Abstract
Ethanol (EtOH)-induced alterations in intestinal homeostasis lead to multi-system pathologies, including liver injury. ω-6 PUFAs exert pro-inflammatory activity, while ω-3 PUFAs promote anti-inflammatory activity that is mediated, in part, through specialized pro-resolving mediators [e.g., resolvin D1 (RvD1)]. We tested the hypothesis that a decrease in the ω-6:ω-3 PUFA ratio would attenuate EtOH-mediated alterations in the gut-liver axis. ω-3 FA desaturase-1 (fat-1) mice, which endogenously increase ω-3 PUFA levels, were protected against EtOH-mediated downregulation of intestinal tight junction proteins in organoid cultures and in vivo. EtOH- and lipopolysaccharide-induced expression of INF-γ, Il-6, and Cxcl1 was attenuated in fat-1 and WT RvD1-treated mice. RNA-seq of ileum tissue revealed upregulation of several genes involved in cell proliferation, stem cell renewal, and antimicrobial defense (including Alpi and Leap2) in fat-1 versus WT mice fed EtOH. fat-1 mice were also resistant to EtOH-mediated downregulation of genes important for xenobiotic/bile acid detoxification. Further, gut microbiome and plasma metabolomics revealed several changes in fat-1 versus WT mice that may contribute to a reduced inflammatory response. Finally, these data correlated with a significant reduction in liver injury. Our study suggests that ω-3 PUFA enrichment or treatment with resolvins can attenuate the disruption in intestinal homeostasis caused by EtOH consumption and systemic inflammation with a concomitant reduction in liver injury.
Keywords: intestine, inflammation, omega-3 fatty acids, diet and dietary lipids, gut microbiome, bile acid metabolism, alcoholic liver disease, polyunsaturated fatty acid
The gastrointestinal tract is the tissue of first contact with ingested ethanol (EtOH), and excessive consumption can lead to numerous deleterious effects, including loss of gut barrier integrity, inflammation, and pathological changes in the gut microbiota composition and function (1). These intestinal alterations induced by EtOH as well as bidirectional communication between the gut and other organs, for example, the liver, play important roles in the pathogenesis of alcohol-induced multi-organ pathology, including alcoholic liver disease (ALD). A large body of both preclinical animal and human studies indicates that in the case of EtOH-mediated intestinal barrier dysfunction and increased permeability, the liver is exposed to a plethora of gut microbiota-derived products that contribute to hepatic inflammation and to the development of alcohol-related liver injury (2). The gut microbiota is also an integral part of the bile acid (BA) enterohepatic circulation, and EtOH-mediated changes in BA metabolism may be a mechanistic link between EtOH-induced alterations in the gut microbiota and impaired host phenotype, including intestinal and liver damage (3).
Previous work from our group and others underscores the importance of different types of dietary fat in EtOH-associated pathologies. For example, a diet high in ω-6 PUFAs exacerbated EtOH-induced intestinal permeability, endotoxemia, and liver injury in mice (4). We also showed that a diet high in ω-6 PUFAs combined with excessive alcohol intake resulted in intestinal inflammation (5) and gut microbiota alterations, specifically a prominent reduction in Bacteroidetes and an increase in the gram-negative Proteobacteria and the gram-positive Actinobacteria phyla (6). However, the effects of dietary ω-3 PUFAs on alcohol-mediated tissue and organ alterations are not clear and are somewhat controversial (7). Recent promising studies demonstrated that both dietary ω-3 PUFA supplementation and an endogenous increase in ω-3 PUFAs with a consequent decrease in the ω-6:ω-3 PUFA ratio in transgenic ω-3 FA desaturase-1 (fat-1) mice (due to endogenous conversion of ω-6 to ω-3 PUFAs by the fat-1 gene, which encodes an ω-3 PUFA desaturase) attenuated acute alcohol-induced liver injury (8, 9). It has also been shown that fat-1 mice have reduced metabolic endotoxemia and systemic low-grade inflammation in response to a diet high in ω-6 PUFAs (10) and are protected from high fat/high sucrose diet-induced intestinal permeability and alterations in intestinal tight junctions (11). Further, endogenous conversion of ω-6 to ω-3 PUFAs in fat-1 mice can modulate the gut microbiota (10), and transplantation of feces from fat-1 mice to WT littermates prevented obesity and associated metabolic disorders (12). Evidence suggests that ω-3 PUFA supplementation can alter gut microbial composition in humans (13) and may induce an increase in several beneficial taxa, including the butyrate-producing Roseburia, Bifidobacterium, and Lactobacillus (14). Studies showing reduced intestinal inflammation by ω-3 PUFA supplementation, both in patients and in animal models, suggest anti-inflammatory and pro-resolving properties of ω-3 PUFAs and their derivatives in the gut (15). Supplementation with ω-3 PUFAs resulted in significant improvement of metabolic risk factors and liver steatosis in patients with nonalcoholic liver disease (16), and substitution of the ω-6 PUFA, linoleic acid, with ω-3 PUFAs prevented Western diet-induced nonalcoholic steatohepatitis (17). Many of the benefits of increased ω-3 PUFAs can be attributed to the class of bioactive metabolites collectively termed specialized pro-resolving mediators (SPMs), which includes resolvins, protectins, and maresins, that have been shown to limit the inflammatory response in a number of models of disease [reviewed in (18)].
Despite promising studies suggesting beneficial effects of ω-3 PUFAs and their metabolites in a variety of pathological conditions (19–21), there is still limited evidence for their beneficial effects on alcohol-mediated tissue/organ damage. Given that the Western diet is heavily reliant on ω-6 PUFA-rich sources (e.g., corn oil and soybean oil) at the expense of ω-3 PUFA-rich food (e.g., oily fish), leading to an increase in the ω-6:ω-3 PUFA ratio, which contributes to numerous deleterious health effects (22), it is essential to investigate the role of modulation/decrease of the ω-6:ω-3 PUFA ratio as a potential dietary preventative/therapeutic approach in different pathological conditions, including alcohol-induced multi-organ pathology. Further compounding the problem of excessive ω-6 PUFA intake is the observation that the rise in obesity rates mirrors that of soybean oil consumption (23). Obesity has been linked to exacerbation of ALD (24). The objective of the current study was to investigate the impact of a decreased ω-6:ω-3 PUFA ratio due to endogenous ω-3 PUFA enrichment on EtOH-induced alterations in intestinal homeostasis, gut microbiota, BA metabolism, and associated liver injury in transgenic fat-1 mice subjected to chronic EtOH administration.
MATERIALS AND METHODS
Animals and experimental setup
Animal studies were approved by and performed in accordance with the guidelines of the University of Louisville Institutional Animal Care and Use Committee (protocol number 15423 to I.A.K.). Transgenic fat-1 mice were maintained as heterozygotes. Expression of the the fat-1 gene is driven by the chicken β-actin promotor and the cytomegalovirus enhancer (25). To produce mice for the experiments described herein, heterozygous fat-1 male mice were mated to female WT C57BL/6J mice that were bred in-house (supplemental Fig. S1A). Age and sex-matched fat-1+/− and WT littermates were used for all experiments. For simplicity, fat-1 will be used throughout to indicate heterozygous mice. Mice were genotyped by PCR (supplemental Fig. S1B) and primers to Gdf5 were used as a positive control (see supplemental Table S1 for sequences). Mice were housed in a temperature-controlled room (23.9°C) with a 12 h light-dark cycle in a specific pathogen-free animal facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
All mice were maintained on autoclaved laboratory rodent chow diet (5010; LabDiet, St. Louis, MO) and had ad libitum access to food and water. At 8–10 weeks of age, male fat-1 mice and their WT littermates were placed on control (maltose dextrin) or EtOH-containing Lieber-DeCarli liquid diets (BioServ, Flemington, NJ; catalog numbers F1259SP and F1258SP, respectively). The mice were fed for 6 weeks with a step-wise increase in EtOH concentration (0, 1, and 2% for 2 days each, 4% and 5% for 1 week each, and then 6% for 3 weeks). Lipopolysaccharide [LPS; LPS-EB Ultrapure from InvivoGen, San Diego, CA (p/n tlrl-3pelps)] was given to a subgroup of EtOH-treated mice 24 h prior to euthanasia (5 mg/kg ip) to mimic human alcoholic hepatitis in which systemic inflammation and infection are major contributors to morbidity and mortality (26). Additionally, a subset of EtOH+LPS-treated mice received resolvin D1 (RvD1; Cayman Chemical, Ann Arbor, MI) treatment (500 ng ip on each of the final 5 days of the experiment). The experimental design is presented in supplemental Fig. S1C. The control and EtOH-containing diets were prepared fresh each day, and food consumption was monitored daily. There was no significant difference in food intake between WT and fat-1 mice. The control pair-fed (PF) groups received the same amount of isocaloric food (maltose dextrin-containing diets) that EtOH-fed animals consumed in the previous day.
Intestinal tissue PUFA analysis
FA profiles in intestinal samples were analyzed by gas chromatography in Dr. J. Kang’s laboratory as described previously (27). Briefly, tissue samples were ground to powder under liquid nitrogen and subjected to total lipid extraction and FA methylation by 14% boron trifluoride-methanol reagent (Sigma-Aldrich, St. Louis, MO) at 100°C for 1 h. FA methyl esters were analyzed using a fully automated HP5890 gas chromatography system equipped with a flame-ionization detector (Agilent Technologies, Palo Alto, CA). The FA peaks were identified by comparing their relative retention times with the commercial mixed standards (NuChek Prep, Elysian, MN), and area percentages for all resolved peaks were analyzed by using a PerkinElmer M1 integrator.
Histopathological and immunohistochemical analysis of the intestinal tissue
Formalin-fixed paraffin-embedded samples of ileum were sectioned, stained with H&E, and evaluated by light microscopy. The ileum was chosen for analysis because it is the region that has the highest degree of gut barrier permeability in response to alcohol feeding (4). For morphometric analysis, complete villus-crypt structures were randomly selected from H&E-stained sections, and villus height, width, and crypt depth were measured using ImageJ software (National Institutes of Health). Villus length was measured from the tip to the base of the villus, villus width was measured at the base of the villus, and the crypt length was measured from the bottom of the crypt to the opening of the crypt (28).
For immunohistological analyses, intestinal sections were stained for the tight junction protein, ZO1 (Invitrogen, Camarillo, CA; catalog number 61-7300), Paneth cells (anti-lysozyme; Abcam, Cambridge, MA; ab36362), and goblet cells (Alcian Blue 8GX; Sigma-Aldrich). Neutrophil accumulation was assayed by chloroacetate esterase (CAE) staining using a commercially available kit (Sigma-Aldrich). Quantification of CAE staining was performed by counting CAE-positive cells in a random series of 20–30 digital images per section (200× field) in a blinded approach; the number of CAE-positive cells were then summed and averaged to obtain an estimate for each mouse (n = 3–8 animals per group).
Isolation of intestinal crypts and crypt-villus organoid culture
Isolation of intestinal crypts and subsequent organoid culture were largely performed as previously described (29, 30) using commercially available reagents (StemCell Technologies, Vancouver, Canada). In brief, mice were humanely euthanized by CO2 asphyxiation and the entire length of the small intestine was removed and the lumen flushed thoroughly with cold PBS. The intestine was then opened longitudinally and the mucosal layer was collected by gently scraping the luminal aspect with a glass microscope slide. The mucosa was washed and processed according to the manufacturer’s instructions (StemCell Technologies). Isolated crypts (200–400 per well) were suspended in Matrigel (BD Biosciences, San Jose, CA) and plated in 24-well plates prewarmed to 37°C. The crypts were grown with crypt niche growth factors in IntestiCult growth media (StemCell Technologies). The organoid cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
For experiments evaluating organoid growth, crypts were isolated from the small intestines of naïve WT and fat-1 mice, and the resulting organoids were cultured in triplicate and treated with 50 mM of EtOH. Growth was analyzed by measuring the cross-sectional area of representative organoids every 24 h (>10 in each condition). In a second series of experiments, intestinal organoids were established from the experimental WT and fat-1 mice fed control or EtOH-containing diets. Total RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA), and cDNA was synthesized and amplified using the Nugen Ovation PicoSL WTA System V2 (San Carlos, CA). Changes in gene expression were analyzed by quantitative (q)RT-PCR using PerfeCTa SYBR Green Fast Mix (Quanta Biosciences, Beverly, MA) on the Applied Biosystems 7900HT platform (Foster City, CA). Primers are presented in supplemental Table S1.
Cytokine measurement in intestinal tissue
Ileum samples were homogenized in PBS + 0.05% Tween-20 supplemented with protease and phosphatase inhibitors (Halt™; Thermo Fisher, Waltham, MA), in tubes filled with 1.5 mm zirconium beads (Benchmark Scientific, Edison, NJ). Following centrifugation at 20,000 g, supernatants were collected and protein concentrations were determined by the BCA method (Pierce™ BCA protein assay kit, Thermo Fisher). Six hundred micrograms were analyzed on the V-PLEX immunoassay pro-inflammatory panel 1 plate and data were collected on the MESO Sector S 600 instrument and analyzed with Discovery Workbench, v. 4.0 (MesoScale Discovery, Rockville, MD).
RNA isolation and real-time qRT-PCR
Total RNA from intestinal and liver tissues was isolated with Trizol reagent (Thermo Fisher) as described by the manufacturer, and any contaminating genomic DNA was removed by digestion with DNase I (Thermo Fisher). cDNA was synthesized with qScript cDNA Supermix (Quanta Biosciences, Beverly, MA) and a cDNA equivalent of 10 ng RNA (or 0.1 ng for 18S rRNA) was analyzed in each quantitative (q)PCR reaction. qRT-PCR assays were performed with PerfeCTa SYBR Green Fast Mix (Quanta Biosciences) on the Applied Biosystems 7900HT platform (Foster City, CA). Primers are presented in supplemental Table S1.
RNA-seq analysis and bioinformatics
RNA-seq analysis was performed in the ileal segments of the intestine by the University of Louisville Center for Genetics in Molecular Medicine Core facility, and bioinformatic analysis was conducted by the National Institutes of Health-funded Kentucky Biomedical Research Infrastructure Network Bioinformatics Core according to the protocols presented in the supplemental Materials and Methods.
Gut microbiota analysis
DNA extraction of fecal samples, bacterial 16S rRNA gene amplification, and sequence analysis was performed as previously described (31, 32) with modifications shown in the supplemental Materials and Methods.
Fecal BA analysis
BA analysis from fecal samples was performed by the Wayne State University Lipidomic Core facility (supported by National Institutes of Health Grant S10RR027926) according to the protocol presented in the supplemental Materials and Methods.
Serum BA analysis
Total serum BA concentrations were determined using the Mouse Total Bile Acids Assay Kit from Crystal Chem (Elk Grove Village, IL).
Blood plasma metabolomic analysis
Blood metabolomic analysis was performed by the University of Louisville Metabolomic Core Facility according to the protocol presented in the supplemental Materials and Methods.
Measurement of blood alcohol levels
Plasma blood alcohol levels were measured using the EnzyChrom™ EtOH assay kit (BioAssay Systems, Hayward, CA).
Assessment of liver injury
Plasma levels of alanine aminotransferase (ALT; a marker of liver injury) were determined using the ALT/GPT Reagent (Thermo Fisher) according to the manufacturer’s instructions and using normal and abnormal serum for controls (Data Trol™; Thermo Fisher). Hepatic triglycerides (TGs) were measured as previously described (33). For histological evaluation of liver injury, 5 μm sections were prepared from formalin-fixed paraffin-embedded liver tissue and were stained with H&E.
Statistical analysis
The data were expressed as mean value ± SEM. Differences among multiple groups were analyzed by two-way ANOVA followed by post hoc multiple comparisons tests. Differences between two groups were tested by the unpaired two-tailed Student’s t-test. A P-value of <0.05 was considered statistically significant. The correlation between various characteristics was assessed using Pearson correlation analysis. Statistical analysis was performed using GraphPad Prism 5.01 software for Windows (GraphPad Software, Inc., La Jolla, CA).
Additional protocols and procedures are described in supplemental Materials and Methods.
RESULTS
Intestinal ω-6:ω-3 PUFA ratio and EtOH-mediated alterations in the FA composition in WT and fat-1 mice
Analysis of intestinal FA composition revealed that fat-1 mice compared with WT littermates showed a markedly decreased ω-6:ω-3 PUFA ratio in the ileal segments with a ratio of ∼8:1 and 7:1 in fat-1 PF and EtOH-fed, respectively, and ∼14:1 and 20:1 in WT PF and EtOH-fed, respectively (Fig. 1A). Interestingly, while total ω-3 PUFAs were elevated in fat-1 as compared with WT in both PF and EtOH-fed animals (Fig. 1B), ω-6 PUFA levels were similar between genotypes in PF animals. However, EtOH consumption markedly increased total ω-6 PUFAs in WT mice, resulting in significantly higher ω-6 PUFA levels in WT-EtOH versus WT-PF and fat-1-EtOH animals (Fig. 1C). Further analysis revealed elevated levels of linoleic acid (an ω-6 PUFA) in WT-EtOH-fed mice, while EPA (an ω-3 PUFA) was below detectible levels in these mice (supplemental Table S2). There were no noticeable effects of EtOH on the ω-3 PUFA DHA in either WT or fat-1 mice, although fat-1 mice had significantly higher DHA levels compared with WT-PF littermates.
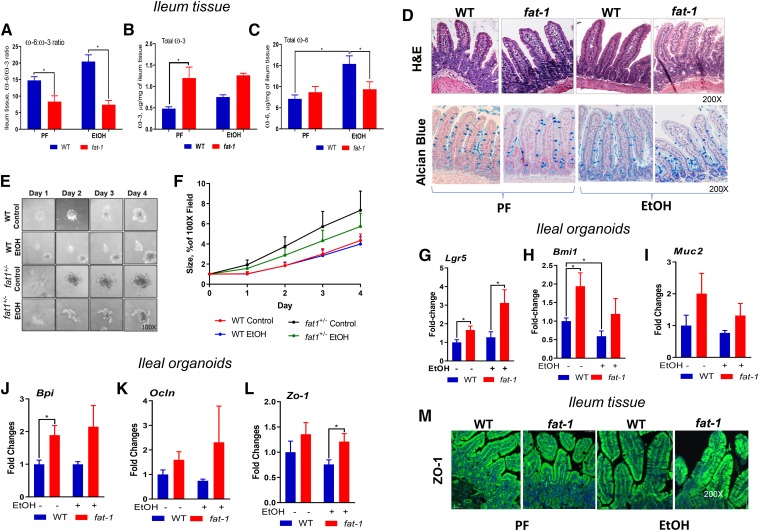
Fig. 1.
Effects of EtOH and endogenous conversion of tissue ω-6 to ω-3 PUFAs on intestinal morphology and homeostasis. A–C: Ileum tissue ω-6:ω-3 PUFA ratio and total ω-6 and ω-3 PUFAs (n = 3–5). The full panel of FAs is presented in supplemental Table S2. D: Representative images of stained intestinal sections with H&E or Alcian blue, 200× magnification. E, F: Effect of EtOH on small intestine-derived organoids from naïve WT and fat-1 mice cultured for 4 days, 100× magnification (n = 5). Expression of growth and proliferation markers (G–H), mucus production (I), bacteriocidal activity (J), and tight junction genes (K–L) in intestinal organoids from WT or fat-1 mice fed control or EtOH diets cultured for 10 days (n = 3). *P < 0.05. M: immunohistochemistry/confocal imaging of ZO-1 expression (200× magnification).
Effects of a decreased ω-6:ω-3 PUFA ratio on intestinal morphology and homeostasis
Histological evaluation of intestinal sections indicated that varying the content of essential FAs in WT and fat-1 mice had no effects on the overall structure of the ileum tissue morphology in PF mice (Fig. 1D). EtOH feeding resulted in increased villus length in WT mice compared with WT-PF and fat1-EtOH littermates, while there were no differences in crypt height between the experimental groups (supplemental Fig. S2A, B).
Using small intestine organoids, we determined whether a decrease in tissue ω-6:ω-3 PUFA ratio would benefit intestinal stem cell (ISC) growth, proliferation, and function. We observed that crypts derived from naïve fat-1 mice yielded substantially larger organoids and increased early organoid growth within 4 days in culture compared with crypts obtained from naïve WT littermates. EtOH treatment did not significantly affect the formation and growth of organoids derived from either fat-1 or WT mice (Fig. 1E, F). Although, it has been previously shown that EtOH can inhibit the growth and budding of mouse ileal organoids after 5 days of culture (29), we did not observe this effect with longer time course (after 7 days in culture, data not shown) possibly due to differences in the establishment of the cultures or growth medium. In organoids derived from the ileum of mice fed either control or EtOH-containing diets, we observed a significant increase in the expression of the ISC markers, Lgr5 and Bmi1, in fat-1 compared with WT mice, both in EtOH- and control diet-fed animals (Fig. 1G, H). Expression of Muc2, a goblet cell and mucus production marker, was elevated in fat-1 mice, both in EtOH-fed and PF mice, compared with WT littermates (although this increase was not significant, Fig. 1I), which was consistent with a similar goblet cell distribution in the ileal tissue of the WT and fat-1 animals (supplemental Fig. S2C). Compared with WT, intestinal organoids derived from fat-1 experimental mice had upregulated bactericidal permeability-increasing protein (Bpi1), an antibacterial and endotoxin-neutralizing molecule (Fig. 1J). Expression of intestinal tight junction proteins, Ocln and Zo1, was similar in fat-1 and WT PF mice, while significantly higher in fat-1-EtOH compared with WT-EtOH littermates (Fig. 1K, L), which was consistent with higher ZO1 protein levels in the ileal tissue of fat-1-EtOH versus WT-EtOH experimental animals (Fig. 1M). However, there were no appreciable EtOH-induced changes in LPS and CD14 (circulating markers of intestinal permeability) in either PF or EtOH-fed fat-1 and WT littermates (supplemental Fig. S3A, B).
The impact of the differential ω-6:ω-3 PUFA ratio on the intestinal mucosa transcriptome responses to EtOH challenge
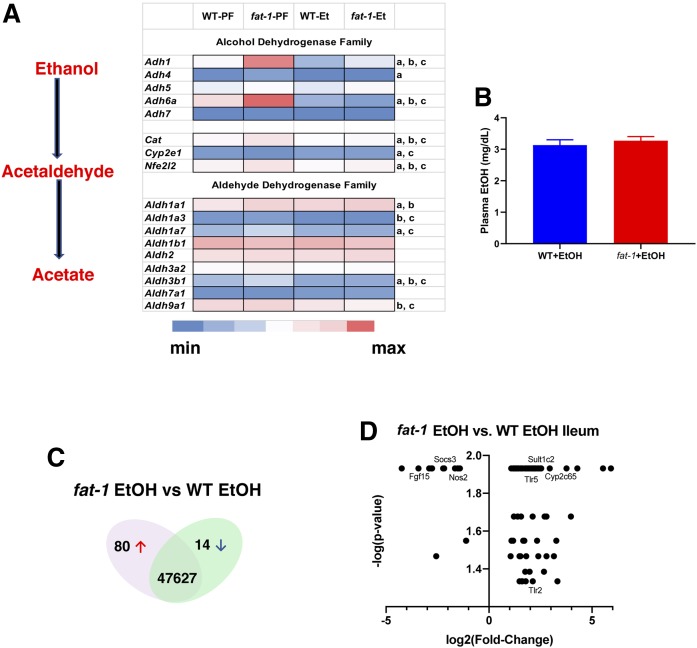
To gain deeper insight into the impact of the ω-6:ω-3 PUFA ratio on intestinal mucosa transcriptome responses to EtOH challenge, we performed an RNA-seq analysis of intestinal mucosal epithelial tissue (ileal segments) isolated from WT and fat-1 mice (n = 3–5). Although the liver is the primary tissue responsible for EtOH metabolism, the gut is also an important, albeit quantitatively lower, first-line location for EtOH detoxification (34). There are several metabolic pathways/enzymes involved in EtOH metabolism, including alcohol dehydrogenases (Adhs), a family of dehydrogenases/reductases responsible for oxidation of EtOH to acetaldehyde. Adh1 (expressed both in the liver and the intestine) and Adh6 [almost exclusively expressed in the small intestine (35)] were the most abundantly expressed genes in both WT and fat-1 PF control animals with significantly higher levels in fat-1 mice (Fig. 2A). EtOH administration resulted in downregulation of these genes to similar levels in both genotypes. The enzymes of the aldehyde dehydrogenase (Aldh) family further catalyze the oxidation and detoxification of aldehydes, including acetaldehyde, to carboxylic acids. The most abundantly expressed Aldhs in the intestines were Aldh1b1, Aldh9a, Aldh2, and Aldh1a1. Several Aldhs were significantly higher in fat-1-PF compared with WT-PF mice, including Aldh1a1, I Aldh1a7, and Aldh3b1. Interestingly, expression of Aldh1a1 was upregulated while Aldh9a was downregulated by EtOH administration in both WT and fat-1 mice. The levels of Aldh1b1 and Aldh2 (the major enzyme for the acetaldehyde oxidation) were similar in all experimental groups. Although we did not evaluate the EtOH-metabolizing system in the liver, it is most likely that there were no significant differences in expression of these genes between WT and fat-1 EtOH-fed animals, as blood alcohol levels were similar in WT and fat-1 mice on the EtOH-supplemented diet (Fig. 2B).
Fig. 2.
Differential gene expression in WT versus fat-1 mice. A: Effects of modulation of the ω-6:ω-3 PUFA ratio on EtOH-metabolizing enzymes in the intestine. Data are presented as gene expression levels of the corresponding group. Lowercase letters indicate statistically significant changes between groups (a: fat-1-PF vs. WT-PF; b: WT-PF vs. WT-EtOH; c: fat-1-PF vs. fat-1-EtOH; P < 0.05). B: Plasma EtOH levels (n = 6–10). Venn diagram (C) and graphical representation (D) of total number of genes differentially expressed in the EtOH groups.
Further analysis revealed that there were a total of 94 genes that were differentially regulated between fat-1 and WT EtOH-fed mice (80 upregulated and 14 downregulated, Fig. 2C, D). The complete list of differentially expressed genes is presented in supplemental Table S3 and organized by biological function as determined from published literature. Twenty-five of the genes were involved in immune function [e.g., Toll-like receptor (Tlr)2, Tlr5, and Nos2], while 14 are important for lipid metabolism or transport (e.g., Abca12, Acot2, and Mgll). There were a number of genes differentially expressed in fat-1 versus WT mice that are involved in cell proliferation and ISC renewal, and are potentially important for tissue repair (e.g., Adra2a, Pla2g10, and Plet1). Interestingly, the expression of tryptophan hydroxylase-1 (Tph-1; which synthesizes serotonin) was significantly increased as well as several other novel genes. For example, the prion protein gene (Prnp), the E3 ubiquitin protein ligase and ulcerative colitis (UC)-linked gene (Rnf186), and cystathionine β synthase (Cbs) were all increased in fat-1 versus WT mice.
Inflammation and antimicrobial response to LPS challenge in mice chronically fed EtOH
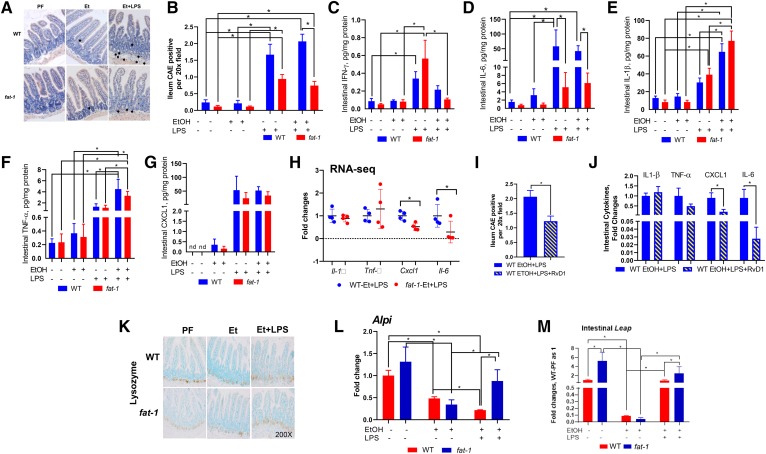
To assess the role of increased ω-3 PUFAs and a concomitant decrease in the ω-6:ω-3 PUFA ratio in intestinal inflammation, we investigated the intestinal mucosa pro-inflammatory responses to LPS challenge in a setting of chronic EtOH pre-exposure. Intestinal inflammation was significantly lower in fat-1 compared with WT mice in response to EtOH+LPS treatment, and was characterized by reduced numbers of neutrophils and mononuclear cells (Fig. 3A, B). This was also accompanied by markedly decreased levels of the pro-inflammatory cytokines, INF-γ (Fig. 3C) and IL-6, in EtOH+LPS fat-1 versus WT mice (Fig. 3D). IL-1β and TNFα levels were similar in both fat-1 and WT EtOH+LPS-treated mice (Fig. 3E, F). Although CXCL1 protein levels were equally induced in fat-1 and WT mice, Cxcl1 gene expression was significantly downregulated in EtOH+LPS fat-1 mice compared WT littermates (Fig. 3G, H). Gene expression levels of Il-1β, Tnf-α, and Il-6 were consistent with their protein levels (Fig. 3H). Given that ω-3 PUFAs are precursors to SPMs that actively promote the resolution of inflammation, we tested the effect of the DHA-derived resolvin, RvD1, on intestinal inflammation caused by EtOH+LPS challenge. RvD1 treatment significantly decreased intestinal neutrophil infiltration induced by EtOH+LPS (Fig. 3I) in parallel with decreased IL-6 and CXCL1 protein levels, while TNFα levels were moderately reduced, but IL-1β levels were unchanged (Fig. 3J).
Fig. 3.
Increased ω-3 PUFAs reduced LPS-mediated intestinal inflammation in mice chronically fed EtOH. A, B: CAE-stained intestine sections and quantification of CAE-positive cells (n = 6–18 mice). C–G: Ileal pro-inflammatory cytokine expression determined by immunoassay: INF-γ (C), IL-6 (D), Il-1β (E), TNF-α (F), and CXCL1 (G). H: Ileal mRNA levels of pro-inflammatory genes (n = 3–11 mice). I: Quantification of CAE-positive cells in the EtOH+LPS group treated with RvD1 (n = 12–14). J: Effects of RvD1 treatment on EtOH+LPS-induced pro-inflammatory cytokines (n = 9–10). K: Immunohistochemistry for lysozyme expression in the intestine. L, M: Ileal expression of antimicrobial genes (n = 3–5).
In order to maintain intestinal homeostasis and promote host defense against pathogens, different epithelial cells (mainly Paneth cells, a highly specialized population of intestinal cells located at the crypt base) produce a diverse array of antimicrobial peptides (AMPs). In our experiment, the number of Paneth cells was not observably different between the experimental groups; however, LPS administration resulted in redistribution of lysozyme staining (a marker of Paneth cells) in both WT and fat-1 EtOH+LPS-treated mice (Fig. 3K). Accordingly, the expression of genes encoding lysozymes, such as Lyz1 and Lyz2, was not altered by any of the experimental conditions. There were also no significant differences between WT and fat-1 mice in the expression of other AMPs, including defensin and C-type lectin families, in response to EtOH or EtOH+LPS treatments (supplemental Fig. S4). In line with a previously reported study (10), expression of intestinal alkaline phosphatase (Alpi), a glycosylphosphatidylinositol-linked protein expressed primarily in mucosal epithelia (36) and an important factor in inflammatory resolution in the mucosa (37), was significantly upregulated in fat-1-PF compared with WT-PF littermates (Fig. 3L). EtOH treatment resulted in Alpi downregulation in both genotypes. Notably, LPS administration elevated Alpi expression in fat-1 but not WT mice, resulting in significantly higher Alpi levels in fat-1 as compared with WT EtOH+LPS-treated littermates. The expression of another AMP, liver-enriched antimicrobial peptide 2 (Leap2), followed a similar trend as that for Alpi (Fig. 3M).
Compositional and structural shift of gut microbiota in WT and fat-1 mice caused by chronic EtOH feeding and LPS challenge
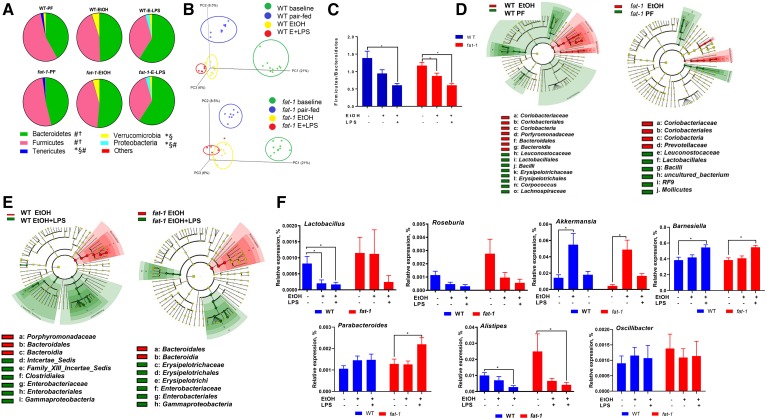
Because dysregulation of intestinal homeostasis and susceptibility to inflammation are often associated with alterations in the gut microbiome, we investigated how bacterial communities were impacted by chronic EtOH administration and evaluated the effects of tissue modulation of the ω-6:ω-3 PUFA ratios on these alterations. At the phylum level, Firmicutes and Bacteroidetes accounted for the major proportion of the bacterial population in all groups regardless of genotype or treatment (Fig. 4A). Six weeks of either control or EtOH-containing diet consumption resulted in significant changes in the gut microbiota in both genotypes. Importantly, the gut microbial communities of EtOH-fed mice clustered separately from naïve (baseline control) or PF mice (experimental control), indicating a unique impact of EtOH on the gut microbiome (Fig. 4B). EtOH treatment resulted in a shift in the Firmicutes:Bacteroidetes ratio (due to increased Bacteroidetes and decreased Firmicutes) in both WT and fat-1 mice compared with control PF littermates (Fig. 4C). WT and fat-1 EtOH-fed mice were differentially enriched with Verruomicrobiaceae, Clostridiaceae, Coriobacteriaceae, Peptostreptococcaceae, Prevotellaceae, and several operational taxonomic units (OTUs) in the Bacteroidales order, whereas Lachnospiraceae, Leuconastocaceae, and Pseudomonadales were depleted in response to EtOH challenge compared with control diet feeding in both genotypes (Fig. 4D, supplemental Fig. S5A, B). Lactobacillaceae and Erysipelotrichaceae were depleted in EtOH-fed mice compared with PF mice in WT but not fat-1 animals. Further analysis revealed several Lachnospiraceae, Ruminococcaceae (short-chain FA producers), and Bacteroidales OTUs that were differentially enriched in EtOH-fed fat-1 mice, although microbiome diversity and richness were comparable between these two groups (data not shown). LPS challenge in EtOH-fed mice induced a modest shift in gut microbial community structure in both WT and fat-1 mice without significant changes in microbial diversity and richness. LEfSe analysis revealed a significant expansion of Enterobacteriaceae and Erysipelotrichaceae in response to LPS challenge in WT mice, whereas several Ruminococcaceae and Bacteroidales OTUs were depleted as a result of LPS challenge in both WT and fat-1 animals (Fig. 4E). Importantly, Porphyromonadaceae:Barnesiella and Lachnospiraceae were differentially enriched (increased) in EtOH-fed fat-1 mice in response to LPS challenge, but not in EtOH-fed WT mice (supplemental Fig. S5C, D).
Fig. 4.
Effects of endogenous ω-6:ω-3 PUFA modulation on EtOH-associated gut microbiota alterations. A: Pie charts of taxonomic composition of bacterial community at the phylum levels in stool samples from WT and fat-1 experimental mice. Statistically significant changes (P < 0.05) between groups are shown: *WT-PF versus WT-EtOH; §fat1-PF versus fat-1-EtOH; #WT-EtOH versus WT-EtOH-LPS; †fat-1-EtOH versus fat-1-EtOH-LPS. B: Principal component analysis of gut microbiota composition based on unweighted UniFrac. P = 0.001, PERMANOVA analysis. C: Firmicutes:Bacteroidetes ratio in experimental groups. D, E: Cladograms showing the taxa most differentially associated with EtOH or post-EtOH LPS treatment in WT and fat-1 mice. Circle sizes in the cladogram plot are proportional to bacterial abundance. The circles represent phyla, genus, class, order, and family going from the inner to the outer circle. F: Selected bacteria at the genus levels in WT and fat-1 experimental mice. For each analysis, n = 5–10 mice per group.
We further analyzed selected bacterial taxa with roles known to be important in EtOH-induced multi-organ pathology. Specifically, Lactobacillus, a beneficial taxa for ALD (38), was significantly reduced in WT mice in response to EtOH alone and in response to EtOH+LPS, while levels of this bacteria were not affected by EtOH in fat-1 mice (Fig. 4F). Considering that metabolites of gut microbiota, such as short-chain FAs, are the primary energy source for intestinal epithelial cells, we analyzed Roseburia, a butyrate-producing bacteria, which was enriched (although not significantly) in fat-1-PF compared with WT-PF animals, and similarly decreased in both genotypes in EtOH- and EtOH+LPS-treated animals. In line with a previous report (39), EtOH feeding resulted in increased abundance of Akkermansia, and this effect was independent of the genotype. The abundance of Parabacteroides was increased by EtOH+LPS in fat-1 mice. Other genera, such as Barnesiella, were significantly increased, while Alistipes were decreased by EtOH+LPS challenge in both genotypes. The abundance of the Oscillibacter genus was not affected in any experimental condition.
Impact of ω-6:ω-3 ratio modulation on the plasma metabolome following EtOH administration and LPS challenge
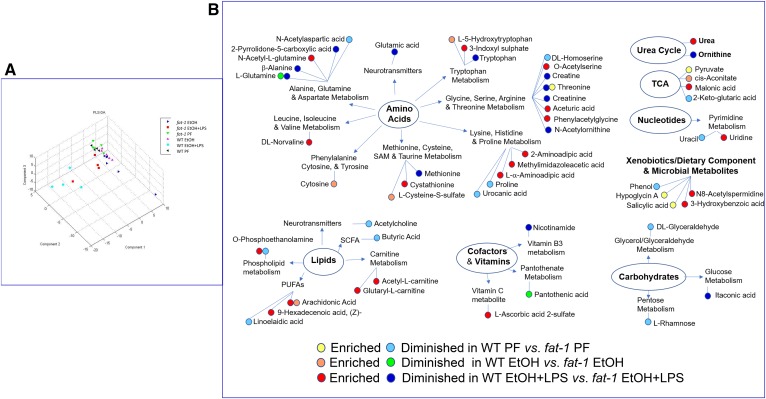
The intestinal microbiota communicates with the host via numerous microbial metabolites (40). The blood metabolome, a net result of metabolic activity of the gut microbiota and metabolic changes in several tissues, communicates signals between multiple organs, including the gut and the liver. Therefore, we conducted a plasma metabolomic analysis to characterize metabolic phenotypes and identify key differences between WT and fat-1 experimental mice. Principal component analysis demonstrated that WT and fat-1 mice differed in overall plasma metabolite composition in PF, EtOH-fed, and EtOH-fed+LPS-challenged groups (Fig. 5A). These changes are schematically presented in Fig. 5B. Overall, 18 plasma metabolites had significant differences between WT and fat-1 PF animals, including butyric acid, which was significantly lower in WT; however, this difference was lost in EtOH-fed mice. Chronic EtOH feeding resulted in minimal differences between WT and fat-1 mice (there were seven differentially expressed metabolites), while LPS administration produced the majority of observed metabolomic differences (37 differentially expressed metabolites). Statistically significant changes belonged to multiple metabolic pathways, including amino acid, carbohydrate, lipid, xenobiotic, and microbial metabolism among others. Specifically, EtOH-treated WT mice compared with fat-1 littermates had higher levels of cytosine, cysteine-S-sulfate, l-5-hydroxy-tryptophan, arachidonic acid (AA), and cis-aconitate, while exhibiting lower levels of glutamine and pantothenic acid. Compared with EtOH+LPS-treated fat-1 mice, WT counterparts revealed increased levels of several lipid compounds such as AA, 9-hexadecenoic acid (a trans-isomer of palmitoleic acid), and a phospholipid O-phosphoethanolamine. Molecules involved in carnitine metabolism, such as acetyl-l-carnitine and glutaryl-l-carnitine, were elevated in WT-EtOH+LPS-treated as compared with fat-1-EtOH+LPS-treated mice. The most prominent alterations between EtOH+LPS-treated fat-1 and WT mice were observed in protein degradation and amino acid metabolism. Compared with fat-1 mice, WT animals treated with EtOH+LPS revealed reduced levels of tryptophan, creatine, threonine, methionine, and glutamic acids. Several acetylated amino acids and dipeptides, including O-acetylserine, phenylacetylglycine, n-acetyl-l-glutamine, and 2-aminoadipic acid, were increased in WT-EtOH+LPS versus fat-1-EtOH+LPS mice, while the levels of n-acetylornithine were reduced. WT-EtOH+LPS mice also exhibited increased levels of urea and uridine, as well as several molecules belonging to dietary components and microbial metabolites, such as N8-acetylspermidine and 3-hydroxybenzoic acid.
Fig. 5.
Impact of ω-6:ω-3 PUFA ratio modulation on plasma metabolome alterations associated with EtOH administration and LPS challenge. A: Partial least squares-discriminant analysis. B: Plasma metabolites significantly differentially enriched in experimental groups.
Alterations in fecal BAs and related metabolic pathways in WT and fat-1 mice
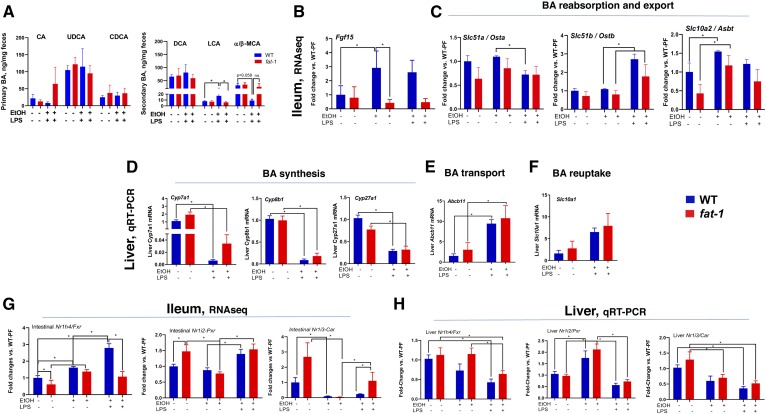
One of the essential functions provided by the gut microbiota is modulation of gastrointestinal metabolites, including their synthesis, digestion, fermentation, and secondary metabolism. Among these metabolites are BAs. EtOH-induced alterations in BA metabolism has been found in humans and in multiple animal models (41–44). We next examined the impact of decreased ω-6:ω-3 PUFA ratio on selected BAs under the EtOH+LPS experimental condition. There were no significant differences in fecal levels of primary BAs [cholic acid, ursodeoxycholic acid (UDCA), or chenodeoxycholic acid (CDCA)] or in the levels of secondary BAs (deoxycholic acid) in either genotype in PF mice or in response to EtOH+LPS treatment. Notably, the secondary BA, lithocholic acid (LCA), a product of microbial metabolism of UDCA and CDCA via 7-dehydroxylation (45, 46), was the only BA markedly elevated by EtOH+LPS in WT compared with WT-PF as well as in fat-1-EtOH+LPS-treated mice (Fig. 6A). Measurement of total BA concentrations in serum revealed a significant increase in both WT and fat-1 EtOH+LPS-treated mice with no differences between genotypes (supplemental Fig. S6).
Fig. 6.
Effects of modulation ω-6:ω-3 PUFA ratio on EtOH- and LPS-mediated alterations in fecal BA profiles and related metabolic pathways. A: Fecal primary and secondary BA concentrations (n = 3–7). Expression of genes related to BA detoxification in the ileum (RNA-seq) (B) and in the liver (C) as determined by qRT-PCR in WT and fat-1 mice. Expression of genes involved in BA synthesis (D), transport (E), and reuptake (F) in the liver. G, H: Expression of Nr1h4/Fxr, Nr1i2/Pxr, and Nr1i3/Car in the ileum and liver, respectively (n = 3–5). CA, cholic acid, DCA, deoxycholic acid.
Given the importance of BA enterohepatic circulation, we measured the relative expression of genes involved in BA metabolism in both the intestine and in the liver. In the intestine, expression of Fgf15 (FGF19 in humans), which is involved in a negative feedback loop of BA synthesis in the liver, was upregulated by EtOH and EtOH-LPS in WT but not in fat-1 mice (Fig. 6B). Intestinal expression of genes of BA reabsorption and export (Osta, Ostb, and Asbt) were similar between genotypes and not affected in any treatment group (Fig. 6C). In the liver, genes associated with the major pathways of BA synthesis (Cyp7a1, Cyp27a1, and Cyp8b) were similarly downregulated in response to EtOH+LPS treatment in both WT and fat-1 mice (Fig. 6D), while markers of BA transport (Abcb11) and reuptake (Slc10a1) were equally overexpressed as compared with control animals (Fig. 6E, F). Several nuclear receptors, including Fxr, pregnane X receptor (Pxr), and constitutive androstane receptor (Car), are all involved in the regulation of BA synthesis and in protection against the cytotoxic effects of BAs by increasing the expression of binding proteins, transporters, and enzymes that detoxify BAs (47). Expression of Fxr, which represses BA synthesis in the liver and activates Fgf15 in the intestine, was upregulated by EtOH in the intestine but not in the liver in either genotype. EtOH+LPS treatment further increased intestinal Fxr in WT but not in fat-1 mice (Fig. 6G, H). Interestingly Pxr, which is an LCA receptor, was differentially modulated by LPS treatment in the intestine (upregulated) and the liver (downregulated) after EtOH treatment. Compared with PF control animals, expression of Car was downregulated in EtOH- and EtOH+LPS-treated groups in both WT and fat-1 genotypes. Notably, expression of Car was significantly higher in the intestines of fat-1 mice compared with EtOH+LPS-treated WT mice.
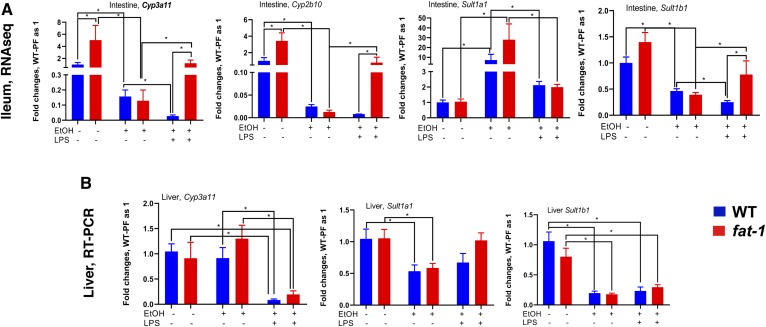
Hydroxylation and sulfation, key pathways for the conversion of BAs to more hydrophilic and less toxic compounds, are important mechanisms to prevent BA-induced tissue/organ damage. Hydroxylation occurs predominantly via Cyp3a11 (CYP3A4 in humans) both in the intestine and in the liver (48, 49), and sulfation via specific sulfotransferases, predominately in the liver. We measured the relative expression of major genes involved in these pathways in the intestine (Fig. 7A) as well as in the liver (Fig. 7B). Expression of Cyp3a11, Cyp2b10, and Sult1b1 was significantly higher in the intestines but not in the livers of fat-1 PF mice compared with WT PF littermates. EtOH feeding significantly decreased intestinal but not liver Cyp3a11 levels in both genotypes. Intestinal and liver Sult1b1 levels were similarly significantly reduced by EtOH in fat-1 and WT animals, while Sult1a1 expression was significantly increased in the intestines and decreased in the livers of both genotypes. Interestingly, compared with EtOH alone, EtOH+LPS administration significantly induced intestinal Cyp3a11, Cyp2b10, and Sult1b1 in fat-1 mice, while expression of these genes was further decreased by EtOH+LPS in WT littermates. This pattern was not observed in the livers, where expression of Cyp3a11 and Sult1b1 were equally decreased in WT and fat-1 mice in response to EtOH+LPS administration.
Fig. 7.
BA detoxification gene expression. Expression of genes involved in the metabolic inactivation of BA in the ileum (A) (RNA-seq, n = 3–5) and liver (B) (qPCR, n = 5–10). *P < 0.05, one-way ANOVA.
Decreased ω-6:ω-3 PUFA ratio resulted in the attenuation of liver injury associated with EtOH and LPS administration
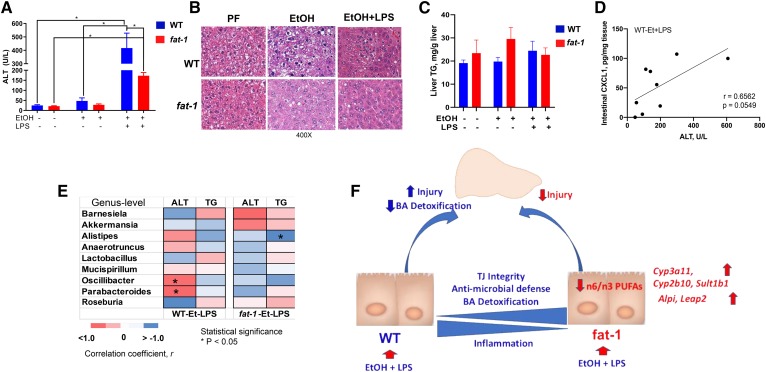
We next sought to determine whether the effects of a decreased ω-6:ω-3 PUFA ratio on the gut was associated with attenuation of liver pathology caused by EtOH consumption and LPS challenge. EtOH alone moderately increased plasma ALT levels (a marker of liver injury) in WT but not in fat-1 mice. Further, compared with WT, fat-1 mice had significantly decreased ALT levels in response to EtOH+LPS treatment (Fig. 8A), but there were no differences in hepatic TG accumulation, and the degree of steatosis was similar in both genotypes (Fig. 8B, C). We next performed multiple correlation analyses to evaluate the potential link between markers of liver injury, intestinal inflammation, and the gut microbiota. ALT levels were positively correlated with intestinal CXCL1 levels (Fig. 8D), and the frequency of the bacteria genera Oscillibacter (Oscillospiraceae family in the phylum Firmicutes) and Parabacteroides (Porphyromonadaceae family in the phylum Bacteroidetes) in WT but not in fat-1 mice subjected to EtOH+LPS treatment (Fig. 8E). Negative correlation was noted between liver TG levels and the genus Alistipes (Rikenellaceae family in the phylum Bacteroidetes) in EtOH+LPS-treated fat-1 mice.
Fig. 8.
Decreased ω-6:ω-3 PUFA ratio resulted in the improvement of liver injury associated with EtOH and LPS administration. Serum ALT levels (A), representative H&E-stained liver sections (400×) (B), liver TGs (C), correlation between intestinal CXCL1 protein expression and ALT (D), and heat map to illustrate Spearman correlations of the relative abundance of microbiome at the genus levels and parameters of liver injury WT and fat-1 mice in the EtOH+LPS group (n = 5, statistical significance indicated as *P < 0.05) (E). The r values are represented by gradient colors, where red and blue cells indicate positive and negative correlations, respectively; *P < 0.05. F: Model showing potential mechanism by which decreased tissue ω-6 PUFA and increased ω-3 PUFA results in a specific changes in intestinal homeostasis, the gut microbiota, and BA metabolism that coordinate to ameliorate liver injury in mice chronically fed EtOH and challenged with LPS. A–E: n = 3–12 mice per group.
DISCUSSION
In this study, we showed that reduction in the ω-6:ω-3 PUFA ratio in fat-1 mice produced changes that attenuated many of the toxic effects of EtOH and EtOH+LPS on the intestine and was associated with reduced liver injury. Because there were no differences in the expression of intestinal EtOH-metabolizing genes between fat-1 and WT mice, the differential effects identified are due to other mechanisms as discussed below. Our results demonstrated that endogenous ω-3 PUFA enrichment and reduction in the ω-6:ω-3 PUFA ratio preserved intestinal tight junction protein expression in both intestinal organoids and as determined in vivo by RNA-seq analysis. One potential mechanism is through downregulation of Nos2 expression. In addition to its well-established role in innate immunity, iNOS has also been demonstrated to disrupt intestinal tight junctions (50). A reduced ω-6:ω-3 PUFA ratio also positively impacted ISC growth and proliferation, shown in organoid cultures and in vivo. The expression of several genes were increased in fat-1 versus WT mice that have been demonstrated to regulate proliferation of intestinal cells [e.g., adrenoceptor α2A (Adra2a)] and renewal of the colonic mucosa in mice [e.g., aldo-keto reductase family 1 B8 (Akr1b8)]. Finally, the expression of Tph1 was also significantly upregulated in fat-1 mice. TPH1 synthesizes 5-hydroxytryptamine (5-HT) (serotonin) in the gut and although the role of 5-HT in gut health is controversial, it was recently shown that mice with enhanced uptake of 5-HT in the intestine had increased mucosal growth (51).
There were several other interesting observations related to intestinal health and disease revealed by RNA-seq analysis of the intestinal mucosa, including Plet1 with ∼46-fold induction in fat-1 mice versus WT. Zepp et al. (52) recently demonstrated that IL-17A-mediated induction of Plet1 expression was necessary for tissue repair following dextran sulfate sodium (DSS)-induced colitis. In a series of reports in mouse models of colitis, a number of other genes have been demonstrated to be beneficial. For example, cellular prion protein (PrPC, encoded by Prnp) and Rnf186, were elevated in fat-1 mice, and each has been demonstrated to protect mice from DSS-induced colitis (53, 54). The expression of Toll-like receptor genes (Tlr2 and Tlr5) was upregulated in fat-1 versus WT mice, and Choteau et al. (55) demonstrated that Tlr2 knockout mice had greater intestinal dysfunction and inflammation when compared with WT mice.
One of the most significant findings from this study is that endogenous ω-3 PUFA enrichment and RvD1 treatment attenuated EtOH-mediated intestinal inflammation. Altering the ω-6:ω-3 PUFA ratio has also proven beneficial in other mouse models. For example, fat-1 mice were protected from DSS-induced colitis (56), and many SPMs, including RvD1, are effective in mouse models of inflammatory bowel disease [reviewed in (57)]. In humans, the role of ω-3 PUFAs in intestinal inflammation is not well-established, but there is evidence for beneficial effects in UC and Crohn’s disease [reviewed in (58)]; however, the role of SPMs has not been investigated, but there is evidence that patients with UC had low levels of DHA-derived SPMs compared with patients in remission (59). Based upon these studies and on our results, treatment with RvD1 and other SPMs may be an effective treatment strategy, alongside ω-3 PUFA supplementation, to treat inflammatory disorders of the intestine.
The intestine, among other organs exposed to environmental factors, produces a number of protective factors, including AMPs. Gut AMPs protect the host from pathogens and control intestinal inflammation by either enzymatic (alkaline phosphatase and soluble phospholipase A2) or nonenzymatic mechanisms [e.g., defensins, cathelecidins, and C-type lectins (REG3 family)] (60). Although there were no differences in the expression of defensins between fat-1 and WT mice challenged with LPS, fat-1 mice had significantly higher intestinal expression of two antimicrobial genes, Leap2 and Alpi. LEAP2, despite the name, is most highly expressed in the small intestine (61), and though there is evidence that LEAP2 has antimicrobial activity in vitro, its in vivo role is less clear. In contrast, much more is known about the function of ALPI. ALPI is an important defense mechanism in the intestine because it can inactivate LPS and regulate the microbiota [reviewed in (62)]. Interestingly, the expression of Alpi can be induced by the EPA-derived SPM, resolvin E1 (37). Another novel finding from this study was the observation of increased expression of a “non-classical” antibacterial and endotoxin-neutralizing molecule, Bpi1, in fat-1 mouse-derived intestinal organoids. Bpi is transcriptionally upregulated by lipoxins (63) and exerts a potent (nanomolar) and selective bioactivity against gram-negative bacterial species.
Our results provide evidence that tissue ω-3 PUFA enrichment favorably modulates intestinal microbiota. It is well-documented that alcohol consumption resulted in alterations of the intestinal microbiota in humans and rodents (64); therefore, modulation of gut microbial composition is an attractive therapeutic approach for alcohol-related pathologies, including liver disease. We previously showed that a diet enriched in saturated FAs prevented gut microbiota changes induced by EtOH and a diet enriched in ω-6 PUFA, linoleic acid, further attenuated intestinal and liver damage (6). In the present study, we observed significant EtOH-induced changes in both WT and fat-1 mice; however, modulation of the ω-6:ω-3 PUFA ratio did not produce marked changes in the intestinal microbiome (in α or β diversity, or phyla composition) between fat-1 and WT mice. Our data are in agreement with studies demonstrating the lack of significant changes in microbial diversity associated with ω-3 PUFA supplementation (14). However, our results are in some disagreement with the study by Kaliannan et al. (10) that demonstrated significant differences in gut microbiota in fat-1 compared with WT mice, including significantly lower levels of Proteobacteria in fat-1 mice, which we did not observe in our study. This discrepancy can potentially be explained by environmental differences in the mouse facilities between our institutions (e.g., animal husbandry practices, bedding, water, etc.). This effect was convincingly demonstrated recently in a study where mice from the same vendor were fed the same Lieber-DeCarli EtOH diet but housed in two different institutions in France, which produced drastically different outcomes attributed to differences in the gut microbiome due to differences in animal husbandry between these two institutions (43). Nevertheless, there were several individual taxa that revealed differences associated with modulation of the ω-6:ω-3 PUFA ratio. Thus, an important observation in our study was that the EtOH-induced drop in Lactobacillus in WT mice was prevented in fat-1 mice. A decrease in Lactobacillus was associated with alcohol consumption in humans and in rodents (65, 66). Lactobacillus species exert multiple beneficial properties [e.g., production of antimicrobial substances (67)], and treatment with Lactobacillus rhamnosus Gorbach Goldin, for example, resulted in improvement of intestinal barrier function and subsequent protection against alcohol-induced endotoxemia and liver injury in mice (38, 68, 69). In addition, many species of lactic acid bacteria, e.g., Lactobacillus plantarum, possess enzymes involved in saturation metabolism of PUFAs generating multiple FA species, such as hydroxy, oxo, conjugated, and partially saturated trans-FAs (70). Some of these unique lipid species exert specific physiological functions. For instance, 10-Hydroxy-cis-12-octadecenoic acid, a gut microbial metabolite of linoleic acid, attenuated intestinal epithelial barrier dysfunction and ameliorated intestinal inflammation in DSS-induced colitis in mice by the suppression of TNFR2 upregulation (71). In line with the observation that ω-3 PUFA supplementation was associated with an increase in abundance of the butyrate-producing genera Roseburia (14), we also observed elevated levels (also not significant) of Roseburia in fat-1 mice as compared with WT littermates. Despite the relatively small changes in Roseburia, it most likely had functional consequences, as the blood butyric acid, an energy source for enterocytes and an important epigenetic modulator of gene expression, was significantly higher in fat-1 mice, but only in the PF animals. Butyrate supplementation attenuated EtOH-induced intestinal barrier and liver injury (72). In addition to butyrate production, Roseburia metabolizes linoleic acid via conjugated linoleic acids to vaccenic acid in humans and animals (73), and both of these compounds are considered to be beneficial for health (74).
Metabolomic analyses of plasma from fat-1 and WT mice revealed several interesting findings. ω-6 PUFA-derived AA was elevated in the blood of WT relative to fat-1 mice (both EtOH alone and EtOH+LPS exposures). AA and its metabolites can be pro-inflammatory, and we have recently demonstrated that many AA metabolites were upregulated in mice fed a diet high in ω-6 PUFAs and EtOH and were associated with increased liver inflammation (75). Tryptophan metabolism is a critical function of the gut microbiota that regulates many host functions, including metabolism and immunity (76). Blood tryptophan was diminished in WT mice relative to fat-1 mice exposed to EtOH+LPS, while the toxic tryptophan metabolite 3-indoxyl sulfate (I3S) was increased. I3S is an aryl hydrocarbon receptor (AhR) ligand (77) that can promote systemic inflammation through immune cell activation (78). Because fat-1 mice had reduced levels of I3S, this may be one mechanism by which fat-1 mice are protected from EtOH+LPS-induced intestinal inflammation. These data also revealed lower concentrations of nicotinamide in WT mice exposed to EtOH+LPS relative to fat-1 mice. Nicotinamide is an NAD+ precursor and the metabolism of EtOH decreases the NAD+:NADH ratio in the liver disrupting energy metabolism leading to fatty liver (79). fat-1 mice maintain levels of NAD+ precursors, which could elicit protection through preservation of tissue REDOX status. We also found decreased levels of methionine but increased levels of cystathionine in the EtOH-fed and LPS-challenged WT mice. Interestingly, a recent study by Yang et al. (80) found increased levels of these two amino acids in human patients with alcoholic cirrhosis.
Important roles for EtOH-mediated alterations in BA profile and metabolism have been reported in clinical and experimental studies (81, 82). Our data demonstrated that the profile of BAs was similar between WT and fat-1 PF and EtOH+LPS-treated mice. Similar expression of genes involved in BA synthesis (e.g., Cyp7a1, Cyp27a1, and Cyp8b1) in the livers of WT and fat-1 mice may explain the comparable levels of BA in these animals. LCA, the most lipophilic secondary BA, was the only BA that was significantly elevated by EtOH+LPS in WT mice but not in fat-1 mice. Increased LCA levels were also observed in actively drinking cirrhotic patients (44). At physiological levels, LCA, may prevent TNF-α release from colonic epithelial cells in vitro and protect against colonic inflammation in a DSS-induced colitis animal model (83). However, high LCA concentrations are toxic, particularly to the liver, as serious liver damage was noted in mice exposed to LCA (84). The expression of a number of genes important for BA detoxification was increased in fat-1 mice, suggesting that they are able to resist the generalized downregulation of the detoxification pathway in response to LPS. Finally, we found that the expression of Nr1h4/Fxr was induced by EtOH; however, LPS challenge did not further increase its expression in fat-1 mice, as was found in WT mice. Conversely, the expression of Nr1i3/Car was decreased by EtOH, but increased to a greater extent in fat-1 mice versus WT mice in the EtOH+LPS group. Interestingly, this pattern was specific to the intestine. It was recently shown that inhibition of intestinal FXR protected mice from nonalcoholic liver disease (85). In addition, NR1I3/CAR has been demonstrated to play a role in reducing intestinal damage in mice treated with DSS (86).
In our study, we used a multi-pronged approach to investigate alterations in the intestinal homeostasis caused by EtOH. While this design has significant strengths and advantages, we acknowledge that there are some limitations in our study. For example, the RNA-seq analysis was performed on total ileal mucosa RNA. Thus, with some of the genes identified, we cannot identify the specific cell type responsible for the differential expression, particularly with genes of the immune system. Therefore, future studies using single-cell sequencing would be useful in elucidating the role of the various cells of the gut. Nevertheless, these data provide a solid foundation upon which to further investigate the effect of ω-3 PUFAs on intestinal homeostasis.
There are several important implications to our findings. First, that modest changes in the ω-6:ω-3 PUFA ratio can lead to significant changes in the response of the intestine to EtOH with respect to attenuation of inflammation suggests that dietary factors can have a profound effect. Second, this effect is mediated not only by changes in the expression of the inflammatory pathway but also through the induction of AMP gene expression and subsequent changes in the microbiota. Indeed, AMPs are emerging as a therapeutic treatment for a number of diseases (87). In addition to these effects seen in fat-1 mice, additional pathways are involved, including that by which some hepatotoxic BAs (e.g., LCA) are detoxified through hydroxylation and conjugation in both the intestine (88) and liver (89). Furthermore, the effects of ω-3 PUFAs on the associated liver injury may be mediated by other factors, including plasma metabolites and intrinsic changes in the ω-6:ω-3 PUFA ratio in the livers of fat-1 mice. It is also possible that some of the changes observed in fat-1 mice are secondary to an overall attenuation in inflammation and protection of the liver from EtOH-induced damage mediated by other mechanisms. In conclusion, our results provide the basis for a model in which a decreased tissue ω-6:ω-3 PUFA ratio leads to specific changes in intestinal homeostasis, the gut microbiota, BA metabolism, and the plasma metabolome that coordinate to reduce intestinal inflammation and associated liver damage in the context of chronic EtOH exposure and LPS challenge (summarized in Fig. 8F).
Supplementary Material
Acknowledgments
The authors thank Marion McClain for manuscript editing. The authors acknowledge Dr. Shubha Gosh-Dastidar and Xinmin Yin for their contributions to this project.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- Adh
- alcohol dehydrogenase
- ALD
- alcoholic liver disease
- Aldh
- aldehyde dehydrogenase
- Alpi
- intestinal alkaline phosphatase
- ALT
- alanine aminotransferase
- AMP
- antimicrobial peptide
- BA
- bile acid
- CAE
- chloroacetate esterase
- Car
- constitutive androstane receptor
- CDCA
- chenodeoxycholic acid
- DSS
- dextran sulfate sodium
- EtOH
- ethanol
- fat-1
- ω-3 FA desaturase-1
- 5-HT
- 5-hydroxytryptamine
- ISC
- intestinal stem cell
- I3S
- 3-indoxyl sulfate
- LCA
- lithocholic acid
- Leap2
- liver-enriched antimicrobial peptide 2
- LPS
- lipopolysaccharide
- OTU
- operational taxonomic unit
- PF
- pair-fed
- Pxr
- pregnane X receptor
- RvD1
- resolvin D1
- SPM
- specialized pro-resolving mediator
- TG
- triglyceride
- Tlr
- Toll-like receptor
- Tph-1
- tryptophan hydroxylase-1
- UC
- ulcerative colitis
- UDCA
- ursodeoxycholic acid
This work was supported by National Institutes of Health Grants R01AA024102-01A1 (I.A.K.), U01AA022489 (C.J.M.), 1U01AA021901-01 (C.J.M.), 1U01AA021893-01 (C.J.M.), R01AA023681 (C.J.M.), S10OD020106 (X.Z.), P20GM103436 (E.C.R.), and P30GM106396 (E.C.R.); U.S. Department of Veterans Affairs Grant I01BX000350 (C.J.M.). Research reported in this publication was also supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grant P20GM113226 (C.J.M.) and the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under Grant P50AA024337 (C.J.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bishehsari F., Magno E., Swanson G., Desai V., Voigt R. M., Forsyth C. B. and Keshavarzian A.. 2017. Alcohol and gut-derived inflammation. Alcohol Res. 38: 163–171. [PMC free article] [PubMed] [Google Scholar]
- 2.Rao R. 2009. Endotoxemia and gut barrier dysfunction in alcoholic liver disease. Hepatology. 50: 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie G., Zhong W., Li H., Li Q., Qiu Y., Zheng X., Chen H., Zhao X., Zhang S., Zhou Z. et al. 2013. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 27: 3583–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirpich I. A., Feng W., Wang Y., Liu Y., Barker D. F., Barve S. S., and McClain C. J.. 2012. The type of dietary fat modulates intestinal tight junction integrity, gut permeability, and hepatic toll-like receptor expression in a mouse model of alcoholic liver disease. Alcohol. Clin. Exp. Res. 36: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirpich I. A., Feng W., Wang Y., Liu Y., Beier J. I., Arteel G. E.,Falkner K. C. Barve S. S.and McClain C. J.. 2013. Ethanol and dietary unsaturated fat (corn oil/linoleic acid enriched) cause intestinal inflammation and impaired intestinal barrier defense in mice chronically fed alcohol. Alcohol. 47: 257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirpich I. A., Petrosino J., Ajami N., Feng W., Wang Y., Liu Y.,Beier J. I., Barve S. S., Yin X., Wei S., et al. 2016. Saturated and unsaturated dietary fats differentially modulate ethanol-induced changes in gut microbiome and metabolome in a mouse model of alcoholic liver disease. Am. J. Pathol. 186: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirpich I. A., Miller M. E., Cave M. C., Joshi-Barve S., and McClain C. J.. 2016. Alcoholic liver disease: update on the role of dietary fat. Biomolecules. 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W., Wang B., Li X., and Kang J. X.. 2015. Endogenously elevated n-3 polyunsaturated fatty acids alleviate acute ethanol-induced liver steatosis. Biofactors. 41: 453–462. [DOI] [PubMed] [Google Scholar]
- 9.Wang M., Zhang X., Ma L. J., Feng R. B., Yan C., Su H., He C., Kang J. X., Liu B. And Wan J. B.. 2017. Omega-3 polyunsaturated fatty acids ameliorate ethanol-induced adipose hyperlipolysis: a mechanism for hepatoprotective effect against alcoholic liver disease. Biochim. Biophys. Acta Mol. Basis Dis. 1863: 3190–3201. [DOI] [PubMed] [Google Scholar]
- 10.Kaliannan K., Wang B., Li X. Y., Kim K. J., and Kang J. X.. 2015. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci. Rep. 5: 11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidu C., Escoula Q., Bellenger S., Spor A., Galan M., Geissler A., Bouchot A., Dardevet D., Morio-Liondor B., Cani P. D., et al. 2019. Erratum. The transplantation of omega3 PUFA-altered gut microbiota of fat-1 mice to wild-type littermates prevents obesity and associated metabolic disorders. Diabetes 2018;67:1512-1523. Diabetes. 68: 235. [DOI] [PubMed] [Google Scholar]
- 12.Bidu C., Escoula Q., Bellenger S., Spor A., Galan M., Geissler A., Bouchot A., Dardevet D., Morio B., Cani P. D., et al. 2018. The transplantation of ω3 PUFA-altered gut microbiota of fat-1 mice to wild-type littermates prevents obesity and associated metabolic disorders. Diabetes. 67: 1512–1523. [DOI] [PubMed] [Google Scholar]
- 13.Costantini L., Molinari R., Farinon B., and Merendino N.. 2017. Impact of omega-3 fatty acids on the gut microbiota. Int. J. Mol. Sci. 18: E2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson H., Mitra S., Croden F. C., Taylor M., Wood H. M., Perry S. L., Spencer J. A., Quirke P., Toogood G. J., Lawton C. L., et al. 2018. A randomised trial of the effect of omega-3 polyunsaturated fatty acid supplements on the human intestinal microbiota. Gut. 67: 1974–1983. [DOI] [PubMed] [Google Scholar]
- 15.Ungaro F., Rubbino F., Danese S., and D’Alessio S.. 2017. Actors and factors in the resolution of intestinal inflammation: lipid mediators as a new approach to therapy in inflammatory bowel diseases. Front. Immunol. 8: 1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musa-Veloso K., Venditti C., Lee H. Y., Darch M., Floyd S., West S., and Simon R.. 2018. Systematic review and meta-analysis of controlled intervention studies on the effectiveness of long-chain omega-3 fatty acids in patients with nonalcoholic fatty liver disease. Nutr. Rev. 76: 581–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeyapal S., Kona S. R., Mullapudi S. V., Putcha U. K., Gurumurthy P., and Ibrahim A.. 2018. Substitution of linoleic acid with alpha-linolenic acid or long chain n-3 polyunsaturated fatty acid prevents Western diet induced nonalcoholic steatohepatitis. Sci. Rep. 8: 10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serhan C. N. 2014. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 510: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zehr K. R., and Walker M. K.. 2018. Omega-3 polyunsaturated fatty acids improve endothelial function in humans at risk for atherosclerosis: a review. Prostaglandins Other Lipid Mediat. 134: 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitas R. D. S., and Campos M. M.. 2019. Protective effects of omega-3 fatty acids in cancer-related complications. Nutrients. 11: E945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K. R., Midgette Y., and Shah R.. 2018. Fish oil derived omega 3 fatty acids suppress adipose NLRP3 inflammasome signaling in human obesity. J. Endocr. Soc. 3: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simopoulos A. P. 2016. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients. 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasbalg T. L., Hibbeln J. R., Ramsden C. E., Majchrzak S. F., and Rawlings R. R.. 2011. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am. J. Clin. Nutr. 93: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart C. L., Morrison D. S., Batty G. D., Mitchell R. J., and Davey Smith G.. 2010. Effect of body mass index and alcohol consumption on liver disease: analysis of data from two prospective cohort studies. BMJ. 340: c1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang J. X., Wang J., Wu L., and Kang Z. B.. 2004. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 427: 504. [DOI] [PubMed] [Google Scholar]
- 26.Gustot T., Fernandez J., Szabo G., Albillos A., Louvet A., Jalan R., Moreau R., Moreno C.. 2017. Sepsis in alcohol-related liver disease. J. Hepatol. 67: 1031–1050. [DOI] [PubMed] [Google Scholar]
- 27.Kang J. X., and Wang J.. 2005. A simplified method for analysis of polyunsaturated fatty acids. BMC Biochem. 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins J. F., Hu Z., Ranganathan P. N., Feng D., Garrick L. M., Garrick M. D., and Browne R. W.. 2008. Induction of arachidonate 12-lipoxygenase (Alox15) in intestine of iron-deficient rats correlates with the production of biologically active lipid mediators. Am. J. Physiol. Gastrointest. Liver Physiol. 294: G948–G962. [DOI] [PubMed] [Google Scholar]
- 29.Lu R., Voigt R. M., Zhang Y., Kato I., Xia Y., Forsyth C. B., Keshavarzian A. and Sun J.. 2017. Alcohol injury damages intestinal stem cells. Alcohol. Clin. Exp. Res. 41: 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsyth C. B., Shaikh M., Bishehsari F., Swanson G., Voigt R. M., Dodiya H., Wilkinson P., Samelco B., Song S. and Keshavarzian A.. 2017. Alcohol feeding in mice promotes colonic hyperpermeability and changes in colonic organoid stem cell fate. Alcohol. Clin. Exp. Res. 41: 2100–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antharam V. C., Li E. C., Ishmael A., Sharma A., Mai V., Rand K. H., and Wang G. P.. 2013. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51: 2884–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozlov A., Bean L., Hill E. V., Zhao L., Li E., and Wang G. P.. 2018. Molecular identification of bacteria in intra-abdominal abscesses using deep sequencing. Open Forum Infect. Dis. 5: ofy025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirpich I. A., Gobejishvili L. N., Bon Homme M., Waigel S., Cave M., Arteel G., Barve S. S., McClain C. J., and Deaciuc I. V.. 2011. Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease. J. Nutr. Biochem. 22: 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seitz H. K., and Poschl G.. 1997. The role of gastrointestinal factors in alcohol metabolism. Alcohol Alcohol. 32: 543–549. [DOI] [PubMed] [Google Scholar]
- 35.Fu Z. D., Selwyn F. P., Cui J. Y., and Klaassen C. D.. 2016. RNA sequencing quantification of xenobiotic-processing genes in various sections of the intestine in comparison to the liver of male mice. Drug Metab. Dispos. 44: 842–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaishnava S., and Hooper L. V.. 2007. Alkaline phosphatase: keeping the peace at the gut epithelial surface. Cell Host Microbe. 2: 365–367. [DOI] [PubMed] [Google Scholar]
- 37.Campbell E. L., MacManus C. F., Kominsky D. J., Keely S., Glover L. E., Bowers B. E., Scully M., Bruyninckx W. J., and Colgan S. P.. 2010. Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc. Natl. Acad. Sci. USA. 107: 14298–14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Kirpich I., Liu Y., Ma Z., Barve S., McClain C. J., and Feng W.. 2011. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am. J. Pathol. 179: 2866–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan A. W., Fouts D. E., Brandl J., Starkel P., Torralba M., Schott E., Tsukamoto H., Nelson K. E., Brenner D. A., and Schnabl B.. 2011. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 53: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastelli M., Knauf C., and Cani P. D.. 2018. Gut microbes and health: a focus on the mechanisms linking microbes, obesity, and related disorders. Obesity (Silver Spring). 26: 792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llopis M., Cassard A. M., Wrzosek L., Boschat L., Bruneau A., Ferrere G., Puchois V., Martin J. C., Lepage P., Le Roy T., et al. 2016. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 65: 830–839. [DOI] [PubMed] [Google Scholar]
- 42.Wu W. B., Chen Y. Y., Zhu B., Peng X. M., Zhang S. W., and Zhou M. L.. 2015. Excessive bile acid activated NF-kappa B and promoted the development of alcoholic steatohepatitis in farnesoid X receptor deficient mice. Biochimie. 115: 86–92. [DOI] [PubMed] [Google Scholar]
- 43.Ferrere G., Wrzosek L., Cailleux F., Turpin W., Puchois V., Spatz M., Ciocan D., Rainteau D., Humbert L., Hugot C., et al. 2017. Fecal microbiota manipulation prevents dysbiosis and alcohol-induced liver injury in mice. J. Hepatol. 66: 806–815. [DOI] [PubMed] [Google Scholar]
- 44.Bajaj J. S., Kakiyama G., Zhao D., Takei H., Fagan A., Hylemon P., Zhou H., Pandak W. M., Nittono H., Fiehn O., et al. 2017. Continued alcohol misuse in human cirrhosis is associated with an impaired gut-liver axis. Alcohol. Clin. Exp. Res. 41: 1857–1865. [DOI] [PubMed] [Google Scholar]
- 45.Hirano S., and Masuda N.. 1982. Enhancement of the 7 alpha-dehydroxylase activity of a gram-positive intestinal anaerobe by Bacteroides and its significance in the 7-dehydroxylation of ursodeoxycholic acid. J. Lipid Res. 23: 1152–1158. [PubMed] [Google Scholar]
- 46.Ishii M., Toda T., Ikarashi N., Kusunoki Y., Kon R., Ochiai W., Machida Y., and Sugiyama W.K.. 2014. Gastrectomy increases the expression of hepatic cytochrome P450 3A by increasing lithocholic acid-producing enteric bacteria in mice. Biol. Pharm. Bull. 37: 298–305. [DOI] [PubMed] [Google Scholar]
- 47.Garcia M., Thirouard L., Sedes L., Monrose M., Holota H., Caira F., Volle D. H., and Beaudoin C.. 2018. Nuclear receptor metabolism of bile acids and xenobiotics: a coordinated detoxification system with impact on health and diseases. Int. J. Mol. Sci. 19: E3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alnouti Y. 2009. Bile Acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 49.Owen B. M., Milona A., van Mil S., Clements P., Holder J., Boudjelal M., Cairns W., Parker M., White R., and Williamson C.. 2010. Intestinal detoxification limits the activation of hepatic pregnane X receptor by lithocholic acid. Drug Metab. Dispos. 38: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han X., Fink M. P., Yang R., and Delude R. L.. 2004. Increased iNOS activity is essential for intestinal epithelial tight junction dysfunction in endotoxemic mice. Shock. 21: 261–270. [DOI] [PubMed] [Google Scholar]
- 51.Tackett J. J., Gandotra N., Bamdad M. C., Muise E. D., and Cowles R. A.. 2017. Enhanced serotonin signaling stimulates ordered intestinal mucosal growth. J. Surg. Res. 208: 198–203. [DOI] [PubMed] [Google Scholar]
- 52.Zepp J. A., Zhao J., Liu C., Bulek K., Wu L., Chen X., Hao Y., Wang Z., Wang X., Ouyang W., et al. 2017. IL-17A-induced PLET1 expression contributes to tissue repair and colon tumorigenesis. J. Immunol. 199: 3849–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin G. R., Keenan C. M., Sharkey K. A., and Jirik F. R.. 2011. Endogenous prion protein attenuates experimentally induced colitis. Am. J. Pathol. 179: 2290–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujimoto K., Kinoshita M., Tanaka H., Okuzaki D., Shimada Y., Kayama H., Okumura R., Furuta Y., Narazaki M., Tamura A., et al. 2017. Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 10: 446–459. [DOI] [PubMed] [Google Scholar]
- 55.Choteau L., Vancraeyneste H., Le Roy D., Dubuquoy L., Romani L., Jouault T., Poulain D., Sendid B., Calandra T., Roger T., et al. 2017. Role of TLR1, TLR2 and TLR6 in the modulation of intestinal inflammation and Candida albicans elimination. Gut Pathog. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudert C. A., Weylandt K. H., Lu Y., Wang J., Hong S., Dignass A., Serhan C. N., and Kang J. X.. 2006. Transgenic mice rich in endogenous omega-3 fatty acids are protected from colitis. Proc. Natl. Acad. Sci. USA. 103: 11276–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwanke R. C., Marcon R., Bento A. F., and Calixto J. B.. 2016. EPA- and DHA-derived resolvins’ actions in inflammatory bowel disease. Eur. J. Pharmacol. 785: 156–164. [DOI] [PubMed] [Google Scholar]
- 58.Mozaffari H., Daneshzad E., Larijani B., Bellissimo N., and Azadbakht L.. Dietary intake of fish, n-3 polyunsaturated fatty acids, and risk of inflammatory bowel disease: a systematic review and meta-analysis of observational studies. Eur. J. Nutr. Epub ahead of print. January 24, 2019; doi:10.1007/s00394-019-01901-0. [DOI] [PubMed] [Google Scholar]
- 59.Ungaro F., Tacconi C., Massimino L., Corsetto P. A., Correale C., Fonteyne P., Piontini A., Garzarelli V., Calcaterra F., Della Bella S., et al. 2017. MFSD2A promotes endothelial generation of inflammation-resolving lipid mediators and reduces colitis in mice. Gastroenterology. 153: 1363–1377.e6. [DOI] [PubMed] [Google Scholar]
- 60.Mukherjee S., and Hooper L. V.. 2015. Antimicrobial defense of the intestine. Immunity. 42: 28–39. [DOI] [PubMed] [Google Scholar]
- 61.Ge X., Yang H., Bednarek M. A., Galon-Tilleman H., Chen P., Chen M., Lichtman J. S., Wang Y., Dalmas O., Yin Y., et al. 2018. LEAP2 is an endogenous antagonist of the ghrelin receptor. Cell Metab. 27: 461–469.e6. [DOI] [PubMed] [Google Scholar]
- 62.Fawley J., and Gourlay D. M.. 2016. Intestinal alkaline phosphatase: a summary of its role in clinical disease. J. Surg. Res. 202: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Canny G., Levy O., Furuta G. T., Narravula-Alipati S., Sisson R. B., Serhan C. N., and Colgan S. P.. 2002. Lipid mediator-induced expression of bactericidal/ permeability-increasing protein (BPI) in human mucosal epithelia. Proc. Natl. Acad. Sci. USA. 99: 3902–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarin S. K., Pande A., and Schnabl B.. 2019. Microbiome as a therapeutic target in alcohol-related liver disease. J. Hepatol. 70: 260–272. [DOI] [PubMed] [Google Scholar]
- 65.Bull-Otterson L., Feng W., Kirpich I., Wang Y., Qin X., Liu Y., Gobejishvili L., Joshi-Barve S., Ayvaz T., Petrosino J., et al. 2013. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 8: e53028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leclercq S., Matamoros S., Cani P. D., Neyrinck A. M., Jamar F., Starkel P., Windey K., Tremaroli V., Bäckhed F., Verbeke K., et al. 2014. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA. 111: E4485–E4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Vuyst L., and Leroy F.. 2007. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 13: 194–199. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y., Liu Y., Sidhu A., Ma Z., McClain C., and Feng W.. 2012. Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am. J. Physiol. Gastrointest. Liver Physiol. 303: G32–G41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forsyth C. B., Farhadi A., Jakate S. M., Tang Y., Shaikh M., and Keshavarzian A.. 2009. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 43: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kishino S., Takeuchi M., Park S. B., Hirata A., Kitamura N., Kunisawa J., Kiyono H., Iwamoto R., Isobe Y., Arita M., et al. 2013. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA. 110: 17808–17813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miyamoto J., Mizukure T., Park S. B., Kishino S., Kimura I., Hirano K., Bergamo P., Rossi M., Suzuki T., Arita M., et al. 2015. A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40-MEK-ERK pathway. J. Biol. Chem. 290: 2902–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cresci G. A., Glueck B., McMullen M. R., Xin W., Allende D., and Nagy L. E.. 2017. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol. 32: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devillard E., McIntosh F. M., Duncan S. H., and Wallace R. J.. 2007. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 189: 2566–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Field C. J., Blewett H. H., Proctor S., and Vine D.. 2009. Human health benefits of vaccenic acid. Appl. Physiol. Nutr. Metab. 34: 979–991. [DOI] [PubMed] [Google Scholar]
- 75.Warner D. R., Liu H., Ghosh Dastidar S., Warner J. B., Prodhan M. A. I., Yin X., Zhang X., Feldstein A. E., Gao B., Prough R. A., et al. 2018. Ethanol and unsaturated dietary fat induce unique patterns of hepatic omega-6 and omega-3 PUFA oxylipins in a mouse model of alcoholic liver disease. PLoS One. 13: e0204119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agus A., Planchais J., and Sokol H.. 2018. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 23: 716–724. [DOI] [PubMed] [Google Scholar]
- 77.Schroeder J. C., Dinatale B. C., Murray I. A., Flaveny C. A., Liu Q., Laurenzana E. M., Lin J. M., Strom S. C., Omiecinski C. J., Amin S., et al. 2010. The uremic toxin 3-indoxyl sulfate is a potent endogenous agonist for the human aryl hydrocarbon receptor. Biochemistry. 49: 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim H. Y., Yoo T. H., Hwang Y., Lee G. H., Kim B., Jang J., Yu H. T., Kim M. C., Cho J. Y., Lee C. J., et al. 2017. Indoxyl sulfate (IS)-mediated immune dysfunction provokes endothelial damage in patients with end-stage renal disease (ESRD). Sci. Rep. 7: 3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purohit V., Gao B., and Song B. J.. 2009. Molecular mechanisms of alcoholic fatty liver. Alcohol. Clin. Exp. Res. 33: 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Z., Kusumanchi P., Ross R. A., Heathers L., Chandler K., Oshodi A., Thoudam T., Li F., Wang L. and Liangpunsakul S.. 2019. Serum metabolomic profiling identifies key metabolic signatures associated with pathogenesis of alcoholic liver disease in humans. Hepatol. Commun. 3: 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brandl K., Hartmann P., Jih L. J., Pizzo D. P., Argemi J., Ventura-Cots M., Coulter S., Liddle C., Ling L., Rossi S. J., et al. 2018. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J. Hepatol. 69: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Donepudi A. C., Ferrell J. M., Boehme S., Choi H. S., and Chiang J. Y. L.. 2017. Deficiency of cholesterol 7alpha-hydroxylase in bile acid synthesis exacerbates alcohol-induced liver injury in mice. Hepatol. Commun. 2: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ward J. B. J., Lajczak N. K., Kelly O. B., O’Dwyer A. M., Giddam A. K., Ni Gabhann J., Franco P., Tambuwala M. M., Jefferies C. A., Keely S., et al. 2017. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 312: G550–G558. [DOI] [PubMed] [Google Scholar]
- 84.Matsubara T., Tanaka N., Patterson A. D., Cho J. Y., Krausz K. W., and Gonzalez F. J.. 2011. Lithocholic acid disrupts phospholipid and sphingolipid homeostasis leading to cholestasis in mice. Hepatology. 53: 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang C., Xie C., Li F., Zhang L., Nichols R. G., Krausz K. W., Cai J., Qi Y., Fang Z. Z., Takahashi S., et al. 2015. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 125: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hudson G. M., Flannigan K. L., Erickson S. L., Vicentini F. A., Zamponi A., Hirota C. L., Alston L., Altier C., Ghosh S., Rioux K. P., et al. 2017. Constitutive androstane receptor regulates the intestinal mucosal response to injury. Br. J. Pharmacol. 174: 1857–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shukla P. K., Meena A. S., Rao V., Rao R. G., Balazs L., and Rao R.. 2018. Human defensin-5 blocks ethanol and colitis-induced dysbiosis, tight junction disruption and inflammation in mouse intestine. Sci. Rep. 8: 16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng J., Fang Z. Z., Kim J. H., Krausz K. W., Tanaka N., Chiang J. Y., and Gonzalez F. J.. 2014. Intestinal CYP3A4 protects against lithocholic acid-induced hepatotoxicity in intestine-specific VDR-deficient mice. J. Lipid Res. 55: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khan A. A., Chow E. C., Porte R. J., Pang K. S., and Groothuis G. M.. 2011. The role of lithocholic acid in the regulation of bile acid detoxication, synthesis, and transport proteins in rat and human intestine and liver slices. Toxicol. In Vitro. 25: 80–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.