Abstract
The Ron proto-oncogene is a human receptor for macrophage-stimulating protein (MSP). The exclusion of exon 11 in alternative splicing generates ΔRON protein that is constitutively activated. Heterogenous ribonucleaoprotein (hnRNP) C1/C2 is one of the most abundant proteins in cells. In this manuscript, we showed that both hnRNP C1 and C2 promoted exon 11 inclusion of Ron pre-mRNA and that hnRNP C1 and hnRNP C2 functioned independently but not cooperatively. Moreover, hnRNP C1 stimulated exon 11 splicing through intron 10 activation but not through intron 11 splicing. Furthermore, we showed that, whereas the RRM domain was required for hnRNP C1 function, the Asp/Glu domain was not. In conclusion, hnRNP C1/C2 promoted exon 11 splicing independently by stimulating intron 10 splicing through RRM but not through the Asp/Glu domain.
Keywords: Exon 11 inclusion, hnRNP C1/C2, Pre-mRNA splicing, Ron proto-oncogene, RRM domain
INTRODUCTION
Pre-mRNA splicing occurs in a large RNA-protein complex called a spliceosome (1). Spliceosome assembly is a stepwise process in which U1 snRNP basepairs with a 5′ splice-site, then U2 snRNP basepairs with a branch-point. Then, the U4/U5/U6 snRNP is loaded into the complex. Through alternative exon inclusion/exclusion, proteins with slightly different, completely different, and opposite functions are produced (2–4).
Ron is a cell surface-located receptor composed of disulfide-linked α and β subunits, which are produced from a 190 kDa single chain precursor by proteolytic cleavage (5). ΔRON is an uncleaved protein isoform lacking 49 amino acids in the extracellular domain, which is produced by the exclusion of exon 11 in alternative spicing procedures (6). For ΔRON is unable to bind the ligand, it must be constitutively activated by tyrosine phosphorylation through intracellular oligomerization (6, 7). SRSF1, SRSF2, and hnRNP A1 have been demonstrated to regulate exon 11 splicing (8–10).
Heterogeneous nuclear ribonucleoprotein (hnRNP) C1/C2 is one of the most abundant proteins in cells (11). hnRNP C1/C2 includes an N-terminal RNA recognition motif (RRM), a basic leucine zipper (bZIP)-like motif (bZLM), and an acidic aspartic acid/glutamic acid-rich (Asp/Glu) domain (UniprotKB -P07910) (12). While RRM plays a minimal role in the overall affinity of hnRNP C1/C2 for RNA, a highly basic 40 amino acid (aa) domain preceding the leucine zipper motif provides high-affinity for RNA (13, 14). An acidic Asp/Glu domain is necessary for homo- or heterotetramer formation of hnRNP C1 and C2 (13). hnRNP C1/C2 inhibits splicing of the Alu element in the human genome by interfering with U2AF65 binding to the cryptic PPT sequence (15).
Here, we show that hnRNP C1/C2 promoted exon 11 inclusion of Ron pre-mRNA through stimulation of intron 10, but not intron 11, splicing. In addition, we found that hnRNP C1 and hnRNP C2 functioned independently, not cooperatively. Importantly, although the RRM domain was required for hnRNP C1/C2 function, the Asp/Glu domain of hnRNP C1 was not necessary for promoting Ron exon 11 splicing.
RESULTS
Reduced hnRNP C1/C2 expression inhibits exon 11 splicing of Ron pre-mRNA
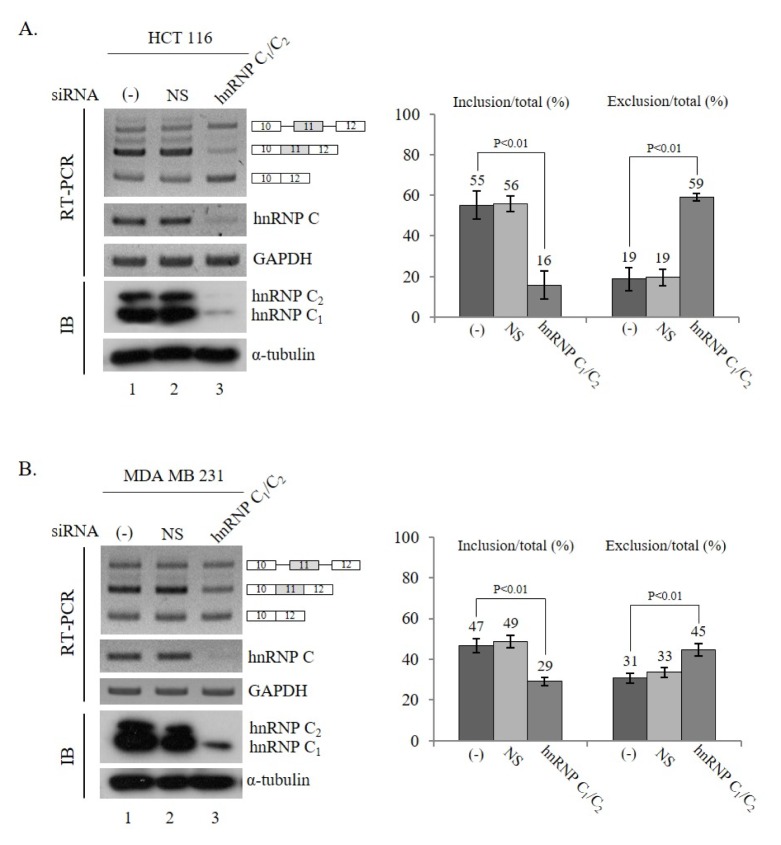
In order to understand the role of hnRNP C1/C2 in the splicing of Ron pre-mRNA, we investigated whether reduced expression of hnRNP C1/C2 affected Ron alternative splicing. Fig. 1A shows that hnRNP C1/C2-targeted shRNA treatment reduced both hnRNP C1 and hnRNP C2 expression analyzed by RT-PCR and immunoblotting analysis (left panel). The results in Fig. 1A show that hnRNP C1/C2-targeting shRNA virus treatment induced a significant decrease in the exon 11-included isoform (~39%) and an increase in the exon 11-skipped isoform (~40%) in HCT 116 cells. Thus, reduced hnRNP C1/C2 expression inhibited exon 11 splicing in HCT 116 cells. Similarly, as shown in Fig. 1B (left panel), shRNA-mediated knockdown also successfully reduced hnRNP C1/C2 expression in MDA MB 231 cells (lane 3) and reduced hnRNP C1/C2 expression suppressed exon 11 inclusion (~18%). Taken together, these results demonstrate that reduced expression of hnRNP C1/C2 inhibited exon 11 inclusion of Ron pre-mRNA.
Fig. 1.
Reduced hnRNP C1/C2 expression inhibits exon 11 splicing of Ron pre-mRNA. RT-PCR analysis of Ron exon 11 splicing was performed with RNAs extracted from untreated, non-silencing shRNA, and hnRNP C1/C2 shRNA lentivirus-treated HCT 116 (A) and MDA MB 231 (B) cells. The exon 11 included, skipped, and unspliced isoforms are shown as indicated. The reduced expression of hnRNP C1/C2 is demonstrated by RT-PCR and immunoblotting using anti-hnRNP C1/C2 antibody, with GAPDH and α-tubulin as controls. The quantitation results of exon 11 inclusion or exclusion in total RNA are shown in the right panel.
hnRNP C1 and C2 promotes exon 11 of Ron pre-mRNA independently
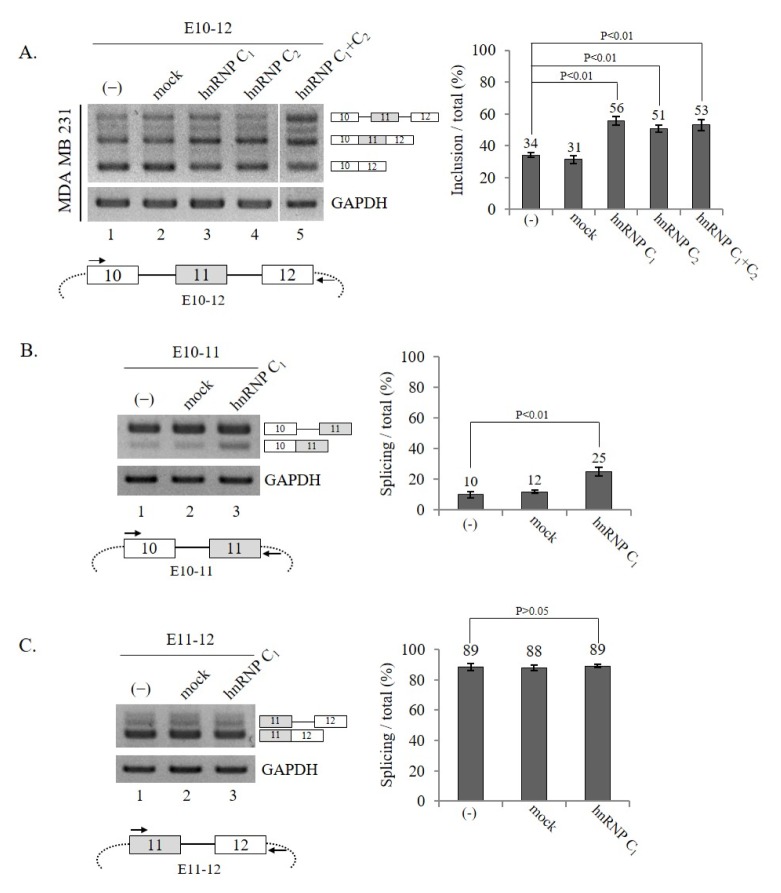
Next, we investigated whether increased expression of hnRNP C1 or hnRNP C2 had the opposite effect of reduced hnRNP C1/C2 expression on exon 11 splicing. In order to address the question, an hnRNP C1 or C2 expression plasmid and Ron exon 10–12 mini-gene were co-transfected into MDA MB 231 cells. Fig. 2A shows that hnRNP C1 and C2 promoted an increase in exon 11 inclusion (~22% or ~17% independently, respectively) to similar levels, which was the opposite effect of hnRNP C1/C2 knockdown (lane 3 and 4). In addition, the results also indicate that the 13 amino acids included only in hnRNP C2 did not play a significant role in this activity.
Fig. 2.
hnRNP C1 and hnRNP C2 promote exon 11 splicing of Ron pre-mRNA independently. (A) (Upper panel) Shown are RT-PCR results of Ron exon 11 splicing from the E10–12 mini-gene in MDA MB 231 cells expressing hnRNP C1, hnRNP C2, or both hnRNP C1 and C2. The quantitation results are shown on the right. (Lower panel) The scheme of Ron mini-gene (E10–12) is shown. The intronic RNA is represented with solid lines and the vector sequence is shown as dot lines. The primers used to detect the mini-gene splicing are shown with arrows. (B, C) hnRNP C1 promotes intron 10 but not intron 11 splicing of Ron pre-mRNA. (B) (Upper panel) RT-PCR analysis of intron 10 splicing from cells expressing hnRNP C1 is shown with GAPDH as a loading control. The quantitation results are shown on the right. (Lower panel) The scheme of the E10–11 mini-gene is shown. The intronic RNA is represented with solid lines and the vector sequence is shown as dotted lines, and the primers used to detect intron 10 splicing are shown as arrows. (C) (Upper panel) RT-PCR analysis of intron 11 splicing is shown, with GAPDH as a loading control. The quantitation results are shown in the right panel. (Lower panel) The scheme of the E11–12 mini-gene is shown. The primers used to detect intron 11 splicing are shown with arrows.
In addition to forming homo-tetramers by themselves, hnRNP C1 and C2 have also been shown to form a hetero-tetramer with a 3:1 ratio (16). We, therefore, asked if hnRNP C1 and C2 could promote Ron exon 11 splicing cooperatively. To answer this question, we introduced both hnRNP C1 and C2 expression plasmids with the Ron mini-gene into cells. The results in Fig. 2A show that the co-expression of hnRNP C1 and C2 had similar effects as the individual expression of either the hnRNP C1 or C2 plasmid (lane 5). Thus, we conclude that these two proteins did not synergistically increase either hnRNP C1 or C2 expression. By combining the results in Fig. 1 and 2A, we conclude that hnRNP C1 and C2 functioned similarly to promote exon 11 inclusion of Ron pre-mRNA and that the 13 amino acids in hnRNP C2 were not required for the function. To simplify our studies, we decided to examine only the roles of hnRNP C1 in this report.
hnRNP C1 promotes intron 10 but not intron 11 splicing of Ron pre-mRNA
We asked if hnRNP C1 affected splicing of intron 10 or intron 11 in Ron pre-mRNA. To detect intron 10 splicing, we applied a mini-gene in which only exon 10 to exon 11 sequences were included (E10–11, lower panel, Fig. 2B). Using this mini-gene, we performed RT-PCR with primer pairs corresponding to exon 10 and the downstream vector sequence. The results in Fig. 2B demonstrate that hnRNP C1 expression increased the intron 10-spliced isoform significantly (~15%, lane 3). Thus, hnRNP C1 promotes intron 10 splicing. We next analyzed intron 11 splicing using another mini-gene, which includes exons 11–12 (E11–12). The primer pairs that corresponded to exon 11 and the vector sequences were used to detect intron 11 splicing (Fig. 2C, lower panel). The results in Fig. 2C show that intron 11 was almost completely spliced in the E11–12 mini-gene (lane 1). Thus, we expect that a further increase in exon 11-spliced products would be hard to detect. The results in Fig. 2C show that intron 11 splicing was not altered by the hnRNP C1 treatment. The results in Fig. 2B and 2C indicate that hnRNP C1 affected intron 10 but not intron 11 splicing. Therefore, we conclude that hnRNP C1 promoted exon 11 inclusion through activation of intron 10 splicing.
Conserved splice-site sequences of exon 11 do not affect hnRNP C1 function
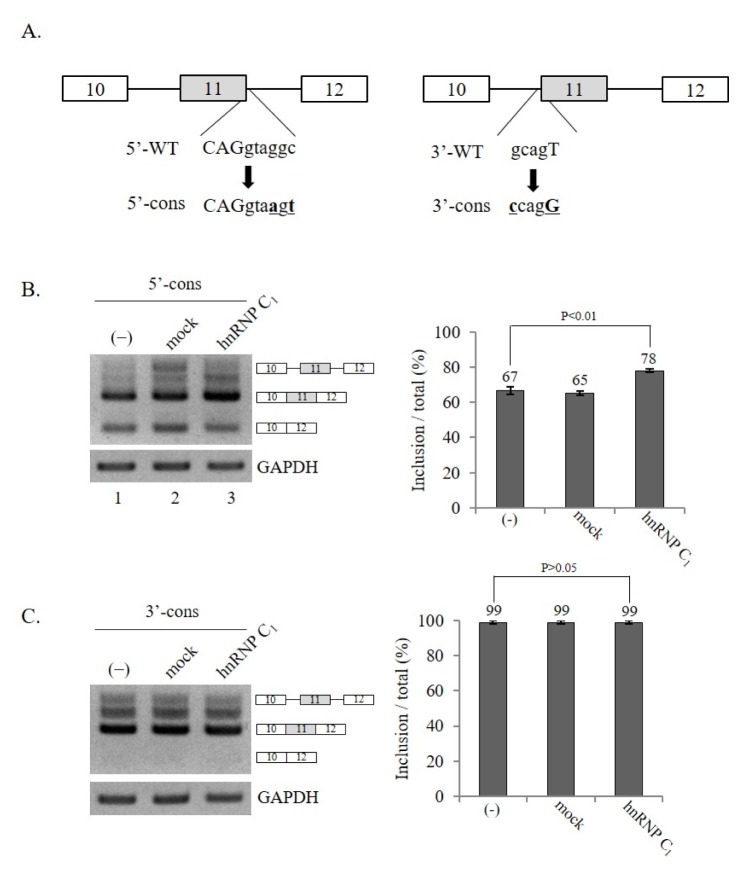
We further asked if splicing sites flanking exon 11 would regulate the effect of hnRNP C1 on the exon 11-inclusion of Ron pre-mRNA. To test this possibility, we used two previously described mutant mini-gene constructs (10) in which either the 5′ splice-site or the 3′ splice-site was mutated into conserved sequences (5′-cons or 3′-cons, Fig. 3A). Consistent with our previous reports, more conserved splice-sites sequences in exon 11 facilitated exon 11 inclusion significantly (10), leading to the predominant production of exon 11-included isoforms by these two mutants (lane 1 of Fig. 3B and 3C). However, the results in Fig. 3B demonstrate that a mutation of the 5′ splice-site in a conserved sequence did not disrupt hnRNP C1 function on Ron exon 11 splicing because hnRNP C1 still promoted exon 11 inclusion in the 5′-cons mutant. In the 3′-cons mutant, since the exon 11-excluded isoform was not detectable, a decrease in the exon 11-skipped form was not observable. Taken together, we conclude that conserved splice-site sequences of exon 11 did not affect the role of hnRNP C1 in exon 11 splicing.
Fig. 3.
Conserved splice-site sequences of exon 11 do not affect hnRNP C1 function. (A) (Left panel) 5′ splice-site sequence of Ron exon 11 (5′-WT) and its conserved splice-site mutant sequence are shown (5′-cons). (Right panel) 3′ splice-site sequence of Ron exon 11 (3′-WT) and its conserved splice-site mutant sequence are shown (3′-cons). (B) RT-PCR analysis of exon 11 splicing using 5′-cons mini-gene in hnRNP C1-expressing cells. GAPDH was used as a loading control. The quantitation of exon 11 inclusion in total RNA is shown. (C) RT-PCR analysis of exon 11 splicing using 3′-cons mini-gene in hnRNP C1-expressing cells. GAPDH was used as a loading control. The quantitation of exon 11 inclusion in total RNA is shown.
The Asp/Glu domain is dispensable for hnRNP C1 function in Ron exon 11 splicing
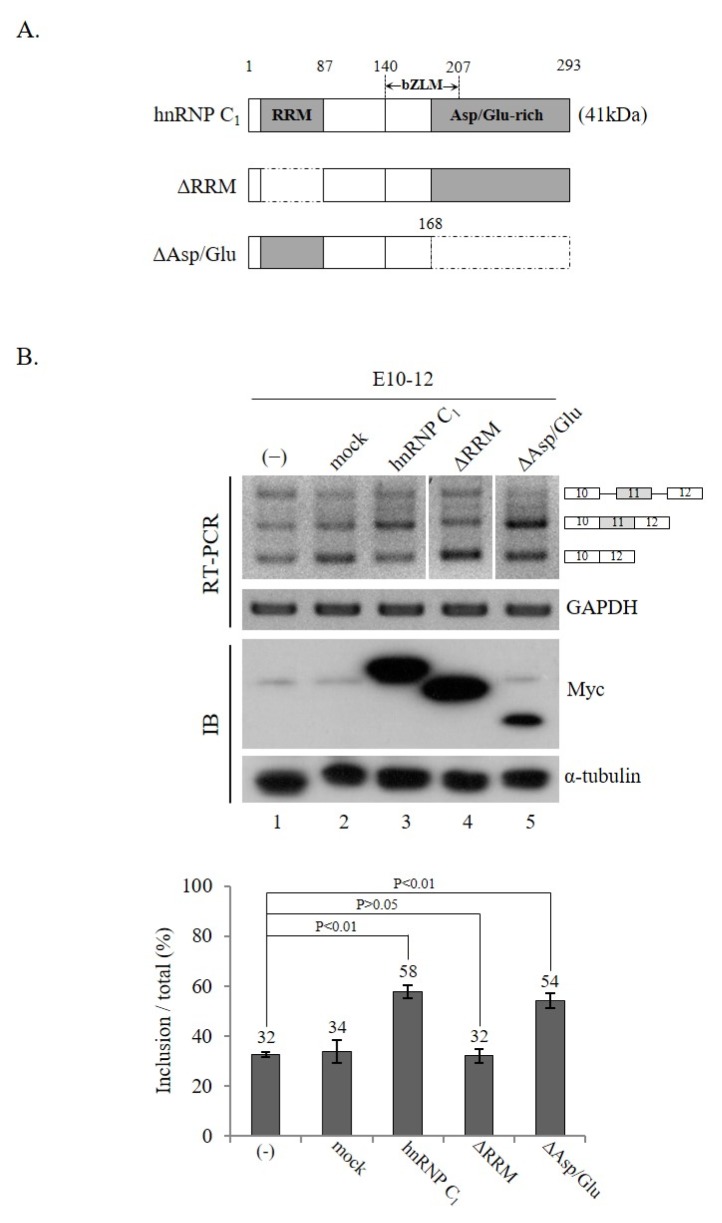
hnRNP C1 includes RRM, bZLM, and Asp/Glu domains. While the RRM domain is required for RNA binding, the Asp/Glu domain is required for the formation of tetramers of hnRNP C1. To determine if these two domains were necessary for hnRNP C1-mediated Ron exon 11 splicing, we produced two hnRNP C1 mutants, in which either the RRM domain or the Asp/Glu domain was deleted (ΔRRM, ΔAsp/Glu) (Fig. 4A). The results in Fig. 4B show that the ΔRRM mutant protein was not able to promote exon 11 inclusion (lane 4). Thus, the RRM domain was required for the role of hnRNP C1 in exon 11 splicing. This was not unexpected because the RRM domain is required for the binding target RNA in exon 10. In contrast, we found that the ΔAsp/Glu mutant of hnRNP C1 was still capable of promoting exon 11 inclusion (lane 5). Therefore, we conclude that the Asp/Glu domain was not needed for the role of hnRNP C1 in Ron exon 11 splicing. The results indicate that tetramer formation was not required for the role of hnRNP C1 in Ron exon 11 alternative splicing.
Fig. 4.
The Asp/Glu domain is dispensable for the function of hnRNP C1 in Ron exon 11 splicing. (A) Scheme of the hnRNP C1 protein domains is shown. RRM and Asp/Glu domain are shown with gray boxes. Parts deleted in the ΔRRM and ΔAsp/Glu mutants are shown in dotted line-framed boxes. (B) RT-PCR analysis of exon 11 splicing using RNAs extracted from cells expressing ΔRRM and ΔAsp/Glu mutant proteins. Expression of myc-tagged wild-type hnRNP C1, ΔRRM, or ΔAsp/Glu mutant protein is confirmed with anti-myc antibody. The quantitation of exon 11 inclusion in total RNA is demonstrated in the lower panel.
DISCUSSION
We previously demonstrated that SRSF2 regulated Ron exon 11 splicing by contacting exon 11 sequences (10). In this study, by using shRNA-mediated knockdown and overexpression, we showed that hnRNP C1/C2 promoted Ron exon 11 splicing by stimulating intron 10 but not intron 11 splicing. Moreover, hnRNP C1 and C2 played roles in Ron exon 11 splicing independently but not synergistically, demonstrated by experiments using cells co-expressing both proteins. Furthermore, the acidic Asp/Glu domain required for tetramer formation was not necessary for the role of hnRNP C1 in Ron splicing.
Ron pre-mRNA splicing is regulated by multiple proteins, including SRSF1, hnRNP A1, and SRSF2, through various sequences on Ron pre-mRNA (8–10). Whereas SRSF1 and hnRNP A1 target exon 12 RNA and SRSF2 targets exon 11 RNA, here, we demonstrated that hnRNP C1 targeted exon 10 sequences. However, in spite of these studies, it is still unclear if SRSF1, hnRNP A1, SRSF2, and hnRNP C1 function cooperatively in Ron exon 11 splicing, spliceosome assembly, and/or splice-site selection. Moreover, how and why these different proteins only target their respective sequences on Ron RNA, but not other potential binding sequences at other locations, need to be answered.
Although the role of hnRNP C1/C2 was demonstrated in an in vitro study (17), their functions in alternative splicing have not yet been elucidated in great detail. In addition, a previous study found that hnRNP C1/C2 was not necessary for viability (18). Thus, it is likely that hnRNP C1/C2 played redundant roles in splicing in the cells. In this study, we presented direct evidence that hnRNP C1/C2 regulated alternative splicing using cells in which hnRNP C1/C2 was either overexpressed or suppressed. Our results, as well as a few other reports, demonstrated that hnRNP C1/C2 was an essential regulatory protein of alternative splicing (19–21). However, previous studies of hnRNP C1/C2 were primarily based on large scale sequencing and screening, therefore, mechanistic insight from those studies was limited. The results in our study showed that reduced hnRNP C1/C2 expression induced a much more significant change in exon 11 splicing compared to hnRNP C1/C2 overexpression. The difference can be explained by the fact that hnRNP C1/C2 is one of the most abundant proteins in cells (11). Thus, shRNA treatment induced a significant decrease in hnRNP C1/C2 expression. Although we used various approaches, we were not able to show that the endogenous Ron exon 11 splicing was affected by hnRNP C1/C2 overexpression. Nonetheless, the transient expression of hnRNP C1/C2, along with the Ron mini-gene, demonstrated that it affected exon 11 splicing but to a much lesser extent than the effect seen from the knockdown.
hnRNP C1 and C2 are able to form homo- or heterotetramers (13). However, it seems that the role of hnRNP C1/C2 in Ron splicing was not required for tetramer formation, based on two pieces of evidence. First, hnRNP C1 and C2 did not function cooperatively in alternative splicing of Ron pre-mRNA, but rather independently. Second, the acidic Asp/Glu domain that is essential for tetramer formation was dispensable in Ron exon 11 splicing. In addition, we showed that the RRM domain was required for the function of hnRNP C1, which was not surprising. However, what was striking is that the long Asp/Glu domain was not necessary for Ron splicing, although it was previously shown to be essential for tetramer formation and that the hnRNP C tetramer was important for mRNA transport (22). Therefore, the role of Asp/Glu in Ron pre-mRNA splicing cannot be established. However, whether the Asp/Glu domain is required for other pre-mRNA splicing is still unknown. It is also possible that the Asp/Glu domain plays regulatory roles in alternative splicing.
MATERIALS AND METHODS
Plasmid construction
The coding region of hnRNP C1, C2 was inserted into a pcDNA6/myc-His A (Invitrogen) plasmid. The ΔAsp/Glu and ΔRRM mutants of hnRNP were produced by overlapping PCR using the hnRNP C1 expression plasmid as a template.
RT-PCR
Total RNA was extracted using RiboEx (GeneAll) as previously described (23). Reverse transcription was performed using 0.5 μg RNA with oligo (dT) primer and ImProm-IITM reverse transcriptase (Promega). The reaction mixture (0.5 μl) was amplified by PCR using G-Taq polymerase (Cosmo Genetech).
Purification of hnRNP C1 protein
Total protein was extracted from HEK293 cells transfected with the pcDNA6/myc-His A-hnRNP C1 plasmid by 30 min incubation with lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 5 mM imidazole, 0.5% Tween-20, and 1 mM PMSF). Prewashed Ni-NTA agarose beads (QIAGEN) were added to the lysates and the mixture was incubated overnight at 4°C in the binding buffer (50 mM NaH2PO4, 500 mM NaCl, 0.5% Tween-20, and 1 mM PMSF). After washing, the hnRNP C1 protein was eluted from the Ni-NTA agarose beads using elution buffer (250 mM imidazole in binding buffer) for 20 min at 4°C.
Knockdown of hnRNP C1/C2 with shRNA
To generate shRNA lentivirus, 293T cells were transfected with an shRNA-harboring plasmid (Open Biosystems) and PSPAX2 and PMD2G helper plasmids using PEI reagent. The media was changed after 12 h and incubated for another 24 h. The lentivirus-containing supernatants were harvested with a 0.45 μm filter. To knock down the hnRNP C1/C2 expression, lentivirus-containing supernatants were added to the cells supplemented with 10 μg/ml polybrene. After 72 h infection, the RNAs were extracted for RT-PCR.
ACKNOWLEDGEMENTS
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (HI17C0196).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Papoff G, Cascino I, Eramo A, Starace G, Lynch DH, Ruberti G. An N-terminal domain shared by Fas/Apo-1 (CD95) soluble variants prevents cell death in vitro. J Immunol. 1996;156:4622–4630. [PubMed] [Google Scholar]
- 3.Droin N, Rebe C, Bichat F, Hammann A, Bertrand R, Solary E. Modulation of apoptosis by procaspase-2 short isoform: selective inhibition of chromatin condensation, apoptotic body formation and phosphatidylserine externalization. Oncogene. 2001;20:260–269. doi: 10.1038/sj.onc.1204066. [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat Med. 1997;3:614–620. doi: 10.1038/nm0697-614. [DOI] [PubMed] [Google Scholar]
- 5.Iwama A, Okano K, Sudo T, Matsuda Y, Suda T. Molecular cloning of a novel receptor tyrosine kinase gene, STK, derived from enriched hematopoietic stem cells. Blood. 1994;83:3160–3169. doi: 10.1182/blood.V83.11.3160.3160. [DOI] [PubMed] [Google Scholar]
- 6.Collesi C, Santoro MM, Gaudino G, Comoglio PM. A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol Cell Biol. 1996;16:5518–5526. doi: 10.1128/MCB.16.10.5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okino T, Egami H, Ohmachi H, et al. Presence of RON receptor tyrosine kinase and its splicing variant in malignant and non-malignant human colonic mucosa. Int J Oncol. 1999;15:709–714. doi: 10.3892/ijo.15.4.709. [DOI] [PubMed] [Google Scholar]
- 8.Ghigna C, Giordano S, Shen H, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Bonomi S, di Matteo A, Buratti E, et al. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013;41:8665–8679. doi: 10.1093/nar/gkt579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon H, Cho S, Loh TJ, et al. SRSF2 promotes splicing and transcription of exon 11 included isoform in Ron proto-oncogene. Biochim Biophys Acta. 20141839:1132–1140. doi: 10.1016/j.bbagrm.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 12.Brunner JE, Nguyen JH, Roehl HH, Ho TV, Swiderek KM, Semler BL. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J Virol. 2005;79:3254–3266. doi: 10.1128/JVI.79.6.3254-3266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAfee JG, Shahied-Milam L, Soltaninassab SR, LeStourgeon WM. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA. 1996;2:1139–1152. [PMC free article] [PubMed] [Google Scholar]
- 14.Gorlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarnack K, Konig J, Tajnik M, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnett SF, Friedman DL, LeStourgeon WM. The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol Cell Biol. 1989;9:492–498. doi: 10.1128/MCB.9.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 18.Williamson DJ, Banik-Maiti S, DeGregori J, Ruley HE. hnRNP C is required for postimplantation mouse development but Is dispensable for cell viability. Mol Cell Biol. 2000;20:4094–4105. doi: 10.1128/MCB.20.11.4094-4105.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konig J, Zarnack K, Rot G, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venables JP, Koh CS, Froehlich U, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izquierdo JM. Heterogeneous ribonucleoprotein C displays a repressor activity mediated by T-cell intracellular antigen-1-related/like protein to modulate Fas exon 6 splicing through a mechanism involving Hu antigen R. Nucleic Acids Res. 2010;38:8001–8014. doi: 10.1093/nar/gkq698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCloskey A, Taniguchi I, Shinmyozu K, Ohno M. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335:1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 23.Moon H, Cho S, Loh TJ, et al. SRSF2 directly inhibits intron splicing to suppresses cassette exon inclusion. BMB Rep. 2017;50:423–428. doi: 10.5483/BMBRep.2017.50.8.103. [DOI] [PMC free article] [PubMed] [Google Scholar]