Abstract
Severe combined immunodeficiency (SCID) is a group of inherited disorders characterized by compromised T lymphocyte differentiation related to abnormal development of other lymphocytes [i.e., B and/or natural killer (NK) cells], leading to death early in life unless treated immediately with hematopoietic stem cell transplant. Functional NK cells may impact engraftment success of life-saving procedures such as bone marrow transplantation in human SCID patients. Therefore, in animal models, a T cell−/B cell−/NK cell+ environment provides a valuable tool for understanding the function of the innate immune system and for developing targeted NK therapies against human immune diseases. In this review, we focus on underlying mechanisms of human SCID, recent progress in the development of SCID animal models, and utilization of SCID pig model in biomedical sciences. Numerous physiologies in pig are comparable to those in human such as immune system, X-linked heritability, typical T−B+NK− cellular phenotype, and anatomy. Due to analogous features of pig to those of human, studies have found that immunodeficient pig is the most appropriate model for human SCID.
Keywords: Genetic mutations, Pig immunodeficient model, Regenerative medicine, SCID, Xenotransplantation
INTRODUCTION
SCID is a group of rare congenital syndromes caused by mutations in genes responsible for the development of both T and B cells. The immune system consists of different types of specialized white blood cells with different roles. These white blood cells including T-lymphocytes (T-cells) can act as mediators of immune response. They can also directly attack viruses. B-lymphocytes (B-cells) responsible for the production of antibodies can attach themselves to invaders and mark them for destruction. They are dependent on T-cells to work efficiently. Natural killer (NK) cells, a distinct subset of lymphocytes, possess cytotoxic activities. Patients with SCID have genetic defects that can affect T cells and at least one other type of immune cells. SCID is categorized into numerous types based on the cellular profile of immune cells. Its primary symptoms are reduced or absence of immune functions. All types of SCID are fatal unless treated appropriately. A naturally occurring SCID in humans was first described in the 1960s. It was subsequently acknowledged in horses, mice, dogs, and recently in pigs (1, 2). SCID was thought to be inherited from a recessive gene linked to either X-chromosome or an autosome. So far, mutation(s) in more than 30 genes that might cause SCID have been discovered. However, evidence suggests that there are likely other causes of SCID that are yet to be discovered. SCID pig is considered to be the most efficient animal model for studying human SCID due to their similar genetics, anatomy, physiology, and lifespan.
SCID REFLECTS DIFFERENT IMMUNOPHENOTYPES
SCID pig models depict numerous types of immune cell profiles due to defects in different signaling pathways and mutations in different genes. Different types of SCID based on the immunophenotype are summarized in Table 1.
Table 1.
Classification of SCID based on the immunophenotype
| Defects | Phenotypes in T/B/NK cells | Gene | Heredity | Disease |
|---|---|---|---|---|
| Defects in cytokine Signaling | T−B+NK− | IL2RG | XL | X-Lined SCID |
| T−B+NK− | JAK3 | AR | ||
| T−B+NK+ | IL7R-A | AR | ||
| Defect in V (D) J recombination | T−B−NK+ | RAG 1 | AR | Omenn Syndrome |
| T−B−NK+ | RAG 2 | AR | Omenn Syndrome | |
| T−B−NK+ | DCLRE1C | AR | ||
| Impaired signaling through the pre-T cell receptor | T−B+NK+ | CD3D | AR | |
| T−B+NK+ | CD3E | AR | ||
| T−B+NK+ | CD3G | AR | ||
| Increased lymphocyte apoptosis | T−B+NK+/NK− | PTPRC | AR | |
| T−B−NK− | ADA | AR | ADA-SCID | |
| T−B−NK− | AK2 | AR | Reticular dysgenesis | |
| Other mechanisms | T−B+NK+ | CORO1A | AR | |
| T−B+NK+ | RMRP | AR | Cartilage hair hypoplasia (CHH) |
ABERRANT CYTOKINE SIGNALING IN SCID
Defects in cytokine signaling can be caused by mutations in either the gene encoding cytokine receptors [IL2RG (γc), IL7-Rα] or a kinase [Janus kinase 3 (JAK3)] mediating the IL2RG signal transduction (5–7). The common gamma chain, IL2RG, is an IL-2 receptor subunit shared by IL-4, IL-7, IL-9, IL-5, and IL-21 receptors (8–12). IL2RG is found on T-cells, NK cells, natural killer T-cells (NK T), and dendritic cells. It plays a vital role in lymphoid growth induced by JAK3 (13).
X-LINKED-SCID BY IL2RG MUTATIONS: T−B+NK−
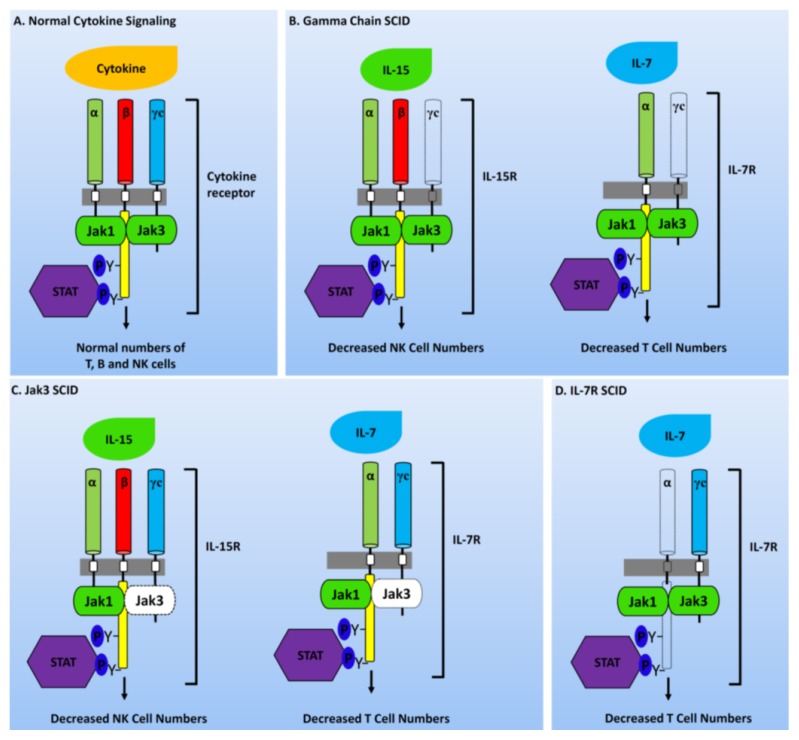
This type of SCID is generally caused by gene mutation that affects the integrity between IL2RG and JAK3 signaling pathway. The most common form of SCID in humans is the X-linked recessive SCID (T−B+NK−). The gamma chain is a part of several cytokine receptors, namely IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 (Fig. 1) (14, 15). All cytokine receptors share a common structure and contain three subunits (α, β, and γ chains) that are not covalently linked. These subunits contribute toward the binding of IL-2. However, only IL-2Rβ and γc are essential for signal transduction and activated T-cell, including regulatory T-cell that expresses a high level of IL-2Rα. This mostly restricts the high-affinity of IL-2R to these cell populations (16). The most common form of SCID affects nearly 45% of cases. It is caused by mutations in IL2RG, resulting in T cell−/B cell+/NK cell− cellular phenotype.
Fig. 1.
Defects in cytokine receptor signaling responsible for SCID phenotypes. (A) Most cytokine receptors are heterodimers or heterotrimers whose cytoplasmic regions are accompanied by JAK kinases which in turn activate STAT transcription factor. Proper functionality of these components leads to the development of normal and functional T, B, and NK cells. (B) Mutation in IL2RG inhibits the production of mature NK and T cells. (C) Mutations in JAK3 also prevent the development of mature NK and T cells. (D) Mutation in IL-7Rα chain affects only IL-7 signaling and blights T cell production (mutated molecules are shown with dashed outline).
AUTOSOMAL RECESSIVE SCID BY JAK3 MUTATIONS: T−B+NK−
Another type of SCID is caused by a mutation in a gene that encodes the JAK3 enzyme found in lymphocytes. More than 50 JAK3 gene mutations have been reported in JAK3-deficient SCID (17). The most common mutations in JAK3 gene either inhibit the production of JAK3 protein and produce an unbalanced protein or inhibit a protein so that it cannot perform its functions. Due to the non-function of JAK3 protein, some signals are not transmitted to the nucleus, thus affecting lymphocyte development. Consequently, individuals with JAK3-deficient SCID have shown T-NK-phenotype and normal count but poorly functioning B cells (Fig. 1).
AUTOSOMAL RECESSIVE SCID BY IL7Rα MUTATIONS: T−B+NK+
Interleukin 7 receptor (IL7R) gene is responsible for encoding the IL-7 receptor alpha chain protein that belongs to both IL-7 receptor and thymic stromal lymph protein (TSLP) receptor. IL-7 receptors are present on B and T cells. TSLP receptors are found on numerous types of immune cells, including B cells, T cells, monocytes and dendritic cells. In individuals with IL7R deficient SCID, T-cells are significantly reduced but counts of B and NK cells are normal (Fig. 1). IL7Rα is a unique alpha chain of IL7R which is essential for the production of T cells. In contrast, the gamma chain of IL7R is shared by receptors for IL-2 and IL-4. Its deficiency causes T−B+NK− phenotypic profile. ILR7 alpha-deficiency is clinically distinct from T−B+NK+ SCID which is caused by mutations in CD45, CD3α, and CD3ɛ genes (18).
DEFECTS IN V(D)J RECOMBINATION
The route of V(D)J recombination leads to the assemblage of genes encoding immunoglobulins and T-cell receptors (TCR). It also leads to the differentiation of lymphocytes into B and T cells which enable the formation of a functional immune system (19–21). RAG1/2 lymphoid specific factors initiates DNA Double Strand breaks (DNA-DSBs). Thereafter, DNA-DSBs are repaired using Non-Homologous End Joining (NHEJ) DNA repair pathway (22). During the initial lymphoid differentiation, diverse gene segments called Variable (V), Diversity (D), and Joining (J) are joined to produce coding sequences for immunoglobulin and T-cell antigen receptor variable regions (23). Mutations in genes involved in the V(D)J rearrangement results in a cellular phenotype like T−B−NK+.
T−B−NK+ OMENN SYNDROME BY RAG1, RAG2, OR DCLRE1C MUTATIONS
Omenn syndrome (OS) is an inherited immunodeficiency disorder caused by mutations in various genes involved in immune system function. The two most prominent reasons for OS are caused by dysregulation of RAG1 and RAG2. These genes provide instructions for the formation of protein complexes that are active in two types of lymphocytes: B-cells and T-cells. As a result, mutations in these genes can lead to the absence of B and T-cells, although they have no effect on NK cells. In humans, mutations in RAG1/2 can lead to the absence of both B and T cells. Previous studies have demonstrated that RAG1/2 KO (KO) mice are immunodeficient with properties similar to the absence of mature T and B cells (Fig. 2) (24, 25). Many other rodent models such as RAG1 KO rats and RAG1/2 KO rabbits have also been used in pre-clinical trials (26–28). Nonetheless, rodent models and other mid-size animals like rabbits are inefficient models for human immune diseases because of their physiological characteristics. Due to pig’s similarity to humans in terms of organ size, life span, anatomical, and physiological properties, it is considered to be the most efficient animal model for biomedical research, particularly for tissue engineering and xenotransplantation (29). The other gene involved in OS is the DCLRE1C gene. This gene encodes a nuclear protein, Artemis, that is involved in V(D)J recombination during DNA repair. There is no difference in survival, early toxicity, or occurrence of tumor between Artemis and RAG-deficient SCID (30). SCID caused by mutations in DCLRE1C results in the absence of both T and B cells, although it has no effect on the number of NK cells (Fig. 2). Immunologically, SCID has been classified into two categories: 1) T-cells are lacking (T−B+), and 2) both T and B cells are absent (T−B−). It has been suggested that 70% of SCID cases are T−B+SCID caused by a T-cell signaling defect. T−B-SCID accounts for 30% of SCID cases. It is caused by defect in V(D)J recombination (31, 32).
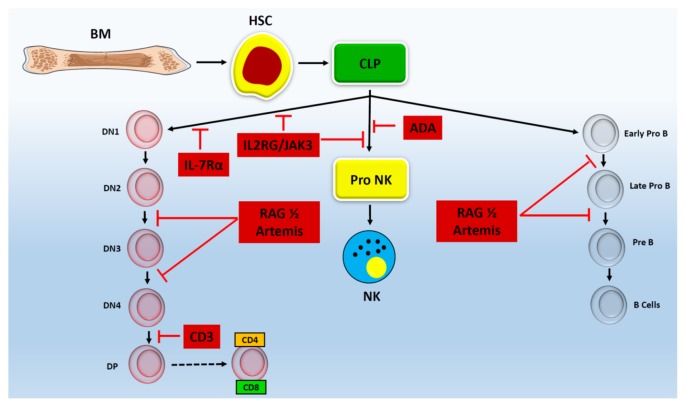
Fig. 2.
Genetic mutations inhibit the development of lymphocyte at different stages. Schematic of lymphocyte development process shows that genetic mutations inhibit which stage of T, B and NK cell development. Mutations in IL2RG and JAK3 block the development of T and NK cells, but not B cell, during early developmental stages whereas IL-7Rα only inhibits T cell development without affecting the development of B or NK cells. Rag complex and Artemis mutations impair the development of B and T cells, but not NK cells, during Pro to pre stages. Mutation in ADA only blocks the development of NK cells without affecting the development of B or T cells. Mutation in CD3 inhibits the development of T cells during DN to DP stage without affecting the development of B or NK cells.
IMPAIRED SIGNALING THROUGH PRE-T CELL RECEPTOR
T-cell differentiation depends on pre-T cell receptor (pTCR) and TCR signaling events that occur at consecutive stages of the pathway (33). The pre-TCR consists of T-cell receptor beta (TCRβ) chain and disulfide-linked pre-T cell receptor alpha (pTCRα) chain. The pTCR complex is encoded by the PTCRA gene. The protein encoded by the PTCRA gene is a single-pass type I membrane protein that is functionally active in immature T-cells, but not in mature T-cells (34). pTCR receptors can transduce signals to mature T lymphocytes by CD3 complex (CD3γ, CD3δ, CD3ɛ, CD3ζ), protein-tyrosine kinases, and protein-phosphotyrosine phosphatases (CD45) (35). The CD3 complex plays an important role in signaling via the T-cell receptor which is needed for normal T-cells development. Mutations in genes encoding CD3 chains (CD3 delta, CD3 epsilon, and CD3 zeta) can cause T−B+NK+/−SCID (Fig. 2). CD45, a common leukocyte antigen, is a transmembrane tyrosine phosphatase involved in T-cell receptor signaling and maturation of T-cell in the thymus. Defect in CD45 can also cause the T−B+NK+/−SCID phenotype, although this type of defect is very rare (36). ZAP70 is a part of the TCR that plays a crucial role in T-cell signaling It belongs to the protein-tyrosine kinase family (37). Defect in ZAP70 gene can also cause SCID wherein CD8+ T-cells are absent. The T lymphocyte-specific protein tyrosine kinase P56lck (LCK) is an important part of the TCR-mediated signal transduction complex (38). Although defect in P56lck is very infrequent, it can lead to an autosomal recessive immunodeficiency syndrome known as idiopathic CD4+ lymphocytopenia, in which CD4+ T cells are reduced (39, 40).
INCREASED LYMPHOCYTE APOPTOSIS
Apoptosis of lymphocyte plays a significant role in proper functioning of immune system. Increased lymphocyte apoptosis has been detected in patients with systemic autoimmune diseases. An autoimmune immune disease is condition where body immune system mistakenly attack and destroy healthy body tissue and organs. Immunodeficiency can be caused due to increased lymphocyte apoptosis. Mutations which causes impairing in survival signals, which travels through cytokine receptors, can result in cell death, causing SCID (41).
T−B− NK− SCID BY ADA MUTATIONS
Adenosine deaminase (ADA) deficiency is an autosomal recessive inheritance disorder which damages the immune system and causes SCID. ADA deficiency affects about 1 in 200,000 newly born babies and accounts for about 1/3 of all SCID cases. Although this enzyme is found throughout the body, it is the most active in lymphocytes (42). ADA is an enzyme of the purine salvage pathway that facilitates alteration of adenosine and 2-deoxyadenosine to inosine (and deoxyinosine). Correspondingly, any type of deficiency in this process can result in metabolic poisoning due to accumulation of adenosine, deoxyadenosine, and deoxyadenosine triphosphate. Excess intra-cellular deoxyadenosine and deoxyadenosine triphosphate can cause comprehensive lymphocyte apoptosis that leads to T−B−NK− SCID phenotype (Fig. 2).
T−B− NK-SCID PHENOTYPE BY AK2 MUTATIONS
Reticular dysgenesis (RD) is a fatal form of SCID. It is categorized by lack of circulation of T-lymphocyte, severe congenital neutropenia and sensorineural hearing loss. This type of SCID typically evident in newborns. In all cases of SCID, less than 2% are associated with RD-SCID. It affects both males and females. RD SCID patients have mutations in both copies of AK2 gene (43, 44). Absence of AK2 protein expression defines RD. AK2 is a member of adenylate kinase family. All hematopoietic cells and majority of tissues contains AK2 expressions. AK2 is located in the intermembrane space of mitochondria. This protein is located in the intermembrane space of mitochondria, rest of the members of adenylate kinase family are confined to cytoplasmic (AK1, 5, 7 and 8), nuclear (AK6), or mitochondrial matrix (AK3 and AK4) (45, 46). RD SCID is characterized by the lack of granulocytes, extreme deficiency of lymphocytes in peripheral blood, and hypoplasia of thymus and other secondary lymphoid organs. Individuals with RD-SCID display low T and NK cell numbers. Their B cell numbers are either absent or low.
DEFICIENCY IN CORONIN-1A GENE
Coronin-1A gene was first discovered by de Hostos and coworkers in 1991. Deficiency in Coronin-1A causes absence of T-cells. However, numbers of B and NK cells are normal (T−B+NK+). The first study on mammalian Coronin-1A was performed with instinctively happening “Peripheral T cell deficient” or ptcd mouse (47). Positional cloning has revealed that Coronin-1A gene mutation primarily causes failure of T cells to leave the thymus in a rodent model, consequently leading to the absence of T cells in the periphery despite intact thymic differentiation. So far, Coronin-1A KO, hypomorphic, and gain-of-function mice models have been developed (48–50). Shiow et al. (51) and others (51–53) have investigated children with Coronin-1A deficiency whose phenotype resonates the that of ptcd mouse (i.e., few peripheral T cells in spite of a normal-sized thymus with normal numbers of B and NK cells).
T−B+ NK+ SCID BY RMRP GENE MUTATION
Cartilage-hair hypoplasia (CHH) is a rare autosomal recessive genetic disorder first reported in old order Amish families of Pennsylvania (54). CHH is caused by mutations in the RMRP gene. Unlike other genes, the RMRP gene does not contain any instruction for coding a protein, hence known as a non-coding RNA. A chemical partner of DNA is generated from the RMRP gene. Several proteins can interact with this RNA molecule and form mitochondrial RNA-processing endoribonuclease or RNase MRP enzyme. Anauxetic dysplasia is the extreme end of the spectrum of mutations in RMRP, with genotype/phenotype correlations. Generally, the degree of rRNA cleavage decreases strongly with the degree of bone dysplasia (55).
GENOME EDITING TECHNIQUES AND SCID PIG MODELS
The gene editing process has led scientists to improve an animal’s genome by identifying and correcting mutant ones. Gene editing is an anticipated, more precise way of rapidly improving genetic makeup in plants and animals. Basic medical research studies are constantly investigating different diseases and striving to introduce novel methods of treatment. Translation of these research findings into captivating therapies is not only difficult, but also expensive. Hence, a need for enhancing transition “from bench to the bedside” lies in the application of a valid animal model. Gene editing techniques like, Zinc-Finger Nucleases (ZFNs), Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated 9 (Cas9) system and transcription activator-like effector nucleases (TALENs), proved pivotal in enhancing the efficacy of genetically engineered (GE) animal models (56).
ZINC FINGER NUCLEASES (ZFNs)
A genome editing method invented in 2011 with engineered nucleases can let researchers pick any specific site of the genome in numerous species and set alteration according to their research purpose, making this a very powerful way to control the function of genes. In clinical research, investigators have utilized gene editing nucleases for therapy. The entire recognition pair site of ZFNs is about 18–24 bp long, allowing precise targeting in human genome (57). Many researchers have used ZFNs to generate mutant animal models such as zebrafish, mice, rats, rabbits, and pig (58–63). The first GE pig generated by using ZFN was reported in 2011. The first step in creating GE pig using ZFNs was to interrupt hemizygous enhanced green fluorescence protein (eGFP) gene. A couple of ZFNs could efficiently deactivate the eGFP gene in pig fibroblast cells via NHEJ. The efficacy of ZFNs was approximately 5%. Whyte et al. in 2011 suggested that DNA repair processes used for gene-editing system were also present in pig somatic cells and that ZFNs could be used for generating pig models. Peroxisome proliferator activated receptor gamma (PPARγ) was the first endogenous gene that was picked and engineered using ZFNs to create a GE pig model for cardiovascular disease. Both alleles in pig somatic cell were interrupted using transfecting ZFNs. Thereafter, phenotypically picking α-Gal negative cells via fluorescence-activated cell sorting (FACS). Alpha-1,3-galactosyltransferase (GGTA1) was meritoriously altered in both alleles in pig fibroblast cells. These cells were developmentally capable via somatic cell nuclear transfer (SCNT). Hauschild et al. in 2013 determined that the effectiveness of targeting was 1%. This was a noteworthy accomplishment in the development of GE pigs since only heterozygous alterations were possible with earlier methods through conventional gene targeting approach. ZFNs can also be used for the generation of double KO pigs without breeding by deactivating two endogenous genes, GGTA1 and CMP-N-glycolylneuraminic acid hydroxylase (CMAH), with ZFN. Initially both alleles of CMAH gene were interrupted in pig somatic cells. Later on, cells obtained from CMAH KO clones were transfected with ZFNs; ZFNs that are targeting GGTA1. SCNT is used to generate double KO pigs with CMAH/GGTA1 double KO cell lines (64). This was an a very useful enhancement in gene editing methods which provided researchers an opportunity to interrupt multiple alleles and genes, reducing the amount of breeding required to create GE pigs appropriate for xenotransplantation. Despite the above mention advantages of ZFN method, it also has some limitations such as very time consuming, off-site target cutting of DNA, and cytotoxicity (65).
TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASES (TALENs)
TALEN is a type of restriction enzyme that can be used to modify specific sequences of DNA. TALENs constitute TAL effector DNA-binding domain and a DNA cleavage domain known as nuclease. TALEs can be altered to attach to any region of interest in a DNA sequence, to combine with nucleases, and to cut a specific DNA region. TALENs were discovered from plant pathogenic bacteria called Xanthomona. TALEN acts by attaching a string of TALEN motif to the region of interest of DNA on the genome. FOK1 enzyme acts as an endonuclease to begin double strand breaks (DSB). TALEN was successfully applied in GE pig production. The first GE pig using this method was produced in 2012 (66). According to that study, utilization of Goldy TALEN set with truncated N- and C-terminal of TALEN worked efficiently for the induction of mutation. A number of pig models genetically modified by TALEN have been used for xenotransplantation and muscle biology. This method gave great breakthrough in the field of gene editing. The significance of this technique has been well documented (67, 68). This technique has also been used for generating a SCID pig model for stem cell transplantation studies. In 2014, Lee K. created RAG2 KO pigs by combining TALEN and SCNT methods and introducing human induced pluripotent stem (hiPS) cells into pigs. Their results indicated that pigs generated by this technique presented prominent signs of SCID phenotype. They could support growth and differentiation of transplanted hiPS cells by teratoma formation. This was the first SCID large animal model with teratoma formation from human stem cells. Most recently, Choi et al. (69) in 2017 created RAG2 biallelic KO pig model where induction of hiPS cells led to the formation of teratoma. Previous studies have indicated that RAG2 biallelic KO pig is athymic. However, Choi et al. (69) demonstrated that RAG2 biallelic KO pig generated by TALEN method had smaller spleen compared to an age-matched control pig.
CRISPR/Cas9
The development of proficient and reliable methods to make appropriate, targeted changes to the genome of organism is a long-awaited goal of researchers in the biomedical field. Advancements in the area of gene editing techniques provide a meaningful advantage for clinical research. CRISPR/Cas9 has been considered as the best invention for the generation of animal models to cure human diseases. CRISPR was first reported in 1987. It is a family of DNA sequences belonging to bacteria and archaea. The CRISPR/Cas9 system is capable of inducing DSBs in any regions of interest in DNA by using RNA-based gene editing technology. The process involves modifying a single guide RNA (sgRNA) combining with tracer-RNA to attach to a region of interest, thereby locating Cas9 protein to the target site on the genome. The function of Cas9 in this procedure is to create DSB in the target site if the protospacer adjacent motif (PAM) is existing at the locus (70). In contrast to ZFN and TALEN that are very time consuming and tedious due to the requirement of an array assembly, the CRISPR/Cas9 method is easy as it needs only the insertion of a 20-bp sgRNA into the targeting vector (71). The CRISPR/Cas9 system is the leading gene-editing system because of its ease of use. Initially, GE pig using the CRISPR/Cas9 method was produced by introducing the CRISPR/Cas9 system into developing zygotes (72). The first CRISPR/Cas9 system efficiently induced precise mutation in pig fibroblast cells for generation of GE pig (73). Recently, a study (74) indicated that the CRISPR/Cas9 system can efficiently interrupt multiple copies of pig endogenous retroviruses (PERV) in somatic cells and that these cells can be used as donor for SCNT to produce pig without PERV. That study has demonstrated that organ transplantation between different species is feasible by using the CRISPR/Cas9 system. CRISPR simplifies the generation of animal models that mimic human disease conditions.
EFFICIENT SCID PIG MODELS THAT MIMIC HUMAN SCID PHENOTYPE
In recent studies, most of SCID phenotypes are artificially attainable by diverse genetic manipulations. To develop SCID pig model, researchers have concentrated on targeting IL2RG and/or the RAG1/2 KOs. Engineered IL2RG SCID pigs have been created using serial nuclear transfer, ZFNs, or CRISPR/cas9 system technologies. Similar to human SCID, IL2RG SCID pig has an X-linked heritability. It also has the typical T−B+NK− cellular phenotype. The RAG targeted pig model generated using TALENs and/or SCNT is appropriate to fill NK cell specific research gaps. Basically, these RAG targeted pigs portray the T−B−NK+ phenotype.
IL2RG KNOCKOUT PIG MODELS
IL2RG gene is typically located on the X-chromosome. Mutation caused in this gene results in X-linked SCID. Herein, we report a pig model with knock-out IL2RG gene. The first IL2Rγ KO model was reported by Suzuki et al. (75) in 2012. They used predictable gene targeting to delete exon 6 of IL2RG, making it non-functional. Male offspring of animals generated via the SCNT technique lacked the thymus and showed reduced numbers of T and NK cells with allogenic (BMT) rescued T−B+NK− immunodeficient phenotype. Female offspring showed a normal T+B+NK+ phenotype (75, 76). Watanabe et al. (77) have attained similar phenotype using ZFNs and showed that their animals are athymic with decreased numbers of T and NK cells. After these two promising pig SCID models, Choi et al. (78) in 2016 reported the generation of monoallelic IL2RG pig using TALENs technique. Their animals were athymic with the T−B+NK− phenotype. Kang et al. (78) in 2016 reported the generation biallelic mutations in IL2RG female offspring by using CRISPR/Cas9 direct injection technique. Their results indicated that IL2Rγ KO female pigs showed the phenotypic characteristic of SCID, with reduced numbers of T, B, and NK cells and absence of thymus.
RAG1/2 KNOCKOUT PIG MODEL
RAG1/2 genes are involved in the production of a group of proteins called the RAG complex involved in immune system cells (lymphocytes) namely B and T cells. These genes constitute V, D, and J segments. Greater than 20 mutations in the RAG1/2 gene that cause Omenn syndrome, an immune system disorder, have been reported. Hence, RAG1/2 genes can be used to create an immunodeficient pig model. Huang et al. (78) have reported the generation and phenotypic characterization of RAG1/2 KO pig model using TALEN. Pig fetal fibroblasts were genetically engineered using TALEN and then used to provide donor nuclei for SCNT. Their results indicated that animals created with biallelic mutation in both RAG1/RAG2 genes had emaciated thymus and spleen and failed to perform V(D)J rearrangement without having mature T and B cells (Fig. 2). Lee et al. (79) have studied RAG2 to generate a KO model by using TALEN technique. According to their research, phenotypic characterization of the RAG2 KO model showed complete absence of T and B cells and functional thymi. Choi et al. (78) have focused on RAG2 to generate SCID pig deficient in two alleles of RAG2, known as RAG-2 biallelic KO pig model. Their results showed phenotypic characteristics like T−B−NK+−SCID. These type of Rag1/2 deficient pig model are very useful for studying human SCID because of similar phenotype.
RESEMBLANCE OF HUMAN TO MOUSE AND PIG
The choice of the model depends on the similarity between the model and humans. In the past, murine models were widely used for human disease modeling. There are many instances that a drug works successfully in these rodent models but fails to produce effect in humans, even genetically altered murine models have significant limitations in that they could not mimic results in humans. The advantage of the pig model is its similarity to humans in genetic makeup, anatomy, and physiology. Previous studies have demonstrated that analyzed parameters of pig immune system are 80% similar to human, while there is only a 10% similarity between human and mouse (2, 80, 81). The SCID mouse is widely used because of the generation of the humanized model in which HSCs cells are induced to a given SCID host to permit the differentiation of elements involved in the immune system (82). This humanized SCID mouse also has some limitations. For example, mouse phagocytes directly involved in the killing of evolving human NK cell ancestors or non-recognition by mouse cytokines are needed for lineage maturation of NK cells. Due to the aforementioned limitations, researchers inclined towards large animals. As previously discussed, many studies have shown that pig is the most efficient model for biomedical research due to its considerable resemblance with humans. Although humanized pig has not been published yet, filling this gap can help develop mature teratomas after injecting human pluripotent stem cells into RAG mutant SCID pig (69, 79). Previously, many successful immunodeficient murine models have been introduced in different areas of research such as xenotransplantation, human immune disease modeling, and regenerative medicine. Researchers have tried to extend the applicability of the pig model due to similarity of pig’s immune system to human. Several SCID pig models have already been generated (2, 83). A humanized pig disease model as a research platform will be a breakthrough that can transform preclinical research because it is the closest animal model to human.
IMPORTANCE OF PIG IMMUNOCOMPROMISED MODEL IN BIOMEDICAL RESEARCH
Animal models are essential resources for the growth in the field of diagnostic and therapeutic procedures for human diseases. Immunodeficient models have attracted great attention because they permit engraftment of human cells and tissue. In addition, they could be beneficial for the authentication of stem cell therapies in cancer research. Understanding the SCID system can provide insight into the generation of SCID animal model, a powerful tool for the development of biomedical treatments. Previously, many researchers have induced SCID condition in mice, rats, and recently in pig. Pig is an extraordinary model with genetics, anatomy, and physiology similar to humans. The potential of stem cell-based treatments is presently not being completely recognized due to no existence of suitable animal models for pre-clinical trials. This leaves the acute necessity of improved animal models that can translate results from preclinical trials in humans. Scientific improvements addressing these hurdles have been realized through the identification of a naturally occurring SCID pig. SCID pig lacks both T and B lymphocytes as well as adaptive immunity. The SCID pig has been identified as the most phenomenal model for human immune diseases. Pig model is widely used in areas of stem cell researches including transplantation of hematopoietic stem cell (HSC) to develop treatment or graft for host disease and regenerative medicine including heart, cartilage, and bone regenerations. Previous studies have demonstrated that SCID pig models can phenocopy human SCID. Thus, the generation of the SCID pig model is very useful for translation human disease modeling. Compared to mouse, pig model has higher sequence similarities with human xenobiotic receptors, thus permitting more precise prediction of pharmacodynamics and pharmacokinetic characteristics of drugs.
CONCLUSION
In this review, we discussed genetics of SCID and the utilization of SCID pig model in different areas of biomedical research such as xenotransplantation model, regenerative medicine, cancer research, and pre-clinical trials. Previously, many rodent animal models have been used for studying different areas of biomedical sciences. However, rodents have numerous limitations. For example, their organ-size, genetic makeup, and pharmaceutical aspects cannot mimic human disease condition. Thus, there are needs for the generation of large animal models thatn can better mimic human disease condition. Main concerns with the utilization of large animals are costs, animal ethics, and social problems. In this review, we provide evidences that pig is the most appropriate animal model for studying human SCID. Different SCID pig models have already been established to develop pigs harboring human organs. These type SCID pig models have prodigious applications in the area of xenotransplantation. The GE SCID pig model plays a vital role in the field of xenotransplantation due to its similarity of organ size with humans. Lastly, based on pervious literatures, we conclude that SCID pig will become the most emerging animal model for regenerative medicine, clinical research, and understanding human immune diseases and physiology.
ACKNOWLEDGEMENTS
This study was supported by a grant (2015R1A5A1009701) from the National Research Foundation funded by the Ministry of Science, ICT & Future Planning, Republic of Korea.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Tasher D, Dalal I. The genetic basis of severe combined immunodeficiency and its variants. Appl Clin Genet. 2012;5:67–80. doi: 10.2147/TACG.S18693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell EJ, Cunnick JE, Tuggle CK. SCID pigs: An emerging large animal NK model. J Rare Dis Res Treat. 2017;2:1–6. doi: 10.29245/2572-9411/2017/3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–655. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 4.Cossu F. Genetics of SCID. Ital J Pediatr. 2010;36:76. doi: 10.1186/1824-7288-36-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macchi P, Villa A, Giliani S, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 6.Noguchi M, Yi H, Rosenblatt HM, et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-O. [DOI] [PubMed] [Google Scholar]
- 7.Russell SM, Tayebi N, Nakajima H, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 8.Takeshita T, Asao H, Ohtani K, et al. Cloning of the gamma chain of the human IL-2 receptor. Science. 1992;257:379–382. doi: 10.1126/science.1631559. [DOI] [PubMed] [Google Scholar]
- 9.Kondo M, Takeshita T, Ishii N, et al. Sharing of the interleukin-2 (IL-2) receptor gamma chain between receptors for IL-2 and IL-4. Science. 1993;262:1874–1877. doi: 10.1126/science.8266076. [DOI] [PubMed] [Google Scholar]
- 10.Noguchi M, Nakamura Y, Russell SM, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880. doi: 10.1126/science.8266077. [DOI] [PubMed] [Google Scholar]
- 11.Giri JG, Ahdieh M, Eisenman J, et al. Utilization of the beta and gamma chains of the IL-2 receptor by the novel cytokine IL-15. EMBO J. 1994;13:2822–2830. doi: 10.1002/j.1460-2075.1994.tb06576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asao H, Okuyama C, Kumaki S, et al. Cutting edge: the common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol. 2001;167:1–5. doi: 10.4049/jimmunol.167.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Colonna M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity. 2018;48:1104–1117. doi: 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol. 2001;1:200–208. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- 15.Sugamura K, Asao H, Kondo M, et al. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu Rev Immunol. 1996;14:179–205. doi: 10.1146/annurev.immunol.14.1.179. [DOI] [PubMed] [Google Scholar]
- 16.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 17.Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter HH, Friedrich W, Dopfer R, et al. NK cell function in severe combined immunodeficiency (SCID): evidence of a common T and NK cell defect in some but not all SCID patients. J Immunol. 1983;131:2332–2339. [PubMed] [Google Scholar]
- 19.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/S0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 20.Oettinger MA. V(D)J recombination: on the cutting edge. Curr Opin Cell Biol. 1999;11:325–329. doi: 10.1016/S0955-0674(99)80044-1. [DOI] [PubMed] [Google Scholar]
- 21.Villa A, Sobacchi C, Notarangelo LD, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.V97.1.81. [DOI] [PubMed] [Google Scholar]
- 22.de Villartay JP. Congenital defects in V(D)J recombination. Br Med Bull. 2015;114:157–167. doi: 10.1093/bmb/ldv020. [DOI] [PubMed] [Google Scholar]
- 23.Wiler R, Leber R, Moore BB, VanDyk LF, Perryman LE, Meek K. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc Natl Acad Sci U S A. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-G. [DOI] [PubMed] [Google Scholar]
- 25.Shinkai Y, Rathbun G, Lam KP, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-C. [DOI] [PubMed] [Google Scholar]
- 26.Menoret S, Fontaniere S, Jantz D, et al. Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J. 2013;27:703–711. doi: 10.1096/fj.12-219907. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Zhong J, Guo X, et al. Generation of RAG 1- and 2-deficient rabbits by embryo microinjection of TALENs. Cell Res. 2013;23:1059–1062. doi: 10.1038/cr.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Fan N, Song J, et al. Generation of knockout rabbits using transcription activator-like effector nucleases. Cell Regen (Lond) 2014;3:3. doi: 10.1186/2045-9769-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang J, Guo X, Fan N, et al. RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193:1496–1503. doi: 10.4049/jimmunol.1400915. [DOI] [PubMed] [Google Scholar]
- 30.Schuetz C, Neven B, Dvorak CC, et al. SCID patients with ARTEMIS vs RAG deficiencies following HCT: increased risk of late toxicity in ARTEMIS-deficient SCID. Blood. 2014;123:281–289. doi: 10.1182/blood-2013-01-476432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckley RH, Schiff RI, Schiff SE, et al. Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J Pediatr. 1997;130:378–387. doi: 10.1016/S0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- 32.Geha RS, Notarangelo LD, Casanova JL, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee. J Allergy Clin Immunol. 2007;120:776–794. doi: 10.1016/j.jaci.2007.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat Rev Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 35.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tchilian EZ, Wallace DL, Wells RS, Flower DR, Morgan G, Beverley PC. A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol. 2001;166:1308–1313. doi: 10.4049/jimmunol.166.2.1308. [DOI] [PubMed] [Google Scholar]
- 37.Otsu M, Steinberg M, Ferrand C, et al. Reconstitution of lymphoid development and function in ZAP-70-deficient mice following gene transfer into bone marrow cells. Blood. 2002;100:1248–1256. doi: 10.1182/blood-2002-01-0247. [DOI] [PubMed] [Google Scholar]
- 38.Legname G, Seddon B, Lovatt M, et al. Inducible expression of a p56Lck transgene reveals a central role for Lck in the differentiation of CD4 SP thymocytes. Immunity. 2000;12:537–546. doi: 10.1016/S1074-7613(00)80205-8. [DOI] [PubMed] [Google Scholar]
- 39.Goldman FD, Ballas ZK, Schutte BC, et al. Defective expression of p56lck in an infant with severe combined immunodeficiency. J Clin Invest. 1998;102:421–429. doi: 10.1172/JCI3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubert P, Bergeron F, Ferreira V, et al. Defective p56Lck activity in T cells from an adult patient with idiopathic CD4+ lymphocytopenia. Int Immunol. 2000;12:449–457. doi: 10.1093/intimm/12.4.449. [DOI] [PubMed] [Google Scholar]
- 41.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell. 2002;109(Suppl):S97–107. doi: 10.1016/S0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 42.Hershfield MS. Genotype is an important determinant of phenotype in adenosine deaminase deficiency. Curr Opin Immunol. 2003;15:571–577. doi: 10.1016/S0952-7915(03)00104-3. [DOI] [PubMed] [Google Scholar]
- 43.Pannicke U, Honig M, Hess I, et al. Reticular dysgenesis (aleukocytosis) is caused by mutations in the gene encoding mitochondrial adenylate kinase 2. Nat Genet. 2009;41:101–105. doi: 10.1038/ng.265. [DOI] [PubMed] [Google Scholar]
- 44.Lagresle-Peyrou C, Six EM, Picard C, et al. Human adenylate kinase 2 deficiency causes a profound hematopoietic defect associated with sensorineural deafness. Nat Genet. 2009;41:106–111. doi: 10.1038/ng.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noma T. Dynamics of nucleotide metabolism as a supporter of life phenomena. J Med Invest. 2005;52:127–136. doi: 10.2152/jmi.52.127. [DOI] [PubMed] [Google Scholar]
- 46.Panayiotou C, Solaroli N, Xu Y, Johansson M, Karlsson A. The characterization of human adenylate kinases 7 and 8 demonstrates differences in kinetic parameters and structural organization among the family of adenylate kinase isoenzymes. Biochem J. 2011;433:527–534. doi: 10.1042/BJ20101443. [DOI] [PubMed] [Google Scholar]
- 47.Yagi H, Matsumoto M, Nakamura M, et al. Defect of thymocyte emigration in a T cell deficiency strain (CTS) of the mouse. J Immunol. 1996;157:3412–3419. [PubMed] [Google Scholar]
- 48.Shiow LR, Roadcap DW, Paris K, et al. The actin regulator coronin 1A is mutant in a thymic egress-deficient mouse strain and in a patient with severe combined immunodeficiency. Nat Immunol. 2008;9:1307–1315. doi: 10.1038/ni.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mueller P, Massner J, Jayachandran R, et al. Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat Immunol. 2008;9:424–431. doi: 10.1038/ni1570. [DOI] [PubMed] [Google Scholar]
- 50.Haraldsson MK, Louis-Dit-Sully CA, Lawson BR, et al. The lupus-related Lmb3 locus contains a disease-suppressing Coronin-1A gene mutation. Immunity. 2008;28:40–51. doi: 10.1016/j.immuni.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiow LR, Paris K, Akana MC, Cyster JG, Sorensen RU, Puck JM. Severe combined immunodeficiency (SCID) and attention deficit hyperactivity disorder (ADHD) associated with a Coronin-1A mutation and a chromosome 16p112 deletion. Clin Immunol. 2009;131:24–30. doi: 10.1016/j.clim.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moshous D, Martin E, Carpentier W, et al. Wholeexome sequencing identifies Coronin-1A deficiency in 3 siblings with immunodeficiency and EBV-associated B-cell lymphoproliferation. J Allergy Clin Immunol. 2013;131:1594–1603. doi: 10.1016/j.jaci.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mace EM, Orange JS. Lytic immune synapse function requires filamentous actin deconstruction by Coronin 1A. Proc Natl Acad Sci U S A. 2014;111:6708–6713. doi: 10.1073/pnas.1314975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McKusick VA, Eldridge R, Hostetler JA, Ruangwit U, Egeland JA. Dwarfism in the amish. Ii. cartilage-hair hypoplasia. Bull Johns Hopkins Hosp. 1965;116:285–326. [PubMed] [Google Scholar]
- 55.Thiel CT, Rauch A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best Pract Res Clin Endocrinol Metab. 2011;25:131–142. doi: 10.1016/j.beem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 56.Ryu J, Prather RS, Lee K. Use of gene-editing technology to introduce targeted modifications in pigs. J Anim Sci Biotechnol. 2018;9:5. doi: 10.1186/s40104-017-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doyon Y, McCammon JM, Miller JC, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci U S A. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geurts AM, Cost GJ, Freyvert Y, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zschemisch NH, Glage S, Wedekind D, et al. Zinc-finger nuclease mediated disruption of Rag1 in the LEW/Ztm rat. BMC Immunol. 2012;13:60. doi: 10.1186/1471-2172-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hauschild J, Petersen B, Santiago Y, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci U S A. 2011;108:12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang D, Yang H, Li W, et al. Generation of PPARgamma mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lutz AJ, Li P, Estrada JL, et al. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose alpha-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2013;20:27–35. doi: 10.1111/xen.12019. [DOI] [PubMed] [Google Scholar]
- 65.Whyte JJ, Zhao J, Wells KD, et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev. 2011;78:2. doi: 10.1002/mrd.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hendriks WT, Jiang X, Daheron L, Cowan CA. TALEN- and CRISPR/Cas9-mediated gene editing in human pluripotent stem cells using lipid-based transfection. Curr Protoc Stem Cell Biol. 2015;34:5B 3 1–25. doi: 10.1002/9780470151808.sc05b03s34. [DOI] [PubMed] [Google Scholar]
- 68.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi YJ, Kim E, Reza A, et al. Recombination activating gene-2(null) severe combined immunodeficient pigs and mice engraft human induced pluripotent stem cells differently. Oncotarget. 2017;8:69398–69407. doi: 10.18632/oncotarget.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitworth KM, Lee K, Benne JA, et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91:78. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niu D, Wei HJ, Lin L, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017;357:1303–1307. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki S, Iwamoto M, Saito Y, et al. Il2rg gene-targeted severe combined immunodeficiency pigs. Cell Stem Cell. 2012;10:753–758. doi: 10.1016/j.stem.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 76.Bauer TR, Jr, Adler RL, Hickstein DD. Potential large animal models for gene therapy of human genetic diseases of immune and blood cell systems. ILAR J. 2009;50:168–186. doi: 10.1093/ilar.50.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe M, Nakano K, Matsunari H, et al. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS One. 2013;8:e76478. doi: 10.1371/journal.pone.0076478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang JT, Cho B, Ryu J, et al. Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod Biol Endocrinol. 2016;14:74. doi: 10.1186/s12958-016-0206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee K, Kwon DN, Ezashi T, et al. Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci U S A. 2014;111:7260–7265. doi: 10.1073/pnas.1406376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dawson HD, Loveland JE, Pascal G, et al. Structural and functional annotation of the porcine immunome. BMC Genomics. 2013;14:332. doi: 10.1186/1471-2164-14-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20:50–57. doi: 10.1016/j.tim.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ito R, Takahashi T, Katano I, Ito M. Current advances in humanized mouse models. Cell Mol Immunol. 2012;9:208–214. doi: 10.1038/cmi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ito T, Sendai Y, Yamazaki S, et al. Generation of recombination activating gene-1-deficient neonatal piglets: a model of T and B cell deficient severe combined immune deficiency. PLoS One. 2014;9:e113833. doi: 10.1371/journal.pone.0113833. [DOI] [PMC free article] [PubMed] [Google Scholar]