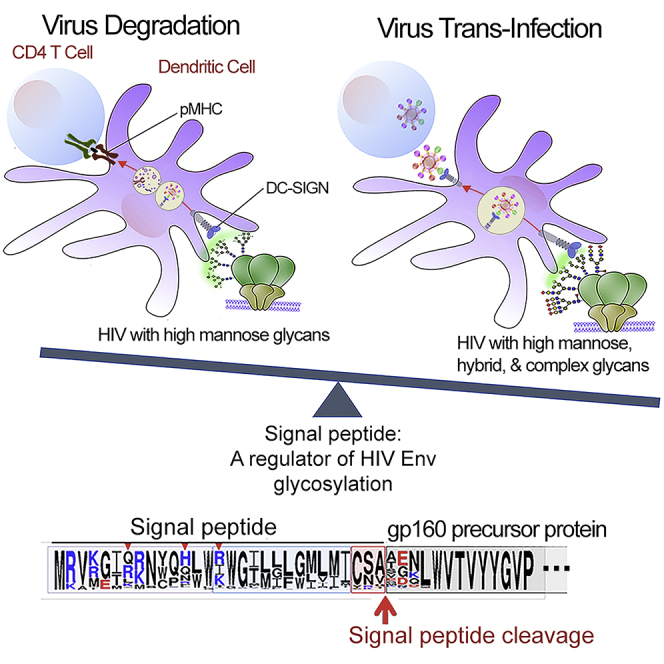
Summary
The HIV-1 envelope (Env) surface is shrouded with an assortment of oligomannose-, hybrid-, and complex-type glycans that enable virus interaction with carbohydrate-recognizing lectins. This study examined the importance of glycan heterogeneity for HIV-1 transmission through the trans-infection pathway by the host mannose-binding lectin DC-SIGN. A diversity of glycan content was observed among HIV-1 strains and associated with varying degrees of trans-infection via DC-SIGN and sensitivity to trans-infection blockage by antiviral lectins. When Env glycans were modified to display only the oligomannose type, DC-SIGN-mediated virus capture was enhanced; however, virus trans-infection was diminished because of increased degradation, which was alleviated by incorporation with hybrid-type glycans. Amino acid changes in the Env signal peptide (SP) modulated the Env glycan content, leading to alterations in DC-SIGN-dependent trans-infection and virus sensitivity to antiviral lectins. Hence, SP variation and glycosylation that confer varied types of oligosaccharides to HIV-1 Env are critical determinants for virus fitness and phenotypic diversity.
Subject Areas: Biological Sciences, Molecular Biology, Microbiology, Virology
Graphical Abstract
Highlights
-
•
HIV-1 Env glycan content determines the fate of virions captured by DC-SIGN
-
•
Viruses with high-mannose glycans are captured by DC-SIGN but are prone to degradation
-
•
Inclusion of hybrid and complex glycans reduces degradation and promotes transmission
-
•
Env glycan content is influenced by amino acid variations in Env signal peptide
Biological Sciences; Molecular Biology; Microbiology; Virology
Introduction
HIV-1 envelope (Env) spikes, the only virus-encoded glycoproteins on the virion surface, mediate virus-host interactions that are crucial for virus propagation and spread. The Env surface is densely shrouded by N-linked glycans, which make up approximately 50% of Env mass (Lee et al., 2016, Rudd and Dwek, 1997, Stewart-Jones et al., 2016). These glycans promote occlusion of immunogenic epitopes, and virus escape from antibody recognition (Binley et al., 2010, Crooks et al., 2015, Kumar et al., 2013, Sanders et al., 2008, Townsley et al., 2016, Wei et al., 2003). N-glycan-induced structural alterations to Env also have been shown to regulate the proteolytic processing of MHCII-restricted CD4 T cell epitopes (Li et al., 2009). In addition, HIV-1 utilizes Env glycans to attach to carbohydrate-binding lectins on host cells, such as the C-type lectin DC-SIGN expressed on dendritic cells and macrophages found at mucosal sites (Garcia-Vallejo and van Kooyk, 2013, Geijtenbeek et al., 2000c). DC-SIGN is a type 2 C-type lectin with a carbohydrate-recognition domain that binds fucose and high-mannose glycans on viruses and bacteria in a Ca2+-dependent manner. DC-SIGN displays enhanced affinity specifically for high-mannose glycans with terminal Manα1-2 structures on HIV Env (Feinberg et al., 2007, Mitchell et al., 2001). Although DC-SIGN serves as a pattern recognition receptor that innate immune cells use to capture and internalize microbial pathogens for destruction and antigen presentation to T cells (Geijtenbeek et al., 2000a, Geijtenbeek et al., 2000c), HIV-1 can evade the cellular degradation machinery and exploit DC-SIGN for its transmission to CD4 T cells, the main cell type that hosts robust, productive HIV-1 replication (Baribaud et al., 2002, Geijtenbeek et al., 2000b, Jan and Arora, 2017, Manel et al., 2010, van Montfort et al., 2011). DC-SIGN interaction with HIV-1 is thus implicated in the initial stages of virus acquisition and spread from the mucosal site of virus entry (Geijtenbeek and van Kooyk, 2003, Hertoghs et al., 2017).
HIV-1 is transmitted from dendritic cells to CD4 T cells through trans- or cis-infection pathways (Lekkerkerker et al., 2006, Tsunetsugu-Yokota and Muhsen, 2013). Virus interaction with DC-SIGN typically leads to trans-infection in the absence of virus replication in dendritic cells (Geijtenbeek et al., 2000b, Kwon et al., 2002). Intact HIV-1 virions captured by mature dendritic cells have been shown to be endocytosed to non-lysosomal tetraspanin-containing multivesicular bodies (Garcia et al., 2005), whereas in a more recent study with immature monocyte-derived dendritic cells, HIV-1 virions were found to accumulate on actin-rich dendrites near the plasma membrane to allow for efficient transfer to CD4 T cells; this process involved the participation of tetraspanin 7 and dynamin 2, which controlled nucleation and cortical stabilization of actin to limit virus endocytosis (Menager and Littman, 2016). Dendritic cells also may support a limited level of HIV-1 replication to produce infectious virions that are efficiently transmitted to CD4 T cells through cis-infection (Burleigh et al., 2006, Tsunetsuguyokota et al., 1995). In addition to DC-SIGN, other C-type lectins, such as Siglec-1 (sialic acid-binding immunoglobulin-like lectin-1) on mature dendritic cells and DCIR (dendritic cell immunoreceptor) on immature dendritic cells, have been reported to participate in the capture and trans-infection of HIV-1 (Izquierdo-Useros et al., 2012, Lambert et al., 2008, Perez-Zsolt et al., 2019). DCIR is also implicated in cis-infection (Lambert et al., 2008). The relative importance of these lectins in HIV-1 transmission remains controversial. In some studies, antibody blockage of DC-SIGN on dendritic cells was found to reduce virus capture only up to 50% (Burleigh et al., 2006, Cambi et al., 2004), but other studies have demonstrated 70%–75% inhibition of virus capture by immature monocyte-derived dendritic cells (MDDCs) and almost complete blockage of transmission to CD4 T cells (van Montfort et al., 2007). siRNA knockdown in immature dendritic cells also decreased HIV-1 transfer to CD4 T cells by 75% (Arrighi et al., 2004). A side-by-side comparison of DC-SIGN and DCIR knockdown further demonstrated a more significant contribution of DC-SIGN versus DCIR in HIV-1 capture and trans-infection by immature dendritic cells (Jin et al., 2014).
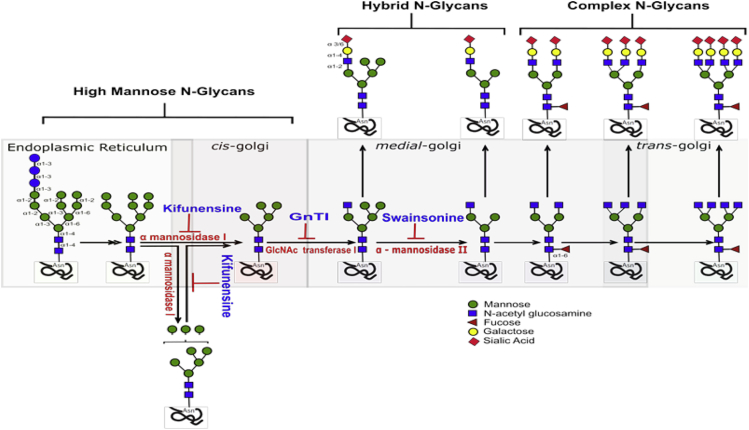
This study was conducted to evaluate HIV-1 trans-infection by DC-SIGN and the capacity of soluble antiviral lectins to block this mode of virus transmission. In particular, we postulated that the oligosaccharide composition of HIV-1 Env glycans is a key factor determining DC-SIGN-mediated virus transmission and virus resistance to antiviral lectins. Glycosylation adds a layer of diversity to the already extremely high level of Env variation found among circulating HIV-1 isolates. At the genetic level, N-linked glycosylation is dictated by the N-X-S/T motif, where X is any amino acid except P. Env proteins have varying numbers of these potential N-glycosylation sites (PNGSs), ranging from 23 to 34 per gp160 protomer; the majority of these are in the gp120 surface subunit, whereas only four to eight are in the external domain of the gp41 subunit (Go et al., 2017). Most PNGSs are not conserved. In fact, only six to eight PNGSs found in Env of clades A, B, C, G, and CRF_01.AE are >90% conserved, and for several PNGSs, conservation is <20% (Go et al., 2017, Pritchard et al., 2015). The PNGSs on HIV-1 Env are not fully occupied (Cao et al., 2018, Struwe et al., 2018). The level of PNGS occupancy is dictated by site accessibility for a series of enzymes participating in the glycan maturation process (Cao et al., 2017). The glycosylation pathway is initiated by the addition of Glc3Man9GlcNAc2 en bloc onto a nascent protein in the ER (Figure 1). As the protein is transported across the ER and the Golgi apparatus, the high-mannose structure is trimmed and subsequently elaborated with hybrid- and complex-type glycans. In contrast to cellular glycoproteins, which are usually adorned with mature complex-type glycans, virus Env carries all three glycan types, including early and intermediate high-mannose, intermediate hybrid, and mature complex glycans (Behrens et al., 2016, Go et al., 2015, Kong et al., 2010). In fact, various glycan types and glycoforms are found in proportions that vary depending on Env strain and host cell type (Bonomelli et al., 2011, Cao et al., 2018, Go et al., 2017, Struwe et al., 2018, Torrents de la Pena et al., 2019). On soluble and membrane-anchored Env mimics, high-mannose-type glycans range from ∼60%–70%, with the most prominent being the least processed Man9GlcNAc2 glycans at 20%–40% (Behrens et al., 2017, Doores et al., 2010, Go et al., 2015). Analysis of soluble, uncleaved, prefusion-optimized BG505 Env gp140 trimers produced in 293F cells similarly has shown that 56% are high-mannose type composed of Man5 (6%), Man6 (3%), Man7 (6%), Man8 (15%), and Man9 (26%) (He et al., 2018). When the same proteins were produced in ExpiCHO cells, the total oligomannose content increased to 64%, with observable changes in glycoform proportions. Site-specific analysis further revealed that each PNGS on gp120 incorporated multiple glycoforms of only the oligomannose type or a mix of oligomannose type and complex type, whereas the gp41 PNGSs had mainly complex-type glycoforms (Cao et al., 2017, He et al., 2018). Consistent with these findings, our previous study of virus-associated Env from a clade B infectious molecular clone REJO.c/2864 identified nine and three unique glycoforms at positions 290 and 446, respectively, each of which included both high-mannose type and fucosylated complex type (Upadhyay et al., 2018). Importantly, the proportion of glycoforms changed when amino acid substitutions were introduced into the Env signal peptide (SP), demonstrating a genetic influence on Env glycan composition (Upadhyay et al., 2018).
Figure 1.
N-linked Glycan Processing
N-linked glycosylation is initiated by the addition of Glc3Man9GlcNAc2en bloc onto a nascent protein in the ER. As the protein is transported across the ER and the Golgi apparatus, the high-mannose structure is trimmed and subsequently elaborated with hybrid- and complex-type glycans. HIV-1 Env is swathed with a dense array of N-glycans composed of high-mannose, hybrid, and complex types. Glycosidase inhibitors and cells lacking a key enzyme for glycan maturation are useful tools to experimentally modify the Env glycans and reduce glycan heterogeneity to certain glycoforms. For example, kifunensine, which inhibits the ER and Golgi mannosidase I, retards glycosylation at the Man9GlcNAc2 stage, enriching for this particular high-mannose glycoform. In GnTI-deficient HEK293S cells (GnTI−/−), glycan processing is arrested at the Man5GlcNAc2 structure. In contrast, treatment with swainsonine, an inhibitor of mannosidase II in the Golgi, generates glycans of high-mannose type bearing Man5-to-9GlcNAc2 and hybrid type carrying GlcNAcMan5GlcNAc2.
Previous studies evaluating the effects of Env glycan composition have revealed the importance of glycan types in modulating virus interactions with antibodies and lectins. In these studies, HIV-1 with Env bearing only high-mannose-type glycans exhibited increased sensitivity to neutralizing antibodies against the crown of the V3 loop (Binley et al., 2010, Kumar et al., 2013, Upadhyay et al., 2014) but increased resistance to antibodies specific for V2i epitopes in the underbelly of the V1V2 beta-barrel domain (Upadhyay et al., 2018). More refined changes of oligosaccharide contents that were induced by substitutions of the basic amino acid residues overrepresented in Env SP also rendered viruses more resistant to V2i-specific antibodies (Upadhyay et al., 2018). Glycans were not part of the V2i and V3 crown epitopes; rather, SP mutations changed the Env oligosaccharide composition, triggering structural alterations that affected the accessibility of these epitopes. Consistent with these results, swapping Env SPs was also reported to change the glycan content of recombinant Env gp120 proteins, with a resultant impact on antibody binding to CD4-induced epitopes (Yolitz et al., 2018). Glycan composition also influences HIV-1 Env interactions with DC-SIGN and other lectins from bacteria, algae, fungi, plants, and animals (Mitchell et al., 2017, Sharon and Lis, 1989). HIV-1 enriched with high-mannose glycans was captured more efficiently by DC-SIGN and shuttled toward a degradation pathway, which augmented MHCII-antigen presentation but impeded trans-infection (van Montfort et al., 2011). Altering the ratio of high-mannose- and complex-type glycans by swapping Env SP caused a dramatic switch in Env affinity for DC-SIGN and for plant lectins specific for α-linked mannose or terminal galactose residues (Upadhyay et al., 2018, Yolitz et al., 2018). DC-SIGN-dependent virus transmission was also reduced, to various extents, by Env SP mutations, although the effect correlated mainly with lower Env incorporation into virions (Upadhyay et al., 2018).
Owing to their exquisite specificity and affinity for unique oligosaccharide configurations on HIV-1 Env glycans, soluble lectins have been investigated for use as antiviral agents to prevent HIV-1 infection (Jan et al., 2018, Jan et al., 2017, Koharudin and Gronenborn, 2014, Mitchell et al., 2017). One such lectin that has advanced to human clinical trials as an anti-HIV-1 microbicide is griffithsin (GRFT), a lectin from the marine red algae Griffithsia species, which is specific for the terminal Manα(1-2)Man residues forming the D1, D2, or D3 arms of Man8/9 glycans (Moulaei et al., 2010, Ziolkowska et al., 2006) and shows efficacy across different HIV-1 isolates and clades (Alexandre et al., 2013, Derby et al., 2018, Girard et al., 2018, Jan et al., 2018, Lal et al., 2018, Mori et al., 2005, Xue et al., 2013). Another lectin with anti-HIV-1 activity, GNA, is derived from snowdrop bulbs of Galanthus nivalis; this lectin also recognizes terminal oligomannoses but is specific for the Manα(1-3)- and Manα(1-6)-linked structures on Man5/6 glycans (Hester et al., 1995, Shibuya et al., 1988). Nonetheless, the inhibitory potency of lectins against HIV-1 isolates varies greatly (Alexandre et al., 2013, Jan et al., 2018, Xue et al., 2013), and the role of glycan heterogeneity found among these viruses warrants further investigation.
The current study evaluated the effects of HIV-1 Env glycan content on virus trans-infection by DC-SIGN and virus sensitivity to transmission inhibition by antiviral lectins GRFT and GNA. Among different HIV-1 isolates, a diversity of Env glycan content was demonstrated by lectin probes and associated with variability in DC-SIGN-dependent transmission and sensitivity to inhibitory lectins. To specifically delineate the contribution of different glycan contents, viruses were generated with modifications specifically in their Env glycan composition, using glycosidase inhibitors or a cell line lacking a key glycosyltransferase in the glycosylation pathway, and introducing mutations to the Env signal peptide. Modifications in Env glycans altered the balance between DC-SIGN-mediated virus degradation and trans-infection to CD4+ cells and also influenced virus sensitivity to transmission inhibition by GRFT and GNA. The results provide direct evidence for the importance of a heterogeneous Env glycan composition for HIV-1 fitness and phenotypic variation. The findings also have important implications for the development of prophylactic vaccines and microbicides, many of which specifically target HIV-1 Env glycans and thus are impacted by the variability of glycan contents found among circulating HIV-1 isolates.
Results
DC-SIGN-Mediated Transmission of HIV-1 Isolates with Varied Oligosaccharide Env Contents
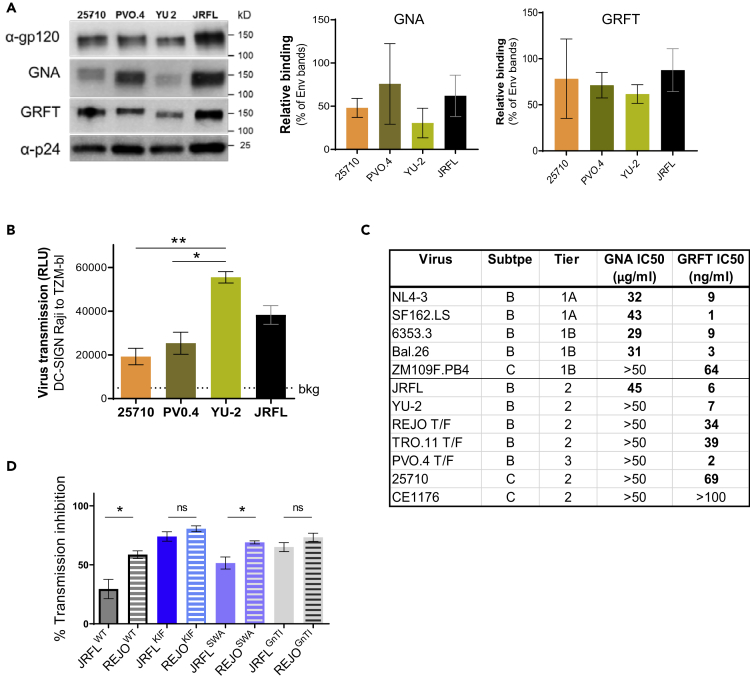
To evaluate the variability in the Env oligomannose composition of different HIV-1 isolates, we determined the binding of mannose-specific lectins GNA and GRFT with Env expressed by viruses that were all produced in transfected 293T cells. Four viruses pseudotyped with Env strains that belong to neutralization-resistant tier 2 or tier 3 were subjected to SDS-PAGE and blotted and probed with GNA, GRFT, or mAbs specific for gp120 or p24. The intensities of lectin bands were measured and normalized to the corresponding Env band. There was a tendency for higher GNA binding to PVO.4 and lower GNA binding to YU-2 and 25710 as compared with JRFL (Figure 2A). In contrast, the relative levels of GRFT binding were more invariant. A separate analysis of subtype B Env strains belonging to tier 1 and tier 2 based on neutralizing antibody sensitivity revealed a greater variability in GNA binding and GRFT binding (Figure S1). Env of tier 1 viruses tended to be more reactive with GNA and GRFT than Env of tier 2 viruses, although the differences between the two groups were not significant.
Figure 2.
HIV-1 Isolates Display Differences in Env Glycan Content that Influence Their Interaction with Lectins and Transmission via DC-SIGN
(A) Oligomannose contents from HIV-1 expressing Env strains of 25710, PV0.4, YU-2, and JRFL as detected by oligomannose-specific lectins GNA and GRFT. Virus lysates were prepared from sucrose-pelleted viruses and analyzed by western blot with anti-gp120 mAb cocktail, GNA, GRFT, and anti-p24 mAb. The intensities of GNA and GRFT bands were calculated as percentages of the corresponding anti-gp120 mAb bands. See also Figure S1.
(B) DC-SIGN-mediated transmission of HIV-1 isolates 25710, PV0.4, YU-2, and JRFL. DC-SIGN+ Raji cells were treated with viruses containing an equivalent infectious dose (i.e., comparable infectivity upon titration in TZM.bl cells: ∼150,000 RLU) for 2 h, washed to remove free virions, and co-cultured with TZM.bl cells. After 48 h, virus transmission to TZM.bl cells was measured by luciferase activity (RLU). Statistical analysis was done using ANOVA (**p < 0.01, *p < 0.05, no asterisk p ≥ 0.05).
(C) Differential sensitivity of HIV-1 strains to transmission inhibition by GRFT and GNA as demonstrated by 50% inhibitory concentration (IC50). To assess transmission inhibition by GRFT or GNA, virus was treated with titrated amounts of lectin for 1 h and then incubated with DC-SIGN+ Raji cells for 2 h.
(D) Comparable sensitivity of HIV-1 JRFL and REJO to transmission inhibition by mannan oligosaccharides when viruses were produced in kifunensine (KIF) or in GnTI−/− cells and expressed Env bearing homogenously high-mannose-type glycans. Statistical analysis was done using unpaired t test (*p < 0.05, ns p ≥ 0.05). For inhibition by mannan, DC-SIGN+ Raji cells were pretreated with mannan (100 μg/mL) for 1 h, washed, and incubated with virus for 2 h. After extensive washing for free virus removal, treated Raji cells were co-cultured with TZM.bl cells, and virus transmission to TZM.bl cells was detected by luciferase activity. All experiments were performed twice; averages and standard deviations of the two experiments are presented.
To determine virus interaction with DC-SIGN and virus trans-infection via this lectin, we evaluated transmission of different HIV-1 strains from DC-SIGN+ Raji cells to CD4+ TZM.bl cells. Expressing no CD4, DC-SIGN+ Raji cells mediated only trans-infection of HIV-1. Treatment of DC-SIGN+ Raji cells with an equivalent infectious dose of 25710, PVO.4, YU-2, and JRFL (∼150,000 RLU) resulted in varying levels of transmission to TZM.bl cells (Figure 2B).
We subsequently evaluated the relative sensitivity of a panel of subtypes B and C HIV-1 strains to transmission inhibition by GNA and GRFT. Viruses known to be relatively sensitive (tier 1) or resistant (tiers 2 and 3) to neutralizing antibodies were tested. GNA was effective mainly against tier 1 viruses and failed to inhibit transmission of most tier 2 viruses except JRFL (IC50 ranging from 29 to >50 μg/mL or 1.45 to >25 nM) (Figure 2C). These inhibitory potencies correlated with GNA binding levels to the respective Env (r2 = 0.77, p = 0.05 by Spearman correlation). GRFT, on the other hand, inhibited transmission of all viruses except CE1176, although IC50 values ranged widely from 1 to 69 ng/mL (0.078–5 nM). CE1176 Env lacks specific PNGSs critical for interaction with GRFT (Alexandre et al., 2013, Xue et al., 2013).
Variability was also observed with inhibition of transmission of different HIV-1 strains by mannan oligosaccharides, which bind DC-SIGN and compete with HIV-1 Env. JRFL was inhibited by 29% at 100 μg/mL, whereas REJO was inhibited by 58% (Figure 2D). We then reduced the heterogeneity of Env glycan by producing JRFL and REJO viruses in the presence of a glycosidase inhibitor (kifunensine [KIF] or swainsonine [SWA]) or in the 293S cell line lacking GlcNAc transferase I (GnTI−/−) (Kumar et al., 2013, Upadhyay et al., 2018). KIF treatment stops the early step of glycan maturation in which α-mannosidase I trims Man9GlcNAc2, thus enriching the virus with high-mannose glycans with Man9GlcNAc2, whereas glycosylation is arrested in GnTI−/− cells at the Man5GlcNAc2 stage (Figure 1). Both high-mannose-enriched KIF and GnTI−/− viruses exhibited increased sensitivity and became equally sensitive to mannan inhibition (>65% inhibition) (Figure 2D). Higher inhibition was also attained when the viruses were produced in the presence of SWA, but a difference in mannan sensitivity between JRFL and REJO was retained. SWA targets α-mannosidase II in the later stage of glycosylation, permitting the accumulation of high-mannose glycans with Man5-9GlcNAc2 and hybrid glycans bearing GlcNAcMan5GlcNAc2, but not of complex-type glycans. Unlike KIF or GnTI−/− cells, treatment with SWA generates viruses that express a mix of high-mannose and hybrid glycans and thus remain relatively heterogeneous in their Env glycan composition. These data demonstrate that heterogeneity in Env sugar composition is a key factor contributing to phenotypic differences in virus sensitivity to trans-infection inhibition by glycan-targeting blockers.
Effect of HIV Envelope Glycan Modification on DC-SIGN-Mediated HIV-1 Capture and Transmission
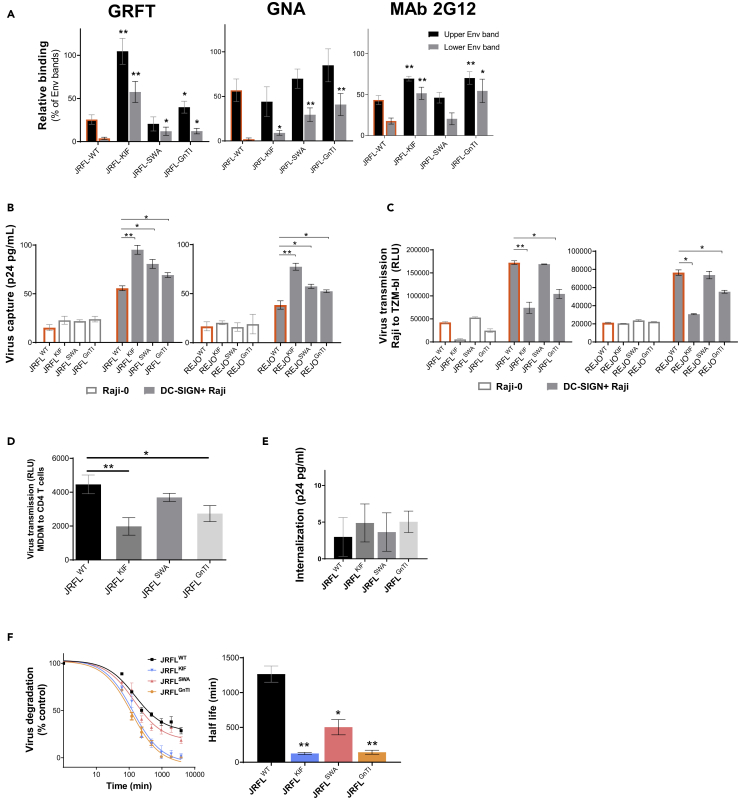
Although DC-SIGN is a lectin with the propensity for binding oligomannose glycans (Mitchell et al., 2001), the composition of HIV-1 Env glycan optimal for virus interaction with DC-SIGN versus trans-infection to CD4+ cells is not fully understood. To address this, JRFL and REJO pseudoviruses with global modifications in their Env glycan contents were produced in the presence of KIF or SWA or in the GnTI−/− cells. Altered glycan contents of JRFL were verified in western blots probed with mannose-binding lectins (GNA and GRFT) and mannose-specific mAb 2G12, along with anti-gp120 and anti-gp41 mAbs (Figures 3A, S2A, and S2C). As shown in Figure 3A, JRFL-KIF was more reactive with GRFT than was untreated JRFL (denoted as WT), consistent with the enrichment of Man9GlcNAc2-bearing terminal Manα1-2, which is recognized by GRFT. GNA showed a minimal change in binding to JRFL-KIF versus JRFL-WT. In contrast, GNA was more reactive with JRFL-SWA and JRFL-GnTI−/− versus JRFL-WT, indicative of an increase in Manα1-3 and Manα1-6 particularly in the lower Env bands of these glycan-modified viruses. Alterations of oligomannose content were also evident from increased reactivity of JRFL-KIF and JRFL-GnTI−/−, but not of JRFL-SWA, with mAb 2G12, further verifying that the Env of these viruses contained a distinct oligomannose composition.
Figure 3.
Glycan Composition Determines the Fate of HIV-1 upon DC-SIGN Interaction
(A) Changes in glycan composition of JRFL Env from sucrose-pelleted virions produced under different conditions (KIF, SWA, GnTI−/−), as detected using oligomannose-specific lectins (GNA specific for terminal Manα1-3 and GRFT specific for terminal Manα1-2) or mAb 2G12. Virus lysates were run on SDS-PAGE under reducing condition, blotted, and probed with an anti-gp120 mAb cocktail (anti-V3 2219, anti-V3 2558, anti-C2 841, anti-C2 1006-30D, anti-C5 722), GNA, GRFT, or 2G12. The reactivity of the upper and lower Env bands with lectins or 2G12 were quantified relative to the anti-gp120 mAb cocktail band for each virus. A set of representative western blots is shown in Figure S2.
(B) DC-SIGN-mediated capture of glycan-modified HIV-1 JRFL and REJO. Raji cells with versus without DC-SIGN were treated with virus (10 ng p24/mL for capture and 150,000 RLU for transmission) for 2 h. After extensive washing to remove free virus, cell-associated p24 protein was measured by ELISA. See also Figure S3.
(C) Virus transmission was measured after Raji cells were co-cultured with TZM.bl cells for 48 h. See also Figure S3B.
(D) Transmission of glycan-modified viruses from MDDCs to primary PHA-stimulated CD4 T cells.
(E) Virus internalization by MDDCs. Cells were incubated with glycan-modified JRFL viruses (20 ng p24/mL) for 4 h at 37°C. Surface-bound virus particles were removed by treatment with 0.05% trypsin for 5 min, and internalized viruses were quantified by ELISA.
(F) Virus degradation by MDDCs. MDDCs were treated with glycan-modified viruses (10 ng p24/mL) for 2 h at 37°C, washed to remove unbound virus, then incubated at 37°C for different time points. The amount of p24 associated with the cells over time was measured by ELISA and calculated relative to the p24 level at t = 0 (100%). The virus half-life (t1/2) was analyzed based on the one-phase decay model (GraphPad Prism 6).
Experiments were repeated independently two to three times. Means and standard deviations from all experiments are shown. Data were analyzed using unpaired t test (*p < 0.05, **p < 0.01 versus untreated WT), except for (F), which was analyzed by two-way ANOVA. Differences with p ≥ 0.05 are left unmarked.
Western blot analysis also demonstrated that each of these glycan-modified viruses expressed Env with lower molecular size as compared with WT in the following order: WT > SWA > KIF > GnTI−/−. The size reduction was apparent for Env bands reactive with anti-gp120 and anti-gp41 mAbs (Figure S2A). Of note, uncleaved gp160 and cleaved gp120 were detected. As compared with the gp120 bands, the gp160 bands reacted more intensely with GNA, GRFT, and mAb 2G12, all of which recognize terminal oligomannoses (Figure 3A). This pattern was seen with glycan-modified and WT viruses, indicating that uncleaved gp160 and cleaved gp120 associated with these viruses displayed distinct glycan profiles. In this study, Env was expressed on pseudotyped viruses under an exogenous CMV promotor; however, similar findings were observed in our earlier study using full-length infectious molecular clones (Upadhyay et al., 2018), demonstrating that uncleaved gp160 bearing more oligomannose glycoforms was also present when Env was expressed under its own endogenous promotor.
The glycan-modified viruses were first compared with WT for DC-SIGN-mediated virus capture using DC-SIGN+ Raji cells. Virus capture was assessed by measuring the total amount of cell-associated Gag p24 after treatment of DC-SIGN+ Raji cells with the same amount of p24 virus input. Virus-treated Raji-0 cells were tested in parallel to establish the background control. Higher levels of virus capture were observed with DC-SIGN+ Raji cells treated with each of the 3 glycan-modified viruses versus WT, with the KIF virus captured at the highest level (Figure 3B). These results were consistently observed with JRFL and REJO (Figure 3B) as well as with JRCSF and NL4-3 strains (Figure S3A). The enhanced capture of glycan-modified viruses was likely due to preferential binding of DC-SIGN for oligomannoses that were enriched in these viruses. It was not associated with increased Env incorporation into these viruses, as the levels of Env incorporated into glycan-modified viruses, measured by Env/p24 ratios, tended to be lower than that of WT Env (Figure S2B (Jan et al., 2018)).
Subsequently, we evaluated transmission of these glycan-modified viruses from DC-SIGN+ Raji cells to TZM.bl cells using an equivalent dose of virus input. Interestingly, although virus capture was enhanced (Figure 3B), virus transmission was reduced, particularly for the high-mannose-enriched KIF and GnTI−/− viruses (Figure 3C). In contrast, transmission of the SWA viruses bearing both high-mannose- and hybrid-type glycans was comparable with that of WT. Similar results were also observed with glycan-modified viruses of JRCSF and NL4.3 (Figure S3B). The reduced transmission efficiency of KIF and GnTI−/− viruses, but not SWA virus, was also apparent when virus transmission was assessed from primary MDDCs to primary CD4 T cells (Figure 3D).
The reduced transmission of glycan-modified viruses despite enhanced virus capture indicated that these viruses were not released to the CD4+ target cells, possibly due to alteration in virus internalization or degradation, as reported previously (van Montfort et al., 2011). To measure virus internalization, trypsin was added to virus-treated MDDCs to remove surface-bound viruses, and the amounts of virus that remained cell associated were detected by p24 ELISA. Figure 3E shows that comparable levels of internalization were seen with glycan-modified versus WT viruses. However, the amounts of virus internalized were very low (<5 pg of 20 ng p24 input), indicating that most WT and glycan-modified virions bound to cells were not internalized.
Next, we examined whether the viruses differed in their sensitivity to degradation based on the amount of p24 that remained over time. Virus-treated MDDCs were incubated at 37°C up to 64 h, and the levels of p24 retention were measured periodically. The data showed that the half-life of JRFL WT was 22.5 h, whereas the half-lives of KIF and GnTI−/− viruses were reduced to ∼2 h (Figure 3F). This remarkably faster degradation of oligomannose-enriched viruses elucidates the poor transmission of the otherwise efficient virus capture by DC-SIGN. In contrast, the SWA virus had a half-life of 8.4 h, demonstrating that resistance to degradation was increased by the incorporation of hybrid-type glycans, although the half-life of the SWA virus remained lower than that of the WT. These results indicate that HIV-1 Env glycosylation that incorporates a mixture of high-mannose, hybrid, and complex glycans is beneficial for the virus; this feature is necessary for HIV-1 evasion from degradation by dendritic cells, while allowing efficient DC-SIGN-mediated virus capture and transmission to CD4 T cells.
DC-SIGN-Mediated Trans-infection of HIV-1 SP Mutants Expressing Env with Altered Glycan Contents
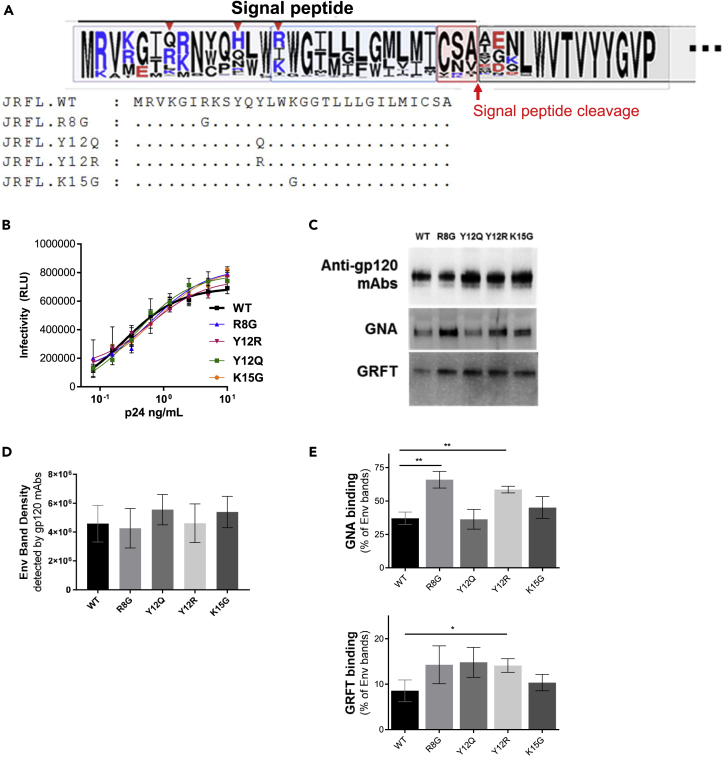
Biosynthesis of HIV-1 Env is initiated by translation of the signal peptide (SP), which transports nascent protein to the ER and Golgi pathways. Notably, HIV-1 Env SP is highly variable and contains several basic residues in its N-terminal region (Figure 4A); these features are unique to Env SPs and are not seen among SPs of other viral or cellular glycoproteins (Li et al., 1994). We previously showed that SP sequences influence HIV-1 Env glycosylation; particularly, mutations at SP positions 8, 12, and 15 were each found to change the oligosaccharide contents of Env glycans, which, in turn, affected virus neutralization by antibody (Upadhyay et al., 2018). Unlike the global shifts of glycan types incurred by glycosidase inhibitors, SP mutations affected Env glycosylation in a more subtle but physiologic way, to result in altered proportions of glycoforms associated with Env. To examine whether such changes are sufficient to influence virus trans-infection via DC-SIGN, HIV-1 JRFL pseudoviruses were generated with R8G, Y12Q, Y12R, and K15G mutations (Figure 4A). In contrast to alterations seen when they were introduced to full-length infectious clones (Upadhyay et al., 2018), the SP mutations did not affect pseudovirus infectivity, as measured in TZM.bl reporter cells (Figure 4B). The mutations also did not change the level of Env incorporation, as indicated by comparable intensities of Env bands detected by anti-gp120 mAbs in all four SP mutants versus WT (Figures 4C and 4D). The intensity of the gp41 band was also similar for all viruses (not shown). To detect changes in Env sugar composition, lectin-probed western blot analyses were performed using GNA (specific for terminal Manα1-3 and Manα1-6) and GRFT (specific for terminal Manα1-2) on virus lysates from SP mutants versus WT (Figure 4C). Distinct patterns of lectin binding were observed among different SP mutants and in those of WT (Figures 4C and 4E). GNA reacted more to R8G and Y12R compared with WT and the other two SP mutants. GRFT, on the other hand, showed slightly more binding with R8G, Y12Q, and Y12R compared with WT, although a significant difference was attained only with Y12R.
Figure 4.
Alterations of HIV-1 Env Glycan Contents by SP Mutations
(A) Sequence logo showing amino acid variability in the HIV-1 Env SPs from the Los Alamos HIV database. Letter size indicates the frequency of each amino acid found at the particular position; basic, acidic, and neutral residues are shown in blue, red, and black, respectively. Red arrows show the positions of amino acid substitutions evaluated in this study; the position numbering is based on alignment with the HXB2 sequence. SP mutations (R8G, Y12Q, Y12R, and K15G) were introduced to the HIV-1 JRFL Env.
(B) Infectivity of SP mutants versus WT in TZM.bl cells. Viruses were produced in transfected HEK293T cells, and viruses with equivalent p24 contents were titrated and used to infect TZM.bl cells.
(C) Alterations in glycan contents of SP mutants versus WT as detected by oligomannose-specific lectins GNA (binding terminal Manα1-3) and GRFT (binding terminal Manα1-2). Virus lysates (20 ng Env/mL) were analyzed by western blot with anti-gp120 mAb cocktail, GNA, or GRFT.
(D and E) Quantification of Env-band density as probed with anti-gp120 mAbs (D) or lectins GNA and GRFT (E). Lectin binding was calculated relative to the Env band density of each virus. All experiments were repeated twice; averages and standard deviations from both experiments are presented. Data were analyzed using unpaired t test (*p < 0.05, **p < 0.01 versus WT; no asterisk indicates p ≥ 0.05).
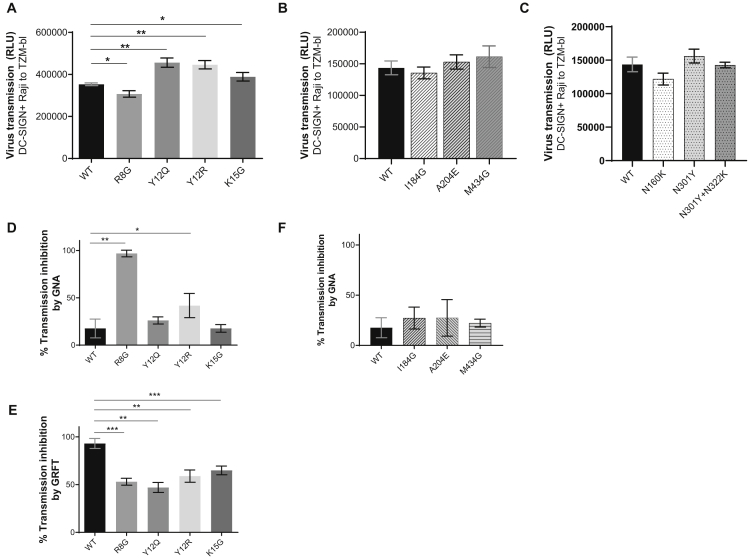
Because GNA and GRFT binding data indicated that SP mutants expressed Env with altered compositions of terminal oligomannoses, we postulated that, similar to the alterations in glycan-modified viruses studied in Figure 3, the SP mutants would also display alterations in DC-SIGN interaction and transmission. The data in Figure 5A demonstrate that, indeed, each of the four SP mutants showed reduced or increased DC-SIGN-dependent trans-infection. Trans-infection of R8G was lower than that of WT, whereas transmission of the remaining three SP mutants was higher. However, virus capture of these SP mutants by DC-SIGN+ cells was lower, except for Y12R, which was captured to a similar extent as WT (Figure S4). Hence, changes in transmission of these viruses did not correlate with virus capture, similar to the lack of correlation seen with glycan-modified viruses (Figures 3B and 3C). Of note, although the levels of alteration were modest (only <30% decrease or increase), the changes were specifically incurred by SP mutations. Mutations in the gp120 subunit outside the SP region (I184G, A204E, and M434G) did not cause any changes in virus trans-infection via DC-SIGN (Figure 5B). These non-SP mutations are located at the protomer Env interfaces (I184G) or in the gp120 hydrophobic core (A204E and M434G). Similarly, non-SP mutations that removed 1 or 2 PNGSs from positions 160, 301, and 322 did not incur significant changes in DC-SIGN-mediated trans-infection (Figure 5C). Each of these non-SP gp120 mutations in the inter-protomer surface, the hydrophobic core, or PNGSs has been shown to expose the occluded Ab epitopes, thus increasing virus sensitivity to neutralization by mAbs against the V3 crown, V2i epitopes, the CD4-binding site, or CD4-induced epitopes (Powell et al., 2017, Zolla-Pazner et al., 2016).
Figure 5.
Effects of SP versus Non-SP Mutations on DC-SIGN-Mediated Virus Transmission and Virus Sensitivity to Antiviral Lectins
(A) DC-SIGN-mediated trans-infection of SP mutants versus WT. Virus transmission was measured using DC-SIGN+ Raji and TZM.bl cells as described in Figure 2. See also DC-SIGN-mediated capture of SP mutants versus WT in Figure S4.
(B) Trans-infection of viruses with gp120 mutations outside SP for comparison with the SP mutants.
(C) Trans-infection of viruses with single or double PNGS knockout mutations.
(D–F) Altered sensitivity of SP mutants but not gp120 mutants to transmission inhibition by oligomannose-binding lectins GNA or GRFT. Viruses were treated with a final concentration of 25 μg/mL of GNA (D and F) or 0.1 μg/mL of GRFT (E) for 1 h and incubated with DC-SIGN+ Raji cells for another 2 h. Raji cells were then washed extensively to remove free virus and lectin and co-cultured with TZM.bl cells. After 48 h, virus infection in TZM.bl cells was measured by luciferase activity (RLU). GRFT was tested only with SP mutants. Percent inhibition was calculated based on co-cultures with no virus (100%) and co-cultures with virus alone (0%).
All experiments were repeated twice; averages and standard deviations from both experiments are presented. Data were analyzed using unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.001 versus WT; no asterisk indicates p ≥ 0.05).
Subsequently, SP mutants were compared with WT for sensitivity to transmission inhibition by GNA and GRFT (Figures 5D and 5E). R8G and Y12R showed increased sensitivity to inhibition by GNA (Figure 5D), consistent with higher binding of GNA to Env of these mutants (Figures 4C and 4E). In contrast, all four SP mutants were more resistant to inhibition by GRFT compared with WT (Figure 5E). Notably, the non-SP gp120 mutants I184G, A204E, and M434G did not display such changes; they were all equally resistant to GNA and inhibited by <25%, similar to WT (Figure 5F). Hence, alterations of virus sensitivity to lectins were induced by SP mutations but not by non-SP mutations examined here. Taken together, the data demonstrate that modest shifts in HIV-1 Env glycoform contents—specifically the terminal oligomannose structures induced as a result of SP mutations—can influence the efficiency by which HIV-1 is transmitted via DC-SIGN and inhibited by antiviral lectins.
Discussion
This study demonstrates the importance of HIV-1 expression of Env with a heterogeneous glycan composition in exploitation of a DC-SIGN-mediated virus trans-infection pathway and promotion of virus resistance to antiviral lectins. By testing viruses with defined glycan modifications, we revealed that virus Env glycan contents determine the balance between DC-SIGN-mediated virus degradation and virus transmission. Viruses with homogenously high-mannose-type glycans, as a result of production in the presence of KIF or in GnTI−/− cells, were found to be more efficiently captured, but they degraded faster and were poorly transmitted to CD4 T cells, consistent with previously reported findings (van Montfort et al., 2011). Such viruses were also more efficiently transcytosed across epithelial cells but displayed impaired infectivity in explanted human intestinal mucosa (Shen et al., 2014). The current study further demonstrates that the addition of hybrid-type glycans, as seen in viruses grown in the presence of SWA, reduced the rate of virus degradation and increased the rate of virus transmission. However, the presence of all three glycan types—high mannose, hybrid, and complex—normally decorating the HIV-1 Env conferred the highest half-life and transmissibility, although requisite proportions for optimal virus transmission are yet to be defined.
The mechanisms by which hybrid and complex glycans help HIV-1 evade degradation are not understood. DC-SIGN, a pattern-recognition receptor with high affinity for oligomannoses, has been shown to shuttle HIV-1 bearing high-mannose glycans to the degradative pathway, similar to other pathogens with such glycans (van Montfort et al., 2011); this was indicated by an increased association with vesicles containing early and late endosomal markers and by more efficient antigen processing and presentation to MHCII-restricted CD4 T cells (Gringhuis et al., 2009, Jan and Arora, 2017, van Montfort et al., 2011). This study demonstrates that HIV-1 evades this degradative pathway by incorporating host-like hybrid and complex glycans into its Env glycoproteins. In fact, addition of the hybrid type alone is sufficient to tip the balance from degradation to trans-infection. The presence of hybrid and complex glycans reduces DC-SIGN affinity for Env but may activate signals that prevent virus transport to endolysosomal compartments and preserve infectious virions on dendrites to promote their transfer to T cells (Menager and Littman, 2016). Indeed, DC-SIGN binding to fucose- versus mannose-expressing ligands has been shown to elicit distinct responses. Interactions with mannose ligands on Mycobacterium tuberculosis and HIV-1 recruited effector proteins to DC-SIGN signalosome and activated Raf-1, whereas fucose-bearing pathogens such as Schistosoma mansoni and Helicobacter pylori dissociated the Raf-1 complex from the signalosome (Gringhuis et al., 2009). Unlike mannose, engagement of fucose ligands by DC-SIGN also favors the induction of Th2-type responses via activation of Bcl3 and IKKϵ (Gringhuis et al., 2014a, Gringhuis et al., 2014b). More recently, removal of sialic acid residues from Env gp120 proteins was found to enhance the uptake of these proteins by bone marrow dendritic cells (Sun et al., 2018), although the effects of asialyation on DC-SIGN-mediated virus uptake and trans-infection are unknown. Hence, successful evasion of DC-SIGN-dependent degradation and preservation of virus for transmission to CD4 T cells may depend on specific monosaccharide or oligosaccharide moieties present on certain hybrid and/or complex glycoforms.
The oligosaccharide composition of N-linked glycan is controlled post-translationally as a nascent glycoprotein traverses through the ER and Golgi compartments; however, our understanding of the mechanisms regulating Env glycan composition is incomplete. Only a fraction of Env glycans is adorned with hybrid or complex oligosaccharides that are normally found on most other viral and mammalian glycoproteins (Behrens et al., 2017, Bonomelli et al., 2011, Doores et al., 2010, Go et al., 2013, Go et al., 2015, Kong et al., 2010, Raska et al., 2010). It is presumed that the dense glycosylation of Env, especially on the gp120 subunit, imposes a steric hindrance to glycosyl transferases and glycosylases involved in sequential attachment, trimming, and addition of specific oligosaccharides, resulting in incomplete maturation and leaving a substantial proportion of gp120 glycans to remain as high-mannose type. It is also unclear how diversity in glycoform composition across different HIV-1 strains is regulated. We previously demonstrated that amino acid substitutions in Env SP caused detectable changes in the proportions of Env glycan types and glycoforms that affect virus neutralization by antibodies against the V1V2 region (Upadhyay et al., 2018). Variability among SPs of circulating HIV-1 isolates further indicates that Env SPs are under selective pressure and contribute to HIV-1 genetic variability (da Silva et al., 2011, Gnanakaran et al., 2011, Upadhyay et al., 2018). In the current study, we confirmed that mutations introduced to Env SP, without any modifications to the mature Env sequence and the rest of the HIV-1 genome, indeed affected the oligomannose contents of Env, as measured by interactions with lectins: three of the four tested SP mutations increased Env binding to Manα(1-3)-specific lectin GNA or to Manα(1-2)-specific lectin GRFT. All four SP mutations also affected DC-SIGN-dependent virus capture and transmission to CD4 T cells and altered virus sensitivity to transmission inhibition by GNA and GRFT. Such alterations were not seen when mutations were introduced to amino acid residues in the non-SP gp120 subunit that increased the propensity of Env to adopt more open conformations; neither were they seen when mutations removed one or two PNGSs from the gp120 subunit (Powell et al., 2017, Zolla-Pazner et al., 2016). Of note, HIV-1 Env SP cleavage occurs late, after the full-length gp160 chain translation is terminated and after a near-native folding of the gp120 subunit is attained, owing to the occlusion of the SP cleavage site by a secondary structure formed by conserved SP residues and residues downstream of the cleavage site (Land and Braakman, 2001, Li et al., 1994, Li et al., 2000, Snapp et al., 2017). Altogether, these data corroborate that Env SP plays a role in regulating the post-translational glycosylation of Env, which, in turn, influences virus interactions not only with antibodies but also with the host lectin DC-SIGN believed to facilitate virus spread from the mucosal entry site. Moreover, Env SP affects virus interactions with soluble lectins such as GNA and GRFT, which are being explored for potential use as anti-HIV microbicides.
We observed a high degree of variability among different HIV-1 isolates in their sensitivity to lectins GNA and GRFT. Interestingly, virus sensitivity to lectins correlated with virus neutralization by antibodies. Comparison of tier 1 with tier 2 and 3 viruses that are classified based on sensitivity to neutralizing antibodies commonly produced by HIV-infected subjects showed that DC-SIGN-dependent transmission of tier 1 viruses was more sensitive to inhibition by GNA and GRFT than that of tier 2 and 3 viruses. Hence, GNA blocked trans-infection of tier 1 viruses with a mean IC50 of 37 μg/mL, whereas tier 2 and 3 viruses were highly resistant to GNA, with IC50 > 50 μg/mL for all except JRFL. GRFT, one of the most potent lectins against HIV-1, was able to inhibit tier 1, tier 2, and tier 3 viruses, but GRFT was on average ∼2x more potent against tier 1 versus tier 2 and tier 3 viruses (IC50: 17.2 versus 36.7 μg/mL). In a recent study (Jan et al., 2018), we tested direct neutralization of two tier 1 and two tier 2 viruses with GNA, GRFT, and other mannose-binding lectins (SVN, CVN, HHA, and ConA) and observed a comparable pattern: tier 1 viruses SF162 and BaL were more sensitive to neutralization by lectins (mean IC50: 0.97–2.4 μg/mL for all lectins tested) than tier 2 viruses JRFL and REJO (mean IC50: 9.25–14.55 μg/mL). The relative levels of virus sensitivity to lectin-mediated inhibition are associated to some extent with the lectin-binding data, which infer that, as compared with Env of tier 2 or tier 3 viruses, Env of tier 1 viruses expressed more terminal Manα1-3 and Manα1-6 targeted by GNA and also more Manα1-2 targeted by GRFT. These terminal mannose configurations are abundant on the less processed high-mannose-type glycans and some hybrid-type glycans, implicating the enrichment of Env from tier 1 with these glycan types compared with that of tier 2 and 3 viruses. It should be noted, however, that the presence of specific PNGSs is also critical for lectin recognition. Many subtype C viruses, including CE1176, are relatively resistant to GRFT and other mannose-specific lectins owing to the absence of critical PNGSs in C2, C3, V4, and/or C4 regions of Env gp120 (Alexandre et al., 2013, Xue et al., 2013). Therefore, comprehensive analyses of both PNGSs and glycan contents from the array of circulating HIV-1 isolates are needed to inform the potential application of GRFT and other lectins as anti-HIV prophylactics for high-risk populations.
Limitations of the Study
This study examined DC-SIGN-mediated trans-infection of HIV-1 in vitro, using a DC-SIGN+ Raji cell line and primary immature MDDCs. The findings are yet to be verified with DC-SIGN+ dendritic cells and other cell types that participate in virus trans-infection in vivo in mucosal or lymphoid tissues. The study is also limited to the evaluation of virus interaction with DC-SIGN, although a variety of C-type lectins, including Siglec-1 and DCIR, are expressed along with DC-SIGN on dendritic cells and have been shown to participate in capture and trans-infection of HIV-1 (Izquierdo-Useros et al., 2012, Lambert et al., 2008, Perez-Zsolt et al., 2019). In addition, HIV-1 pseudoviruses generated from transfected HEK293T cells were used throughout the study. Although this approach removed the confounding effects of the host cell and production system variability, the glycan contents incorporated into HIV-1 Env can vary depending on how the virions are produced (Bonomelli et al., 2011, Go et al., 2017, Struwe et al., 2018). Further investigation is thus warranted to examine replication-competent HIV-1 isolates produced in primary cells from different donors for their trans-infection efficiency and antiviral lectin sensitivity.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors would like to thank Dr. Susan Zolla-Pazner and Ms. Vincenza Itri for providing HIV-1-specific mAbs, Ms. Roya Feyznezhad and Ms. Xiaomei Liu for general laboratory supports, Ms. Alisa Fox for assistance with flow cytometry and cell cultures, Dr. Rajnish Kumar for reagents, and Drs. Svenja Weiss and Lili Wang for helpful discussion. The authors also thank Ms. Rina Kleege for reviewing the manuscript.
This work is supported by VA Merit Review Award (I01BX003860 to C.E.H.), VA Research Career Scientist Award (IK6BX004607 to C.E.H.), NIH R21 grant (AI124863 to C.U. and C.E.H.), and NIH R01 grant (AI140909 to C.U.).
Author Contributions
Conceptualization, M.J. and C.E.H.; Methodology M.J. and C.U.; Investigation, M.J.; Resources, M.J. and C.U.; Writing – Original Draft, M.J. and C.E.H.; Writing – Review & Editing, M.J., C.U., and C.E.H.; Funding Acquisition, C.U. and C.E.H.
Declaration of Interests
The authors declare no competing interests.
Published: November 22, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.10.030.
Supplemental Information
References
- Alexandre K.B., Moore P.L., Nonyane M., Gray E.S., Ranchobe N., Chakauya E., McMahon J.B., O'Keefe B.R., Chikwamba R., Morris L. Mechanisms of HIV-1 subtype C resistance to GRFT, CV-N and SVN. Virology. 2013;446:66–76. doi: 10.1016/j.virol.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.F., Pion M., Garcia E., Escola J.M., van Kooyk Y., Geijtenbeek T.B., Piguet V. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J. Exp. Med. 2004;200:1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F., Doms R.W., Pohlmann S. The role of DC-SIGN and DC-SIGNR in HIV and Ebola virus infection: can potential therapeutics block virus transmission and dissemination? Expert Opin. Ther. Targets. 2002;6:423–431. doi: 10.1517/14728222.6.4.423. [DOI] [PubMed] [Google Scholar]
- Behrens A.J., Vasiljevic S., Pritchard L.K., Harvey D.J., Andev R.S., Krumm S.A., Struwe W.B., Cupo A., Kumar A., Zitzmann N. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A.J., Struwe W.B., Crispin M. Glycosylation profiling to evaluate glycoprotein immunogens against HIV-1. Expert Rev. Proteomics. 2017;14:881–890. doi: 10.1080/14789450.2017.1376658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley J.M., Ban Y.E., Crooks E.T., Eggink D., Osawa K., Schief W.R., Sanders R.W. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J. Virol. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C., Doores K.J., Dunlop D.C., Thaney V., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh L., Lozach P.Y., Schiffer C., Staropoli I., Pezo V., Porrot F., Canque B., Virelizier J.L., Arenzana-Seisdedos F., Amara A. Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J. Virol. 2006;80:2949–2957. doi: 10.1128/JVI.80.6.2949-2957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M.H.P., de Bakker B.I., Fransen J.A.M., Bovee-Geurts P.H.M., van Leeuwen F.N. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 2004;164:145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Diedrich J.K., Kulp D.W., Pauthner M., He L., Park S.R., Sok D., Su C.Y., Delahunty C.M., Menis S. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat. Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Pauthner M., Andrabi R., Rantalainen K., Berndsen Z., Diedrich J.K., Menis S., Sok D., Bastidas R., Park S.R. Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat. Commun. 2018;9:3693. doi: 10.1038/s41467-018-06121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks E.T., Tong T., Chakrabarti B., Narayan K., Georgiev I.S., Menis S., Huang X., Kulp D., Osawa K., Muranaka J. Vaccine-elicited tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the CD4 binding site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby N., Lal M., Aravantinou M., Kizima L., Barnable P., Rodriguez A., Lai M., Wesenberg A., Ugaonkar S., Levendosky K. Griffithsin carrageenan fast dissolving inserts prevent SHIV HSV-2 and HPV infections in vivo. Nat. Commun. 2018;9:3881. doi: 10.1038/s41467-018-06349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores K.J., Bonomelli C., Harvey D.J., Vasiljevic S., Dwek R.A., Burton D.R., Crispin M., Scanlan C.N. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc. Natl. Acad. Sci. U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H., Castelli R., Drickamer K., Seeberger P.H., Weis W.I. Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J. Biol. Chem. 2007;282:4202–4209. doi: 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallejo J.J., van Kooyk Y. The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Garcia E., Pion M., Pelchen-Matthews A., Collinson L., Arrighi J.F., Blot G., Leuba F., Escola J.M., Demaurex N., Marsh M. HIV-1 trafficking to the dendritic cell-T-cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic. 2005;6:488–501. doi: 10.1111/j.1600-0854.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B.H., van Kooyk Y. DC-sign: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr. Top. Microbiol. 2003;276:31–54. doi: 10.1007/978-3-662-06508-2_2. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Krooshoop D.J., Bleijs D.A., van Vliet S.J., van Duijnhoven G.C., Grabovsky V., Alon R., Figdor C.G., van Kooyk Y. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 2000;1:353–357. doi: 10.1038/79815. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Kwon D.S., Torensma R., van Vliet S.J., van Duijnhoven G.C., Middel J., Cornelissen I.L., Nottet H.S., KewalRamani V.N., Littman D.R. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Torensma R., van Vliet S.J., van Duijnhoven G.C., Adema G.J., van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- Girard L., Birse K., Holm J.B., Gajer P., Humphrys M.S., Garber D., Guenthner P., Noel-Romas L., Abou M., McCorrister S. Impact of the griffithsin anti-HIV microbicide and placebo gels on the rectal mucosal proteome and microbiome in non-human primates. Sci. Rep. 2018;8:8059. doi: 10.1038/s41598-018-26313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanakaran S., Bhattacharya T., Daniels M., Keele B.F., Hraber P.T., Lapedes A.S., Shen T., Gaschen B., Krishnamoorthy M., Li H. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Liao H.X., Alam S.M., Hua D., Haynes B.F., Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. J. Proteome Res. 2013;12:1223–1234. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Ding H., Zhang S., Ringe R.P., Nicely N., Hua D., Steinbock R.T., Golabek M., Alin J., Alam S.M. Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven Env trimers. J. Virol. 2017;91 doi: 10.1128/JVI.02428-16. pii: e02428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E.P., Herschhorn A., Gu C., Castillo-Menendez L., Zhang S., Mao Y., Chen H., Ding H., Wakefield J.K., Hua D. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. J. Virol. 2015;89:8245–8257. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis S.I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T.B. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 2009;10:1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- Gringhuis S.I., Kaptein T.M., Wevers B.A., Mesman A.W., Geijtenbeek T.B. Fucose-specific DC-SIGN signalling directs T helper cell type-2 responses via IKKepsilon- and CYLD-dependent Bcl3 activation. Nat. Commun. 2014;5:3898. doi: 10.1038/ncomms4898. [DOI] [PubMed] [Google Scholar]
- Gringhuis S.I., Kaptein T.M., Wevers B.A., van der Vlist M., Klaver E.J., van Die I., Vriend L.E., de Jong M.A., Geijtenbeek T.B. Fucose-based PAMPs prime dendritic cells for follicular T helper cell polarization via DC-SIGN-dependent IL-27 production. Nat. Commun. 2014;5:5074. doi: 10.1038/ncomms6074. [DOI] [PubMed] [Google Scholar]
- He L., Kumar S., Allen J.D., Huang D., Lin X., Mann C.J., Saye-Francisco K.L., Copps J., Sarkar A., Blizard G.S. HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 2018;4:eaau6769. doi: 10.1126/sciadv.aau6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertoghs N., van Pul L., Geijtenbeek T.B.H. Mucosal dendritic cells in HIV-1 susceptibility: a critical role for C-type lectin receptors. Future Virol. 2017;12:373–388. [Google Scholar]
- Hester G., Kaku H., Goldstein I.J., Wright C.S. Structure of mannose-specific snowdrop (Galanthus nivalis) lectin is representative of a new plant lectin family (vol 2, pg 472, 1995) Nat. Struct. Biol. 1995;2:704. doi: 10.1038/nsb0695-472. [DOI] [PubMed] [Google Scholar]
- Izquierdo-Useros N., Lorizate M., Puertas M.C., Rodriguez-Plata M.T., Zangger N., Erikson E., Pino M., Erkizia I., Glass B., Clotet B. Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol. 2012;10:e1001448. doi: 10.1371/journal.pbio.1001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Arora S.K. Innate sensing of HIV-1 by dendritic cell-specific ICAM-3 grabbing nonintegrin on dendritic cells: degradation and presentation versus transmission of virus to T cells is determined by glycan composition of viral envelope. AIDS Res. Hum. Retroviruses. 2017;33:765–767. doi: 10.1089/aid.2016.0290. [DOI] [PubMed] [Google Scholar]
- Jan M., Upadhyay C., Sharma A., Hioe C.E., Arora S.K. Short communication: manalpha1-2man-binding anti-HIV lectins enhance the exposure of V2i and V3 crown neutralization epitopes on the V1/V2 and V3 hypervariable loops of HIV-1 envelope. AIDS Res. Hum. Retroviruses. 2017;33:941–945. doi: 10.1089/aid.2016.0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M., Upadhyay C., Alcami Pertejo J., Hioe C.E., Arora S.K. Heterogeneity in glycan composition on the surface of HIV-1 envelope determines virus sensitivity to lectins. PLoS One. 2018;13:e0194498. doi: 10.1371/journal.pone.0194498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Li C., Du T., Hu K., Huang X., Hu Q.X. DC-SIGN plays a stronger role than DCIR in mediating HIV-1 capture and transfer. Virology. 2014;458:83–92. doi: 10.1016/j.virol.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Koharudin L.M., Gronenborn A.M. Antiviral lectins as potential HIV microbicides. Curr. Opin. Virol. 2014;7:95–100. doi: 10.1016/j.coviro.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Sheppard N.C., Stewart-Jones G.B.E., Robson C.L., Chen H., Xu X., Krashias G., Bonomelli C., Scanlan C.N., Kwong P.D. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J. Mol. Biol. 2010;403:131–147. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Tuen M., Liu J., Nadas A., Pan R., Kong X., Hioe C.E. Elicitation of broadly reactive antibodies against glycan-modulated neutralizing V3 epitopes of HIV-1 by immune complex vaccines. Vaccine. 2013;31:5413–5421. doi: 10.1016/j.vaccine.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D.S., Gregorio G., Bitton N., Hendrickson W.A., Littman D.R. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity. 2002;16:135–144. doi: 10.1016/s1074-7613(02)00259-5. [DOI] [PubMed] [Google Scholar]
- Lal M., Lai M., Ugaonkar S., Wesenberg A., Kizima L., Rodriguez A., Levendosky K., Mizenina O., Fernandez-Romero J., Zydowsky T. Development of a vaginal fast-dissolving insert combining griffithsin and carrageenan for potential use against sexually transmitted infections. J. Pharm. Sci. 2018;107:2601–2610. doi: 10.1016/j.xphs.2018.06.002. [DOI] [PubMed] [Google Scholar]
- Lambert A.A., Gilbert C., Richard M., Beaulieu A.D., Tremblay M.J. The C-type lectin surface receptor DICIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood. 2008;112:1299–1307. doi: 10.1182/blood-2008-01-136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land A., Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83:783–790. doi: 10.1016/s0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Ozorowski G., Ward A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekkerkerker A.N., van Kooyk Y., Geijtenbeek T.B.H. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr. HIV Res. 2006;4:169–176. doi: 10.2174/157016206776055020. [DOI] [PubMed] [Google Scholar]
- Li Y., Luo L., Thomas D.Y., Kang C.Y. Control of expression, glycosylation, and secretion of HIV-1 gp120 by homologous and heterologous signal sequences. Virology. 1994;204:266–278. doi: 10.1006/viro.1994.1531. [DOI] [PubMed] [Google Scholar]
- Li Y., Luo L., Thomas D.Y., Kang C.Y. The HIV-1 Env protein signal sequence retards its cleavage and down-regulates the glycoprotein folding. Virology. 2000;272:417–428. doi: 10.1006/viro.2000.0357. [DOI] [PubMed] [Google Scholar]
- Li H., Xu C.F., Blais S., Wan Q., Zhang H.T., Landry S.J., Hioe C.E. Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes. J. Immunol. 2009;182:6369–6378. doi: 10.4049/jimmunol.0804287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N., Hogstad B., Wang Y., Levy D.E., Unutmaz D., Littman D.R. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menager M.M., Littman D.R. Actin dynamics regulates dendritic cell-mediated transfer of HIV-1 to T cells. Cell. 2016;164:695–709. doi: 10.1016/j.cell.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.A., Fadden A.J., Drickamer K. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 2001;276:28939–28945. doi: 10.1074/jbc.M104565200. [DOI] [PubMed] [Google Scholar]
- Mitchell C.A., Ramessar K., O'Keefe B.R. Antiviral lectins: selective inhibitors of viral entry. Antiviral Res. 2017;142:37–54. doi: 10.1016/j.antiviral.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort T., Nabatov A.A., Geijtenbeek T.B.H., Pollakis G., Paxton W.A. Efficient capture of antibody neutralized HIV-1 by cells expressing DC-SIGN and transfer to CD4(+) T lymphocytes. J. Immunol. 2007;178:3177–3185. doi: 10.4049/jimmunol.178.5.3177. [DOI] [PubMed] [Google Scholar]
- van Montfort T., Eggink D., Boot M., Tuen M., Hioe C.E., Berkhout B., Sanders R.W. HIV-1 N-glycan composition governs a balance between dendritic cell-mediated viral transmission and antigen presentation. J. Immunol. 2011;187:4676–4685. doi: 10.4049/jimmunol.1101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., O'Keefe B.R., Sowder R.C., 2nd, Bringans S., Gardella R., Berg S., Cochran P., Turpin J.A., Buckheit R.W., Jr., McMahon J.B. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 2005;280:9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- Moulaei T., Shenoy S.R., Giomarelli B., Thomas C., McMahon J.B., Dauter Z., O'Keefe B.R., Wlodawer A. Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity. Structure. 2010;18:1104–1115. doi: 10.1016/j.str.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Zsolt D., Cantero-Perez J., Erkizia I., Benet S., Pino M., Serra-Peinado C., Hernandez-Gallego A., Castellvi J., Tapia G., Arnau-Saz V. Dendritic cells from the cervical mucosa capture and transfer HIV-1 via siglec-1. Front. Immunol. 2019;10:825. doi: 10.3389/fimmu.2019.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell R.L.R., Totrov M., Itri V., Liu X., Fox A., Zolla-Pazner S. Plasticity and epitope exposure of the HIV-1 envelope trimer. J. Virol. 2017;91 doi: 10.1128/JVI.00410-17. pii: e00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard L.K., Spencer D.I., Royle L., Bonomelli C., Seabright G.E., Behrens A.J., Kulp D.W., Menis S., Krumm S.A., Dunlop D.C. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat. Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska M., Takahashi K., Czernekova L., Zachova K., Hall S., Moldoveanu Z., Elliott M.C., Wilson L., Brown R., Jancova D. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J. Biol. Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P.M., Dwek R.A. Glycosylation: heterogeneity and the 3D structure of proteins. Crit. Rev. Biochem. Mol. Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- Sanders R.W., van Anken E., Nabatov A.A., Liscaljet I.M., Bontjer I., Eggink D., Melchers M., Busser E., Dankers M.M., Groot F. The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology. 2008;5:10. doi: 10.1186/1742-4690-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N., Lis H. Lectins as cell recognition molecules. Science. 1989;246:227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- Shen R., Raska M., Bimczok D., Novak J., Smith P.D. HIV-1 envelope glycan moieties modulate HIV-1 transmission. J. Virol. 2014;88:14258–14267. doi: 10.1128/JVI.02164-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N., Goldstein I.J., Vandamme E.J.M., Peumans W.J. Binding-properties of a mannose-specific lectin from the snowdrop (Galanthus nivalis) bulb. J. Biol. Chem. 1988;263:728–734. [PubMed] [Google Scholar]
- da Silva J.X., Franco O.L., Lemos M.A., Gondim M.V., Prosdocimi F., Arganaraz E.R. Sequence variations of Env signal peptide alleles in different clinical stages of HIV infection. Peptides. 2011;32:1800–1806. doi: 10.1016/j.peptides.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Snapp E.L., McCaul N., Quandte M., Cabartova Z., Bontjer I., Kallgren C., Nilsson I., Land A., von Heijne G., Sanders R.W. Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide. Elife. 2017;6 doi: 10.7554/eLife.26067. pii: e26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones G.B., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe W.B., Chertova E., Allen J.D., Seabright G.E., Watanabe Y., Harvey D.J., Medina-Ramirez M., Roser J.D., Smith R., Westcott D. Site-specific glycosylation of virion-derived HIV-1 Env is mimicked by a soluble trimeric immunogen. Cell Rep. 2018;24:1958–1966.e5. doi: 10.1016/j.celrep.2018.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Ishihara M., Middleton D.R., Tiemeyer M., Avci F.Y. Metabolic labeling of HIV-1 envelope glycoprotein gp120 to elucidate the effect of gp120 glycosylation on antigen uptake. J. Biol. Chem. 2018;293:15178–15194. doi: 10.1074/jbc.RA118.004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Pena A., Rantalainen K., Cottrell C.A., Allen J.D., van Gils M.J., Torres J.L., Crispin M., Sanders R.W., Ward A.B. Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog. 2019;15:e1007920. doi: 10.1371/journal.ppat.1007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley S., Mohamed Z., Guo W., McKenna J., Cleveland B., LaBranche C., Beaumont D., Shen X., Yates N.L., Pinter A. Induction of heterologous tier 2 HIV-1-Neutralizing and cross-reactive V1/V2-specific antibodies in rabbits by prime-boost immunization. J. Virol. 2016;90:8644–8660. doi: 10.1128/JVI.00853-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunetsugu-Yokota Y., Muhsen M. Development of human dendritic cells and their role in HIV infection: antiviral immunity versus HIV transmission. Front. Microbiol. 2013;4:178. doi: 10.3389/fmicb.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunetsuguyokota Y., Akagawa K., Kimoto H., Suzuki K., Iwasaki M., Yasuda S., Hausser G., Hultgren C., Meyerhans A., Takemori T. Monocyte-derived cultured dendritic cells are susceptible to human-immunodeficiency-virus infection and transmit virus to resting T-cells in the process of nominal antigen presentation. J. Virol. 1995;69:4544–4547. doi: 10.1128/jvi.69.7.4544-4547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay C., Mayr L.M., Zhang J., Kumar R., Gorny M.K., Nadas A., Zolla-Pazner S., Hioe C.E. Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope. J. Virol. 2014;88:12853–12865. doi: 10.1128/JVI.02125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay C., Feyznezhad R., Yang W., Zhang H., Zolla-Pazner S., Hioe C.E. Alterations of HIV-1 envelope phenotype and antibody-mediated neutralization by signal peptide mutations. PLoS Pathog. 2018;14:e1006812. doi: 10.1371/journal.ppat.1006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Xue J., Hoorelbeke B., Kagiampakis I., Demeler B., Balzarini J., Liwang P.J. The griffithsin dimer is required for high-potency inhibition of HIV-1: evidence for manipulation of the structure of gp120 as part of the griffithsin dimer mechanism. Antimicrob. Agents Chemother. 2013;57:3976–3989. doi: 10.1128/AAC.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolitz J., Schwing C., Chang J., Van Ryk D., Nawaz F., Wei D., Cicala C., Arthos J., Fauci A.S. Signal peptide of HIV envelope protein impacts glycosylation and antigenicity of gp120. Proc. Natl. Acad. Sci. U S A. 2018;115:2443–2448. doi: 10.1073/pnas.1722627115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska N.E., O'Keefe B.R., Mori T., Zhu C., Giomarelli B., Vojdani F., Palmer K.E., McMahon J.B., Wlodawer A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure. 2006;14:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S., Cohen S.S., Boyd D., Kong X.P., Seaman M., Nussenzweig M., Klein F., Overbaugh J., Totrov M. Structure/Function studies involving the V3 region of the HIV-1 envelope delineate multiple factors that affect neutralization sensitivity. J. Virol. 2016;90:636–649. doi: 10.1128/JVI.01645-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.