Abstract
Gastrointestinal cancers account for more cancer-related deaths than any other organ system, owing in part to difficulties in early detection, treatment response assessment, and post-treatment surveillance. Circulating biomarkers hold the promise for noninvasive liquid biopsy platforms to overcome these obstacles. Although tumors shed detectable levels of degraded genetic material and cellular debris into peripheral blood, identifying reproducible and clinically relevant information from these analytes (eg, cell-free nucleotides, exosomes, proteins) has proven difficult. Cell-based circulating biomarkers also present challenges, but have multiple advantages including allowing for a more comprehensive tumor analysis, and communicating the risk of metastatic spread. Circulating tumor cells have dominated the cancer cell biomarker field with robust evidence in extraintestinal cancers; however, establishing their clinical utility beyond that of prognostication in colorectal and pancreatic cancers has remained elusive. Recently identified novel populations of tumor-derived cells bring renewed potential to this area of investigation. Cancer-associated macrophage-like cells, immune cells with phagocytosed tumor material, also show utility in prognostication and assessing treatment responsiveness. In addition, circulating hybrid cells are the result of tumor–macrophage fusion, with mounting evidence for a role in the metastatic cascade. Because of their relative abundance in circulation, circulating hybrid cells have great potential as a liquid biomarker for early detection, prognostication, and surveillance. In all, the power of the cell reaches beyond enumeration by providing a cellular source of tumor DNA, RNA, and protein, which can be harnessed to impact overall survival.
Keywords: Fusion Hybrid, CAML, CHC, Liquid Biopsy, Macrophage
Abbreviations used in this paper: BMT, bone marrow transplant; CAML, cancer-associated macrophage-like cell; CHC, circulating hybrid cell; CK, cytokeratin; CRC, colorectal cancer; CTC, circulating tumor cell; ctDNA, cell-free tumor DNA; EMT, epithelial-to-mesenchymal transition; EpCAM, epithelial cellular adhesion molecule; GFP, green fluorescent protein; GI, gastrointestinal; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; RFP, red fluorescent protein; TAM, tumor-associated macrophage; TME, tumor microenvironment
Summary.
Circulating cell-based biomarkers, a source of tumor DNA, RNA, and proteins, can be enumerated or provide in-depth tumor analyses to aid in cancer detection and disease monitoring. Here, we review the progress toward this goal and highlight future directions.
Cancers of the gastrointestinal (GI) tract account for more cancer-related deaths in the United States than any other organ site, including pulmonary.1 Each GI cancer has unique challenges in early diagnosis, staging, and treatment that could benefit from improved noninvasive biomarkers to diagnose and track disease evolution. Specifically, as the second leading cause of cancer-related deaths in the United States, colorectal cancer (CRC) accounts for more than 150,000 cancer diagnoses and more than 51,000 deaths annually.1 Despite advances in screening regimens for adults older than age 50 years, new CRC diagnoses in younger adults has increased 1.4% annually since 2004.2 CRC diagnosed after a symptom-initiated work-up often portends an advanced burden of disease and a dramatic decrease in expected survival; Surveillance, Epidemiology and End Results data report 5-year survival for CRC diagnosed as locoregional disease at 80%–90%, compared with 14% in distantly metastatic disease.1, 3, 4 Late-stage diagnosis is even more common in pancreatic ductal adenocarcinoma (PDAC) owing to absent or nonspecific symptoms during the early stages of disease and contributes to its dismal prognosis. Although CRC has multiple effective screening regimens, PDAC currently lacks effective early detection modalities or validated biologic biomarkers,5 however, both cancers would benefit from additional noninvasive modalities for early detection and surveillance.
Noncellular Circulating Biomarkers
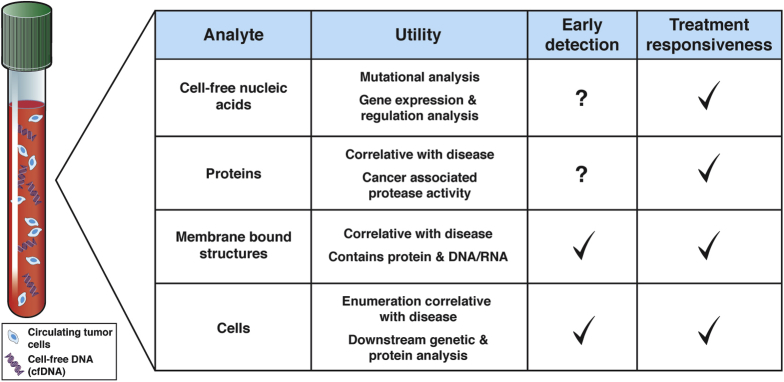
The holy grail of early cancer detection is the development of noninvasive biomarkers that elucidate both the presence of cancer and tumor progression. Current screening methods fall short of this goal. Screening colonoscopies for CRC are recommended for average-risk adults aged 50–75 years and are effective at detecting cancer with the added benefit of removing premalignant adenomas.6 However, colonoscopy is not universally accessible owing to high cost and the need for trained staff with specialized equipment. The fecal occult blood test and fecal immunochemical test are Food and Drug Administration–approved stool assays that expand accessibility but reduce the specificity of CRC detection.7 In addition, gold standard serum biomarkers available for PDAC and CRC, including carcinoembryonic antigen and cancer antigen 19-9, fall far short of reliable usage for diagnosis. Today, these tests are used primarily for surveillance and to monitor disease response during treatment.5, 8 To improve the sensitivity and specificity of cancer detection through noninvasive methods, a new generation of blood-based analytes with correlative or biologic value are in development, including exosomes, cell-free tumor DNA (ctDNA) or nucleic acids, and proteins (Figure 1).9, 10
Figure 1.
Circulating biomarkers in cancer. Summary of analytes detectible in peripheral blood, including cell-free nucleic acids (both DNA and RNA), proteins, membrane-bound structures, and cells. Functional use of each analyte, as well as their suitability for use in early detection and treatment responsiveness, is reported.
ctDNA is hypothesized to arise from tumor cell death, whether by necrosis, cell lysis, or apoptosis, resulting in the release of naked DNA into circulation and creating a residual fingerprint. ctDNA was first detected in healthy individuals in the late 1940s. However, it was not until the 1970s–1980s that neoplastic characteristics were discovered, leading to the realization that cancer patients had higher concentrations of ctDNA relative to healthy controls.11, 12 Quantification of ctDNA is useful in some disease states when used alongside more established blood assays,13 but is better suited for detecting the mutational evolution of cancer. However, despite major technological advances, ctDNA is not readily detectable in all cancers and is scarce in early disease.14 In addition, ctDNA does not always reflect tumor cell biology15 and further complicates its utility as a noninvasive biomarker.
Exosomes are another noncellular analyte sparking excitement in cancer research as an emerging biomarker with the potential to forecast the presence of malignancy, treatment response, and tumor progression. First described in the late 1980s, these small membrane-bound vesicles range in size between 50 and 140 nm and carry cargo that include proteins, DNA, RNA, and various lipid types.16 Detected in a myriad of cancers, exosomes are described to mediate angiogenesis,17 establish a premetastatic niche, and contribute to tumor progression.18 As a liquid biomarker, they remain a promising prognostic and diagnostic analyte, carrying an array of microRNAs that differ significantly between healthy controls and patients with various different cancer types, including glioblastoma multiforme,19, 20 pancreatic,21, 22 colorectal,23 lung,24 and breast cancer.25 The wide variety of cargo carried by exosomes points to their functional relevance in intercellular communication, with the potential to inform tumor state and response to treatment.
Cellular Circulating Biomarkers
Beyond noncellular markers, circulating tumor cells (CTCs) were first identified by Ashworth26 in 1869 in a metastatic cancer patient. CTCs are cells shed into peripheral blood directly from tumors. Although extremely rare in circulation,27 evidence that CTCs correlate with poor prognoses exists in a number of disease sites, including CRC28, 29, 30 and PDAC.31, 32 Conventionally defined CTCs are identified based on the presence of epithelial or tumor markers, typically cytokeratin (CK) or epithelial cellular adhesion molecule (EpCAM), and the absence of the panleukocyte marker CD45. Platforms for CTC detection leverage various features of these cells, including density,33, 34 charge,35, 36, 37 size,38, 39, 40 and associated antigens41, 42, 43, 44, 45 (Table 1), which allow for investigations in early46, 47 and late-stage cancers.48, 49 More recently, stem cell identities in CTCs were shown to correlate with the presence of brain metastases in breast cancer patients,48 supporting that circulating cells harbor important biologic information. This and other evidence suggest that CTCs may be useful in defining discrete differentiation states50 and drug susceptibility.51
Table 1.
CTC Isolation Methods
| Isolation method | Platforms | Advantages | Disadvantages |
|---|---|---|---|
| Density gradient33, 34 | Accucyte (RareCyte, Inc, Seattle, WA) OncoQuick (Greiner Bio-One, Monroe, NC) |
Relatively fast and inexpensive Independent of cellular antigens |
Low specificity |
| Electrophoresis35, 36, 37 | DEPArray (Menarini-Silicon Biosystems, Castel Maggiore, Italy) ApoStream (Precision for Medicine, Frederick, MD) |
Independent of cellular antigens | Pre-enrichment required High cell loss during sample preparation |
| Size-based38, 39, 40 | ISET (Rarecells Diagnostics, Paris France) ScreenCell (Westford, MA) CellSieve |
Relatively fast and inexpensive Captures CTC aggregates Independent of cellular antigens |
Small CTCs not captured Large leukocytes captured |
| Immunoaffinity41, 42, 43, 44, 45 | CellSearch MagSweeper (Stanford University, Stanford, CA) Isoflux (IsoFlux, Inc, Pittsford, NY) Microfluidic chips |
FDA approved (CellSearch) Some platforms are semi-automated Good reproducibility |
Cost Specialized equipment required Dependent on cellular antigens |
FDA, Food and Drug Administration; ISET, isolation-by-size of epithelial tumor cells.
In theory, a cell-based assay for early cancer detection would provide the greatest possible diversity of information, including DNA mutation status, tissue of origin, protein expression for signaling pathway activation, stem cell phenotypes, and gene expression. CTCs are the only cell population with commercially available assays approved for use in cancer-related treatment decisions. Unfortunately, CTCs are rare entities in circulation, even in patients with metastatic cancer. This rarity impedes their utility as a basis for routine transcriptomic or robust protein assessment. To date CTCs have not been shown to provide biologic insights to inform therapeutic decision making, despite initially promising results.52 However, CTCs represent the first identified cell population in an exciting new field, specifically that of circulating cells in cancer patients that have either tumor identity or characteristics that may have utility in a cell-based assay. This review recapitulates the advances made in the field of circulating cellular biomarkers, including a review of CTCs and the discovery of cancer-associated macrophage-like cells (CAMLs),53 culminating in the newly described tumor-derived circulating hybrid cell (CHC).54
CTCs: Prognostic Tumor Dandruff
Since their discovery in the mid-19th century, convincing correlation of CTC detection with disease burden has led to validated commercially available assays (Table 1). The Food and Drug Administration–approved CellSearch system (Menarini Silicon Biosystems Inc, Huntingdon Valley, PA) enriches cells using magnetic ferrofluid-coated antibodies targeting EpCAM for initial separation. Cells screened for the expression of CK8/18/19 and lack of CD45 expression identify EpCAM+/CK+/CD45- cells as CTCs.55 However, it should be noted that the CellSearch approach misses CTC subpopulations that express CD45 (ie, CHCs),54 or that lack EpCAM expression, which can result from an epithelial-to-mesenchymal transition (EMT) and has important prognostic implications.56 Platforms such as the isolation-by-size of epithelial tumor cells technique uses size-based filtration to capture a range of CTC types for downstream analysis and enumeration.57 Although the clinical utility of CTC assays is undisputed in certain organ sites, there remains controversy over the causative role of CTCs in cancer metastasis, as well as their practical applications in early cancer diagnosis and treatment guidance. To this end, CTCs have been studied in a variety of malignancies; the subsequent sections review advances in CTC utilization.
Advances in CTC Utilization
Although CTCs are of interest in GI-derived cancers, they are perhaps best studied in the setting of breast and prostate cancers. Higher levels of CTCs portend poorer survival in locally advanced breast cancer patients undergoing adjuvant treatment, as well as in patients with distant metastatic disease.29, 58, 59 Similarly, CTCs are a valuable biomarker in metastatic, castrate-resistant prostate cancer because levels correlate with overall survival (OS)60 and outperforms prostate-specific antigen (PSA) as a marker of early response after the initiation of chemotherapy.61 In addition, patients with a decrease in CTCs after systemic therapy have improved OS.62 The integration of CTCs into decision making during cancer treatment has remained an elusive goal. To date, the only clinical trial investigating CTC enumeration to measure the effectiveness of chemotherapy and inform a switch to alternative regimens was the SWOG S0500 trial, which did not show a survival difference when using CTC levels to guide chemotherapy regimens in breast cancer patients.63 Upcoming trials investigating the utility of CTCs for this purpose, including the CirCe01 trial (NCT01349842) and the STIC CTC trial (NCT 01710605), may provide additional insight.
CTCs in GI Cancers
There is significant interest in the development of a CTC-based liquid biopsy platform for GI cancers; however, efforts have been stymied by lower CTC detection rates in many GI-derived cancers compared with breast and prostate cancers.64 CellSearch and other platforms detect as low as a single CTC in 7.5 mL of blood, reflecting poor sensitivity, especially at early cancer stages, and have limited the clinical utility of CTCs to impact outcomes based on earlier detection.65 However, improved detection of PDAC has been shown when CTCs are used in combination with other circulating biomarkers, such as cancer antigen 19-9 and doublecortin-like kinase 1.18, 66, 67 Although CTC enumeration has been shown to correlate with disease stage32, 68, 69, 70 and prognosticate future development of metastasis in PDAC,32, 71 some studies have found no correlation.72, 73, 74 Advances in isolation techniques in more recent studies may account for this discrepancy; however, the clinical role of CTC enumeration in PDAC staging remains unclear.
Beyond CTC Enumeration
Although poorer survival has been associated with detectable CTC levels, the reliance on a minute number of cells has led many investigators to look beyond enumeration to molecular and phenotypic characteristics of CTCs for an enhanced prognostic readout in CRC and PDAC. In addition, as with the CellSearch system, the sole reliance on epithelial markers fail to capture subpopulations, which provide prognostic insight.18, 75, 76, 77 By expanding marker profiling, associations between cyclooxygenase-2 (COX2), Leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5), or caudal-type homeobox-2 (CDX2), expression in CTCs have been reported to convey poor OS in CRC patients.78, 79, 80 However, detection of mesenchymal CTCs through the co-expression of CK and mesenchymal markers, such as vimentin or Twist family BHLH Transcription Factor 1 (TWIST1), predict aggressive tumor biology and earlier cancer recurrence in PDAC.69, 70, 75, 77 This supports the theory that EMT facilitates distant metastasis through tumor cell acquisition of mesenchymal properties that are essential for migration and motility.81, 82 Furthermore, the Circulating Tumor Cells in Pancreatic Cancer (CLUSTER) trial (NCT02974764) showed that alterations in total CTC levels and the ratio of mesenchymal CTCs may provide a marker of treatment response and disease progression to help guide therapy decisions.77 There is also evidence that EMT encourages the formation of CTC clusters,83 which have been shown to possess greater metastatic potential than individual CTCs84 and predict worse survival in patients with PDAC,85 as well as metastatic breast cancer patients.86 Although the mechanisms are not fully understood, multicellular clustering in the portal vein may contribute to liver metastasis in PDAC patients by promoting CTC growth and immune cell evasion.87 In addition, a recent study by Szczerba et al88 showed that CTC clusters with associated neutrophils in the peripheral blood of breast cancer patients drives cell-cycle progression, and may be a potential therapeutic target.
A major strength of cellular-based circulating biomarkers is their potential to harness the entire tumor genome and assess treatment-related changes without the need for multiple invasive biopsies. Although KRAS mutations have been detected in CTCs using polymerase chain reaction,73, 89, 90, 91 in-depth genetic analysis at the single-cell level has been challenging.92 Newer technologies that leverage unique CTC features, such as glycoprotein surface expression, can facilitate their isolation for downstream analyses of DNA mutations and gene expression.93 These advances could be developed for the monitoring of CTC subtypes across therapeutic treatment for a real-time readout of tumor burden.
The prognostic value of CTCs is established in a number of malignancies, robust data demonstrating their ability to inform management decisions or detect early disease is lacking. These limitations have prompted investigations for cellular alternatives, leading to the discovery and characterization of new populations of circulating cells in the past decade, including CAMLs and CHCs.
CAMLs: A Circulating Sampler Platter of the Tumor Microenvironment
Adams et al53 reported the isolation of large cells with atypical nuclei and vacuoles of tumor material from blood samples of patients with breast, pancreatic, and prostate cancers in 2014 using low-flow microfiltration. These circulating cells predominantly express CD14, a typical macrophage antigen, thus the name cancer-associated macrophage-like cells. Macrophage and other monocyte-derivative cells are a multifaceted immune population with key roles in maintaining tissue homeostasis through direct cellular functions, such as phagocytosis, and the transmission of immune regulatory cell signals.94 Macrophages are recruited to the tumor microenvironment (TME) through cancer cell–derived cytokines,95 where they can account for up to 50% of a tumor’s mass and become known as tumor-associated macrophages (TAMs).96 It is hypothesized that CAMLs originate from TAMs that have undergone macrophage–macrophage fusion and phagocytose dying neoplastic cells before ultimately disseminating back into circulation with internalized tumor fragments. Although cellular fusion of monocyte-derived giant cells has been well-described in inflammatory diseases, the exact mechanism of CAML formation is largely unknown. However, a portion of their life cycle may derive from ongoing interaction with CTCs in circulation because 10% of studied patients with metastatic cancer have CAMLs bound to CTCs.53
CAMLs are enlarged, highly differentiated cells with atypical or multiple nuclei and phagocytosed tumor matter. As an immune cell population, they are differentiated from CTCs by their CD45 positivity and detection of internalized tumor markers (CK or EpCAM) within the cytoplasm. Notably, CAMLs are large cells that range from 25 to 300 μm and have highly variable morphology,53 although CHCs and CTCs generally are round, with sizes ranging from 5 to 20 μm and 12 to 25 μm, respectively (Table 2).54, 97 In addition, as immune cells, CAMLs are not thought to be directly tumorigenic but may play a role in facilitating metastatic seeding when bound to CTCs by providing a mechanism of immune evasion.53
Table 2.
Characteristics of Circulating Cellular Biomarkers
| Identity | Function | Relative abundancea |
Marker expression |
Morphology | Size, μm | |||
|---|---|---|---|---|---|---|---|---|
| Early stage | Late stage | Cell surface | Cytoplasmic | |||||
| CTC52 | Disseminated tumor cell in circulation | Metastatic seeding | ↑ | ↑↑ | EpCAM | Cytokeratin | Shape: round Nuclei: ≥50% of cytoplasm |
15–25 |
| CAML53 | Macrophage with phagocytosed tumor material | Immune cell functions | ↑↑ | ↑↑↑ | CD45 | Cytokeratin EpCAM |
Shape: variable (amorphous, round, oblong) Nuclei: large or multiple |
25–300 |
| CHC54 | Product of tumor–macrophage fusion Characteristics of both macrophage and tumor cells | Metastatic seeding | ↑↑↑ | ↑↑↑↑ | CD45 EpCAM MUC4 |
Cytokeratin | Shape: round Nuclei: round or binucleated |
5–15 |
MUC4, mucin 4.
Relative abundance based on pancreatic adenocarcinoma data.
CAMLs as a Diagnostic and Prognostic Tool
By using the CellSieve (Creatv MicroTech, Inc., Potomac, MD) microfiltration system, CAMLs have been isolated from peripheral blood in patients with a wide range of cancer types, including esophageal, liver, pancreatic, colorectal, breast, and prostate.98, 99 CAMLs are not detected in healthy patients, but are identified in patients with noninvasive conditions, including benign breast conditions.100 Similar to CTCs, CAMLs have high detection rates in late-stage disease, but lower sensitivity in stage I patients, which ultimately limits their utility for early detection.98 However, CAML enumeration does show promise as an indicator of treatment responsiveness, shown by increased levels in patients after initiating chemotherapy.53 In addition, both increased CAML size and higher levels in untreated breast cancer patients correlated with shorter progression-free survival and worse OS.98, 101
Ultimately, CAMLs provide a promising method for cancer detection, prognostication, and treatment response. However, more research is needed to understand the mechanisms of their formation, function in circulation, and contribution to the metastatic progression of cancer. Interestingly, CAMLs have some similarities with CHCs, another newly described circulating cell population with tumor characteristics. Despite their shared CD45 positivity and tumor marker expression, the large size, variable morphology, and cytoplasmic staining of EpCAM distinguish CAMLs from CHCs. In addition, although both CHCs and CAMLs derive from macrophages, CAMLs retain their immune cell identity, while CHCs are a distinct product of macrophage fusion with tumor cells that imbues properties essential for the initiation of the metastatic cascade.54
CHCs: Chimeras of Groundbreaking Significance
Cellular fusion is a phenomenon by which cells from identical (homotypic) or distinct (heterotypic) lineage combine into a single cell with shared nuclear and cytoplasmic contents, and is known to occur during both homeostatic and inflammatory states.102, 103, 104 Recent investigations in both murine and human models have described fusion in malignant states, where cell fusion hybrids may play an important role in cancer progression and metastasis.105, 106, 107, 108, 109, 110, 111, 112, 113 The concept that fusion between leukocytes and tumor cells may promote cancer metastasis was first postulated by Aichel114 more than a century ago. However, evidence directly linking cellular fusion to phenotypic diversity and cancer metastasis only recently has come to light through the discovery of tumor-derived hybrid cells within the TME and in circulation.54 CHCs represent a possible mechanism of cancer metastasis, and CHC enumeration and analysis may have utility as a diagnostic and prognostic biomarker in human malignancies.
Cellular Fusion in Solid Tumors
Direct observation through in vitro live imaging provides the most compelling evidence of cancer–macrophage fusion. By co-culturing MC-38 colorectal cancer cells expressing red fluorescent protein (RFP) with macrophages expressing green fluorescent protein (GFP), cellular fusion and the formation of hybrid cells harboring cytoplasmic GFP and nuclear RFP has been observed.54 These GFP+/RFP+ hybrid cells retain mitotic activity, producing daughter cells with identical fluorescence expression. Fusion hybrids are identified using similar methods in a murine breast cancer model and in vitro human breast cancer cell lines.115, 116 Together, these data distinguish cellular fusion from other immune cell functions such as phagocytosis and trogocytosis, a process by which leukocytes can extract and express surface antigens from antigen-presenting cells.117
Although all leukocytes may be capable of cellular fusion, the macrophage is the principal fusogenic leukocyte in solid tumors, as evident in mouse models of bone marrow transplants (BMTs) and mouse models of breast cancer.106, 115 Notably, the fusion process involves the up-regulation of genes in pathways linked to metastatic spread, including activated leukocyte cell adhesion molecule (ALCAM) FMS related tyrosine kinase 4 (FLT4), and runt-related transcription factor 1 (RUNX1).106 Further, fusion hybrids show enhanced migratory and invasive properties relative to unfused cancer cells.54 In addition to the acquisition of macrophage properties, hybrids retain tumorigenicity, as evidenced by tumor growth after injection into recipient mice in models of colon and ovarian carcinoma.54, 118
In human malignancies, the most compelling evidence of fusion in solid tumors is reported in female patients who previously received a sex-mismatched BMT and subsequently developed PDAC. When tumor specimens were interrogated with pan-CK antibodies and Y chromosome fluorescence in situ hybridization (FISH) probes, Y chromosomes were discovered within the nuclei of CK+ cancer cells, indicating cellular fusion between male donor leukocytes and recipient pancreatic cells.54 In this way, fusion hybrid cells are identified in several other solid tumors of sex-mismatched BMT patients, including renal cell carcinoma, head and neck squamous cell carcinoma, lung adenocarcinoma, and ovarian carcinoma.54, 118, 119 Fusion hybrids also have been discovered in non–small-cell lung cancer, melanoma, and prostate cancers using other methodologies.120, 121, 122, 123 The full extent to which cellular fusion plays a role in the metastatic cascade remains unknown, but it has been theorized to contribute to the development of chemotherapy resistance, and may induce the Warburg effect.124, 125
Fusion Hybrids: From Tumor to Circulation
In addition to their role in CAML formation, TAMs also serve as the reservoir for intratumoral cellular fusion, and fusion-derived hybrid cells retain properties of both TAMs and tumor cells.54 Subsequently, fusion hybrids are hypothesized to disseminate into circulation, where they are detectable as CHCs. The prevalence of CHCs in peripheral blood of murine cancer models and human cancer patients, combined with their robust tumorigenic capacity, underline their potential role as effectors of cancer metastasis.54 Further supporting this, experimental assays of metastasis using in vitro–derived hybrid cells resulted in pulmonary metastases with notably higher seeding and growth when compared with unfused-MC-38 cells (analogous to CTCs).54 Furthermore, in a spontaneous metastasis model, detectible tumor cells at both the primary and distant metastatic sites were of fusion origin,54 thus providing compelling evidence for the role of CHCs in disease progression.
CHCs appear to be more prevalent in circulation than conventional CTCs. In a murine model of melanoma, CHCs derived from orthotopic injection of RFP+ B16F10 melanoma cells into GFP+ mice were the predominant tumor-derived cell in circulation (identified as RFP+/GFP+).54 Conventionally defined CTCs (RFP+/GFP-) comprised only 10% of all RFP+ circulating cells. Furthermore, of key importance is the fact that the majority of CHCs were shown to express the pan-leukocyte antigen CD45, indicating that dual expression of CD45 and a tumor marker could be translated to human patients for CHC detection.
Reports of atypical circulating macrophage-like cells are surprisingly common in the literature, and it is likely that CHCs have gone unrecognized and unappreciated for some time (Table 3).18, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135 Toyoshima et al127 described atypical cells in immunodeficient mice injected with CD45+/EpCAM+ or CD45-/EpCAM+ tumor cells isolated from patients with advanced gastric cancer. The CD45+ fraction had significantly enhanced tumorigenicity compared with the CD45- fraction, an unanticipated finding.127 Similarly, a CD45+/CK+ cell population isolated from breast cancer patients was reported to prognosticate worse OS, although their significance was not investigated.130
Table 3.
Unappreciated CD45+/CK+ Cells in Prior Publications
| Study | Cancer site | Explanation for CD45+/CK+ cells | Relevant findings |
|---|---|---|---|
| Zhang et al18 | PDAC | Unknown | ND |
| Toyoshima et al127 | Gastric | Unknown | Increased tumorigenicity |
| de Wit et al126 | NSCLC | Unknown | ND |
| Nel et al129 | NSCLC | Unknown | ND |
| Stott et al133 | NSCLC, prostate | Trogocytosis | CD45+/CK+ more prevalent than CTCs |
| Sajay et al128 | NSCLC, breast | Unknown | ND |
| Takao and Takeda132 | NSCLC, breast | False positive | ND |
| Lustberg et al131 | HNSCC, breast | Unknown | ND |
| Lustberg et al130 | Breast | Artifact | ND |
| Riethdorf et al134 | Breast | Artifact | ND |
| Allan et al135 | Breast | Artifact | ND |
HNSCC, head and neck squamous cell carcinoma; ND, not determined; NSCLC, non–small-cell lung cancer.
Clawson et al136 suggested that CHCs occur in human patients. Cells cultured from peripheral blood of melanoma patients expressed macrophage (CD204, CD206, CD163), epithelial (CK, EpCAM), and melanocyte (MLANA, ALCAM) markers and grew tumors when injected into immunodeficient mice. These cells were presumed to be fusion-derived because they were also found in the primary tumors of the melanoma patients.136 Similar findings were reported in PDAC patients137 and by Gast et al,54 who identified CHCs in peripheral blood from the female PDAC patient with a prior sex-mismatched BMT by the expression of CD45, EpCAM, and donor-derived Y chromosome that were also carrying macrophage-specific epitopes and tumor-specific mucin 4 expression. Studies that have reported, but not characterized, the CD45+ fraction are listed in Table 3.
Implications of CHCs on Cancer Diagnosis and Treatment
The existence of CHCs in human malignancies invites questions as to their diagnostic and prognostic potential as a liquid biopsy. Promising early data have shown potential for CHC enumeration to discriminate between PDAC disease stage, with high levels significantly correlating with advanced disease states, and correlating with OS regardless of cancer stage.54 In this patient population, CHC level showed improved performance compared with CTCs, which were not correlated significantly with either stage or OS.54
In summary, CHCs are a newly described circulating cell population in cancer patients, and compelling evidence from multiple investigators has characterized their tumorigenicity, spontaneous formation in murine and human cancers, as well as their relative abundance in circulation. Additionally, CHCs correlate with disease presence, stage, and prognosis. Despite promising early results, CHCs are still a newly described circulating neoplastic cell population with only a few descriptions in the literature54, 136 and therefore further validation of these cells is needed. Furthermore, the utility of CHCs in predicting advanced disease states and in prognostication has yet to be established in larger cohorts of patients with PDAC or across a wider range of GI malignancies. Similarly, the relative prognostic and diagnostic utility of CHCs compared with CTCs across multiple malignancies is untested and requires further study. Nonetheless, there is clearly great promise for this novel cell type.
Discussion
Prognostic biomarkers with enhanced sensitivity and specificity have promise to transform survival from cancer. Thus, the ideal effective biomarker will provide precise and actionable information about tumor location, stage, mutational status, therapeutic vulnerability, and extent of tumor heterogeneity. This is a tall order for a single biomarker. Certainly, dual-analyte biomarkers have enhanced potential for providing the most comprehensive information. For example, a newly developed platform, CancerSEEK (Johns Hopkins Kimmel Cancer Center, Baltimore, MD), combines ctDNA and protein biomarkers to impart tissue localization and a cancer diagnoses in 8 different cancers. Although promising, the ctDNA-based assay showed poor sensitivity for PDAC and other organ sites, and the study lacked inclusion of high-risk cohorts with premalignant pathology.138 Novel multi-analyte platforms could use biomarkers that deliver on multiple fronts: DNA, protein, and cellular information.
Assays built around cells that originate directly from the tumor or the TME have the potential to provide genomic information, mutational analysis, tumor-associated protein identity, the ability to survey epigenetic alterations, and cellular heterogeneity that all derive from the same source. The Achilles’ heel of CTCs is their low prevalence in circulation, which presents a challenge to extend analyses beyond enumeration or protein expression. CHCs show immense promise as a plentiful biomarker for the early detection, diagnosis, and surveillance of a wide range of cancers, and have potential applicability beyond that of CTCs and CAMLs. Further study of this cell population may show an untapped resource for the development of effective biomarkers to impact cancer treatment and survival.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by CA172334, CA2111134, and the Knight Cancer Institute grant P30 CA069533 (M.H.W.).
References
- 1.Noone AM, Howlader N, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER cancer statistics review, 1975-2015. Bethesda, MD: National Cancer Institute. https://seer.cancer.gov/csr/1975_2015/ Available from:
- 2.Dozois E.J., Boardman L.A., Suwanthanma W., Limburg P.J., Cima R.R., Bakken J.L., Vierkant R.A., Aakre J.A., Larson D.W. Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine. 2008;87:259–263. doi: 10.1097/MD.0b013e3181881354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Connell M.J., Campbell M.E., Goldberg R.M., Grothey A., Seitz J.F., Benedetti J.K., Andre T., Haller D.G., Sargent D.J. Survival following recurrence in stage II and III colon cancer: findings from the ACCENT data set. J Clin Oncol. 2008;26:2336–2341. doi: 10.1200/JCO.2007.15.8261. [DOI] [PubMed] [Google Scholar]
- 4.Ryuk J.P., Choi G.S., Park J.S., Kim H.J., Park S.Y., Yoon G.S., Jun S.H., Kwon Y.C. Predictive factors and the prognosis of recurrence of colorectal cancer within 2 years after curative resection. Ann Surg Treat Res. 2014;86:143–151. doi: 10.4174/astr.2014.86.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooperman A.M., Iskandar M.E., Wayne M.G., Steele J.G. Prevention and early detection of pancreatic cancer. Surg Clin North Am. 2018;98:1–12. doi: 10.1016/j.suc.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman D.A., Weiss D.G., Bond J.H., Ahnen D.J., Garewal H., Chejfec G. Use of colonoscopy to screen asymptomatic adults for colorectal cancer. Veterans Affairs Cooperative Study Group 380. N Engl J Med. 2000;343:162–168. doi: 10.1056/NEJM200007203430301. [DOI] [PubMed] [Google Scholar]
- 7.Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit Rev Environ Sci Technol. 2015;45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locker G.Y., Hamilton S., Harris J., Jessup J.M., Kemeny N., Macdonald J.S., Somerfield M.R., Hayes D.F., Bast R.C., Jr. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313–5327. doi: 10.1200/JCO.2006.08.2644. [DOI] [PubMed] [Google Scholar]
- 9.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Dalerba P., Cho R.W., Clarke M.F. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 11.Leon S.A., Shapiro B., Sklaroff D.M., Yaros M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 12.Stroun M., Anker P., Maurice P., Lyautey J., Lederrey C., Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46:318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro B., Chakrabarty M., Cohn E.M., Leon S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::aid-cncr2820511127>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 14.de Kok J.B., van Solinge W.W., Ruers T.J., Roelofs R.W., van Muijen G.N., Willems J.L., Swinkels D.W. Detection of tumour DNA in serum of colorectal cancer patients. Scand J Clin Lab Invest. 1997;57:601–604. doi: 10.3109/00365519709055283. [DOI] [PubMed] [Google Scholar]
- 15.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., Antonarakis E.S., Azad N.S., Bardelli A., Brem H., Cameron J.L., Lee C.C., Fecher L.A., Gallia G.L., Gibbs P., Le D., Giuntoli R.L., Goggins M., Hogarty M.D., Holdhoff M., Hong S.M., Jiao Y., Juhl H.H., Kim J.J., Siravegna G., Laheru D.A., Lauricella C., Lim M., Lipson E.J., Marie S.K., Netto G.J., Oliner K.S., Olivi A., Olsson L., Riggins G.J., Sartore-Bianchi A., Schmidt K., Shih l M., Oba-Shinjo S.M., Siena S., Theodorescu D., Tie J., Harkins T.T., Veronese S., Wang T.L., Weingart J.D., Wolfgang C.L., Wood L.D., Xing D., Hruban R.H., Wu J., Allen P.J., Schmidt C.M., Choti M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Papadopoulos N., Diaz L.A., Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnstone R.M., Adam M., Hammond J.R., Orr L., Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 17.Maji S., Chaudhary P., Akopova I., Nguyen P.M., Hare R.J., Gryczynski I., Vishwanatha J.K. Exosomal annexin II promotes angiogenesis and breast cancer metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Wang F., Ning N., Chen Q., Yang Z., Guo Y., Xu D., Zhang D., Zhan T., Cui W. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. Int J Cancer. 2015;136:1228–1233. doi: 10.1002/ijc.29070. [DOI] [PubMed] [Google Scholar]
- 19.Gourlay J., Morokoff A.P., Luwor R.B., Zhu H.J., Kaye A.H., Stylli S.S. The emergent role of exosomes in glioma. J Clin Neurosci. 2017;35:13–23. doi: 10.1016/j.jocn.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Shao N., Wang L., Xue L., Wang R., Lan Q. Plasma miR-454-3p as a potential prognostic indicator in human glioma. Neurol Sci. 2015;36:309–313. doi: 10.1007/s10072-014-1938-7. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong E.A., Beal E.W., Chakedis J., Paredes A.Z., Moris D., Pawlik T.M., Schmidt C.R., Dillhoff M.E. Exosomes in pancreatic cancer: from early detection to treatment. J Gastrointest Surg. 2018;22:737–750. doi: 10.1007/s11605-018-3693-1. [DOI] [PubMed] [Google Scholar]
- 22.Li W., Li C., Zhou T., Liu X., Liu X., Li X., Chen D. Role of exosomal proteins in cancer diagnosis. Mol Cancer. 2017;16:145. doi: 10.1186/s12943-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., Gunji T., Ohta H., Okamoto H., Sonoda H., Watanabe M., Nakagama H., Yokota J., Kohno T., Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabinowits G., Gercel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 25.Eichelser C., Stuckrath I., Muller V., Milde-Langosch K., Wikman H., Pantel K., Schwarzenbach H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashworth T. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J. 1869;1869:146–147. [Google Scholar]
- 27.Miller M.C., Doyle G.V., Terstappen L.W. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen S.J., Punt C.J., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y., Picus J., Morse M., Mitchell E., Miller M.C., Doyle G.V., Tissing H., Terstappen L.W., Meropol N.J. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 29.Cristofanilli M., Budd G.T., Ellis M.J., Stopeck A., Matera J., Miller M.C., Reuben J.M., Doyle G.V., Allard W.J., Terstappen L.W., Hayes D.F. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 30.Krebs M.G., Sloane R., Priest L., Lancashire L., Hou J.M., Greystoke A., Ward T.H., Ferraldeschi R., Hughes A., Clack G., Ranson M., Dive C., Blackhall F.H. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 31.Torphy R.J., Tignanelli C.J., Kamande J.W., Moffitt R.A., Herrera Loeza S.G., Soper S.A., Yeh J.J. Circulating tumor cells as a biomarker of response to treatment in patient-derived xenograft mouse models of pancreatic adenocarcinoma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Court C.M., Ankeny J.S., Sho S., Winograd P., Hou S., Song M., Wainberg Z.A., Girgis M.D., Graeber T.G., Agopian V.G., Tseng H.R., Tomlinson J.S. Circulating tumor cells predict occult metastatic disease and prognosis in pancreatic cancer. Ann Surg Oncol. 2018;25:1000–1008. doi: 10.1245/s10434-017-6290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg R., Gertler R., Friederichs J., Fuehrer K., Dahm M., Phelps R., Thorban S., Nekarda H., Siewert J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 34.Campton D.E., Ramirez A.B., Nordberg J.J., Drovetto N., Clein A.C., Varshavskaya P., Friemel B.H., Quarre S., Breman A., Dorschner M., Blau S., Blau C.A., Sabath D.E., Stilwell J.L., Kaldjian E.P. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360. doi: 10.1186/s12885-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elvington E.S., Salmanzadeh A., Stremler M.A., Davalos R.V. Label-free isolation and enrichment of cells through contactless dielectrophoresis. J Vis Exp. 2013 doi: 10.3791/50634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez S.V., Bingham C., Fittipaldi P., Austin L., Palazzo J., Palmer G., Alpaugh K., Cristofanilli M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 2014;16:445. doi: 10.1186/s13058-014-0445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta V., Jafferji I., Garza M., Melnikova V.O., Hasegawa D.K., Pethig R., Davis D.W. ApoStream, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freidin M.B., Tay A., Freydina D.V., Chudasama D., Nicholson A.G., Rice A., Anikin V., Lim E. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer. 2014;85:182–185. doi: 10.1016/j.lungcan.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Farace F., Massard C., Vimond N., Drusch F., Jacques N., Billiot F., Laplanche A., Chauchereau A., Lacroix L., Planchard D., Le Moulec S., Andre F., Fizazi K., Soria J.C., Vielh P. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams D.L., Stefansson S., Haudenschild C., Martin S.S., Charpentier M., Chumsri S., Cristofanilli M., Tang C.M., Alpaugh R.K. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch(®) CTC test. Cytometry A. 2015;87:137–144. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 41.Raimondi C., Nicolazzo C., Gradilone A., Giannini G., De Falco E., Chimenti I., Varriale E., Hauch S., Plappert L., Cortesi E., Gazzaniga P. Circulating tumor cells: exploring intratumor heterogeneity of colorectal cancer. Cancer Biol Ther. 2014;15:496–503. doi: 10.4161/cbt.28020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talasaz A.H., Powell A.A., Huber D.E., Berbee J.G., Roh K.H., Yu W., Xiao W., Davis M.M., Pease R.F., Mindrinos M.N., Jeffrey S.S., Davis R.W. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009;106:3970–3975. doi: 10.1073/pnas.0813188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winer-Jones J.P., Vahidi B., Arquilevich N., Fang C., Ferguson S., Harkins D., Hill C., Klem E., Pagano P.C., Peasley C., Romero J., Shartle R., Vasko R.C., Strauss W.M., Dempsey P.W. Circulating tumor cells: clinically relevant molecular access based on a novel CTC flow cell. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagrath S., Sequist L.V., Maheswaran S., Bell D.W., Irimia D., Ulkus L., Smith M.R., Kwak E.L., Digumarthy S., Muzikansky A., Ryan P., Balis U.J., Tompkins R.G., Haber D.A., Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brychta N., Drosch M., Driemel C., Fischer J.C., Neves R.P., Esposito I., Knoefel W., Mohlendick B., Hille C., Stresemann A., Krahn T., Kassack M.U., Stoecklein N.H., von Ahsen O. Isolation of circulating tumor cells from pancreatic cancer by automated filtration. Oncotarget. 2017;8:86143–86156. doi: 10.18632/oncotarget.21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Dalum G., Stam G.J., Scholten L.F., Mastboom W.J., Vermes I., Tibbe A.G., De Groot M.R., Terstappen L.W. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int J Oncol. 2015;46:1361–1368. doi: 10.3892/ijo.2015.2824. [DOI] [PubMed] [Google Scholar]
- 47.van Dalum G., van der Stam G.J., Tibbe A.G., Franken B., Mastboom W.J., Vermes I., de Groot M.R., Terstappen L.W. Circulating tumor cells before and during follow-up after breast cancer surgery. Int J Oncol. 2015;46:407–413. doi: 10.3892/ijo.2014.2694. [DOI] [PubMed] [Google Scholar]
- 48.Boral D., Vishnoi M., Liu H.N., Yin W., Sprouse M.L., Scamardo A., Hong D.S., Tan T.Z., Thiery J.P., Chang J.C., Marchetti D. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat Commun. 2017;8:196. doi: 10.1038/s41467-017-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giuliano M., Giordano A., Jackson S., De Giorgi U., Mego M., Cohen E.N., Gao H., Anfossi S., Handy B.C., Ueno N.T., Alvarez R.H., De Placido S., Valero V., Hortobagyi G.N., Reuben J.M., Cristofanilli M. Circulating tumor cells as early predictors of metastatic spread in breast cancer patients with limited metastatic dissemination. Breast Cancer Res. 2014;16:440. doi: 10.1186/s13058-014-0440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jordan N.V., Bardia A., Wittner B.S., Benes C., Ligorio M., Zheng Y., Yu M., Sundaresan T.K., Licausi J.A., Desai R., O'Keefe R.M., Ebright R.Y., Boukhali M., Sil S., Onozato M.L., Iafrate A.J., Kapur R., Sgroi D., Ting D.T., Toner M., Ramaswamy S., Haas W., Maheswaran S., Haber D.A. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537:102–106. doi: 10.1038/nature19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu M., Bardia A., Aceto N., Bersani F., Madden M.W., Donaldson M.C., Desai R., Zhu H., Comaills V., Zheng Z., Wittner B.S., Stojanov P., Brachtel E., Sgroi D., Kapur R., Shioda T., Ting D.T., Ramaswamy S., Getz G., Iafrate A.J., Benes C., Toner M., Maheswaran S., Haber D.A. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pimienta M., Edderkaoui M., Wang R., Pandol S. The potential for circulating tumor cells in pancreatic cancer management. Front Physiol. 2017;8:381. doi: 10.3389/fphys.2017.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams D.L., Martin S.S., Alpaugh R.K., Charpentier M., Tsai S., Bergan R.C., Ogden I.M., Catalona W., Chumsri S., Tang C.M., Cristofanilli M. Circulating giant macrophages as a potential biomarker of solid tumors. Proc Natl Acad Sci U S A. 2014;111:3514–3519. doi: 10.1073/pnas.1320198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gast C.E., Silk A.D., Zarour L., Riegler L., Burkhart J.G., Gustafson K.T., Parappilly M.S., Roh-Johnson M., Goodman J.R., Olson B., Schmidt M., Swain J.R., Davies P.S., Shasthri V., Iizuka S., Flynn P., Watson S., Korkola J., Courtneidge S.A., Fischer J.M., Jaboin J., Billingsley K.G., Lopez C.D., Burchard J., Gray J., Coussens L.M., Sheppard B.C., Wong M.H. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv. 2018;4 doi: 10.1126/sciadv.aat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millner L.M., Linder M.W., Valdes R., Jr. Circulating tumor cells: a review of present methods and the need to identify heterogeneous phenotypes. Ann Clin Lab Sci. 2013;43:295–304. [PMC free article] [PubMed] [Google Scholar]
- 56.Andree K.C., van Dalum G., Terstappen L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol. 2016;10:395–407. doi: 10.1016/j.molonc.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen F., Wang S., Fang Y., Zheng L., Zhi X., Cheng B., Chen Y., Zhang C., Shi D., Song H., Cai C., Zhou P., Xiong B. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application. Oncotarget. 2017;8:3029–3041. doi: 10.18632/oncotarget.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lucci A., Hall C.S., Lodhi A.K., Bhattacharyya A., Anderson A.E., Xiao L., Bedrosian I., Kuerer H.M., Krishnamurthy S. Circulating tumour cells in non-metastatic breast cancer: a prospective study. Lancet Oncol. 2012;13:688–695. doi: 10.1016/S1470-2045(12)70209-7. [DOI] [PubMed] [Google Scholar]
- 59.Bidard F.C., Michiels S., Riethdorf S., Mueller V., Esserman L.J., Lucci A., Naume B., Horiguchi J., Gisbert-Criado R., Sleijfer S., Toi M., Garcia-Saenz J.A., Hartkopf A., Generali D., Rothe F., Smerage J., Muinelo-Romay L., Stebbing J., Viens P., Magbanua M.J.M., Hall C.S., Engebraaten O., Takata D., Vidal-Martinez J., Onstenk W., Fujisawa N., Diaz-Rubio E., Taran F.A., Cappelletti M.R., Ignatiadis M., Proudhon C., Wolf D.M., Bauldry J.B., Borgen E., Nagaoka R., Caranana V., Kraan J., Maestro M., Brucker S.Y., Weber K., Reyal F., Amara D., Karhade M.G., Mathiesen R.R., Tokiniwa H., Llombart-Cussac A., Meddis A., Blanche P., d'Hollander K., Cottu P., Park J.W., Loibl S., Latouche A., Pierga J.Y., Pantel K. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J Natl Cancer Inst. 2018;110:560–567. doi: 10.1093/jnci/djy018. [DOI] [PubMed] [Google Scholar]
- 60.Danila D.C., Heller G., Gignac G.A., Gonzalez-Espinoza R., Anand A., Tanaka E., Lilja H., Schwartz L., Larson S., Fleisher M., Scher H.I. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–7058. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 61.Scher H.I., Jia X., de Bono J.S., Fleisher M., Pienta K.J., Raghavan D., Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lorente D., Olmos D., Mateo J., Bianchini D., Seed G., Fleisher M., Danila D.C., Flohr P., Crespo M., Figueiredo I., Miranda S., Baeten K., Molina A., Kheoh T., McCormack R., Terstappen L.W., Scher H.I., de Bono J.S. Decline in circulating tumor cell count and treatment outcome in advanced prostate cancer. Eur Urol. 2016;70:985–992. doi: 10.1016/j.eururo.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smerage J.B., Barlow W.E., Hortobagyi G.N., Winer E.P., Leyland-Jones B., Srkalovic G., Tejwani S., Schott A.F., O'Rourke M.A., Lew D.L., Doyle G.V., Gralow J.R., Livingston R.B., Hayes D.F. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32:3483–3489. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allard W.J., Matera J., Miller M.C., Repollet M., Connelly M.C., Rao C., Tibbe A.G., Uhr J.W., Terstappen L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 65.Nagrath S., Jack R.M., Sahai V., Simeone D.M. Opportunities and challenges for pancreatic circulating tumor cells. Gastroenterology. 2016;151:412–426. doi: 10.1053/j.gastro.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 66.Xu Y., Qin T., Li J., Wang X., Gao C., Xu C., Hao J., Liu J., Gao S., Ren H. Detection of circulating tumor cells using negative enrichment immunofluorescence and an in situ hybridization system in pancreatic cancer. Int J Mol Sci. 2017;18:E622. doi: 10.3390/ijms18040622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qu D., Johnson J., Chandrakesan P., Weygant N., May R., Aiello N., Rhim A., Zhao L., Zheng W., Lightfoot S., Pant S., Irvan J., Postier R., Hocker J., Hanas J.S., Ali N., Sureban S.M., An G., Schlosser M.J., Stanger B., Houchen C.W. Doublecortin-like kinase 1 is elevated serologically in pancreatic ductal adenocarcinoma and widely expressed on circulating tumor cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ankeny J.S., Court C.M., Hou S., Li Q., Song M., Wu D., Chen J.F., Lee T., Lin M., Sho S., Rochefort M.M., Girgis M.D., Yao J., Wainberg Z.A., Muthusamy V.R., Watson R.R., Donahue T.R., Hines O.J., Reber H.A., Graeber T.G., Tseng H.R., Tomlinson J.S. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br J Cancer. 2016;114:1367–1375. doi: 10.1038/bjc.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao X.H., Wang Z.R., Chen C.L., Di L., Bi Z.F., Li Z.H., Liu Y.M. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: potential role in clinical practice. World J Gastroenterol. 2019;25:138–150. doi: 10.3748/wjg.v25.i1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao R., Cai Z., Li S., Cheng Y., Gao H., Liu F., Wu S., Liu S., Dong Y., Zheng L., Zhang W., Wu X., Yao X. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget. 2017;8:9293–9302. doi: 10.18632/oncotarget.14065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsai W.S., Chen J.S., Shao H.J., Wu J.C., Lai J.M., Lu S.H., Hung T.F., Chiu Y.C., You J.F., Hsieh P.S., Yeh C.Y., Hung H.Y., Chiang S.F., Lin G.P., Tang R., Chang Y.C. Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci Rep. 2016;6:24517. doi: 10.1038/srep24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Albuquerque A., Kubisch I., Breier G., Stamminger G., Fersis N., Eichler A., Kaul S., Stolzel U. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. Oncology. 2012;82:3–10. doi: 10.1159/000335479. [DOI] [PubMed] [Google Scholar]
- 73.Kulemann B., Pitman M.B., Liss A.S., Valsangkar N., Fernandez-Del Castillo C., Lillemoe K.D., Hoeppner J., Mino-Kenudson M., Warshaw A.L., Thayer S.P. Circulating tumor cells found in patients with localized and advanced pancreatic cancer. Pancreas. 2015;44:547–550. doi: 10.1097/MPA.0000000000000324. [DOI] [PubMed] [Google Scholar]
- 74.Bessa X., Elizalde J.I., Boix L., Pinol V., Lacy A.M., Salo J., Pique J.M., Castells A. Lack of prognostic influence of circulating tumor cells in peripheral blood of patients with colorectal cancer. Gastroenterology. 2001;120:1084–1092. doi: 10.1053/gast.2001.23245. [DOI] [PubMed] [Google Scholar]
- 75.Poruk K.E., Valero V., 3rd, Saunders T., Blackford A.L., Griffin J.F., Poling J., Hruban R.H., Anders R.A., Herman J., Zheng L., Rasheed Z.A., Laheru D.A., Ahuja N., Weiss M.J., Cameron J.L., Goggins M., Iacobuzio-Donahue C.A., Wood L.D., Wolfgang C.L. Circulating tumor cell phenotype predicts recurrence and survival in pancreatic adenocarcinoma. Ann Surg. 2016;264:1073–1081. doi: 10.1097/SLA.0000000000001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poruk K.E., Blackford A.L., Weiss M.J., Cameron J.L., He J., Goggins M., Rasheed Z.A., Wolfgang C.L., Wood L.D. Circulating tumor cells expressing markers of tumor-initiating cells predict poor survival and cancer recurrence in patients with pancreatic ductal adenocarcinoma. Clin Cancer Res. 2017;23:2681–2690. doi: 10.1158/1078-0432.CCR-16-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gemenetzis G., Groot V.P., Yu J., Ding D., Teinor J.A., Javed A.A., Wood L.D., Burkhart R.A., Cameron J.L., Makary M.A., Weiss M.J., He J., Wolfgang C.L. Circulating tumor cells dynamics in pancreatic adenocarcinoma correlate with disease status: results of the prospective CLUSTER study. Ann Surg. 2018;268:408–420. doi: 10.1097/SLA.0000000000002925. [DOI] [PubMed] [Google Scholar]
- 78.Cai J., Huang L., Huang J., Kang L., Lin H., Huang P., Zhu P., Wang J., Dong J., Wang L., Xian C.J. Associations between the cyclooxygenase-2 expression in circulating tumor cells and the clinicopathological features of patients with colorectal cancer. J Cell Biochem. 2019;120:4935–4941. doi: 10.1002/jcb.27768. [DOI] [PubMed] [Google Scholar]
- 79.Messaritakis I., Sfakianaki M., Papadaki C., Koulouridi A., Vardakis N., Koinis F., Hatzidaki D., Georgoulia N., Kladi A., Kotsakis A., Souglakos J., Georgoulias V. Prognostic significance of CEACAM5mRNA-positive circulating tumor cells in patients with metastatic colorectal cancer. Cancer Chemother Pharmacol. 2018;82:767–775. doi: 10.1007/s00280-018-3666-9. [DOI] [PubMed] [Google Scholar]
- 80.Wang W., Wan L., Wu S., Yang J., Zhou Y., Liu F., Wu Z., Cheng Y. Mesenchymal marker and LGR5 expression levels in circulating tumor cells correlate with colorectal cancer prognosis. Cell Oncol (Dordr) 2018;41:495–504. doi: 10.1007/s13402-018-0386-4. [DOI] [PubMed] [Google Scholar]
- 81.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 83.Haeger A., Krause M., Wolf K., Friedl P. Cell jamming: collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim Biophys Acta. 2014;1840:2386–2395. doi: 10.1016/j.bbagen.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 84.Cheung K.J., Ewald A.J. A collective route to metastasis: seeding by tumor cell clusters. Science. 2016;352:167–169. doi: 10.1126/science.aaf6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chang M.C., Chang Y.T., Chen J.Y., Jeng Y.M., Yang C.Y., Tien Y.W., Yang S.H., Chen H.L., Liang T.Y., Wang C.F., Lee E.Y., Chang Y.C., Lee W.H. Clinical significance of circulating tumor microemboli as a prognostic marker in patients with pancreatic ductal adenocarcinoma. Clin Chem. 2016;62:505–513. doi: 10.1373/clinchem.2015.248260. [DOI] [PubMed] [Google Scholar]
- 86.Wang C., Mu Z., Chervoneva I., Austin L., Ye Z., Rossi G., Palazzo J.P., Sun C., Abu-Khalaf M., Myers R.E., Zhu Z., Ba Y., Li B., Hou L., Cristofanilli M., Yang H. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res Treat. 2017;161:83–94. doi: 10.1007/s10549-016-4026-2. [DOI] [PubMed] [Google Scholar]
- 87.Arnoletti J.P., Fanaian N., Reza J., Sause R., Almodovar A.J., Srivastava M., Patel S., Veldhuis P.P., Griffith E., Shao Y.P., Zhu X., Litherland S.A. Pancreatic and bile duct cancer circulating tumor cells (CTC) form immune-resistant multi-cell type clusters in the portal venous circulation. Cancer Biol Ther. 2018;19:887–897. doi: 10.1080/15384047.2018.1480292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szczerba B.M., Castro-Giner F., Vetter M., Krol I., Gkountela S., Landin J., Scheidmann M.C., Donato C., Scherrer R., Singer J., Beisel C., Kurzeder C., Heinzelmann-Schwarz V., Rochlitz C., Weber W.P., Beerenwinkel N., Aceto N. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566:553–557. doi: 10.1038/s41586-019-0915-y. [DOI] [PubMed] [Google Scholar]
- 89.Brychta N., Krahn T., von Ahsen O. Detection of KRAS mutations in circulating tumor DNA by digital PCR in early stages of pancreatic cancer. Clin Chem. 2016;62:1482–1491. doi: 10.1373/clinchem.2016.257469. [DOI] [PubMed] [Google Scholar]
- 90.Timme-Bronsert S., Bronsert P., Werner M., Kulemann B., Hoppner J. [Circulating tumor cells in pancreatic cancer: results of morphological and molecular analyses and comparisons with the primary tumor] Der Pathologe. 2018;39:311–314. doi: 10.1007/s00292-018-0550-7. [DOI] [PubMed] [Google Scholar]
- 91.Kidess-Sigal E., Liu H.E., Triboulet M.M., Che J., Ramani V.C., Visser B.C., Poultsides G.A., Longacre T.A., Marziali A., Vysotskaia V., Wiggin M., Heirich K., Hanft V., Keilholz U., Tinhofer I., Norton J.A., Lee M., Sollier-Christen E., Jeffrey S.S. Enumeration and targeted analysis of KRAS, BRAF and PIK3CA mutations in CTCs captured by a label-free platform: comparison to ctDNA and tissue in metastatic colorectal cancer. Oncotarget. 2016;7:85349–85364. doi: 10.18632/oncotarget.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Court C.M., Ankeny J.S., Sho S., Hou S., Li Q., Hsieh C., Song M., Liao X., Rochefort M.M., Wainberg Z.A., Graeber T.G., Tseng H.R., Tomlinson J.S. Reality of single circulating tumor cell sequencing for molecular diagnostics in pancreatic cancer. J Mol Diagn. 2016;18:688–696. doi: 10.1016/j.jmoldx.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neves M., Azevedo R., Lima L., Oliveira M.I., Peixoto A., Ferreira D., Soares J., Fernandes E., Gaiteiro C., Palmeira C., Cotton S., Mereiter S., Campos D., Afonso L.P., Ribeiro R., Fraga A., Tavares A., Mansinho H., Monteiro E., Videira P.A., Freitas P.P., Reis C.A., Santos L.L., Dieguez L., Ferreira J.A. Exploring sialyl-Tn expression in microfluidic-isolated circulating tumour cells: a novel biomarker and an analytical tool for precision oncology applications. N Biotechnol. 2019;49:77–87. doi: 10.1016/j.nbt.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 94.Malyshev I., Malyshev Y. Current concept and update of the macrophage plasticity concept: intracellular mechanisms of reprogramming and m3 macrophage "switch" phenotype. Biomed Res Int. 2015;2015:341308. doi: 10.1155/2015/341308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morita Y., Zhang R., Leslie M., Adhikari S., Hasan N., Chervoneva I., Rui H., Tanaka T. Pathologic evaluation of tumor-associated macrophage density and vessel inflammation in invasive breast carcinomas. Oncol Lett. 2017;14:2111–2118. doi: 10.3892/ol.2017.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J., Bae J.S. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediat Inflamm. 2016;2016:6058147. doi: 10.1155/2016/6058147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hao S.J., Wan Y., Xia Y.Q., Zou X., Zheng S.Y. Size-based separation methods of circulating tumor cells. Adv Drug Deliv Rev. 2018;125:3–20. doi: 10.1016/j.addr.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 98.Tang C.M., Zhu P., Li S., Makarova O.V., Amstutz P.T., Adams D.L. Blood-based biopsies-clinical utility beyond circulating tumor cells. Cytometry A. 2018;93:1246–1250. doi: 10.1002/cyto.a.23573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mu Z., Benali-Furet N., Uzan G., Znaty A., Ye Z., Paolillo C., Wang C., Austin L., Rossi G., Fortina P., Yang H., Cristofanilli M. Detection and characterization of circulating tumor associated cells in metastatic breast cancer. Int J Mol Sci. 2016;17:E1665. doi: 10.3390/ijms17101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adams D.L., Adams D.K., Alpaugh R.K., Cristofanilli M., Martin S.S., Chumsri S., Tang C.M., Marks J.R. Circulating cancer-associated macrophage-like cells differentiate malignant breast cancer and benign breast conditions. Cancer Epidemiol Biomarkers Prev. 2016;25:1037–1042. doi: 10.1158/1055-9965.EPI-15-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mu Z., Wang C., Ye Z., Rossi G., Sun C., Li L., Zhu Z., Yang H., Cristofanilli M. Prognostic values of cancer associated macrophage-like cells (CAML) enumeration in metastatic breast cancer. Breast Cancer Res Treat. 2017;165:733–741. doi: 10.1007/s10549-017-4372-8. [DOI] [PubMed] [Google Scholar]
- 102.Singec I., Snyder E.Y. Inflammation as a matchmaker: revisiting cell fusion. Nat Cell Biol. 2008;10:503–505. doi: 10.1038/ncb0508-503. [DOI] [PubMed] [Google Scholar]
- 103.Johansson C.B., Youssef S., Koleckar K., Holbrook C., Doyonnas R., Corbel S.Y., Steinman L., Rossi F.M., Blau H.M. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat Cell Biol. 2008;10:575–583. doi: 10.1038/ncb1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nygren J.M., Liuba K., Breitbach M., Stott S., Thoren L., Roell W., Geisen C., Sasse P., Kirik D., Bjorklund A., Nerlov C., Fleischmann B.K., Jovinge S., Jacobsen S.E. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat Cell Biol. 2008;10:584–592. doi: 10.1038/ncb1721. [DOI] [PubMed] [Google Scholar]
- 105.Rizvi A.Z., Swain J.R., Davies P.S., Bailey A.S., Decker A.D., Willenbring H., Grompe M., Fleming W.H., Wong M.H. Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci U S A. 2006;103:6321–6325. doi: 10.1073/pnas.0508593103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Powell A.E., Anderson E.C., Davies P.S., Silk A.D., Pelz C., Impey S., Wong M.H. Fusion between intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011;71:1497–1505. doi: 10.1158/0008-5472.CAN-10-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davies P.S., Powell A.E., Swain J.R., Wong M.H. Inflammation and proliferation act together to mediate intestinal cell fusion. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Silk A.D., Gast C.E., Davies P.S., Fakhari F.D., Vanderbeek G.E., Mori M., Wong M.H. Fusion between hematopoietic and epithelial cells in adult human intestine. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Duelli D., Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 110.Pawelek J.M. Tumour-cell fusion as a source of myeloid traits in cancer. Lancet Oncol. 2005;6:988–993. doi: 10.1016/S1470-2045(05)70466-6. [DOI] [PubMed] [Google Scholar]
- 111.Pawelek J.M. Tumour cell hybridization and metastasis revisited. Melanoma Res. 2000;10:507–514. doi: 10.1097/00008390-200012000-00001. [DOI] [PubMed] [Google Scholar]
- 112.Pawelek J.M., Chakraborty A.K. Fusion of tumour cells with bone marrow-derived cells: a unifying explanation for metastasis. Nat Rev Cancer. 2008;8:377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 113.Pawelek J.M., Chakraborty A.K. The cancer cell--leukocyte fusion theory of metastasis. Adv Cancer Res. 2008;101:397–444. doi: 10.1016/S0065-230X(08)00410-7. [DOI] [PubMed] [Google Scholar]
- 114.Aichel O. Über zellverschmelzung mit qualitativ abnormer chromosomenverteilung als ursache der geschwulstbildung [About cell fusion with qualitatively abnormal. chromosome distribution as cause for tumor formation] Vorträge Aufsätze Entvickelungsmechanik Organismen. 1911;XIII:92–111. [Google Scholar]
- 115.Lizier M., Anselmo A., Mantero S., Ficara F., Paulis M., Vezzoni P., Lucchini F., Pacchiana G. Fusion between cancer cells and macrophages occurs in a murine model of spontaneous neu+ breast cancer without increasing its metastatic potential. Oncotarget. 2016;7:60793–60806. doi: 10.18632/oncotarget.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rappa G., Mercapide J., Lorico A. Spontaneous formation of tumorigenic hybrids between breast cancer and multipotent stromal cells is a source of tumor heterogeneity. Am J Pathol. 2012;180:2504–2515. doi: 10.1016/j.ajpath.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joly E., Hudrisier D. What is trogocytosis and what is its purpose? Nat Immunol. 2003;4:815. doi: 10.1038/ni0903-815. [DOI] [PubMed] [Google Scholar]
- 118.Ramakrishnan M., Mathur S.R., Mukhopadhyay A. Fusion-derived epithelial cancer cells express hematopoietic markers and contribute to stem cell and migratory phenotype in ovarian carcinoma. Cancer Res. 2013;73:5360–5370. doi: 10.1158/0008-5472.CAN-13-0896. [DOI] [PubMed] [Google Scholar]
- 119.Yilmaz Y., Lazova R., Qumsiyeh M., Cooper D., Pawelek J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005;35:1021–1024. doi: 10.1038/sj.bmt.1704939. [DOI] [PubMed] [Google Scholar]
- 120.Xu M.H., Gao X., Luo D., Zhou X.D., Xiong W., Liu G.X. EMT and acquisition of stem cell-like properties are involved in spontaneous formation of tumorigenic hybrids between lung cancer and bone marrow-derived mesenchymal stem cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.LaBerge G.S., Duvall E., Grasmick Z., Haedicke K., Pawelek J. A melanoma lymph node metastasis with a donor-patient hybrid genome following bone marrow transplantation: a second case of leucocyte-tumor cell hybridization in cancer metastasis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0168581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lazova R., Laberge G.S., Duvall E., Spoelstra N., Klump V., Sznol M., Cooper D., Spritz R.A., Chang J.T., Pawelek J.M. A melanoma brain metastasis with a donor-patient hybrid genome following bone marrow transplantation: first evidence for fusion in human cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo F., Liu T., Wang J., Li J., Ma P., Ding H., Feng G., Lin D., Xu Y., Yang K. Bone marrow mesenchymal stem cells participate in prostate carcinogenesis and promote growth of prostate cancer by cell fusion in vivo. Oncotarget. 2016;7:30924–30934. doi: 10.18632/oncotarget.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dittmar T., Zanker K.S. Tissue regeneration in the chronically inflamed tumor environment: implications for cell fusion driven tumor progression and therapy resistant tumor hybrid cells. Int J Mol Sci. 2015;16:30362–30381. doi: 10.3390/ijms161226240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lazova R., Chakraborty A., Pawelek J.M. Leukocyte-cancer cell fusion: initiator of the warburg effect in malignancy? Adv Exp Med Biol. 2011;714:151–172. doi: 10.1007/978-94-007-0782-5_8. [DOI] [PubMed] [Google Scholar]
- 126.de Wit S., Zeune L.L., Hiltermann T.J.N., Groen H.J.M., Dalum G.V., Terstappen L. Classification of cells in CTC-enriched samples by advanced image analysis. Cancers. 2018;10:377. doi: 10.3390/cancers10100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Toyoshima K., Hayashi A., Kashiwagi M., Hayashi N., Iwatsuki M., Ishimoto T., Baba Y., Baba H., Ohta Y. Analysis of circulating tumor cells derived from advanced gastric cancer. Int J Cancer. 2015;137:991–998. doi: 10.1002/ijc.29455. [DOI] [PubMed] [Google Scholar]
- 128.Sajay B.N., Chang C.P., Ahmad H., Khuntontong P., Wong C.C., Wang Z., Puiu P.D., Soo R., Rahman A.R. Microfluidic platform for negative enrichment of circulating tumor cells. Biomed Microdevices. 2014;16:537–548. doi: 10.1007/s10544-014-9856-2. [DOI] [PubMed] [Google Scholar]
- 129.Nel I., Jehn U., Gauler T., Hoffmann A.C. Individual profiling of circulating tumor cell composition in patients with non-small cell lung cancer receiving platinum based treatment. Transl Lung Cancer Res. 2014;3:100–106. doi: 10.3978/j.issn.2218-6751.2014.03.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lustberg M.B., Balasubramanian P., Miller B., Garcia-Villa A., Deighan C., Wu Y., Carothers S., Berger M., Ramaswamy B., Macrae E.R., Wesolowski R., Layman R.M., Mrozek E., Pan X., Summers T.A., Shapiro C.L., Chalmers J.J. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014;16:R23. doi: 10.1186/bcr3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lustberg M., Jatana K.R., Zborowski M., Chalmers J.J. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 2012;195:97–110. doi: 10.1007/978-3-642-28160-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takao M., Takeda K. Enumeration, characterization, and collection of intact circulating tumor cells by cross contamination-free flow cytometry. Cytometry A. 2011;79:107–117. doi: 10.1002/cyto.a.21014. [DOI] [PubMed] [Google Scholar]
- 133.Stott S.L., Hsu C.H., Tsukrov D.I., Yu M., Miyamoto D.T., Waltman B.A., Rothenberg S.M., Shah A.M., Smas M.E., Korir G.K., Floyd F.P., Jr., Gilman A.J., Lord J.B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L.V., Lee R.J., Isselbacher K.J., Maheswaran S., Haber D.A., Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riethdorf S., Fritsche H., Muller V., Rau T., Schindlbeck C., Rack B., Janni W., Coith C., Beck K., Janicke F., Jackson S., Gornet T., Cristofanilli M., Pantel K. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 135.Allan A.L., Vantyghem S.A., Tuck A.B., Chambers A.F., Chin-Yee I.H., Keeney M. Detection and quantification of circulating tumor cells in mouse models of human breast cancer using immunomagnetic enrichment and multiparameter flow cytometry. Cytometry A. 2005;65:4–14. doi: 10.1002/cyto.a.20132. [DOI] [PubMed] [Google Scholar]
- 136.Clawson G.A., Matters G.L., Xin P., Imamura-Kawasawa Y., Du Z., Thiboutot D.M., Helm K.F., Neves R.I., Abraham T. Macrophage-tumor cell fusions from peripheral blood of melanoma patients. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Clawson G.A., Matters G.L., Xin P., McGovern C., Wafula E., dePamphilis C., Meckley M., Wong J., Stewart L., D'Jamoos C., Altman N., Imamura Kawasawa Y., Du Z., Honaas L., Abraham T. "Stealth dissemination" of macrophage-tumor cell fusions cultured from blood of patients with pancreatic ductal adenocarcinoma. PLoS One. 2017;12 doi: 10.1371/journal.pone.0184451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., Hruban R.H., Wolfgang C.L., Goggins M.G., Dal Molin M., Wang T.L., Roden R., Klein A.P., Ptak J., Dobbyn L., Schaefer J., Silliman N., Popoli M., Vogelstein J.T., Browne J.D., Schoen R.E., Brand R.E., Tie J., Gibbs P., Wong H.L., Mansfield A.S., Jen J., Hanash S.M., Falconi M., Allen P.J., Zhou S., Bettegowda C., Diaz L.A., Jr., Tomasetti C., Kinzler K.W., Vogelstein B., Lennon A.M., Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]