 1 |
ai.e. at the time of first-line treatment start or, for candidates to BSC only, at the first visit for metastatic disease
The vast majority of patients (89%) received at least one treatment for metastatic disease: first-line treatment was monochemotherapy ± a biologic agent (anti-EGFR monoclonal antibody or bevacizumab) in 9.4% of treated patients, doublet ± a biologic agent in 55.8%, triplet ± a biologic agent in 24.6%, immunotherapy in 7.3% and anti-BRAF treatment in 2.2%.
Distribution of molecular variables analysed in whole population is shown in Table 2, correlation data between paired single parameters are reported in Supplementary Table 1. For 46 patients, paired primary and metastasis samples were available: data obtained from IHC analyses were concordant in most cases, as shown in Supplementary Table 2.
Table 2.
Distribution of molecular variables analysed in whole population
| Characteristic | TOT = 155 |
|---|---|
| N (%) | |
| CDX2 | |
| Low | 47 (32.6%) |
| Intermediate | 50 (34.8%) |
| High | 47 (32.6%) |
| NEa | 11 |
| CK7 | |
| Low | 81 (87.1%) |
| High | 12 (12.9%) |
| NE | 9 |
| Not tested | 53 |
| CK20 | |
| Low | 11 (11.8%) |
| High | 82 (88.2%) |
| NEa | 9 |
| Not tested | 53 |
| CMS | |
| 1 | 44 (39.7%) |
| 2–3 | 47 (42.3%) |
| 4 | 20 (18.0%) |
| NEa | 44 |
| TILs | |
| Low | 61 (39.6%) |
| High | 93 (60.4%) |
| NEa | 1 |
| Barras et al. subtypes | |
| BM1 | 51 (49%) |
| BM2 | 53 (51%) |
| NEa | 51 |
| Clinical prognostic scoreb | |
| Low | 69 (44.8%) |
| Intermediate | 59 (38.3%) |
| High | 26 (16.9%) |
| NEa | 1 |
aNot evaluable
bSimplified version
After a median follow-up of 27.9 months (95%CI 20.3–35.5), 104 patients (67.1%) died. Median OS of the whole population was 18.5 months (95%CI 13.3–23.7), median PFS from the beginning of the first line treatment was 7.6 months (95%CI 5.2–10.0).
Univariate analyses
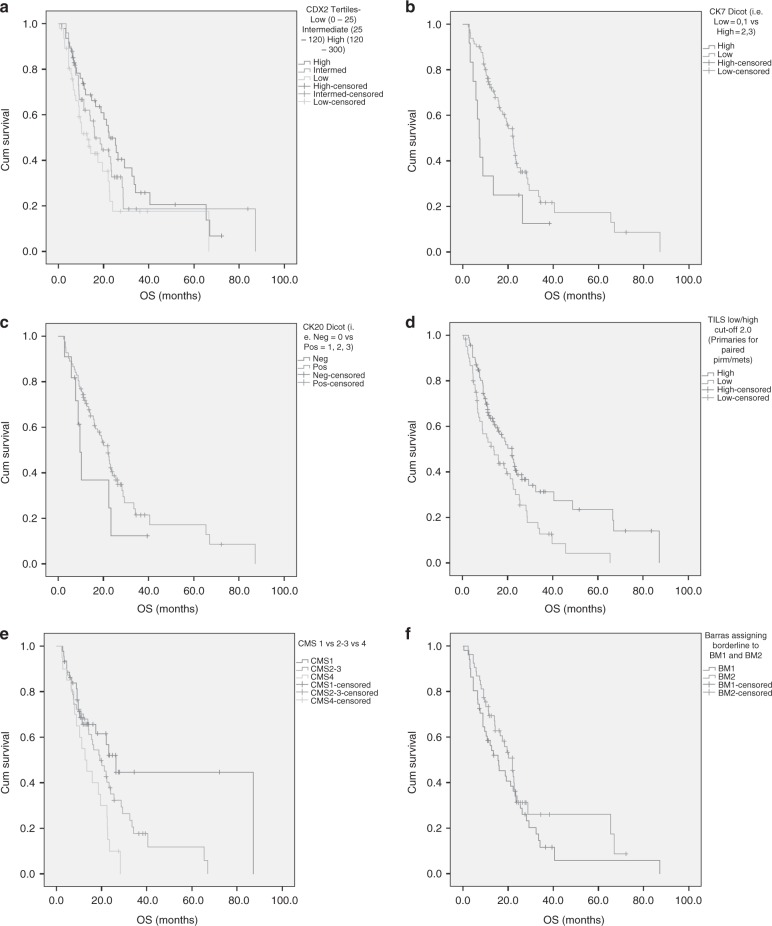
Results on OS and PFS are reported in Table 3 and Supplementary Table 3 and graphically represented in Fig. 1 and Supplementary Fig. 1, respectively and described below for each single variable.
Table 3.
Univariate analysis for overall survival
| Characteristics | Median OS (months) | Overall survival | ||
|---|---|---|---|---|
| HR | 95% CI | p | ||
| CDX2 | ||||
| High | 22.3 | 1 | – | – |
| Intermediate | 16 | 1.72 | 1.03–2.86 | 0.036 |
| Low | 12.7 | |||
| CK7 | ||||
| Low | 22.3 | 1 | – | – |
| High | 7.2 | 2.17 | 1.10–4.29 | 0.026 |
| CK20 | ||||
| Pos | 22 | 1 | – | |
| Neg | 9.7 | 1.75 | 0.83–3.69 | 0.14 |
| CMS | ||||
| 1 | 26.3 | 1 | – | – |
| 2–3 | 19.2 | 2.7 | 1.41–5.26 | 0.003 |
| 4 | 12.7 | |||
| TILs | ||||
| High | 22 | 1 | – | – |
| Low | 13.9 | 1.72 | 1.16– 2.56 | 0.007 |
| BM | ||||
| 2 | 22 | 1 | – | – |
| 1 | 15.6 | 1.37 | 0.87–2.17 | 0.177 |
| Simplified score | ||||
| Low | 23.3 | 1 | – | – |
| Intermediate | 19.5 | |||
| High | 6.6 | 2.61 | 1.53–4.48 | <0.001 |
Bold values indicate statistical significance p < 0.05
Fig. 1.
Kaplan–Meier curves for Overall survival (OS). a CDX2 tertiles expression (low vs intermediate and high). b CK7 expression (high vs low). c CK20 expression (negative vs positive). d TILs expression (low vs high). e CMS classification (CMS2/3 and CMS4 vs CMS1). f Barras classification (BM1 vs BM2)
CDX2
Patients with low or intermediate expression had a shorter OS compared to patients with high expression (HR = 1.72, 95%CI 1.03–2.86, p = 0.036). Similar trend, but no significant differences were detected in terms of PFS (HR = 1.41, 95%CI 0.86–2.30, p = 0.169).
CK7–CK20
Patients with higher CK7 expression had a shorter OS compared to patients with lower CK7 expression (HR = 2.17, 95%CI 1.10–4.29, p = 0.026). No significant differences were detected in terms of PFS (HR = 1.13, 95%CI 0.56–2.29, p = 0.74). Patients with negative CK20 had a shorter OS compared to patients with positive CK20 (HR = 1.75, 95%CI 0.83–3.69, p = 0.14). Similar trend, but no significant differences were detected in terms of PFS (HR = 1.72, 95%CI 0.73–4.05, p = 0.21).
CMS
CMS2-3 or CMS4 patients had a shorter OS compared to CMS1 (HR = 2.70, 95%CI 1.41–5.26, p = 0.003), similar results were reported for PFS (HR = 2.22, 95%CI 1.14–4.35, p = 0.02).
TILs
Patients with low TILs levels had a shorter OS compared to patients with high levels (HR = 1.72, 95%CI 1.16–2.56, p = 0.007), results confirmed also in PFS (HR = 1.72, 95%CI 1.18–2.56, p = 0.005).
BM1/BM2
No significant differences between BM1 and BM2 patients were detected in terms of OS (HR = 1.37, 95%CI 0.87–2.17, p = 0.18), or PFS (HR = 1.27, 95%CI 0.80–2.03, p = 0.31).
Clinical score
As expected, patients with high score had a shorter OS compared to patients with intermediate or low score (HR = 2.61, 95%CI 1.53–4.48, p < 0.001), similar results were obtained in terms of PFS (HR = 2.13, 95%CI 1.21–3.75, p = 0.009).
Multivariate analysis
Since the clinical score was built on OS data, it was included to adjust for integrating clinical and molecular prognostication. At multivariable analysis, CK7 overexpression was independently associated with worse OS with a HR of 2.11 (95%CI 1.03–4.34, p = 0.041), as was CMS2-3 and 4 over CMS1, with a HR of 2.22 (95%CI 1.03–5.02, p = 0.049). Complete data are reported in Table 4. The poor prognostic score that was determined clinically retained an independent prognostic impact (HR = 2.42, 95%CI 1.16–5.05, p = 0.019).
Table 4.
Multivariate analysis for overall survival
| Characteristics | Overall survival | ||
|---|---|---|---|
| HR | 95%CI | p | |
| CDX2 | |||
| High | 1 | – | – |
| Low + Intermediate | 1.92 | 0.94–4.00 | 0.07 |
| CK7 | |||
| Low | 1 | – | – |
| High | 2.11 | 1.03–4–34 | 0.041 |
| CMS | |||
| 1 | 1 | – | – |
| 2–3 + 4 | 2.22 | 1.03–5.02 | 0.049 |
| TILs | |||
| High | 1 | – | – |
| Low | 1.19 | 0.66–2.17 | 0.55 |
| Simplified score | |||
| Intermediate + Low | 1 | – | – |
| High | 2.42 | 1.16–5.05 | 0.019 |
Bold values indicate statistical significance p < .05
Similarly, in multivariate analysis for PFS, CMS2-3 and 4 were significant determinants of worse outcome over CMS1 (HR 2.17, 95%CI 1.01–4.76, p = 0.049). Complete data are reported in Supplementary Table 4
Discussion
In the present study, we explored and clarified the prognostic role of CDX2, CK7 and 20, TILs, CMS and BM1/BM2 subtypes in V600EBRAF mutated mCRC with a modern multivariate model including a validated clinical prognostic score as covariate. For each variable, we verified the impact on OS and secondarily on PFS. Moreover, level of concordance between primary tumours and metastatic sites was studied for each parameter.
Major findings were: CDX2 loss, high CK7 expression, less TILs, CMS2-3 or CMS4 (compared to CMS1) conferred worse OS. Progression-free survival analyses led to similar results. At multivariate analysis, CK7 and CMS subgrouping retained their significant correlation with OS.
CRC is usually associated to a CK7 negative and CK20/CDX2 positive profile.12,13 Notwithstanding, some evidence in literature suggests that in V600EBRAF mutated mCRC this profile may be different, with a higher prevalence of CK20/CDX2 negative tumours.14,15 On the other hand, it has never been properly explored how the cytokeratins’ profile affects prognosis among BRAF mutated patients, moreover none of the survival analyses conducted so far included important covariates such as MSI status.11 In 2014 Landau et al. documented lower expression of CDX2 in V600EBRAF mutated mCRC compared to V600EBRAF wild type mCRC, irrespective of MSI status: these results suggest that loss of expression of CDX2 could depend on BRAF mutations more than on microsatellite status. Of note, in the same study a higher prevalence of CK7 expression was found in BRAF mutated MSS CRC. Unfortunately, no survival analyses were planned in that study.25 Recently, another work described the relationship between CDX2 and prognosis in CRC: in this study, loss of CDX2 expression was associated with significantly worse OS, but specific analyses for stage IV V600EBRAF patients were not possible due to small sample size.26 According to our data, low CDX2 expression was associated to worse OS (HR of intermediate/low vs high expression = 1.72, 95%CI 1.03–2.86, p = 0.036).
We were also able to report on the prognostic role of CK7 expression, emerging as a strong determinant of outcome even in the multivariate model (HR of CK7 positive vs negative = 2.11, 95%CI 1.03–4–34, p = 0.041). From a mechanistic perspective, this finding is in line with what Harbaum et al. described earlier: a high prevalence of CK7 positive cells at the invasive front of tumour buds in samples of CRC. The same authors recorded a trend toward a higher risk of disease progression or mortality in patients with high CK7 expression.27 Again, no specific information was available regarding V600EBRAF mutated patients in this study. The authors argued that CK7 could be associated with epithelial-to-mesenchymal transition. In our study we did not find significant correlations between CDX2/CK7/CK20 expression results and grading (Supplementary Table 5), nor with CMS classification (Supplementary Table 1).
In our study, CMS2-3 and 4 were associated with significantly worse OS and PFS (HR = 2.70, 95%CI 1.41–5.26, p = 0.003 and HR = 0.22, 95%CI 1.14–4.35, p = 0.02, respectively) when compared to CMS1 also in multivariate analyses. Given that we arbitrarily assigned MSI-H tumours to the CMS1 subgroup as previously described,22,23 our data clarify the effect of MSI-H phenotype on V600EBRAF mutated mCRC. This finding provides an answer to an open issue still unsolved from studies conducted so far.2,19 It is unlikely that treatment with anti-PD1 could have influenced these results since the percentage of patients treated with those agents is below 10% in our series. Given the rapid development of anti-BRAF and immunotherapy with checkpoint inhibitor-based treatments in mCRC, our data would be quite useful to inform the design of future clinical trials by evaluating the adoption of new stratification factors. It should be noted that while IHC assessment for CMS subgrouping may lead to different results than standard transcriptome-based classification a concordance of nearly 90% was reported in specific studies23 and the intrinsic advantages of IHC in terms of costs and ease of reproducibility are obvious.
Another interesting observation is that presence of TILs was related to better OS and PFS outcome (respectively, HR = 0.58, 95%CI 0.39–0.86 for OS and HR = 0.58, 95%CI 0.39–0.85 for PFS). Recently, Shibutani et al. described a correlation between high immune infiltrate in primary tumour and response to chemotherapy in a cohort of 57 mCRC patients, with higher response rate to chemotherapy (79.3 vs. 48.1%, p = 0.025) and better PFS (10.1 months vs. 7.3 months, p = 0.013) in the high-TILs group. Of note, in the high-TILs group a significantly better OS was observed compared to low-TILs group (35.5 months vs. 22.4 months, p = 0.022).28 Taken together, data on TILs and CMS suggest a role for tumour-immune system interaction in affecting prognosis of V600EBRAF mutated mCRC.
We also explored the reproducibility of BM1/BM2 categorisation with IHC based surrogate markers and correlated those results with outcome. Similarly, to what Barras et al. previously reported,5 no differences were found in PFS and OS between the two groups.
Major points of strength of our data rely on: (a) clinical homogeneity (i.e. all metastatic patients), (b) large numbers (V600EBRAF mutated mCRC constitutes around 8% and 155 patients with detailed clinical data and biologic material constitute one the biggest cohort ever studied), (c) adjustment with a modern, robust and validated clinical prognostic score, (d) real world data. The latter is a two-faced point, with its intrinsic pros and cons. From one side, being BRAF a determinant of extremely bad prognosis, it has been reported how patients with available biologic material or enrolled in trials do not resemble exactly in terms of incidence and specific characteristics. From this may obviously originate a dangerous selection bias when analysing patients and samples from trials.1 From the other side, we should admit that quality and detail of real-world data are certainly limited compared to prospective controlled trials. In fact, the most relevant limitation of our study is its retrospective nature. Patients have been selected according to stage, but not for type or number of previous lines of treatment received: in this way, a slight selection bias could not be excluded. Additional limitations reside in not having adjusted for multiple testing and not having enough biologic material for testing all the markers in the totality of the population included.
In conclusion, present data provide new, original and informative observations. These findings would deserve external confirmatory studies but give already a guide to interpret clinical heterogeneity within the subgroup of V600EBRAF mCRC, being therefore useful to inform further translational research, for patients’ stratification in clinical trials and in routine clinical practice to better estimate patients’ prognosis.
Supplementary information
Acknowledgments
Author contributions
Study conception and design: F.L., M.D.M. Provision of patient data: all authors. Data analysis and interpretation: F.L., M.D.M. Paper writing: P.B., A.A.P., F.L., M.D.M. Paper revision and final approval: all authors.
Competing interests
F.L. had roles as consultant or advisor for Roche, Bayer, Amgen, Genentech. F.P. received honoraria and had roles as consultant or advisor for Amgen, Merck, Roche, Sanofi, Bayer, Servier, Lilly. S.L. had roles as consultant or advisor for Amgen, Bayer, Merck Serono, Lilly. She received research funding from Amgen, Merck Serono and she is part of speakers’ bureau of Lilly, BMS. Vittorina Zagonel received honoraria and had roles as consultant or advisor for Bristol-Mayers Squibb, Bayer, Roche, Pfizer, Janssen, Novartis, Astellas, Servier. She had roles as consultant or advisor for Celgene, Merck. M.D.M. received honoraria and had roles as consultant or advisor for AstraZeneca, Bristol Myers Squibb, MSD, Takeda and Janssen; received a research grant from Tesaro. M.F. received a research grant from Astellas Pharma. The remaining authors declare no competing interest.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Veneto Institute of Oncology (ID CESC IOV 186/2017) and was conducted according to ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Patients provided additional Informed Consent when appropriate
Funding
Regione Veneto – RP-2014-00000395., DOR Funds University of Padua.
Consent for publication
No individual person’s data were reported in any form in the paper.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fotios Loupakis, Phone: +39 049/8215919, Email: fotios.loupakis@iov.veneto.it.
Matteo Fassan, Phone: +39 049 821 1312, Email: matteo.fassan@unipd.it.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0560-0.
References
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin. Cancer Res. 2014;20:5322–5330. doi: 10.1158/1078-0432.CCR-14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88–99. doi: 10.1053/j.gastro.2014.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goey KKH, Sørbye H, Glimelius B, Adams RA, André T, Arnold D, et al. Consensus statement on essential patient characteristics in systemic treatment trials for metastatic colorectal cancer: supported by the ARCAD Group. Eur. J. Cancer. 2018;100:35–45. doi: 10.1016/j.ejca.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Barras D, Missiaglia E, Wirapati P, Sieber OM, Jorissen RN, Love C, et al. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin. Cancer Res. 2017;23:104–115. doi: 10.1158/1078-0432.CCR-16-0140. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loupakis F, Intini R, Cremolini C, Orlandi A, Sartore-Bianchi A, Pietrantonio F, et al. A validated prognostic classifier for V600E BRAF mutated metastatic colorectal cancer: the “BRAF BeCool” study. Eur. J. Cancer. 2019;118:121–130. doi: 10.1016/j.ejca.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto N, Feng Y, Stolfi C, Kurosu Y, Green M, Lin J, et al. BRAF V600E cooperates with CDX2 inactivation to promote serrated colorectal tumorigenesis. Elife. 2017;6:e20331. doi: 10.7554/eLife.20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong K, Pellón-Cárdenas O, Sirihorachai VR, Warder BN, Kothari OA, Perekatt AO, et al. Degree of tissue differentiation dictates susceptibility to BRAF-driven colorectal cancer. Cell Rep. 2017;21:3833–3845. doi: 10.1016/j.celrep.2017.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalerba P, Sahoo D, Paik S, Guo X, Yothers G, Song N, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N. Engl. J. Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirripa M, Loupakis F, Lenz HJ. CDX2 as a prognostic biomarker in colon cancer. N. Engl. J. Med. 2016;374:2183. doi: 10.1056/NEJMc1602584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moll R, Zimbelmann R, Goldschmidt MD, Keith M, Laufer J, Kasper M, et al. The human gene encoding cytokeratin 20 and its expression during fetal development and in gastrointestinal carcinomas. Differentiation. 1993;53:75–93. doi: 10.1111/j.1432-0436.1993.tb00648.x. [DOI] [PubMed] [Google Scholar]
- 13.Werling RW, Yaziji H, Bacchi CE, Gown AM. CDX2, a highly sensitive and specific marker of adenocarcinomas of intestinal origin: an immunohistochemical survey of 476 primary and metastatic carcinomas. Am. J. Surg. Pathol. 2003;27:303–310. doi: 10.1097/00000478-200303000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Rhee YY, Bae JM, Cho NY, Kang GH. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am. J. Surg. Pathol. 2013;37:1532–1541. doi: 10.1097/PAS.0b013e31829ab1c1. [DOI] [PubMed] [Google Scholar]
- 15.Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J. Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 16.Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J. Clin. Oncol. 2012;30:1288–1295. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 17.Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, et al. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int. J. Cancer. 2011;128:2075–2084. doi: 10.1002/ijc.25555. [DOI] [PubMed] [Google Scholar]
- 18.Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–4632. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein J, Tran B, Ensor J, Gibbs P, Wong HL, Wong SF, et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H) Ann. Oncol. 2014;25:1032–1038. doi: 10.1093/annonc/mdu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remo A, Fassan M, Lanza G. Immunohistochemical evaluation of mismatch repair proteins in colorectal carcinoma: the AIFEG/GIPAD proposal. Pathologica. 2016;108:104–109. [PubMed] [Google Scholar]
- 22.Ten Hoorn S, Trinh A, de Jong J, Koens L, Vermeulen L. Classification of colorectal cancer in molecular subtypes by immunohistochemistry. Methods Mol. Biol. 2018;1765:179–191. doi: 10.1007/978-1-4939-7765-9_11. [DOI] [PubMed] [Google Scholar]
- 23.Trinh A, Trumpi K, De Sousa E, Melo F, Wang X, de Jong JH, Fessler E, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin. Cancer Res. 2017;23:387–398. doi: 10.1158/1078-0432.CCR-16-0680. [DOI] [PubMed] [Google Scholar]
- 24.Williams DS, Mouradov D, Jorissen RN, Newman MR, Amini E, Nickless DK, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68:465–474. doi: 10.1136/gutjnl-2017-315664. [DOI] [PubMed] [Google Scholar]
- 25.Landau MS, Kuan SF, Chiosea S, Pai RK. BRAF-mutated microsatellite stable colorectal carcinoma: an aggressive adenocarcinoma with reduced CDX2 and increased cytokeratin 7 immunohistochemical expression. Hum. Pathol. 2014;45:1704–1712. doi: 10.1016/j.humpath.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Bruun J, Sveen A, Barros R, Eide PW, Eilertsen I, Kolberg M, et al. Prognostic, predictive, and pharmacogenomic assessments of CDX2 refine stratification of colorectal cancer. Mol. Oncol. 2018;12:1639–1655. doi: 10.1002/1878-0261.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbaum Lars, Pollheimer Marion J, Kornprat Peter, Lindtner Richard A, Schlemmer Andrea, Rehak Peter, Langner Cord. Keratin 7 expression in colorectal cancer - freak of nature or significant finding? Histopathology. 2011;59(2):225–234. doi: 10.1111/j.1365-2559.2011.03694.x. [DOI] [PubMed] [Google Scholar]
- 28.Shibutani M, Maeda K, Nagahara H, Fukuoka T, Iseki Y, Matsutani S, et al. Tumor-infiltrating lymphocytes predict the chemotherapeutic outcomes in patients with stage IV colorectal cancer. In Vivo. 2018;32:151–158. doi: 10.21873/invivo.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.