Abstract
Recently, emerging evidence shows that dysregulation of circadian genes is closely associated with liver fibrosis. However, how dysregulation of circadian genes promotes liver fibrosis is unknown. In this study, we show that neuronal PAS domain protein 2 (NPAS2), one of the core circadian molecules that has been shown to promote hepatocarcinoma cell proliferation, significantly contributed to liver fibrogenesis. NPAS2 is upregulated in hepatic stellate cells (HSCs) after fibrogenic injury, which subsequently contributes to the activation of HSCs. Mechanistically, NPAS2 plays a profibrotic role via direct transcriptional activation of hairy and enhancer of split 1 (Hes1), a critical transcriptor of Notch signaling for the fibrogenesis process, in HSCs. Our findings demonstrate that NPAS2 plays a critical role in liver fibrosis through direct transcriptional activation of Hes1, indicating that NPAS2 may serve as an important therapeutic target to reverse the progression of liver fibrosis.
Keywords: neuronal PAS domain protein 2, circadian gene, liver fibrosis, hepatic stellate cell, Hes1
Introduction
Progressive liver fibrosis (LF) is characterized by the deposition of collagen, usually caused by hepatitis virus infection, as well as cholestatic/biliary and metabolic dysregulation.1, 2, 3, 4 As liver fibrosis progresses, it disrupts the architecture and function of liver tissue via replacing hepatocytes with fibrotic tissues, ultimately leading to cirrhosis, hepatic dysfunction, and even hepatocellular carcinoma (HCC).2,5 Recent experimental research has advanced our knowledge on the pathogenesis of LF. However, the translation of basic exploration into clinical practice has been impeded due to a lack of understanding of the disease, which thus emphasizes the urgency of clarifying the detailed mechanism of LF.
LF is modulated by dynamic communication between hepatocytes and other non-parenchymal cells, among which the extracellular matrix (ECM)-producing myofibroblasts play key roles in this process.1,6 Hepatic stellate cells (HSCs), located in the hepatic sinusoid, are the major source of fibrogenic cells and play a hub role in liver fibrogenesis.7 Upon liver injury, quiescent HSCs transdifferentiate from lipid-storing cells into proliferating, contractile, pro-inflammatory, and fibrogenic myofibroblasts, which leads to enhanced profibrotic signals and suppressed anti-fibrotic pathways.7,8 In recent years, we have witnessed significant progress in exploring the mechanisms underlying LF. Also, many signaling molecules and pathways involved in HSC activation have been identified.9,10 So far, however, our understanding of the regulation of HSCs remains unclear.
Circadian rhythms occur around a 24-h oscillation in behavior and physiology associated with the solar day.11 These daily rhythms are driven by a network of transcriptional-translational feedback loops that exist in essentially all tissue and cell types of the organism.12,13 Previous research reports that dysregulation of some circadian genes, such as Bmal1, Clock, Per2, Cry2, and RORα, contributes to liver fibrogenesis.14, 15, 16, 17 Neuronal PAS domain protein 2 (NPAS2), one of the core circadian molecules, was proven to be a critical oncogene in hepatocellular carcinogenesis by our previous study, which showed that NPAS2 significantly facilitated cell survival both in vitro and in vivo by promoting cell proliferation and inhibiting apoptosis.18 Nevertheless, it is still unknown whether and how NPAS2 affects LF progression, the pre-stage of HCC.
In this study, we found that NPAS2 was exclusively expressed in activated HSCs (aHSCs) and played an essential role in LF, suggesting that NPAS2 is a novel biomarker for aHSCs and a potential target for antifibrotic therapy.
Results
NPAS2 Expression Is Markedly Increased in aHSCs of Fibrotic Liver
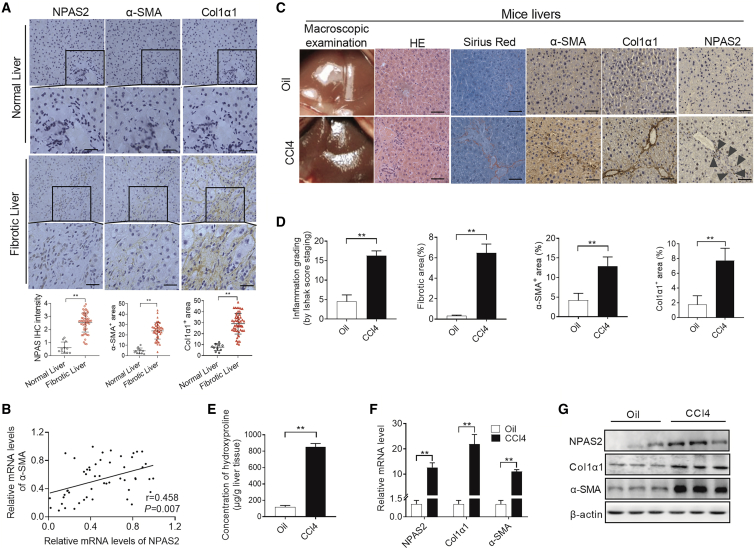
To explore the role of NPAS2 in LF, we first examined the expression of NPAS2 in human liver tissues. As shown in Figure 1A, immunohistochemical (IHC) analyses revealed that NPAS2 expression was higher in fibrotic liver tissues as compared to normal liver tissues. Also, NPAS2 mRNA and protein levels were increased in the fibrotic livers (Figures S1A and S1B). Notably, there was a positive correlation between the increased NPAS2 expression and LF progression (r = 0.727, p < 0.001) (Figure S1C). Importantly, we showed that the expression pattern of NPAS2 coincides with that of alpha-smooth muscle actin (α-SMA), a well-established marker of aHSCs in human fibrotic livers, implying the relatively restricted expression of NPAS2 in aHSCs (Figure 1A). Also, NPAS2 mRNA was increased in the fibrotic livers, correlating with the induction of α-SMA mRNA (r = 0.458, p = 0.007) (Figure 1B) and suggesting that NPAS2 may play vital roles in HSCs. Next, as expected, increased hydroxyproline content and the upregulation of NPAS2, α-SMA, and Col1α1 were also observed in carbon tetrachloride (CCl4)-induced murine LF (Figure 1C–1G). Moreover, similar results were observed in bile duct ligation (BDL)-induced fibrotic models (Figures S1D–S1H). Importantly, we investigated that varieries of cell types in the liver after inducing LF. As shown in Figure S2G, only NPAS2 expression in HSCs significantly increased after liver injury, compared with no change in liver sinusoidal endothelial cells (LSECs), Kupffer cells, and hepatocytes, further suggesting that NPAS2 in HSCs contributes to liver fibrogenesis. Collectively, these data suggest that NPAS2 expression in HSCs is positively correlated with LF progression.
Figure 1.
NPAS2 Expression Is Upregulated in aHSCs of Fibrotic Liver
(A) Representative results by IHC staining in normal and fibrotic liver tissues. (B) Pearson’s correlation analysis of NPAS2 mRNA with α-SMA mRNA was measured in liver lysate with treatment as indicated. (C) Representative results to evaluate liver fibrosis progression. (D) Semiquantification results to statistically analyze fibrosis progression indicated in (C). (E) Liver hydroxyproline content was analyzed to compare the fibrosis progression stage of different groups. Data shown are the means ± SEM from three independent experiments. (F and G) Classical fibrosis-related genes in fibrotic livers were more than that in control group at the mRNA level and protein level. *p < 0.05; **p < 0.01.
NPAS2-KO Mice Are Protected against Hepatic Fibrosis
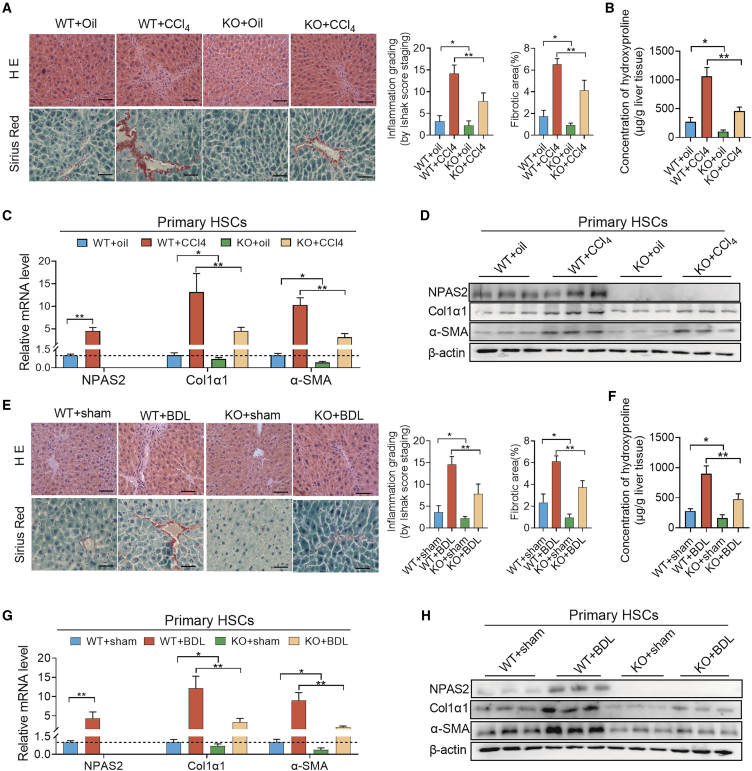
Then, we focused on the contribution of NPAS2 to LF, using NPAS2-knockout (KO) mice. As shown in Figure S2A (arrowheads point to HSCs), S2C, and S2E, IHC and western blotting analyses were carried out to assess the efficiency of NPAS2 KO. H&E and picrosirius red staining revealed the attenuated liver-bridging fibrosis and collagen deposition in NPAS2-KO mice in the CCl4-induced fibrosis model (Figure 2A). Hydroxyproline content in the liver tissue of NPAS2−/− mice, as well as α-SMA and Col1α1 expression, showed a decrease in the CCl4-induced fibrosis model, compared with wild-type (WT) mice. These results suggest that NPAS2 may contribute to the mechanism of LF (Figures 2B–2D). Consistently, in BDL-induced murine fibrotic liver tissues, liver-bridging fibrosis, and collagen deposition, hydroxyproline content, α-SMA, and Col1α1 expression decreased in mice with NPAS2 abrogation, compared with WT mice (Figures 2E–2H). The results demonstrated that NPAS2 KO could attenuate liver injury and alleviate LF in vivo, and NPAS2 may contribute to molecular mechanisms in the pathogenesis of LF.
Figure 2.
NPAS2-KO Mice Are Protected against Hepatic Fibrosis
(A) H&E and picrosirius red staining revealed the attenuated liver-bridging fibrosis and collagen deposition in NPAS2-KO mice in the CCl4-induced fibrosis model. (B) Hydroxyproline content in the CCl4-induced liver tissues. (C and D) Col1α1 and α-SMA expression were downregulated in NPAS-KO mice compared with that in WT mice in CCl4-induced fibrosis model. (E) H&E and picrosirius red staining revealed the attenuated liver-bridging fibrosis and collagen deposition in NPAS2-KO mice in the BDL-induced fibrosis model. (F) Hydroxyproline content in BDL-induced liver tissues. (G and H) Col1α1 and α-SMA expression were downregulated in NPAS-KO mice compared with that in WT mice in BDL-induced fibrosis model. (n = 8–10 for each group). *p < 0.05; **p < 0.01.
Overexpression of NPAS2 Aggravates Hepatic Fibrogenesis in Both CCl4 and BDL Models
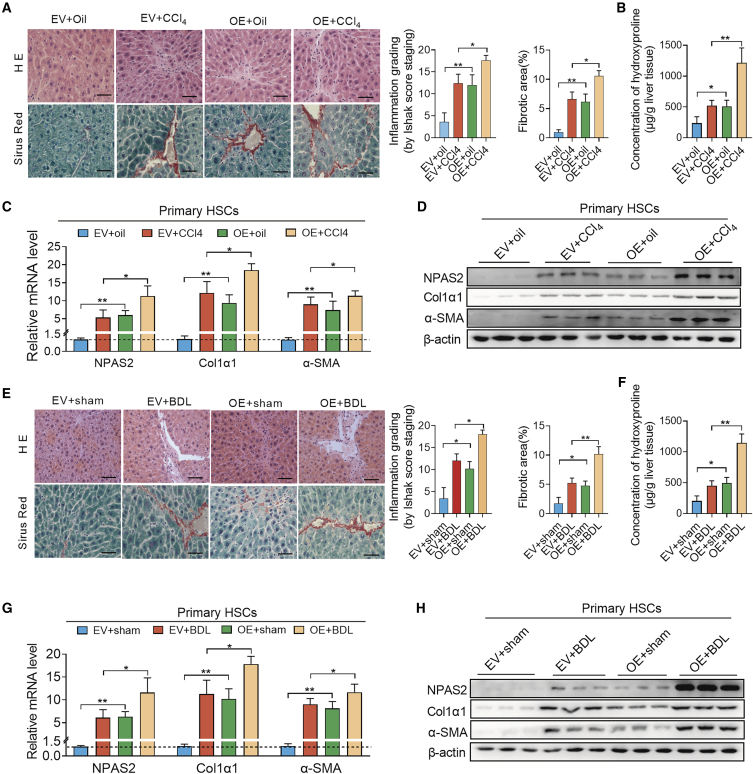
Additionally, we constructed lentiviruses carrying an NPAS2-overexpressing lentiviral vector to confirm the essential role of NPAS2 in hepatic fibrosis progression. As shown in Figure S2B (arrowheads point to HSCs), 2D, and 2F, IHC staining and western blotting were carried out to assess the efficiency of NPAS2 overexpression (OE). Then, as shown in Figure 3A, H&E and picrosirius red staining revealed the considerable liver-bridging fibrosis and collagen deposition in NPAS2-OE mice in the CCl4-induced fibrosis model. Hydroxyproline content in the liver tissue of lenti-NPAS2 mice, as well as α-SMA and Col1α1 expression, showed an increase in the CCl4-induced fibrosis model, compared with WT mice. These results also suggest that NPAS2 may contribute to the mechanism of liver fibrosis (Figures 3B–3D). Consistently, in BDL-induced murine fibrotic liver tissues, liver-bridging fibrosis, and collagen deposition, hydroxyproline content, α-SMA, and Col1α1 expression were augmented in OE mice, compared with WT mice (Figures 3E–3H). The results demonstrated that NPAS2 OE in HSCs aggravates liver fibrogenesis in vivo.
Figure 3.
Overexpression of NPAS2 Aggravates Hepatic Fibrogenesis in Both the CCl4 and BDL Models
(A) H&E and picrosirius red staining revealed the increased liver-bridging fibrosis and collagen deposition in lenti-NPAS2 mice in the CCl4-induced fibrosis model. (B) Hydroxyproline content in the CCl4-induced liver tissues. (C and D) Col1α1 and α-SMA expression were upregulated in NPAS-OE mice compared with that in control mice in CCl4-induced fibrosis model. (E) H&E and picrosirius red staining revealed the aggravated liver-bridging fibrosis and collagen deposition in lenti-NPAS2 mice in the BDL-induced fibrosis model. (F) Hydroxyproline content in BDL-induced liver tissues. (G and H) Col1α1 and α-SMA expression were increased in NPAS-OE mice compared with that in control mice in BDL-induced fibrosis model (n = 8–10 for each group). *p < 0.05; **p < 0.01.
NPAS2 Is Involved in the Activation of HSCs
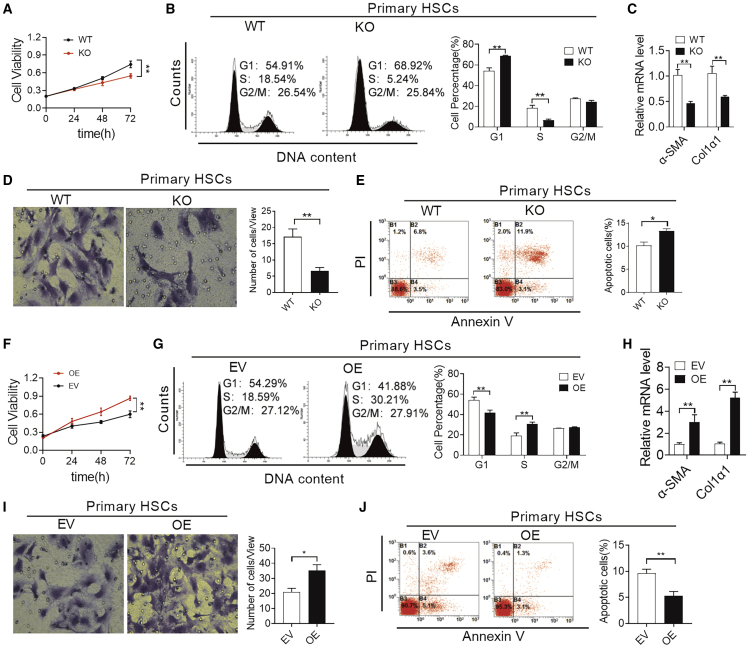
HSC activation is characterized by key phenotypes such as enhanced proliferation, cell migration, actin remodeling, resistance to apoptosis, and collagen production.1,8 Next, we explored whether NPAS2 was required for some or most of these phenotypes, and thereby for HSC activation. First, we analyzed NPAS2 expression in cultured primary murine HSCs acquired from NPAS2-WT mice at different days. Importantly, NPAS2 expression was undetectable in the isolated quiescent primary HSCs from WT mice, whereas evident NPAS2 expression was noted in the culture-activated HSCs since the third day of incubation on plastic plates (Figure S2H), which further supported the specific expression of NPAS2 in aHSCs and that the upregulation of NPAS2 might be an important event in HSC activation. As shown in Figures 4A–4D, NPAS2 abrogation attenuated the proliferation, migration, and collagen production capability of primary HSCs. In addition, compared to primary HSCs from WT mice, increased cell apoptosis was determined in primary HSCs from NPAS2-KO mice by flow cytometry assay (Figure 4E). Additionally, the opposite results have been achieved in lenti-NPAS2 mice. Specifically, the enhanced abilities of proliferation, migration, collagen production, and apoptotic resistance were observed in OE mice, compared with empty vector (EV)-treated mice (Figures 4F–FJ). Moreover, we also investigated the ability of NPAS2 to activate HSCs in LX2 cells, immortal human HSCs.19,20 As shown in Figures S3A and S3B, the efficiency of siNPAS2 and NPAS2 OE was analyzed by qPCR and western blotting assays. Also, similar results were achieved in LX2 cells with treatment as indicated, compared with the results from primary HSCs (Figures S3C–S3L). Taken together, these results demonstrate that NPAS2 function is required for several key phenotypes that are critical for transdifferentiation and sustained activation of HSCs during progressive fibrosis.
Figure 4.
NPAS2 Contributes to HSC Activation
(A) MTS assay for primary HSC proliferation from WT and KO mice as indicated. (B) Cell cycle analysis by flow cytometry in primary HSC proliferation from WT and KO mice as indicated. (C) Col1α1 and α-SMA mRNA expression in primary HSCs from WT and KO mice. (D) Transwell Matrigel invasion assay for cell invasion ability of primary HSCs from WT and KO mice. (E) Flow cytometry analysis of apoptosis by annexin V and propidium iodide (PI) staining in primary HSCs from WT and KO mice. (F) MTS assay for primary HSC proliferation from EV and OE mice as indicated. (G) Cell cycle analysis by flow cytometry in primary HSC proliferation from EV and OE mice as indicated. (H) Col1α1 and α-SMA mRNA expression in primary HSCs from WT and KO mice. (I) Transwell Matrigel invasion assay for cell invasion ability of primary HSCs from EV and OE mice. (J) Flow cytometry analysis of apoptosis by annexin V and PI staining in primary HSCs from EV and OE mice. Data shown are the means ± SEM from three independent experiments. *p < 0.05; **p < 0.01.
Hes1 Is Essential for the Effect of NPAS2 in Activating HSCs
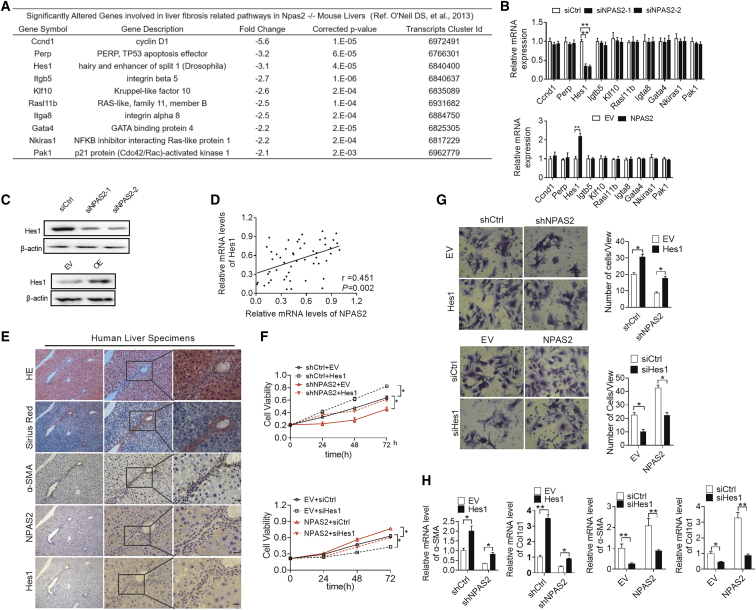
Recently, a study demonstrated that a series of genes might be NPAS2-targeted genes.21 We investigated all of these genes and selected LF-related genes from components of the transforming growth factor β (TGF-β) pathway, Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling, Wnt signaling, Hippo signaling, Notch signaling, and Hedgehog signaling19,22,23 (see Data S1). The top 10 significantly downregulated genes in NPAS2-KO mice liver were identified as genes that might be regulated by NPAS2 (Figure 5A). Next, to elucidate the molecular mechanism of NPAS2-mediated liver fibrosis, the mRNA levels of selected genes were evaluated with treatment of lenti-NPAS2, while only the expressions of hairy and enhancer of split 1 (Hes1) was upregulated by qPCR and western blot analysis (Figures 5B and 5C). As expected, only Hes1 expression was significantly decreased at the mRNA and protein levels when NPAS2 was knocked down in LX2 cells (Figures 5B and 5C). To provide further support, we examined both the mRNA and protein levels of Hes1 in LF tissue samples by qPCR and IHC analysis. Scatterplot analysis (Figure 5D) showed a positive correlation between the mRNA levels of NPAS2 and Hes1 (r = 0.451, p = 0.002). Representative images are shown in Figure 5E, which demonstrates that regions with strong NPAS2 IHC staining also had strong Hes1 staining in HSCs. Furthermore, we explored whether Hes1 was involved in NPAS2-promotive fibrosis. As shown in Figure 5F, forced expression of Hes1 significantly increased proliferation ability as downregulated by NPAS2 knockdown in LX2 cells. Also as expected, overexpression of Hes1 reversed cell migration and collagen production inhibited by NPAS2 knockdown in LX2 cells (Figures 5G and 5H). In contrast, knockdown of Hes1 exhibited opposite effects (Figures 5F–5H). In addition, since Hes1 is one component of the Notch signaling target gene,22 we next explored whether the NPAS2-promotive effect needs Notch signaling. As shown in Figure S4A, Notch signaling-related genes, Notch, Jag1, and Hey2 remained unchanged after NPAS2 upregulation or downregulation. Moreover, we utilized γ-secretase inhibitor (GSI) to inhibit Notch signaling in LX2 cells.24 As shown in Figure S4B, although the Notch pathway was attenuated, Hes1 was upregulated after NPAS2 overexpression. The results seem to show that the NPAS2 profibrotic effect might need intrinsic transcriptional activation of Hes1, independent of Notch signaling. However, this needs further exploration. In addition, NPAS2 and Hes1 expression levels were detected using primary HSC lysates (Figures 4D–4G). Taken these results together, we recognize that Hes1 might be essential for NPAS2-promoted liver fibrogenesis.
Figure 5.
Hes1 Is Essential for the Effect of NPAS2 in Activation of HSCs
(A) Possible candidate transcriptional target genes of NPAS2 as referred from O’Neil et al.21 (B) Among the ten target genes, only Hes1 was dowregulated or upregulated after downregulating or upregulating NPAS2 expression. (C) Hes1 expression was downregulated by siRNA against NPAS2 and upregulated by overexpression of NPAS2 using western blotting assay. (D) Scatterplot analysis of correlation between mRNA expression levels of NPAS2 and Hes1. (E) Representative immunohistochemical (IHC) staining images of NPAS2 and HES1 in LF tissues. (F) MTS analysis for LX2 cell proliferation with treatment as indicated. (G) Transwell Matrigel invasion assay for cell invasion ability of LX2 cells with treatment as indicated. (H) Col1α1 and α-SMA mRNA expression in LX2 cells with treatment as indicated. Data shown are the means ± SEM from three independent experiments. *p < 0.05; **p < 0.01.
Pro-fibrotic Effects of NPAS2 Directly Depends on the Transcriptional Activation of Hes1 Signaling in HSCs In Vivo
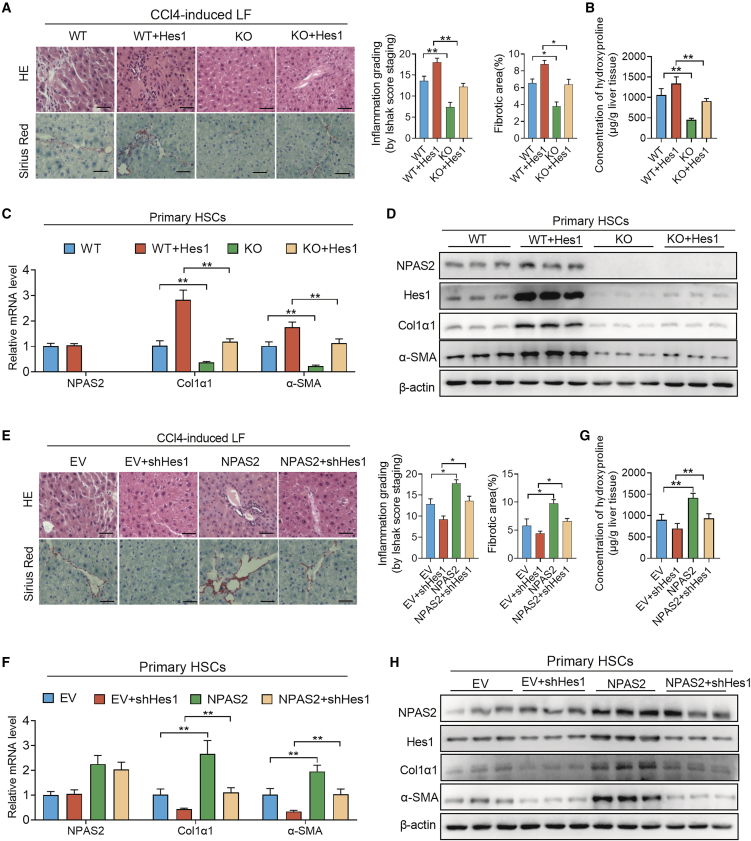
We hypothesized that NPAS2 could promote LF progression by transcriptional activation of Hes1, an indicated LF-promotive molecule.25. To further investigate whether NPAS2 could promote LF via Hes1 transcriptional activation in vivo, pcDNA3.1-Hes1 or small interfering RNA (siRNA) against Hes1 was respectively transfected into LX2 cells with NPAS2 expression stably knocked down or overexpressed. We found that forced expression of Hes1 significantly restored the attenuated inflammation effect and ECM deposition in NPAS2 knockout mice after CCl4-induced (Figure 6A) and BDL-induced (Figure S5A) injuries. Taken together, the hydroxyproline content, mRNA, and protein levels of Col1α1 and α-SMA showed similar results (Figures 6B–6D; Figures S5B–S5D). Meanwhile, Hes1 knockdown significantly reversed LF progression induced by NPAS2 overexpression in CCl4-induced (Figure 6E) and BDL-induced (Figure S5E) LF models. As expected, we also observed that Hes1 downregulation alleviated the aggravated inflammation effect and ECM deposition in NPAS2-OE mice (Figures 6F–6H; Figures S5F–S5H). These results indicate that the fibrosis-promotive function of NPAS2 is dependent on Hes1-mediated profibrotic progression.
Figure 6.
The Liver Fibrosis-Promotive Function of NPAS2 Is Dependent on Transcriptional Activation of Hes1 In Vivo
(A) H&E and picrosirius red staining revealed the reversed liver-bridging fibrosis and collagen deposition in NPAS2-KO mice together with lenti-Hes1 mice in the CCl4-induced liver fibrosis (LF) model. (B) Hydroxyproline content in CCl4-induced liver tissues. (C and D) Lenti-Hes1 significantly restored the attenuated Col1α1 and α-SMA expression in NPAS2 knock-out mice after CCl4-induced LF. (E) H&E and picrosirius red staining revealed that lenti-shHes1 injection inhibits the aggravated liver-bridging fibrosis and collagen deposition in NPAS2-OE mice in the CCl4-induced LF model. (F) Hydroxyproline content in CCl4-induced liver tissues. (G and H) Lenti-shHes1 significantly decreased the augmented Col1α1 and α-SMA expression in NPAS2-OE mice after CCl4-induced LF. *p < 0.05; **p < 0.01.
NPAS2 Transcriptionally Upregulates Hes1 Expression by Heterodimerization with BMAL1 in HSCs
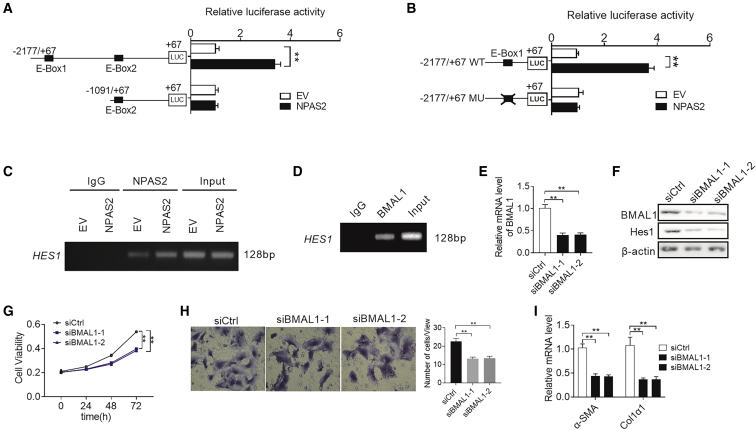
Recently, O’Neil et al.21 identified 352 intersecting genes that are candidates for direct regulation by the NPAS2/BMAL1 complex. In their results, Hes1 was also targeted by the NPAS2/BMAL1 complex. Accordingly, we hypothesized that Hes1 may be a transcriptional target of NPAS2. To test this idea, we developed a series of truncated promoter constructs and determined their transcriptional activity in LX2 cells transfected with EV or NPAS2. A luciferase reporter assay showed that all Hes1 promoter constructs truncated from −2177 to −1091 exhibited obvious NPAS2-related transcriptional activities, and the construct (from −1091 to +67) completely abolished the activity of the reporter gene in all transfected cells (Figure 7A). These data strongly suggest that the Hes1 promoter region may contain a binding site of NPAS2. To test this finding, we analyzed the DNA sequence of Hes1 promoter and identified an E-box (nucleotide −1875 to nucleotide −1869), which is a putative DNA-binding site for NPAS2. Site-directed mutagenesis analyses showed that the E-box we identified was critical for NPAS2-induced Hes1 transcriptional activation in LX2 cells (Figure 7B). Moreover, the chromatin immunoprecipitation (ChIP) assay confirmed that NPAS2 binds directly to the E-box region of Hes1 promoter (Figure 7C).
Figure 7.
NPAS2 Transcriptionally Upregulates Hes1 Expression in HSCs
(A) LX2 cells were transfected with constructs with serially truncated Hes1 promoter constructs, and the cells were treated with or without ectopic expression of NPAS2 as indicated. Twenty-four hours after treatment, the luciferase activity was determined. (B) LX2 cells were transfected with −2177/+67 Hes1-WT or −2177/+67 Hes1-MU (selective mutation) promoter constructs. Twenty-four hours after treatment, the luciferase activity was determined. (C) Amplification of the Hes1 promoter sequence from ChIP DNA was performed. Input and immunoglobulin G (IgG) served as positive and negative controls, respectively. The agarose gel electrophoresis was then used for analysis of the PCR products. (D) Amplification of the Hes1 promoter sequence from ChIP DNA. Input and IgG served as positive and negative controls, respectively. (E and F) qRT-PCR and western blot analysis for expression levels of Hes1 at both mRNA and protein levels. (E and F) Hes1 expression was decreased after knockdown of BMAL1 and upregulated by overexpression of BMAL1. (G) MTS assay for LX2 cells with treatment as indicated. (H) Transwell Matrigel invasion assay for cell invasion ability of LX2 cells with treatment as indicated. (I) Col1α1 and α-SMA mRNA expression in LX2 cells with treatment as indicated. Data shown are the means ± SEM from three independent experiments. *p < 0.05; **p < 0.01.
Alternatively, NPAS2 generally forms heterodimers with BMAL1, another core circadian rhythm transcription factor, to transcriptionally regulate the expression of numerous target genes.26,27 We thus explored whether BMAL1 is involved in the promotion of liver fibrosis by NPAS2. ChIP-PCR assay indicated that BMAL1 directly binds to the promoter of Hes1 (Figure 7D). As expected, knockdown of BMAL1 robustly decreased the expression of Hes1 at both the mRNA and protein levels (Figures 7E and 7F). Additionally, decreased cell proliferation and migration, as well as Col1α and α-SMA mRNA levels, were also observed when BMAL1 was knocked down in LX2 cells (Figures 7G–7I), which are similar to the effects of NPAS2 knockdown. Collectively, these results demonstrate that NPAS2 directly binds to the E-box site in the Hes1 promoter region to transactivate Hes1 expression by heterodimerization with BMAL1.
Overexpression of NPAS2 in HSCs Is Mainly Mediated by Downregulation of miR-19b-3p
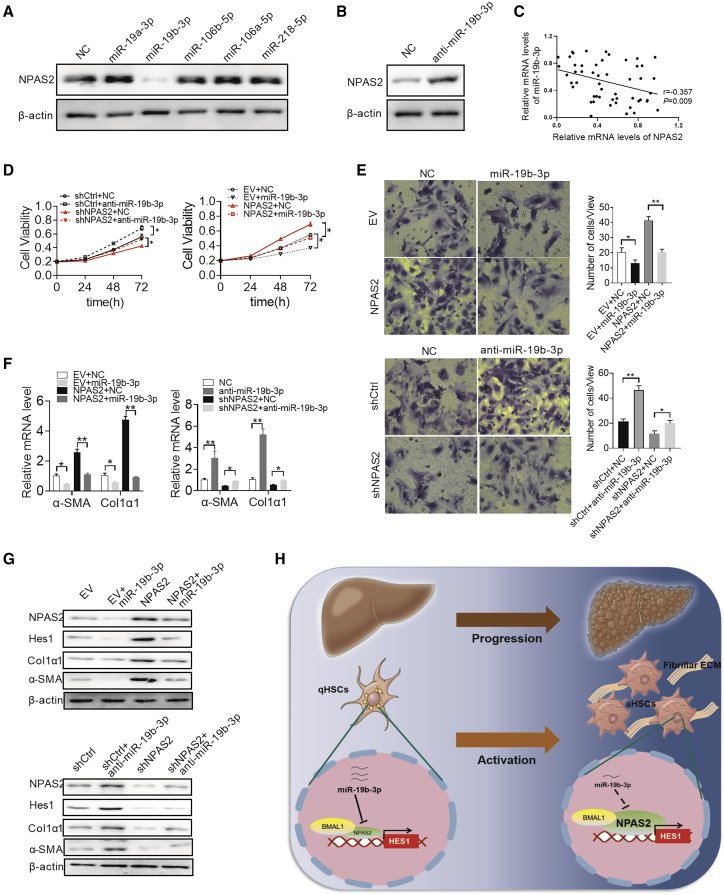
MicroRNA plays an important modulating role in the gene-expression network.28 To identify potential microRNAs involved in the overexpression of NAPS2 in LF, microRNA data integration portal (mirDIP)29-based target prediction programs were performed. Among the top 10 predicted microRNAs targeting NPAS2 represented in Figure S6, miR-19a-3p, miR-19b-3p, miR-106a-5p, miR-106b-5p, and miR-218-5p were reported to be frequently downregulated in aHSCs.30, 31, 32, 33 We thus explored which microRNA might contribute to upregulate the expression of NPAS2 in LX2 cells. As shown in Figure 8A, only miR-19b-3p-transfected LX2 cells show significant downregulation of NPAS2. To further test this possibility, miR-19b-3p inhibition enhanced NPAS2 protein expression (Figure 8B). These results suggest that miR-19b-3p represses NPAS2 expression in LX2 cells. In addition, a significant negative correlation between miR-19b-3p and NPAS2 was found in liver fibrosis patients (Figure 8C). Moreover, enforced expression of miR-19b-3p greatly attenuated the ability of NPAS2 to induce the activation of HSCs (Figures 8D–8G), indicating that the miR-19b-3p/NPAS2 axis might contribute to hepatic fibrogenesis.
Figure 8.
Overexpression of NPAS2 Is Mainly Mediated by Downregulation of miR-19b-3p
(A) Western blot analysis for NPAS2 expression in LX2 cells transfected with the miR-19a-3p, miR-19b-3p, miR-106a-5p, miR-106b-5p, and miR-218-5p mimics. (B) Western blot analysis for NPAS2 expression in LX2 cells transfected with the miR-19b-3p inhibitor. (C) The correlation between mRNA levels of NPAS2 and miR-19b-3p was determined. (D–G) MTS assay for cell proliferation (D), Transwell Matrigel invasion assay (E), Col1α1 and α-SMA mRNA expression (F), and Hes1, Col1α1, and α-SMA protein expression (G) were measured in LX2 cells with treatment as indicated. (H) Graphic abstract of this study. Data shown are the means ± SEM from three independent repeats. *p < 0.05; **p < 0.01.
Discussion
A determined effort to uncover the cellular and molecular basis of hepatic fibrosis is now yielding some success, which recognizes that cumulative hepatic injury induces an attenuated wound-healing response, eventually resulting in LF and later progression.1 Importantly, accumulative ECM deposition in turn promotes the progression of hepatic fibrogenesis.34 Usually, the resident HSCs consist of the primary source of ECM-producing myofibroblasts in liver, and they undergo a well-established process to be activated.7,8 However, the in-depth mechanism of HSC activation remains largely unknown. Alternatively, reports have demonstrated inseparable links between circadian genes and LF. Chen et al.16 first used Per−/− mice to demonstrate that circadian dysregulation critically affects liver fibrosis. Also, reversed melatonin administration may inhibit liver fibrogenesis.17,35 Nonetheless, the mechanism of aberrant circadian rhythm promoting LF remains scarce so far. Our study was the first to address the crucial role of circadian gene NPAS2 in LF. First, NPAS2 expression was significantly elevated in human fibrotic specimens and in murine fibrotic models, and it was positively correlated with LF progression stage in patients and in mice. Second, NPAS2 silencing alleviated LF in vivo, whereas OE of NPAS2 aggravates LF progression. Third, NPAS2 functioned as a contributor to HSC activation by transcriptionally activating Hes1. Therefore, NPAS2 is a novel regulator of HSC activation and therapeutic target in LF.
Our research previously reported that circadian gene NPAS2, a component of clock genes, could promote HCC progression by enhancing hepatocarcinoma cell proliferation.18 Also, these results were observed in acute myeloid leukemia.36 Nevertheless, the role of NPAS2 in LF, the pre-stage of HCC, remains unreported. In the current study, we detected the NPAS2 transcript and protein expression in whole liver, and we found that NPAS2 expression was elevated after liver injuries. Next, we found that NPAS2 protein was undetectable in normal liver or in cell types other than HSCs in fibrotic liver by western blot or histological examination. More importantly, the abrogation of NPAS2 attenuated HSC activation and liver fibrosis, suggesting that the upregulated NPAS2 is an intrinsic response of HSC and is involved in HSC activation. Except for producing ECM, aHSCs might also affect liver injury, inflammation, or regeneration via dynamic communication with hepatocytes and non-parenchymal cells.6 However, more studies revealed that KO of HSC fibrosis-associated gene could reverse the progression of LF without involving the signals of liver inflammation.9,10,37,38 Considering the limitation of whole-body KO of NPAS2, we cannot rule out terms that NPAS2 in other types of liver cells contributes to HSC activation. Thus, we considered examining the effects of NPAS2 in hepatocytes, the parenchymal cells in the liver, on the activation of HSCs. On the one hand, from our results (Figures S7A–S7D), we found that NPAS2 in hepatocytes could also affect cell proliferation, consistent with previous studies.18,39 However, further mechanisms need to be investigated. On the other hand, it seems that NPAS2 abrogation or OE in hepatocytes does not affect the activation of HSCs from our results (Figures S7E–S7H). Also, this phenomenon needs intensive studies in vivo and in vitro. Taken together, it is worth considering whether other mechanisms involve the NPAS2 profibrotic effect in HSCs; for example, apart from NPAS2 promoting hepatocyte proliferation, whether inflammation signaling, microRNA release,40 and NPAS2 in other non-parenchymal cells41,42 contribute to HSCs activation. Accordingly, the results imply that NPAS2 depletion attenuated HSC activation and experimental LF. However, further studies are needed to illustrate more mechanisms underlying the profibrotic effects of NPAS2.
Accumulating evidence has illustrated that the TGF-β/Smad pathway, JAK/STAT3 pathway, Notch pathway, and others play a vital role in HSC activation,22 but the detailed machinery has not been identified so far. In our study, we analyzed 10 LF-relevant genes to explore the possible mechanism of NPAS2 profibrotic effects.21 Our data showed that NPAS2 directly bound to the Hes1 promoter and enhanced Hes1 transcription. Previous studies have demonstrated that Hes1 could enhance the expression of α-SMA and Col1α1 in HSCs and be a novel anti-fibrotic target.25,43 In this study, we observed a significant decrease in α-SMA and Col1α1 expression in the livers and primary HSCs of NPAS2-depleted mice compared with WT mice that were included in the experimental fibrosis models. Importantly, considering that Hes1 is a key target gene of Notch signaling, we explored whether NPAS2 relies on Notch signaling to promote Hes1 expression. Strikingly, our results indicate that NPAS2 transcriptional activation of Hes1 is independent of Notch signaling. In addition, as expected, our study demonstrates that the liver profibrotic function of NPAS2 is dependent on Hes1-mediated ECM synthesis both in vivo and in vitro, indicating that NPAS2 may serve as an important therapeutic target to reverse liver fibrosis. Taken together, our results also suggest that Hes1 is a liver fibrogenic factor. Meanwhile, the present study also needs further investigations. Additionally, normal and abnormal circadian rhythms of NPAS2 and Hes1 are required for better understanding of the profibrotic role of NPAS2.44 Alternatively, considering that both NPAS2 and Hes1 contain basic helix-loop-helix (bHLH) domains,45,46 another important question concerning whether the two proteins could form a heterodimer to play a profibrotic role remains to be explored.
A series of microRNAs have been demonstrated as frequently downregulated in aHSCs. For example, six microRNAs, such as miR-30c, miR-93, and miR-106 and others, showed downregulation in chronic hepatitis C Egyptian patients.30 Brandon-Warner et al.31 reported that miR-19b-3p is downregulated in aHSCs. As predicted by the mirDIP program, we investigated which microRNA might contribute to the upregulation of NPAS2 in aHSCs. As shown in our results, only miR-19b-3p antagonizes NPAS2 expression. Collectively, these results suggest that miR-19b-3p functions as an important suppressor of NPAS2 in aHSCs. However, the question as to which region is targeted in endogenous and ectopically expressed NPAS2 transcripts needs further investigation. Furthermore, we still cannot rule out the possibility that other factors may also contribute to the upregulation of NPAS2 in aHSCs and liver fibrosis, such as genetic and epigenetic aberrations. Therefore, the possibility of involvement of other factors in NPAS2 overexpression in LF still needs further confirmation.
In conclusion, these findings broaden the regulatory mechanism of HSC activation and deepen our understanding of the molecular mechanism of LF. In the present study, we demonstrate a pivotal regulatory role of NPAS2 in LF promotion, which provides novel insights to help understand the mechanisms of circadian dysfunction in LF progression and find new therapeutic targets for treatment of this disease.
Materials and Methods
Human Tissue Specimens
Liver tissues were obtained from the liver tissue bank of the Tangdu Hospital and Xijing Hospital, The Fourth Military Medical University (Xi’an, China). The healthy control (n = 11) was normal liver or liver with angioma. Fibrotic liver (n = 52) was from patients with hepatic fibrosis or cirrhosis. Informed consent was obtained from all subjects. The study protocol was approved by the Scientific Investigation Board of The Fourth Military Medical University.
Animals and Treatments
Male C57BL/6 (WT) mice (25–30 g) were provided by the animal experiment center of The Fourth Military Medical University. Constitutive NPAS2−/− mice were generated using the CRISPR/Cas9 approach at Cyagen Biosciences (Suzhou, China), and a 561-bp DNA fragment containing NPAS2 was deleted to produce the null allele. The primer sequences for mouse genotyping are as follows: primer 1 (product size, 561 bp): forward 1, 5′-CTTGTTGAGTAGTGAGTGTTCTG-3′; reverse 1, 5′-CAACTCTGCTTGTGTGGTCTTC-3′; primer 1 (product size, 603 bp): forward 2, 5′-CTTGTTGAGTAGTGAGTGTTCTG-3′; reverse 2, 5′-TAACTAGAACACTCCACGTCACG-3′. Animals were maintained in a temperature-controlled environment (20°C–22°C) with 12-h light/12-h dark cycles and fed ad libitum standard chow with free access to drinking water. Male mice aged 6–8 weeks were intraperitoneally injected with CCl4 (0.5 mL/kg body weight, dissolved in olive oil, 1:4; Sigma) or vehicle (olive oil) three times per week for 4 weeks to induce fibrosis and were sacrificed 48 hours after the last injection.47 The mice subjected to BDL were anesthetized with chloral hydrate followed by midline laparotomy. The common bile duct was ligated twice with 6-0 silk sutures and cut through between the ligations. Sham-operated mice were subjected to laparotomy without BDL. The mice that received BDL or the sham operation were sacrificed 2 weeks later. To perform NPAS2 overexpression, a single dose of 1 × 109 viral particles lentivirus (Shanghai GenePharma, Shanghai, China) was delivered via tail vein 7 days before CCl4 or BDL treatment. Simultaneous administration of lenti-NPAS2 and lenti-shHes1 (Shanghai GenePharma, Shanghai, China) to liver injury mice was performed to investigate the role of Hes1 on the profibrotic effect of NPAS2. The mouse livers and serum were collected for subsequent experiments. All animal procedures were conducted with the approval of The Fourth Military Medical University.
Histology and Immunohistological Analysis
Liver tissues were processed for IHC and H&E staining as previously described.48 The extent of scarring was assessed in livers using picrosirius red staining (Beyotime Biotechnology) and performed as previously described. Areas of positive-stained sites were measured using image analyses software Image-Pro Plus 6.0 (Media Cybernetics). The percentage of positive area in the corresponding field of liver tissue was calculated to show the intensity of collagen deposition or protein expression.
Measurement of Liver Hydroxyproline Content
Wet liver samples (100 mg) were subjected to a commercial kit (A030-2, Jiancheng, Nanjing, China) to determine total hepatic hydroxyproline level.
Isolation and Culture of Primary Cells
Primary murine HSCs were isolated from the livers of male NPAS2−/− or WT mice aged 6–10 weeks using Nycodenz density gradient centrifugation. Briefly, after cannulation of the inferior vena cava, the portal vein was cut, allowing retrograde stepwise perfusion with pronase (Roche, Basel, Switzerland) and collagenase (Roche, Basel, Switzerland)-containing solutions, and isolated by differential centrifugation, respectively. Nycodenz used in density gradient centrifugation for isolation was purchased from Sigma-Aldrich (St. Louis, MO, USA). The viability of HSCs was observed by trypan blue staining, and the purity of HSCs was >98%, as assessed by vitamin A autofluorescence under an BX51 fluorescence microscope (Olympus, NY, USA) and by flow cytometry. The HSCs were then cultured on plastic culture plates in DMEM with 10% fetal bovine serum (FBS). Considering that LSECs attach poorly to a plastic culture dish, LSECs were removed from Kupffer cell (KC) fractions by selective adherence.
qRT-PCR and Western Blotting
RNA extraction, cDNA synthesis, and qRT-PCR reactions and western blotting were performed as previously described.49 Primer sequences used were provided in Table S2. Proteins (30 mg) were subjected to SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane. Native PAGE was utilized for Col1α1 protein detection.50 The antibodies used for western blot are listed in Table S2.
Cell Migration and Proliferation Assays
Cell proliferation was determined by an 3-(4,5-dimethylthiazol-2-yl)-5-(3- carboxymethoxyphe nyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, G3581) according to the manufacturer’s protocol. Briefly, primary HSCs or LX2 cells were plated in 96-well plates (Costar) at 1,000 cells per well. After 12 h, cell viability and growth were measured by addition of 20 μL of MTS (0.2%)-PMS (0.092%; phenazine methosulfate, 20:1) solution and incubation for 2 h at 37°C. The microplates were read in a spectrophotometer at a wavelength of 490 nm. Each sample was analyzed in triplicate. For the cell migration assay, 2 × 105 cells were cells were seeded in the upper chamber that was precoated with 10 mg/mL growth factor-reduced Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). After incubation for 48 h, cells that migrated onto the lower surface of the filter were fixed and stained with crystal violet. The cell counts were expressed as the mean number of cells per field of view.
Luciferase Assay
Four micrograms of Hes1 promoter constructs was co-transfected with Renilla luciferase-expressing control vector into 1 × 106 LX2 cells. Transfected cells were cultured for 48 h. Cells were lysed and the luciferase activates were determined using the Dual-Luciferase reporter assay kit (Promega, E1910) according to the manufacturer’s instructions. The relative light units were measured by a Luminoskan Ascent microplate luminometer (Thermo Scientific). The firefly luciferase activity corresponding to a specific promoter construct was normalized to renilla luciferase activity.
ChIP Assays
The ChIP assays were carried out using a ChIP assay kit (Cell Signaling Technology, 9005), according to the manufacturer’s instructions. Briefly, the cells were cross-linked with 1% formaldehyde for 10 min and then disrupted in cell lysate buffer. The supernatant was collected after the samples were sonicated to break nuclear membrane. Subsequently, the chromatin was immunoprecipitated with anti-NPAS2 antibody (1:100, Abcam, ab157165) or equal amounts of negative control normal rabbit immunoglobulin G (IgG). Finally, DNA extractions were PCR amplified using primer pairs within the regulatory region of the Hes1. The specific PCR primers are listed in Table S1.
Statistical Analysis
Experiments were repeated three times, where appropriate. SPSS 23.0 software (SPSS, Chicago, IL, USA) was used for all statistical analyses, and p < 0.05 was considered significant. Unpaired t tests were used for comparisons between two groups where appropriate. Correlations between measured variables were tested by Spearman’s rank correlation analyses.
Author Contributions
T.Y. and H.L. Chang designed the project. T.Y., P.Y. and Y.Y. carried out most of the experiments, analyzed the data, and wrote the manuscript. J.M. and Z.Y. Yan acquired, analyzed, and interpreted data and drafted the manuscript. N.L., Q.W., J.L., R.L. and H.X. Zhang were responsible for the concept, analyzed and interpreted data, and performed critical revision of the manuscript. Z.Y. Yan, H.X. Zhang and H.L. Chang helped to design and coordinate the experiment.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81600478, 81802345, 81772934) and by the State Key Laboratory of Cancer Biology Project (CBSKL2017Z01)
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.025.
Contributor Information
Jiao Mu, Email: fzzxmj@126.com.
Zhaoyong Yan, Email: yanzytdtt@163.com.
Hulin Chang, Email: changhulin_spph@163.com.
Supplemental Information
References
- 1.Friedman S.L. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuppan D., Afdhal N.H. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao B., Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohinc B.N., Diehl A.M. Mechanisms of disease progression in NASH: new paradigms. Clin. Liver Dis. 2012;16:549–565. doi: 10.1016/j.cld.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Zhang D.Y., Friedman S.L. Fibrosis-dependent mechanisms of hepatocarcinogenesis. Hepatology. 2012;56:769–775. doi: 10.1002/hep.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seki E., Schwabe R.F. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashi T., Friedman S.L., Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv. Drug Deliv. Rev. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 9.Du K., Hyun J., Premont R.T., Choi S.S., Michelotti G.A., Swiderska-Syn M., Dalton G.D., Thelen E., Rizi B.S., Jung Y., Diehl A.M. Hedgehog-YAP signaling pathway regulates glutaminolysis to control activation of hepatic stellate cells. Gastroenterology. 2018;154:1465–1479.e13. doi: 10.1053/j.gastro.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishio T., Hu R., Koyama Y., Liang S., Rosenthal S.B., Yamamoto G., Karin D., Baglieri J., Ma H.Y., Xu J. Activated hepatic stellate cells and portal fibroblasts contribute to cholestatic liver fibrosis in MDR2 knockout mice. J. Hepatol. 2019;71:573–585. doi: 10.1016/j.jhep.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Eckel-Mahan K., Sassone-Corsi P. Metabolism and the circadian clock converge. Physiol. Rev. 2013;93:107–135. doi: 10.1152/physrev.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver R., Kriegsfeld L.J. Circadian rhythms have broad implications for understanding brain and behavior. Eur. J. Neurosci. 2014;39:1866–1880. doi: 10.1111/ejn.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurley J.M., Loros J.J., Dunlap J.C. Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem. Sci. 2016;41:834–846. doi: 10.1016/j.tibs.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kudo T., Tamagawa T., Shibata S. Effect of chronic ethanol exposure on the liver of Clock-mutant mice. J. Circadian Rhythms. 2009;7:4. doi: 10.1186/1740-3391-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P., Kakan X., Zhang J. Altered circadian rhythm of the clock genes in fibrotic livers induced by carbon tetrachloride. FEBS Lett. 2010;584:1597–1601. doi: 10.1016/j.febslet.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Chen P., Han Z., Yang P., Zhu L., Hua Z., Zhang J. Loss of clock gene mPer2 promotes liver fibrosis induced by carbon tetrachloride. Hepatol. Res. 2010;40:1117–1127. doi: 10.1111/j.1872-034X.2010.00695.x. [DOI] [PubMed] [Google Scholar]
- 17.González-Fernández B., Sánchez D.I., Crespo I., San-Miguel B., de Urbina J.O., González-Gallego J., Tuñón M.J. Melatonin attenuates dysregulation of the circadian clock pathway in mice with CCl4-induced fibrosis and human hepatic stellate cells. Front. Pharmacol. 2018;9:556. doi: 10.3389/fphar.2018.00556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yuan P., Li J., Zhou F., Huang Q., Zhang J., Guo X., Lyu Z., Zhang H., Xing J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017;8:e2704. doi: 10.1038/cddis.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang D.M., Sun W., Ning B.F., Zhou T.F., Li X.F., Zhong W., Cheng Z., Xia M.Y., Wang X., Deng X. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 2018;67:1704–1715. doi: 10.1136/gutjnl-2016-313392. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Sanchez I., Dunkel Y., Roh Y.S., Mittal Y., De Minicis S., Muranyi A., Singh S., Shanmugam K., Aroonsakool N., Murray F. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat. Commun. 2014;5:4451. doi: 10.1038/ncomms5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neil D., Mendez-Figueroa H., Mistretta T.A., Su C., Lane R.H., Aagaard K.M. Dysregulation of Npas2 leads to altered metabolic pathways in a murine knockout model. Mol. Genet. Metab. 2013;110:378–387. doi: 10.1016/j.ymgme.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang K., Han X., Zhang Z., Zheng L., Hu Z., Yao Q., Cui H., Shu G., Si M., Li C. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat. Commun. 2017;8:144. doi: 10.1038/s41467-017-00204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang J.X., Mikami K., Venugopal S., Li Y., Török N.J. Apoptotic body engulfment by hepatic stellate cells promotes their survival by the JAK/STAT and Akt/NF-κB-dependent pathways. J. Hepatol. 2009;51:139–148. doi: 10.1016/j.jhep.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu C., Kim K., Wang X., Bartolome A., Salomao M., Dongiovanni P., Meroni M., Graham M.J., Yates K.P., Diehl A.M. Hepatocyte Notch activation induces liver fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aat0344. eaat0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K., Zhang Y.Q., Ai W.B., Hu Q.T., Zhang Q.J., Wan L.Y., Wang X.L., Liu C.B., Wu J.F. Hes1, an important gene for activation of hepatic stellate cells, is regulated by Notch1 and TGF-β/BMP signaling. World J. Gastroenterol. 2015;21:878–887. doi: 10.3748/wjg.v21.i3.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haque R., Ali F.G., Biscoglia R., Abey J., Weller J., Klein D., Iuvone P.M. CLOCK and NPAS2 have overlapping roles in the circadian oscillation of arylalkylamine N-acetyltransferase mRNA in chicken cone photoreceptors. J. Neurochem. 2010;113:1296–1306. doi: 10.1111/j.1471-4159.2010.06698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf D., Wang L.L., Diemer T., Welsh D.K. NPAS2 compensates for loss of CLOCK in peripheral circadian oscillators. PLoS Genet. 2016;12:e1005882. doi: 10.1371/journal.pgen.1005882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kittelmann S., McGregor A.P. Modulation and evolution of animal development through microRNA regulation of gene expression. Genes (Basel) 2019;10:e321. doi: 10.3390/genes10040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirdel E.A., Xie W., Mak T.W., Jurisica I. NAViGaTing the micronome—using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE. 2011;6:e17429. doi: 10.1371/journal.pone.0017429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Hefny M., Fouad S., Hussein T., Abdel-Hameed R., Effat H., Mohamed H., Abdel Wahab A.H. Circulating microRNAs as predictive biomarkers for liver disease progression of chronic hepatitis C (genotype-4) Egyptian patients. J. Med. Virol. 2019;91:93–101. doi: 10.1002/jmv.25294. [DOI] [PubMed] [Google Scholar]
- 31.Brandon-Warner E., Feilen N.A., Culberson C.R., Field C.O., deLemos A.S., Russo M.W., Schrum L.W. Processing of miR17-92 cluster in hepatic stellate cells promotes hepatic fibrogenesis during alcohol-induced injury. Alcohol. Clin. Exp. Res. 2016;40:1430–1442. doi: 10.1111/acer.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Y., Pu R., Han X., Zhao J., Li W., Yin J., Zhang Y., Shen Q., Xie J., Zhang Q. Association of a potential functional pre-miR-218 polymorphism and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Liver Int. 2014;34:728–736. doi: 10.1111/liv.12313. [DOI] [PubMed] [Google Scholar]
- 33.Bae M., Lee Y., Park Y.K., Shin D.G., Joshi P., Hong S.H., Alder N., Koo S.I., Lee J.Y. Astaxanthin attenuates the increase in mitochondrial respiration during the activation of hepatic stellate cells. J. Nutr. Biochem. 2019;71:82–89. doi: 10.1016/j.jnutbio.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S.W. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y.R., Hong R.T., Xie Y.Y., Xu J.M. Melatonin ameliorates liver fibrosis induced by carbon tetrachloride in rats via inhibiting TGF-beta1/Smad signaling pathway. Curr. Med. Sci. 2018;38:236–244. doi: 10.1007/s11596-018-1871-8. [DOI] [PubMed] [Google Scholar]
- 36.Song B., Chen Y., Liu Y., Wan C., Zhang L., Zhang W. NPAS2 regulates proliferation of acute myeloid leukemia cells via CDC25A-mediated cell cycle progression and apoptosis. J. Cell. Biochem. 2018;120:8731–8741. doi: 10.1002/jcb.28160. [DOI] [PubMed] [Google Scholar]
- 37.Wilhelm A., Aldridge V., Haldar D., Naylor A.J., Weston C.J., Hedegaard D., Garg A., Fear J., Reynolds G.M., Croft A.P. CD248/endosialin critically regulates hepatic stellate cell proliferation during chronic liver injury via a PDGF-regulated mechanism. Gut. 2016;65:1175–1185. doi: 10.1136/gutjnl-2014-308325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan J., Tung H.C., Li S., Niu Y., Garbacz W.G., Lu P., Bi Y., Li Y., He J., Xu M. Aryl hydrocarbon receptor signaling prevents activation of hepatic stellate cells and liver fibrogenesis in mice. Gastroenterology. 2019;157:793–806.e14. doi: 10.1053/j.gastro.2019.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crumbley C., Wang Y., Kojetin D.J., Burris T.P. Characterization of the core mammalian clock component, NPAS2, as a REV-ERBα/RORα target gene. J. Biol. Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian H., Deng X., Huang Z.W., Wei J., Ding C.H., Feng R.X., Zeng X., Chen Y.X., Ding J., Qiu L. An HNF1α-regulated feedback circuit modulates hepatic fibrogenesis via the crosstalk between hepatocytes and hepatic stellate cells. Cell Res. 2015;25:930–945. doi: 10.1038/cr.2015.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Araújo Júnior R.F., Garcia V.B., Leitão R.F., Brito G.A., Miguel Ede.C., Guedes P.M., de Araújo A.A. Carvedilol improves inflammatory response, oxidative stress and fibrosis in the alcohol-induced liver injury in rats by regulating Kuppfer cells and hepatic stellate cells. PLoS ONE. 2016;11:e0148868. doi: 10.1371/journal.pone.0148868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan J.L., Ruan B., Yan X.C., Liang L., Song P., Yang Z.Y., Liu Y., Dou K.F., Han H., Wang L. Endothelial Notch activation reshapes the angiocrine of sinusoidal endothelia to aggravate liver fibrosis and blunt regeneration in mice. Hepatology. 2018;68:677–690. doi: 10.1002/hep.29834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie G., Karaca G., Swiderska-Syn M., Michelotti G.A., Krüger L., Chen Y., Premont R.T., Choi S.S., Diehl A.M. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice. Hepatology. 2013;58:1801–1813. doi: 10.1002/hep.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dardente H., Cermakian N. Molecular circadian rhythms in central and peripheral clocks in mammals. Chronobiol. Int. 2007;24:195–213. doi: 10.1080/07420520701283693. [DOI] [PubMed] [Google Scholar]
- 45.Axelson H. The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 2004;204:171–178. doi: 10.1016/S0304-3835(03)00453-1. [DOI] [PubMed] [Google Scholar]
- 46.Bersten D.C., Sullivan A.E., Peet D.J., Whitelaw M.L. bHLH-PAS proteins in cancer. Nat. Rev. Cancer. 2013;13:827–841. doi: 10.1038/nrc3621. [DOI] [PubMed] [Google Scholar]
- 47.Sahreen S., Khan M.R., Khan R.A. Ameliorating effect of various fractions of Rumex hastatus roots against hepato- and testicular toxicity caused by CCl4. Oxid. Med. Cell. Longev. 2013;2013:325406. doi: 10.1155/2013/325406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J., Huang Q., Long X., Zhang J., Huang X., Aa J., Yang H., Chen Z., Xing J. CD147 reprograms fatty acid metabolism in hepatocellular carcinoma cells through Akt/mTOR/SREBP1c and P38/PPARα pathways. J. Hepatol. 2015;63:1378–1389. doi: 10.1016/j.jhep.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 49.Li J., Huang Q., Long X., Guo X., Sun X., Jin X., Li Z., Ren T., Yuan P., Huang X. Mitochondrial elongation-mediated glucose metabolism reprogramming is essential for tumour cell survival during energy stress. Oncogene. 2017;36:4901–4912. doi: 10.1038/onc.2017.98. [DOI] [PubMed] [Google Scholar]
- 50.Kao Y.H., Chen C.L., Jawan B., Chung Y.H., Sun C.K., Kuo S.M., Hu T.H., Lin Y.C., Chan H.H., Cheng K.H. Upregulation of hepatoma-derived growth factor is involved in murine hepatic fibrogenesis. J. Hepatol. 2010;52:96–105. doi: 10.1016/j.jhep.2009.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.