Abstract
Background
Luminal B cancers show much worse outcomes compared to luminal A. This present study aims to screen key lncRNAs and mRNAs correlated with luminal-B breast cancer.
Methods
Luminal-B breast cancer tissue samples and adjacent tissue samples were obtained from 4 patients with luminal-B breast cancer. To obtain differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) between luminal-B breast cancer tumor tissues and adjacent tissues, RNA-sequencing and bioinformatics analysis were performed. Functional annotation of DEmRNAs and protein-protein interaction networks (PPI) construction were performed. DEmRNAs transcribed within a 100 kb window up- or down-stream of DElncRNAs were searched, which were defined as cis nearby-targeted DEmRNAs of DElncRNAs. DElncRNA-DEmRNA co-expression networks were performed. The mRNA and lncRNA expression profiles were downloaded from The Cancer Genome Atlas (TCGA) database to validate the expression patterns of selected DEmRNAs and DElncRNAs.
Results
A total of 1178 DEmRNAs and 273 DElncRNAs between luminal-B breast cancer tumor tissues and adjacent tissues were obtained. Hematopoietic cell lineage, Cytokine-cytokine receptor interaction, Cell adhesion molecules (CAMs) and Primary immunodeficiency were significantly enriched KEGG pathways in luminal-B breast cancer. FN1, EGFR, JAK3, TUBB3 and PTPRC were five hub proteins of the PPI networks. A total of 99 DElncRNAs-nearby-targeted DEmRNA pairs and 1878 DElncRNA-DEmRNA co-expression pairs were obtained. Gene expression results validated in TCGA database were consistent with our RNA-sequencing results, generally.
Conclusion
This study determined key genes and lncRNAs involved in luminal-B breast cancer, which expected to present a new avenue for the diagnosis and treatment of luminal-B breast cancer.
Keywords: Luminal-B breast cancer, mRNA, Long non-coding RNA (lncRNA), RNA-sequencing
Background
Breast cancer is the leading cause of cancer-related death in women, both overall and in less developed countries (1). It is a heterogeneous disease with regard to molecular alterations, cellular composition, and clinical outcome, both between tumor subtypes and within a single tumor, which were commonly defined by gene expression profiling as four main subtypes including luminal A, luminal B, HER-2 enriched and basal-like, (2–4). Luminal B breast cancer is unique with regard to somatic point mutations, the profile of gene copy number alterations (CNAs), and DNA methylation (5). Expression profiles and gene sets, with prognostic, predictive functions, or both for patients with breast cancer, have been identified in multiple studies (6). Although both luminal-A and luminal-B breast cancers are ER-positive, luminal-B cancers showed worse outcomes as compared to luminal-A cancers (7, 8). Therefore, it is urgent to discover novel biomarkers with prognostic and predictive functions for luminal B breast cancer that can be therapeutically targeted.
With advances in high-throughput technology, it is discovered that human transcriptome mainly consists of non-coding RNAs (ncRNAs) with limited or no protein-coding capacity (9, 10). Long non-coding RNAs (lncRNAs), with over 200 nucleotides base long, attracts more attention and has been widely linked with various diseases, including cancers (11–14). The lncRNAs exert momentous roles in multiple cellular processes at transcriptional and post-transcriptional regulation level through transcriptional interference and histone modifications (15, 16). The higher expression of SPRY4-IT1 was reported to modulate apoptosis and invasion in melanoma (17). LncRNA UCA1a (CUDR) may promote proliferation and tumorigenesis in human bladder cancer (18).
In this study, differentially expressed lncRNAs (DElncRNA) and mRNAs (DEmRNAs) in tumor tissues of patients with luminal-B breast cancer were identified by RNA-sequencing. Subsequently, protein-protein interaction (PPI) networks of DEmRNAs were conducted. Identification of cis nearby-targeted DEmRNAs of DElncRNAs and construction of DElncRNA-DEmRNA co-expression networks were performed. In this light, we expect this study could represent a new avenue to improve the understanding of the pathogenesis and be helpful for treatment of luminal B breast cancer.
Methods
Patients and samples
Luminal-B breast cancer tissue samples and adjacent tissue samples were obtained from 4 patients with luminal-B breast cancer in People’s Hospital of Deyang City, which were free of treatment. The detailed characteristics of patients are displayed in Table 1. Written informed consent about the use of these samples was obtained from each patient. All procedures performed in this study was in accordance with the ethical standards of the ethics committee of People’s Hospital of Deyang City (2017–045) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Table 1.
Patient characteristics
| Index | Age Range |
TNM stage | ER | PR | HER-2 | ||
|---|---|---|---|---|---|---|---|
| T | N | M | |||||
| Case 1 | 46–56 | 3 | 0 | 0 | 30% | 10% | – |
| Case 2 | 2 | 1 | 0 | 80% | 60% | + | |
| Case 3 | 2 | 1 | 0 | 40% | 30% | – | |
| Case 4 | 2 | 1 | 0 | 40% | 20% | – | |
TNM stage Tumor-node-metastasis stage, ER ertrogen receptor, PR progestrone receptor, HER-2 human epidermal growth factor receptor-2
RNA isolation and sequencing
According to the manufacturer’s protocol, RNA was extracted with PAXgene blood RNA kit (PreAnalytiX GmbH, Hombrechtikon, CH, Switzerland). With Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit), the concentration, integrity and RNA integrity number (RIN) values of RNA were assessed. Sequencing was performed based on the Illumina Hiseq X-ten platform (Illumina, Inc., San Diego, CA, USA) with PE150 bp sequencing mode. The sequencing was done with paired-ends and 10G depth. With Base Calling version 0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), the FASTQ sequence data were acquired from the RNA-sequencing data. Read QC tool in FastQC version 0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for the quality control of FASTQ data with Q > 30. Trimming of raw data was performed with cutadapt version 1.16 (http://cutadapt.readthedocs.io). Reads with low quality (adaptor sequences, sequences with a quality score < 20, and sequences with an N base rate of raw reads > 10%) were removed to obtain the clean reads.
Identification of DEmRNAs and DElncRNAs
In order to align the clean reads with the human reference genome, Ensemble GRCh38.p7 (ftp://ftp.ncbi.nlm.nih.gov/genomes/Homo_sapiens), HISAT2 version 2.1.0 (https://ccb.jhu.edu/software/hisat2/index.shtml) was applied. Expression of mRNAs and lncRNAs was normalized and outputted with StringTie version 1.3.3b (http://ccb.jhu.edu/software/stringtie/). Fragments per Kilobase of exon per million fragments mapped (FPKM) of lncRNAs and mRNAs were calculated with StringTie. With edgeR version 3.24 (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html), both DEmRNAs and DElncRNAs were obtained with |log2FC| > 1 and p-value < 0.05. By using R package “pheatmap”, hierarchical clustering analysis of DElncRNAs and DEmRNAs were conducted.
Functional annotation of DEmRNAs
Functional annotation, including Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses was performed with Metascape (http://metascape.org/gp/index.html). A value of p < 0.05 was set as the cut-off for significance.
Protein-protein interaction (PPI) networks construction
Top 100 up- and down-regulated DEmRNAs were scanned with the BioGrid (http://www.uniprot.org/database/DB-0184). Then, PPI networks were visualized with Cytoscape software (version 3.5.0, http://www.cytoscape.org).
Cis nearby-targeted DEmRNAs of the DElncRNAs
DEmRNAs transcribed within a 100-kb window upstream or downstream of DElncRNAs were searched, which were defined as cis nearby-targeted DEmRNAs of DElncRNAs, to obtain the targeted DEmRNAs of DElncRNAs with cis-regulatory effects. The networks were visualized by Cytoscape software. Functional annotation of the cis nearby-targeted DEmRNAs of the DElncRNAs was performed with Metascape. A value of p < 0.05 was set as the cut-off for significance.
DElncRNA-DEmRNA co-expression networks
To further examine the potential roles of DElncRNAs and DEmRNAs in luminal-B breast cancer, the DElncRNA-DEmRNA co-expression networks were constructed. DElncRNA-DEmRNA pairs with an absolute value of PCC > 0.95 and p < 0.01 were defined as co-expressed DElncRNA-DEmRNA pairs. By using Cytoscape, the co-expressed DElncRNA-DEmRNA networks were visualized. Functional annotation of the DEmRNAs co-expressed with DElncRNAs was performed with Metascape. A value of p < 0.05 was set as the cut-off for significance.
Validation in the Cancer genome atlas (TCGA) database
The mRNA and lncRNA expression profiles of 171 patients with luminal B breast cancer and 59 normal tissues were downloaded from TCGA database to validate the expression patterns of selected DEmRNAs and DElncRNAs.
Results
DEmRNAs and DElncRNAs between luminal-B breast cancer tumor tissues and adjacent tissues
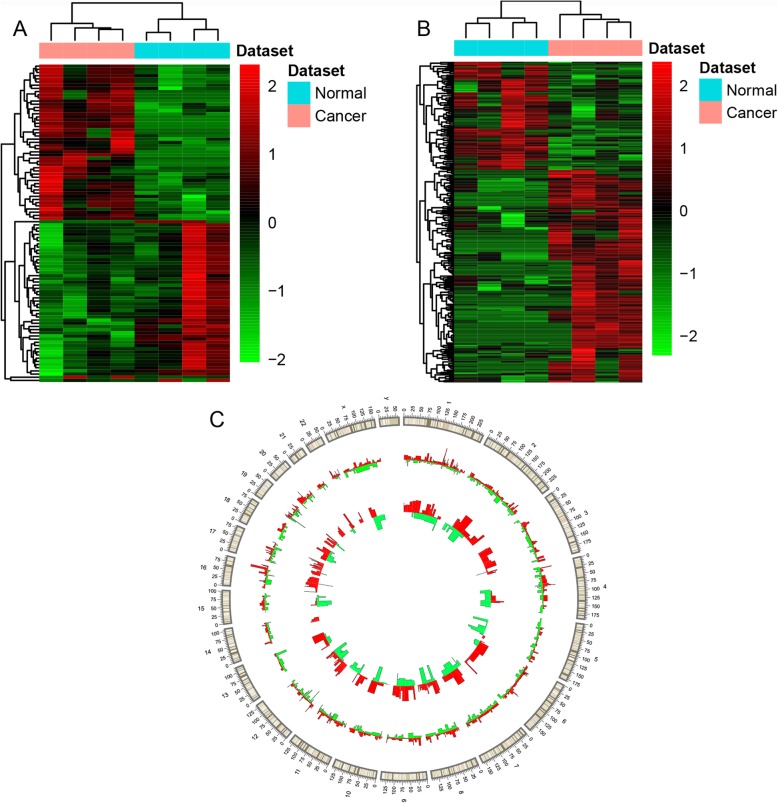
A total of 1178 DEmRNAs (666 up-regulated and 512 down-regulated DEmRNAs) and 273 DElncRNAs (181 up-regulated and 92 down-regulated DElncRNAs) were obtained. The top 10 up- and down-regulated DEmRNAs and DElncRNAs were exhibited in Table 2 and Table 3, respectively. Hierarchical clustering analysis of top 100 10 up- and down-regulated DEmRNAs and DElncRNAs was showed in Fig. 1a and Fig. 1b, respectively. Furthermore, the distribution of DElncRNAs and DEmRNAs on all chromosomes was showed in Fig. 1c. The raw-data have been uploaded to Gene Expression Omnibus (GEO) (GSE139274, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139274).
Table 2.
Top 10 up- and down-regulated DEmRNAs between luminal-B breast cancer tumor tissues compared with adjacent tissues
| ID | Symbol | log2FC | p-value | Regulation |
|---|---|---|---|---|
| ENSG00000143556 | S100A7 | 8.127250867 | 7.08E-10 | up |
| ENSG00000171951 | SCG2 | 4.46199863 | 7.47E-09 | up |
| ENSG00000163993 | S100P | 5.865266947 | 2.43E-08 | up |
| ENSG00000169245 | CXCL10 | 5.755282864 | 2.57E-08 | up |
| ENSG00000188404 | SELL | 4.205586942 | 2.63E-08 | up |
| ENSG00000180549 | FUT7 | 3.310463733 | 3.42E-08 | up |
| ENSG00000138755 | CXCL9 | 4.319459635 | 4.19E-08 | up |
| ENSG00000099953 | MMP11 | 4.120231544 | 1.44E-07 | up |
| ENSG00000184937 | WT1 | 2.831855381 | 2.43E-07 | up |
| ENSG00000143546 | S100A8 | 5.625786123 | 3.05E-07 | up |
| ENSG00000269711 | AC008763.3 | −8.573808999 | 3.62E-11 | down |
| ENSG00000272414 | FAM47E-STBD1 | −4.255105776 | 3.94E-07 | down |
| ENSG00000109846 | CRYAB | −2.409617655 | 1.98E-06 | down |
| ENSG00000109107 | ALDOC | −2.539792463 | 3.92E-06 | down |
| ENSG00000134548 | SPX | −7.064583075 | 5.21E-06 | down |
| ENSG00000135447 | PPP1R1A | −4.272501282 | 8.38E-06 | down |
| ENSG00000120049 | KCNIP2 | −3.142698521 | 1.76E-05 | down |
| ENSG00000270181 | BIVM-ERCC5 | −5.716264875 | 1.93E-05 | down |
| ENSG00000162433 | AK4 | −1.720478242 | 1.94E-05 | down |
| ENSG00000159387 | IRX6 | −4.343462068 | 3.97E-05 | down |
DEmRNAs, differentially expressed mRNAs. FC, fold change
Table 3.
Top 10 up- and down-regulated DElncRNAs between luminal-B breast cancer tumor tissues compared with adjacent tissues
| ID | Symbol | log2FC | p-value | Regulation |
|---|---|---|---|---|
| ENSG00000235123 | DSCAM-AS1 | 6.99539501 | 2.71E-06 | up |
| ENSG00000273445 | AC133644.2 | 7.081588171 | 3.65E-06 | up |
| ENSG00000261039 | LINC02544 | 7.125899953 | 1.29E-05 | up |
| ENSG00000225783 | MIAT | 2.369853869 | 3.75E-05 | up |
| ENSG00000224950 | AL390066.1 | 2.333212245 | 7.65E-05 | up |
| ENSG00000270120 | AC007728.3 | 5.314205083 | 8.62E-05 | up |
| ENSG00000279930 | AL032819.2 | 5.526214467 | 1.62E-04 | up |
| ENSG00000247774 | PCED1B-AS1 | 1.82134551 | 1.78E-04 | up |
| ENSG00000261218 | AC099524.1 | 4.377268771 | 2.60E-04 | up |
| ENSG00000234261 | AL138720.1 | 4.488627977 | 2.87E-04 | up |
| ENSG00000253434 | LINC02237 | −6.553462635 | 1.55E-04 | down |
| ENSG00000250961 | AC025470.2 | −5.149166872 | 1.76E-04 | down |
| ENSG00000236333 | TRHDE-AS1 | −4.862423419 | 4.38E-04 | down |
| ENSG00000261441 | AC124068.2 | −4.394333958 | 7.43E-04 | down |
| ENSG00000261888 | AC144831.1 | −3.00198307 | 1.98E-03 | down |
| ENSG00000251660 | AC007036.3 | −2.997221226 | 2.05E-03 | down |
| ENSG00000260947 | AL356489.2 | −3.611436753 | 2.33E-03 | down |
| ENSG00000235033 | AL590999.1 | −2.980398474 | 2.69E-03 | down |
| ENSG00000230333 | AC004160.1 | −3.679435378 | 2.71E-03 | down |
| ENSG00000272701 | MESTIT1 | −3.528818654 | 2.85E-03 | down |
DElncRNAs, differentially expressed lncRNAs. FC, fold change
Fig. 1.
DElncRNAs and DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues. a and b displayed hierarchical clustering results of top 100 DEmRNAs and DElncRNAs between luminal-B breast cancer tumor tissues and adjacent tissues, respectively. Row and column represented DEmRNAs/DElncRNAs and tissue samples, respectively. The color scale represented the expression levels. c displayed distribution of DElncRNAs and DEmRNAs on chromosomes. The outer layer cycle was the chromosome map of the human genome hg19 (GRCh37). The larger inner layer and smaller inner layer represented the distribution of DEmRNAs and DElncRNAs on different chromosome, respectively. The red and green color represented the up- and down-regulated
Functional annotation
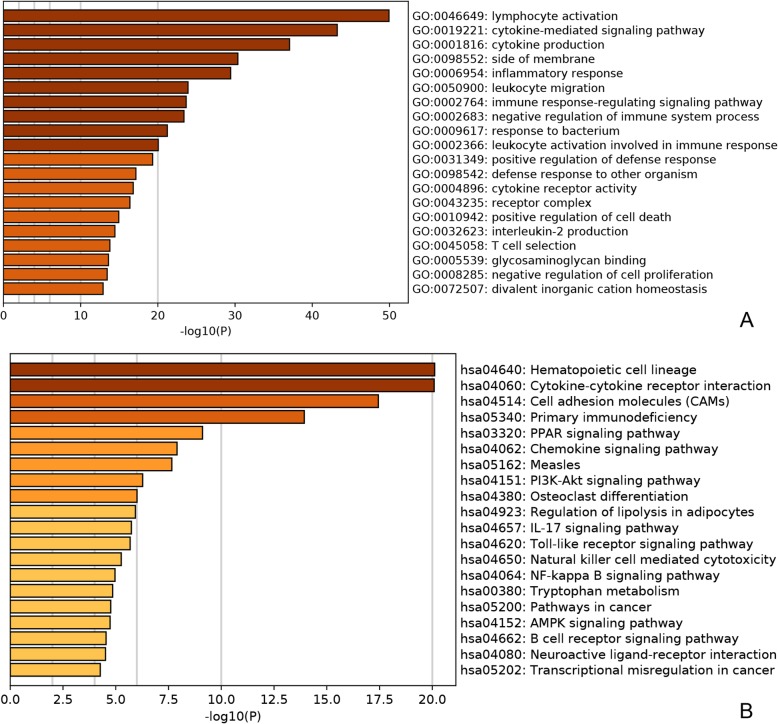
Lymphocyte activation (p = 1.14E-50), cytokine-mediated signaling pathway (p = 5.74E-44) and cytokine production (p = 8.78E-38) were significantly enriched GO terms in luminal-B breast cancer (Fig. 2a). Hematopoietic cell lineage (p = 7.75E-21), Cytokine-cytokine receptor interaction (p = 8.19E-21), Cell adhesion molecules (CAMs) (p = 3.61E-18) and Primary immunodeficiency (p = 1.17E-14) were significantly enriched KEGG pathways in luminal-B breast cancer (Fig. 2b).
Fig. 2.
Significantly enriched GO (a) terms and KEGG (b) pathways of DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues. The x-axis represented -lg p-value and the y-axis shows GO terms or KEGG pathways
Protein-protein interaction (PPI) networks
The PPI networks included 281 nodes and 263 edges. FN1 (degree = 28), EGFR (degree = 14), JAK3 (degree = 11), TUBB3 (degree = 11) and PTPRC (degree = 10) were five hub proteins of the PPI networks (Fig. 3).
Fig. 3.
Protein-protein interaction (PPI) networks. The red and blue ellipses represented proteins encoded by up- and down-regulated DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues. Ellipses with black border were DEmRNAs derived from top 10 down- and up-regulated DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues
Cis-nearby-targeted DEmRNAs of DElncRNAs
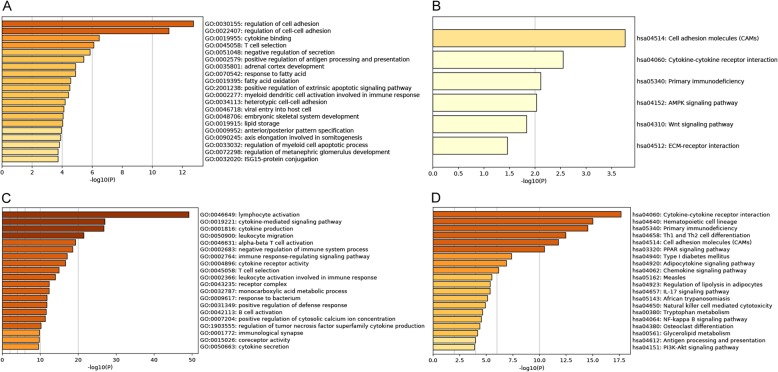
A total of 99 DElncRNAs-nearby-targeted DEmRNA pairs, involving in 78 DElncRNAs and 86 DEmRNAs, were detected (Fig. 4). Top two DElncRNAs with most nearby DEmRNAs were AL121985.1 and AL031316.1, which owned 5 and 4 nearby DEmRNAs, respectively. Regulation of cell adhesion (p = 1.84E-13), regulation of cell-cell adhesion (p = 7.86E-12), cytokine binding (p = 3.41E-07), T cell selection (p = 7.75E-07) and negative regulation of secretion (p = 1.38E-06) were significantly enriched GO terms (Fig. 6a). Cell adhesion molecules (CAMs) (p = 1.72E-04), Cytokine-cytokine receptor interaction (p = 2.81E-03), Primary immunodeficiency (p = 7.71E-03), AMPK signaling pathway (p = 9.30E-03) and Wnt signaling pathway (p = 1.46E-02) were significantly enriched KEGG pathways (Fig. 6b).
Fig. 4.
DElncRNA-nearby DEmRNA interaction networks. The inverted triangles and ellipses represent DElncRNAs and their nearby DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues, respectively. Red and blue color represent up- and down-regulation in luminal-B breast cancer tumor tissues compared to adjacent tissues, respectively. Inverted triangles and ellipses with black border were DElncRNAs/DEmRNAs derived from top 10 up- and down-regulated DElncRNAs/DEmRNAs
Fig. 6.
Significantly enriched GO terms and KEGG pathways of DElncRNA-nearby DEmRNAs (a-b) and DEmRNAs co-expressed with DElncRNAs (c-d). The x-axis represented -lg p-value and the y-axis shows GO terms or KEGG pathways
DElncRNA-DEmRNA co-expression networks
A total of 1878 DElncRNA-DEmRNA co-expression pairs including 225 DElncRNAs and 737 DEmRNAs were obtained with an absolute value of PCC > 0.95 and p < 0.01 (Fig. 5).
Fig. 5.
DElncRNA-DEmRNA co-expression networks. The inverted triangles and ellipses represent DElncRNAs and their nearby DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues, respectively. Red and blue color represent up- and down-regulation in luminal-B breast cancer tumor tissues compared to adjacent tissues, respectively. Inverted triangles and ellipses with black border were DElncRNAs/DEmRNAs derived from top 10 up- and down-regulated DElncRNAs/DEmRNAs
Lymphocyte activation (p = 7.08E-50), cytokine-mediated signaling pathway (p = 9.35E-28), cytokine production (p = 1.68E-27), leukocyte migration (p = 3.20E-22) and alpha-beta T cell activation (p = 4.89E-20) were significantly enriched GO terms (Fig. 6c). Cytokine-cytokine receptor interaction (p = 2.18E-18), Hematopoietic cell lineage (p = 9.66E-16), Primary immunodeficiency (p = 2.85E-15), Th1 and Th2 cell differentiation (p = 3.38E-13) and Cell adhesion molecules (CAMs) (p = 1.66E-12) were significantly enriched KEGG pathways (Fig. 6d).
Validation in TCGA database
With mRNA and lncRNA expression profiles downloaded from TCGA database, the expression patterns of four DEmRNAs, including S100A7, CCL5, MIAT and WT1-AS, were verified. As shown in Fig. 7, compared to normal controls, MIAT was down-regulated in luminal B breast cancer tumor tissues which were inconsistent with our results, while S100A7, CCL5 and WT1-AS were up-regulated in luminal B breast cancer tumor tissues which were consistent with our results.
Fig. 7.
Validation in TCGA database. The x-axis shows luminal-B breast cancer tumor tissues and adjacent tissues, and the y-axis shows expression levels, respectively. A) S100A7; B) CCL5; C) MIAT; D) WT1-AS
Discussion
Breast cancer, as the most common non-cutaneous type of cancer, is the leading cause of cancer-related mortality among female globally (19). As luminal-B cancers showed poorer prognosis as compared to luminal-A cancers, we performed this study and identified abundant DElncRNAs and DEmRNAs between luminal-B breast cancer tumor tissues and adjacent tissues.
S100A7 is a member of the S100 protein family, which have been associated with preinvasive ductal carcinoma in situ (DCIS) (20). During breast tumorigenesis and/or progression, several S100 s, including S100A2, S100A4 and S100A7, exhibit altered expression levels based on molecular analysis of breast tumors (21). Cancemi et al. suggested that S100A7 was involved in critical phases of the breast cancer growth and progression (22). Mayama et al. proposed that S100A7 was linked to an aggressive phenotype of ER-positive breast carcinoma, and was potent marker for distant metastasis of ER-positive breast cancer patients (23). In current study, S100A7 was the most significant up-regulated DEmRNAs in luminal B breast cancer tumor tissues, which may indicated that S100A7 exert momentous roles in luminal B breast cancer.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA, and its over-expression in multiple cancerous tissues has been linked to the proliferation and metastasis of tumor cells. It was first identified as being up-regulated in lung tumors, and a prognostic marker for metastasis and patient survival in non-small cell lung cancer (NSCLC), specifically in early stages of lung adenocarcinoma (24). Subsequently, MALAT1 was shown to be up-regulated in a broad spectrum of tumor types, such as endometrial stromal sarcoma and hepatocellular carcinomas (25, 26). Additional, it has been found that MALAT1 gene mutations frequently occurred in luminal-type breast tumors (27). Besides, MALAT1 was one of the top 10 up-regulated DElncRNAs in this study, and co-expressed with S100A7, which emphasized the critical role the MALAT1 in luminal B breast cancer and suggested that MALAT1 may involve in luminal B breast cancer by regulating S100A7.
Chemokines, small-molecular-weight cytokines involved in the physiological control of immune cell migrationare, were reported to perform a crucial function in breast cancer tumorigenesis and progression (19). Recent years, the chemokine C-C motif ligand 5 (CCL5), also known as RANTES, is a member of the CC subfamily, has been associated with aggressive breast cancer (28). Svensson et al. identified CCL2 and CCL5 as two therapeutic targets for estrogen-dependent breast cancer (29). Previous study suggested that endothelial cells (ECs) enhance endothelial-mesenchymal transition (EMT)-induced triple-negative breast cancer (TNBC) cell metastasis through PAI-1 and CCL5 signaling (30). Zhang et al. found that CCL5-mediated Th2 polarization of CD4+ T cells promotes metastasis in luminal breast cancer (31). In our analysis, CCL5 was significant up-regulated in luminal B breast cancer tumor tissues, which indicated the key role of CCL5 in luminal B breast cancer.
LncRNA myocardial infarction-associated transcript (MIAT) is primarily expressed in heart and fetal brain tissue (32). Dysregulated MIAT was first reported to correlated with myocardial infarction and involved in cardiac hypertrophy and diabetic cardiomyopathy (32–35). Recent studies suggested that MIAT promoted gastric cancer growth and metastasis by regulation of miR-141/DDX5 pathway, promoted proliferation and metastasis of non-small cell lung cancer via MMP9 activation, promoted hepatocellular carcinoma cells proliferation and invasion through sponging miR-214 (36–38). Luan et al. proposed that overexpression of MIAT was related to the TNM stage and lymphnode metastasis of breast cancer (39). MIAT was found to be overexpressed in both ER-positive breast cancer tissues and ER-positive breast cancer cell line MCF-7, and play a pivotal role in ER-positive breast cancer cell growth (40). Almnaseer-M et al. suggested that MIAT performs a critical function in breast tumorigenesis (41). MIAT was detected to be one of the top 10 up-regulated DElncRNAs, and co-expressed with CCL5. All these findings suggest that MIAT may involve in luminal B breast cancer by regulating the expression level of CCL5.
The Wilms’ tumor 1 (WT1) was first cloned in 1990 as a suppressor in Wilms’ tumor, which was located at chromosome 11p13 (42). WT1 gene mutations are linked with a subset of Wilm’s tumors, the most common pediatric renal cancer (43). Substantial evidence has linked WT1 with the pathogenesis of breast cancer. WT1 is linked to the progression of breast cancer, including migration, invasion and angiogenesis. Knockdown of WT1 was demonstrated to lead to mitochondrial damage and then inhibit malignant cell growth (44). Highly expressed WT1 was linked to poor prognosis of patients with breast cancer (45). Over expression of WT1 was detected in TNBC (46). In agreement with previous studies, the expression of WT1 was observed significant up-regulated in luminal B breast cancer tumor tissues in present study.
Wilms tumor 1 Antisense RNA (WT1-AS) is located upstream of the WT1 gene, and these two genes are bi-directionally transcribed from the same promoter region. Down-regulation of WT1-AS was related to a poorer prognosis in ovarian clear cell adenocarcinoma (47). Lv et al. suggested that WT1-AS promoted cell apoptosis in hepatocellular carcinoma (HCC) and may function as a tumor suppressor in HCC (48). It is reported that WT1-AS was significantly down-regulated in gastric cancers and may correlates with tumor progression (49). In addition, WT1-AS was detected to be was under-expressed in cervical carcinoma and suppress cervical cancer cell growth and aggressiveness (50, 51). In the current study, we found that WT1-AS was a DElncRNA and WT1 was a nearby-targeted DEmRNA of WT1-AS, which reminded us to explore the role of WT1-AS-WT1 in luminal B breast cancer.
Conclusion
In conclusion, a total of 1178 DEmRNAs and 273 DElncRNAs between luminal-B breast cancer tumor tissues and adjacent tissues were obtained. We discussed and emphasized the importance role of three DElncRNA-DEmRNA pairs, including MALAT1-S100A7, MIAT-CCL5 and WT1-AS-WT1, involved in luminal B breast cancer, which expected to provide new insight into understanding the mechanism underlying pathogenesis of luminal B breast cancer. The small sample size was a limitation of our study. Although the validation results in TCGA database indicated that our RNA-sequencing results were generally reliable, larger cohorts of patients and further experimental validation studies are needed to conduct to verify this conclusion.
Acknowledgements
We are appreciated to Pro. Xin-Jian Jia for supplying the experimental samples.
Abbreviations
- CAMs
Cell adhesion molecules
- CCL5
Chemokine C-C motif ligand 5
- CNAs
Copy number alterations
- DCIS
Ductal carcinoma in situ
- DElncRNA
Differentially expressed lncRNA
- DEmRNA
Differentially expressed mRNA
- ECs
Endothelial cells
- EMT
Endothelial-mesenchymal transition
- FPKM
Per million fragments mapped
- GO
Gene Ontology
- HCC
Hepatocellular carcinoma
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNAs
Long non-coding RNAs
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- MIAT
Myocardial infarction-associated transcript
- ncRNAs
Non-coding RNAs
- NSCLC
Non-small cell lung cancer
- PCC
Pearson’s correlation coefficient
- PPI
Protein-protein interaction network
- RIN
RNA integrity number
- TNBC
Triple-negative breast cancer
- WT1
Wilms’ tumor 1
- WT1-AS
Wilms tumor 1 Antisense RNA
Authors’ contributions
CY and NZ2 made substantial contributions to conception and design. CY, XJ and YY performed the experiment. JE, NZ1 and XL collected and analyzed the data. WW and YL interpreted the data. All authors were involved in drafting and revising the manuscript and gave final approval of the manuscript.
Funding
This study was supported by Scientific Research Project of Sichuan Provincial Department of Health (090163). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The raw-data have been uploaded to Gene Expression Omnibus (GEO) (GSE139274, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139274) and is publicly available until October 2020 due to data confidentiality.
Ethics approval and consent to participate
Written informed consent about the use of these samples was obtained from each patient. This study was approved by the ethical committee of People’s Hospital of Deyang City (2017–045).
Consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Parker JS, Michael M, Cheang MCU, Samuel L, David V, Tammi V, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornelia P. Heterogeneity in breast cancer. J Clin Investig. 2011;121(10):3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Untch M, Gerber B, Harbeck N, Jackisch C, Marschner N, Mobus V, et al. 13th st. Gallen international breast cancer conference 2013: primary therapy of early breast cancer evidence, controversies, consensus - opinion of a german team of experts (zurich 2013). Breast care (Basel, Switzerland). 2013;8(3):221–9. [DOI] [PMC free article] [PubMed]
- 5.Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Kyle E, et al. The Cancer genome atlas Pan-Cancer analysis project. Chin J Lung Cancer. 2015;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng F, Oh DS, Lodewyk W, Britta W, Nuyten DSA, Nobel AB, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 7.SøRlie T, ., Perou CM, Tibshirani R, ., Aas T, ., Geisler S, ., Johnsen H, ., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci 2001. [DOI] [PMC free article] [PubMed]
- 8.Christos S, Lajos P. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz JE, Hongjae S, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Francesco C, Akira W, Luca Q, Hui X, Larissa P, Abhijit P, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448(3):315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wu N, Liu J, Wu Z, Dong D. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol. 2015;10:131. doi: 10.1186/s13000-015-0310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Li Y, Yang B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem Biophys Res Commun. 2016;478(1):323–329. doi: 10.1016/j.bbrc.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 17.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71(11):3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X, et al. Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int J Oncol. 2012;41(1):276–284. doi: 10.3892/ijo.2012.1443. [DOI] [PubMed] [Google Scholar]
- 19.Marco VV, Wendy X, Pestell RG. The potential to target CCL5/CCR5 in breast cancer. Expert Opin Ther Targets. 2014;18(11):1265–1275. doi: 10.1517/14728222.2014.949238. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Wang L, Wang X, Cao Z, Yang Q, Zhang K. S100A7 enhances invasion of human breast cancer MDA-MB-468 cells through activation of nuclear factor-kappaB signaling. World J Surg Oncol. 2013;11:93. doi: 10.1186/1477-7819-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emberley ED, Murphy LC, Watson PH. S100A7 and the progression of breast cancer. Breast Cancer Res. 2004;6(4):153–159. doi: 10.1186/bcr816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancemi P, Di Cara G, Albanese NN, Costantini F, Marabeti MR, Musso R, et al. Differential occurrence of S100A7 in breast cancer tissues: a proteomic-based investigation. Proteomics Clin Appl. 2012;6(7–8):364–373. doi: 10.1002/prca.201100072. [DOI] [PubMed] [Google Scholar]
- 23.Mayama A, Takagi K, Suzuki H, Sato A, Onodera Y, Miki Y, et al. OLFM4, LY6D and S100A7 as potent markers for distant metastasis in estrogen receptor-positive breast carcinoma. Cancer Sci. 2018;109(10):3350–3359. doi: 10.1111/cas.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 25.Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97(2):106–112. doi: 10.1111/j.1349-7006.2006.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 27.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ayesha K, Joy W, Ilaria F, Chaofeng M, Junhua M, Zhizhou Y, et al. Recent Advances in Discovering the Role of CCL5 in Metastatic Breast Cancer. Mini Reviews in Medicinal Chemistry. 2016;15(13). [DOI] [PMC free article] [PubMed]
- 29.Svensson S, Abrahamsson A, Rodriguez GV, Olsson AK, Jensen L, Cao Y, et al. CCL2 and CCL5 are novel therapeutic targets for estrogen-dependent breast Cancer. Clin Cancer Res. 2015;21(16):3794–3805. doi: 10.1158/1078-0432.CCR-15-0204. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Wenwen, Xu Jing, Fang Hehui, Tang Lin, Chen Weiwei, Sun Qian, Zhang Qun, Yang Fang, Sun Zijia, Cao Lulu, Wang Yucai, Guan Xiaoxiang. Endothelial cells promote triple-negative breast cancer cell metastasisviaPAI-1 and CCL5 signaling. The FASEB Journal. 2018;32(1):276–288. doi: 10.1096/fj.201700237RR. [DOI] [PubMed] [Google Scholar]
- 31.Qianfei Z, Jilong Q, Lin Z, Lei G, Bing Z, Yan Z, et al. CCL5-mediated Th2 immune polarization promotes metastasis in luminal breast Cancer. Cancer Res. 2015;75(20):4312–4321. doi: 10.1158/0008-5472.CAN-14-3590. [DOI] [PubMed] [Google Scholar]
- 32.Zhu XH, Yuan YX, Rao SL, Wang P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Euro Rev Med Pharmacol Sci. 2016;20(17):3653. [PubMed] [Google Scholar]
- 33.Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8(7):e2929. doi: 10.1038/cddis.2017.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Wang J, Sun L, Zhu S. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol. 2017;818:508–517. doi: 10.1016/j.ejphar.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 35.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT , that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 36.Sha M, Lin M, Wang J, Ye J, Xu J, Xu N, et al. Long non-coding RNA MIAT promotes gastric cancer growth and metastasis through regulation of miR-141/DDX5 pathway. Journal of experimental & clinical cancer research : CR. 2018;37(1):58. doi: 10.1186/s13046-018-0725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai I, Yang CA, Lin PC, Chan WL, Lee YT, Yen JC, et al. Long noncoding RNA MIAT promotes non-small cell lung cancer proliferation and metastasis through MMP9 activation. Oncotarget. 2017;8(58):98148–98162. doi: 10.18632/oncotarget.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang X, Gao Y, Qin J, Lu S. lncRNA MIAT promotes proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2018;314(5):G559–Gg65. doi: 10.1152/ajpgi.00242.2017. [DOI] [PubMed] [Google Scholar]
- 39.Luan T, Zhang X, Wang S, Song Y, Zhou S, Lin J, et al. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget. 2017;8(44):76153–76164. doi: 10.18632/oncotarget.19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Jiang B, Wu X, Huang Q, Chen W, Zhu H, et al. Long non-coding RNA MIAT is estrogen-responsive and promotes estrogen-induced proliferation in ER-positive breast cancer cells. Biochem Biophys Res Commun. 2018;503(1):45–50. doi: 10.1016/j.bbrc.2018.05.146. [DOI] [PubMed] [Google Scholar]
- 41.Almnaseer ZA, Mourtada-Maarabouni M. Long noncoding RNA MIAT regulates apoptosis and the apoptotic response to chemotherapeutic agents in breast cancer cell lines. Biosci Rep. 2018;38(4). [DOI] [PMC free article] [PubMed]
- 42.Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms' tumor locus. Cell. 1990;60(3):509–520. doi: 10.1016/0092-8674(90)90601-A. [DOI] [PubMed] [Google Scholar]
- 43.Silberstein GB, Van Horn K, Strickland P, Roberts CT, Jr, Daniel CW. Altered expression of the WT1 wilms tumor suppressor gene in human breast cancer. Proc Natl Acad Sci U S A. 1997;94(15):8132–8137. doi: 10.1073/pnas.94.15.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tatsumi N, Oji Y, Tsuji N, Tsuda A, Higashio M, Aoyagi S, et al. Wilms' tumor gene WT1-shRNA as a potent apoptosis-inducing agent for solid tumors. Int J Oncol. 2008;32(3):701–711. [PubMed] [Google Scholar]
- 45.Yasuo M, Akiko A, Chiyomi E, Tetsuya T, Yasuhiro T, Hiroya T, et al. High expression of Wilms' tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin Cancer Res. 2002;8(5):1167. [PubMed] [Google Scholar]
- 46.Xiao-Wei Q, Fan Z, Xin-Hua Y, Lin-Jun F, Yi Z, Yan L, et al. High Wilms' tumor 1 mRNA expression correlates with basal-like and ERBB2 molecular subtypes and poor prognosis of breast cancer. Oncol Rep. 2012;28(4):1231–1236. doi: 10.3892/or.2012.1906. [DOI] [PubMed] [Google Scholar]
- 47.Kaneuchi M, Sasaki M, Tanaka Y, Shiina H, Yamada H, Yamamoto R, et al. WT1 and WT1-AS genes are inactivated by promoter methylation in ovarian clear cell adenocarcinoma. Cancer. 2005;104(9):1924–1930. doi: 10.1002/cncr.21397. [DOI] [PubMed] [Google Scholar]
- 48.Lv L, Chen G, Zhou J, Li J, Gong J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J Exp Clin Cancer Res : CR. 2015;34:119. doi: 10.1186/s13046-015-0233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J, et al. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta. 2016;1862(1):12–19. doi: 10.1016/j.bbadis.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Cui L, Nai M, Zhang K, Li L, Li R. lncRNA WT1-AS inhibits the aggressiveness of cervical cancer cell via regulating p53 expression via sponging miR-330-5p. Cancer Manag Res. 2019;11:651–667. doi: 10.2147/CMAR.S176525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai SG, Guo LL, Xia X, Pan Y. Long non-coding RNA WT1-AS inhibits cell aggressiveness via miR-203a-5p/FOXN2 axis and is associated with prognosis in cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23(2):486–495. doi: 10.26355/eurrev_201901_16860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw-data have been uploaded to Gene Expression Omnibus (GEO) (GSE139274, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139274) and is publicly available until October 2020 due to data confidentiality.