Abstract
The development and progression of diabetic kidney disease (DKD), a highly prevalent complication of diabetes mellitus, is influenced by both genetic and environmental factors. DKD is an important contributor to the morbidity of patients with diabetes mellitus, indicating a clear need for an improved understanding of disease aetiology to inform the development of more efficacious treatments. DKD is characterized by an accumulation of extracellular matrix, hypertrophy, and fibrosis in kidney glomerular and tubular cells. Increasing evidence shows that genes associated with these features of DKD are regulated not only by classical signalling pathways, but also by epigenetic mechanisms, involving chromatin histone modifications, DNA methylation, and non-coding RNAs. These mechanisms can respond to changes in the environment and importantly, might mediate the persistent long-term expression of DKD-related genes and phenotypes induced by prior glycaemic exposure, despite subsequent glycaemic control, a phenomenon called metabolic memory. Detection of epigenetic events during the early stages of DKD could be valuable for timely diagnosis and prompt treatment to prevent progression to end-stage renal disease. Identification of epigenetic signatures of DKD via epigenome-wide association studies might also inform precision medicine approaches. Here, we highlight the emerging role of epigenetics and epigenomics in DKD, and the translational potential of candidate epigenetic factors and non-coding RNAs as biomarkers and drug targets for DKD.
Introduction
The worldwide prevalence of both type 1 mellitus (T1DM) and type 2 diabetes mellitus (T2DM) has rapidly increased over the past 30 years1,2, with consequential increases in the prevalence of microvascular and macrovascular diabetes complications3-7. Epidemiological studies show that improvements in diabetes management have led to reductions in diabetes-associated cardiovascular morbidity over recent decades; however, the impact of these strategies on rates of end-stage renal disease (ESRD) have been minimal8. Approximately 30–40% of patients with diabetes mellitus (T1DM and T2DM) develop DKD, and about 50% of them can progress to ESRD9-12. Current therapies to manage DKD include control of blood pressure and glucose levels, and treatment with angiotensin-receptor blockers (ARBs) and angiotensin-converting-enzyme inhibitors; however, these modalities show limited efficacy in preventing the progression of DKD. Genetic factors and signal transduction pathways affect the expression of genes and phenotypes associated with DKD. However, the escalating rates of DKD indicate the need for a more in-depth understanding of the underlying molecular mechanisms to identify better therapies for this disease13-16.
Epigenetic mechanisms can affect gene expression and function without changing the underlying DNA sequence and can mediate crosstalk between genes and the environment.17,18 As the development and progression of diabetes mellitus and DKD are strongly influenced by environmental factors, such as nutritional status, which in turn alters epigenetic states, it is likely that epigenetic mechanisms have a key role in the pathogenesis of DKD.14,19,20 In this Review we describe our current understanding of the role of epigenetics and epigenomics in DKD. We discuss how epigenetic factors and non-coding (nc)RNAs could be used as biomarkers and drug targets for DKD diagnosis, prognosis and treatment, and highlight how our rapidly expanding knowledge of epigenetic variations opens a window of opportunity for improved clinical management of DKD.
Pathogenesis of DKD
DKD is characterized by an accumulation of extracellular matrix (ECM) proteins such as collagen and fibronectin in renal compartments, resulting in tubular interstitial fibrosis, glomerular mesangial hypertrophy and expansion, thickening of the glomerular basement membrane, podocyte foot process effacement, and inflammation due to the infiltration of monocytes and macrophages. All of these factors contribute to renal dysfunction and can ultimately lead to ESRD.13,21-23 Diabetogenic stimuli, including high blood glucose levels (> 15 mM); advanced glycation end products (AGEs); growth factors such as transforming growth factor beta 1 (TGF-β1), angiotensin II (AngII) and platelet-derived growth factor; and inflammatory cytokines have been implicated in the pathogenesis of DKD owing to their adverse effects on multiple renal cell types.24-29 Upregulation of the above-mentioned growth factors and cytokines, activates signal transduction pathways, including protein kinase C and Akt kinase cascades, leading to the activation of key effector transcription factors such as SMADs, nuclear factor kappa B (NF-κB) and upstream stimulatory factors (USFs) (Fig. 1). Together, these mechanisms promote the expression of genes associated with fibrosis, hypertrophy, apoptosis, inflammation, oxidative stress, endoplasmic reticulum (ER) stress, mitochondrial dysfunction, and autophagy (Fig. 1).13-16,22,28-36 In particular, TGF-β1, which is overexpressed in several renal cell types in diabetes mellitus, has been widely studied in DKD progression due to its pro-fibrotic effects.13,21,26,27 Identification of these pathways and the use of unbiased transcriptomic profiling approaches to uncover additional pathogenic factors have led to the initiation of clinical trials to assess new therapeutic targets in DKD.37-42 However, the escalating rates of DKD underscore the need to explore the contribution of other mechanisms, such as epigenetics, to DKD, particularly for patients with long-standing diabetes mellitus who experience persistent complications even after glucose normalization, a phenomenon attributed to metabolic (or hyperglycaemic) memory.
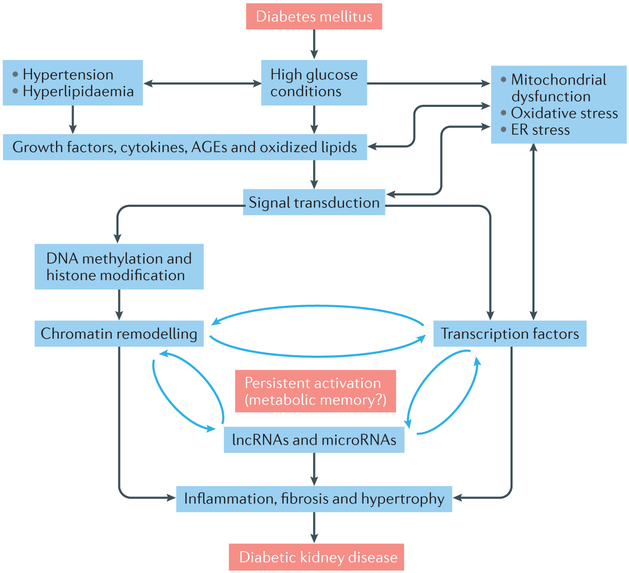
Fig. 1. Molecular mechanisms of diabetic kidney disease.
The pathophysiology of diabetic kidney disease (DKD) is complex and involves interactions between genetic factors, epigenetic factors, and the environment. High glucose levels in the context of diabetes mellitus, together with hypertension, and hyperlipidaemia induce the expression of growth factors, angiotensin II (AngII), cytokines, advanced glycation end products (AGEs), and produce oxidized lipids, mitochondrial dysfunction, and oxidative and endoplasmic reticulum (ER) stress. Growth factors, such as such as transforming growth factor β1 (TGF-β1), stimulate signal transduction pathways, including those involving MAPK and AKT kinases that activate downstream transcription factors, such as upstream stimulatory factors, SMADs, AP1 and the ER stress-related transcription factor, CHOP. They also affect epigenetic processes, including DNA methylation, histone modifications, and the expression of non-coding RNAs (microRNAs and long non-coding RNAs), resulting in altered chromatin accessibility and/or altered expression of target genes. Interactions between transcription and epigenetic factors lead to the persistent activation of signalling pathways, which might contribute to the phenomenon of metabolic memory, whereby exposure to high glucose levels leads to continued detrimental effects, even after glycaemic control has been established. Together, these conditions promote inflammation, fibrosis, and glomerular hypertrophy, which are hallmarks of DKD.
Metabolic memory in DKD
Most complications in patients with diabetes mellitus are associated with hyperglycaemia. Although glucose levels can be controlled through diet, exercise, and medications including insulin, many patients continue to experience numerous life-threatening complications following glucose normalization, suggesting the existence of a ‘memory’ of the prior glucose exposure in target cells, which leads to persistence of its detrimental effects long after glycaemic control has been established.43-46 The phenomenon of hyperglycaemic memory has been observed in clinical trials and experimental models. The landmark Diabetes Control and Complications Trial (DCCT) showed that intensive glycaemic control delayed progression of microvascular complications, including nephropathy and neuropathy, compared to conventional therapy in patients with T1DM. After cessation of the DCCT in 1993, both groups of patients received intensive glycaemic control and were followed long-term (1994 to present) as part of the observational Epidemiology of Diabetes Intervention and Complications (EDIC) study47. During EDIC, both groups achieved similar mean haemoglobin A1c (HbA1c) levels of approximately 8%.43,44 However, patients in the original DCCT intensive treatment group demonstrated a significantly lower risk of developing microvascular and macrovascular complications relative to those of patients in the original conventional treatment arm of DCCT.43-46 This difference in risk between the two groups was attributed to prolonged hyperglycaemia in the conventional therapy group during DCCT. The UK Prospective Diabetes Study reported similar long-term benefits of intensive glycaemic control in patients with T2DM. These benefits lasted well after cessation of the intervention, suggesting a ‘legacy effect’ of glycaemic control.48
The persistent effects of high glucose through metabolic memory remain a major hurdle in the effective management of diabetic complications. An increasing body of evidence supports a role for epigenetic mechanisms in metabolic memory, suggesting that targeting these mechanisms might offer opportunities for the development of novel approaches to prevent and treat persistent diabetic complications, including DKD.
Epigenetics: new insights into DKD
Genome-wide association studies (GWAS) have identified only a few potential genes, loci, and single nucleotide polymorphisms (SNPs) associated with DKD, and few of these have achieved genome-wide significance in replication studies.49-52 By contrast, a growing body of literature implicates a role for epigenetic modifications in DKD, particularly because they can induce changes in the expression of genes in response to environmental factors14,53-56. Epigenetic modifications enable mitotically and/or meiotically heritable changes in gene function to occur without altering the underlying DNA sequence17,18. Of note, most disease-associated SNPs are present in non-coding regions of the genome, including regulatory regions such as promoters and enhancers, and in ncRNAs,57 which can affect gene expression by altering transcription factor binding, chromatin accessibility, and through other epigenetic mechanisms. Thus, findings from GWAS and epigenome studies show convergence on shared pathways, suggesting that combined analyses of genetic and epigenetic susceptibility loci might provide a powerful approach to identify pathogenic pathways in DKD, as has been done in CKD58. In order to understand the mechanisms and consequences of epigenetic changes, it is important to first understand the organizational structure of nuclear DNA and chromatin.
Nuclear DNA is packaged into a histone–protein complex called chromatin (Fig. 2). The basic subunit of chromatin is the nucleosome, which comprises an octamer of two copies of each of the core histone proteins H2A, H2B, H3 and H4, wrapped by 147 base pairs of chromosomal DNA.59,60 Epigenetic modifications affect chromatin structure and the expression of genes by either covalently modifying DNA or histone proteins to affect the binding of transcriptional regulators to DNA, or through actions of non-coding (nc)RNAs [G]. The most well-studied epigenetic marks include DNA methylation [G] of cytosines, histone post-translational modifications (PTMs), and (nc)RNAs. Whereas ‘epigenetics’ refers to epigenetic modifications that occur in single genes or sets of genes, ‘epigenomics’ refers to the broader, genome-wide profiles and effects of these modifications. The epigenome therefore collectively dictates cell identity and the gene expression and phenotypes of specific cells. Epigenetic control of gene expression is important in several important biological processes, including development, cellular identity, genomic imprinting [G] and cell differentiation. In the context of diabetes mellitus, epigenetic control of gene expression can mediate cellular responses to environmental factors and contributes to the differential susceptibility of monozygotic twins to diabetes mellitus and its complications.
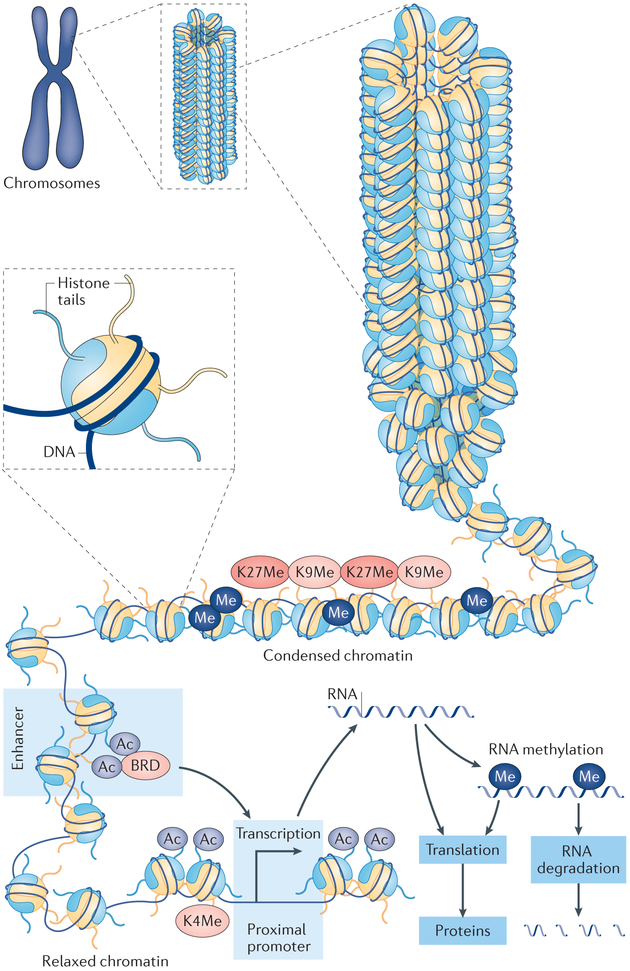
Fig. 2. The effects of epigenetic modifications on chromatin structure.
Chromosomes are composed of DNA–protein complexes called chromatin. The basic subunit of chromatin is the nucleosome, which comprises an octamer of two copies of each of the core histone proteins H2A, H2B, H3 and H4, wrapped by 147 base pairs of chromosomal DNA. Condensed chromatin is generally characterized by promoter DNA methylation (Me) and histone Me depending on the site of modification on histone (for example, lysine 9 methylation (K9Me) and lysine 27 methylation (K27Me) on histone H3), resulting in repressed gene expression. Relaxed chromatin is marked by histone lysine acetylation (Ac) and methylation of lysine 4 on histone 3 (K4Me) in the promoter regions, which allows efficient gene expression. Histone lysine acetylation at the enhancer region also enhances gene expression by interacting with the promoter region through bromodomain-containing proteins (BRD) and other proteins. RNA methylation (Me) is a form of epigenetic regulation related to the translation and degradation of RNAs.
Changes in the environment, including nutritional changes, can either transiently or chronically affect epigenetic states.54,61,62 Furthermore, as epigenetic marks are heritable, maternal behaviour and nutritional status (that is, undernutrition or overnutrition) can induce intrauterine and early postnatal epigenetic changes that might predispose offspring to metabolic diseases.54,62 A number of studies have also highlighted a connection between low birth weight, poor nutrition, low nephron endowment and the development of renal disease — a series of events most likely mediated by epigenetic factors.63 Notably, the role of epigenetics in mediating the effects of maternal nutritional status on kidney development was elegantly demonstrated in a 2018 study64, which showed that in a rat model of intrauterine growth restriction, kidneys of postnatal day 1 pups had reduced weight as well as pronounced DNA hypomethylation relative to controls. In mice, deletion of Dnmt1, which encodes the maintenance DNA methyltransferase 1, in nephron progenitor cells, phenocopied the effects of intrauterine growth restriction, with reduced nephron number at birth and reduced expression of genes critical for nephrogenesis,64 supporting a role for DNA methylation in mediating the effects of altered nutritional status on renal programming.
Epigenetics has also been strongly associated with risk of both T1DM and T2DM, 65-67but this aspect is not discussed in this Review.
DNA methylation in DKD
DNA methylation is mediated by DNA methyltransferases DNMT1, DNMT2 and the DNMT3 family members DNMT3a, DNMT3b and DNMT3-like (DNMT3L). DNMT3a/3b mediate de novo DNA methylation (that is, the establishment of new DNA methylation patterns), whereas DNMT1 is a maintenance methyltransferase that copies the methylation pattern of parental DNA strands. 60 On the other hand, the function of DNMT2 is relatively less clear with data suggesting it serves more as a tRNA methyltransferase.68 DNA methylation can be reversed by ten-eleven translocation (TET) proteins, which convert 5-methylcytosine to 5-hydroxymethylcytosine.69 Methylation of DNA at gene promoter regions can repress gene expression through various mechanisms, including the recruitment of transcriptional repressors and by interfering with transcription factor binding, whereas methylation of DNA at gene bodies/transcribed regions can regulate the elongation [G] phase of transcription and alternative splicing [G].60
Several experimental and clinical studies indicate the existence of links between CpG DNA methylation and DKD. For example, experimental models support the involvement of DNA methylation in glomerular and proximal tubular epithelial cell functions, including filtration, glucose and solute handling;70 dysregulation of critical genes associated with these processes has a critical role in the pathogenesis of CKD and DKD. In models of TGF-β1-induced renal fibrosis, upregulation of TGF-β1 induced hypermethylation of Rasal1promoter, leading to reduced expression of this Ras-GTP suppressor and increased Ras-GTP signalling, increased fibroblast activation, and fibrosis.71 Changes in Rasal1 promoter DNA methylation were reversed by TET3-mediated hydroxymethylation, with concomitant reduction in fibrosis.72 In another study, mice with streptozotocin-induced albuminuria showed decreased expression of the histone deacetylase [G] SIRT1. Interestingly, tubule-specific overexpression of SIRT1 prevented albuminuria by inducing hypermethylation of the Cldn1 gene (as SIRT1 deacetylates and activates DNMT1), leading to downregulation of the tight junction protein Claudin-1, in podocytes, suggesting crosstalk between podocytes and proximal tubules through epigenetic mechanisms.73 On the other hand, mice with proximal tubule-specific deletion of Sirt1 showed aggravation of albuminuria in association with reduced Cldn1methylation, enhanced histone acetylation, and upregulation of Claudin-1.73 These and other studies showing the protective effects of SIRT1 in DKD have prompted further evaluation of SIRT1 as a therapeutic target for DKD.73 In the db/db mouse model of diabetic kidney disease, transcriptional repression of the transcription factor KLF4 was associated with increased DNA methylation at the promoter region of the gene that encodes nephrin, along with podocyte apoptosis and proteinuria.74 Another in vivo study showed that the promoter region of Agt, which encodes angiotensinogen, and is associated with hypertension and CKD, exhibited DNA hypomethylation and acetylation of histone H3 at lysine 9 (H3K9) along with increased expression in early stages of DKD in db/db mice. These epigenetic marks were resistant to treatment with the antidiabetic drug pioglitazone, suggesting that conventional therapies might be unable to reverse epigenetic marks (as discussed later).75 A 2017 study demonstrated that cultured proximal tubule cells exposed to high glucose, and kidneys from streptozotocin-treated mice exhibited DNA hypomethylation of MIOX, associated with enhanced expression of its encoded protein, myo-inositol oxygenase. Hypomethylation of MIOX was associated with enhanced binding of the transcription factor, Sp1, to its promoter and the expression of genes involved in oxidative stress, hypoxia and fibrosis.76 In a further study, levels of SP1, NFκB, and DNMT11 were markedly increased in podocytes from diabetic db/db mice; inhibition of DNMT1 with 5-azacytidine, 5-aza-2'-deoxycytidine, or Dnmt1 small interfering RNA, induced renal protection, suggesting that epigenetic regulation of the SP1–NFκB–DNMT1 signalling pathway might be involved in podocyte injury in DKD.77 Increased expression of the DNA methylation eraser, TET2, increased expression of TGF-β1 by demethylating CpG islands in the TGF-β1 regulatory region.78 Demethylation of USF1 binding sites in the Tgfb1 promoter, associated with increased USF1 binding, decreased DNMT1 binding and increased expression of Tgfb1 has also been demonstrated in primary mesangial cells from diabetic db/db mice.79 In this study, treatment of mesangial cells with hydrogen peroxide also induced demethylation of the Tgfb1 promoter, leading to upregulation of Tgfb1 expression, suggesting that reactive oxygen species might mediate mesangial fibrosis in DKD through aberrant DNA methylation of the Tgfb1 locus.
DNA methylation EWAS
An increased appreciation of the contribution of epigenetic mechanisms to human disease, including complications of diabetes, has led to a surge in epigenome-wide association studies (EWAS) of DNA methylation status in populations of patients with CKD and DKD (Table 1).
Table 1.
Summary of EWAS studies in patients with diabetic kidney disease
| Study design | Population | Tissue | Method | Gene (s) modified |
Main findings | Ref |
|---|---|---|---|---|---|---|
| DKD in T1DM | 192 patients of Irish descent with T1DM (with and without DKD) | Whole blood | Illumina HumanMethylation27 BeadChip (~27,000 CpGs) | UNC13B | 19 CpGs correlated with time to DKD development, including one near UNC13B, which contains a SNP associated with DKD. | 80 |
| Microdissected tubuli from subjects with DKD and CKD and controls without | 87 microdissected human kidney tubule samples: 21 from patients with DKD (cases) and 66 from controls for 450K arrays; 12 CKD cases and 14 controls for the HELP assay | Microdissected tubuli | Illumina 450K Bead ChIP; and HpaII fragment enrichment by ligation mediated PCR (HELP) assay and hybridization to oligonucleotide arrays | COLIV4A1/A2, SIX2, HNF, and TCFAP EZR TGFBR3, SMAD3, SMAD6 SMAD3 RUNX1 and others | Differentially methylated regions localized to enhancers and were enriched in binding sites for key renal TFs. Key genes associated with renal fibrosis, such as collagens, exhibited differential methylation that correlated with gene expression. | 84 |
| CKD versus controls including DKD | 255 cases with CKD including 113 with DKD; 152 controls without renal disease including 113 T1DM | Whole blood | Illumina HumanMethylation450 BeadChip (~450,000 CpG features) | CUX1, ELMO1, FKBP5, INHBA-AS1, PTPRN2, PRKAG2 ELMO1 and PRKAG2 | 23 genes with significant methylation changes associated with CKD. | 85 |
| Mitochondrial function in T1DM with versus without DKD | 196 patients with T1DM and DKD ; 246 patients with T1DM without renal disease | Whole blood | A combination of Illumina HumanMethylation 27K and 450K BeadChips | TAMM41, PMPCB, TSFM, and AUH at multiple CpGs | 54 probes were associated with DKD; 46 of the top-ranked variants for DKD, including genes related to mitochondrial function, showed differential methylation in subjects with ESRD. | 86 |
| eGFR decline in CKD including DKD | Chronic Renal Insufficiency Cohort (CRIC): 20 (including 10 with DKD) with the lowest and 20 (including 10 DKD) with the highest rates of eGFR decline | Whole blood | Illumina HumanMethylation450 BeadChip (~450,000 CpG features) | NPHP4, IQSEC1 and TCF3 NOS3, NFKBIL2, CLU, NFKBIB, TGFB3 and TGFBI | CpG islands in certain genes associated with renal fibrosis (NPHP4, IQSEC1, and TCF3) showed greater DNAme in participants with stable renal function. | 87 |
| Offspring exposed in utero to T1DM | 29 adult non-diabetic offspring of mothers with T1DM (cases) and 28 non-diabetic offspring of fathers with T1DM (controls) | Blood leukocytes | Illumina HumanMethylation27 BeadChip (~27,000 CpGs) | DNMT1 | DNMT1, a DNA methyltransferase involved in gene regulation in development, showed reduced DNAme in cases. | 89 |
| eGFR in CKD including DKD | Patients from the Atherosclerosis Risk in Communities (n=2,264 including 586 DM) and Framingham Heart (n=2,595 including 394 DM) studies | Whole blood | Illumina HumanMethylation450 BeadChip (~450,000 CpG features) | PTPN6/PHB2 , ANKRD11, and TNRC18 | Of 19 CpG sites significantly associated with eGFR and/or CKD five also associated with renal fibrosis in biopsies from CKD patients and showed concordant DNAme changes in kidney cortex. | 90 |
| Pima Indians with T2DM and DKD | 181 diabetic Pima Indians | Blood leukocytes | Illumina HumanMethylation450 BeadChip (~450,000 CpG features) | CDGAP, FKBPL and ATF6B | Methylation at 77 sites were associated with eGFR decline; three of these 77 probes showed directionally consistent and significant associations with fibrosis in matched kidney tissue. | 91 |
| DCCT / and EDIC cohort of T1DM | Patients who received conventional therapy in DCCT and showed retinopathy or albuminuria progression by EDIC year 10 (cases) versus patients who received intensive therapy in DCCT who did not develop retinopathy or DKD progression by EDIC year 10 (controls). | Whole blood (sampled in ~1993), Monocytes (sampled in ~2010) | Illumina HumanMethylation450 BeadChip (~450,000 CpG features) | GATAD1, BSN, PRKCE, TXNIP, LHX6, MAP7, CDH3, SMYD1, CLCN7, ZNF167, CUED1 | 12 differentially methylated loci were common in both whole blood and monocytes, i.e., sustained over 16 years. Loci included TXNIP, known to be associated with hyperglycaemia and related complications, including DKD. Demonstrate a connection between DNAme and human metabolic memory. | 127 |
ChIP, chromatin immunoprecipitation; CKD, chronic kidney disease; DCCT, Diabetes Control and Complications Trial; DKD, diabetic kidney disease; DNAme, DNA methylation; EDIC; Epidemiology of Diabetes Intervention and Complications; eGFR, estimated glomerular filtration rate; ESRD, end-stage renal disease; HELP, HpaII fragment enrichment by ligation mediated PCR; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus; TFs, transcription factors
In a 2010 case-control study of 192 patients with T1DM,80 researchers used the Illumina HumanMethylation27 BeadChip (which assesses ~27,000 CpGs across the genome) to compare DNA methylation levels from genomic DNA of patients with and without DKD. Nineteen CpGs correlated with time to DKD development, including one near UNC13B, which contains a SNP associated with DKD. In another early study, DNA from saliva was used to compare DNA methylation levels in >14,000 genes in African American and Hispanic patients with diabetes mellitus (3 T1DM and 21 T2DM) and ESRD, with those of patients with diabetes mellitus but no DKD.81 Interestingly, 21% of the 187 differentially methylated genes had been previously associated with CKD or DKD, suggesting potential functional relevance of the findings to disease pathogenesis. Another study that used the Illumina HumanMethylation27 BeadChip to assess epigenetic markers of renal function in 972 African Americans (including 298 with DKD) from the Genetic Epidemiology Network of Arteriopathy (GENOA) study82 found that 24 of the top 30 DNA methylation markers significantly associated with estimated glomerular filtration rate (eGFR) were related to aging, inflammation, or cholesterol pathways.
The development of improved DNA methylation arrays, high-throughput sequencing technologies, and powerful approaches for data analysis have prompted additional EWAS in DKD and CKD populations over the past ~ 10 years with the aim of improving disease prognosis and patient classification.83 In the first EWAS study of kidney biopsy samples, DNA methylation profiling of microdissected tubuli from patients with DKD or nondiabetic CKD compared with those of control individuals without renal disease revealed significant differences between the two groups.84 Notably, the differentially methylated regions localized mostly to enhancers and were enriched in binding sites for important kidney specific transcription factors. Key genes associated with renal fibrosis, such as collagens, showed differential methylation patterns that correlated with gene expression. Another study, used the Illumina HumanMethylation450 BeadChip (which assesses ~450,000 CpG features across the genome) to profile DNA methylation in 255 T1DM patients with CKD (including 113 with DKD) and 152 controls (including 113 T1DM) without renal disease.85 The researchers then evaluated the biological and functional relevance of the differentially methylated genes using in silico analyses based on the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database of pathways. These analyses led to the identification of 23 genes that had significant methylation changes associated with CKD, and suggested a potential functional role for DNA methylation changes in influencing CKD development.
Mitochondrial function has an important role in the pathogenesis of DKD. A study of DNA methylation alterations in genes related to mitochondrial function in patients with DKD compared the methylation of blood DNA from 196 patients with T1DM and DKD to that of 246 patients with T1DM but with no renal disease.86 The researchers identified 54 CpG sites where methylation changes were significantly associated with DKD using both the HumanMethylation27 and HumanMethylation450 BeadChips; from a further sub-analyis, 46 of the top-ranked identied variants for DKD, including several genes related to mitochondrial function, were found to be differentially methylated in patients with ESRD relative to those without renal disease.
An analysis of DNA methylation levels in participants from the Chronic Renal Insufficiency Cohort (CRIC) with the lowest and highest rates of eGFR decline (that is, those with stable renal function versus rapid decliners) using the HumanMethylation450 BeadChip 87 identified associations between methylation at CpG islands in certain genes and renal fibrosis. Participants (20 patients including 10 with DKD) with stable renal function had greater DNA methylation of NPHP4, IQSEC1, and TCF3 than rapid decliners (20 patients including 10 with DKD). Genes associated with oxidative stress and inflammation were also differentially methylated between the two groups. These data support a role for epigenetic changes in renal function decline in patients with CKD. Replication studies are needed to validate these findings and to determine whether these key epigenetic differences exist in patients with DKD, although these CKD cohorts had a significant number of subjects with DKD.
The role of DNA methylation in the trans-generational inheritance of metabolic diseases has been well studied because of the ability of diet and the environment to alter epigenetic marks at genes associated with metabolic functions.54,62,88 A study of the effects of foetal exposure to hyperglycaemia on kidney development and function89 used the HumanMethylation27 BeadChip to examine leukocyte DNA methylation in 29 adult non-diabetic offspring of mothers with T1DM (the case group) and 28 non-diabetic offspring of fathers with T1DM (the control group). Differentially methylated CpGs were not located in any gene directly related to kidney development or function, but DNMT1, a key DNA methyltransferase involved in gene regulation in early development, showed reduced DNA methylation in the case group, prompting the researchers to conclude that T1DM and exposure to hyperglycaemia in utero might predispose offspring to changes in kidney function via pathways related to imprinting. Additional studies in larger cohorts are needed to strengthen these conclusions and determine whether kidney-specific processes are also involved.
Many of the early EWAS were limited by small sample sizes, cross-sectional design, lack of replication samples, and/or the presence of comorbidities in the patient population under study. A 2017 EWAS 90 assessed whole blood DNA methylation profiles associated with kidney function in patients from the Atherosclerosis Risk in Communities (n=2,264 including 586 with diabetes) and Framingham Heart (n=2,595 including 394 with diabetes) studies, representing the largest EWAS of a CKD cohort to date. Using the HumanMethylation450 BeadChip, the researchers identified 19 CpGs associated with eGFR and CKD, five of which associated with renal fibrosis and showed concordant DNA methylation changes in renal cortical biopsy samples. The top CpGs mapped to enhancer regions; moreover, the researchers identified methylation at a CpG site cg19942083 at PTPN6 that was associated with low renal PTNP6 expression, higher eGFR, and reduced renal fibrosis, implying that PTPN6 could be further evaluated as an epigenetic biomarker for fibrosis/CKD because locus-specific DNA methylation changes may cause CKD through alteration in the expression of the related functionally relevant genes. Notably, the data also shows the utility of non-invasive blood based DNA methylation profiling to screen for kidney disease specific targets. They also observed significant enrichment of eGFR-associated CpGs in regions that bind to the transcription factors EBF1, EP300, and CEBPB. ,In addition, they used epigenome data from ENCODE [G] to determine putative functions of these CpGs which showed the presence of active enhancers around them implying that they are associated with gene expression. This well-designed EWAS has several key strengths, including its large sample size, use of appropriate methodologies to correct for white blood cell subtype composition, the evaluation of important kidney phenotypes, use of an ethnically diverse population, its replication of findings in an independent cohort, and its rigorous analytical methods. Future studies are needed to stratify data obtained by this study according to diabetes status to identify methylation changes that are also specific to DKD. Nevertheless, many of the data analyses and covariate correction approaches used in this study will likely inform future EWAS efforts to establish epigenetic markers of renal function in patients with CKD and DKD.
The Pima Indians of Southwestern USA have among the highest incidence of T2DM of any population in the world, and also very high incidence of DKD. A 2018 study that examined leukocyte DNA methylation patterns at ~400,000 CpG sites in 181 Pima Indians with T2DM91 identified an association between methylation at 77 CpG sites and eGFR decline over a 6-year period. Furthermore, three of these 77 probes showed directionally consistent associations with fibrosis in matched kidney tissue, adding to the growing number of reports supporting a role of DNA methylation and epigenetic variations as potential biomarkers of DKD progression.
EWAS: challenges and opportunities
A limitation of EWAS of renal phenotypes is the paucity of available renal biopsy material. In one approach, to circumvent this limitation, a 2018 study used a network in silico analytic approach to identify renal compartment-specific gene expression and methylated sites related to DKD through mining publicly available gene expression omnibus (GEO) database for glomerular and tubular datasets, enriched function analyses by gene ontology (GO), and KEGG analyses, and integrated the expression data with Illumina 27k methylation ChIP data. This endeavour enabled the authors to generate glomerular and tubular specific networks of genes associated with related tissue-specific functions in DKD pathogenesis, including mitochondrial function and acid-base regulation.92 With increasing availability of transcriptome, epigenome and other datasets from numerous clinical trials and consortia, as well as more powerful data integrative tools and reference datasets, it is likely that such in silico approaches will help accelerate new discoveries in DKD epigenome research.
Since epigenetic marks affect gene expression in a cell-specific manner, early EWAS were often confounded by the cellular heterogeneity of the samples. This challenge has been addressed to some extent by the adoption of computational deconvolution approaches that use published reference DNA methylation datasets to adjust for differences in the cell composition between samples from different participants, so that methylation changes can be corrected for any variation caused by cell type composition.93 In addition, efforts have been made to compare EWAS findings from whole blood genomic DNA with candidate DNA methylation and gene expression changes in matched renal biopsy samples in the same patient to determine the extent to which changes in blood reflect those in the kidney.90
Another challenge is the limited genome coverage provided by the HumanMethylation27 and HumanMethylation450 BeadChips. Although these BeadChips have been used extensively for population-based studies, they cover only a small fraction of the genome with little coverage of enhancer regions, and might therefore miss important epigenetic changes. The more recently developed Illumina EPIC arrays cover nearly 850,000 CpG sites, and have many improved features, including coverage at many enhancers.94 Sequencing methods, including methylated DNA immunoprecipitation sequencing (Me-DIP-seq) [G], reduced representation bisulfite sequencing (RRBS-seq) [G] and whole genome bisulfite sequencing [G], are also increasingly being used to identify more numbers of differentially methylated sites across the genome, facilitated by steadily decreasing costs. These sequencing approaches, especially whole genome bisulfite, are powerful and can not only uncover differential changes genome-wide, but can also be combined with in silico bioinformatics and data analysis packages, as well as publicly available omics datasets for integrative analyses to obtain unprecedented new biological data. This information in turn can accelerate discovery of disease relevant loci and drug targets through associations with clinical phenotypes, biomarkers in biofluids (serum and urine) as well as subject-specific variations for personalized medicine.95 As mentioned earlier, given that most disease-associated SNPs identified by GWAS are present in non-coding regions of the genome, identification of epigenotype [G] variations using EWAS can complement genotype variations uncovered by GWAS and provide powerful information about disease susceptibility and causality. In addition, advances in single-cell (transcriptome and epigenome) sequencing of multiple kidney cell types will greatly aid future EWAS efforts.96 Exciting reports of single-cell and single-nucleus sequencing of mouse and human kidney cells have already uncovered cell types related to specific kidney diseases because epigenetic changes and their effects on gene expression and DKD can be renal cell type-specific , for example, podocytes, endothelial cells, tubular cells and mesangial cells in the kidney.97-100 Thus, a combination of GWAS, EWAS, and single-cell or single-nucleus sequencing, along integrative analyses with other omics datasets, has potential to uncover novel new causal genes or loci associated with DKD and determine the role of epigenetic changes in regulating the expression of these genes. Furthermore, controlling the epigenetic changes at these newly identified genes and loci might be a novel therapeutic approach for treating DKD and metabolic memory.
Histone modifications
PTMs of nucleosomal histone proteins in chromatin regulate gene expression and are an important component of epigenetic regulation59 (Fig. 2). Genome-wide profiling of modified histones using chromatin immunoprecipitation (ChIP) sequencing [G] (ChIP-seq) has shown that specific different types of histone modifications are enriched in specific genomic locations such as regulatory regions (enhancers and promoters), transcriptionally-repressed regions, transcriptional start sites, and actively transcribed regions.101 The major histone PTMs are lysine acetylation, methylation, and ubiquitination; serine and threonine phosphorylation; and arginine methylation.59,102
Histone acetylation can unwind and relax chromatin to enhance gene expression by various mechanisms which facilitate accession of transcription factors and cofactors, leading to transcription initiation and elongation through nucleosome remodelling59 (Fig. 2, Fig. 3). In general, histone acetylation is enriched at promoters and enhancers of actively transcribed genes and reduced in repressed genomic regions. By contrast, the location of histone methylation varies. For example, monomethylation, dimethylation or trimethylation of H3 at lysine 4 (H3K4me1/2/3) and dimethylation or trimethylation of H3 at lysine 36 (H3K36me2/3) are enriched at transcriptionally active genome regions, whereas trimethylation of H3 at lysine 9 (H3K9me3), trimethylation of H3 at lysine 27 (H3K27me3), and trimethylation of H4 at lysine 20 (H4K20me3) are enriched in repressed regions,59 and histone monomethylation of H3 at lysine 4 (H3K4me1 is enriched at enhancer regions.103 Active enhancers are marked by enrichment of acetylated H3 at lysine 27 (H3K27ac) along with mediator complexes [G] and transcription factors. Super-enhancers, which are long stretches of enhancers enriched for binding motifs of transcription factors, are cell type-specific; hence, examination of their differential regulation by epigenetic marks could provide a wealth of information about cell-type function alterations in disease states.104 Poised promoters [G] in developmental genes are enriched with bivalent chromatin modifications, for example, both active (H3K4me3) and repressive (H3K27me3) marks which allow dynamic changes in expression (silenced to highly expressed) of developmentally regulated genes in a stage-specific manner.101,102 Although the role of DNA methylation in epigenetic transmission and heredity is generally better accepted than that of histone modifications, some repressive histone PTMs can be epigenetically inherited.105
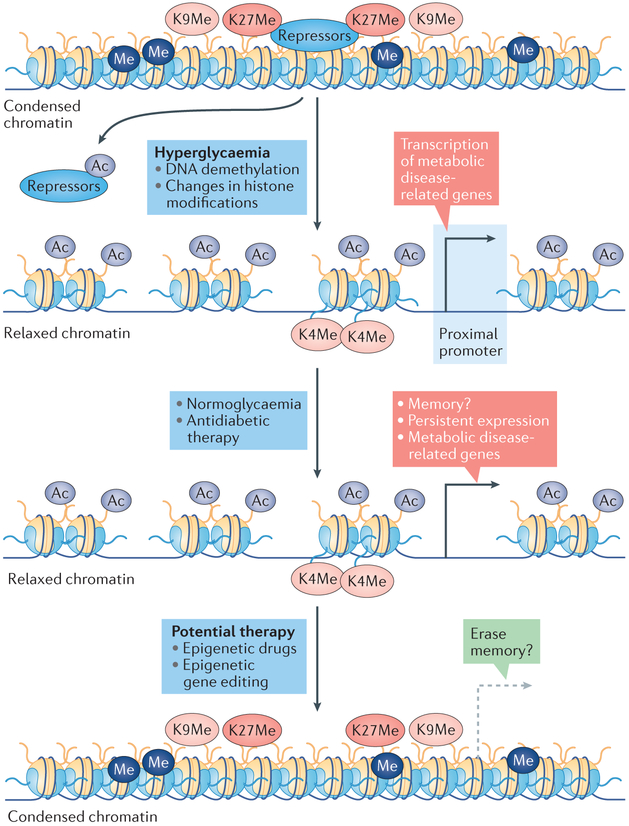
Fig. 3. Epigenetic mechanisms of metabolic memory.
Hyperglycaemia alters histone modifications and DNA methylation status, resulting in a change in chromatin structure from condensed (marked by DNA methylation (Me), methylation of lysine 9 on histone 3 (K9Me), and methylation of lysine 27 on histone 3 (K27Me)), which is associated with repressed gene expression, to relaxed (marked by histone lysine acetylation (Ac) and methylation of lysine 4 on histone 3 (K4Me)) to induce the expression of pathogenic genes. Acetylation relaxes chromatin and enhances gene expression through various mechanisms, including by facilitating the release of transcriptional repressors from condensed chromatin. Once established, the chromatin status (including DNA methylation and histone modification status) is maintained, even after normoglycaemia has been established by administration of insulin or antidiabetic drugs, with persistent expression of pathogenic genes, in a phenomenon called ‘metabolic memory’. This epigenetic memory could potentially be erased using epigenetic drugs or by inducing locus-specific changes in histone modifications or DNA methylation through approaches such as gene editing directed by guide RNAs complementary to a specific locus.
The histone acetyltransferases p300 and CREB-binding protein (CBP) are transcriptional coactivators and catalyse histone acetylation; histone deacetylases (HDACs) and sirtuins can act as transcriptional corepressors and remove acetylation marks from histones. 59,102 Histone lysine methyltransferases (HMTs) catalyse lysine methylation whereas histone lysine demethylases (KDMs) remove methyl groups from histones.59 HMTs and KDMs use specific lysine residues as substrates106 and can also target non-histone proteins. Acetyltransferases and methylases are called epigenetic writers; deacetylases and demethylases are called epigenetic erasers; and proteins such as the bromodomain-containing proteins (for example, bromodomain-containing protein 4), which recognize acetylated proteins at promoters and enhancers and have an active role in gene transcription, are called epigenetic readers.107-109
Studies of histone PTMs at key metabolic genes suggest that their epigenetic regulation have a critical role in the pathogenesis of T1DM, T2DM, obesity, and the metabolic syndrome.65,110,111 These findings indicate that histone PTMs are also involved in diabetic complications — a proposal supported by preclinical studies of DKD. For example, induction of the pro-fibrotic genes Pai1 (which encodes plasminogen activator inhibitor 1) and p21 in cultured rat mesangial cells in response to TGFβ or high glucose is associated with enrichment of H3K9ac and H3K14ac and the presence of p300 and CBP at Smad and SP1 binding sites in the promoters of these genes.112 ChIP assays of glomeruli of diabetic mice showed that increases in Pai1 and p21 expression in vivo were associated with a similar enrichment of H3K9ac and H3K14ac at their promoters. Furthermore, increased expression of Pai1, Col1a1 (which encodes the collagen α−1(I) chain), and Ctgf (which encodes connective tissue growth factor) induced by TGF-β1 was accompanied by enrichment of active histone methylation (H3K4me1/2/3) and decreased repressive marks (H3K9me2/3), at their promoters, together with increased occupancy of the HMT SET7.113 Consistent with these findings, exposure of cultured rat mesangial cells to 12(S)-hydroxyeicosatetraenoic acid, which is increased in DKD, induced similar effects to TGF-β1 on these histone PTMs and pro-fibrotic genes, and the effects of this oxidised lipid on fibrosis were also mediated in part via SET7.114
Changes in the expression of key DKD-associated non-coding RNAs can also be regulated by histone PTMs. TGF-β1 induced histone acetylation (H3K9ac, H3K14ac and H3K27ac) of the promoter region of micro RNA (miR)-192 in renal cells is induces the expression of Smads and the proto-oncogene Ets-1.30,115,116 As discussed below, miR-192 is upregulated by TGF-β1 and has an important role in renal fibrosis and DKD pathogenesis.
In vivo studies in the db/db model of T2DM have demonstrated that several chromatin histone PTMs, occur in the promoters of Pai1 and Ager (which encodes the AGE-specific receptor, RAGE) in glomeruli, together with increased recruitment of the RNA polymerase II suggesting that both active and repressive epigenetic marks cooperate to promote chromatin unwinding and enhance the recruitment of transcription factors to these DKD-associated genes. Of note, administration of the ARB losartan ameliorated key indices of DKD and reduced H3K9ac enrichment at Pai1 and Ager promoters, but did not reverse other diabetes-induced histone PTMs in these mice.117 This incomplete amelioration of epigenetic changes following treatment could be a reason why some patients do not respond well to routine DKD drugs such as ARBs. Combination therapy with conventional therapies and drugs that target epigenetic processes could be a viable option for such patients.24 In another study, increased recruitment of RNA polymerase II and enriched H3K4me2, but diminished H3K27me3, correlated with the expression of DKD-related genes in mouse and rat models of T1DM. However, there were some differences in the expression of a few genes (Mcp-1 and Lamc1) and epigenetic regulators (Ezh2 K27 methylase and the corresponding demethylase) between rats and mice, suggesting some epigenetic changes could be species-specific.118 This aspect should be taken into consideration while translating findings of epigenetic changes/mechanisms in animals to human DKD. Podocyte-specific deletion of the HMT EZH2 decreased H3K27me3 levels at the promoter region of Jag1, which encodes the Notch ligand Jagged1, to increase Jag1 expression, and sensitized mice to glomerular disease in three injury models, including diabetes (db/db T2DM mice) because Jag1 can activate Notch signalling and glomerular disease. 119 By contrast, inhibition of the KDMs Jmjd3 (also known as Kdm6b) and Utx (also known as Kdm6a) increased H3K27me3 levels and attenuated glomerular disease in these models. Lower H3K27me3 levels and higher UTX levels were also detected in glomerular podocytes from humans with DKD, suggesting that JAG1 induction by histone demethylation through UTX1 and to activate Notch signalling in podocytes may also be involved in the development of glomerular dysfunction in human DKD.119
Increased renal inflammation as a result of macrophage infiltration contributes to the pathogenesis of DKD. Histone modifications have been implicated in mediating the activation of NF-κB and other inflammatory factors in vascular cells and monocytes in response to high glucose.120,121 Epigenome profiling studies have demonstrated differences in H3K4me2 and H3K9me2 marks at genes associated with diabetes and inflammation in high glucose-treated monocytes, and in blood monocytes and lymphocytes from patients with T1DM diabetes compared to those in normoglycaemic controls.121-123 The HMT SET7 promotes methylation of H3K4 and coactivates inflammatory genes downstream of NFκB in monocytes.124 Given the above-described role of SET7 in promoting the expression of pro-fibrotic genes in mesangial cells exposed to TGFβ, these findings suggest that this HMT might have a general role in regulating the expression of key pathological genes in various cell types including monocytes, endothelial and mesangial cells associated with diabetic complications, and supports efforts to explore small molecule inhibitors of SET7.125
Together, these data from cell, animal, and clinical models clearly suggest that epigenetic regulation of DKD-related genes through histone PTMs in addition to DNA methylation has a critical role in the pathogenesis and progression of DKD (Fig. 1). Population-based EWAS of histone PTM profiles are less common than those of DNA methylation status because they require relatively large amounts of high quality ChIP-grade antibodies, fresh tissues or blood samples, and larger sample sizes. However, the increased availability of online datasets of enhancers with histone PTM profiles and of the chromatin states of multiple cell types should prove useful for integration with data from RNA sequencing and DNA methylation. Having such integrative and imputed data can significantly aid in interpreting the functional relevance of epigenome changes to the etiology and pathogenesis of DKD for future development of biomarkers and drug targets.
Epigenetics in metabolic memory
Epigenetic mechanisms, including DNA methylation and histone PTMs, have been implicated in the metabolic memory of diabetic complications, including DKD (Fig. 3). Analysis of chromatin and DNA in blood monocytes and lymphocytes of patients with T1DM who participated in the DCCT and EDICs trials126,127 showed significant enrichment of H3K9ac at key inflammatory genes in monocytes from patients who received conventional therapy during DCCT and experienced progression of nephropathy or retinopathy (cases) during subsequent EDIC follow-up, compared with levels in patients who received intensive glycaemic control during DCCT and who did not experience progression of nephropathy and retinopathy (controls) during EDIC.126 Furthermore, this study identified a significant association between HbA1c level and H3K9ac, suggesting that H3K9ac might contribute to metabolic memory in patients with T1DM126 owing to hyperglycemia induced sustained chromatin relaxation at susceptible genomic regions via histone hyperacetylation.
Another study of the DCCT and EDIC cohort127 used the HumanMethylation450 BeadChip to profile DNA methylation in genomic DNA from whole blood taken at DCCT baseline and from blood monocytes obtained ~17 years later during the EDIC phase from the same set of T1DM patients. After adjustment for multiple co-variates, including age, gender, and cell-type composition, the researchers identified a set of 150–250 regions that were differentially hypomethylated or hypermethylated in cases (patients who received conventional therapy during DCCT and experienced progression of nephropathy or retinopathy by EDIC year 10) and controls (patients who received intensive glycaemic control during DCCT and did not experience progression of nephropathy or retinopathy during EDIC). Notably, 12 differentially methylated loci were common in both whole blood and monocytes, including hypomethylation in cases at the 3’untranslated region (UTR) of TXNIP, which encodes thioredoxin-interacting protein, and is associated with hyperglycaemia and related complications including DKD.127,128 Similarly, exposure of cultured THP1 human monocytes to high glucose induced persistent hypomethylation at the 3’-UTR of TXNIP. Furthermore, association analyses were performed between DNAme in whole blood and monocytes from each DCCT patient (samples collected ~ 17 years apart) and their individual mean HbA1c levels measured at multiple time points from the start of DCCT to the time of monocyte collection during EDIC years 16/17. This led to the identification of a set of differentially methylated loci (including TXNIP 3’UTR) that depicted similar association with prior HbA1c in both whole blood and monocyte DNA, i.e the association persisted for ~17 years in the same patient. These findings of a persistence of memory in an epigenetic signature and association with prior history of hyperglycemia strongly support a role for epigenetic mechanisms in metabolic memory. Interestingly, hypomethylation of TXNIP at the same site (cg19693031) has also been observed in other cohorts, in association with higher fasting blood glucose levels and lipidaemia129, indicating that metabolic abnormalities and diabetes might promote similar DNA methylation changes at specific genomic loci that in turn lead to sustained complications.
Studies of experimental models also support a role for epigenetic mechanisms in metabolic memory. One study showed that, compared to VSMCs from aortas of non-diabetic heterozygous db/+ mice, VSMCs from diabetic db/db mice showed high expression of inflammatory genes, associated with lower levels of the HMT SUV39H1 and lower levels of promoter H3K9me3 — a repressive mark.130 This difference in histone methylation persisted even after the VSMCs had been cultured for several passages. VSMCs from diabetic db/db mice also showed upregulation of miR-125b, which targets SUV39H1, demonstrating that miRNAs are key regulators of epigenetic factors in metabolic memory.130,131 In another study, endothelial cells cultured for short periods under conditions of high glucose exhibited persistent induction of the NF-κB subunit p65, indicative of inflammation, accompanied by sustained enrichment of H3K4me1 and the corresponding HMT SET7 at the p65 promoter, long after normalization of glucose levels.132,133 In parallel, the researchers observed sustained and persistent increases in the expression of NF-κB-dependent inflammatory genes Ccl2 (also known as Mcp-1) and Vcam-1. These cell culture data demonstrate the existence of metabolic memory mediated by epigenetic mechanisms in cultured cells, and are supported by in vivo findings from studies of glucose-infused normal mice and in diabetic mice prone to atherosclerosis.132-135 Similar evidence of metabolic memory have also been reported in experimental models of diabetic retinopathy and cardiovascular disease, 136 suggesting that metabolic memory is an important component of the epigenetic changes that occur in diabetes, and that it has consequences for disease outcomes and response to treatment.
RNA modifications in DKD
In addition to DNA, RNA can also be epigenetically modified (Fig. 2). RNA methylation regulates gene expression by altering RNA stability and degradation. The methyltransferase complex METTL3–METTL14–WTAP converts adenosine to N6-methyladenosine (m6A) on RNA.137 In human stem cells, activation of SMAD2/3 by TGF-β signalling promotes binding of the m6A methyltransferase complex to transcripts involved in early cell-fate decisions: the rapid downregulation (that is, degradation) of these transcripts enables the timely exit of cells from pluripotency.138 Given the major role of TGF-β1 in DKD, similar epigenetic crosstalk between TGF-β, SMAD2/3, and m6A methyltransferase could be involved in DKD. Indeed, according to the Nephroseq database (https://www.nephroseq.org), the expression of METTL3, METTL14, and WTAP is lower in glomeruli from diabetic mice, patients with CKD, and patients with focal segmental glomerulosclerosis than in healthy control samples, suggesting that negative feedback of m6A methylation could be disrupted resulting in sustained expression of TGF-β and SMAD2/3. This mechanism might also contribute to the persistent expression of genes associated with DKD.
Non-coding RNAs
Data from ENCODE and other consortia show that the majority of the genome is transcribed into RNA, much of which is non-coding.101,139,140 ncRNA includes not only small ncRNAs, such as miRNAs, but also long ncRNAs (lncRNAs), both of which are part of the epigenome and have regulatory functions.
MicroRNAs in DKD
miRNAs are well established epigenetic regulators and modulators of gene expression. These endogenous small ncRNAs are 20–25 nucleotides in length, and typically silence expression of target genes by hybridizing to the 3’UTR of target mRNAs, leading to translational repression and/or mRNA degradation.141 miRNAs are thought to regulate up to 60% of human protein-coding genes. They are generated as relatively long primary transcripts (pri-miRNAs) following transcription by RNA polymerase II in the nucleus, which are then processed to ~70 nucleotide stem-loop structures called precursor miRNAs (pre-miRNAs) by the Drosha–DGCR8 complex. Pre-miRNAs are transported to the cytoplasm and processed to mature miRNAs by Dicer. Genetic deletion of Dicer or Drosha in mice results in severe renal phenotypes, including proteinuria, underscoring the functional relevance of miRNAs in the kidney.142-146 The first miRNA shown to have a functional role in DKD was miR-192, which acts by targeting key repressors to promote the expression of ECM and collagen, and to augment the pro-fibrotic effects of TGF-β1.30,116,147,148 Numerous miRNAs are now thought to regulate key features of DKD, such as podocyte apoptosis, the accumulation of ECM, glomerular and tubular hypertrophy, and fibrosis. The roles of miRNAs in DKD have been reviewed elsewhere.14,149-151 Of note, MeCP2 (methyl CpG binding protein 2) supresses miRNA processing through Drosha–DGCR8 complex152 and modulates miR-25-NOX4 expression related to oxidant stress in diabetic kidney,153 suggesting an interesting connection from DNA methylation to miRNA regulation.
Long non-coding RNAs in DKD.
LncRNAs are long transcripts (>200 nucleotides and up to ~100kb in length) that have many similarities with mRNAs but lack protein-coding (translation) potential.139,154 Increasing evidence suggests that lncRNAs have distinct cellular roles that affect various biological mechanisms and processes, including gene transcription, splicing, mRNA stability, epigenetic regulation, cell cycle control, differentiation, and the immune response.155,156 lncRNAs regulate the expression of local and distal genes through various mechanisms, which include binding and targeting histone modifying complexes to various loci to affect chromatin states and modulate transcription.155-157 lncRNAs also serve as host RNAs for miRNAs, and act as modular molecules with individual domains that enable them to specifically associate with DNA, RNA, and/or proteins to affect gene expression.28,30,31,156-158 Thus, lncRNAs form an integral part of the epigenome. Interest in lncRNAs increased following the discovery that many lncRNAs are dysregulated in human diseases; furthermore, many GWAS studies have observed SNPs in lncRNA loci,155,159 underscoring the potential connection of lncRNAs to disease states, including diabetes and its complications.
Reports showing the involvement of lncRNAs in DKD have emerged only in the past few years.160 A 2009 report showed that key miRNAs (miR-216a and miR-217) with DKD-related functions were induced by TGF-β1 together with their host lncRNA RP23–298H6. 1–001 in mesangial cells.28 Another study in mouse mesangial cells treated with TGF-β1 (mentioned earlier) demonstrated that miR-192 is co-regulated with its host lncRNA CJ241444 through histone acetylation of the lncRNA promoter and the proto-oncogene Ets-1, leading to the enrichment of Smads and up-regulation of both the lncRNA and miR-192.30 Administration of AngII — a peptide hormone that is associated with renal dysfunction, hypertension, and DKD — to cultured VSMCs induced several lncRNAs; including lnc-Ang362, which serves as a host for miR-221 and miR-222, known to promote VSMC proliferation. Findings from this study showed that lnc-Ang362 might drive VSMC proliferation through these miRNAs.161 Another AngII-induced novel lncRNA called Giver is regulated by the nuclear receptor NR4A3 and promotes oxidant stress and inflammation in VSMCs.162 Expression of Giver was increased in AngII infused mice with hypertension, and expression of a human orthologue GIVER was elevated in arteries from patients with hypertension. A further study showed that key lncRNAs induced by AngII overlap with enhancers regulated by AngII in VSMCs; CRISPR–Cas9-mediated deletion of candidate lncRNA-associated enhancers reduced the expression of nearby AngII-regulated genes, demonstrating crosstalk between these two epigenetic systems.163
The lncRNA plasmacytoma variant translocation 1 (PVT1) is also involved in fibrosis and DKD pathogenesis.164 The PVT1 locus was first implicated in diabetes-associated ESRD in a GWAS in 2007.165 Several miRNAs that map to the PVT1 locus, including miRs1204–1208, were subsequently shown to be upregulated in human mesangial cells in response to high glucose conditions, inducing the expression of ECM-related genes.166 Another study found that 21 lncRNAs were upregulated in two models of renal fibrosis but downregulated in Smad3-knockout mice, indicating a role for TGFβ–SMAD signalling in the regulation of these lncRNAs.167 A 2016 study identified lnc-megacluster (lnc-MGC), a lncRNA host for nearly 40 miRNAs in the miR-379 cluster, to promote mesangial cell ECM accumulation and hypertrophy in mouse models of early DKD.31 Expression of lnc-MGC and several of its resident miRNAs were increased in glomeruli of mouse models of T1DM and T2DM, and in cultured mouse mesangial cells exposed to high glucose and TGF-β1 through mechanisms involving the endoplasmic reticulum (ER) stress-related transcription factor, CHOP (C/EBP homologous protein)). miR-379 targets (and therefore represses) Edem3, which encodes an inhibitor of ER stress, whereas miR-494, another miRNA in the miR-379 cluster, targets Atf3, a repressor of CHOP. Thus, upregulation of lnc-MGC and this miR-379 cluster can augment ER stress in DKD, which in turn might lead to persistent expression of lnc-MGC and miR-379 cluster miRNAs, continuing the circuits and potentially contributing to metabolic memory (Fig. 4).31 Notably, a human orthologue of lnc-MGC is also upregulated in human mesangial cells following exposure to high glucose and TGF-β1.31 This finding is important because unlike miRNAs, lncRNAs are generally not well conserved across species, so it suggests that this lncRNA cluster could also be relevant to human DKD.
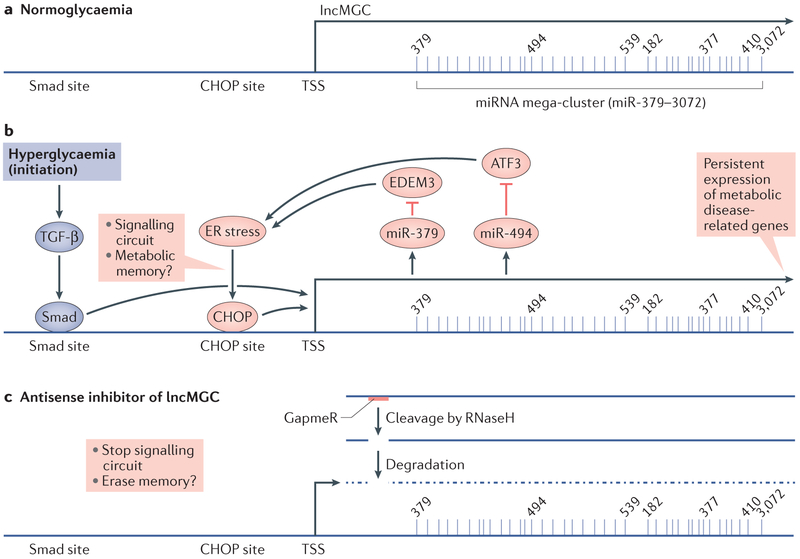
Fig. 4. Contribution of signalling circuits to persistent long non-coding RNA expression.
a ∣ The long noncoding RNA megacluster, lnc-MGC hosts nearly 40 miRNAs within a microRNA cluster called the miR-379 cluster. Smad site and CHOP site are binding sites for SMAD and CHOP transcription factors in the promoter region of lnc-MGC. b ∣ Expression of lnc-MGC, is induced by hyperglycaemia through the actions of TGF-β, which induces the binding of transcription factors such as SMADs. The resultant induction of miR-379 and miR-494 (which are located in the miR-379 cluster) targets and reduces the expression of negative regulators of endoplasmic reticulum (ER) stress, such as EDEM3 and ATF3, which in turn induces activation of the transcription factor CHOP, which in turn increases the expression of lnc-MGC and the miR-379 cluster. This signalling circuit might facilitate the persistent expression of lnc-MGC and the miR-379 cluster even after blood glucose levels have been controlled by antidiabetic agents, providing a potential mechanism for metabolic memory. c ∣ Erasure of this mechanism of metabolic memory might be achieved with use of a locked nucleic acid (LNA)-modified chimeric antisense oligonucleotide (a GapmeR) that targets lnc-MGC. Such an approach would result in cleavage of the lnc-MGC RNA by RNaseH and stop the persistent expression of lnc-MGC and the miR-379 cluster induced by the signalling circuit. TSS, Transcription start site.
Podocyte foot process effacement, apoptosis, and dysfunction are key features of proteinuria and DKD. In mouse podocytes, the lncRNA Tug1 was shown to regulate changes in mitochondrial bioenergetics associated with DKD via interactions with PPAR gamma coactivator α (PGC-1α), and subsequent effects on key PGC-1α targets that are important for mitochondrial biogenesis. Mice with diabetes showed downregulation of Tug1; however, overexpression of Tug1 in mice was sufficient to reverse the negative impact of diabetes on mitochondrial function and ameliorate DKD168. These data suggest that decrease of Tug1 in podocytes in diabetes reduces mitochondrial function and biogenesis due to down-regulation of PGC-1α target genes.
Several reports have described crosstalk between lncRNAs and miRNAs in the pathogenesis of DKD.169,170 For example, the lncRNA Erbb4-IR promotes diabetic kidney injury by hybridizing and inhibiting miR-29b, which targets genes involved in collagen production and ECM accumulation.169 Another lncRNA, downregulation of Lin01619 promotes oxidative stress and podocyte injury in DKD through reduced sequestering of miR-27a, which inhibits forkhead transcriptional factor Foxo1 and induces ER stress.170 The lncRNA 1700020I14Rik reduces cell proliferation and fibrosis in DKD through interactions with the miR-34a-5p–Sirt1–HIF-1α signaling pathway.171 Finally, the lncRNA MALAT1 controls renal tubular epithelial pyroptosis [G] in experimental models of DKD by inhibiting the expression of miR-23c, leading to increased expression of its gene target Elavl1 which induce pro-inflammatory programmed cell death in tubular epithelial cells related to tubular injury, 172 further highlighting the numerous mechanisms by which lncRNAs induce tissue injury in DKD.
As mentioned earlier, infiltration of inflammatory cells and inflammation have important roles in DKD. Transcriptional profiling of blood cell samples from patients with T2DM has revealed several dysregulated lncRNAs with potential roles in inflammation and insulin resistance.173 RNA sequencing of bone marrow macrophages from obese, diabetic db/db mice showed differential expression of several lncRNAs compared to those of control non-diabetic db/+ mice, including increased expression of E330013P06 , a pro-inflammatory lncRNA that enhances the formation of foam cells [G].174 Levels of E330013P06 were also higher in macrophages from mice fed a high-fat diet and in monocytes from humans with T2DM than in chow-fed and non-diabetic counterparts, respectively. A 2018 study showed that another pro-inflammatory lncRNA (Dnm3os) was similarly upregulated in macrophages of obese and diabetic mice, and in monocytes of humans with T2DM.175 Dnm3os binds to nucleolin, a major nucleolar protein of eukaryotic cells; the available evidence indicates this interaction is disrupted under diabetic conditions.175 Disruption of Dnm3os –nucleolin binding enables Dnm3os to enhance histone acetylation (H3K9ac) at promoters of key inflammatory genes, increasing their transcription and promoting macrophage dysfunction.175 Human macrophage lncRNAs have also been associated with cardiometabolic disorders,176 indicating that macrophages in the circulation might represent a non-invasive source for the identification of lncRNAs associated with inflammation and DKD.
Together, these reports highlight not only the involvement of lncRNAs in the pathogenesis of DKD, but also illustrate crosstalk between epigenetic layers (that is, between lncRNAs, miRNAs and chromatin), and provide insights into the signalling circuits these factors trigger to mediate fibrotic events, inflammation, and changes in mitochondrial bioenergetics associated with DKD.15,28,31,151,168,177. We expect that future studies will uncover additional lncRNAs related to DKD, including causal SNPs in lncRNA loci.
Epigenetic biomarkers
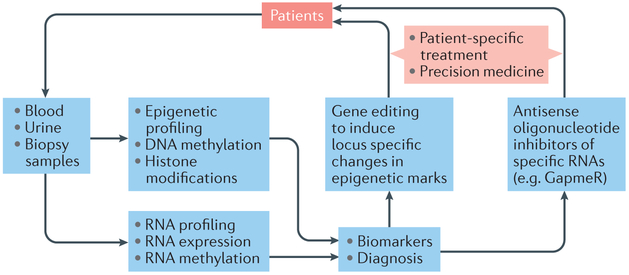
Early detection of DKD is important for preventing progression to renal failure; thus, predictive biomarkers could have a major clinical impact in the diagnosis and treatment of DKD. Several biomarkers of DKD progression have been reported178 with most being proteins, peptides, growth factors, and cytokines. As discussed earlier, EWAS have also identified epigenetic signatures of specific DNA methylation sites and histone PTMs that are associated with key DKD clinical phenotypes, including eGFR, albuminuria, and aberrant renal morphology. Such signatures could be developed as predictive biomarkers of DKD development as has been done in other fields. For example, the methylation status of the gene that encodes insulin has been exploited as a biomarker to examine pancreatic beta cell loss,179 and tracking methylation patterns in circulating DNA can be used to identify tissue-specific cell death.180 As podocytes are observed in the urine of patients with DKD, methylation of podocyte-specific genes in urine could serve as a biomarker of podocyte dysfunction and death. Combining traditional biomarkers (such as urinary albumin, serum cystatin C, serum and urine neutrophil gelatinase associated lipocalin (NGAL), urine kidney injury molecule 1 (KIM-1), and urine transferrin ) with novel biomarkers, including circulating proteins, DNA and ncRNAs, could greatly enhance the early detection of renal function decline in patients with diabetes mellitus.
miRNAs in biofluids (that is, urine, serum and plasma) are already being investigated as non-invasive epigenetic biomarkers of various diseases including DKD, and remain a very active area of translational research.14,151 These ncRNAs are of particular interest as diagnostic biomarkers of DKD owing to their inherent stability in exosomes [G] and biofluids and availability of methods for their detection and quantification. A number of reports have characterized profiles of key miRNAs in urine, urinary sediment, and plasma of patients with various kidney diseases, showing varying levels of correlation with nephropathy or fibrosis.181-187 Urinary exosomes, which originate from most renal cells, can be extremely valuable for miRNA profiling in renal disorders.186,188 Urinary exosomes from patients with T1DM and microalbuminuria, and mesangial cell-derived exosomes from cultured mesangial cells exposed to high glucose levels showed enrichment for miR-145 which is associated with inflammatory pathways and thus might be a potential biomarker for DKD.189 Of note, however, the heterogeneity of extracellular vesicles, including exosomes, and the diversity of circulating extracellular RNAs are poorly understood, as are the mechanisms by which they elicit cellular signalling and communication. These limitations have hampered progress in this field and caution must therefore be taken when evaluating the potential of exosomes and their contents as biomarkers of disease.190
A study that profiled over 700 miRNAs in the urine of 40 patients with T1DM and various stages of DKD provided strong support for the use of miRNA profiles as molecular predictors of DKD.191 Other studies have profiled miRNAs in urine, urinary sediment, and serum from patients with differing degrees of DKD, fibrosis, renal function decline and albuminuria, 181,184,187,189,192-194. For example, one report suggested that TGF-β1-regulated circulating miRNAs can be used as biomarkers of renal function decline and rapid progression to ESRD in patients with T1DM.193 Another study reported tissue-specific miRNA expression patterns for four types of kidney disease —namely, DKD, focal segmental glomerulosclerosis, IgA nephropathy, and membranoproliferative glomerulonephritis, suggesting such patterns could be used as biomarkers for these diseases and provide clues as to the underlying pathological mechanisms.195
Of note, as miRNAs and DNA are relatively stable in paraffin-embedded sections, archived pathology sections represent a rich resource for the future development of epigenetic biomarkers. Methods to assess circulating lncRNAs in biofluids are also under development and the utility of lncRNAs as biomarkers of disease is expected to be another active area of research. For example, one study demonstrated the expression of the lncRNA NR_033515 to be significantly higher in- serum of patients with DKD than in controls and to correlate with the severity of DKD and other diagnostic markers of kidney injury such as KIM-1.196
Translating epigenetic findings in DKD
Targeting epigenetic factors
Currently available therapies for DKD, such as ARBs, show limited efficacy in preventing the progression of renal disease in many patients24. Studies in animal models show that these agents are limited in their ability to inhibit or reverse epigenetic changes that occur in the diabetic kidney14,117, suggesting that future therapeutic approaches might require a combination of conventional therapy plus epigenetic agents. The potential reversibility of epigenetic marks provides exciting opportunities for therapeutic intervention.
Some epigenetic drugs are already being used in oncology, neuronal diseases, inflammation, and fibrosis197, including HDAC inhibitors and DNA methylation inhibitors198, suggesting their efficacy could be assessed in DKD. A 2017 computational systems-level network analysis of microarray data from 33 independent HDAC inhibitor studies, revealed enrichment of pathways involved in the metabolic syndrome and diabetes mellitus. 199 Integration of these data with ENCODE ChIP-seq datasets suggested that these HDAC inhibitors suppressed gene targets of the transcription factor EP300, which had previously been implicated in the pathogenesis of diabetes. These in silico data were tested experimentally by treating vascular endothelial cells from a diabetic patient with HDAC inhibitors or EP300 inhibitors which blocked diabetes associated EP300 target genes.199 This type of in silico computational method could be useful to identify new drugs for diabetes and DKD, and also repurpose drugs that are already in use for other diseases. Although early generation HDAC inhibitors have been tested in animal models of DKD with some showing beneficial effects, 200 these agents were generally non-specific; thus, it may be of value to study the efficacy of newer, more selective inhibitors or activators of HDACs. The class III HDAC protein, SIRT1, suppresses AGE-induced expression of pro-fibrotic genes via upregulation of antioxidant gene in glomerular mesangial cells.201 Administration of the SIRT1 agonist BF175 to diabetic OVE26 mice attenuated kidney injury, suggesting that targeting (upregulating) this HDAC might be of therapeutic value.202 Inhibitors of the HMT EZH2 have also been studied, mostly in models of renal fibrosis200,203,204.
As described earlier, AngII-regulated enhancers overlap with key AngII-regulated genes in VSMCs. This study also showed that an inhibitor of the epigenetic reader bromodomain-containing protein 4, called JQ1, which also inhibits enhancer function, inhibited AngII-regulated genes in vitro, as well as AngII-induced hypertension in mice,163 again highlighting the translational potential of targeting epigenetic marks for diabetic vascular complications.
Targeting non-coding RNAs
The finding that several miRNAs are dysregulated in DKD suggests they could represent valuable targets for epigenetic therapies.186 miRNA levels have traditionally been manipulated using antisense miRNAs or miRNA mimics; however, technical advances have led to the development of stable modified nuclease-resistant oligonucleotides that can act as miRNA inhibitors and mimics. These chemical modifications to the oligonucleotides enhance target affinity, nuclease resistance in biofluids and bioavailability, with reduced toxicity. miRNAs that are upregulated can be inactivated using miRNA sponges, antisense oligonucleotides, or through gene editing. One widely used modification is LNA (locked nucleic acid), a high-affinity RNA analogue in which the ribose ring is chemically locked in a relatively rigid conformation by a 2′-O,4′-C methylene bridge. LNA-modified oligonucleotides possess high thermal stability when hybridized with their complementary mRNA targets, as well as superior sensitivity and specificity in detecting corresponding miRNAs.205 Locked nucleic acid (LNA) [G] -modified anti-miRNAs (antagomirs [G]) (antisense oligonucleotides that hybridize with target miRNAs can inhibit specific miRNAs.28,147,206 One such antagomir is being evaluated in clinical trials for the treatment of hepatitis C virus infection.205
In the context of diabetes, administration of a LNA-modified inhibitor of miR-192 to mice with streptozotocin-induced diabetes inhibited both miR-192 and its downstream miRNAs (miR-216a, miR-217, and the miR-200 family), as well as p53 expression in the renal cortex, and reduced key parameters of DKD.28,147,148,207 Several studies have also reported efficacy of miR-21 inhibitors in mouse models of DKD, 208-211 while another study showed that injection of 2’-O-methyl antisense oligonucleotides, which target miR-29c, reduced rates of DKD in diabetic db/db mice.212 The upstream mechanisms that control the expression and/or transcription of miRNAs could also be targeted to prevent or treat DKD. For example, the induction of miR-192 by TGF-β1 is mediated by Smad2/3 and acetylation of Ets-1 and histones by the by the histone acetyltransferase p300 (which is activated by Akt)30, suggesting that miR-192 expression could be therapeutically targeted by inhibiting Akt or histone acetyltransferases to prevent DKD.
Despite the potential of miRNA-targeted therapies, it will be a challenge to avoid undesired off-target effects, especially in a heterogeneous organ such as the kidney. Stable, synthetic, and biodegradable miRNA carriers, as well as optimal delivery methods, are being assessed to ensure efficiency without toxicity.213-216
Beyond miRNAs, a number of studies have demonstrated the value of targeting lncRNAs in DKD. Subcutaneous injection of an LNA-modified chimeric antisense oligonucleotide (GapmeR) that targets the DKD-associated lncRNA megacluster, lnc-MGC, led to accumulation of the GapmeR in the kidneys of diabetic mice, led to reduced expression not only of lnc-MGC, but also key cluster miRNAs hosted within it, and attenuated key features of early DKD. 31A similar GapmeR that targets human lnc-MGC also reduced fibrotic gene expression in human mesangial cells cultured with high glucose or TGF-β mimicking diabetic conditions, suggesting translational potential of this agent for human DKD.31 As described above, miR-379–Edem3 and miR-494–Atf3 signalling can activate circuits that augment ER stress to induce persistent expression of lnc-MGC and miR-379 cluster miRNAs as a potential mechanism of metabolic memory; thus, GapmeR-mediated inhibition of lnc-MGC might confer additional benefits by erasing metabolic memory (Fig. 4).
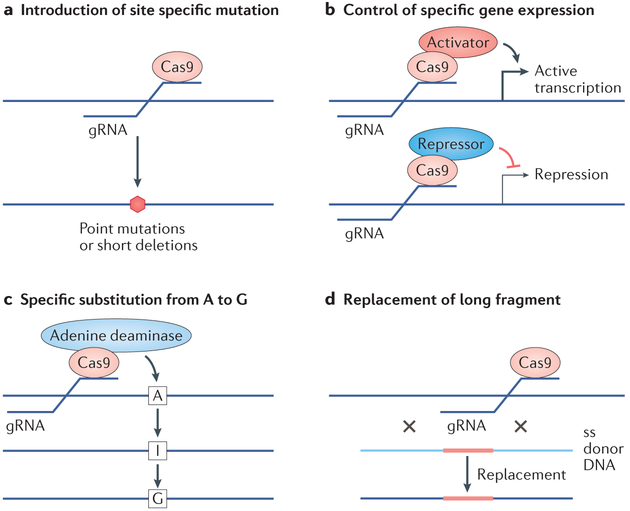
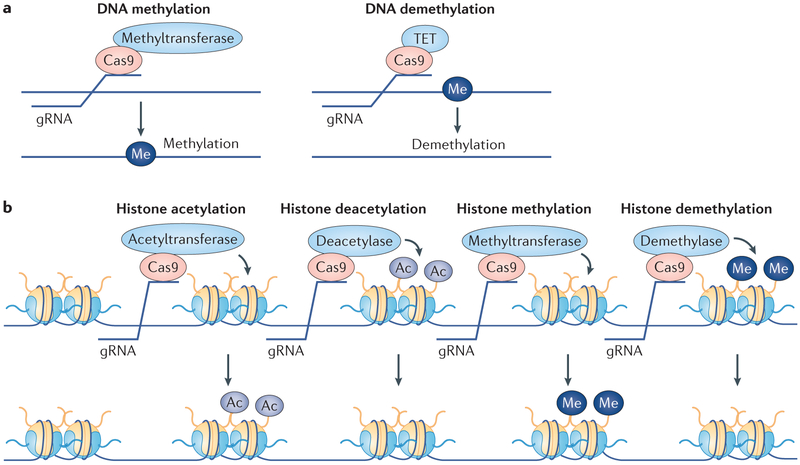
Epigenetic editing
Genome editing via CRISPR–Cas9 and other methods217-220 is a powerful approach to modify genetic and epigenetic changes. The CRISPR–Cas9 system is relatively easier and faster than other genome editing methods. It can be used to introduce site-specific mutations, large deletions and replacements, even in mammalian cells, and to create animal models harbouring such mutations (Fig. 5)221-223. By fusing Cas9 proteins with transcriptional activators or repressors, this system can also be used for the site-specific control of gene activation or repression.224,225 Similarly, fusion of Cas9 with DNA methyltransferases, TETs, or histone modifying enzymes, enables locus-specific alteration of epigenetic marks, DNA methylation, or histone acetylation or methylation, thus enabling erasure or reversal of epigenetic memory (Figs. 3 & 6).226-231In the future, such site-specific epigenetic modifications could be used improve efficacy of pharmacological treatments, and overcome the broad off-target, non-specific genomic effects of currently available epigenetic-modifying drugs.
Fig. 5. Approaches to generate locus-specific genetic changes as a therapy for diabetic kidney disease.
CRISPR-Cas9 is a tool that enables targeted editing of the genome. Briefly, the Cas9 endonuclease is delivered into cells with a guide RNA (gRNA) that is designed to bind Cas9 and the target gene of interest. Following targeted Cas9 cleavage, the DNA is repaired by the cellular repair machinery (through non-homologous end joining or homology-directed repair). The system can be modified to achieve various outcomes, including (a) locus-specific induction of point mutations or short deletions, (b) the control of expression of specific genes (by fusing Cas9 with transcriptional activators or transcriptional repressors), (c) specific substitutions (for example by fusing Cas9 to adenine deaminase, which converts adenine (A) to inosine (I), which pairs with cytosine (C) and will be eventually converted to guanine (G) during replication), and (d) replacement of a long fragment of the genome by homology-directed repair. DNA damage (double-strand breaks) induced by CRISPR-Cas9 system can be repaired by homology-directed repair if the homologous template is close to the site. In this process, if an extra sequence is inserted between left arm and right arm of the homologous template (including extra sequences), the targeted sites can be replaced with the desired extra sequence.
Fig. 6. Locus-specific epigenetic changes as potential therapy for diabetic kidney disease.
In addition to introducing locus-specific genetic changes, the CRISPR-Cas9 system can also be used to introduce locus-specific epigenetic changes, including (a) the induction of DNA methylation (by fusing Cas9 with DNA methyltransferase) or demethylation at specific sites (by fusing Cas9 with ten-eleven translocation (TET) proteins that convert 5-methylcytosine into 5-hydroxymethylcytosine) and (b) the induction of histone modifications, including histone acetylation (by fusing Cas9 with acetyltransferase), histone deacetylation (by fusing Cas9 with histone deacetylase), histone methylation (by fusing Cas9 with histone methyltransferase), and histone demethylation (by fusing Cas9 with histone demethylase). Such changes of epigenetic marks might facilitate the erasure of epigenetic memory and improve long-term outcomes for patients with diabetic kidney disease. gRNA, guide RNA.
Conclusions
DKD pathogenesis is driven by complex interactions between metabolic and haemodynamic factors. Epigenetic mechanisms that might integrate these interactions include DNA and chromatin modifications, miRNAs and lncRNAs. Epigenetics therefore offers new insights into DKD that will complement GWAS data. Long-term persistence of epigenetic modifications and ncRNAs triggered by diabetic stimuli could be key mechanisms underlying metabolic memory. Several HMTs and corresponding histone PTMs, DNA methylation as well as multiple miRNAs and lncRNAs, have been implicated in the expression of fibrotic and inflammatory genes associated with DKD. Approaches for controlling these molecules using small molecules or genetic interventions, including CRISPR-Cas9 editing, might lead to the development of new therapeutics for DKD and could potentially even erase metabolic memory. On-going efforts to evaluate epigenetic changes and ncRNA profiles in archived or freshly obtained biological samples, blood cells, and renal tissues from clinical cohorts of patients with DKD and CKD will lead to the discovery of non-invasive biomarker signatures of renal function decline and DKD. Integrating epigenetic datasets with available genetic patient data using experimental and in silico methods is expected to accelerate the identification of new mediators of DKD progression.
As epigenetics mostly affects gene expression in a cell type-specific manner, challenges exist to the interpretation of epigenome data from heterogenous white blood cell samples and renal biopsy samples. Computational deconvolution approaches to correct for cell-type composition have partially overcome these hurdles,127,232 but additional reference datasets are needed. Research in the area of EWAS in DKD is highly active and expected to further escalate, as better computational deconvolution approaches, multi-omics reference datasets, and human kidney single-cell sequencing datasets become available. This rapidly expanding field is aided by unprecedented technological and scientific advances in the fields of high-throughput sequencing, sophisticated bioinformatics, and gene editing, as well as the wealth of publicly available datasets from the NIH Roadmap Epigenetics Project, ENCODE, and other international consortia. EWAS coupled with GWAS and other omics profiles, including data from enhancer and chromatin accessibility studies,83 will help determine the functional relevance or causality of genetic variants, many of which occur in non-coding regulatory regions, including lncRNAs and enhancers.
Unlike genetics, epigenetic changes are mostly reversible generating exciting opportunities for therapeutic intervention through the use of emerging approaches that enable locus-specific control of epigenetic marks. In addition, epigenetic biomarkers could support precision medicine approaches to optimally stratify patients for effective treatment (Fig. 7). Thus, evaluation of epigenetic and epigenomic alterations is expected to yield new insights into the pathogenesis of DKD and metabolic memory that, in turn, can be translated into novel biomarkers and therapeutic modalities.
Fig. 7. Epigenetic and RNA profiling to support precision medicine.
Epigenetic changes such as DNA methylation and histone modifications in patients represent potential biomarkers for particular renal diseases, including diabetic kidney disease (DKD). Locus-specific correction of such epigenetic changes using the CRISPR-Cas9 system could be a future avenue for patient-specific treatment. Circulating RNAs (including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs)) in biological fluids, such as blood and urine, or biopsy tissue are also potential diagnostic biomarkers for DKD. Synthetic chemical inhibitors of specific RNAs (for example, antisense oligonucleotides (for example, GapmeRs) could also facilitate patient-specific treatment.
Key Points.
Diabetic kidney disease (DKD), involves the accumulation of extracellular matrix, hypertrophy, and fibrosis in kidney glomerular and tubular cells, can lead to end-stage renal disease (ESRD).
Genes associated with DKD are regulated not only by classical signalling pathways and transcription factors, but also by epigenetic mechanisms, including chromatin histone modifications, DNA methylation, and non-coding RNAs.
Metabolic memory is a phenomenon characterized by the persistent expression of DKD-related genes and phenotypes induced by glycaemic exposure, despite subsequent glycaemic control.
Persistent epigenetic changes (DNA methylation and histone modifications) and signalling circuitry mediated by non-coding RNAs may be involved in metabolic memory.
Targeting epigenetic factors (using small molecules or locus-specific epigenetic editing via the CRISPR–Cas9 system) or non-coding RNAs (using chemically-modified antisense oligonucleotides) are potential approaches to prevent or treat DKD and erase metabolic memory.
Epigenetic profiling through epigenome-wide association studies (EWAS) and RNA (transcriptome) profiling of patient samples (blood, urine, and biopsy samples) might facilitate a precision (personalized) medicine approach for the treatment of DKD.
Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health (NIDDK and NHLBI), the Wanek Family Project to Cure Type 1 Diabetes at City of Hope, and the Juvenile Diabetes Research Foundation. The authors thank Sarah T. Wilkinson and Kerin Higa (Office of Faculty and Institutional Support, Beckman Research Institute of City of Hope) for help editing the manuscript before submission.
Glossary
- Non-coding RNA (ncRNA)
An RNA molecule that is not translated into protein; ncRNAs can regulate gene expression at the transcriptional and post-transcriptional levels
- DNA methylation
An epigenetic modification that adds a methyl (CH3) group to DNA, typically at the 5’-cytosine (C) of CpG dinucleotides
- Histone post-translational modifications (PTMs)
Covalent modifications (e.g. methylation, phosphorylation, and acetylation) on histone residues that facilitate epigenetic regulation of gene expression by altering chromatin structure or recruiting chromatin modifiers
- Genomic imprinting
A phenomenon that maternal and paternal chromosomes (or genes) are differentially marked and either maternal or paternal genes is selectively suppressed. DNA methylation is a key mark for genomic imprinting
- Genome-wide association study (GWAS)
A cataloguing of genetic variants across the genomes of multiple individuals that are associated with a specific phenotype
- Epigenome-wide association study (EWAS)
A cataloguing of epigenetics marks (typically focused on DNA methylation) across the genomes of multiple individuals that are associated with a specific phenotype
- Enhancers
Short (50–1500 bp) regions of DNA that can be bound by transcriptional activators to increase the transcription of a particular gene. An enhancer may be located upstream or downstream of the gene it regulates, and does not have to be close to the transcription initiation site
- Epigenotype
Encompasses a set of epigenetic marks (DNA methylation and histone modifications) unique to specific cells or individuals which determines their cell fate or phenotypes (disease susceptibility) respectively
- Expression quantitative trait loci
(eQTLs) are genomic loci that explain the variation in expression levels of mRNAs
- Elongation
A process of copying DNA into the growing RNA chain by RNA polymerase during the elongation phase of transcription
- Alternative splicing
A process which allows multiple mRNAs to be transcribed from a single gene. Usually certain exons of a gene are included or excluded from the final processed mRNA
- Histone deacetylase
Enzymes which remove the acetyl group from histones
- ENCODE (Encyclopedia of DNA Elements)
A public research consortium funded by the National Human Genome Research Institute (NHGRI) to systematically identify all functional elements in the human and mouse genomes
- Methylated DNA immunoprecipitation sequencing (Me-DIP-seq)
A method to identify the profile of genome wide DNA methylation,by sequencing DNA immunoprecipitated with antibody to 5-methylcytosine (5mC)
- Reduced representation bisulfite sequencing (RRBS-seq)
A technique for analyzing the genome-wide DNA methylation profiles with single nucleotide resolution at relatively low cost. It combines restriction enzyme and bisulfite sequencing to enrich (~ 1% of genome) for areas of the genome with a high CpG content
- Whole genome bisulfite sequencing
A gold standard for analyses of DNA methylation of single cytosines at base resolution in the whole genome. It involves next-generation sequencing of bisulfite converted DNA. Sodium bisulfite converts unmethylated cytosine into uracil which eventually will appear as thymine after sequencing. Methylated cytosines appear as cytosine
- Chromatin immunoprecipitation (ChIP) sequencing
A method to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) using specific antibodies to target protein followed high throughput DNA sequencing to identify the binding sites of DNA-associated proteins in a genome-wide fashion.
- Mediator complex
A multiprotein complex that is required for gene transcription by RNA polymerase II and functions as a transcriptional coactivator in eukaryotes. Subunits of the complex relay information from signals and transcription factors to the RNA polymerase II machinery to control the expression of specific genes
- Poised promoters
Promoters which are defined by the simultaneous presence of histone modifications associated with both gene activation and repression (bivalent). They are frequently found at the promoters and enhancers of developmental and lineage specific genes
- Pyroptosis
A process of pro-inflammatory programmed cell death which is also a form of regulated necrosis, a lytic type of cell death inherently associated with inflammation
- Foam cells
Lipid-laden macrophages that serve as the hallmark of early stage atherosclerotic lesion formation
- Exosomes
Cell-derived vesicles that are present in biofluids including blood, urine, cerebrospinal fluid and conditioned medium of cultured cells. The diameter of exosomes is between 30 and 100 nm
- Locked nucleic acid (LNA)
A modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2′ oxygen and 4′ carbon. The bridge “locks” the ribose in the 3′-endo conformation, which is often found in the A-form duplexes. LNA nucleotides are very stable and have higher affinity to the target DNA or RNA residues, hybridizing with them according to Watson-Crick base-pairing rules
- Antagomirs
A small synthetic RNA that is designed to silence endogenous miRNAs
Footnotes
Competing interests
The authors declare no competing interests.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Databases
Nephroseq database (https://www.nephroseq.org)
References:
- 1.Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4·4 million participants. The Lancet 387, 1513–1530, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmet PZ, Magliano DJ, Herman WH & Shaw JE Diabetes: a 21st century challenge. The Lancet Diabetes & Endocrinology 2, 56–64, (2014). [DOI] [PubMed] [Google Scholar]
- 3.Nathan DM Long-term complications of diabetes mellitus. N Engl J Med 328, 1676–1685, (1993). [DOI] [PubMed] [Google Scholar]
- 4.Rask-Madsen C & King GL Vascular Complications of Diabetes: Mechanisms of Injury and Protective Factors. Cell Metabolism 17, 20–33, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes JM & Cooper ME Mechanisms of Diabetic Complications. Physiol Rev 93, 137–188, (2013). [DOI] [PubMed] [Google Scholar]
- 6.Shah MS & Brownlee M Molecular and Cellular Mechanisms of Cardiovascular Disorders in Diabetes. Circ Res 118, 1808–1829, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathan DM Diabetes: Advances in Diagnosis and Treatment. JAMA 314, 1052–1062, (2015). [DOI] [PubMed] [Google Scholar]
- 8.Gregg EW et al. Changes in Diabetes-Related Complications in the United States, 1990–2010. New England Journal of Medicine 370, 1514–1523, (2014). [DOI] [PubMed] [Google Scholar]
- 9.Alicic RZ, Rooney MT & Tuttle KR Diabetic Kidney Disease. Challenges, Progress, and Possibilities 12, 2032–2045, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fineberg D, Jandeleit-Dahm KAM & Cooper ME Diabetic nephropathy: diagnosis and treatment. Nat Rev Endocrinol 9, 713–723, (2013). [DOI] [PubMed] [Google Scholar]
- 11.Jones CA et al. Epidemic of end-stage renal disease in people with diabetes in the United States population: do we know the cause? Kidney Int 67, 1684–1691, (2005). [DOI] [PubMed] [Google Scholar]
- 12.Park CW Diabetic kidney disease: from epidemiology to clinical perspectives. Diabetes & metabolism journal 38, 252–260, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanwar YS, Sun L, Xie P, Liu FY & Chen S A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annual review of pathology 6, 395–423, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato M & Natarajan R Diabetic nephropathy[mdash]emerging epigenetic mechanisms. Nat Rev Nephrol 10, 517–530, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng X.-m., Nikolic-Paterson DJ & Lan HY TGF-β: the master regulator of fibrosis. Nature Reviews Nephrology 12, 325, (2016). [DOI] [PubMed] [Google Scholar]
- 16.Reidy K, Kang HM, Hostetter T & Susztak K Molecular mechanisms of diabetic kidney disease. The Journal of clinical investigation 124, 2333–2340, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo VE, Martienssen RA & Riggs AD Epigenetic mechanisms of gene regulation. (Cold Spring Harbor Laboratory Press, 1996). [Google Scholar]
- 18.Berger SL, Kouzarides T, Shiekhattar R & Shilatifard A An operational definition of epigenetics. Genes & Development 23, 781–783, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keating ST, van Diepen JA, Riksen NP & El-Osta A Epigenetics in diabetic nephropathy, immunity and metabolism. Diabetologia 61, 6–20, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Susztak K Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol 25, 10–17, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S, Jim B & Ziyadeh FN Diabetic nephropathy and transforming growth factor-beta: transforming our view of glomerulosclerosis and fibrosis build-up. Seminars in nephrology 23, 532–543, (2003). [DOI] [PubMed] [Google Scholar]
- 22.Qian Y, Feldman E, Pennathur S, Kretzler M & Brosius FC 3rd. From fibrosis to sclerosis: mechanisms of glomerulosclerosis in diabetic nephropathy. Diabetes 57, 1439–1445, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagtalunan ME et al. Podocyte loss and progressive glomerular injury in type II diabetes. The Journal of clinical investigation 99, 342–348, (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruggenenti P, Cravedi P & Remuzzi G The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol 6, 319–330, (2010). [DOI] [PubMed] [Google Scholar]
- 25.Abboud HE Role of platelet-derived growth factor in renal injury. Annual review of physiology 57, 297–309, (1995). [DOI] [PubMed] [Google Scholar]
- 26.Sharma K & Ziyadeh FN Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes 44, 1139–1146, (1995). [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T, Nakamura T, Noble NA, Ruoslahti E & Border WA Expression of transforming growth factor beta is elevated in human and experimental diabetic nephropathy. Proc Natl Acad Sci U S A 90, 1814–1818, (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato M et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11, 881–889, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M et al. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol 17, 3325–3335, (2006). [DOI] [PubMed] [Google Scholar]
- 30.Kato M et al. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal 6, ra43, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato M et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nature communications 7, 12864, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sedeek M et al. Oxidative stress, Nox isoforms and complications of diabetes--potential targets for novel therapies. J Cardiovasc Transl Res 5, 509–518, (2012). [DOI] [PubMed] [Google Scholar]
- 33.Inagi R, Ishimoto Y & Nangaku M Proteostasis in endoplasmic reticulum[mdash]new mechanisms in kidney disease. Nat Rev Nephrol 10, 369–378, (2014). [DOI] [PubMed] [Google Scholar]
- 34.Kropski JA & Blackwell TS Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. The Journal of clinical investigation 128, 64–73, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brownlee M Biochemistry and molecular cell biology of diabetic complications. Nature 414, 813–820, (2001). [DOI] [PubMed] [Google Scholar]
- 36.Deshpande S et al. Reduced Autophagy by a microRNA-mediated Signaling Cascade in Diabetes-induced Renal Glomerular Hypertrophy. Scientific reports 8, 6954, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komorowsky CV, Brosius FC 3rd, Pennathur S & Kretzler M Perspectives on systems biology applications in diabetic kidney disease. J Cardiovasc Transl Res 5, 491–508, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brosius FC 3rd & Alpers CE New targets for treatment of diabetic nephropathy: what we have learned from animal models. Current opinion in nephrology and hypertension 22, 17–25, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woroniecka KI et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 60, 2354–2369, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brosius FC, Tuttle KR & Kretzler M JAK inhibition in the treatment of diabetic kidney disease. Diabetologia 59, 1624–1627, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuttle KR et al. JAK1/JAK2 inhibition by baricitinib in diabetic kidney disease: results from a Phase 2 randomized controlled clinical trial. Nephrology Dialysis Transplantation, gfx377–gfx377, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pena MJ et al. The effects of atrasentan on urinary metabolites in patients with type 2 diabetes and nephropathy. Diabetes, Obesity and Metabolism 19, 749–753, (2017). [DOI] [PubMed] [Google Scholar]
- 43.Nathan DM et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med 353, 2643–2653, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. Jama 290, 2159–2167, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Boer IH et al. Intensive Diabetes Therapy and Glomerular Filtration Rate in Type 1 Diabetes. New England Journal of Medicine 365, 2366–2376, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Boer IH & Group, f. t. D. E. R. Kidney Disease and Related Findings in the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study. Diabetes care 37, 24–30, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nathan DM The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study at 30 Years: Overview. Diabetes care 37, 9–16, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colagiuri S, Cull CA & Holman RR Are Lower Fasting Plasma Glucose Levels at Diagnosis of Type 2 Diabetes Associated With Improved Outcomes?: U.K. Prospective Diabetes Study 61. Diabetes care 25, 1410–1417, (2002). [DOI] [PubMed] [Google Scholar]
- 49.Thomas MC, Groop PH & Tryggvason K Towards understanding the inherited susceptibility for nephropathy in diabetes. Current opinion in nephrology and hypertension 21, 195–202, (2012). [DOI] [PubMed] [Google Scholar]
- 50.Sandholm N et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS genetics 8, e1002921, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandholm N et al. The Genetic Landscape of Renal Complications in Type 1 Diabetes. Journal of the American Society of Nephrology 28, 557–574, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Zuydam NR et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes 67, 1414–1427, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper ME & El-Osta A Epigenetics: mechanisms and implications for diabetic complications. Circulation research 107, 1403–1413, (2010). [DOI] [PubMed] [Google Scholar]
- 54.Jirtle RL & Skinner MK Environmental epigenomics and disease susceptibility. Nat Rev Genet 8, 253–262, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feinberg A The Key Role of Epigenetics in Human Disease. N Engl J Med 379, 400–401, (2018). [DOI] [PubMed] [Google Scholar]
- 56.Rakyan VK, Down TA, Balding DJ & Beck S Epigenome-wide association studies for common human diseases. Nature Reviews Genetics 12, 529, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu C et al. Renal compartment–specific genetic variation analyses identify new pathways in chronic kidney disease. Nature medicine 24, 1721–1731, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kouzarides T Chromatin Modifications and Their Function. Cell 128, 693–705, (2007). [DOI] [PubMed] [Google Scholar]
- 60.Jones PA Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13, 484–492, (2012). [DOI] [PubMed] [Google Scholar]
- 61.Portela A & Esteller M Epigenetic modifications and human disease. Nat Biotech 28, 1057–1068, (2010). [DOI] [PubMed] [Google Scholar]
- 62.Simmons R Epigenetics and maternal nutrition: nature v. nurture. The Proceedings of the Nutrition Society 70, 73–81, (2011). [DOI] [PubMed] [Google Scholar]
- 63.Woroniecki R, Gaikwad AB & Susztak K Fetal environment, epigenetics, and pediatric renal disease. Pediatric nephrology (Berlin, Germany) 26, 705–711, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wanner N et al. DNA Methyltransferase 1 Controls Nephron Progenitor Cell Renewal and Differentiation. Journal of the American Society of Nephrology, ASN.2018070736, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosen ED et al. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes 67, 1923–1931, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rakyan VK et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS genetics 7, e1002300, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davegardh C, Garcia-Calzon S, Bacos K & Ling C DNA methylation in the pathogenesis of type 2 diabetes in humans. Molecular metabolism 14, 12–25, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schaefer M et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 24, 1590–1595, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tahiliani M et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 324, 930–935, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wanner N & Bechtel-Walz W Epigenetics of kidney disease. Cell and tissue research 369, 75–92, (2017). [DOI] [PubMed] [Google Scholar]
- 71.Bechtel W et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med 16, 544–550, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tampe B et al. Tet3-Mediated Hydroxymethylation of Epigenetically Silenced Genes Contributes to Bone Morphogenic Protein 7-Induced Reversal of Kidney Fibrosis. Journal of the American Society of Nephrology 25, 905–912, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hasegawa K et al. Renal tubular Sirt1 attenuates diabetic albuminuria by epigenetically suppressing Claudin-1 overexpression in podocytes. Nature medicine 19, 1496–1504, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayashi K et al. KLF4-dependent epigenetic remodeling modulates podocyte phenotypes and attenuates proteinuria. The Journal of clinical investigation 124, 2523–2537, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marumo T et al. Diabetes Induces Aberrant DNA Methylation in the Proximal Tubules of the Kidney. J Am Soc Nephrol 26, 2388–2397, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma I, Dutta RK, Singh NK & Kanwar YS High Glucose-Induced Hypomethylation Promotes Binding of Sp-1 to Myo-Inositol Oxygenase: Implication in the Pathobiology of Diabetic Tubulopathy. The American journal of pathology 187, 724–739, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L et al. DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int 92, 140–153, (2017). [DOI] [PubMed] [Google Scholar]
- 78.Yang L et al. Effect of TET2 on the pathogenesis of diabetic nephropathy through activation of transforming growth factor beta1 expression via DNA demethylation. Life Sci 207, 127–137, (2018). [DOI] [PubMed] [Google Scholar]
- 79.Oba S et al. Aberrant DNA methylation of Tgfb1 in diabetic kidney mesangial cells. Scientific reports 8, 16338, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bell CG et al. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC medical genomics 3, 33, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sapienza C et al. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics 6, 20–28, (2011). [DOI] [PubMed] [Google Scholar]
- 82.Bomotti SM et al. Epigenetic Markers of Renal Function in African Americans. Nursing Research and Practice 2013, 687519, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dirks RA, Stunnenberg HG & Marks H Genome-wide epigenomic profiling for biomarker discovery. Clinical epigenetics 8, 122, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ko YA et al. Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol 14, R108, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smyth LJ, McKay GJ, Maxwell AP & McKnight AJ DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics 9, 366–376, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Swan EJ, Maxwell AP & McKnight AJ Distinct methylation patterns in genes that affect mitochondrial function are associated with kidney disease in blood-derived DNA from individuals with Type 1 diabetes. Diabetic Medicine 32, 1110–1115, (2015). [DOI] [PubMed] [Google Scholar]
- 87.Wing MR et al. DNA methylation profile associated with rapid decline in kidney function: findings from the CRIC study. Nephrol Dial Transplant 29, 864–872, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Block T & El-Osta A Epigenetic programming, early life nutrition and the risk of metabolic disease. Atherosclerosis 266, 31–40, (2017). [DOI] [PubMed] [Google Scholar]
- 89.Gautier JF et al. Kidney Dysfunction in Adult Offspring Exposed In Utero to Type 1 Diabetes Is Associated with Alterations in Genome-Wide DNA Methylation. PloS one 10, e0134654, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chu AY et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nature communications 8, 1286, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qiu C et al. Cytosine methylation predicts renal function decline in American Indians. Kidney International 93, 1417–1431, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y-Z et al. Specific expression network analysis of diabetic nephropathy kidney tissue revealed key methylated sites. Journal of Cellular Physiology 233, 7139–7147, (2018). [DOI] [PubMed] [Google Scholar]
- 93.Houseman EA et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics 13, 86, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu MC & Kuan PF A Guide to Illumina BeadChip Data Analysis. Methods Mol Biol 1708, 303–330, (2018). [DOI] [PubMed] [Google Scholar]
- 95.Goodwin S, McPherson JD & McCombie WR Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics 17, 333, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stuart T & Satija R Integrative single-cell analysis. Nat Rev Genet, (2019). [DOI] [PubMed] [Google Scholar]
- 97.Park J et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu H, Kirita Y, Donnelly EL & Humphreys BD Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. Journal of the American Society of Nephrology, ASN.2018090912, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karaiskos N et al. A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. Journal of the American Society of Nephrology 29, 2060–2068, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu H et al. Single-Cell Transcriptomics of a Human Kidney Allograft Biopsy Specimen Defines a Diverse Inflammatory Response. Journal of the American Society of Nephrology 29, 2069–2080, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.EncodeProjectConsortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou VW, Goren A & Bernstein BE Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet 12, 7–18, (2011). [DOI] [PubMed] [Google Scholar]
- 103.Jin F, Li Y, Ren B & Natarajan R Enhancers: multi-dimensional signal integrators. Transcription 2, 226–230, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hnisz D et al. Super-Enhancers in the Control of Cell Identity and Disease. Cell 155, 934–947, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reinberg D & Vales LD Chromatin domains rich in inheritance. Science 361, 33–34, (2018). [DOI] [PubMed] [Google Scholar]
- 106.Klose RJ & Zhang Y Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol 8, 307–318, (2007). [DOI] [PubMed] [Google Scholar]
- 107.Filippakopoulos P & Knapp S Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13, 337–356, (2014). [DOI] [PubMed] [Google Scholar]
- 108.Marmorstein R & Zhou MM Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harbor perspectives in biology 6, a018762, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao S et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science 327, 1000–1004, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang H & Pollin TI Epigenetics Variation and Pathogenesis in Diabetes. Current diabetes reports 18, 121, (2018). [DOI] [PubMed] [Google Scholar]
- 111.Smith CJ & Ryckman KK Epigenetic and developmental influences on the risk of obesity, diabetes, and metabolic syndrome. Diabetes, metabolic syndrome and obesity : targets and therapy 8, 295–302, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan H et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol 304, F601–613, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun G et al. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 21, 2069–2080, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuan H et al. Epigenetic Histone Modifications Involved in Profibrotic Gene Regulation by 12/15-Lipoxygenase and Its Oxidized Lipid Products in Diabetic Nephropathy. Antioxidants & redox signaling 24, 361–375, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chung AC, Huang XR, Meng X & Lan HY miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21, 1317–1325, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kato M et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104, 3432–3437, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reddy MA et al. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int 85, 362–373, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Komers R et al. Epigenetic changes in renal genes dysregulated in mouse and rat models of type 1 diabetes. Laboratory investigation; a journal of technical methods and pathology 93, 543–552, (2013). [DOI] [PubMed] [Google Scholar]
- 119.Majumder S et al. Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. The Journal of clinical investigation 128, 483–499, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giacco F & Brownlee M Oxidative stress and diabetic complications. Circ Res 107, 1058–1070, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Miao F, Gonzalo IG, Lanting L & Natarajan R In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem 279, 18091–18097, (2004). [DOI] [PubMed] [Google Scholar]
- 122.Miao F et al. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem 282, 13854–13863, (2007). [DOI] [PubMed] [Google Scholar]
- 123.Miao F et al. Lymphocytes from patients with type 1 diabetes display a distinct profile of chromatin histone H3 lysine 9 dimethylation: an epigenetic study in diabetes. Diabetes 57, 3189–3198, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 283, 26771–26781, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Okabe J et al. Endothelial transcriptome in response to pharmacological methyltransferase inhibition. ChemMedChem 9, 1755–1762, (2014). [DOI] [PubMed] [Google Scholar]
- 126.Miao F et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 63, 1748–1762, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen Z et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proceedings of the National Academy of Sciences 113, E3002–E3011, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shalev A Minireview: Thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Molecular endocrinology (Baltimore, Md 28, 1211–1220, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Walaszczyk E et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case–control sample of the Lifelines study. Diabetologia 61, 354–368, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Villeneuve LM et al. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci U S A 105, 9047–9052, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Villeneuve LM et al. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes 59, 2904–2915, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.El-Osta A et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. The Journal of experimental medicine 205, 2409–2417, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tonna S, El-Osta A, Cooper ME & Tikellis C Metabolic memory and diabetic nephropathy: potential role for epigenetic mechanisms. Nat Rev Nephrol 6, 332–341, (2010). [DOI] [PubMed] [Google Scholar]
- 134.Brasacchio D et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58, 1229–1236, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okabe J et al. Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circulation research 110, 1067–1076, (2012). [DOI] [PubMed] [Google Scholar]
- 136.Reddy MA, Zhang E & Natarajan R Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 58, 443–455, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heyn H & Esteller M An Adenine Code for DNA: A Second Life for N6-Methyladenine. Cell 161, 710–713, (2015). [DOI] [PubMed] [Google Scholar]
- 138.Bertero A et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guttman M et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Carninci P et al. The Transcriptional Landscape of the Mammalian Genome. Science 309, 1559–1563, (2005). [DOI] [PubMed] [Google Scholar]
- 141.Bartel DP Metazoan MicroRNAs. Cell 173, 20–51, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Harvey SJ et al. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19, 2150–2158, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ho J et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol 19, 2069–2075, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi S et al. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19, 2159–2169, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nagalakshmi VK et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int 79, 317–330, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhdanova O et al. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int 80, 719–730, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Putta S et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 23, 458–469, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deshpande SD et al. Transforming growth factor-beta-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes 62, 3151–3162, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kato M & Natarajan R MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Annals of the New York Academy of Sciences 1353, 72–88, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Trionfini P & Benigni A MicroRNAs as Master Regulators of Glomerular Function in Health and Disease. J Am Soc Nephrol 28, 1686–1696, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Trionfini P, Benigni A & Remuzzi G MicroRNAs in kidney physiology and disease. Nat Rev Nephrol 11, 23–33, (2015). [DOI] [PubMed] [Google Scholar]
- 152.Cheng TL et al. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev Cell 28, 547–560, (2014). [DOI] [PubMed] [Google Scholar]
- 153.Oh HJ et al. Inhibition of the processing of miR-25 by HIPK2-Phosphorylated-MeCP2 induces NOX4 in early diabetic nephropathy. Scientific reports 6, 38789, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cabili MN et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wapinski O & Chang HY Long noncoding RNAs and human disease. Trends in cell biology 21, 354–361, (2011). [DOI] [PubMed] [Google Scholar]
- 156.Taft RJ, Pang KC, Mercer TR, Dinger M & Mattick JS Non-coding RNAs: regulators of disease. The Journal of pathology 220, 126–139, (2010). [DOI] [PubMed] [Google Scholar]
- 157.Moran VA, Perera RJ & Khalil AM Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40, 6391–6400, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wilusz JE, Sunwoo H & Spector DL Long noncoding RNAs: functional surprises from the RNA world. Genes & development 23, 1494–1504, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Moran I et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell metabolism 16, 435–448, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Leung A, Amaram V & Natarajan R Linking diabetic vascular complications with LncRNAs. Vascular pharmacology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leung A et al. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circulation research 113, 266–278, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Das S et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circulation research 123, 1298–1312, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Das S et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nature communications 8, 1467, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alvarez ML & DiStefano JK Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PloS one 6, e18671, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hanson RL et al. Identification of PVT1 as a Candidate Gene for End-Stage Renal Disease in Type 2 Diabetes Using a Pooling-Based Genome-Wide Single Nucleotide Polymorphism Association Study. Diabetes 56, 975–983, (2007). [DOI] [PubMed] [Google Scholar]
- 166.Alvarez ML, Khosroheidari M, Eddy E & Kiefer J Role of MicroRNA 1207–5P and Its Host Gene, the Long Non-Coding RNA Pvt1, as Mediators of Extracellular Matrix Accumulation in the Kidney: Implications for Diabetic Nephropathy. PloS one 8, e77468, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhou Q et al. Identification of Novel Long Noncoding RNAs Associated with TGF-beta/Smad3-Mediated Renal Inflammation and Fibrosis by RNA Sequencing. The American journal of pathology 184, 409–417, (2014). [DOI] [PubMed] [Google Scholar]
- 168.Long J et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. The Journal of clinical investigation 126, 4205–4218, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sun SF et al. Novel lncRNA Erbb4-IR Promotes Diabetic Kidney Injury in db/db Mice by Targeting miR-29b. Diabetes, (2017). [DOI] [PubMed] [Google Scholar]
- 170.Bai X et al. Long Noncoding RNA LINC01619 Regulates MicroRNA-27a/Forkhead Box Protein O1 and Endoplasmic Reticulum Stress-Mediated Podocyte Injury in Diabetic Nephropathy. Antioxidants & redox signaling 29, 355–376, (2018). [DOI] [PubMed] [Google Scholar]
- 171.Li A et al. LincRNA 1700020I14Rik alleviates cell proliferation and fibrosis in diabetic nephropathy via miR-34a-5p/Sirt1/HIF-1α signaling. Cell death & disease 9, 461, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li X et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Experimental cell research 350, 327–335, (2017). [DOI] [PubMed] [Google Scholar]
- 173.Wang X et al. Aberrant Expression of Long Non-Coding RNAs in Newly Diagnosed Type 2 Diabetes Indicates Potential Roles in Chronic Inflammation and Insulin Resistance. Cell Physiol Biochem 43, 2367–2378, (2017). [DOI] [PubMed] [Google Scholar]
- 174.Reddy MA et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes 63, 4249–4261, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Das S et al. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arterioscler Thromb Vasc Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang H et al. Deep RNA Sequencing Uncovers a Repertoire of Human Macrophage Long Intergenic Noncoding RNAs Modulated by Macrophage Activation and Associated With Cardiometabolic Diseases. Journal of the American Heart Association 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lorenzen JM & Thum T Long noncoding RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol 12, 360–373, (2016). [DOI] [PubMed] [Google Scholar]
- 178.Fassett RG et al. Biomarkers in chronic kidney disease: a review. Kidney Int 80, 806–821, (2011). [DOI] [PubMed] [Google Scholar]
- 179.Husseiny MI, Kaye A, Zebadua E, Kandeel F & Ferreri K Tissue-Specific Methylation of Human Insulin Gene and PCR Assay for Monitoring Beta Cell Death. PloS one 9, e94591, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lehmann-Werman R et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proceedings of the National Academy of Sciences 113, E1826–E1834, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Szeto CC et al. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Disease markers 33, 137–144, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Neal CS et al. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant 26, 3794–3802, (2011). [DOI] [PubMed] [Google Scholar]
- 183.Luk CC et al. Urinary biomarkers for the prediction of reversibility in acute-on-chronic renal failure. Disease markers 34, 179–185, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang Y et al. Urine miRNAs: potential biomarkers for monitoring progression of early stages of diabetic nephropathy. Medical hypotheses 81, 274–278, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ichii O et al. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int 81, 280–292, (2012). [DOI] [PubMed] [Google Scholar]
- 186.DiStefano JK, Taila M & Alvarez ML Emerging roles for miRNAs in the development, diagnosis, and treatment of diabetic nephropathy. Current diabetes reports 13, 582–591, (2013). [DOI] [PubMed] [Google Scholar]
- 187.Cai X et al. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatric nephrology (Berlin, Germany) 28, 1797–1801, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karpman D, Ståhl A. l. & Arvidsson I Extracellular vesicles in renal disease. Nature Reviews Nephrology 13, 545, (2017). [DOI] [PubMed] [Google Scholar]
- 189.Barutta F et al. Urinary Exosomal MicroRNAs in Incipient Diabetic Nephropathy. PloS one 8, e73798, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li K et al. Advances, challenges, and opportunities in extracellular RNA biology: insights from the NIH exRNA Strategic Workshop. JCI insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Argyropoulos C et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PloS one 8, e54662, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Higuchi C et al. Identification of Circulating miR-101, miR-375 and miR-802 as Biomarkers for Type 2 Diabetes. Metabolism: clinical and experimental 64, 489–497, (2015). [DOI] [PubMed] [Google Scholar]
- 193.Pezzolesi MG et al. Circulating TGF-beta1-Regulated miRNAs and the Risk of Rapid Progression to ESRD in Type 1 Diabetes. Diabetes 64, 3285–3293, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Satake E et al. Circulating miRNA Profiles Associated With Hyperglycemia in Patients With Type 1 Diabetes Mellitus. Diabetes 67, 1013–1023, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baker MA et al. Tissue-Specific MicroRNA Expression Patterns in Four Types of Kidney Disease. Journal of the American Society of Nephrology 28, 2985–2992, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gao J, Wang W, Wang F & Guo C LncRNA-NR_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting miR-743b-5p in diabetic nephropathy. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 106, 543–552, (2018). [DOI] [PubMed] [Google Scholar]
- 197.Arrowsmith CH, Bountra C, Fish PV, Lee K & Schapira M Epigenetic protein families: a new frontier for drug discovery. Nature Reviews Drug Discovery 11, 384, (2012). [DOI] [PubMed] [Google Scholar]
- 198.Jones PA, Issa JP & Baylin S Targeting the cancer epigenome for therapy. Nat Rev Genet 17, 630–641, (2016). [DOI] [PubMed] [Google Scholar]
- 199.Rafehi H et al. Systems approach to the pharmacological actions of HDAC inhibitors reveals EP300 activities and convergent mechanisms of regulation in diabetes. Epigenetics 12, 991–1003, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Advani A et al. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. The American journal of pathology 178, 2205–2214, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Huang K et al. Sirt1 resists advanced glycation end products-induced expressions of fibronectin and TGF-beta1 by activating the Nrf2/ARE pathway in glomerular mesangial cells. Free Radic Biol Med 65, 528–540, (2013). [DOI] [PubMed] [Google Scholar]
- 202.Hong Q et al. Increased podocyte Sirtuin-1 function attenuates diabetic kidney injury. Kidney International 93, 1330–1343, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhou X et al. Enhancer of Zeste Homolog 2 Inhibition Attenuates Renal Fibrosis by Maintaining Smad7 and Phosphatase and Tensin Homolog Expression. J Am Soc Nephrol 27, 2092–2108, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhou X et al. Targeting histone methyltransferase enhancer of zeste homolog-2 inhibits renal epithelial-mesenchymal transition and attenuates renal fibrosis. Faseb j, fj201800237R, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lindow M & Kauppinen S Discovering the first microRNA-targeted drug. J Cell Biol 199, 407–412, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Krutzfeldt J et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689, (2005). [DOI] [PubMed] [Google Scholar]
- 207.Kato M et al. A microRNA circuit mediates transforming growth factor-beta1 autoregulation in renal glomerular mesangial cells. Kidney Int 80, 358–368, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhong X et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 56, 663–674, (2013). [DOI] [PubMed] [Google Scholar]
- 209.Chau BN et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Science translational medicine 4, 121ra118, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kölling M et al. Therapeutic miR-21 Silencing Ameliorates Diabetic Kidney Disease in Mice. Molecular Therapy 25, 165–180, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dey N et al. MicroRNA-21 Orchestrates High Glucose-induced Signals to TOR Complex 1, Resulting in Renal Cell Pathology in Diabetes. J Biol Chem 286, 25586–25603, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Long J, Wang Y, Wang W, Chang BH & Danesh FR MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem 286, 11837–11848, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dowdy SF Overcoming cellular barriers for RNA therapeutics. Nature Biotechnology 35, 222, (2017). [DOI] [PubMed] [Google Scholar]
- 214.Crooke ST, Witztum JL, Bennett CF & Baker BF RNA-Targeted Therapeutics. Cell metabolism 27, 714–739, (2018). [DOI] [PubMed] [Google Scholar]
- 215.Rupaimoole R & Slack FJ MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16, 203–222, (2017). [DOI] [PubMed] [Google Scholar]
- 216.Zhou J & Rossi J Aptamers as targeted therapeutics: current potential and challenges. Nature Reviews Drug Discovery 16, 181, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Doudna JA & Charpentier E The new frontier of genome engineering with CRISPR-Cas9. Science 346, (2014). [DOI] [PubMed] [Google Scholar]
- 218.Dominguez AA, Lim WA & Qi LS Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nature Reviews Molecular Cell Biology 17, 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liao H-K et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1–13, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, (2016). [DOI] [PubMed] [Google Scholar]
- 222.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nature Biotechnology 35, 371, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mali P et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology 31, 833, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gilbert Luke A. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kearns NA et al. Functional annotation of native enhancers with a Cas9–histone demethylase fusion. Nature Methods 12, 401, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu XS et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247.e217, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hilton IB et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology 33, 510, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Verma N et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nature Genetics 50, 83–95, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Morita S et al. Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv–TET1 catalytic domain fusions. Nature Biotechnology 34, 1060, (2016). [DOI] [PubMed] [Google Scholar]
- 231.WareJoncas Z et al. Precision gene editing technology and applications in nephrology. Nature Reviews Nephrology 14, 663–677, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cardenas A et al. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics 11, 773–779, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]